Abstract
Fertilization is fundamental for sexual reproduction, yet its molecular mechanisms are poorly understood. We found that an oocyte-expressed Ly6/uPAR protein, which we call Bouncer, is a crucial fertilization factor in zebrafish. Membrane-bound Bouncer mediates sperm-egg binding and is thus essential for sperm entry into the egg. Remarkably, Bouncer not only is required for sperm-egg interaction but is also sufficient to allow cross-species fertilization between zebrafish and medaka, two fish species that diverged more than 200 million years ago. Our study thus identifies Bouncer as a key determinant of species-specific fertilization in fish. Bouncer’s closest homolog in tetrapods, SPACA4, is restricted to the male germline in internally fertilizing vertebrates, which suggests that our findings in fish have relevance to human biology.
Keywords: fertilization, species-specificity, sperm-egg interaction, zebrafish
Introduction, results and discussion
Fertilization, whereby two gametes fuse to form the single-cell zygote in sexually reproducing organisms, is highly efficient, yet species-restricted. This strategy ensures reproductive success and the survival of distinct species. However, the means by which nature has fulfilled these seemingly contradictory requirements, particularly at the molecular level, have remained a mystery. The only vertebrate proteins known so far to be essential for sperm-egg binding are the sperm-expressed IZUMO1 (1, 2) and the egg membrane proteins JUNO (3) and CD9 (4–6). Binding of IZUMO1 to JUNO mediates adhesion between sperm and egg in mammals (1–3, 7), whereas the role of CD9 in this process remains unclear. Although in vitro binding assays show that human IZUMO1 binds more efficiently to human JUNO than to mouse JUNO (8), an in vivo function in mediating species-specificity has not been identified for any of these factors.
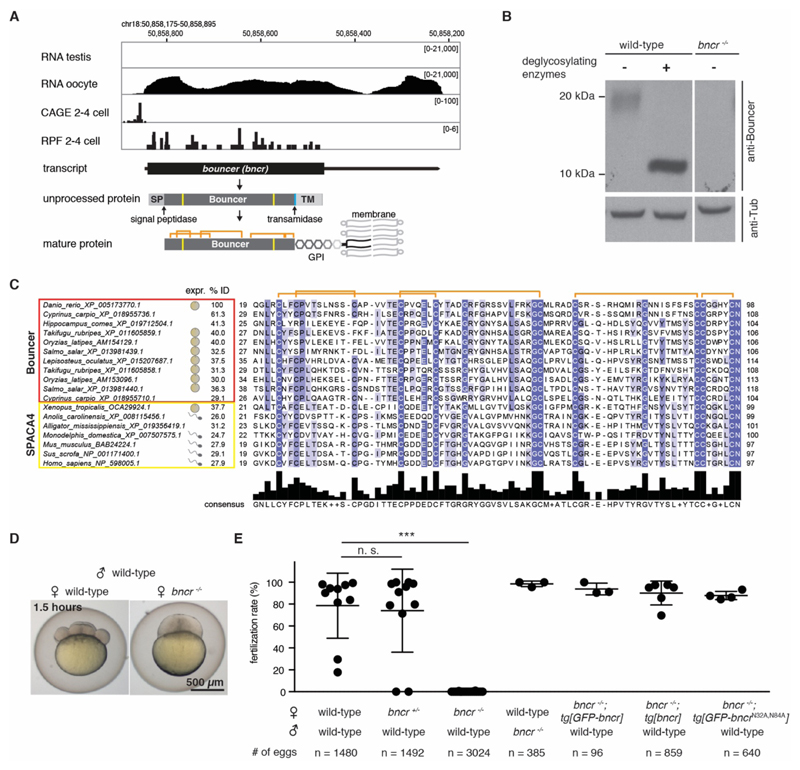
To identify factors required for fertilization in vertebrates, we examined our collection of predicted protein-coding genes (9) that are expressed in zebrafish oocytes and/or testis. A single-exon gene stood out because of its high expression in zebrafish oocytes (Fig. 1A) and the presence of homologous sequences in other vertebrates. On the basis of its loss-of-function phenotype (see below), we named this gene bouncer (bncr) in reference to the colloquial name of a security guard at a bar. Although bouncer lacks any gene annotation in the newest zebrafish genome release (GRCz11), our RNA sequencing (RNA-seq) and in situ hybridization analyses (fig. S1A, B), ribosome profiling data (9–11), and cap analysis gene expression (CAGE)-based transcription start site analysis (12) suggested that bouncer is a maternal transcript that generates a mature 80-amino acid glycosylphosphatidylinositol (GPI)-anchored protein (Fig. 1A). Consistent with two predicted N-glycosylation sites (Fig. 1A), a Bouncer-specific antibody detected glycosylated Bouncer in the egg (Fig. 1B).
Figure 1. Identification of Bouncer in fish.
(A) Expression and genomic features of Bouncer. Coverage tracks for RNA sequencing, ribosome profiling (RPF) (10), and CAGE data (12) are shown. Genomic coordinates are based on GRCz10. SP, signal peptide; TM, transmembrane region; orange, predicted disulfide bonds; yellow, predicted N-glycosylation sites; turquoise, predicted transamidase cleavage site. (B) Endogenous Bouncer protein is glycosylated. Endogenous Bouncer is detected in the zebrafish egg by a Bouncer-specific antibody at a higher molecular weight than predicted (~20 kDa) but shifts down to the expected size (10 kDa) after treatment with deglycosylating enzymes. No Bouncer signal is detected in eggs from bncr-/- mutant females. (C) Protein sequence alignment of the mature domain of Bouncer/SPACA4 protein family members. Apart from the well conserved cysteines (orange denotes predicted disulfide bonds), Bouncer/SPACA4 shows high amino acid divergence among different species (% ID, percent sequence identity to the mature domain of zebrafish Bouncer). The extent of the mature domain displayed here is based on the prediction for zebrafish Bouncer. For all species for which expression data were available [(29–33): human expression based on GTEx Portal; expressed sequence tags based on NCBI], bouncer/Spaca4 RNA is restricted to either the male (symbol: sperm) or female (symbol: egg) germline. For sequences and accessions see data S1 and S2 and table S1. Amino acid abbreviations: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; Y, Tyr. (D and E) Lack of Bouncer causes near-sterility in female zebrafish. (D) Representative images of a developing, eight-cell stage embryo derived from a wild-type female, and an arrested, one-cell stage egg derived from a bncr-/- female 1.5 hours after mating. (E) Transgenically expressed, ubiquitin promoter driven untagged, GFP-tagged, and non-glycosylatable Bouncer rescue the mutant phenotype. Data are means ± SD; n = number of eggs. ***P < 0.0001 (Kruskal-Wallis test with Dunn multiple comparisons test); n.s., not significant.
A protein domain search classified Bouncer as a member of the Ly6/uPAR (Ly6/urokinase-type plasminogen activator receptor) protein superfamily, which includes proteins as diverse as toxins, immunoregulators, and cell surface receptors (13). This protein family is characterized by a 60- to 80-amino acid domain containing 8 to 10 highly conserved cysteines that form a three-finger structure (Fig. 1A, C; fig. S2A). Apart from the cysteines, other amino acids have diverged substantially within this protein superfamily (Fig. 1C; fig. S2A and B). BLASTP searches with zebrafish Bouncer and phylogenetic sequence analyses suggested that SPACA4 is the closest homolog in mammals, reptiles, and amphibians (Fig. 1C; fig. S2A-C; data S1 and 2 and table S1). Human SPACA4/SAMP14 (sperm acrosome membrane-associated protein 4/sperm acrosomal membrane protein 14) was originally identified in a proteomics study as a sperm acrosomal protein, and in vitro experiments implied a possible function in fertilization (14). However, the in vivo function and importance of SPACA4 are unknown.
Intrigued by our finding that zebrafish Bouncer is expressed in oocytes and that its closest homolog in humans was reported to be expressed in sperm (14), we analyzed the expression patterns of other Bouncer/SPACA4 homologs. We found that externally fertilizing vertebrates (e.g. fish and amphibians) show oocyte-restricted expression, whereas all internally fertilizing vertebrates analyzed (e.g. reptiles and mammals) show testis-specific expression (Fig. 1C; Fig. S2A). Together, these results identified Bouncer and SPACA4 as homologs with opposing sex-specific, germline-restricted expression patterns in externally versus internally fertilizing vertebrates.
To investigate the function of Bouncer, we used CRISPR/Cas9 to generate bouncer knockout zebrafish. We established a stable mutant line with a bouncer allele carrying a 13-nucleotide deletion, which abolishes the production of mature Bouncer protein (Fig. 1B; fig. S3A). Incrosses of bouncer heterozygous (bncr+/-) fish gave rise to homozygous mutant adults (bncr-/-) at a Mendelian ratio of ~25%, which suggests that Bouncer is not essential for development. However, in vivo mating experiments showed that only 7 of 3024 eggs (0.11%) derived from bncr-/- females developed into cleavage-stage embryos, as opposed to the majority of eggs from wild-type or bncr+/- females or from wild-type females fertilized by bncr-/- males (Fig. 1D, E; fig. S3A and B). Notably, female near-sterility was fully rescued by ubiquitous expression of transgenic untagged or green fluorescent protein (GFP)-tagged Bouncer (Fig. 1D, E; fig. S3A and C), which confirms that the observed defect was indeed due to the lack of Bouncer protein. Ubiquitous expression of a Bouncer mutant that cannot be glycosylated (GFP-BncrN32A,N84A; fig. S3A and D) also fully rescued female near-sterility (Fig. 1E); this finding demonstrates that glycosylation of Bouncer does not contribute to its function. Thus, oocyte-expressed Bouncer protein is necessary for efficient reproduction in zebrafish.
Zebrafish eggs are activated upon contact with spawning medium, independently of the presence of sperm. Egg activation appeared unaffected in eggs from bncr-/- females, as was evident by normal elevation of the chorion (the outer protective envelope of fish embryos), polar body extrusion, and cytoplasmic streaming (fig. S4A-C; movie S1). Moreover, the micropyle, an opening in the chorion that serves as the sole entry point for sperm into zebrafish eggs, is present in bncr-/- eggs, and its size is similar to that of wild-type eggs (fig. S4D). These results suggest that Bouncer is not required for egg activation and micropyle formation.
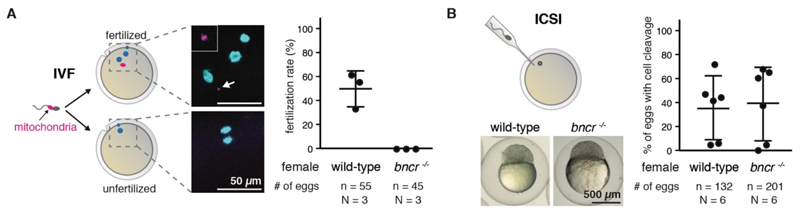
Because eggs from bncr-/- females lack any apparent morphological defects, yet do not develop beyond the one-cell stage, Bouncer might be required for fertilization and/or the initiation of early cleavage cycles. To distinguish between these possibilities, we first asked whether sperm can enter eggs lacking Bouncer. In vitro fertilization (IVF) of wild-type and Bouncer-deficient eggs with MitoTracker-labeled sperm allowed us to detect sperm only in wild-type eggs (50%), but never in Bouncer-deficient eggs (Fig. 2A), which suggests that Bouncer might play a role in sperm entry during fertilization. Consistent with the idea that Bouncer’s sole function is to allow sperm to enter the egg, delivery of sperm into Bouncer-deficient eggs by intra-cytoplasmic sperm injection (ICSI) bypassed the requirement for Bouncer and restored embryonic development beyond the one-cell stage (Fig. 2B and fig. S3B). Bouncer’s key function is therefore in enabling sperm entry during fertilization.
Figure 2. Bouncer is required for sperm entry into the egg.
(A) Sperm does not enter bncr-/- eggs. Left: Experimental setup. Wild-type sperm was stained with MitoTracker label and used for IVF of wild-type and bncr-/- eggs. Representative images are shown (arrow: MitoTracker signal, enlarged in white box). Right: Percentage of fertilized eggs, as indicated by the presence of one MitoTracker-labeled sperm, enlarged three DAPI signals (male nucleus, female nucleus, polar body). Means ± SD are indicated. (B) ICSI is able to rescue bncr-/- eggs. Top left: Experimental setup. Wild-type sperm was injected into wild-type or bncr-/- eggs. Cell cleavage was scored after 3 hours. Bottom left: Representative images. Right: Percentage of eggs that show cell cleavage. Means ± SD are indicated. n = total number of eggs; N = number of biological replicates.
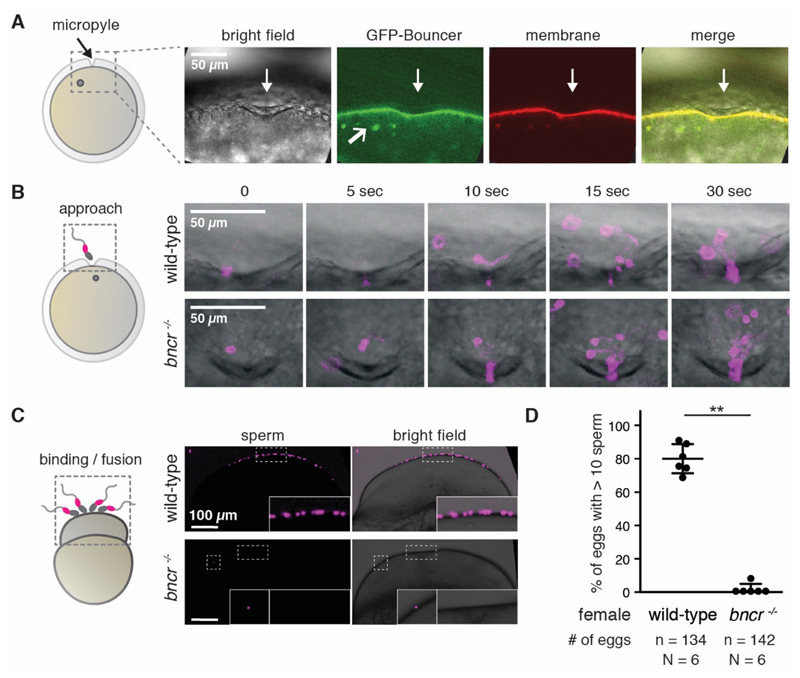
To gain further insight into Bouncer’s function, we assessed its localization. Consistent with its predicted GPI-anchorage, confocal imaging revealed that the fully functional, GFP-tagged Bouncer (Fig. 1E) localized to the egg membrane and to vesicles within the egg (Fig. 3A). Further, ubiquitous expression of a version of Bouncer lacking the C-terminal membrane anchor (GFP-BncrnoTM) did not rescue the near-sterility of bncr-/- females (fig. S3A and C), which suggests that membrane localization of Bouncer is required for its function.
Figure 3. Bouncer mediates binding between sperm and egg.
(A) Bouncer localizes to the egg membrane and to vesicles. Confocal images of eggs expressing GFP-tagged Bouncer (green) and lyn-Tomato (membrane, red) during egg activation show that Bouncer localizes to the egg membrane around the micropyle (downwards pointing white arrow) and to vesicles (angled white arrow in GFP-Bouncer panel). Gray circle, egg nucleus. (B) Bouncer is not required for sperm approach. Left: Experimental setup. Right: Representative time series of multiple MitoTracker-labeled wild-type sperm (magenta) approaching the micropyle area of wild-type (top) and bncr-/- (bottom) eggs. (C) bncr-/- eggs are impaired in sperm-egg binding. Left: Experimental setup. Activated and dechorionated wild-type and bncr-/- eggs were incubated with MitoTracker-labeled wild-type sperm and gently washed. Right: Representative images of a wild-type egg (top, scored as >10) and a bncr-/- egg (bottom) with a single bound sperm. Boxed areas are also shown at higher magnification. (D) Quantification of sperm-egg binding. Eggs were classified as either >10 sperm bound or <10 sperm bound. Data are means ± SD (P < 0.002, Mann-Whitney test; n = number of eggs; N = number of biological replicates).
The requirement for Bouncer at the egg membrane implies that it could promote the approach of sperm to the egg or sperm-egg binding/fusion. Live cell imaging revealed that multiple MitoTracker-labeled sperm are recruited to the micropyle independently of Bouncer (Fig. 3B; movie S2). Thus, Bouncer does not provide an essential attractive cue that guides sperm towards the egg/micropyle.
During live imaging, multiple sperm entered the narrow opening of the micropyle simultaneously, rendering a more detailed analysis of sperm-egg binding capability infeasible. To investigate Bouncer’s potential role in sperm-egg binding, we exposed the entire egg surface to sperm by removing the chorion. MitoTracker-labeled sperm remained bound to the surface of wild-type eggs in large clusters (>10 sperm) (Fig. 3C and D). In contrast, only a few individual sperm (<10) remained attached to the majority of Bouncer-deficient eggs (P < 0.002) (Fig. 3C and D). These results suggest that Bouncer promotes sperm-egg binding.
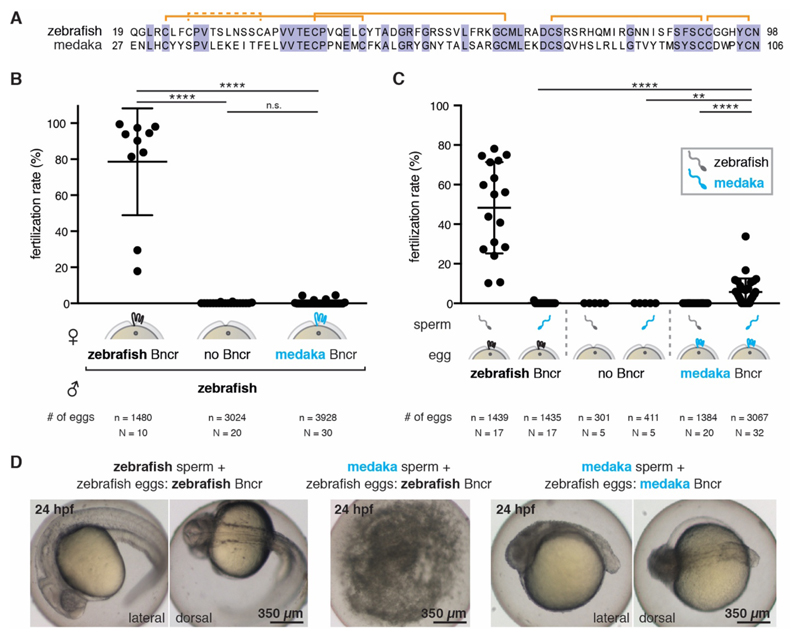
Bouncer shows a high degree of amino acid sequence divergence among different fish species, similar to other proteins involved in species-specificity in mammals (fig. S5A). This raised the interesting possibility that Bouncer might contribute to the species-specificity of fertilization in fish. To test this hypothesis, we generated bncr-/- zebrafish that ubiquitously express medaka Bouncer (bncr-/-; tg[ubi:medaka-bncr]), henceforth called transgenic medaka Bouncer fish. Medaka was chosen because of its large evolutionary distance from zebrafish (~200 million years), its inability to cross-hybridize with zebrafish, and the low (40%) amino acid identity between the mature zebrafish and medaka Bouncer proteins (Fig. 4A). Expression of medaka Bouncer in bncr-/- females did not efficiently rescue fertility when crossed to wild-type male zebrafish (average fertilization rate of 0.45%, versus 0.11% for bncr-/- and 78.6% for wild-type females) (Fig. 4B), supporting our hypothesis that Bouncer might influence species-specific gamete interaction. To directly test this possibility, we performed a series of IVF experiments. As expected, wild-type zebrafish eggs exhibited high fertilization rates with zebrafish sperm (average rate of 48.3%) but were not fertilized by medaka sperm (Fig. 4C). Moreover, zebrafish bncr-/- eggs were fertilized by neither sperm (Fig. 4C). Remarkably, eggs from transgenic medaka Bouncer females were fertilized by medaka, but not zebrafish, sperm (Fig. 4C), and eggs from females expressing both medaka Bouncer and zebrafish Bouncer could be fertilized by both sperm (fig. S5B). The average fertilization rate of all transgenic medaka Bouncer females (15) tested was 3.9% (fig. S5B). Whereas 6 of 15 females were infertile (average fertilization rate < 0.5%; fig. S5C), eggs from the remaining nine females were fertilized by medaka sperm at an average rate of 5.7% (Fig. 4C). Fertility rates of individual transgenic medaka Bouncer females were found to correlate with expression levels of medaka bouncer mRNA in eggs (fig. S5C), supporting a causal link between medaka Bouncer expression and medaka sperm entry. The resulting embryos were zebrafish-medaka hybrids and not haploid zebrafish embryos (fig. S5D). Hybrid embryos underwent cell cleavage and gastrulation (fig. S5E) and displayed anterior-posterior axis formation after 24 hours (Fig 4D; fig. S5F) but did not survive past 48 hours. These results demonstrate that Bouncer is necessary and sufficient for mediating species-specific fertilization in fish. Like a security guard, Bouncer is decisive for allowing conspecific sperm to enter, while keeping heterospecific sperm out. This is the first report of a protein in any organism allowing entry of another species’ sperm.
Figure 4. Bouncer mediates species-specific fertilization.
(A) Mature zebrafish and medaka Bouncer have only 40% amino acid sequence identity. Orange denotes predicted disulfide bonds; the dashed orange line denotes a disulfide bond predicted in zebrafish but not in medaka. (B) Medaka Bouncer does not efficiently rescue the fertilization defect of zebrafish bncr-/- females. Means ± SD are indicated [Kruskal-Wallis test with Dunn multiple-comparisons test: wild-type (wt) x wt versus bncr-/- x wt, adj. P**** < 0.0001; wt x wt versus medaka Bouncer x wt, adj. P**** < 0.0001; bncr-/- x wt versus medaka Bouncer x wt, n.s.; n = number of eggs; N = number of biological replicates]. (C) Medaka Bouncer is sufficient to allow entry of medaka sperm into zebrafish eggs. Medaka sperm did not fertilize wild-type zebrafish eggs, but medaka Bouncer expressing zebrafish eggs had an average fertilization rate of 5.7% in IVF experiments. Data are shown for the subset of medaka Bouncer expressing females (9 of 15) that were fertile (see fig. S5B for data of all 15 females tested). Data are means ± SD (Kruskal-Wallis test with Dunn multiple-comparisons test: medaka sperm on zebrafish versus medaka Bouncer eggs, adj. P**** < 0.0001; zebrafish sperm on medaka Bouncer eggs versus medaka sperm on medaka Bouncer eggs, adj. P**** < 0.0001; medaka sperm on bncr-/- zebrafish eggs versus medaka Bouncer eggs, adj. P** = 0.0052; n = number of eggs; N = number of biological replicates). (D) Fertilization of zebrafish eggs expressing only medaka Bouncer yields medaka-zebrafish hybrid embryos. Left: Wild-type zebrafish embryos fertilized by zebrafish sperm. Center: Wild-type zebrafish embryos are not fertilized by medaka sperm and decompose within 24 hours. Right: Zebrafish eggs expressing only medaka Bouncer are fertilized by medaka sperm and develop into hybrid embryos. hpf, hours post-fertilization.
Our finding that ectopic expression of another species’ Bouncer is sufficient to allow cross-species fertilization strongly suggests that Bouncer has a direct, species-specific interaction partner on sperm. Additionally, the low average fertilization rate (5.7%) of medaka Bouncer-expressing zebrafish eggs by medaka sperm implies that other factors likely contribute to species-specific sperm-egg interaction. The identification of these factors and Bouncer’s interaction partner on sperm will be crucial to unraveling the mechanism of species specificity of fertilization.
Thus far, the only known interacting membrane-bound proteins on vertebrate sperm and egg are IZUMO1 and JUNO in mammals (1–3, 7). Whether these two proteins also play a role in mediating species-specific fertilization in vivo is, however, still unclear (8). In many organisms, species-specificity of fertilization is mediated between proteins on the sperm membrane and those localized to the egg coat (15, 16). For example, in sea urchin, the vitelline envelope protein EBR1 (egg bindin receptor protein-1) binds specifically to the sperm membrane protein bindin (17–19). Similarly, the egg coat protein VERL of abalone is species-specifically bound by lysin, a small secreted protein from sperm (20–23). Whereas lysin has no known homolog in vertebrates, VERL shows structural homology to the mammalian zona pellucida protein ZP2 (22), which was shown to be involved in species-specific binding of sperm to the zona pellucida in mouse and humans (24). Furthermore, from the side of the sperm, the mammalian sperm acrosomal protein zonadhesin binds species-specifically to the zona pellucida, even though it is not required for fertility (25). In contrast to these proteins, Bouncer mediates species-specific binding of sperm to the egg membrane, not to the egg coat.
Bouncer and SPACA4, both members of the large Ly6/uPAR superfamily, have opposing germline-specific expression patterns in externally versus internally fertilizing organisms. The underlying reason for this is unclear, but one can speculate that in externally fertilizing species, oocyte expression of Bouncer contributes to post-copulatory female mate choice (also called cryptic female mate choice) (26). Vertebrates performing external fertilization cannot guarantee that only conspecific sperm reaches the egg by pre-copulatory mate choice (27, 28). Oocyte-expressed proteins such as Bouncer could therefore support the selection of conspecific sperm. Our work on Bouncer also raises the intriguing possibility that SPACA4 might play an important role in mammalian fertilization, albeit from the side of the male. Although a knockout for murine Spaca4 has not yet been reported, this idea is consistent with the localization of SPACA4 to the inner acrosomal membrane of sperm and the observed reduction of sperm-egg binding and fusion in vitro by incubation of sperm with antibody to SPACA4 (14). Future experiments that address the in vivo function of mammalian SPACA4 during fertilization will therefore be of interest. Given that both genes are restricted to the germline, our findings in fish may have direct relevance for fertilization in mammals.
Supplementary Material
Acknowledgements
We thank K. Tessmar-Raible and B.M. Fontinha for generously providing medaka fish and expertise; A. Schier for his generous support during the start of this project and for valuable feedback on the manuscript; J. Gagnon for his help in generating the bouncer mutant and obtaining germline RNA-seq data; M. Novatchkova and L.E. Cabrera Quio for help with RNA-seq data mapping and gene expression analyses; K. Panser for help with genotyping; the IMP animal facility personnel, especially J. König and F. Ecker, for their excellent care of our fish; P. Pasierbek from the BioOptics core facility for support in microscopy; T. Heuser and N. Fellner from the VBCF EM facility for help with EM; M. Madalinski for synthesizing Bouncer peptides for antibody production; J. Farrell for providing the sfGFP plasmid; C.-P. Heisenberg for providing the tg[lyn-tdTomato] fish line; the Pauli lab for discussions; the unidentified scientist at the 20. Anniversary Symposium of the Zebrafish Course at the Marine Biological Labs in Woods Hole, MA, for suggesting the name ‘Bouncer’ to us; and A. Anderson (Life Science Editors), A. Stark, C.-P. Heisenberg, L. Cochella, E. Tanaka, M. Ikawa, Y. Fujihara and B. Podbilewicz for helpful comments on the manuscript.
Funding
Supported by the Research Institute of Molecular Pathology (IMP), Boehringer Ingelheim, and the Austrian Academy of Sciences; a DOC Fellowship from the Austrian Academy of Sciences (S.H.); and the HFSP Career Development Award CDA00066/2015 and the FWF START program (A.P.).
Footnotes
Author contributions
S.H. and A.P. conceived the study; S.H. performed most experiments except experiments regarding species-specificity, which were performed by K.R.G., and generation of RNA-seq data and bncr-/- fish, which were performed by A.P; S.H., K.R.G., and A.P. analyzed the data. A.S. and A.P. performed the phylogenetic analysis; and S.H. and A.P. wrote the manuscript with input from K.R.G. and A.S.
Competing interests
The authors declare no competing interests.
Data and materials availability
RNA-seq data first reported here were deposited at the Gene Expression Omnibus (GEO) and are available under GEO acquisition number GSE111882. All other data are available in the manuscript or the supplementary material.
References
- 1.Inoue N, Ikawa M, Isotani A, Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- 2.Satouh Y, Inoue N, Ikawa M, Okabe M. Visualization of the moment of mouse sperm-egg fusion and dynamic localization of IZUMO1. J Cell Sci. 2012;125:4985–4990. doi: 10.1242/jcs.100867. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi E, Doe B, Goulding D, Wright GJ. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature. 2014;508:483–7. doi: 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaji K, et al. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat Genet. 2000;24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- 5.Miyado K, et al. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287:321–324. doi: 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- 6.Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287:319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- 7.Kato K, et al. Structural and functional insights into IZUMO1 recognition by JUNO in mammalian fertilization. Nat Commun. 2016;7:12198. doi: 10.1038/ncomms12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi E, Wright GJ. Cross-species fertilization: the hamster egg receptor, Juno, binds the human sperm ligand, Izumo1. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0101. 20140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pauli A, et al. Toddler: an embryonic signal that promotes cell movement via apelin receptors. Science. 2014;343 doi: 10.1126/science.1248636. 1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew G-L, et al. Ribosome profiling reveals resemblance between long non-coding RNAs and 5’ leaders of coding RNAs. Development. 2013;140:2828–2834. doi: 10.1242/dev.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chew G-L, Pauli A, Schier AF. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun. 2016;7:11663. doi: 10.1038/ncomms11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haberle V, et al. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature. 2014;507:381–385. doi: 10.1038/nature12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loughner CL, et al. Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Hum Genomics. 2016;10:10. doi: 10.1186/s40246-016-0074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shetty J, et al. SAMP14, a novel, acrosomal membrane-associated, glycosylphosphatidylinositol-anchored member of the Ly-6/Urokinase-type plasminogen activator receptor superfamily with a role in sperm-egg interaction. J Biol Chem. 2003;278:30506–30515. doi: 10.1074/jbc.M301713200. [DOI] [PubMed] [Google Scholar]
- 15.Wassarman PM, Litscher ES. A Bespoke Coat for Eggs: Getting Ready for Fertilization. ed.1. Vol. 117. Elsevier Inc.; 2016. [DOI] [PubMed] [Google Scholar]
- 16.Wassarman PM, Litscher ES. Mammalian fertilization: the egg’s multifunctional zona pellucida. Int J Dev Biol. 2008;52:665–676. doi: 10.1387/ijdb.072524pw. [DOI] [PubMed] [Google Scholar]
- 17.Stapper AP, Beerli P, Levitan DR. Assortative Mating Drives Linkage Disequilibrium between Sperm and Egg Recognition Protein Loci in the Sea Urchin Strongylocentrotus purpuratus. Mol Biol Evol. 2015;32:859–870. doi: 10.1093/molbev/msv010. [DOI] [PubMed] [Google Scholar]
- 18.Kamei N, Glabe CG. The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes Dev. 2003;17:2502–2507. doi: 10.1101/gad.1133003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vacquier VD, Moy GW. Isolation of Binding: The Protein Responsible for Adhesion of Sperm To Sea-Urchin Eggs. Proc Natl Acad Sci U S A. 1977;74:2456–2460. doi: 10.1073/pnas.74.6.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyon JD, Vacquier VD. Interspecies Chimeric Sperm Lysins Identify Regions Mediating Species-Specific Recognition of the Abalone Egg Vitelline Envelope. Dev Biol. 1999;214:151–159. doi: 10.1006/dbio.1999.9411. [DOI] [PubMed] [Google Scholar]
- 21.Swanson WJ, Vacquier VD. The abalone egg vitelline envelope receptor for sperm lysin is a giant multivalent molecule. Proc Natl Acad Sci U S A. 1997;94:6724–6729. doi: 10.1073/pnas.94.13.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raj I, et al. Structural Basis of Egg Coat-Sperm Recognition at Fertilization. Cell. 2017;169:1315–1326.e17. doi: 10.1016/j.cell.2017.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis CA, Talbot CF, Vacquier VD. A protein from abalone sperm dissolves the egg vitelline layer by a nonenzymatic mechanism. Dev Biol. 1982;92:227–239. doi: 10.1016/0012-1606(82)90167-1. [DOI] [PubMed] [Google Scholar]
- 24.Avella MA, Baibakov B, Dean J. A single domain of the ZP2 zona pellucida protein mediates gamete recognition in mice and humans. J Cell Biol. 2014;205:801–809. doi: 10.1083/jcb.201404025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tardif S, et al. Zonadhesin is essential for species specificity of sperm adhesion to the egg zona pellucida. J Biol Chem. 2010;285:24863–24870. doi: 10.1074/jbc.M110.123125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firman RC, Gasparini C, Manier MK, Pizzari T. Postmating Female Control: 20 Years of Cryptic Female Choice. Trends Ecol Evol. 2017;32:368–382. doi: 10.1016/j.tree.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levitan DR. Do Sperm Really Compete and Do Eggs Ever Have a Choice? Adult Distribution and Gamete Mixing Influence Sexual Selection, Sexual Conflict, and the Evolution of Gamete Recognition Proteins in the Sea. Am Nat. 2018;191:88–105. doi: 10.1086/694780. [DOI] [PubMed] [Google Scholar]
- 28.Levitan DR, Ferrell DL. Selection on Gamete Recognition proteins depends on sex, density, and genotype frequency. Science. 2006;312:267–270. doi: 10.1126/science.1122183. [DOI] [PubMed] [Google Scholar]
- 29.Li B, et al. A Comprehensive Mouse Transcriptomic BodyMap across 17 Tissues by RNA-seq. Sci Rep. 2017;7:4200. doi: 10.1038/s41598-017-04520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marin R, et al. Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage. Genome Res. 2017;27:1974–1987. doi: 10.1101/gr.223727.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasquier J, et al. Gene evolution and gene expression after whole genome duplication in fish: The PhyloFish database. BMC Genomics. 2016;17:368. doi: 10.1186/s12864-016-2709-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Session AM, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, et al. Comparative RNA-Seq analysis of differentially expressed genes in the testis and ovary of Takifugu rubripes. Comp Biochem Physiol - Part D Genomics Proteomics. 2017;22:50–57. doi: 10.1016/j.cbd.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7 doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen TN, Brunak S, Von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 37.Hofmann K, Stoffel W. A Database of Membrane Spanning Protein Segments. Biol Chem Hoppe-Seyler. 1993;374 [Google Scholar]
- 38.Eisenhaber B, Bork P, Eisenhaber F. Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng. 1998;11:1155–1161. doi: 10.1093/protein/11.12.1155. [DOI] [PubMed] [Google Scholar]
- 39.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 41.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 42.Wernersson R, Pedersen AG. RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2014;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minh BQ, Nguyen MAT, Von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gagnon JA, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 50.Westerfield M. The Zebrafish Book. A guide for the laboratory use of zebrafish (Danio rerio) Univ Oregon Press; Eugene: 2000. [Google Scholar]
- 51.Kamei Y, et al. Development of a convenient in vitro fertilization method using interspecific hybrids between Oryzias latipes and Oryzias curvinotus. Dev Growth Differ. 2007;49:721–730. doi: 10.1111/j.1440-169X.2007.00966.x. [DOI] [PubMed] [Google Scholar]
- 52.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nair S, Lindeman RE, Pelegri F. In vitro oocyte culture-based manipulation of zebrafish maternal genes. Dev Dyn. 2013;242:44–52. doi: 10.1002/dvdy.23894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meeker ND, Hutchinson SA, Ho L, Trede NS. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques. 2007;43:610–614. doi: 10.2144/000112619. [DOI] [PubMed] [Google Scholar]
- 55.Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:10–11. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pauli A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berthelot C, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5 doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.