Abstract
Cas9 (from S. pyogenes) in complex with a guide-RNA targets complementary DNA for cleavage. Here, we developed single molecule FRET analysis to study the mechanisms of specificity enhancement of two engineered Cas9s (eCas9 and Cas9-HF1). DNA unwinding assay showed that mismatches affect cleavage reactions through rebalancing the unwinding-rewinding equilibrium. Increasing PAM-distal mismatches facilitate rewinding, and the associated cleavage impairment shows that cleavage proceeds from the unwound state. Engineered Cas9s depopulate the unwound state more readily with mismatches. Intrinsic cleavage rate is much lower for engineered Cas9s, preventing cleavage from transiently unwound off-targets. Engineered Cas9s require about one additional base-pair match for stable binding, freeing them from sites that would otherwise sequester them. Therefore, engineered Cas9s achieve their improved specificity (1) by inhibiting stable DNA binding to partially matching sequences, (2) by making DNA unwinding more sensitive to mismatches, and (3) by slowing down intrinsic cleavage reaction.
In bacteria and archaea, CRISPR (clustered regularly interspaced short palindromic repeats)–Cas systems impart adaptive defense against phages and plasmid1. In type II CRISPR-Cas systems, the Cas9 endonuclease in complex with two RNAs, a CRISPR guide-RNA (crRNA) and a trans-activating RNA (tracrRNA), targets complementary 20 base pair (bp) sequences (protospacers) in foreign DNA for double-stranded cleavage, with a requirement that they be followed by a motif called PAM (protospacer adjacent motif, 5’-NGG-3’ for S. pyogenes Cas9)2–4. Programmable DNA binding and cleavage by Cas9 has revolutionized life sciences, where Cas9 cleavage is used for genome editing and Cas9 binding is used for tagging genomic sites with markers or effectors for wide-ranging applications5,6. Minimizing off-target effects of both binding and cleavage7,8 remains an active area of study. In vitro and in vivo investigations have shown that Cas9-RNA specificity is most affected by PAM plus an 8–10 PAM-proximal seed region and concentrations of both Cas9 and RNA7,9. Rationally designed and engineered S. Pyogenes Cas9s (EngCas9s), namely enhanced Cas9 (eCas9; K848A/K1003A/R1060A mutations) and high fidelity Cas9 (Cas9-HF1; N497A/R661A/Q695A/Q926A mutations), led to remarkable specificity improvements in unbiased genome wide CRISPR-Cas9 specificity measurements10,11.
The stability of Cas9-RNA-DNA complex is in part a function of the specific RNA-DNA base-pairing and sequence-independent interactions between Cas9 residues and DNA, the latter being a potential source of promiscuity. Cas9-RNA uses binding energy to initially melt DNA near PAM, and DNA unwinding can continue downstream in the presence of sequence complementary with guide-RNA. The motivation behind the mutations in EngCas9s was to destabilize the Cas9-RNA-DNA complex by diminishing the favorable sequence-independent interactions between Cas9 residues and the DNA backbone (Fig. 1b). Single molecule FRET (Fluorescence Resonance Energy Transfer) measurements showed that Cas9 conformational changes that bring a nuclease domain to the cleavage site can be disrupted by fewer mismatches for the EngCas9, leading to the proposal that the nuclease movement is the conformational checkpoint against off-target cleavage12,13. Here, we report a comparative analysis of DNA interrogation and rejection, DNA unwinding and rewinding dynamics, and cleavage activation among three Cas9s (wild type, eCas9 and Cas9-HF1) using single molecule imaging methods that can detect multiple conformations and transient intermediates14 and have been used previously to study CRISPR systems in vitro9,12,13,15–21. We found evidence that DNA unwinding is the checkpoint that guard against off-target cleavage and could determine the equilibrium between unwinding and rewinding that allowed us to estimate the intrinsic cleavage rate from the unwound state. Based on our findings, we propose that engineered Cas9s achieve their improved specificity (1) by inhibiting stable DNA binding to partially matching sequences, (2) by making DNA unwinding more sensitive to mismatches, and (3) by slowing down intrinsic cleavage reaction.
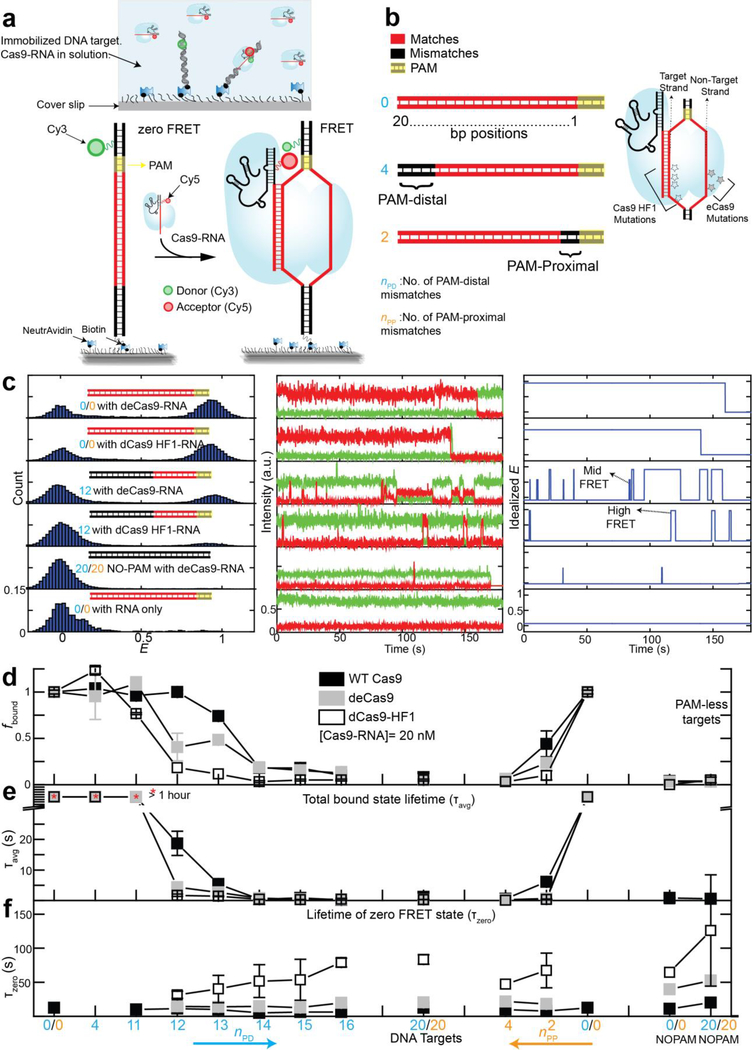
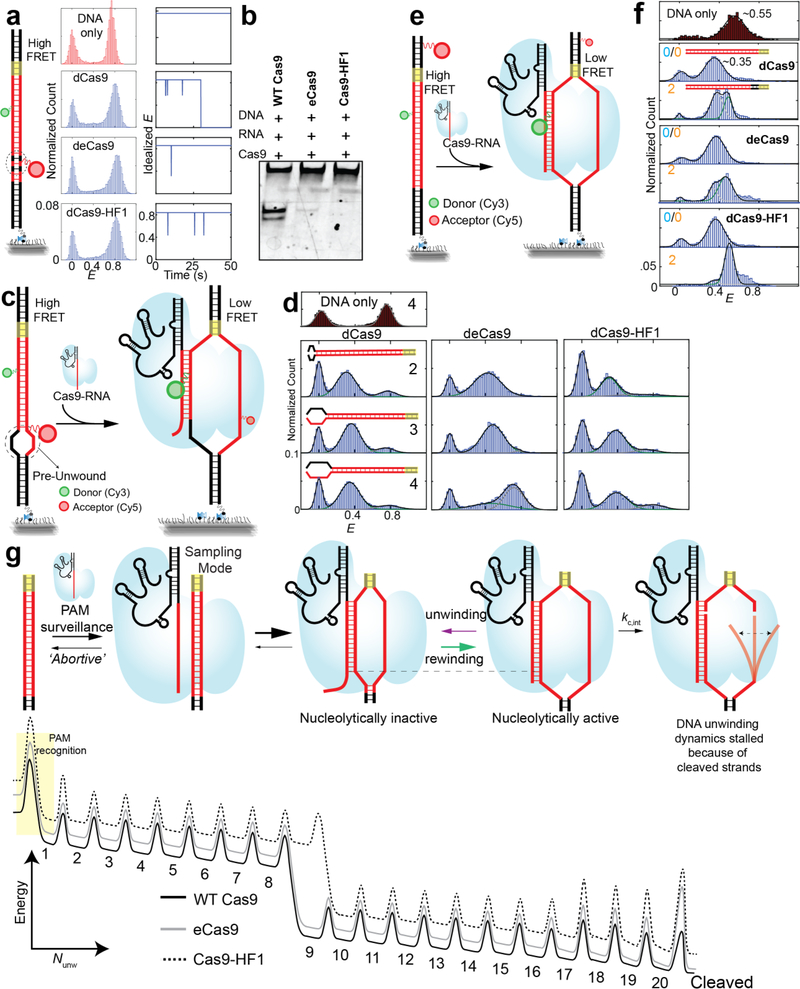
Figure 1. smFRET assay to study DNA interrogation by engineered Cas9-RNA.
(a) Schematic of smFRET assay. Cas9 in complex with an acceptor labeled guide-RNA binds a donorlabeled cognate DNA target. (b) DNA targets with mismatches in the protospacer region against the guide-RNA. The number of mismatches PAM-distal (nPD) and PAM-proximal (nPP) are shown in cyan and orange, respectively. Also shown are locations of EngCas9 mutations in dCas9-RNA-DNA complex (PDB ID: 4UN3). (c) E histograms (left) at 20 nM EngCas9-RNA or RNA only. Representative single molecule intensity time traces of donor (green) and acceptor (red) are shown (middle), along with E values idealized (right) by hidden Markov modeling. (d) Normalized fraction (fbound) of DNA molecules bound with Cas9-RNA at 20 nM Cas9-RNA. Fractions were normalized relative to the bound fraction of cognate DNA target. (e) τavg, obtained from dwell times of E >0.2 states. (f) Unbound state lifetimes at [Cas9RNA] = 20 nM. The mean of τavg over various DNA targets was used to calculate kon. Number of PAMdistal (nPD) and PAM-proximal mismatches (nPP) are shown in cyan and orange respectively. Error bars represent standard deviation (s.d.) from n= 2 or 3. Data for WT Cas9-RNA is taken from a previous study9.
Results
Real-time single molecule FRET assay for DNA interrogation
We employed a single-molecule FRET (smFRET) assay22 to investigate DNA interrogation by EngCas9-RNA. DNA targets (donor-labeled, 82 bp long) were immobilized on polyethylene glycol (PEG)-passivated flow chambers and EngCas9 pre-complexed with acceptor-labeled guide-RNA (EngCas9-RNA) was added to observe their interactions in real time. The guide-RNA was generated by hybridizing an acceptorlabeled crRNA to a tracrRNA. Locations of donor (Cy3) and acceptor (Cy5) were chosen such that specific interaction between DNA target and EngCas9-RNA would lead to high FRET efficiency (Fig. 1a and Supplementary Fig. 1). Labeling at these locations do not affect Cas9 cleavage activity9. Unless mentioned otherwise, we used catalytically dead versions of all Cas9s (denoted with the prefix d) in order to focus on the properties prior to cleavage. Wild-type (WT) Cas9 and dCas9 showed similar behavior in DNA interrogation experiments9.
We examined different DNA targets containing mismatches relative to the guide-RNA. As the naming convention, we used nPD (the number of PAM-distal mismatches) and nPP (the number of PAM-proximal mismatches) (Fig. 1b). The cognate DNA target gave two distinct populations centered at FRET efficiency (E) of 0.9 and 0 in the presence of 20 nM EngCas9-RNA in solution. The E=0.9 population was negligible if only the labeled RNA was added without Cas9 or if a fully mismatched DNA without PAM was used. Therefore, we assigned the E=0.9 species to a sequence-specific EngCas9-RNA-DNA complex (Fig. 1c). The E=0 population is a combination of unbound states and bound states with inactive or missing acceptor. Cas9-RNA titration gave the apparent dissociation constant (Kd) of 0.50 nM (dCas9), 0.5 nM (deCas9) and 2.7 nM (dCas9-HF1) (Supplementary Fig. 1).
To quantify the impact of mismatches, we determined the apparent Cas9-RNA bound fraction fbound, defined as the normalized fraction of DNA molecules with E > 0.75 (20 nM Cas9-RNA) (Fig. 1d and Supplementary Fig. 2). (i) fbound remained unchanged when nPD increased from 0 to 10 or 11, but precipitously decreased with additional PAM-distal mismatches, (ii) even very few PAM-proximal mismatches, nPP of 2 or 4, caused a > 50 or > 95 % drop in fbound, respectively, (iii) binding is ultrastable with >8–9 PAM-proximal matches because fbound remained high even one hour after washing away free Cas9-RNA (Supplementary Fig. 3).
With DNA targets with too many mismatches to bind Cas9 stably, smFRET time traces showed repetitive transitions between E = 0 and E ~ 0.45 states in addition to transitions between E = 0 and E = 0.9, suggesting that there are multiple bound states distinguishable based on E 9 (Fig. 1c). We used a hidden Markov modeling analysis23 to determine τavg as a fraction-weighted average of the high (E = 0.9) and mid (E ~ 0.45) FRET state lifetimes. τavg was over one hour for DNA targets with at least m PAM-proximal matches (m=9 for WT Cas9 and deCas9, 10 for dCas9-HF1) but decreased to 0.5 – 15 s with fewer PAMproximal matches or any PAM-proximal mismatches (Fig. 1e). Dwell times of the unbound state (E < 0.2) were only weakly dependent on sequence (Fig. 1f) (Supplementary Fig. 3)
We list below qualitative features common between EngCas9s and WT Cas9 as well as quantitative differences:
(1) All Cas9s interrogate and bind DNA in two distinct modes. Sequence-independent sampling of DNA target in search of PAM results in transient mid FRET events (Fig. 1c) as reported previously for WT Cas99. The high FRET state results upon PAM detection and RNA-DNA heteroduplex formation, and its lifetime increases with increasing base-pairing between guide-RNA and DNA target. (2) The binding frequency is independent of sequence. The bimolecular association rate constant kon decreased for EngCas9s (2.9×106 M−1 s−1 for deCas9 and 1×106 M−1 s−1 for dCas9-HF1, compared to 5.4×106 M−1 s−1 for WT Cas9), possibly as a result of alteration in electrostatic and polar interaction upon removal of positively charged and polar residues (Fig. 1f). (3) PAM-proximal mismatches are much more deleterious for stable binding compared to PAM-distal mismatches, and mismatches in the middle of the protospacer renders PAM-distal matches inconsequential (Supplementary Fig. 2)9, suggesting that all Cas9s extend RNA-DNA heteroduplex unidirectionally from PAM-proximal to PAM-distal end. (4) PAM-proximal mismatches are more deleterious for EngCas9 binding than WT Cas9, and (5) deCas9 and WT Cas99,24 require 9 PAMproximal matches for ultrastable (> 1 hour) binding whereas dCas9-HF1 requires 10 (Fig. 1e, Supplementary Fig. 3).
Overall, EngCas9 are similar to WT Cas9 in their sequence-dependent binding properties except for the higher ability to reject both PAM-proximal and PAM-distal mismatches which may help reduce Cas9-RNA sequestration by partially matching targets.
Cas9-RNA induced DNA unwinding
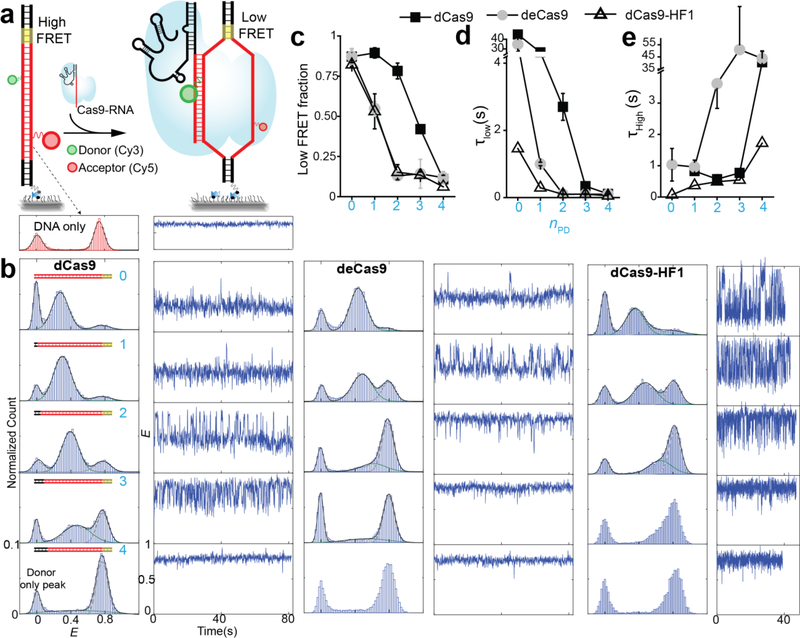
Genome wide characterization of EngCas9 cleavage specificity showed marked improvements over WT Cas910,11. Yet, our binding study shows that the EngCas9s still stably bind to DNA with nPD as large as 10. We reasoned that EngCas9s may differ from WT Cas9 in their mismatch dependence of DNA unwinding which, if promoted by annealing to guide-RNA, may be a determinant of cleavage action25. Therefore, we probed the internal unwinding of PAM-distal region of the protospacer by labeling the target and non-target strands with a donor and an acceptor, respectively, with 9 bp spacing (Supplementary Fig. 4). Labeling at these locations did not perturb cleavage or binding (Supplementary Fig. 4). The DNA by itself showed high FRET (E ~ 0.75) (Fig. 2a-b), and upon addition of 100 nM WT dCas9-RNA to cognate DNA, we observed a shift to a stable low FRET state (E ~ 0.30), likely because of DNA unwinding (Fig. 2a-b). A similar FRET change was observed when the locations of the donor and acceptor were swapped (Supplementary Fig. 4). A DNA with nPD = 1 showed a similarly stable unwinding but with a slightly higher E of ~ 0.35 and occasional short-lived transitions to the high FRET state, likely due to one bp fewer unwinding and increased frequency to rewind, respectively. The trend continued upon increasing nPD to 2 and 3; E value for the unwound state increased, and the relative population of the rewound state increased. With nPD = 4, the unwound state is rarely populated. deCas9 and dCas9-HF1 showed a similar behavior but their unwound state population and lifetime were smaller and decreased more quickly with increasing nPD (Fig. 2b). Therefore, EngCas9s are more sensitive to PAM-distal mismatches in their ability to unwind DNA. The rewound state must still have at least 8 PAM-proximal bp unwound because we did not see stable binding with fewer PAM-proximal matches (Supplementary Fig. 3), and can have up to 16 PAM-proximal bp unwound.
Figure 2. Internal DNA unwinding-rewinding dynamics modulated by mismatches and Cas9 mutations.
(a) Schematic of smFRET assay for Cas9-RNA induced unwinding of surface-tethered DNA. (b) E histograms for nPD = 0, 1, 2, 3 and 4 (cyan numbering) and representative time traces. (c-e) Relative population of the unwound state, average lifetime of the unwound state and average lifetime of the rewound state vs. nPD. Error bars represent standard deviation (s.d.) from n=2. n=1 in absence of error bar.
We observed two distinct E populations even at a higher time resolution (35 ms) (Supplementary Fig. 5), indicating that Cas9-RNA induces primarily two states, unwound and rewound, without spending appreciable time in between. We used the Hidden Markov modeling analysis to segment single molecule time traces into two states (Supplementary Fig. 6). All Cas9s showed a reduction in the relative population and lifetime of the unwound state with increasing nPD (Fig. 2c-e). Combined with the gradual increase of E values for the unwound state (Fig. 2b), we can conclude that PAM-distal mismatches reduce the time spent in the unwound state in addition to reducing the maximal extent of unwinding. For a given nPD, EngCas9s showed lower occupancy and shorter lifetime of the unwound state (Fig. 2c-e), suggesting that their mutations destabilize the maximally unwound states. We observed a similar unwinding behavior from catalytically active Cas9 but E distribution was broader and state transitions were less frequent (Supplementary Fig. 6), suggesting a change in unwinding dynamics after cleavage, possibly due to disordered non-target strand25,26.
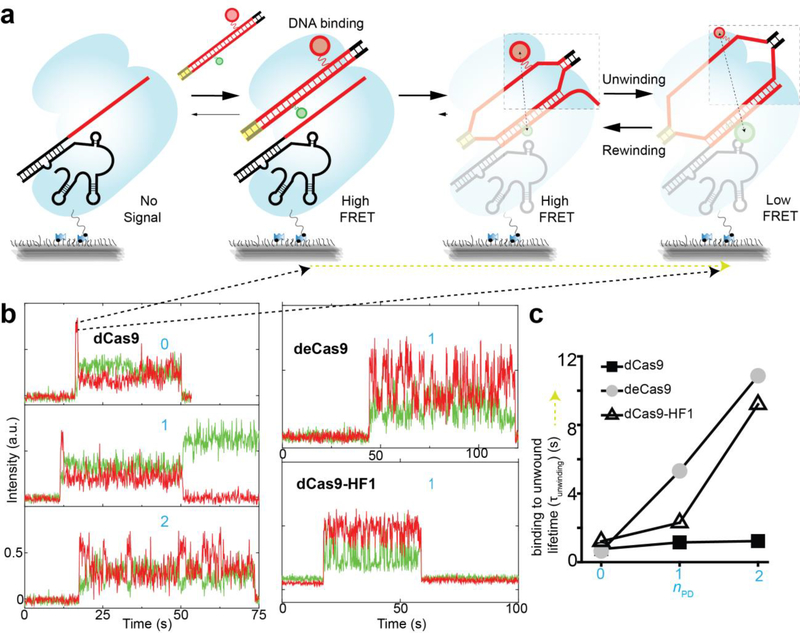
In order to capture the initial DNA unwinding event, we added labeled DNA to surface-immobilized Cas9-RNA molecules (Fig. 3a) (Supplementary Fig. 7). DNA binding is detected as a sudden appearance of fluorescence signal in a high E state, followed by a single step change to a low E (unwound state) (Fig. 3b). Subsequently, we observed unwinding-rewinding dynamics similar to what we observed in the steady state (Fig. 2b) (Supplementary Fig. 7). The average dwell time of the initial high E state which we attribute to the time it takes to unwind DNA for the first time (τunwinding) remained constant (~1 s) for dCas9 when nPD changed from 0 to 2. In contrast, τunwinding increased ~9 folds for EngCas9s when nPD changed from 0 to 2 (Fig. 3c), suggesting that EngCas9s take longer to unwind in the presence of PAMdistal matches.
Figure 3. smFRET assay to investigate initial DNA unwinding upon binding of DNA to Cas9-RNA.
(a) Schematic of smFRET assay for DNA unwinding by surface-tethered Cas9-RNA. (b) Representative single molecule fluorescence intensity time traces of donor (green) and acceptor (red) show abrupt signal increase upon DNA binding followed by FRET changes. (c) Time taken to go from initial binding to its first unwound state configuration (τunwinding) as denoted by a dark yellow line (a). Error bars represent standard deviation (s.d.) from n=2. n=1 in absence of error bar.
DNA cleavage vs. mismatches
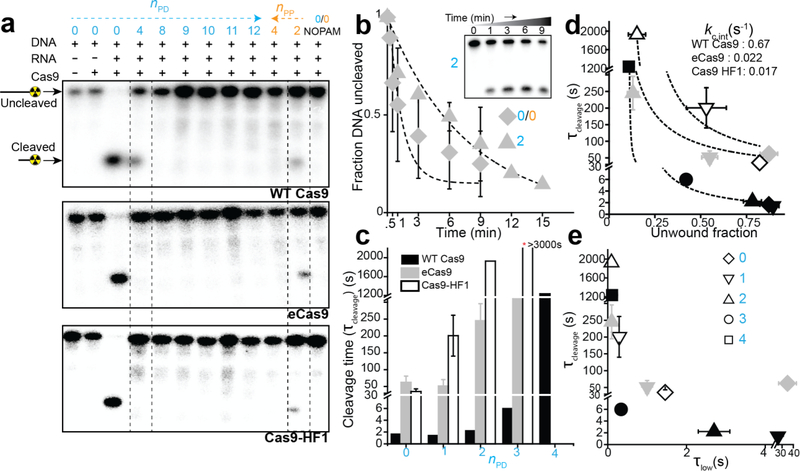
Next, we probed how mismatches influence target cleavage through modulating DNA unwindingrewinding dynamics. Cleavage was more specific for EngCas9s because they cleaved only up to nPD = 3 compared to nPD = 4 for WT Cas9 (Fig. 4a & Supplementary Fig. 4). EngCas9s did not release cleavage products under physiological conditions (Supplementary Fig. 3), as was the case for WT Cas99. In terms of the cleavage reaction, EngCas9s were much slower than WT Cas9 for cognate or PAM-distal mismatched DNA (Fig. 4b-c) but were faster for PAM-proximal mismatched DNA (Fig. 4a). Overall, we observed a clear correlation between the cleavage time scale, τcleavage, and DNA unwinding signatures; τcleavage decreased with increasing population (Fig. 4d) and lifetime (Fig. 4e) of the unwound state. Therefore, cleavage must proceed from the unwound state.
Figure 4. Cleavage vs mismatches and its relation with DNA unwinding.
(a) Cas9-RNA induced cleavage pattern analyzed by 15% denaturing polyacrylamide gel electrophoresis of target strand 5’-labeled with 32-P. Dashed boxes highlight DNA targets with nPD =4 and nPP =2 that show differences between EngCas9s and WT Cas9. (b) Fraction of uncleaved DNA vs time for eCas9 as a function of time and single exponential decay fits to obtain cleavage time τcleavage. Inset shows a representative gel image. [DNA] = 1 nM. [Cas9-RNA] = 100 nM. (c) τcleavage vs nPD. Data for WT Cas9-RNA is taken from a previous study33. (d) τcleavage vs unwound fraction as shown in Fig. 2c. Fits are made to determine kc,int (see methods). (e) τcleavage vs. unwound state lifetime as shown in Fig 2d. nPD and nPP are shown in cyan and orange, respectively. Error bars represent standard deviation (s.d.) from n=2 or 3. n=1 in the absence of error bar.
HNH and RuvC nuclease domains cleave target and non-target strand respectively. Additional 3’−5’ exonuclease activity of RuvC3,25,27, which normally causes multiple bands of cleaved non-target strand, decreased with increasing nPD, and more efficiently so for EngCas9s (Supplementary Fig. 4).
Armed with the kinetic information on DNA binding, interrogation, unwinding-rewinding and cleavage, we can clarify in which reaction steps sequence mismatches and Cas9 mutations exert their influence. Because cleavage likely occurs from the unwound state, we estimated the lower limits of intrinsic rate of cleavage (kc,int) from the apparent cleavage time τcleavage and unwound fraction by fitting τcleavage vs unwound fraction using τcleavage = 1/([unwound fraction]*kc,int + C) where C is a constant. Because a single value of kc,int was able to fit the data for each Cas9, we conclude that changes in τcleavage caused by mismatches can be attributed to changes in the unwound fraction (Fig. 3d). Therefore, although DNA molecules with different nPD values show different extents of unwinding, their unwound states have similar intrinsic cleavage rates. kc,int was ~0.67 s−1 for WT Cas9 but decreased to 0.025–0.015 s−1 for EngCas9s.
Inserting two single bp mismatches in the PAM-distal region gave a predominantly rewound state with only brief excursions to the unwound state for all Cas9s (Fig. 5a and Supplementary Fig. 8), indicating that there is little difference in unwound fraction among them. Nevertheless, WT Cas9 cleaved the DNA whereas EngCas9s did not, further confirming the much higher kc,int for WT Cas9 which in turn allows cleavage even from a transiently populated unwound state, likely contributing to higher off-target cleavage yields (Fig. 5b).
Figure 5. Cas9-RNA induced unwinding of various DNA and mechanisms of increased specificity by EngCas9s.
(a) Unwinding and cleavage of DNA with internal single base mismatches at positions 16th and 18th. E histograms were obtained at [Cas9-RNA] =100 nM or in its absence. Idealized E traces show transient unwinding observed in a subset of molecules. E drop to 0 for dCas9 is due to acceptor photobleaching. (b) Gel image of non-target strand shows that WT Cas9, but not EngCas9s, cleave the DNA. (c) Schematics of Cas9-RNA induced unwinding of pre-unwound DNA. (d) E histograms obtained at [Cas9-RNA]=100 nM or in its absence. Black numbers denote the number of base pairs pre-unwound and are mismatched. (e) Schematics of Cas9-RNA induced unwinding of PAM-proximal region. (f) E histograms obtained at [Cas9-RNA] =100 nM or in its absence. nPP is shown in orange digits. (g) Proposed model of DNA targeting, unwinding, rewinding, and cleavage (top). Energy diagram as a function of Nunw, the number of unwound base pairs.
Mechanism of mismatch sensitivity of EngCas9s
The changing balance between unwinding and rewinding caused by mismatches reflect the energetic competition between target strand annealing with RNA vs with non-target strand. Mismatches disrupt RNA-DNA duplex and help recover the parental duplex through rewinding. Mutations in EngCas9s further promote rewinding, likely contributing to improved cleavage specificity. In order to gain insights into how EngCas9s more readily rewind in the presence of mismatches, we disrupted the parental DNA duplex by pre-unwinding the PAM-distal mismatched portion (Fig. 5c). Pre-unwinding shifted the balance toward unwinding for WT Cas9 and Cas9-HF1 even with nPD = 4, but not for eCas9 (Fig. 5d & Supplementary Fig. 9), likely because the residues that help sequester the unwound non-target strand are mutated in eCas9 but not in Cas9-HF1 (Fig. 1b & Supplementary Fig. 9).
PAM-proximal DNA unwinding
To investigate if EngCas9-RNA mutations also affect PAM-proximal DNA unwinding we designed a DNA construct with a donor and an acceptor at the PAM proximal site separated by 12 bp (Fig. 5e & Supplementary Fig. 9). Cas9-RNA binding to cognate DNA shifted E from 0.55 to 0.35 for all Cas9s, indicating a stable DNA unwinding at PAM-proximal site. With nPP = 2, the fraction of unwound population decreased to 50 % for dCas9, 40 % for deCas9 and < 5 % for dCas9-HF1 (Fig. 5f), mirroring the decreases in fbound. However, this decrease did not correlate with cleavage efficiency of DNA target with nPP = 2, where EngCas9s were more efficient than WT Cas9 (Fig. 4a). Therefore, EngCas9s can cleave DNA better once the initial barrier caused by PAM-proximal mismatches is overcome.
Discussion
Like WT Cas9, EngCas9s also require only 9–10 PAM-proximal matches for ultrastable binding and thus will likely not confer significant specificity advances for applications utilizing Cas9 binding, for example transcription regulation28 or imaging29. EngCas9s, however, do show a modest increase in binding specificity. For example, Cas9-HF1 requires one additional basepair match for ultrastable binding compared to WT Cas9. Ultrastable binding to partially matching sequences can sequester Cas9-RNA, potentially limiting the speed of genome editing9. To overcome sequestration, one would need higher Cas9-RNA concentrations which may in turn lead to an increase in off-target cleavage. One base pair difference would mean about four-fold reduction in such off-target sinks for Cas9-HF1, and even such a modest improvement in binding specificity may improve cleavage specificity by reducing the total concentration of Cas9-RNA required.
Activation and dynamics of proofreading (REC2) and HNH domains in response to PAM-distal mismatches and Cas9 mutations has been studied using single molecule FRET 12,13. But what changes in the DNA target conformation trigger these movements of Cas9 domains were not known. Cas9’s cleavage activity requires many more base pair matches than stable binding3,4 and we found that DNA unwinding, instead of DNA binding, is strongly correlated with the cleavage rate when we vary the DNA sequence or Cas9 mutations. Unwound DNA configuration is likely verified by REC3 domain and linker connecting HNH and RuvC nuclease domains in Cas913,30, guiding HNH nuclease movement for cleavage. Here, we show that PAM-distal mismatches impair DNA unwinding, consequently this also results in impairments in REC2 (coupled to REC3) proofreading and HNH movements12,13. In fact, Chen et al showed that the cleavage active conformation is more readily depopulated with mismatches for EngCas9s compared to WT Cas9, raising the possibility that HNH movement and DNA unwinding are concurrent13.To test this possibility, we utilized the observation that HNH movement requires divalent cations and is inhibited by EDTA12. We found that DNA unwinding occurs even in the absence of divalent cations (Supplementary Fig. 10), suggesting that DNA unwinding occurs before HNH movement and not with or because of the HNH movement as had been suggested in a simulation study31. All these results suggest that DNA unwinding is the critical step that verifies sequence matching, which then subsequently triggers a series of steps toward cleavage including HNH movement. The unwound state (mid FRET state) can likely be a time-averaged ensemble of degenerate states, which can be uncovered in experiments with higher spatio-temporal resolution and/or with additional fluorescent geometries. ~15 % of Cas9-RNA-DNA underwent no unwinding dynamics, consistent with 15 % of Cas9-RNA-DNA that exist in cleavage-incompetent state31 (Supplementary Fig. 10).
The impact of PAM-distal matches in opposing DNA unwinding is even greater for EngCas9 because their mutations destabilize the unwound DNA configuration, and because cleavage likely proceeds from the unwound state, the increased sensitivity to mismatches in DNA unwinding is likely to contribute to the increased specificity of cleavage by EngCas9s. Any perturbation that destabilizes unwound DNA or fully extended R-loop will delay or fully impair the cleavage action and is also likely the reason why Cas9 with truncated guide-RNAs are more specific in cleavage32.
DNA targets with only a few interspersed single base-pair mismatches at PAM-distal site can prevent stably unwound states by all Cas9 but even transiently unwound states appear to allow cleavage by WT Cas9, but not by EngCas9s which we showed here to have much lower intrinsic cleavage rate. Therefore, the large enhancement in cleavage specificity for EngCas9s may in part be due to the lower intrinsic cleavage rate that prevents cleavage from transiently unwound states that are populated in partially matching sequences.
Fig. 5g summarizes our findings on the three possible mechanisms of specificity increase for EngCas9 in the form of free energy diagram along the axis of Nunw, the number of bp unwound, leading toward cleavage. Once the initial barrier of PAM detection is overcome, DNA unwinding proceeds in the direction from PAM-proximal to PAM-distal. There is a sudden drop in energy at Nunw=9 for WT Cas9 and eCas9, marking a threshold for stable binding. Because the threshold is shifted to Nunw=10 for Cas9-HF1, Cas9-HF1 can achieve higher cleavage specificity by reducing the number of off-target sites which can otherwise sequester Cas9-RNA (Mechanism 1). EngCas9s also tilt the energetic balance toward rewinding in the PAM-distal region such that PAM-distal mismatches readily depopulate the fully unwound state needed for cleavage (Mechanism 2). Finally, EngCas9s have lower intrinsic cleavage rates from the unwound state, represented as higher energetic barrier against cleavage, preventing cleavage from transiently unwound off-targets (Mechanism 3).
EngCas9s still cleave DNA targets with ~3 PAM-distal mismatches thus there is room for further improvement. Cas9-HF1 and eCas9 mutations eliminated some of the favorable sequence-independent interactions for target and non-target strands separately and may act in an independent manner and thus, their mutations may be combined. EngCas9 mutations may also be combined with truncated RNA to improve the specificity even further. But, combining a number of such strategies that destabilize unwound state and decrease the intrinsic cleavage rate from the unwound state as we showed here could lead to substantially reduced on-target cleavage as well, as has been observed when Cas9-HF1 was used with the truncated RNA10.
METHODS
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of the paper.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
ONLINE METHODS
Preparation of DNA targets.
DNA targets used in smFRET assay for DNA interrogation by Cas9-RNA are the same as what were used previously for WT Cas9 studies9 and Supplementary Figure 1 shows the overall schematics. The schematic of DNA targets used for smFRET assay of Cas9-RNA induced DNA unwinding is shown in Supplementary Figure 4, 7, 10. All DNA oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA 52241). A thymine modified with an amine group through a C6 linker (amino-dT) was used to label DNA with Cy3 or Cy5 N-hydroxysuccinimido (NHS). Non-target strand, target strand and a 22 nt biotinylated adaptor strand were assembled by mixing them in buffered solution with 10 mM Tris-HCl, pH 8 and 50 mM NaCl and heating to 90 °C followed by cooling to room temperature over 3 hrs. For DNA unwinding by surface-tethered Cas9-RNA, the biotin adaptor strand was omitted. Full sequence and modifications of DNA targets used in smFRET assays for DNA interrogation and DNA unwinding are shown in Supplementary Table 1 and Supplementary Table 2, respectively. For radio-labeled gel electrophoresis cleavage experiments, the target strand was phosphorylated with P32 using T4 polynucleotide kinase reaction and was annealed with the non-target strand as described above. Sequences are the same as in Supplementary Table 1 and Supplementary Table 2 but without the biotinylated adaptor strand. For fluorescently-labeled gel electrophoresis cleavage experiments, the DNA targets used are the same as those used in smFRET assays.
Expression and purification of Cas9.
All Cas9 were expressed and purified as described previously2,34. A pET-based expression vector was used for protein expression which consisted of sequence encoding Cas9 (Cas9 residues 1–1368 from S. pyogenes) and an N-terminal decahistidine-maltose binding protein (His10-MBP) tag, followed by a peptide sequence containing a tobacco etch virus (TEV) protease cleavage site.
Mutations for cleavage impairment, enhanced mutations (eCas9), HF1 mutations or the desired combinations of them were introduced by site-directed mutagenesis (QuickChange Lightning; Agilent Technologies, Santa Clara, CA 95050). Proteins were expressed in E. coli strain BL21 Rosetta 2 (DE3) (EMD Biosciences), grown in TB (Terrific Broth) or 2YT medium (higher expression obtained for TB) at 37 °C for a few hours. When the optical density at 600 nm (OD600) reached 0.6, protein expression was induced with 0.5 mM IPTG and the temperature was lowered to 18 °C. The induction was then continued for 12–16 h. The medium was then discarded and cells were harvested. The harvested cells were lysed in 50 mM Tris pH 7.5, 500 mM NaCl, 5% glycerol, 1 mM TCEP, supplemented with protease inhibitor cocktail (Roche) and with/without Lysozyme (Sigma Aldrich), and then homogenized in a microfluidizer (Avestin) or homogenized with Fisher Model 500 Sonic Dismembrator (Thermo Fisher Scientific) at 30% amplitude in 3 one minute cycles, each consisting of series of 2 s sonicate-2 s repetitions. The lysed solution was then ultra-centrifuged at 15,000 g for 30–45 minutes, supernatant of lysate was collected and cellular debris was discarded. The supernatant was added to Ni-NTA agarose resin (Qiagen). The resin was washed extensively with 50 mM Tris pH 7.5, 500 mM NaCl, 10 mM imidazole, 5% glycerol, 1 mM TCEP and the bound protein was eluted in a single-step with 50 mM Tris pH 7.5, 500 mM NaCl, 300 mM imidazole, 5% glycerol, 1 mM TCEP. Dialysis of Cas9 into Buffer A (20 mM Tris-Cl pH 7.5, 125 mM KCl, 5% glycerol, 1 mM TCEP) and cleavage of TEV-protease site by TEV protease was simultaneously carried out overnight at 4 °C. Deconstitution of 10-His-MBP-TEV-Cas9 by TEV protease resulted in 10-His-MBP and Cas9 constituents in the solution. Another round of Ni-NTA agarose column was performed to arrest 10-His-MBP out of the solution and obtain free Cas9. Cas9 was then further purified by size-exclusion chromatography on a Superdex 200 16/60 column (GE Healthcare) in Cas9 Storage Buffer (20 mM Tris-Cl pH 7.5, 100 mM KCl, 5 % glycerol and 5 mM MgCl2) and stored at −80 °C. All the purification steps were performed at 4 °C. In some preparations, TEV protease was first added to the elutant and cleavage of the protein fusion was carried out overnight.
Following TEV protease cleavage, Cas9 was then dialyzed into Buffer A (20 mM Tris-Cl pH 7.5, 125 mM KCl, 5% glycerol, 1 mM TCEP) for 3 h at 4 °C, before being applied onto a 5 ml HiTrap SP HP sepharose column (GE Healthcare). After washing with Buffer A for three column volumes, Cas9 was eluted using a linear gradient from 0–100% Buffer B (20 mM Tris-Cl pH 7.5, 1 M KCl, 5% glycerol, 1 mM TCEP) over 20 column volumes. The protein was further purified by gel filtration chromatography on a Superdex 200 16/60 column (GE Healthcare) in Cas9 Storage Buffer (20 mM Tris-Cl pH 7.5, 200 mM KCl, 5% glycerol, 1 mM TCEP). Cas9 was stored at −80 °C.
Preparation of guide-RNA and Cas9-RNA.
Guide-RNA for Cas9 is a combination of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA). For smFRET assay for DNA interrogation by Cas9-RNA, crRNA with an amino-dT was purchased from IDT and labeled with Cy5-NHS. Location of the Cy5 in the crRNA is shown in Supplementary Figure 1 The tracrRNA was in vitro transcribed as described previously4. For DNA unwinding smFRET assay, both crRNA and tracrRNA were unlabeled and transcribed in vitro. Guide-RNA was assembled by mixing crRNA and tracrRNA at 1:1.2 ratio in buffer containing 10 mM Tris HCl (pH 8) and 50 mM NaCl, heated to 90 °C and slowly cooled to room temperature. Cas9-RNA was assembled by mixing guide-RNA and Cas9 at a ratio of 1:3 in Cas9-RNA activity buffer (20 mM Tris HCl (pH 8), 100 mM KCl, 5 mM MgCl2, 5% v/v glycerol). Cas9-RNA cleavage activity on cognate sequence used for smFRET assay for DNA interrogation was characterized previously4. We used a slightly difference sequence for optical placement of amino-dTs for DNA unwinding smFRET assay and Cas9RNA cleavage activity on that sequence was also confirmed (Supplementary Fig. 4). RNA sequences are available in Supplementary Table 1 & 2. For smFRET assay of DNA unwinding by surface-tethered Cas9-RNA, a biotin adaptor DNA strand was annealed to a complementary extension on guide-RNA (Supplementary Figure 7).
Single-molecule fluorescence imaging and data analysis.
DNA targets were immobilized on the polyethylene glycol-passivated flow chamber surface (purchased from Johns Hopkins University Microscope Supplies Core or prepared following protocols reported previously 22 using neutrAvidin-biotin interaction and imaged in the presence of Cas9-RNA at the stated concentration using two-color total internal reflection fluorescence microscopy. For DNA unwinding by surface-tethered Cas9-RNA smFRET assay, 20 nM of biotin-labeled Cas9-RNA was immobilized on surface before adding FRET pair labeled DNA targets. All the imaging experiments were done at room temperature in a Cas9-RNA activity buffer with oxygen scavenging for extending photostability (20 mM Tris-HCl, 100 mM KCl, 5 mM MgCl2, 5% (v/v) glycerol, 0.2 mg ml−1 BSA, 1 mg ml−1 glucose oxidase, 0.04 mg ml−1 catalase, 0.8% dextrose and saturated Trolox (>5 mM)35. Time resolution was 100 ms unless stated otherwise. Detailed methods of single-molecule FRET (smFRET) data acquisition and analysis have been described previously22. Video recordings obtained using EMCCD camera (Andor) were processed to extract single molecule fluorescence intensities at each frame and custom written scripts were used to calculate FRET efficiencies. Data acquisition and analysis software can be downloaded from https://cplc.illinois.edu/software/. FRET efficiency (E) of the detected spot was approximated as E = IA/(ID+IA), where ID and IA are background and leakage corrected emission intensities of the donor and acceptor, respectively. For DNA unwinding experiments, only the fluorescent spots in the acceptor channels were used for the analysis to avoid DNA molecules with inactive or missing acceptor.
E histograms, Cas9-RNA bound DNA fraction and unwound fraction.
Unless stated otherwise, first five frames of each molecule’s E value time traces were used as data points to construct E histograms. At least 2,500 molecules were included in each histogram. Cas9-RNA bound DNA fraction was calculated as a fraction of data points with E > 0.75 and was then normalized relative to Cas9-RNA bound DNA fraction for the cognate DNA target, which is not 100% because missing or inactive acceptor fluorophore, and is referred to as fbound. To estimate Kd, fbound vs Cas9-RNA concentration (c) was fit using
fbound= M × c / (Kd + c) where M is the maximum observable fbound. M is typically less than 1 because inactive or missing acceptors or because not all of the DNA on the surface are capable of binding Cas9RNA.
For DNA unwinding smFRET assay, unwound fraction was calculated as the ratio of data points with 0.2 <E <0.6 and total number of data points with E>0.2.
Kinetic analysis of DNA interrogation by Cas9-RNA
A bimolecular association-dissociation kinetics was used for the analysis of DNA binding by Cas9-RNA.
A hidden Markov model analysis of smFRET traces of DNA targets that showed real-time reversible association-dissociation of Cas9-RNA yielded three pre-dominant FRET states, of zero, mid and high E values. Dwell times of high FRET (E>0.6) states before their transition to zero FRET (E<0.2) states were used to calculate lifetime of high FRET (τhigh) (Supplementary Fig. 3) binding events by fitting their distribution using a double-exponential decay. Dwell times of mid FRET (0.2 <E <0.6) states before their transition to zero FRET (E<0.2) states were used to calculate lifetime of mid FRET (τmid) (Supplementary Fig. 3) binding events by fitting their distribution using a single exponential decay.
To calculate the total bound state lifetime (τavg) (Fig. 1d), mid and high FRET states were taken as bound states and zero FRET state as the unbound state. The survival probability of all the bound state events (mid and high FRET states taken as a single state, FRET > 0.2) vs time was fit using a double exponential decay profile (A1 exp(−t/τ1)+A2 exp(−t/τ2)).τavg is an amplitude weighted average of two distinct lifetimes τ1 and τ2.
A1 values are also used as the fraction of mid FRET state binding events (fmid FRET). Dwell time distribution of zero FRET state (E<0.2) was fit to a single exponential decay to calculate lifetime of unbound state (τunbound) (Fig. 1f). Cy5 labeling efficiency of guide-RNA was 88.7% and thus τunbound was appropriately corrected to account for it. Inverse of τunbound was used to calculate kbinding and kon. Since Cas9-RNA DNA association were sequence independent, a mean of kon for different DNA target was used to determine kon for each Cas9 (Fig. 1f)
Kinetic analysis of Cas9-RNA induced DNA unwinding and rewinding
Cas9-RNA induced DNA unwinding was modeled as a two-state system (Fig. 2b):
A hidden Markov model analysis segmented single molecule time traces into unwound and rewound states (low and high FRET states). Dwell time histograms were fitted using single exponential decay to determine the average lifetime of the high (τhigh) and low FRET (τlow) states respectively (Fig. 2d-e). A small fraction of its smFRET time trajectories had a constant high FRET without any fluctuations, likely representing DNA molecules unable to bind Cas9-RNA and were excluded from kinetic analysis.
Unwinding rate (ku) was calculated as inverse of τhigh and rewinding rate (kr) was calculated as inverse of τlow. Intrinsic cleavage rate kc,int was determined by fitting cleavage lifetime vs unwound fraction using τcleavage = 1/([unwound fraction]*kc,int + C), where C=0 for eCas9 but small values of C had to be used for WT Cas9 and Cas9-HF1.
In smFRET experiments to capture the initial DNA unwinding event, distribution of dwell times of the initial high FRET state before the first transition to the low FRET state was fit with a single exponential to obtain τunwinding.
Code availability.
Any custom code used in the study are available from the corresponding author upon request.
Gel-electrophoresis to investigate Cas9-RNA induced cleavage.
FRET pair labeled DNA targets used in the smFRET DNA unwinding assays were also used in fluorescence-based gel electrophoresis experiments. The DNA targets were incubated with Cas9-RNA in Cas9-RNA activity buffer (20 mM Tris HCl (pH 8), 100 mM KCl, 5 mM MgCl2, 5% v/v glycerol) at the specified concentrations and for specified durations. The reaction samples were then denatured by formamide loading buffer (95% formamide, 5 mM EDTA) and resolved via polyacrylamide gel electrophoresis (PAGE) using 15% Polyacrylamide TBE Urea pre-cast gels (Bio-rad laboratories) and imaged via Cy5/Cy3 excitation (GE Amersham Imager 600). Sequences of DNA targets and guide-RNA are shown in Supplementary Table 2.
For radio-labeled PAGE experiments, P32 -labeled DNA targets at 1 nM concentration were incubated with 100 nM Cas9-RNA in Cas9-RNA activity buffer (20 mM Tris HCl (pH 8), 100 mM KCl, 5 mM MgCl2, 5% v/v glycerol) at for 10 minutes. The reaction samples were then denatured by formamide loading buffer (95% formamide, 5 mM EDTA), resolved via PAGE and imaged via phosphorimaging (GE Healthcare). For the rate of cleavage experiments, aliquots of Cas9-RNA DNA running reaction were taken at different time points for the PAGE analysis. Sequences of DNA targets and guide-RNA are the same as in Supplementary Table 2 except that the biotinylated adaptor strand is omitted. τcleavage for WT Cas9 is taken from a study33 which used a slightly different protospacer, shown in Supplementary Table 1. In a control, the τcleavage for WT Cas9 were similar between the two protospacers and much lower than τcleavage for EngCas9. For all experiments reported in the manuscript, error bars represent s.d. from n=2 or 3. n=1 in absence of error bar. Uncropped gel images are shown in Supplementary Data Set 1.
Supplementary Material
ACKNOWLEDGEMENTS
The project was supported by grants from National Science Foundation (PHY-1430124 to T.H.) and National Institutes of Health (GM065367; GM112659 to T.H and GM097330 to S.B.); T.H. is an investigator with the Howard Hughes Medical Institute. J.M is supported by the Nation Institutes of Health Chemical Biology Interface training program (T32GM080189). We would like to thank Jennifer A. Doudna and Samuel H. Sternberg for useful early discussions about the design of experiments. We would also like to thank Samuel H. Sternberg and Janice S. Chen of Doudna lab for generously providing some Cas9 stocks and EngCas9 expression plasmids, respectively.
Footnotes
COMPETING FINANCIAL INTERESTS
Authors declare no competing financial interests.
References
- 1.Marraffini LA & Sontheimer EJ CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nature reviews. Genetics 11, 181–190 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gasiunas G, Barrangou R, Horvath P & Siksnys V Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America 109, E2579–E2586 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sternberg SH, Redding S, Jinek M, Greene EC & Doudna JA DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, La Russa M & Qi LS CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem 85, 227–64 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Barrangou R & Doudna JA Applications of CRISPR technologies in research and beyond. Nat Biotechnol 34, 933–941 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Kriz AJ & Sharp PA Target specificity of the CRISPR-Cas9 system. Quant Biol 2, 59–70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Geen H, Yu AS & Segal DJ How specific is CRISPR/Cas9 really? Curr Opin Chem Biol 29, 72–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh D, Sternberg SH, Fei J, Doudna JA & Ha T Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat Commun 7, 12778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinstiver BP et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slaymaker IM et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagdas YS, Chen JS, Sternberg SH, Doudna JA & Yildiz A A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci Adv 3(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen JS et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joo C, Balci H, Ishitsuka Y, Buranachai C & Ha T Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem 77, 51–76 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Sternberg SH, Redding S, Jinek M, Greene EC & Doudna JA DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szczelkun MD et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A 111, 9798–803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutkauskas M et al. Directional R-Loop Formation by the CRISPR-Cas Surveillance Complex Cascade Provides Efficient Off-Target Site Rejection. Cell Rep (2015). [DOI] [PubMed] [Google Scholar]
- 18.Redding S et al. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell 163, 854–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blosser TR Two distinct DNA binding modes guide dual roles Of a CRISPR-Cas protein complex. 58, 60–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Josephs EA et al. Structure and specificity of the RNA-guided endonuclease Cas9 during DNA interrogation, target binding and cleavage. Nucleic Acids Res 43, 8924–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim Y et al. Structural roles of guide RNAs in the nuclease activity of Cas9 endonuclease. Nat Commun 7, 13350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy R, Hohng S & Ha T A practical guide to single-molecule FRET. Nat Methods 5, 507–16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKinney SA, Joo C & Ha T Analysis of Single-Molecule FRET Trajectories Using Hidden Markov Modeling. Biophys J 91, 1941–51 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyle EA et al. High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc Natl Acad Sci U S A 114, 5461–5466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang F et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson CD, Ray GJ, DeWitt MA, Curie GL & Corn JE Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34, 339–44 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Zuo Z & Liu J Cas9-catalyzed DNA Cleavage Generates Staggered Ends: Evidence from Molecular Dynamics Simulations. Sci Rep 5, 37584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert LA et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 154, 442–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen B et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 155, 1479–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palermo G, Miao Y, Walker RC, Jinek M & McCammon JA CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc Natl Acad Sci U S A 114, 7260–7265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong S, Yu HH, Johnson KA & Taylor DW DNA Unwinding Is the Primary Determinant of CRISPR-Cas9 Activity. Cell Rep 22, 359–371 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Y, Sander JD, Reyon D, Cascio VM & Joung JK Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32, 279–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sternberg SH, LaFrance B, Kaplan M & Doudna JA Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 527, 110–3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jinek M et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasnik I, McKinney SA & Ha T Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods 3, 891–3 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.