Summary
ER membrane contact sites (MCSs) mark positions where endosomes undergo fission for cargo sorting. To define the role of ER at this unique MCS, we targeted a promiscuous biotin ligase to cargo sorting domains on endosome buds. This strategy identified the ER membrane protein TMCC1, a member of a conserved protein family. TMCC1 concentrates at the ER-endosome MCSs that are spatially and temporally linked to endosome fission. When TMCC1 is depleted, endosome morphology is normal, buds still form, but ER-associated bud fission and subsequent cargo sorting to the Golgi are impaired. We find that the endosome-localized actin regulator Coronin1C is required for ER-associated fission of actin-dependent cargo sorting domains. Coronin1C is recruited to endosome buds independently of TMCC1, while TMCC1/ER recruitment requires Coronin1C. This link between TMCC1 and Coronin1C suggests the timing of TMCC1-dependent ER recruitment is tightly regulated to occur after cargo has been properly sequestered into the bud.
Graphical Abstract

ETOC
The timing and location of endosome fission is tightly regulated by TMCC1-mediated ER recruitment to ER-membrane contact to ensure that budding happens only cargo has been properly sequestered.
Introduction
The endoplasmic reticulum (ER) spreads throughout the cytoplasm as an interconnected network of membrane tubules and cisternae. ER tubules are highly dynamic and move in a motor-dependent fashion along the microtubule (MT) cytoskeleton (Lee and Chen, 1988; Terasaki et al., 1984; Waterman-Storer and Salmon, 1998). A striking feature of these dynamic ER tubules is that they are linked to other cytoplasmic organelles through membrane contact sites (MCS) (for reviews see (Phillips and Voeltz, 2016; Prinz, 2014; Salvador-Gallego et al., 2017) (Friedman et al., 2010; Zajac et al., 2013)We have recently described a surprising new function for MCSs between ER tubules and endosomes. Dynamic ER tubules form MCS at the base of endosome budding domains and define the position and timing of endosome fission (Rowland and Chitwood et al., 2014).
Membrane fission is essential to early and late endosome function. Endosomes sort signaling receptors or nutrient cargos for recycling or degradation. Cargo destined for recycling to the plasma membrane (PM) or trans Golgi network (TGN) is packaged into a tubular structure that buds from the endosome vacuolar domain. The cargo that remains behind in the vacuolar domain is later degraded when late endosomes fuse with lysosomes. Several well-characterized factors regulate formation of the budding domain and the partitioning of cargoes into these distinct sorting domains. Microtubules and their motor proteins aid in creating membrane tension by pulling out the budding membrane, which is subsequently stabilized and shaped by the membrane shaping proteins, sorting nexins (SNXs). Cargo is then sorted into the newly formed bud and retained by the combined action of the cargo-binding retromer and Arp2/3 branched actin networks generated by WASH (Derivery et al., 2009; Gautreau et al., 2014; Hunt et al., 2013; Puthenveedu et al., 2010). While much is known about the process of bud formation and cargo sequestration into the bud, it is not clear what machinery drives ER MCS formation with the bud to regulate the timing of ER-associated fission.
We have observed that dynamic ER tubules form MCSs with the endosome bud to drive ER-associated fission after the cargo domain is established (Rowland and Chitwood et al., 2014). This sequence of events suggests that sorting domains must be properly established before the ER can be recruited. We set out to identify the mechanism by which a primed budding domain would signal ER recruitment, as well as the molecular machineries required to establish this contact. Our strategy was to localize the promiscuous biotin ligase, BioID, to the endosome budding domain in order to identify factors involved in ER-associated endosome fission. With this strategy, we have discovered a role for the ER protein TMCC1 in driving MCS formation between dynamic ER tubules and endosome sorting domains, which orchestrates ER-associated bud fission. TMCC1 specifically regulates ER recruitment to endosome buds primed with the actin regulating protein Coronin 1C. Additionally, we have found that TMCC1 and Coronin 1C coordinate to facilitate the timing of ER recruitment for ER-associated endosome fission.
Results
Identification of ER-endosome MCS proteins by proximity biotinylation
Our goal was to identify ER proteins specifically localized to MCSs between dynamic ER tubules and the position of endosome fission. A challenging feature of these contacts is that they are extremely transient and occupy a relatively small surface area of the endosome. Towards this goal, we took advantage of a proximity-based labeling system, BioID, a biotin ligase mutated to promiscuously biotinylate proteins within approximately 10-30 nm (Kim et al., 2014; Roux et al., 2012). We previously determined that Fam21, a component of the WASH complex, localizes discretely to the endosome bud at the position of ER contact and fission (Rowland and Chitwood et al., 2014). Thus, we generated a fusion protein using Fam21 to target BioID to the endosome bud and biotinylate nearby ER MCS proteins. To confirm Fam21 localization was unaffected, Cos7 cells were transiently co-transfected with a general ER marker (BFP-Sec61β), a late endosome marker (mCh-Rab7), and GFP-BioID-Fam21, then imaged live by confocal fluorescence microscopy. The GFP-BioID-Fam21 correctly localized to the endosome bud and these domains successfully execute fission (Figure 1A-B).
Figure 1. A targeted BioID strategy to identify the ER-endosome MCS protein TMCC1.
(A) Merged image of a live Cos7 cell expressing BFP-Sec61β (ER in green), mCh-Rab7 (late endosome, LE in red) and GFP-BioID-Fam21 (bud in cyan). (B) Magnified time-lapse images of the region boxed in (A) shows an example of endosome bud fission. Note that GFP-BioID-Fam21 (yellow arrowheads) correctly localizes to the sorting domain at the position of ER-associated fission. (C) Merged image of a Cos7 cell expressing BFP-KDEL (ER in gray), mCh-Rab7 (red) and GFP-BioID-Fam21 (green) treated with 50μM biotin for 1 hour and then probed for biotinylation using Streptavidin-647 (cyan). (D) Magnified images of the region boxed in (C). Note that GFP-BioID-Fam21 localizes to neck of the bud at an ER MCS labeled by biotin (white arrowhead). (E) Strategy used in HeLa cells to biotinylate ER-endosome bud MCS proteins following transfection with GFP-BioID-Fam21 versus controls: GFP-Fam21 “mock” condition or myc-BioID-Rab7 (F) Differential centrifugation yielded a final 20,000×g pellet that was enriched for ER proteins and was depleted of nuclei, mitochondria, and cytoplasm (fractions were analyzed by immuno-blot (IB) for Rtn-3 (ER), tubulin (cytosol), LE (Rab7), and Tom20 (mitochondria). (G) The 20,000×g pellets were solubilized, bound and eluted from streptavidin beads, and probed with Streptavidin HRP to reveal biotinylation profiles for each sample. Asterisks denote a band size indicative of construct self biotinylation. (H) List of eluted proteins identified using mass spectrometry. Note: TMCC1 was enriched in the GFP-BioID-Fam21 elution. (I) The relative fluorescence intensity of internalized anti-CI-MPR antibody by immuno-staining reveals reduced trafficking of internalized anti-CI-MPR to TGN in Cos7 cells treated with TMCC1 siRNA, FAM21 siRNA, and VPS35 siRNA relative to control siRNA. Smaller insets show Golgi localization by staining with anti-Giantin antibody. (J) The ratio between fluorescence intensity in vesicular structures (outside of the square) relative to that of the TGN (inside the square) was measured for control siRNA, TMCC1 siRNA, FAM21 siRNA, and VPS35 siRNA treated cells (averages 2.1, 4.4, 4.9, 3.6 for n=36, 37, 32, and 35 cells, respectively). Scale bars = 5μm in A and C; 1μm in B and D; 10 μm in I. In the graph in J, median=black line, mean=red line. ***p<0.0005, two-tailed test.
We also tested whether the fusion protein was enzymatically active and capable of biotinylating proteins at ER MCSs. Cos7 cells were co-transfected with GFP-BioID-Fam21, BFP-Sec61β and mCh-Rab7. Biotin was added (5μM for 1hr), the cells were fixed, and biotinylation was visualized with a streptavidin probe conjugated to a fluorescent label (Figure 1C-D). Discrete patches of biotin were observed at MCS between ER tubules and Fam21 puncta on the endosome sorting domain (Figure 1C-D). Similar results were obtained in HeLa cells (Figure S1A-B).
To purify biotinylated ER MCS proteins, we transfected HeLa cells with GFP-BioID-Fam21 and incubated with biotin (as before). We performed two parallel control experiments where cells were transfected with a GFP-Fam21 construct (mock, without BioID fused) or with myc-BioID-Rab7 (designed to biotinylate proteins on the entire surface of late endosomes and may also biotinylate ER proteins found at various endosome MCSs) (Figure 1E). We then lysed the cells and enriched for ER membrane proteins by performing a series of differential centrifugation steps. Extracts were depleted of cell debris, nuclei, mitochondria and cytosol, while isolating a 20,000×g pellet (light membrane fraction). Immunoblot analysis confirmed the presence of both ER and endosomal proteins within this final fraction (Figure 1F). To purify biotinylated proteins, the light membrane fraction was solubilized and applied to streptavidin-conjugated beads. Bound proteins were eluted in SDS-Biotin buffer (as described in M&M). The elutions were analyzed by SDS-PAGE and probed with streptavidin-HRP, which confirmed that the GFP-BioID-Fam21 sample had a unique biotinylation profile compared to the two controls (Figure 1G, S1C). The eluted biotinylated samples were analyzed using mass spectrometry. The sample containing BioID fused to either Rab7 or Fam21 contained peptide spectral matches (Old et al., 2005) to the known ER-endosome MCS proteins VAPA and VAPB (Figure 1H, middle row) (Alpy et al., 2013a; Dong et al., 2016; Rocha et al., 2009). The GFP-BioID-Fam21 sample was further enriched with peptides for known Fam21 interacting proteins (Figure 1H, top rows) (Freeman et al., 2014; Harbour et al., 2012; Jia et al., 2012), as well as a single ER membrane protein candidate, TMCC1, which was identified with four unique peptides and was not found in controls (Figure 1H bottom row, Figure S1D).
We validated whether the TMCC1 candidate functions during retromer-mediated cargo sorting from endosome buds. We efficiently depleted TMCC1 versus known retromer-mediated cargo sorting factors (WASH component FAM21 or the retromer component VPS35) using siRNAs (Figure S1E) and scored whether CI-MPR cargo is still trafficked from endosomes to the TGN as previously described (Arighi et al., 2004; Bonifacino and Rojas, 2006; Burd and Cullen, 2014; Seaman, M N et al., 1998). To measure CI-MPR trafficking, we incubated control versus siRNA treated cells with an antibody that binds the extracellularly-exposed portion of CI-MPR at the plasma membrane (Dong et al., 2016). After 1hr incubation, the intracellular distribution of CI-MPR antibody was detected by immuno-fluorescence. The overall levels of CI-MPR in control cells vs TMCC1 KD cells remained consistent (Figure S1E). In control cells, the CI-MPR antibody was internalized with the receptor and accumulated at the Golgi (Figure 1I, left panel). In contrast, cells that were depleted for TMCC1, Fam21 or VPS35 all had a significant reduction in CI-MPR fluorescence at the Golgi, concomitant with increased levels of fluorescence at peripheral vesicles (Figure 1I-J). These data demonstrate that depletion of the ER resident protein TMCC1 produces a similar defect in endosome retromer-mediated cargo sorting to that observed upon depletion of known cargo sorting components.
TMCCs localize to dynamic domains on ER tubules and all regulate endosome cargo sorting
TMCC1 is an ER membrane protein conserved throughout metazoans, with two additionally ER-localized paralogs in mammalian cells (Zhang et al., 2014) (Figure 2A). Sequence analysis predicts that all TMCC family members contain three conserved coiled coil (CC) and two transmembrane (TM) domains (Zhang et al., 2014) (Figure 2A). We cloned TMCC1, TMCC2, and TMCC3 from HeLa cDNA and created GFP fusions to analyze their subcellular localization. Cos7 cells were co-transfected with TMCC1, TMCC2 or TMCC3 and a general ER marker Sec6β. The general ER protein localizes to all domains of the ER including the nuclear envelope (NE) and the peripheral ER network of tubules and cisternae. In contrast, TMCC family members do not localize homogenously throughout the ER, rather they each concentrate in discrete patches along peripheral ER tubules and are depleted from the flat sheet of the nuclear envelope (Figure 2B-D, S2A-C). These peripheral TMCC patches are dynamic, with concentrations of the protein accumulating and then dissipating at certain places on the tubular network (Movie S1). When TMCC1 was expressed as either an mCh- or a 3xFlag-tagged fusion protein, it showed the same distribution (Figure S2D-E). In contrast, a truncated TMCC1 mutant (TMCC1Δ1-570) that lacks the conserved N-terminal cytoplasmic domain, does not localize to dynamic domains (Figure S2F) and is instead distributed throughout the entire ER.
Figure 2. TMCC protein family members localize to tubules and rescue cargo sorting.
(A) Primary structure of TMCC protein family members and predicted coiled coil (CC) domains (gray) and trans membrane (TM) domains (black). (B-D) Merged images of Cos7 cells expressing a general ER marker (mCh-Sec61β, in red) and low levels of (B) GFP-TMCC1, (C) GFP-TMCC2, or (D) GFP-TMCC3 (in green). Magnified images of 10 μm × 10 μm boxed regions reveal that compared to a general ER marker, TMCC proteins localize to dynamic domains throughout the tubular ER (box2 and movie S1) and is absent from the nuclear envelope (box 1). (E) Representative images from CI-MPR trafficking assay (as described in Figure 1) shows that re-expression of siRNA-resistant GFP-TMCC1, or family members: GFP-TMCC2 or GFP-TMCC3 rescues the cargo sorting defect caused by TMCC1 siRNA depletion. Lower images show expression of GFP vector or expression of the GFP-TMCC family member fluorescence (green) and anti-Giantin staining of the Golgi (red). (F) Graph of experiment described in (E) (GFP, average 4.5, n=23 cells; GFP-TMCC1 FL, average 3.0, n=24 cells; GFP-TMCC2, average 3.3, n=24; GFP-TMCC3, average 3.0 n=22). Median=black line, mean=red line. *p<0.05, two-tailed test. Scale bars =10 μm.
Based on the strikingly similar localization of the TMCC protein family, we next asked if TMCC1-3 have redundant functions in cargo trafficking from late endosomes to the TGN. Only TMCC1 can be detected by immuno-blot analysis in both Cos7 and HeLa cells, and presumably TMCC2 and TMCC3 proteins are not abundant in our cell systems (Figure SG-I). We probed TMCC1-3 redundancy by analyzing whether the CI-MPR sorting defect due to TMCC1 siRNA treatment could be rescued by reintroduction of siRNA-resistant TMCC1, TMCC2 or TMCC3 (Figure 2E-F, S2G). Considering that TMCC1 was specifically identified by our proximity labeling system and the three TMCCs appear to function redundantly, we decided to probe the role of the TMCC family in endosome biogenesis using TMCC1 as a proxy.
TMCC1 localizes to the timing and position of ER-associated endosome fission
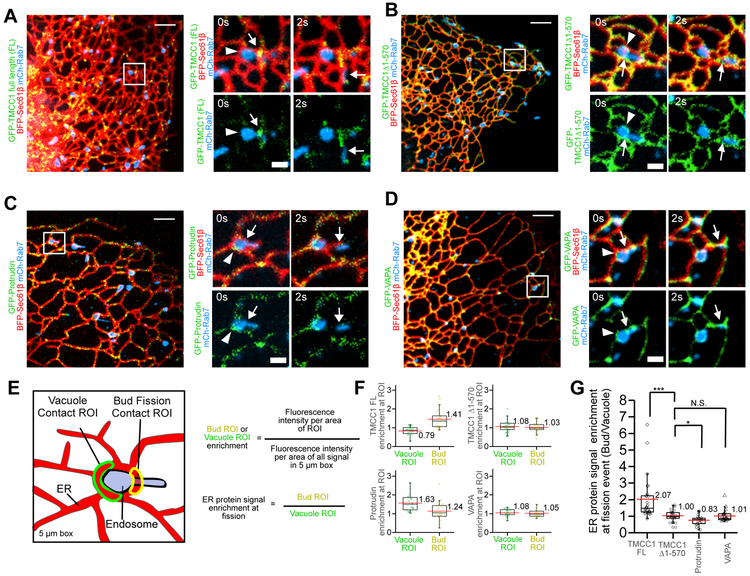
TMCC1’s localization to dynamic domains that move along ER tubules is compelling, so we therefore wondered whether these TMCC1 dynamic domains accumulate at ER MCSs coincident with the position and timing of endosome fission. To test this, we monitored the enrichment of several ER resident proteins relative to the position of ER contact during fission (Figure 3A-D, arrow marks position). Qualitatively and quantitatively TMCC1 concentrates at the position of fission when compared to the general ER protein Sec61β (Figure 3A, S3A-C, 3E-G). Importantly, TMCC1 does not accumulate on ER surrounding the vacuolar compartment of the endosome (arrowhead marks vacuolar contact, Figure 3A, S3A-B). In contrast, the truncated TMCC1 mutant protein (TMCC1Δ1–570) lacking the N-terminus (Figure S2F) which does not localize to dynamic domains, does not accumulate at ER MCSs with the position of fission (Figure 3B, 3E-F, and S3D). We next compared TMCC1 distribution to the localization of Protrudin, an ER protein that has previously been shown to localize at ER-endosome MCS to load endosomes on kinesin for anterograde trafficking (Matsuzaki et al., 2011; Raiborg et al., 2015). Protrudin has a reversed accumulation pattern compared to TMCC1–it is enriched at ER MCSs with the vacuolar compartment but it is not enriched at MCSs with the bud (Figure 3C, 3E-F and S3E). Finally, we measured the distribution of the ER MCS protein VAP, which is known to localize homogenously throughout ER tubules to regulate PI4P metabolism at a variety of MCSs (ER-PM, ER-Golgi, and ER-endosome) (Alpy et al., 2013; Manford et al., 2012; Mesmin et al., 2013; Peretti et al., 2008; Rocha et al., 2009; Salvador-Gallego et al., 2017). The localization of VAP is similar to the general ER protein and does not appear to concentrate at MCSs with either the endosome vacuolar compartment or the bud (Figure 3D, 3E-F and S3F). Together these data demonstrate that TMCC1 has a unique localization to dynamic domains that accumulate at the position and timing of bud fission (Figure 3G). This unique localization suggests TMCC1 could function directly in ER recruitment during ER-associated endosome fission.
Figure 3. Dynamic TMCC1 domains accumulate at the position and timing of endosome bud fission.
(A-D) Merged images of Cos7 cells expressing BFP-Sec61β (ER in red), mCh-Rab7 (LE in blue) and low levels of (A) GFP-TMCC1, (B) GFP-TMCC1 (Δ1-570), (C) GFP-Protrudin, or (D) GFP-VAPA (green). Magnified images of the boxed regions show examples of endosome pre (t=0s) and post (t=2s) fission. White arrowheads highlight vacuolar ER-LE MCS and white arrows highlight the bud ER-LE MCS where fission occurs. (A) Note example of a GFP-TMCC1 dynamic domain (green) that accumulates at a MCS marking the site of fission (arrow). (B) In contrast, GFP-TMCC1(Δ1-570) signal (green) localizes throughout the ER and does not accumulate at the MCS with the bud. (C) Conversely, GFP-Protrudin domains (green) accumulate at vacuolar ER-LE MCS but not at the bud MCS. (D) GFP-VAPA distribution (green) resembles that of a general ER protein. (E) Cartoon diagrams how vacuolar ER-LE MCS and bud ER-LE MCS regions of interest (ROIs) were drawn on images like those shown in (A-D) in order to quantitatively analyze and graph relative protein distribution as shown in (F). (F) TMCC1, TMCC1(Δ1-570), Protrudin and VAPA enrichments quantified at each separate ROI by comparing each fluorescent signal to the general ER marker fluorescent signal in the pre fission frame of multiple fission events (n=41, n=32, n=34, and n=32 fission events respectively). (G) Quantification of TMCC1, TMCC1(Δ 1-570), Protrudin and VAPA protein signal enrichment at LE fission events described in (F) by comparing bud fission ROI to the vacuole ROI (Bud/Vacuole). TMCC1 protein gives a higher Bud/Vacuole enrichment ratio (2.07). Protrudin has a significantly lower Bud/Vacuole ratio (0.83) emphasizing more contact with the vacuole than with the bud. Mutant TMCC1 and VAPA have an equal ratio of Bud/Vacuole suggesting no MCS enrichment. Scale bars in A-D = 5 μm and 1μm in zooms. In the graphs in F-G, median=black line, mean=red line. *p<0.05, *** p<0.0005, N.S. not significant, two-tailed test.
TMCC1 depletion disrupts endosome fission
We next tested whether TMCC1 regulates late endosome fission. Cells were co-transfected with control or TMCC1 siRNAs along with fluorescent markers to visualize late endosomes (GFP-Rab7), the ER (BFP-KDEL), and retromer buds (mCh-FAM21). TMCC1 depletion did not alter ER morphology or dynamics and did not alter endosome diameter or bud length (n=51 cells, 405 buds) when compared to control siRNA treated cells (n=47 cells, 493 buds) (Figure 4A-F). We used live confocal fluorescence microscopy to visualize whether TMCC1 depletion altered the efficiency of endosome fission during 2-minute time-lapse movies (see examples of successful vs. unsuccessful fission events, Movie S3A and S3B, respectively). When we scored fission of all budding domains that formed, we saw a significant decrease in the efficiency of endosome fission in TMCC1 depleted cells compared to control cells (from 23.5% in control to 12.7% in the TMCC1 knockdown, n=47 cells, 493 buds and n=51 cells, 405 buds respectively, Figure 4G). This fission defect could be rescued by re-introduction of an siRNA resistant GFP-TMCC1 protein (Figure S4). We next asked whether TMCC1 depletion would have a more pronounced effect on the efficiency of buds that were labeled with Fam21 because TMCC1 was identified specifically with BioID-Fam21 (Figure 1). Also, Fam21 labeling is indicative of budding domains primed for the retromer mediated cargo sorting pathway (Gomez and Billadeau, 2009; Jia et al., 2012). When we score the efficiency of endosome bud fission for Fam21-labeled buds, we saw a much more dramatic reduction in the efficiency of endosome fission (from 30.4% to 10.4%, for n=47 cells, 380 buds and n=51 cells, 340 buds, respectively, Figure 4H). This significant, 3-fold reduction suggests TMCC1 specifically regulates the process of ER-associated endosome fission for retromer-mediated cargo sorting.
Figure 4. TMCC1 regulates ER-associated endosome fission.
(A) Images of a live Cos-7 cell expressing BFP-KDEL (ER in green), GFP-Rab7 (LE in red), and mCh-Fam21 (sorting domain, cyan) and treated with control siRNA. (B) Magnified image from boxed region in (A) shows ER tubule co-localized with Fam21 on the endosome bud. (C) As in (A), but treated with TMCC1 siRNA. Note the ER morphology is not altered upon siRNA treatment. (D) Magnified images of boxed region in (C) shows reduced co-localization between ER tubules and Fam21. (E-F) Graphs of endosome diameter (E) and bud length (F) reveal similar endosome morphology for control versus TMCC1 siRNA treated cells (control n=47 cells, 493 buds, TMCC1 KD n=51 cells, 405 buds). (G) Graph of endosome bud fission rates reveals all bud fission is reduced following TMCC1 siRNA depletion (from 23.5% to 12.7%). (H) Graph of only Fam21 primed endosome bud fission events reveals a significantly greater reduction in fission efficiency following TMCC1 siRNA depletion (from 30.4% to 10.4% in the TMCC1 knockdown, control n=47 cells, 380 buds, TMCC1 KD n=51 cells, 340 buds). Scale bars = 5μm in A, C and 1μm in B, D. Error bars represent SEM in E-H, **p<0.005 and ***p<0.0005, N.S. not significant, two-tailed test.
Coronin 1C at the endosome bud recruits the ER and regulates bud fission
We have shown that TMCC1 accumulates at MCS with endosome buds coincident with fission (Figure 3). Additionally, TMCC1 depletion has a dramatic effect on fission of buds labeled by the WASH complex component Fam21, a factor that provides the bridge between retromer-mediated cargo sorting and branched actin nucleation (Arighi et al., 2004; Derivery et al., 2009; Gautreau et al., 2014; Gomez and Billadeau, 2009; Harbour et al., 2010; Hunt et al., 2013; Maxfield and McGraw, 2004; Puthenveedu et al., 2010; Seaman, 2004, 2008). Therefore, we wondered what factor(s) on the bud regulate TMCC1 recruitment in order to drive ER-associated bud fission. Such a factor would need to recruit the ER machinery after cargo has been successfully sorted, so this factor should be at the bud with sequestered cargo prior to ER recruitment. We initially tested the WASH complex. However, Wash1/Fam21 depletion did not affect ER recruitment to the budding domain nor did it effect fission of endosome tubules (Rowland and Chitwood et al., 2014)(Figure S5A-D) We therefore searched a recently published protein interactome database (Bioplex 2.0)(Huttlin et al., 2017) for TMCC-interacting factors that have been reported to localize to endosome buds and regulate retromer-mediated cargo sorting and/or branched actin assembly. Interestingly, TMCC2 and TMCC3 both revealed interactions with the actin cytoskeleton regulatory protein, Coronin 1A, Bioplex 2.0/3.0 ((Huttlin et al., 2017); personal communication). This interaction could fit into our model of TMCC1’s role in recruiting the ER to actin patches at the bud. Another member of the Coronin family, Coronin1C, was previously shown to localize to actin-regulated cargo sorting domains on endosome buds in HEK 293T cells (Puthenveedu et al., 2010). Actin-stabilized microdomains are required for cargo sorting and sequestration into the budding domain (Puthenveedu et al., 2010), making Coronin 1C an attractive candidate for regulating the timing of ER recruitment to this site. Importantly, initial experiments in Cos7 cells revealed that Coronin 1C (and not Coronin 1A) localizes to discrete patches on endosome buds (Figure S5F-G).
We tested whether Coronin 1C depletion disrupts MCS formation between dynamic ER tubules and Fam21-labeled endosome buds. Cells transfected with control or Coronin 1C siRNAs (to efficiently deplete Coronin 1C, Figure S5G) were co-transfected with BFP-KDEL (red), GFP-Rab7 (blue), and with mCh-Fam21 (green) to visualize co-localization between ER tubules, and Fam21 domains on LE buds (compare Figure 5A-C to 5D-F, respectively). We monitored the stability of ER MCS with Fam21-labeled endosome buds over time in live cell time-lapsed images (Figure 5G). When Fam21/ER crossing was quantified frame by frame, the ER spent 79.0% of the time contacting Fam21 buds in control cells (n=20) versus 41.4% of the time in the Coronin 1C depleted cells (n=21) (Figure 5H). Thus, Coronin 1C depletion disrupts ER recruitment to the endosome bud.
Figure 5. Coronin 1C regulates ER recruitment to the bud for ER-associated fission.
The effect of CORO1C depletion on ER contact with endosome buds was scored in live Cos7cells. (A-F) Merged images from control cells (A-C) versus CORO1C siRNA-depleted cells (D-F) expressing BFP-KDEL (ER in red), GFP-Rab7 (LEs in blue), and mCh-Fam21 (sorting domain in green). (B and E) Magnified images of the regions boxed in (A and D) shows successful ER crossing with the Fam21 bud in the control cell (B) but no ER contact in the CORO1C KD cell (E) (white arrowheads). However, ER-LE vacuole MCSs are present in both the control cell and the CORO1C KD cell (yellow arrowheads). (C and F) The corresponding line-scan analysis of the relative fluorescence intensity of Rab7, ER, and Fam21 from the lines drawn in (B) and (E). The control cell line scan (C) shows a peak of fluorescence intensity for both Fam21 and ER positioned at the saddle between the endosome and the bud. The CORO1C KD cell line scan (F) reveals there is no peak of ER fluorescence coincident with the peak for Fam21 at the base of the bud. (G) Representative time-lapse images from control versus CORO1C siRNA-depleted cells were used to assess whether ER tubules maintain contact with Fam21 buds over time (white arrowheads). (H) Graph of frame by frame analysis shows a significant reduction in ER contact with the Fam21-marked bud following CORO1C depletion (78.9% in control n=20 cells, 134 buds versus 41.4% in CORO1C knockdown n=21 cells, 159 buds). (I) Graph of Fam21 primed endosome bud fission events reveals a significant reduction in fission efficiency following CORO1C siRNA depletion (from 28.0% to 11.1%; control n=20 cells, 286 buds, CORO1C knockdown n=21 cells, 316 buds). Scale bars = 5μm in A,D and 1μm in B, E, G. In the graph in H, median=black line, mean=red line. Error bars represent SEM in I, ***p<0.0005, two-tailed test.
We next tested whether Coronin 1C depletion affects endosome fission. Similar to TMCC1 depletion, Coronin 1C siRNA treatment dramatically reduced the efficiency of fission for Fam21-labeled buds (from 28.0% to 11.1%; n= 20 cells, 286 buds and n=21 cells, 316 buds,, respectively, Figure 5I). Therefore, we have identified two completely novel factors which directly impact endosome fission. One localizes discretely to mature endosome budding domains and when depleted, impairs endosome fission and disrupts the formation of ER contact (Coronin 1C). The other is ER localized, accumulates at the position and timing of endosome fission, and is also required for fission (TMCC1). Taken together, these data suggest that the actions of TMCC1 and Coronin 1C are coupled during the fission of endosome sorting domains.
TMCC1 regulates ER recruitment to endosome buds
Finally, we measured whether TMCC1 plays a direct role in regulating dynamic ER tubule recruitment to endosome buds to drive ER-associated endosome fission. Since Coronin 1C regulates ER recruitment to the bud, we conversely asked whether TMCC1 functions to regulate ER recruitment to Coronin 1C domains. Cells were co-transfected with control or TMCC1 siRNAs and with BFP-KDEL, mCh-Rab7 and GFP-Coronin1C (Figure 6A-B, D-E, S6C). In the absence of TMCC1, Coronin 1C-labeled buds still form (Figure 6D-E). As before (in Figure 5C-D), we monitored the stability of ER MCSs with Coronin 1C puncta on endosome buds over time in TMCC1 siRNA depleted cells compared to control cells. In control cells, ER tubules maintain contact with both the vacuolar compartment and with Coronin puncta at the base of endosome buds (Figure 6A-C, G, S6A). By comparison, in TMCC1 depleted cells, ER tubules still maintain contact with the vacuolar compartment, but the ER rarely crosses the Coronin 1C domains at the bud (Figure 6D-F, G. S6B, SUP Movie). These analyses revealed a ~2-fold reduction (69.2% to 35.2%) in the percent of time the ER contacts Coronin puncta following TMCC1 depletion (Figure 6H). These data suggest TMCC1 directly regulates the recruitment of dynamic ER tubules to form MCS with Coronin 1C domains on the endosome bud. Coronin 1C or TMCC1 depletion yield strikingly similar defects in ER MCS formation and ER-associated endosome bud fission, suggesting this interaction regulates a precise timing mechanism to ensure successful receptor-mediated cargo sorting before ER recruitment and bud fission.
Figure 6. TMCC1 regulates ER MCS formation with the endosome bud.
The effect of TMCC1 depletion on ER contact with endosome buds was scored in live Cos7 cells. (A-F) Merged images from control cells (A-C) versus TMCC1 siRNA-depleted cells (D-F) expressing BFP-KDEL (ER in red), mCh-Rab7 (LEs in blue), and CORO1C-GFP (green). (B and E) Magnified images of the region boxed in (A and D) shows successful ER crossing with the CORO1C punctum in the control cell (B) but no ER contact with the CORO1C punctum in the TMCC1 KD cell (E) (white arrowheads). However, ER-LE vacuole MCSs are present in both the control cell and the TMCC1 KD cell (yellow arrowheads). (C and F) The corresponding line-scan analysis of the relative fluorescence intensity of Rab7, ER, and CORO1C from the lines drawn in (B) and (E). The control cell line scan (C) shows a peak of fluorescence intensity for both CORO1C and ER positioned at the saddle between the endosome and the bud. The TMCC1 KD cell line scan (F) reveals there is no peak of ER fluorescence coincident with the peak for CORO1C at the base of the bud. (G) Representative images from 2 min time-lapse with images taken every 5 seconds of control cells and TMCC1 KD cells from A-C and D-F used to assess ER tubule tracking with CORO1C over time (white arrowheads). (H) Graph shows a significant reduction in the time ER MCS form with the CORO1C -marked bud following TMCC1 depletion (69.2% in control n=18 cells, 37 buds versus 35.2% in TMCC1 knockdown n=24 cells, 38 buds). Scale bars = 5μm in A, D and 1μm in B, E, G. In the graph in H, median=black line, mean=red line. *** p<0.0005, two-tailed test.
Discussion
We have used a targeted promiscuous biotin ligase strategy to identify TMCC1 as an ER membrane protein that functions at ER MCSs to regulate ER-associated endosome fission. TMCC1 has an unusual localization in that it is partitioned to peripheral ER tubules and is excluded from the nuclear envelope. In addition, it accumulates into dynamic domains throughout the tubular network of the ER. TMCC1 dynamic domains are spatially and temporally linked to the position of ER-associated endosome fission. TMCC1 depletion caused a defect in efficient cargo delivery from late endosomes to the Golgi. Importantly, we observed no abnormalities in endosome morphology or the biogenesis of sorting domains upon TMCC1 depletion, just a defined role in fission of endosome buds. We show that TMCC1 functions to stabilize ER MCS with cargo sorting domains on endosome buds. Together, these data argue a specific role for TMCC1 in the process of ER-associated endosome fission.
TMCC1 specifically regulates ER recruitment to cargo sorting domains that are labelled with the actin regulatory complex WASH. At these cargo sorting domains, the retromer pulls cargo into the bud, the retromer recruits the WASH complex, and WASH promotes Arp2/3 to nucleate branched actin assembly (Derivery et al., 2009; Gautreau et al., 2014; Gomez and Billadeau, 2009; Harbour et al., 2010; Jia et al., 2012; Puthenveedu et al., 2010). Branched Arp2/3 actin has been shown to stabilize the bud to give time for cargo sorting before the bud undergoes fission(Puthenveedu et al., 2010). In the absence of actin, buds still form and undergo fission, but they are likely to only contain cargo that passively diffuses by bulk flow into the bud, like transferrin receptor (Puthenveedu et al., 2010). Interestingly, we found that buds labelled with the WASH complex (promotes actin polymerization at the bud) required both TMCC1 and Coronin 1C for fission.
An important question is what signals the timing of TMCC1-dependent ER recruitment to ensure that it is recruited to drive bud fission only after actin-mediated cargo sequestration has occurred. We would argue that Coronin 1C regulates the timing of ER recruitment. Coronin 1C has previously been shown to label the Arp2/3-mediated actin patches present on endosome buds (Puthenveedu et al., 2010). Our data demonstrates that Coronin 1C regulates TMCC1 recruitment, ER MCS formation with the bud, and ER-associated fission of WASH-labeled buds. Coronins have been shown to regulate actin disassembly by displacing Arp2/3(Chan et al., 2012; Gandhi et al., 2009; Humphries et al., 2002; Liu et al., 2011; Wu et al., 2006). It would thus function to both disassemble branched actin and recruit TMCC1/ER for ER-associated fission (see model in Figure 7).
Figure 7. Model of ER recruitment to endosome sorting and fission factors.
Following retromer and actin mediated sorting of factors into the endosome tubules, Coronin 1C at the endosome bud is required for TMCC1-mediated ER recruitment and the interplay between Coronin 1C and TMCC1 promote fission of the endosome bud.
Work from the De Camilli lab indicates that the efficiency of cargo sorting also depends on the interaction of the ER resident protein VAP and two proteins localized to the endosome: OSBP and SNX2 (Dong et al., 2016). The interaction between VAP and OSBP is required to down-regulate the levels of PI4P on the endosome and in turn reduce aberrant actin accumulations. In the absence of VAP or OSBP, PI4P and actin levels increase around the endosome -the outcome of which is a reduction in cargo trafficking (Dong et al., 2016). It is not known whether depletion of VAP or OSBP also blocks bud formation or alters the rate of ER-associated endosome bud fission. It will be interesting to determine whether VAP-OSBP mediates a reduction in PI4P and actin levels at the bud before or after the process of TMCC1-dependent ER tubule recruitment. It also remains to be determined what machinery drives bud fission. One promising candidate is IST1, which is regulated by an ER localized M1-Spastin. Spastin does not function to create the ER-endosome MCS formation (depleting Spastin actually results in more ER-endosome contact) but Spastin regulation of IST1 contributes to the efficiency of bud fission (Allison et al., 2013, 2017).
TMCC1 has several interesting properties that could further inform its function that will guide future experiments. TMCC1 has been shown to homo and hetero-oligomerize with the other TMCC family members (Zhang et al., 2014). TMCC contains predicted coiled coil (CC) domains in the N terminal cytoplasmic domain that are required for function (Figure S7A). We used the structural prediction program Phyre2, to gain further insight into the functional role of the CC domains of TMCC1. The Phyre2 programs predicts that the two short CCs (aas 215-320) are structurally similar (84% similarity) to the Cdc42 binding domain of cip4 (Figure S7B). It is tempting to speculate that the short coiled coils could function to recruit ER tubules to the same position where actin is also nucleated by a RhoGTPase. RhoGTPases are often used as molecular switches and in this case, a RhoGTPase could provide an additional timer for TMCC1 binding to occur and link the ER to this site when retromer components have successfully been established. A goal of our future work will be to determine whether a RhoGTPase or Cdc42 itself is involved. TMCC1 also contains a longer predicted CC. The Phyre2 structural prediction program also predicts a high similarity (98.5%) between the long CC and a rigid CC found within fibrinogen, a protein that provides structural separation (Figure S7C). We speculate that this long CC would give a predicted length of ~18nm (an ideal spacing for most MCSs, Alpy et al., 2013; Friedman et al., 2011). Furthermore, TMCC1 may form a rigid SNARE-like MCS spacer -perhaps to prevent the two membranes from undergoing heterotypic membrane fusion and/or to establish an important signaling platform that drives and regulates endosome fission.
STAR Methods:
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gia Voeltz (gia.voeltz@colorado.edu)
Method Details
Plasmid Construction
GFP-Sec61β (Shibata et al., 2008), BFP-Sec61β (Zurek et al., 2011), mCh-Sec61β (Zurek et al., 2011), BFP-KDEL (Friedman et al., 2011), GFP-Rab7A (Rowland and Chitwood et al., 2014), mCh-Rab7 (Rowland and Chitwood et al., 2014), and mCh-Fam21 (Rowland and Chitwood et al., 2014) were previously described. GFP-Fam21 was created by PCR amplifying FAM21 out of the shFAM21/HA-YFP-FAM21 rescue vector given to us by Dr. Daniel Billadeau (Gomez and Billadeau, 2009; Jia et al., 2012), and ligating into Kpn1/BamH1 sites of pAcGFP1-C1 (Clontech, Mountain View, CA). GFP-BioID-Fam21 was created by PCR amplifying BioID from myc-BioID ((Roux et al., 2012)Addgene #35700) and ligating into Xho1/Kpn1 sites of GFP-Fam21 vector. Myc-BioID-Rab7A was made by PCR amplifying Rab7A from GFP-Rab7A and ligating into Xho1/BamH1 sites of myc-BioID vector. GFP-TMCC1, GFP-TMCC2 and GFP-TMCC3 were generated by PCR amplifying human TMCC1 (National Center for Biotechnology Information [NCBI] accession number NM_001336192.1), human TMCC2 (NM_001297611.1), and human TMCC3 (NM_020698.3) from a HeLa cell derived cDNA library and ligating into Xho1/Kpn1 sites of pAcGFP1-C1. mCh-TMCC1 and 3xFlag-TMCC1 were generated by cloning mCh or 3xFlag into the NheI/XhoI sites of GFP-TMCC1, replacing the GFP. siRNA resistant GFP-TMCC1 was created using a series of site directed mutagenesis PCR reactions to mutate the base pairs (bps) in the four siRNA binding regions for the four TMCC1 siRNA oligonucleotides. GFP-TMCC1 truncations were created by PCR amplifying the the GFP-TMCC1 vector with forward primers starting 1711 bps or 958 bp into the TMCC1 gene for Δ1-570 or Δ1-319 respectively, and then ligating back into the Xho1/Kpn1 sites of pAcGFP1-C1. Δ420-575 was created using a Gibson reaction procedure with primers that aligned adjacent to but skipped bps 1378-1725 in the GFP-TMCC1 construct. GFP-VAPA was generated by Jonathan Friedman by PCR amplifying human VAPA (NCBI accession number NM_003574.5) from a HeLa cell derived cDNA library and ligating into BglII/BamH1 sites of pAcGFP1-C1. GFP-Protrudin was generated by PCR amplifying human Protrudin1C (NCBI accession number NM_001002262) from a HeLa cell derived cDNA library and ligating into Kpn1/BamH1 sites of pAcGFP1-C1. Coronin1A-GFP was generated by PCR amplifying human Coronin 1A (NCBI accession number NM_001193333) from a HeLa cell derived cDNA library and ligating into Xho1/Kpn1 sites of pAcGFP1-N1. Coronin1C-GFP was gift from Dr. Manojkumar Puthenveedu and Dr. Mark von Zastrow (Puthenveedu et al., 2010).
Cell growth and plasmid DNA transfections
Cos-7 cells (ATCC, Manassas, Virginia) were grown in DMEM media (Invitrogen, Carlsbad, CA) supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were seeded at 2×105 cells per well of a 6-well dish ~16 hours prior to transfection. Cells were transfected ~24 hours prior to imaging with plasmid DNA in Opti-MEM media (Invitrogen) with 5 μl of Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. After ~5 hours of transfection, cells were seeded in 35mm glass-bottom microscope dishes (MatTek, Ashland, MA) with 2.0×105 cells. Live cells were imaged in 37°C Flourobrite supplemented with 10% FBS. For all microscopy experiments, the following amounts of plasmid DNA were transfected into cells for experiments: 500ng/mL BFP-Sec61β; 250ng/mL mCh-Sec61β and GFP-Sec61β; 100ng/mL BFP-KDEL; 20ng/mL mCh-Rab7 and GFP-Rab7; 250ng/mL mCh-Fam21; 25ng/mL Coronin1A-GFP; 25ng/mL Coronin1C-GFP; 50ng/mL myc-BioID-Rab7; 50ng/mL GFP-Fam21; 50ng/mL GFP-BioID-Fam21; 50ng/mL for low expression of TMCC1 and TMCC2, 25ng/mL for low expression of TMCC3; 166.5ng/mL for high expression of TMCC1 and truncation variants. For immunofluorescence experiments, cells were fixed with 4% Paraformaldhyde, solubilized in 0.1% Triton-X, PBS solution and then immunostained. Streptavidin-647 was used at 5 μg/mL to visualize biotinylation. Anti-Flag 1:1,000; and all Alexa-Fluor conjugated antibodies (ThermoFisher) used at 1:300 to visualize 3xFlag-TMCC1.
Biotinylation, ER isolation, and biotinylated protein collection
HeLa cells (ATCC-CCL-2, Manassas, Virginia) were grown in complete media (DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS and 1% penicillin/streptomycin). Cells were seeded in T-75 flasks with 15.0×105 cells about ~16 hours prior to transfection. Cells were transfected ~24 hours prior to biotinylation with plasmid DNA in Opti-MEM media (Invitrogen) with 12.5μL of Lipofectamine 3000 reagent per T-75 of cells (Invitrogen). After ~5 hours of transfection, cells were washed and replenished with complete media and recovered overnight until reaching ~90-100% confluency. Then, cells were incubated in 50μM biotin for 1 hour. After biotinylation, cells were rinsed and collected using PBS at 37°C. Each cell pellet from 2, T-75 flasks was resuspended in 1 mL isolation buffer (225 mM mannitol, 75 mM sucrose, 30 mM Tris-HCl pH 7.4, 0.1 mM EGTA) at 4°C and lysed using a 1mL dounce homogenizer with pestle on ice until ~90-95% of cells were lysed as visualized with tryphan blue (~100 strokes). Isolation of the light membrane fraction containing ER occurred as follows: two sequential 600×g spins (pellet nuclei and cell debris, continue with supernatants), three 7,000×g spins (pellet mitochondria, continue with supernatants), and then 20,000×g spin to pellet light membranes/ enrich for ER membranes, collect supernatant as cytosol, add 1 mL isolation buffer-2 (225 mM mannitol, 75 mM sucrose, 30 mM Tris-HCl pH 7.4) to rinse pellet, spin 20,000×g again to re-pellet final ER light membrane fraction (all spins occurred at 4°C). For each step, pellets were resuspended in solubilization buffer (50 mM HEPES, 2.5 mM MgCl2, 200 mM KCl, 5% glycerol, 1% Triton X-100), protein levels were normalized by BCA assay, 10% of sample was diluted in 2X Laemmli sample buffer, and assayed using standard SDS-PAGE/Western Blotting: Rtn3 (YenZym Antibodies) 1:1000; TOM20 1:1000 (sc-11415, Santa Cruz Biotechnologies); α-tubulin (ab18251, Abcam) 1:10,000; and Rab7 (Cell Signaling 99367S, Danvers, MA) 1:500. The remaining 90% of ER light membrane fraction samples were diluted in 1mL solubilization buffer, nutated for 30min at 4°C, and insoluble proteins were spun down at 15,000×g. Solubilized samples were incubated overnight, nutating at 4°C with 90μL of Dynabeads MyOne Streptavidin C1 (Invitrogen). Beads were washed as according to Roux et al., and replicates were pooled during the final wash. Recovered proteins were eluted by boiling the beads at 95°C in 2x Laemmli sample buffer in the presence of 2mM biotin.
Methods for BioID proteomics
Purified proteins were precipitated from Laemli sample buffer by adding 100ng/mL insulin (as a carrier protein), 0.1% (w/v) Deoxycholic acid and 20% (w/v) trichloroacetic acid. Samples were maintained at 4°C throughout the precipitation. Precipitated proteins were pelleted by centrifugation at 21,130×g for 30 minutes. Protein pellets were washed twice with −20°C acetone, air dried and stored at −80°C until further processing. Samples were proteolyzed using the filter-aided sample preparation (FASP) method (Wiśniewski, 2016). Briefly, dried protein pellets were solubilized in 4% (w/v) SDS, 10mM TCEP, 40mM 2-chloroacetamide and 0.1M Tris pH8.5, then boiled for 5 minutes. Samples were allowed to cool to room temperature for 30 minutes, diluted 10-fold with 8M Urea, 0.1M Tris pH8.5 and loaded onto an Amicon Ultra 0.5mL 30kD NMWL cutoff (Millipore) ultrafiltration device. Samples were washed in the filters three time with 8M Urea, 0.1M Tris pH8.5, and again three times with 2M Urea, 0.1M Tris pH8.5. Endoproteinase Lys-C (Wako) was added and incubated 2 hours rocking at room temperature, followed by trypsin (Pierce) which was incubated overnight rocking at room temperature. Tryptic peptides were eluted via centrifugation and desalted using a C-18 spin column (Pierce) according to the manufacture instructions.
Liquid Chromatography/Mass spectrometry analysis.
Samples were suspended in 5% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid and direct injected onto a 1.7um, 130Å C18, 75um X 250mm M-class column (Waters), with a Waters M-class UPLC. Tryptic peptides were gradient eluted at 300nL/minute, from 3% acetonitrile to 20% acetonitrile in 100 minutes, into an Orbitrap Fusion mass spectrometer (Thermo Scientific). Precursor mass spectrums (MS1) were acquired at 120,000 resolution from 380-1500 m/z with an AGC target of 2.0E5 and a maximum injection time of 50ms. Dynamic exclusion was set for 20 seconds with a mass tolerance of +/− 10 ppm. Isolation for MS2 scans was 1.6Da using the quadrupole, and the most intense ions were sequenced using Top Speed for a 3 second cycle time. All MS2 sequencing was performed using higher energy collision dissociation (HCD) and scanned in the Orbitrap with 30,000 resolution. An AGC target of 5.0E4 and 60 second maximum injection time was used. Rawfiles were converted to the mzML format and searched against the Uniprot Human database downloaded April 1, 2015 using Mascot v2.5 with Cysteine Carbamidomethylation as a fixed modification. Methionine oxidation, protein N-terminal and Lysine sidechain Biotinylation were searched as variable modifications. Peptide mass tolerance was 20ppm for MS1 and 20mDa for MS2. All peptides were thresholded at a 1% false discovery rate (FDR).
RNAi transfection and Western blot
TMCC1, FAM21, VPS35, and CORO1C were depleted using ON-TARGETplus Human siRNA SMARTpools (Dharmacon/GE Lifesciences). Cells were seeded at 2×105 cells per well of a 6-well dish ~16 hours prior to transfection. Cells were first transfected ~72 hours prior to imaging with 5 μL Dharmafect (Dharmacon/GE Lifesciences) in DMEM with 10% FBS and 25nM of the siRNA SMARTpool or 25nM Silencer Negative Control #1 siRNA (Ambion AM4635). After ~6 hours of transfection, cells were washed and replenished with DMEM media supplemented with 10% FBS and 1% penicillin/streptomycin. After ~48hrs cells were transfected again ~24 hours prior to imaging with plasmid DNA as described before with the addition of 12.5 nM siRNA SMARTpool or 12.5 nM Silencer Negative Control #1 siRNA. After ~5 hours of transfection, cells were seeded in 35mm glass-bottom microscope dishes (MatTek) at 2.0×105 cells. Cells were imaged in 37° Flourobrite supplemented with 10% FBS.
Cos-7 cells were harvested and TMCC1, FAM21, VPS35, and CORO1C protein levels assayed using standard SDS-PAGE/Western protocols. Primary antibody concentrations were used as follows: TMCC1 (rabbit) (Zhang et al., 2014) 1:1000; FAM21 (rabbit) 1:1000 (gift from Billadeau, Gomez and Billadeau 2009); VPS35 (rabbit) 1:3000 (Abcam ab97545), and CORO1C (rabbit) 1:2000 (Proteintech 14749-1-Ap); M6PR 2G11 (mouse) (ThermoFisher Ma 1-066), 1:1000; GAPDH (rabbit) 1:30,000 (Sigma-Aldrich G9545, St. Louis, MO). Cos-7 cells and HeLa cells were harvested and TMCC1-3 were probed using TMCC1 (rabbit) (Zhang et al., 2014), TMCC2 Ab serum from rabbit injected with TMCC2 aas 442-554 antigen and TMCC3 Ab serum from a rabbit injected with full length TMCC3 antigen. HRP-conjugated goat anti-rabbit secondary antibody (Sigma-Aldrich) was used at 1:3000 for all rabbit antibodies HRP-conjugated goat anti-mouse secondary antibody (Sigma-Aldrich) was used at 1:3000 for all mouse antibodies. Signal was detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo).
Confocal microscopy
Fixed cells were imaged at room temperature. Live cells were kept at 37°C in a live-cell incubation chamber (Pathology Devices, Westminster, MD). Cell imaging was performed with an inverted fluorescence microscope (TE2000-U; Nikon, Melville, NY) equipped with a Yokogawa spinning-disk confocal system (CSU-Xm2; Nikon). Images were taken with a 100× NA 1.4 oil objective or 60X NA 1.4 oil objective (Nikon) on electron-multiplying charge-coupled device (CCD) camera 50X50 (Cascade II; Photometrics), 50×50 (Andor) or 1TB (Andor). Images were acquired with MetaMorph 7.0 (MDS Analytical Technologies, Sunnyvale, CA), with Nikon Elements, or with Micromanager and then analyzed, merged, and contrasted using Fiji (ImageJ), as well as converted to 400dpi using Photoshop (Adobe, San Jose, CA). Scale bars were generated using Fiji. Supplemental videos were generated using ImageJ, Adobe Photoshop, and Quicktime.
CI-MPR uptake and trafficking analysis
Cells were incubated with 10μg/ml mouse anti-CI-MPR monoclonal antibody (ThermoFisher MA1-006) in serum-free DMEM for 1 hr, rinsed with PBS, fixed with 4% Paraformaldhyde, solubilized with Triton-X and immunostained with 1:500 anti-Giantin (rabbit) antibody (BioLegend cat 924302) and then fluorescent anti-rabbit and anti-mouse antibodies 1:300. Using Fiji, images were background corrected. An area encompassing the whole cell was selected manually and the fluorescence intensity of this region was measured. A 10 × 10 μm2 region was selected around the Golgi. Vesicular fluorescence was obtained by subtracting the Golgi region fluorescence intensity from whole cell fluorescence. A ratio of Vesicular fluorescence/Golgi fluorescence was calculated for each cell, control and knockdown populations were compared using a two-tailed t-test.
TMCC1 accumulation at fission site analysis
Cos-7 cells expressing markers for ER, endosomes, and low expression levels of ER proteins (TMCC1, TMCC1Δ1–570, Protrudin, and VAPA) were visualized live by confocal fluorescence microscopy and imaged for 2minute movies with 2 second intervals. Fission events were analyzed if endosomes were in regions of the cell where the ER occupied a single focal plane, the endosome budding domain divided from the vacuolar portion and trafficked away, and the general ER marker labelled ER tubule crossed clearly over the pre fission bud. The frame just before fission was then analyzed. Using only the general ER (Sec61β) and endosome (Rab7) channels regions of interest (ROIs, lines of 5 point thickness) were drawn along the ER adjacent to the vacuolar part of the endosome (vacuole contact ROI) and then drawn along the ER that crossed the endosome bud about to undergo fission (bud fission contact ROI). These ROIs were then used to measure TMCC1, TMCC1Δ1–570, Protrudin, and VAPA fluorescence at the pre fission frames. The general ER marker (Sec61β) fluorescence intensity was also measured at these ROIs. Fluorescence intensity per area for TMCC1, TMCC1Δ1-570, Protrudin, and VAPA compared to the general ER marker was calculated for each ROI. The fluorescence intensity per area was also measured for the entire 5micron × 5micron area around the endosome to assess the fluorescence distribution throughout the entire ER. Bud and vacuole enrichment was then quantified by comparing each ROI fluorescence per area to the 5micron × 5micron area fluorescence per area. Finally, bud enrichment was compared to vacuole enrichment for each event. These ratios were compared for TMCC1Δ1–570 vs TMCC1 full length, protrudin, or VAPA using two tailed tests.
Percent endosome fission analysis
Cos-7 cells expressing markers for ER and endosomes were visualized live by confocal fluorescence microscopy and imaged for 2 minute movies with 5 second intervals. Endosomes were analyzed if they had a resolvable lumen and were in regions of the cell where the ER occupied a single focal plane. If the endosome formed a budding domain, the endosome diameter and the bud length were measured. Successful fission was recorded only if the budding domain divided from the vacuolar portion and trafficked away. The percent of buds that underwent fission was calculated for each cell. Percent fission was compared in control and knockdown cells using a two-tailed t-test. To narrow the analysis to FAM21 primed buds specifically, each bud was assessed to see if FAM21 signal was present at the bud for two or more consecutive frames in the 2 minute time-lapse. If the bud was Fam21 positive, it was counted into the total number of buds in that cell, and if a Fam21 positive bud underwent fission, it was counted in the number of buds that underwent fission in that cell. Each cell’s percent fission was quantified under these conditions and percent fission was compared in control and knockdown cells using a two-tailed t-test.
ER contact with CORO1C puncta
Cos-7 cells expressing markers for ER, endosome, and CORO1C were visualized live by confocal fluorescence microscopy during a 2 minute period, capturing one image every 5 seconds. CORO1C puncta were assayed if they were localized in the cell periphery where the ER occupies a single focal plane, and they remained spatially and temporally associated with a Rab7-marked endosomes over the entire course of the 2 minute movie. Each cell was quantified manually, in every individual frame the acute spatial localization of ER relative to the endosome-associated CORO1C puncta was recorded. Using this information, we calculated the percent of two-tailed t-test. frames within the 2min time course that the ER was spatially overlapping with CORO1C puncta. The percent of time ER crossing Coronin puncta for each bud was averaged per cell, yielding percent of time ER crossing Coronin puncta per cell. Significance testing was done using data from control vs. TMCC1 KD cells, using a two-tailed t-test.
ER contact with FAM21 budding domains
Similar to ER-CORO1C association, ER-FAM21 association was assessed in Cos-7 cells using confocal fluorescence microscopy 2 minute time-lapses, capturing one image every 5 seconds in Cos-7 cells expressed markers for ER, endosome, and Fam21. Fam21 labeled buds were assayed if they were localized in the cell periphery where the ER occupies a single focal plane, and the Fam21 remained spatially and temporally associated with the Rab7 marked endosome over at least 1 min of the movie. Each cell was quantified manually; in every individual frame the acute spatial localization of ER relative to the Fam21 bud was recorded. Using this information, we calculated the percent of frames the ER was spatially overlapping with the Fam21 bud. The percent of time ER crossing the Fam21 bud for each bud was averaged per cell, yielding percent time ER crossing the Fam21 bud per cell. Significance testing was done using data from control vs. KD cells, using a two-tailed t-test.
Amino acid sequence analysis with structure prediction software
Two regions of TMCC1 protein sequence were analyzed using Phyre2 prediction software. TMCC1 aas 457-575 were used to predict the two short CC structure. Confidence in the model: 84.3% and % i.d. 31 with template c2ke4A. TMCC1 aas 457-575 were used to predict the long CC structure. Confidence in the model: 98.5% and % i.d. 14 with template c2wpgA.
Quantification and Statistical Analysis
Statistical parameters including the exact value of n, the mean, median, dispersion and precision measures (mean ± SEM) and statistical significance are reported in the Figures and Figure Legends. Data is judged to be statistically significant when p < 0.05 by two-tailed Student’s t test. In figures, asterisks denote statistical significance as calculated by Student’s t test (*, p < 0.05; **, p < 0.005; ***, p < 0.0005). Statistical analysis was performed using both Origin and Excel unpaired t-test.
Supplementary Material
Related to Figure 1. GFP-BioID-Fam21 biotinylates proteins in both HeLa and Cos7 cells, related to Figure 1. (A) Endosomes from HeLa cells expressing mCh-Rab7 (red) and GFP-BioID-Fam21 (green) that were incubated with 50μM biotin for 0hr or 1hr and then fixed and stained to probe for biotinylated proteins using Streptavidin-647 (cyan). White arrowheads show GFP-BioID-Fam21 localization to the endosome bud in both the 0hr and 1hr cells but only biotinylation at the endosome bud in the 1 hr cell. Scale bars =1μm. Background biotin is from endogenously biotinylated proteins found in mitochondria in 0hr images (B) Western probing biotinylation (streptavidin-HRP) of whole cell lysates from either Cos7 cells or HeLa cells expressing myc-BioID-Rab7 or GFP-BioID-Fam21 and treated with biotin for 0hr or 1hr. Asterisks denote a band size indicative of construct self biotinylation. Note that myc-BioID-Rab7 and GFP-BioID-Fam21 both biotinylated proteins to a level that can be seen above endogenously biotinylated proteins (0hr vs. 1hr for each), myc-BioID-Rab7 and GFP-BioID-Fam21 have unique biotinylation profiles (compare 1hr for each condition) and the myc-BioID-Rab7 and GFP-BioID-Fam21 biotinylation profiles are consistent in Cos7 and HeLa cells. (C) A longer exposure of biotinylation profiles than that shown in Figure 1G. (D) TMCC1 protein sequence with BioID-MS-identified peptides in red with Orbitrap MS2 spectra of each identified peptide. (E) Western blot showing TMCC1 siRNA, FAM21 siRNA, and VPS35 siRNAs sufficiently depleted these endogenous proteins in Cos7 cells.
Related to Figure 2. Localization and expression of TMCC family proteins in animal cells. (A-C) Live Cos7 cells expressing mCh-Sec61β (ER, in red in merged panels) and low levels of GFP-TMCC1, 2, or 3 (green in merge). Magnified images of boxed regions reveal that the general ER marker mCh-Sec61β (left panels) localizes to the NE (box 1) and throughout the tubules of the peripheral ER (box 2). In contrast, TMCC1 proteins (middle panels) localize to dynamic domains throughout the tubular ER network (box 2, movie S1) but are excluded from the NE (box 1 merge, right panel) (D-E) Live Cos7 cell expressing GFP-Sec61β with low levels of mCh-TMCC1 (D) or a fixed Cos7 cell expressing mCh-Sec61β with low levels of 3xFlag-TMCC1 (E). The general ER marker GFP-Sec61β or mCh-Sec61β (left panels) localizes throughout the ER in the nuclear envelope and in peripheral ER tubules. mCh-TMCC1 or 3xFlag-TMCC1(middle panels) does not localize strongly in the nuclear envelope and is primarily in the tubular ER network, as shown in the zoom from the boxed regions. (F) Live Cos7 cell expressing mCh-Sec61β (red in merges) with low levels of GFP-TMCC1(Δ1-570) (green in merges). Note TMCC1(Δ1-570) localizes throughout the ER like the general ER marker (G). Immuno-blot shows TMCC1 siRNA efficiently depleted endogenous TMCC1 and re-expression of siRNA resistant GFP-TMCC1, GFP-TMCC2, and GFP-TMCC3 in Cos7 cells. (H-I). Immuno-blot shows TMCC2 and TMCC3 antibodies detect exogenously expressed GFP-TMCC2 (H) and GFP-TMCC3 (I) (yellow asterisks) but not endogenous TMCC2 and TMCC3 (red arrowheads) in both HeLa and Cos7 cells. Boxes in A-F 10μm.
Related to Figure 3. ER-endosome MCS proteins have differing localization at endosome fission sites. (A-B) Merged image of Cos7 cells expressing BFP-Sec61β (ER in red), mCh-Rab7 (LE in blue) and low levels of GFP-TMCC1 (green). Magnified images of the boxed regions show examples of endosomes pre (t=0s) and post (t=2s) fission. White arrowheads highlight vacuolar ER-LE MCSs and white arrows highlight bud ER-LE MCSs where fission occurs. Note GFP-TMCC1 dynamic domains (green) accumulate at MCSs with the site of bud fission (arrow). (C-F) Examples of GFP-tagged ER protein localization at pre fission frames in Cos7 cells expressing BFP-Sec61β (ER in red), mCh-Rab7 (LE in blue) and low levels of (C) GFP-TMCC1, (D) GFP-TMCC1 (Δ1-570), (E) GFP-Protrudin, or (F) GFP-VAPA (green) from Figure 5 A-D. (C) GFP-TMCC1 enrichments are at the bud fission MCS but not at the vacuole MCS. (D) GFP-TMCC1(Δ1-570) signal is at both bud fission MCS and vacuole MCS. (E) GFP-Protrudin accumulations do not enrich at the bud fission MCS but enrichments mark the vacuole MCS. (F) GFP-VAPA signal is at both bud fission MCS and vacuole MCS. In grayscale images in (C-F), the green ROI marks vacuole contact and the yellow ROI marks bud contact.
Related to Figure 4. siRNA resistant GFP-TMCC1 rescues ER-associated endosome fission. (A-D) Live TMCC1 siRNA treated Cos-7 cells expressing BFP-KDEL (ER in blue) and mCh-Rab7 (LE in red) with re-expression of either (A-B) GFP or (C-D) siRNA resistant GFP-TMCC1 (green). Note that ER and endosome morphology and distribution appear normal under both conditions. (B) Magnified images of boxed regions in (A) shows an endosome with a budding domain and cytosolic GFP. (D) Magnified images from boxed regions in (C) shows an example of endosome fission of a budding domain and GFP-TMCC1 localizes to the ER tubule that crosses the budding domain. (E-G) Endosomes were analyzed in TMCC-depleted versus TMCC1-rescued cells (GFP n=32 cells, 277 buds, GFP-TMCC1 n=29 cells, 261 buds). (E) The mean endosome diameter was not affected by TMCC1 rescue but (F) mean bud length was reduced in the GFP-TMCC1 re-expression cells. (G) Rate of endosome fission increased upon TMCC1 rescue (11.6% in GFP versus 21.5% in GFP-TMCC1). (H) Western blot showing TMCC1 siRNA sufficiently depleted endogenous TMCC1 in Cos7 cells but transfected, siRNA resistant GFP-TMCC1 was expressed. Scale bars = 5μm A, C and 1μm in B, D. Error bars represent SEM in E-G and N.S. not significant, **p<0.005 by two-tailed test.
Related to Figure 5. CORO1C is a candidate for TMCC1-meadiated ER recruitment. (A) Merged image of a live Cos-7 expressing BFP-KDEL (ER in green), mCh-Rab7 (LE in red), and GFP-Vps35 (retromer in cyan) following siRNA depletion of FAM21. (B) Magnified time-lapse images of the region boxed in (A) shows an endosome undergoing fission at a Vps35-marked sorting domain. Upon fission, an ER tubule is positioned over Vps35. (C) Percent of buds undergoing fission was measured following siRNA depletion as indicated, but there was no significant decrease in fission upon FAM21 depletion (control 21.5 % n=47 cells, 493 buds, FAM21 KD 21.8 % n=51 cells, 405 buds). (D) Immuno-blot analysis shows efficient depletion of FAM21 following targeted siRNA treatment. (E-F) Live Cos7 cells expressing BFP-KDEL (ER, in blue), mCh-Rab7 (LE in red) and Coronin1A-GFP (E) or Coronin1C-GFP (F) (green). Magnified images of boxed regions show Coronin1A-GFP does not localize to the LE bud as strongly as Coronin1C-GFP in Cos7 cells. (G) Western blot showing CORO1C siRNA depleted endogenous CORO1C in Cos7 cells. Boxes 5μm in A; 10μm in E-F. Scale bars = 5μm in A, 1μm in E-F. In graph C, error bars represent SEM, N.S. not significant, two-tailed test.
Related to Figure 6. ER tracking with Coronin 1C puncta in control versus TMCC1 KD cells. (A-B) Contact between ER tubules and endosome buds was scored in live Cos7 cells expressing BFP-KDEL (ER in red), mCh-Rab7 (LEs in blue), and Coronin1C-GFP (CORO1C labels actin at the base of the bud in green). Cells were either treated with control siRNA (A) or with TMCC1 KD siRNA (B). Magnified time lapse images were used to score co-localization between ER tubules and CORO1C puncta at the base of the bud over time (white arrowheads). Vacuole contact was also monitored (yellow arrowheads) (C) Immuno-blot analysis shows efficient depletion of TMCC1 following targeted siRNA treatment.
Related to Figure 7. Phyre2 prediction to analyze TMCC1 domains. (A) Primary structure of TMCC1 predicted coiled coil (CC) and trans membrane (TM) domain regions used for Phyre2 analysis. (B) Predicted Phyre2 structure of region containing two short CCs. (C) Predicted Phyre2 structure of region containing the longer CC.
Related to Figure 2. TMCC1 accumulations are dynamic on ER tubules. A 10μm region of a Cos7 cell expressing the general ER marker mch- Sec6β (ER in red) and GFP-TMCC1 (in green) imaged every 5 s for 2min. Left panel mCh- Sec61β, middle panel GFP-TMCC1, right panel merge.
Related to Figure 3. TMCC1 accumulates at the site of endosome fission. A 5μm region of a Cos7 cell expressing mCh-Rab7 (LE in light blue), the general ER marker BFP-8Sec6β (ER in red) and GFP-TMCC1 (in green) imaged every 2s, showing 10 seconds pre fission, the fission event, and then 10 seconds post fission.
Related to Figure 4. (A) Examples of a successful fission event in a control siRNA treated Cos7 cell versus (B) an unsuccessful fission event in a TMCC1 knockdown siRNA treated Cos7 cell.
Related to Figure 6. ER crossing Coronin 1C puncta at endosome bud in Cos7 cells transfected with GFP-Coronin 1C (green), mch-Rab7 (LE in blue), BFP-KDEL (ER in red) in control siRNA conditions (A) versus TMCC1 knockdown siRNA conditions (B).
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Reticulon 3 (rabbit) | YenZym Antibodies | Injected purified N terminal fragment and affinity purified from rabbit |
| Tom20 (rabbit) | Santa Cruz Biotechnologies | Cat# sc-11415 |
| α-tubulin (rabbit) | Abcam | Cat# ab 18251 |
| Rab7 (rabbit) | Cell Signaling | Cat# 99367S |
| TMCC1 (rabbit) | Zhang et. al 2014 | Affinity purified from rabbit injected with TMCC1 antigen |
| Fam21 (rabbit) | gift from Billadeau | Gomez and Billadeau 2009 |
| VPS35 (rabbit) | Abcam | Cat# ab97545 |
| CORO 1C (rabbit) | Proteintech | Cat# 1749-1-Ap |
| M6PR 2G11 (mouse) | ThermoFisher | Cat# MAI-066 |
| GAPDH (rabbit) | Sigma-Aldrich | Cat# G9545 |
| Giantin (rabbit) | BioLegend | Cat# 924302 |
| Flag M2 (mouse) | Sigma-Aldrich | Cat# F1804 |
| Streptavidin-HRP | Invitrogen | Cat# 434323 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lipofectamine 2000 | Invitrogen (now ThermoFisher) | cat# 11668-019 |
| Lipofectamine 3000 | Invitrogen (now ThermoFisher) | cat# 3000-015 |
| Dharmafect | Dharmacon (now ThermoFisher) | cat# T-2001-03 |
| Dynabead MyOne Streptavidin C1 | Invitrogen (now ThermoFisher) | cat# 65001 |
| Critical Commercial Assays | ||
| BCA Assay | Pierce (now ThermoFisher) | cat# 23227 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | ThermoFisher | cat# 34580 |
| Experimental Models: Cell Lines | ||
| Cos-7 cells | ATCC | CRL-1615 |
| HeLa cells | ATCC | CCL-2 |
| Oligonucleotides | ||
| ON-TARGETplus Human TMCC1 (23023) siRNA SMARTpool | Dharmacon | cat# L-023594 |
| ON-TARGETplus Human WASHC2C (253725) siRNA - SMARTpool | Dharmacon | cat# L-029678 |
|
ON-TARGETplus Human VPS35 (55737) siRNA SMARTpool |
Dharmacon | cat# L-010894 |
|
ON-TARGETplus Human CORO1C (23603) siRNA SMARTpool |
Dharmacon | cat# L-017331 |
| Recombinant DNA | ||
| GFP-Sec61β | Voeltz lab, Shibata et al., 2008 | |
| BFP-Sec61β | Voeltz lab, Zurek et al., 2011 | |
| mCh-Sec61β | Voeltz lab, Zurek et al., 2011 | |
| BFP-KDEL | Voeltz lab, Friedman et al., 2011 | |
| GFP-Rab7A | Voeltz lab, Rowland and Chitwood et al., 2014 | |
| mCh-Rab7A | Voeltz lab, Rowland and Chitwood et al., 2014 | |
| mCh-FAM21 | Voeltz lab, Rowland and Chitwood et al., 2014 | |
| shF AM21/HA-YFP-FAM21 | gift from Dr. Billadeau, Gomez and Billadeau et al., 2009 | |
| myc-BioID | Addgene, Roux et al., 2012 | cat# 35700 |
| GFP-BioID-FAM21 | PCR amplify BioID from myc-BioID and ligate into Xho1/Kpn1 sites of GFP-Fam21 | |
| myc-BioID-Rab7 | PCR amplify Rab7A from GFP-Rab7A and ligating it into the Xho1/BamH1 sites of myc-BioID | |
| GFP-TMCC1 | PCR amplify TMCC1 NM_001336192.1, ligate into Xho1/Kpn1 sites of pAcGFP-C1 | |
| GFP-TMCC2 | PCR amplify TMCC2 NM_001297611.1, ligate into Xho1/Kpn1 sites of pAcGFP-C1 | |
| GFP-TMCC3 | PCR amplify TMCC3 NM 020698.3, ligate into Xho1/Kpn1 sites of pAcGFP-C1 | |
| mCh-TMCC1 | PCR amplify mCherry, remove GFP and ligate into Nhe1/Xho1 sites of GFP-TMCC1 | |
| 3xFlag-TMCC1 | PCR amplify 3x-Flag, remove GFP and ligate into Nhe1/Xho1 sites of GFP-TMCC1 | |
| siRNA resistant GFP-TMCC1 | Used GFP-TMCC1 and series of site directed mutagenesis reactions to mutate base pairs in the 4 siRNA binding regions | |
| GFP-TMCC1 D1-570 | PCR amplify GFP-TMCC1 with forward primer start at bp 1711 | |
| GFP-TMCC1 D1-319 | PCR amplify GFP-TMCC1 with forward primer start at bp 958 | |
| GFP-TMCC 1 D420-575 | Created with GFP-TMCC1 using Gibson reaction with primers aligned with part of sequence but skipped bps 1378-1725 | |
| GFP-VAPA | Jonathan Friedman PCR amplify VAPA NM_003574.5, ligate into BglII/BamH1 sites of pAcGFP-C1 | |
| GFP-Protrudin | PCR amplify Protrudin NM_001002262, ligate into Kpn1/BamH1 sites of pAcGFP-C1 | |
| Coronin1C-GFP | gift from Dr. Puthenveedu and Dr. von Zastrow, Putenveedu et al 2010 | |
| Coronin1A-GFP | PCR amplify Coronin 1A NM_00119333, ligate into Xho1/Kpn1 sites of pAcGFP-N1 | |
| pAcGFP-C1 | Clonetech | Cat# 632470 |
| pAcGFP-N1 | Clonetech | Cat# 632469 |
| Software and Algorithms | ||
| Fiji | Schindelin et al., 2012 | https://fiji.sc/ |
| Origin | OriginLab, Northampton, MA | https://www.originlab.com/ |
| Phyre2 prediction program | Kelley LA et al., 2015 | http://www.sbg.bio.ic.ac.uk/phyre2/ |
Hoyer et al. Highlights.
Targeted BioID identifies new family of ER-endosome MCS proteins termed TMCC. (78)
TMCC1 localizes to dynamic ER domains that define the position of endosome fission. (84)
TMCC1 regulates ER recruitment to endosome sorting domains for bud fission. (76)
Coronin1C at buds is a receptor for TMCC1 dependent ER recruitment to endosomes. (81)
Acknowledgments:
We thank A. Rowland, J. Friedman and L. Westrate for helpful discussions. We thank W. Harper for sharing data from his Bioplex Interactome prior to publication. We thank M. Puthenveedu, D. Billadeau, J. Friedman, and K. Heuer for generating plasmids used here. M.J.H. was supported by an NSF Graduate Research Fellowship (DGE 1144083) and a pre-doctoral training grant from the NIH (T32 GM08759). This work is also supported by a Research Scholar Grant from the American Cancer Society to G.K.V., by grants from the NIH to G.K.V. (GM083977 and GM120998), and by a DARPA cooperative agreement to W.M.O. (13-34-RTA-FP-007).
Footnotes
Declaration of Interests:
The authors declare that there are no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Allison R, Lumb JH, Fassier C, Connell JW, Ten Martin D, Seaman MNJ, Hazan J, and Reid E (2013). An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J. Cell Biol 202, 527–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison R, Edgar JR, Pearson G, Rizo T, Newton T, Günther S, Berner F, Hague J, Connell JW, Winkler J, et al. (2017). Defects in ER-endosome contacts impact lysosome function in hereditary spastic paraplegia. J. Cell Biol 216, 1337–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpy F, Rousseau A, Schwab Y, Legueux F, Stoll I, Wendling C, Spiegelhalter C, Kessler P, Mathelin C, Rio M-C, et al. (2013). STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J. Cell Sci 126, 5500–5512. [DOI] [PubMed] [Google Scholar]
- Arighi CN, Harmell LM, Aguilar RC, Haft CR, and Bonifacino JS (2004). Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol 165, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, and Rojas R (2006). Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol 7, 568–579. [DOI] [PubMed] [Google Scholar]
- Burd C, and Cullen PJ (2014). Retromer: a master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol 6, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KT, Roadcap DW, Holoweckyj N, and Bear JE (2012). Coronin 1C harbours a second actin-binding site that confers co-operative binding to F-actin. Biochem. J 444, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordás G, Renken C, Várnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella C. a, and Hajnóczky G. (2006). Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol 174, 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, and Gautreau A (2009). The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712–723. [DOI] [PubMed] [Google Scholar]
- Dong R, Saheki Y, Swarup S, Lucast L, Harper JW, and De Camilli P (2016). Endosome-ER Contacts Control Actin Nucleation and Retromer Function through VAP-Dependent Regulation of PI4P. Cell 166, 408–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman CL, Hesketh G, and Seaman MNJ (2014). RME-8 coordinates the activity of the WASH complex with the function of the retromer SNX dimer to control endosomal tubulation. J. Cell Sci 127, 2053–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, and Voeltz GK (2010). ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol 190, 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, and Voeltz GK (2011). ER tubules mark sites of mitochondrial division. Science 334, 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M, Achard V, Blanchoin L, and Goode BL (2009). Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol. Cell 34, 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A, Oguievetskaia K, and Ungermann C (2014). Function and regulation of the endosomal fusion and fission machineries. Cold Spring Harb. Perspect. Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, and Billadeau DD (2009). A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour ME, Breusegem S.Y. a, Antrobus R, Freeman C, Reid E, and Seaman MNJ (2010). The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J. Cell Sci 123, 3703–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour ME, Breusegem SY, and Seaman MNJ (2012). Recruitment of the endosomal WASH complex is mediated by the extended “tail” of Fam21 binding to the retromer protein Vps35. Biochem. J 442, 209–220. [DOI] [PubMed] [Google Scholar]
- Humphries CL, Balcer HI, D’Agostino JL, Winsor B, Drubin DG, Barnes G, Andrews BJ, and Goode BL (2002). Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J. Cell Biol 159, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SD, Townley AK, Danson CM, Cullen PJ, and Stephens DJ (2013). Microtubule motors mediate endosomal sorting by maintaining functional domain organization. J. Cell Sci 126, 2493–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Billadeau DD, and Rosen MK (2012). Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol. Biol. Cell 23, 2352–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LA, Mezulis S, Yates C, Wass M, and Sternberg M (2015). The Phyre2 web portal for protein modelling, prediction, and analysis. Nat. Protoc 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, and Roux KJ (2014). Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. U. S. A 111, E2453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, and Chen LB (1988). Dynamic behavior of endoplasmic reticulum in living cells. Cell 54, 37–46. [DOI] [PubMed] [Google Scholar]
- Liu S-L, Needham KM, May JR, and Nolen BJ (2011). Mechanism of a concentration-dependent switch between activation and inhibition of Arp2/3 complex by coronin. J. Biol. Chem 286, 17039–17046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford AG, Stefan CJ, Yuan HL, Macgurn J. a, and Emr SD (2012). ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell 23, 1129–1140. [DOI] [PubMed] [Google Scholar]
- Matsuzaki F, Shirane M, Matsumoto M, and Nakayama KI (2011). Protrudin serves as an adaptor molecule that connects KIF5 and its cargoes in vesicular transport during process formation. Mol. Biol. Cell 22, 4602–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, and McGraw TE (2004). Endocytic recycling. Nat. Rev. Mol. Cell Biol 5, 121–132. [DOI] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, and Antonny B (2013). A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 155, 830–843. [DOI] [PubMed] [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, and Ahn NG (2005). Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502. [DOI] [PubMed] [Google Scholar]
- Peretti D, Dahan N, Shimoni E, Hirschberg K, and Lev S (2008). Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell 19, 3871–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, and Voeltz GK (2016). Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol 17, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA (2014). Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol 205, 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M. a, Lauffer B, Temkin P, Vistein R, Carlton P, Thorn K, Taunton J, Weiner OD, Parton RG, and von Zastrow M (2010). Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell 143, 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Wenzel EM, Pedersen NM, Olsvik H, Schink KO, Schultz SW, Vietri M, Nisi V, Bucci C, Brech A, et al. (2015). Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature 520, 234–238. [DOI] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, and Neefjes J (2009). Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol 185, 1209–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, and Burke B (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland AA, Chitwood PJ, Phillips MJ, and Voeltz GK (2014). ER contact sites define the position and timing of endosome fission. Cell 159, 1027–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador-Gallego R, Hoyer MJ, and Voeltz GK (2017). SnapShot: Functions of Endoplasmic Reticulum Membrane Contact Sites. Cell 171, 1224–1224. e1. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ (2004). Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol 165, 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ (2008). Endosome protein sorting: motifs and machinery. Cell. Mol. Life Sci 65, 2842–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN McCaffery M Emr S (1998). Determination of plasma catecholamine and vasoactive peptide concentrations: clinical usefulness. J. Cell Biol 142, 665–681.9700157 [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, and Voeltz GK (2008). The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem 283, 18892–18904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M, Song J, Wong JR, Weiss MJ, and Chen LB (1984). Localization of endoplasmic reticulum in living and glutaraldehyde-fixed cells with fluorescent dyes. Cell 38, 101–108. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, and Salmon ED (1998). Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol 8, 798–806. [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR (2016). Quantitative Evaluation of Filter Aided Sample Preparation (FASP) and Multienzyme Digestion FASP Protocols. Anal. Chem 88, 5438–5443. [DOI] [PubMed] [Google Scholar]
- Wu MM, Buchanan J, Luik RM, and Lewis RS (2006). Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol 174, 803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac AL, Goldman YE, Holzbaur ELF, and Ostap EM (2013). Local cytoskeletal and organelle interactions impact molecular-motor- driven early endosomal trafficking. Curr. Biol 23, 1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Kho Y-S, Wang Z, Chiang YT, Ng GKH, Shaw P-C, Wang Y, and Qi RZ (2014). Transmembrane and coiled-coil domain family 1 is a novel protein of the endoplasmic reticulum. PLoS One 9, e85206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek N, Sparks L, and Voeltz G (2011). Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic 12, 28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 1. GFP-BioID-Fam21 biotinylates proteins in both HeLa and Cos7 cells, related to Figure 1. (A) Endosomes from HeLa cells expressing mCh-Rab7 (red) and GFP-BioID-Fam21 (green) that were incubated with 50μM biotin for 0hr or 1hr and then fixed and stained to probe for biotinylated proteins using Streptavidin-647 (cyan). White arrowheads show GFP-BioID-Fam21 localization to the endosome bud in both the 0hr and 1hr cells but only biotinylation at the endosome bud in the 1 hr cell. Scale bars =1μm. Background biotin is from endogenously biotinylated proteins found in mitochondria in 0hr images (B) Western probing biotinylation (streptavidin-HRP) of whole cell lysates from either Cos7 cells or HeLa cells expressing myc-BioID-Rab7 or GFP-BioID-Fam21 and treated with biotin for 0hr or 1hr. Asterisks denote a band size indicative of construct self biotinylation. Note that myc-BioID-Rab7 and GFP-BioID-Fam21 both biotinylated proteins to a level that can be seen above endogenously biotinylated proteins (0hr vs. 1hr for each), myc-BioID-Rab7 and GFP-BioID-Fam21 have unique biotinylation profiles (compare 1hr for each condition) and the myc-BioID-Rab7 and GFP-BioID-Fam21 biotinylation profiles are consistent in Cos7 and HeLa cells. (C) A longer exposure of biotinylation profiles than that shown in Figure 1G. (D) TMCC1 protein sequence with BioID-MS-identified peptides in red with Orbitrap MS2 spectra of each identified peptide. (E) Western blot showing TMCC1 siRNA, FAM21 siRNA, and VPS35 siRNAs sufficiently depleted these endogenous proteins in Cos7 cells.
Related to Figure 2. Localization and expression of TMCC family proteins in animal cells. (A-C) Live Cos7 cells expressing mCh-Sec61β (ER, in red in merged panels) and low levels of GFP-TMCC1, 2, or 3 (green in merge). Magnified images of boxed regions reveal that the general ER marker mCh-Sec61β (left panels) localizes to the NE (box 1) and throughout the tubules of the peripheral ER (box 2). In contrast, TMCC1 proteins (middle panels) localize to dynamic domains throughout the tubular ER network (box 2, movie S1) but are excluded from the NE (box 1 merge, right panel) (D-E) Live Cos7 cell expressing GFP-Sec61β with low levels of mCh-TMCC1 (D) or a fixed Cos7 cell expressing mCh-Sec61β with low levels of 3xFlag-TMCC1 (E). The general ER marker GFP-Sec61β or mCh-Sec61β (left panels) localizes throughout the ER in the nuclear envelope and in peripheral ER tubules. mCh-TMCC1 or 3xFlag-TMCC1(middle panels) does not localize strongly in the nuclear envelope and is primarily in the tubular ER network, as shown in the zoom from the boxed regions. (F) Live Cos7 cell expressing mCh-Sec61β (red in merges) with low levels of GFP-TMCC1(Δ1-570) (green in merges). Note TMCC1(Δ1-570) localizes throughout the ER like the general ER marker (G). Immuno-blot shows TMCC1 siRNA efficiently depleted endogenous TMCC1 and re-expression of siRNA resistant GFP-TMCC1, GFP-TMCC2, and GFP-TMCC3 in Cos7 cells. (H-I). Immuno-blot shows TMCC2 and TMCC3 antibodies detect exogenously expressed GFP-TMCC2 (H) and GFP-TMCC3 (I) (yellow asterisks) but not endogenous TMCC2 and TMCC3 (red arrowheads) in both HeLa and Cos7 cells. Boxes in A-F 10μm.
Related to Figure 3. ER-endosome MCS proteins have differing localization at endosome fission sites. (A-B) Merged image of Cos7 cells expressing BFP-Sec61β (ER in red), mCh-Rab7 (LE in blue) and low levels of GFP-TMCC1 (green). Magnified images of the boxed regions show examples of endosomes pre (t=0s) and post (t=2s) fission. White arrowheads highlight vacuolar ER-LE MCSs and white arrows highlight bud ER-LE MCSs where fission occurs. Note GFP-TMCC1 dynamic domains (green) accumulate at MCSs with the site of bud fission (arrow). (C-F) Examples of GFP-tagged ER protein localization at pre fission frames in Cos7 cells expressing BFP-Sec61β (ER in red), mCh-Rab7 (LE in blue) and low levels of (C) GFP-TMCC1, (D) GFP-TMCC1 (Δ1-570), (E) GFP-Protrudin, or (F) GFP-VAPA (green) from Figure 5 A-D. (C) GFP-TMCC1 enrichments are at the bud fission MCS but not at the vacuole MCS. (D) GFP-TMCC1(Δ1-570) signal is at both bud fission MCS and vacuole MCS. (E) GFP-Protrudin accumulations do not enrich at the bud fission MCS but enrichments mark the vacuole MCS. (F) GFP-VAPA signal is at both bud fission MCS and vacuole MCS. In grayscale images in (C-F), the green ROI marks vacuole contact and the yellow ROI marks bud contact.
Related to Figure 4. siRNA resistant GFP-TMCC1 rescues ER-associated endosome fission. (A-D) Live TMCC1 siRNA treated Cos-7 cells expressing BFP-KDEL (ER in blue) and mCh-Rab7 (LE in red) with re-expression of either (A-B) GFP or (C-D) siRNA resistant GFP-TMCC1 (green). Note that ER and endosome morphology and distribution appear normal under both conditions. (B) Magnified images of boxed regions in (A) shows an endosome with a budding domain and cytosolic GFP. (D) Magnified images from boxed regions in (C) shows an example of endosome fission of a budding domain and GFP-TMCC1 localizes to the ER tubule that crosses the budding domain. (E-G) Endosomes were analyzed in TMCC-depleted versus TMCC1-rescued cells (GFP n=32 cells, 277 buds, GFP-TMCC1 n=29 cells, 261 buds). (E) The mean endosome diameter was not affected by TMCC1 rescue but (F) mean bud length was reduced in the GFP-TMCC1 re-expression cells. (G) Rate of endosome fission increased upon TMCC1 rescue (11.6% in GFP versus 21.5% in GFP-TMCC1). (H) Western blot showing TMCC1 siRNA sufficiently depleted endogenous TMCC1 in Cos7 cells but transfected, siRNA resistant GFP-TMCC1 was expressed. Scale bars = 5μm A, C and 1μm in B, D. Error bars represent SEM in E-G and N.S. not significant, **p<0.005 by two-tailed test.
Related to Figure 5. CORO1C is a candidate for TMCC1-meadiated ER recruitment. (A) Merged image of a live Cos-7 expressing BFP-KDEL (ER in green), mCh-Rab7 (LE in red), and GFP-Vps35 (retromer in cyan) following siRNA depletion of FAM21. (B) Magnified time-lapse images of the region boxed in (A) shows an endosome undergoing fission at a Vps35-marked sorting domain. Upon fission, an ER tubule is positioned over Vps35. (C) Percent of buds undergoing fission was measured following siRNA depletion as indicated, but there was no significant decrease in fission upon FAM21 depletion (control 21.5 % n=47 cells, 493 buds, FAM21 KD 21.8 % n=51 cells, 405 buds). (D) Immuno-blot analysis shows efficient depletion of FAM21 following targeted siRNA treatment. (E-F) Live Cos7 cells expressing BFP-KDEL (ER, in blue), mCh-Rab7 (LE in red) and Coronin1A-GFP (E) or Coronin1C-GFP (F) (green). Magnified images of boxed regions show Coronin1A-GFP does not localize to the LE bud as strongly as Coronin1C-GFP in Cos7 cells. (G) Western blot showing CORO1C siRNA depleted endogenous CORO1C in Cos7 cells. Boxes 5μm in A; 10μm in E-F. Scale bars = 5μm in A, 1μm in E-F. In graph C, error bars represent SEM, N.S. not significant, two-tailed test.
Related to Figure 6. ER tracking with Coronin 1C puncta in control versus TMCC1 KD cells. (A-B) Contact between ER tubules and endosome buds was scored in live Cos7 cells expressing BFP-KDEL (ER in red), mCh-Rab7 (LEs in blue), and Coronin1C-GFP (CORO1C labels actin at the base of the bud in green). Cells were either treated with control siRNA (A) or with TMCC1 KD siRNA (B). Magnified time lapse images were used to score co-localization between ER tubules and CORO1C puncta at the base of the bud over time (white arrowheads). Vacuole contact was also monitored (yellow arrowheads) (C) Immuno-blot analysis shows efficient depletion of TMCC1 following targeted siRNA treatment.
Related to Figure 7. Phyre2 prediction to analyze TMCC1 domains. (A) Primary structure of TMCC1 predicted coiled coil (CC) and trans membrane (TM) domain regions used for Phyre2 analysis. (B) Predicted Phyre2 structure of region containing two short CCs. (C) Predicted Phyre2 structure of region containing the longer CC.
Related to Figure 2. TMCC1 accumulations are dynamic on ER tubules. A 10μm region of a Cos7 cell expressing the general ER marker mch- Sec6β (ER in red) and GFP-TMCC1 (in green) imaged every 5 s for 2min. Left panel mCh- Sec61β, middle panel GFP-TMCC1, right panel merge.
Related to Figure 3. TMCC1 accumulates at the site of endosome fission. A 5μm region of a Cos7 cell expressing mCh-Rab7 (LE in light blue), the general ER marker BFP-8Sec6β (ER in red) and GFP-TMCC1 (in green) imaged every 2s, showing 10 seconds pre fission, the fission event, and then 10 seconds post fission.
Related to Figure 4. (A) Examples of a successful fission event in a control siRNA treated Cos7 cell versus (B) an unsuccessful fission event in a TMCC1 knockdown siRNA treated Cos7 cell.
Related to Figure 6. ER crossing Coronin 1C puncta at endosome bud in Cos7 cells transfected with GFP-Coronin 1C (green), mch-Rab7 (LE in blue), BFP-KDEL (ER in red) in control siRNA conditions (A) versus TMCC1 knockdown siRNA conditions (B).