Abstract
The extent to which early events shape tumor evolution is largely uncharacterized, even though a better understanding of these early events may help identify key vulnerabilities in advanced tumors. Here, using genetically defined mouse models of small cell lung cancer (SCLC), we uncovered distinct metastatic programs attributable to the cell-type of origin. In one model, tumors gain metastatic ability through amplification of the transcription factor Nfib and a widespread increase in chromatin accessibility, while in the other model, tumors become metastatic in the absence of Nfib-driven chromatin alterations. Gene expression and chromatin accessibility analyses identify distinct mechanisms as well as markers predictive of metastatic progression in both groups. Underlying the difference between the two programs was the cell-type of origin of the tumors, with Nfib-independent metastases arising from mature neuroendocrine cells. Our findings underscore the importance of the identity of cell-type of origin in influencing tumor evolution and metastatic mechanisms.
Keywords: SCLC, cancer metastasis, cell-of-origin, tumor heterogeneity, Nfib
INTRODUCTION
Most cancer patients die from metastatic disease, however many key aspects of metastatic progression remain poorly understood. In particular, the nature of the changes that drive metastasis and the potential impact of early events in tumorigenesis upon the direction of this evolution are largely unexplored. A better understanding of these mechanisms could help diagnose and treat patients more effectively (1, 2).
Small cell lung cancer (SCLC) is one of the most metastatic and lethal of all major cancer types (3, 4). SCLC is thought to acquire metastatic ability early in the course of tumor progression, but emerging evidence suggests that this metastatic ability is not inherent. Rather, genetic events such as upregulation of CXCR4 and NeuroD1, and/or amplification of Nfib are critical for SCLC invasion and metastasis in at least a subset of patients (5–9). Recent data from genetically engineered mouse models and human tumors have uncovered multiple levels of heterogeneity in SCLC; however, how this heterogeneity pertains to metastatic progression is largely unexplored (10–19).
SCLC is a neuroendocrine cancer, thus it made sense when deletion of the key SCLC tumor suppressor genes Rb1 and Trp53 in pulmonary neuroendocrine cells uncovered these cells as one cell-type of origin for SCLC in mouse models ((20–22), reviewed in (4)). Interestingly, while pulmonary neuroendocrine cells are quite rare, induction of these same genetic alterations in the much more prevalent lung epithelial cell types expressing either Scgb1a1 (coding for CC10, a marker of Club cells) or Sftpc (coding for surfactant protein C (SPC), a marker of alveolar type II cells) demonstrated that these cells have very little, if any ability to serve as the cell-of-origin of SCLC (20, 21). Nonetheless, the lung epithelium contains many diverse cell types (23–25), and whether SCLC can be initiated from other cell types is unknown.
Here, through detailed molecular characterization of primary tumors and metastases, we identify two discrete paths by which SCLC gains metastatic ability. Our data indicate that the same genomic alterations in different cell types gives rise to distinct subtypes of SCLC, and that the founding cell-type of origin can define the trajectory of tumor progression.
RESULTS
Mouse SCLC initiated from adult neuroendocrine cells gains metastatic ability without upregulation of Nfib
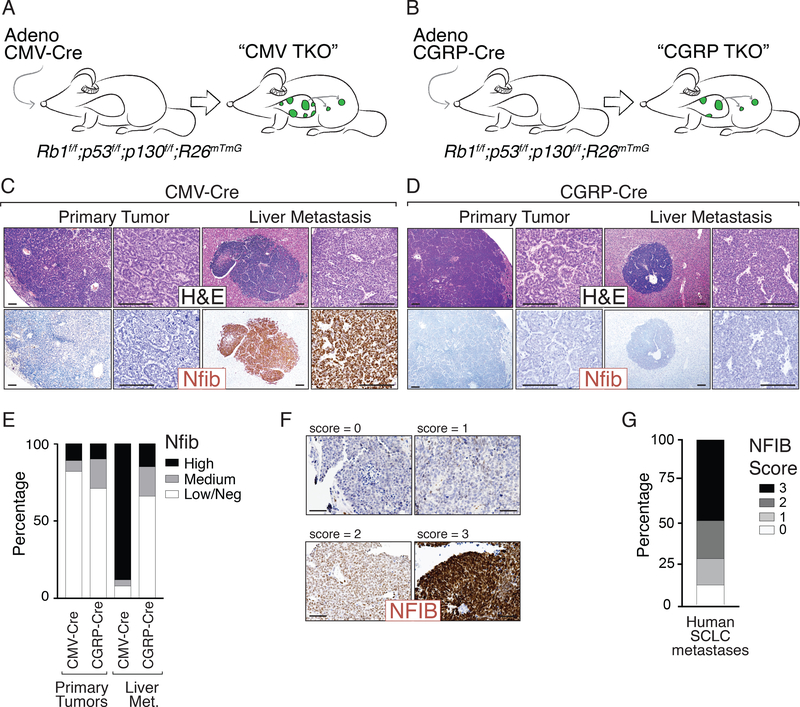
To study the mechanisms underlying SCLC metastasis, we initially used the well characterized Rb1flox/flox;p53flox/flox;p130flox/flox;R26mTmG (TKO;mTmG) mouse model, in which we initiated tumors via intratracheal delivery of an adenoviral vector expressing Cre recombinase under the control of the broadly-expressed CMV promoter (Ad-CMV-Cre) (7, 26). Thus, in this model p53, Rb1, and p130 are inactivated in many different cell types in the lung. We initially uncovered that in this “CMV TKO” model, amplification of the Nfib genomic locus and high expression of the Nfib transcription factor in primary tumors is an important step during metastatic progression (7). To further investigate SCLC metastatic progression in a model in which the tumors arise from a defined cell-type, we subsequently initiated tumors in TKO;mTmG mice with Ad-CGRP-Cre, which specifically directs Cre expression to mature, CGRP-expressing neuroendocrine cells (21, 22, 27) (Fig. 1A-B). These “CGRP TKO” mice developed many fewer SCLCs than CMV TKO mice even when transduced with a 10- to 20-fold higher titer of Ad-CGRP-Cre (Supplementary Fig. S1A). Nonetheless, both CMV TKO and CGRP TKO mice developed SCLC and widespread metastatic disease with metastasis to multiple organs, including the lymph nodes and liver, 6–9 months after tumor initiation. (Fig. 1A-D, Supplementary Fig. S1B-H). Previous studies showed that Ad-CMV-Cre and Ad-CGRP-Cre can each also initiate metastatic SCLC in Rb1flox/flox;p53flox/flox mice, but again, 15-fold more Ad-CGRP-Cre was used to initiate tumors (21, 28).
Figure 1: SCLC initiated from pulmonary neuroendocrine cells metastasizes without upregulating Nfib.
A-B. Mouse models of SCLC. Rb1flox/flox;p53 flox/flox;p130 flox/flox;R26mTmG (TKO;mTmG) mice were transduced with either Adeno-CMV-Cre (“CMV TKO”) or Adeno-CGRP-Cre (“CGRP TKO”) to initiate SCLC.
C-D. Representative H&E images of SCLC tumors from CMV TKO and CGRP TKO mice. Immunostaining for Nfib on primary tumors and metastases is shown. Scale bars = 100μm.
E. Quantification of Nfib expression in primary tumors and liver metastases from CMV TKO and CGRP TKO mice. Most of the metastases in CMV TKO mice are Nfibpositive, while most of the in CGRP TKO mice are Nfiblow/negative.
F. Representative immunostaining for NFIB on human SCLC brain metastases. Expression score for NFIB is indicated. Scale bars = 100μm.
G. Quantification of NFIB expression in human SCLC metastases (including lymph node and brain metastases, N=43).
As part of our characterization of primary tumors and metastases in CMV TKO and CGRP TKO mice, we performed immunostaining for the pro-metastatic transcription factor Nfib. Surprisingly, in contrast to CMV TKO metastases, Nfib was undetectable in the vast majority of CGRP TKO metastases (Fig. 1C-E, Supplementary Fig. S1C-H). In general, primary tumors in CMV TKO mice were characterized by a “rosette” growth pattern with more differentiated glandular structures intermixed with less well differentiated areas, whereas primary tumors in CGRP TKO mice were predominantly characterized by a “solid-nested” growth pattern. Notably, metastases in both models uniformly exhibited solid-nested growth and no rosette formation. Primary tumors with rosette histology were uniformly Nfibnegative/low in both models (Supplementary Table S1). Most of the solid-nested areas in CMV TKO mice were Nfibhigh, while the solid-nested areas in CGRP TKO were mostly Nfibnegative/low (Supplementary Fig. S1G-H). The majority of the macro-metastases in the lymph nodes and liver, as well as most of the cancer cells growing within the pulmonary lymphatics, were Nfibhigh in CMV TKO mice. In contrast, most metastases and lymphatic invasive cells in CGRP TKO mice were Nfibnegative/low (Fig. 1C-E and Supplementary Fig. S1E-F). Similarly, across multiple other genotypes (including Rb1flox/flox;p53flox/flox, Rb1flox/flox;p53flox/flox;Ptenflox/+, and Rb1flox/flox;p53flox/flox;Ptenflox/flox mice), SCLC tumors initiated by Ad-CMV-Cre generally gave rise to Nfibhigh metastases while SCLC tumors initiated by Ad-CGRP-Cre generally gave rise to Nfibnegative/low metastases (5, 29) (Supplementary Fig. S2A). Importantly, the expression of NFIB is also heterogeneous in human SCLC lymph node and brain metastases, suggesting that diverse mechanisms of tumor progression also exist in human SCLC (Fig. 1F-G, Supplementary Fig. S2D-E) (6, 7, 30). Collectively, these data indicate that SCLC initiated from CGRPpositive cells in the mouse lung can metastasize in the absence of Nfib upregulation and this may recapitulate tumor progression in a subset of SCLC patients.
Murine SCLC requires progression prior to dissemination
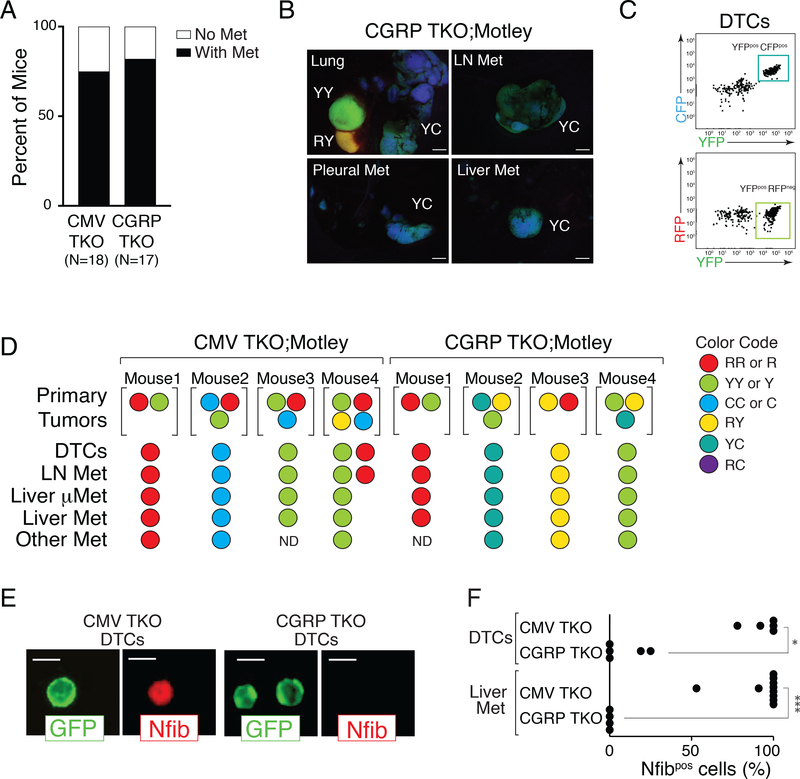
In the CMV TKO model, genomic amplification of Nfib precedes dissemination, consistent with tumor evolution as a prerequisite for metastasis (5–7). The absence of Nfib upregulation in most tumors in CGRP TKO mice prompted us to investigate whether SCLC in this model also progresses to gain metastatic ability or if these tumors were inherently metastatic. Even at a late time point (7–11 months after tumor initiation), when they harbored multiple large primary tumors, not every CMV TKO or CGRP TKO mouse had detectable macro-metastases, micro-metastases, or even disseminated tumor cells (DTCs) (Fig. 2A). These initial data suggested that not all CMV-Cre or CGRP-Cre TKO tumors have the ability to disseminate and metastasize.
Figure 2: Tumor progression is required in both CMV TKO and CGRP TKO mice.
A. Occurrence of metastasis in CMV TKO and CGRP TKO mice. Not every mouse developed metastatic disease at the time of analysis even though each mouse in this analysis had several large primary tumors (7–11 months after initiation).
B. Representative images of tumors and metastases from TKO;Motley mice with CGRP-Cre-initiated SCLC. Primary tumors existed in multiple colors, while all metastases from the same mouse had the same color, suggesting that these metastases are clonally related to one primary tumor. Scale bar = 2mm. Color code, R = RFP, Y = YFP, C = CFP.
C. Representative FACS analysis on disseminated tumor cells (DTCs) from a CGRP TKO;Motley mouse. All The DTCs of this mouse have the same color (CFPpositive and YFPpositive).
D. Summary of results from 4 CMV TKO;Motley mice and 4 CGRP TKO;Motley mice. The colors of the largest primary tumors, disseminated tumors cells (DTCs), and metastases are shown. mMet, micrometastases. ND, not detected.
E-F. Immunofluorescence staining for Nfib on FACS-isolated GFPpositive DTCs from the pleural cavity and cancer cells from liver metastases from CMV TKO and CGRP TKO mice. (E) Representative images of DTCs from CMV TKO and CGRP TKO mice. Membrane GFP staining is encoded by the recombined R26mTmG allele. Scale bars = 10 μm. (F) Quantification of Nfib expression in DTCs and cancer cells from liver metastases from CMV TKO and CGRP TKO mice. CMV TKO data is from Denny, Yang et al, 2016. Each dot represents one mouse. * P = 0.0097, *** P=0.001, Mann-Whitney test.
To further investigate whether only some tumors possess metastatic ability, we incorporated a multi-color reporter allele (R26LSL-Motley) (31) into the Rb1flox/flox;p53flox/flox;p130flox/flox TKO model. In these TKO;Motley mice, clonally-derived tumors are labeled with different combinations of fluorescent markers (Supplementary Fig. S3A). Importantly, in individual TKO;Motley mice with Ad-CMV-Cre- or Ad-CGRP-Cre-initiated SCLC, the metastases were almost always of one color, suggesting that they originated from a single primary tumor. Not only were metastases clonal in origin, but DTCs in the pleural cavity were also almost always of one color (Fig. 2B-D, Supplementary Fig. S3B). Thus, our data are consistent with not all tumors in CGRP TKO mice containing cells with the ability to readily disseminate and metastasize.
While Nfib levels are low in the CGRP TKO model, we considered whether Nfib might be transiently upregulated during metastatic dissemination, then downregulated to generate Nfibnegative/low metastases. To investigate this possibility, we examined Nfib expression by immunofluorescence staining of FACS-isolated SCLC cells. DTCs and cancer cells from metastases from CGRP TKO mice had low Nfib expression, while DTCs and metastatic cancer cells in the CMV TKO model nearly always had high Nfib expression (Fig. 2E-F). This argues against transient expression of Nfib during metastasis in the CGRP TKO model. Collectively, these data indicate that metastatic ability is an acquired property of cancer cells in both models of SCLC, and that SCLC can take different paths during metastatic progression (Supplementary Fig. S3C).
Absence of widespread chromatin changes during metastatic progression of SCLC in CGRP TKO mice
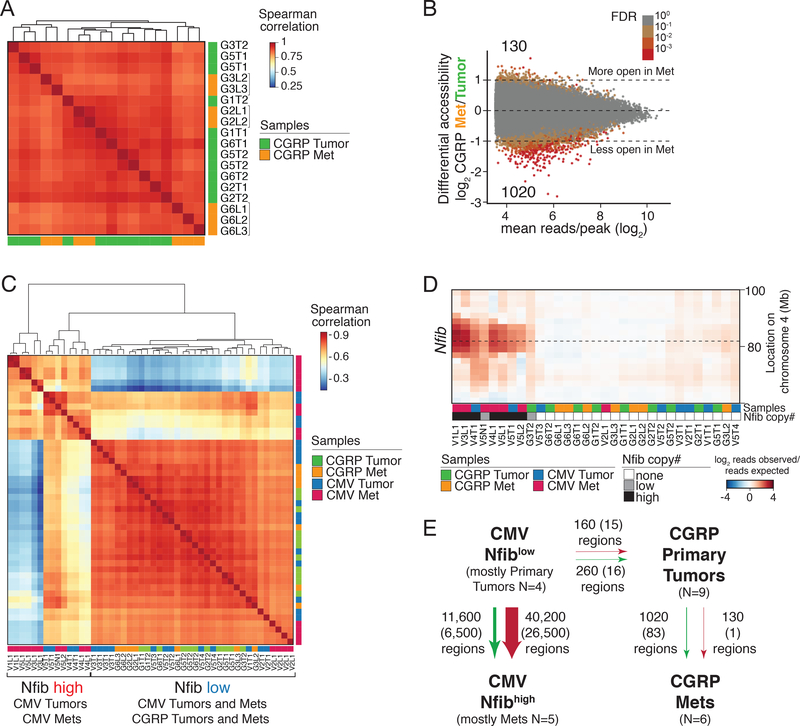
During metastatic progression of CMV TKO tumors, upregulation of Nfib drives a dramatic global increase in chromatin accessibility (7). Metastasis in CGRP-Cre mice could either be driven by widespread chromatin accessibility changes that are independent of Nfib upregulation or by mechanisms that are independent of changes in chromatin accessibility. To characterize the changes in chromatin accessibility during metastasis, we performed ATAC-seq on primary tumors and metastases from CGRP TKO mice (Supplementary Fig. S4-S5, Supplementary Table S2 and Methods) (32). In contrast to our previous analysis of CMV TKO tumors and metastases (7), unsupervised hierarchical clustering based on chromatin accessibility did not clearly separate the primary tumors and liver metastases from CGRP TKO mice (Fig. 3A). Comparison of the accessible chromatin regions in CGRP-Cre primary tumors and liver metastases uncovered only 1020 genomic regions that were less accessible in metastases and 130 genomic regions that were more accessible in metastases (Fig. 3B and Supplementary Fig. S6A-C). Hierarchical clustering of chromatin accessibility of samples from both the CMV TKO and CGRP TKO models showed that CGRP TKO primary tumors, CGRP TKO liver metastases, and Nfiblow CMV TKO primary tumors clustered together but were separate from the Nfibhigh CMV TKO samples (Fig. 3C-D). Direct comparison between CGRP TKO and Nfiblow CMV TKO primary tumors uncovered very few regions with differential chromatin accessibility (260 regions more open in CMV TKO samples and 160 regions more open in CGRP TKO samples; Fig. 3E). Collectively, these data indicate that the chromatin state alterations during metastatic progression of SCLC in CGRP TKO mice are categorically different from those in CMV TKO mice, not just by virtue of Nfib expression, but also at the level of global differences in chromatin accessibility.
Figure 3: Limited chromatin accessibility changes and lack of Nfib amplification during metastatic progression of CGRP TKO tumors.
A. Correlation of chromatin accessibility of 9 primary tumors and 7 liver metastases from CGRP TKO mice. Replicate samples from the same tumors have the same name. Metastases from the same mice (indicated with brackets) correlate most closely with one another.
B. Differential accessibility between CGRP primary tumors and metastases. Log2 fold change in reads per peak between groups plotted against the mean number of reads per peak is shown. Number of regions that have significant accessibility changes (FDR < 0.1, |log2fold change| > 0) is indicated. Color of each point represents the false discovery rate (FDR) that the absolute value of the log2 fold change was greater than 0.
C. Correlation of chromatin accessibility between primary tumors and metastases from CMV TKO and CGRP TKO mice.
D. DNA copy number analysis at the Nfib locus on Chromosome 4 in CMV TKO and CGRP TKO primary tumors and metastases. Heatmap shows copy number variation (CNV) at the Nfib locus. none = CNV < 1, low= CNV >1 and CNV <1.5, high = CNV >1.5.
E. Summary of the chromatin accessibility changes during metastatic progression of CMV TKO and CGRP TKO tumors. Numbers of peaks with changed accessibility are listed (|lfc|>0 at FDR<0.1, no peak changed < 2 fold), numbers in parentheses give the number of peaks that change in accessibility (|lfc|>0.5 at FDR<0.1, no peak changed < 2 fold).
SCLC subtypes have different gene expression programs
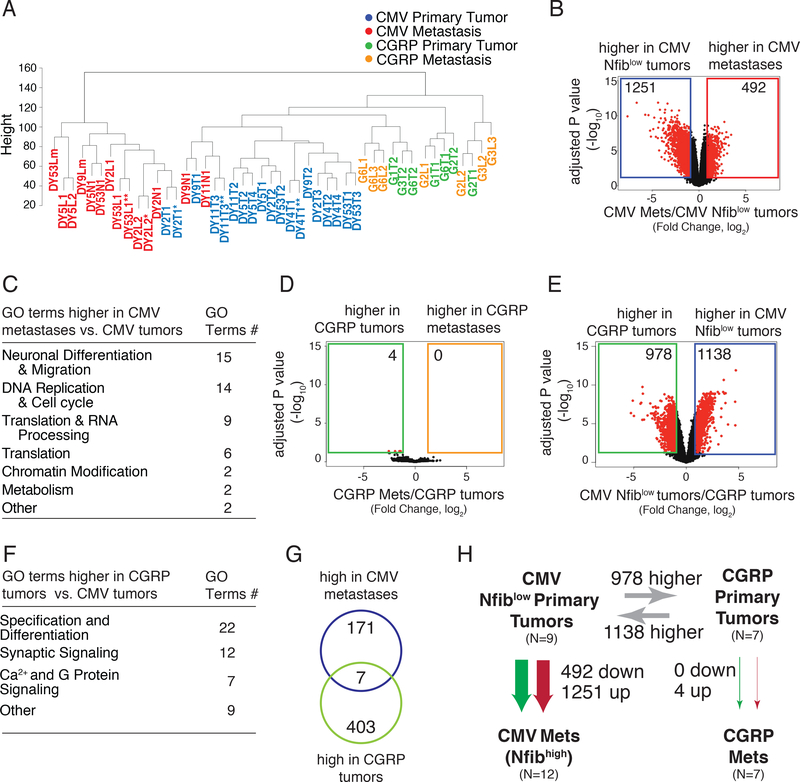
The overall similarity of the chromatin landscape in CMV TKO and CGRP TKO primary tumors suggests that inherent differences in chromatin accessibility likely do not explain the different trajectories of tumor progression in these models. To further understand the basis for the different metastatic paths of SCLC in CMV TKO versus CGRP TKO mice, we next performed RNA-seq on FACS-isolated cancer cells from both models. Unsupervised clustering and Principal Component Analysis (PCA) showed a clear separation of CMV TKO primary tumors from CMV TKO liver metastases. CGRP TKO primary tumors and metastases clustered together and more closely with CMV TKO primary tumors (Fig. 4A and Supplementary Fig. S7A).
Figure 4: Molecularly distinct SCLC subtypes.
A. Unsupervised hierarchical clustering of all CMV TKO and CGRP TKO tumors and metastases. * technical replicates, ** batch replicates.
B. Differential gene expression in CMV TKO Nfiblow primary tumors and CMV TKO metastases. (|Fold change| > 2, adj. p < 0.05)
C. A summary of top 50 GO terms (FDR q value < 0.05) enriched in CMV TKO metastasis in comparison to CMV Nfiblow primary tumor.
D. Differential gene expression in CGRP TKO primary tumors and CGRP TKO metastases. (|Fold change| > 2, adj. p < 0.05)
E. Differential gene expression in CMV TKO Nfiblow tumor and CGRP TKO tumor (|Fold change| > 2, adj. p < 0.05)
F. A summary of top 50 GO terms (FDR q value < 0.05) enriched in CGRP TKO tumor in comparison to CMV Nfiblow primary tumor.
G. Genes that are significantly upregulated (|Log2(Fold change)| > 1.5) in CMV TKO metastasis and CGRP TKO tumors comparing to CMV TKO Nfiblow tumors are mostly mutually exclusive.
H. Summary of RNA expression differences between the 4 different tumor states. Numbers of genes with changed expression are listed (|Fold change| > 2, adj.p < 0.05).
Some metastatic primary tumors in the CMV TKO model express high levels of Nfib (Supplementary Fig. S7B and (7)) and Nfib expression correlated with the clustering of CMV TKO primary tumors and metastases (Supplementary Fig. S7C). To uncover gene expression changes during metastatic progression of CMV TKO tumors, we thus performed differential gene expression analysis between Nfiblow CMV TKO primary tumors and CMV TKO metastases. This analysis revealed widespread changes in gene expression programs during metastatic progression of this subtype of SCLC (Fig. 4B and Supplementary Table S3). Genes that were upregulated in CMV TKO metastases were enriched for gene ontology annotations associated with neuronal differentiation and cell cycle, consistent with what we and others have observed upon ectopic expression of Nfib in SCLC cells (6, 7) (Fig. 4C, Supplementary Fig. S8 and Supplementary Table S4).
Surprisingly, very few genes were significantly differentially expressed by more than two-fold between CGRP TKO primary tumors and metastases (Fig. 4D). As expected from our immunostaining analysis, Nfib expression was not increased in CGRP TKO metastases. In contrast, more than 2,000 genes were differentially expressed between CMV TKO and CGRP TKO primary tumors (Fig. 4E and Supplementary Table S3). Genes that were more highly expressed in CGRP TKO tumors than in Nfiblow CMV TKO tumors were enriched for gene ontology annotations associated with neuronal differentiation, including synaptic signaling (Fig. 4F, Supplementary Fig. S9 and Supplementary Table S4). Notably, while neuronal gene sets were enriched in both CMV TKO metastases and CGRP TKO primary tumors, the genes driving these annotations had little overlap and the neuronal gene programs enriched in CGRP TKO primary tumors relative to Nfiblow CMV TKO primary tumors were specifically related to a more mature neuronal state (Fig. 4G and Supplementary Fig. S9). Together, these data (Fig. 4H) underscore the distinct molecular paths taken by SCLC tumors in the CMV and CGRP TKO models.
To relate the gene expression states of CMV TKO and CGRP TKO tumors to human SCLC, we merged human SCLC datasets (13) and our mouse SCLC datasets and performed principal component analysis. Many principal components seem to be driven primarily by the CMV versus CGRP tumors types, and human tumors were distributed broadly with some being more similar to CGRP TKO tumors and other being more similar to CMV TKO tumors (Supplementary Fig. S10).
Non-metastatic CMV TKO primary tumors contain cells with diverse lung epithelial lineage markers
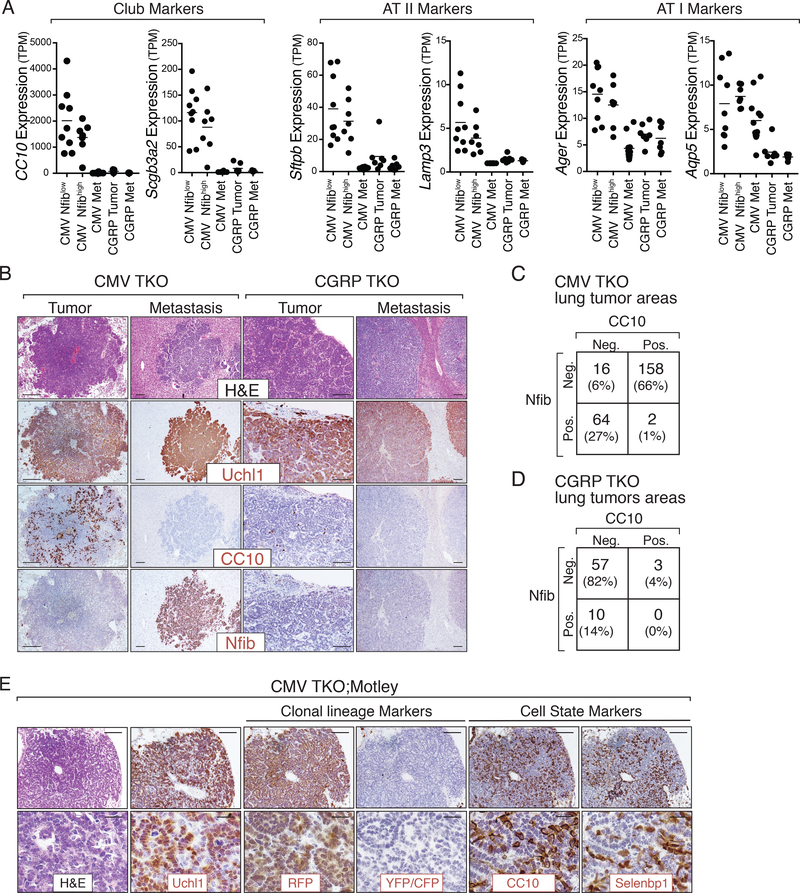
While tumors and metastases in both the CMV and the CGRP TKO models express neuroendocrine genes, CGRP TKO primary tumors and metastases had a slight overall higher expression of neuroendocrine programs compared to CMV TKO primary tumors or metastases (Supplementary Fig. S11). In contrast, compared to CGRP TKO primary tumors, CMV TKO primary tumors specifically had higher expression of lineage markers of several diverse lung epithelial cell types, including club cells (e.g. Scgb1a1/CC10, Scgb3a2), alveolar type II cells (e.g. Sftpb, Lamp3), and alveolar type I cells (e.g. Aqp5, Ager) (Fig. 5A and Supplementary Table S3). Neither SCLC subtypes highly expressed canonical markers of basal cells or lineage negative epithelial progenitors (LNEP) (Krt5, Trp63) or ciliated cells (Foxj1) (Supplementary Fig. S12A). Thus, while CMV TKO tumors still express neuroendocrine markers, they also express a wider range of lung epithelial markers.
Figure 5: Multilineage differentiation in CMV TKO Nfiblow primary tumors.
A. Gene expression of multilineage markers in tumors and metastases from CMV TKO and CGRP TKO mice. CMV TKO primary tumors express markers of Club cells, alveolar type II cells (AT II), and alveolar type I (AT I) cells.
B. Immunohistochemical staining for Uchl1, Nfib, and CC10. Representative images show the presence of CC10pos cells specifically in Nfiblow primary tumors in CMV TKO mice. Scale bars = 100μm.
C-D. Quantification of CC10 and Nfib expression. CC10 and Nfib are mostly mutually exclusive in CMV TKO lung tumors (C), while CC10 and Nfib are only rarely expressed in CGRP TKO lung tumors (D).
E. Immunohistochemical staining for tumor clonal lineage markers (RFP, YFP/CFP) as well as CC10 and Selenbp1 on tumors from CMV TKO;Motley mice. The uniform staining of tumor lineage markers in individual tumors is consistent with their clonal origin. CC10positive and Selenbp1positive cells are cancer cells that are clonally related to the CC10negative and Selenbp1negative cells. Scale bars top = 100 μm, bottom = 20 μm.
Many of these lineage genes have diverse expression in human SCLC, consistent with the inter-tumoral heterogeneity uncovered in the mouse SCLC models (Supplementary Fig. S12B). At the protein level, primary tumors and metastases from both mouse models expressed neuroendocrine markers, including Ascl1 and Uchl1 (Supplementary Fig. S1B and Fig. 5B). However, as anticipated from our RNA-seq analysis, other markers including CC10 and Selenbp1 (a marker of bronchiolar and alveolar non-neuroendocrine cells) were specifically expressed in CMV TKO non-metastatic primary tumors (Fig. 5B-D and Supplementary Fig. S12A and S13A-C). Cells expressing these non-neuroendocrine markers were uniquely present in the Nfibnegative areas within primary CMV TKO SCLC (Supplementary Fig. S13D).
Interestingly, CC10 and Selenbp1 were highly expressed in only a subset of cells in CMV TKO primary tumors (Fig. 5B). Since CC10 and Selenbp1 are also expressed in normal lung epithelial cells (33, 34), we determined whether these cells were cancer cells or normal cells within the tumors. In CMV-Cre initiated tumors in TKO;Motley mice, CC10positive and Selenbp1positive existed within the clonal cancer cell population, indicating that these cells are cancer cells (Fig. 5E and Supplementary Fig. S13E).
Together, these analyses further underscore the difference between CMV TKO and CGRP TKO primary tumors and uncover the presence of subsets of cancer cells expressing markers of multiple lung epithelial lineages in CMV TKO primary tumors.
The two distinct subtypes of mouse SCLC arise from different cell types of origin
The simplest explanation for the differences observed during tumor progression of the CMV TKO and CGRP TKO models is that the tumors are initiated from different cell types. Ad-CMV-Cre and Ad-CGRP-Cre both lead to expression of Cre in neuroendocrine cells but Ad-CMV-Cre also leads to the expression of Cre in many other cell types in the lung (20, 21). The development of fewer tumors in CGRP TKO mice than in CMV TKO mice, even with a higher titer of Ad-CGRP-Cre (Supplementary Fig. S1A), supports the idea that SCLC is initiated from rare neuroendocrine cells in the CGRP TKO model but from a larger pool of cells in the CMV TKO model.
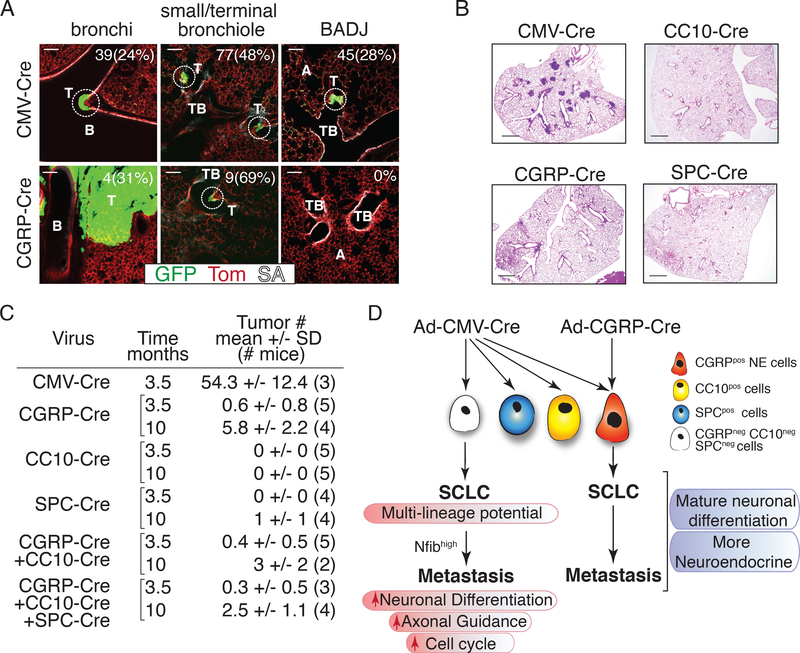
Neuroendocrine cells are often present at bifurcation points in bronchioles as well as in larger airways but are not normally found in alveoli (27, 35, 36). Accordingly, CGRP TKO tumors grow predominantly within the proximal airways, in both the large and small bronchioles. In contrast, CMV TKO tumors grew both in the proximal and distal lung with early stage CMV TKO lesions located at large and small bronchioles, terminal bronchioles, and at bronchial-alveolar duct junctions (Fig. 6A and Supplementary Fig. S14).
Figure 6: SCLC in CMV TKO mice arises from an alternative cell of origin.
A. Representative immunofluorescent images of whole-mount staining and confocal imaging of tumors in CMV TKO and CGRP TKO mice. Tumors are circled. Number and percent of lesions at each location is indicated. T = tumor, B = bronchi, TB = terminal bronchiole, A = alveolar space, SA = streptavidin (which labels lung epithelial cells). Scale bars = 200 μm.
B. Representative H&E images of the lungs of mice 3.5 months after transduction by same titer of adenoviral vectors with distinct promoters driving Cre expression. Scale bars = 1 mm.
C. Quantification of SCLC tumor number 3.5 months and 10 months after transduction with same total titer of different types and combinations of adenoviral vectors with distinct promoters driving Cre expression. Tumor number is quantified from H&E lung sections. The number of mice in each cohort is indicated in parentheses.
D. Model of SCLC development and progression from two distinct cell types of origin.
In addition to the spatial analysis, we also quantitatively re-evaluated the potential of the major lung cell types to give rise to SCLC. Cancer initiation with Ad-CMV-Cre is more than 50-fold more efficient than adenoviral vectors with tissue specific promoters targeting CGRPpositive NE cells, SPCpositive cells (mostly alveolar type II cells), and CC10positive cells (mostly club cells). This is further confirmed by analysis at late stage (10 months after tumor initiation). None of the cell types that we targeted recapitulate the high tumorigenic efficiency of Ad-CMV-Cre (Fig. 6B-C and Supplementary Fig. S15A-B).
Collectively, while our data support the idea that different cells-types of origin provide the most likely explanation for the difference in tumor number and metastatic programs between the CMV TKO and CGRP TKO mouse models, we carefully investigated other variables. First, the time of tumor development was not a major determinant of the acquisition of Nfib-driven metastatic ability, as CMV TKO and CGRP TKO mice analyzed at the same time point after tumor initiation still had Nfibpositive and Nfibnegative metastases, respectively (Supplementary Fig. S16A-B). Second, incomplete recombination of the conditional alleles due to lower Cre expression from one of the viral vectors was largely ruled out by PCR genotyping of the floxed alleles in CMV TKO and CGRP TKO tumors (Supplementary Fig. S4B). Third, while 10- to 20-fold higher titers of Ad-CGRP-Cre were used to initiate tumors, a non-specific effect of overall adenoviral titer did not explain our findings: mice with tumors initiated with Ad-CMV-Cre combined with high titers of control Ad-CMV-GFP or Ad-CC10-Cre still developed CC10positive primary tumors and Nfibhigh metastases (Supplementary Fig. S16C-E). Finally, in the CMV TKO model, Cre is expressed broadly in the lung leading to the generation of many “normal” epithelial cells that lack the floxed tumor suppressor genes. The co-existence of many other cells with these genetic alteration did not contribute to the high number of tumors initiated by CMV-Cre, as TKO;mTmG mice transduced with Ad-CGRP-Cre with or without the addition of Ad-CC10-Cre or Ad-CC10-Cre and Ad-SPC-Cre still developed comparable and low numbers of tumors (Fig. 6C and Supplementary Fig. S15C).
Thus, the most likely explanation for the difference in tumor number and progression remains that Ad-CMV-Cre leads to tumor suppressor inactivation in one or more cell types that do not express Cre after transduction with Ad-CGRP-Cre. These cells ultimately give rise to SCLC tumors that are sufficiently molecularly distinct to result in strikingly different evolutionary paths towards metastasis (Fig. 6D).
DISCUSSION
SCLC is an aggressive cancer type and patients often present or relapse with widespread metastatic disease. Our data from defined genetically engineered mouse models indicate that SCLC is not inherently metastatic and that key genetic and epigenetic changes occur during cancer progression (this study and (5–7, 37)). Furthermore, we provide evidence that SCLC can arise from different cell types, which has profound influences on the course of tumor evolution.
NFIB is highly expressed in more than 50% of human SCLC metastases (6, 7, 13) (Fig. 1G and Supplemental Fig. 2), suggesting that upregulation of this transcription factor could influence metastatic ability of a large fraction of human SCLC. Deletion of Rb/p53 or Rb/p53/p130 in mice after delivery of Ad-CMV-Cre to the lungs may provide an accurate model for this subset of human tumors. SCLC initiated specifically from mature neuroendocrine cells using Ad-CGRP-Cre may model a different subset of human SCLC that does not amplify or upregulate Nfib as often and thus may metastasize via different mechanisms. We cannot exclude, however, that specific combinations of genetic alterations in mature neuroendocrine cells may affect the frequency of Nfib upregulation during SCLC progression. Notably, while SCLC in humans often develops within the lobar or main bronchi and is identified by imaging within the central aspect of the chest, 5–15% of cases SCLC are more peripheral or even subpleural (38–41). These peripheral tumors are histologically similar to central SCLC and are also highly metastatic (42). The more peripheral location of tumors in the CMV TKO model suggests that CMV TKO mice may model this poorly characterized subset of human tumors.
Why CGRP TKO tumors and a fraction of human SCLC tumors metastasize without upregulation of Nfib is unknown, especially when this factor can be such a strong oncogene and pro-metastatic driver. We did not observe large-scale differences in genome-wide chromatin accessibility between CMV TKO and CGRP TKO primary tumors, suggesting that cancer cells in both models could be responsive to Nfib. However, small changes in accessibility at specific loci and/or other chromatin differences may render cancer cells in CGRP TKO tumors less responsive to Nfib upregulation. SCLC derived from mature neuroendocrine cells may be in a more differentiated state that is less amenable to epigenetic transformation by Nfib. The Nfib locus itself could be less frequently amplified or the expression of Nfib could be regulated in a different manner in SCLC derived from mature neuroendocrine cells. Finally, it is possible that co-factors required for Nfib action are not present in SCLC derived from CGRPpositive mature neuroendocrine cells. Interestingly, some primary tumors in the CGRP TKO model do express high levels of Nfib (Fig. 1E). The expansion of Nfibhigh cancer cells in these tumors could suggest that Nfib still has oncogenic properties in this cellular context, but without promoting metastasis.
Whether the molecular paths taken by distinct SCLC tumors affect their response to specific treatments remains an important question. In culture, we did not identify significant differences in the responses of cell lines derived from Nfibhigh CMV-Cre and Nfiblow CGRP-Cre tumors to three different drugs: cisplatin, etoposide, and a Chk1 inhibitor. These cells lines had large differences in their sensitivity to these inhibitors, CGRP TKO cell lines had a trend towards higher IC50 values for each drug (Fig. S17). Future studies, including in vivo studies on the genetically-engineered mouse models and/or in clinical trials will be required to determine whether different SCLC subtypes define therapeutic responses to immunotherapy, epigenetic therapies, or other treatments.
In addition to recent observations that heterogeneity within individual tumors can play a role in the SCLC progression (14, 16, 18, 19), accumulating evidence supports the notion that SCLC is not one disease, but rather many diseases that are distinguishable only at the molecular level (12, 13, 17). Across many cancer types, the genetic and epigenetic changes that occur during tumor evolution are a major focus of efforts to understand this inter-tumoral heterogeneity. However, the impact of the cell-type of origin on cancer initiation and progression has garnered less interest (43–47). Distinct cell types may give rise to different tumor subtypes with distinct histological features; however, in the case of SCLC, although the two subtypes metastasize using different mechanisms, the resulting metastases were histologically indistinguishable.
Our results indicate that fundamental differences in tumor development between the two mouse models may be endowed upon the tumor by the cell-type of origin. The identity of the cell-type(s) of origin in the CMV TKO model remains unknown. Previous work using Rb1flox/flox;p53flox/flox DKO mice and our analysis of Rb1flox/flox;p53flox/flox;p130 flox/flox TKO mice suggests that lung cell types expressing CC10 or SPC (including club cells and alveolar type II cells) are unlikely to be cell-types of origin for SCLC (4, 20, 21). Based on the expression of classic markers of several lung epithelial lineages specifically in CMV TKO primary tumors, including markers of club cells and alveolar type II and type I cells, we speculate that SCLC in this model arises from stem/progenitor cells or facultative stem/progenitor cells that gain multi-lineage potential upon inactivation of Rb1 and p53. Indeed, RB1 loss promotes cellular plasticity (48) and can lead to neuroendocrine differentiation/transdifferentiation in lung adenocarcinoma and prostate cancer (49–51). The formal identification of this cell type or types will require new tools to express Cre specifically in defined subpopulations of lung epithelial cells (23, 24, 52).
SCLC patients continue to have one of the worst survival rates of all cancer patients. Our work indicates that multiple cell types in the lung can serve as the cell of origin of SCLC and suggests that biomarkers of these different subsets of tumors may help predict their evolutionary trajectories towards malignancy. Understanding this diversity may ultimately have value in enabling better patient stratification and more precise treatment plans.
METHODS
Ethics statement
Mice were maintained according to practices prescribed by the NIH at Stanford’s Research Animal Facility accredited by the American Association for Accreditation of Laboratory Animal Care (AAALAC). All animal studies were conducted following approval from the Stanford Animal Care and Use Committee (IACUC) (protocol 13565).
Mice, adenoviral infections, and cancer cell isolation
Rb1 flox, p53 flox, p130 flox, R26mTmG, and R26Motley alleles have been described (7, 26, 31, 53, 54). The SCLC mouse model bearing deletions of p53, Rb, and p130 was previously described (26). Ad-CMV-Cre, Ad-CGRP-Cre, Ad-SPC-Cre, Ad-CC10-Cre and Ad-CMV-GFP were purchased from University of Iowa viral vector core. Intratracheal administration was performed as previously described (7, 26). Unless specified in the text, for long-term experiments, we used Ad-CMV-Cre at 4×107 Pfu. Ad-CGRP-Cre or Ad-CC10-Cre were used at 4×108 or 8×108 Pfu per mouse, respectively. Primary tumors and metastases were harvested when mice became moribund. Mice were maintained at the Stanford Research Animal Facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. Cancer cells are isolated and purified by FACS as previously described (7).
Cell culture
SCLC cell lines were passaged as described before (7). For drug response assays, cells were plated in a 96-well plate at 10,000 cells per well, in triplicate, in 100 μl of media. After 24 hours, drugs or vehicle control were added to the cells at a 2x concentration in 100 μl, for a total volume of 200 μl per well. For each drug, seven different concentrations were tested at 10-fold dilutions. Forty-eight hours after the drugs were added, 20 μl of Alamar Blue was added to each well, and fluorescence values were measured 4 hours later. IC50 values were calculated using GraphPad Prism7. Cisplatin and etoposide were obtained from the Lucile Packard Children’s Hospital at Stanford. The Chk1 inhibitor LY2606368 was a gift from the lab of Dr. Lauren Byers (MD Anderson Cancer Center) and reconstituted in DMSO.
Immunoassays
Protein levels of Nfib and Ascl1 for the tumor-derived cell lines used in IC50 experiments were determined using the Simple Western™ quantitative immunoassay and the Compass software, according to the manufacturer’s protocol. Cells were lysed in TNESV lysis buffer (50 mM Tris·HCl at pH 7.6, 1% IGEPAL®, 20 mM EDTA at pH 8.0, 100mM NaCl), supplemented with proteasome and phosphatase inhibitors, and lysates were cleared by centrifugation at maximum speed for 10 minutes. Total protein was quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher, Cat. #23277). Whole-cell lysates were diluted to a final concentration of 0.2 μl/ml. The antibodies and dilutions used were as follows: Nfib (Abcam, ab186738) 1:20000; Ascl1 (BD Pharmingen, 556604) 1:1000; Hsp90 (Cell Signaling Technology, 4877S) 1:2000.
Histology and immunohistochemistry
Tumor samples were fixed in 4% formalin and paraffin embedded. Hematoxylin and Eosin staining was performed using standard methods. Images were quantified using ImageJ. For IHC, we used antibodies to Nfib (1:1000, Abcam ab186738), Uchl1 (1:500, sigma HPA005993), CC10 (CCSP) (1:1000, Millipore 07–623), Selenbp1 (1:200, Abcam), GFP (1:500, Abcam ab6673), Ascl1 (1:200, BD Biosciences, 556604) and RFP (1:500, Rockland).
SCLC patient brain metastases sections and IHC staining score
Human SCLC brain metastases tissue sections were collected by Anna S. Berghoff and Matthias Preusser from the Medical University of Vienna, Austria. Immunohistochemical staining for NFIB (1:1000, Abcam ab186738) was performed on 4 μM formalin-fixed paraffin-embedded sections. NFIB expression was scored by a board-certified pathologist C.S.K. on a three point scale as follows: 0 = negative or weak staining of less than 10% cells, 1 = weak staining of more than 10% cells, 2 = moderate intensity staining, 3 = strong intensity staining.
Immunofluorescence
FACS-sorted cancer cells from primary tumors, DTCs, and liver metastases were cytospun onto glass slides at 500 rpm for 5 minutes. Cells were fixed with 4% PFA for 15 minutes, and stained for Nfib (Abcam, ab186738) 1:2000 and with a goat anti-rabbit secondary antibody (Invitrogen). For imaging, membrane GFP staining was confirmed to indicate a DTC and Nfib expression was checked through the far-red channel using fluorescence scope (Leica). Nfib staining was quantified by counting directly under the scope. On average, we quantified 30–100 cells per sample based on how many cells were harvested.
Whole mount immunofluorescence staining
Lungs were dissected from mice after perfusion with ~5ml PBS into the right cardiac ventricle and intratracheal inflation with ~2ml 2% low melting point agarose (ThermoFisher cat. #16520) in PBS. The lungs were fixed in 4% paraformaldehyde (EMS cat. #15714) for six hours at 4°C then sectioned with a vibrating blade microtome (Leica Biosystems cat. #VT1000S) at 500μm thickness. Lung slices were stained with rabbit anti-CGRP (Enzo Life Sciences cat. #BML-CA1137, 1:1000 dilution) for 5–7 days at 4°C, then with Alexa Fluor 647-conjugated anti-rabbit IgG (ThermoFisher cat. #A-21244, 1:500 dilution) and streptavidin-conjugated Alexa Fluor 405 (ThermoFisher cat. #S32351, 4 μg/ml final concentration) as a pan-lung epithelial identifier (55) for 3–4 days at 4°C. Finally, stained sections were optically cleared using the CUBIC method (56), comprised of a three-hour incubation at room temperature in CUBIC 1 reagent and long-term storage in CUBIC 2 at 4°C. Sections were imaged using a Zeiss LSM 780 laser scanning confocal microscope with an inverted 5X air objective (Carl Zeiss AG, NA=0.45), and optical sections were collected at 10 μm resolution.
ATAC-seq library preparation, sequencing, and analysis
ATAC-seq library preparation was performed as described (7).
Calling peaks
Accessible regions were called by performing peak calling to obtain peak summits, merging peak calls obtained in different sequencing batches, and subsequently filtering to remove any overlapping regions. Macs2 (v 2.1.0.20140616) was used to call peak summits on the merged bam file of each sequencing batch of samples (three total) using the command “macs2 callpeak --nomodel --call-summits --keep-dup all”. Peak summits were expanded by 250 bps on each side to form 500 bp windows. Each set of accessible regions was filtered to remove blacklist regions (mm9; https://sites.google.com/site/anshulkundaje/projects/blacklists) and copy number amplification (any peak that overlapped a region that differed from expected for any sample by more than 2 fold (up or down) was removed; copy number amplification was determined as described below). Finally, the three peak sets were concatenated, while retaining overlapping peak windows.
Initially, all accessible regions were filtered to obtain only regions with sufficient read depth to compare across samples. The number of overlapping reads per peak was found for all samples using the bedtools multicov module (v2.25.0), using only reads with high mapping quality (-q 30). The read counts per peak for each sample was normalized by the mean read counts per peak for that sample, such that all samples have the same mean reads per peak. Peaks were kept if they fulfilled either one of two criteria: (1) the peak had at least 1 reads for all samples, or (2) the peak had at least one sample with large number of normalized reads (>1 standard deviation above the mean). This operation was performed to filter out peaks with low counts while retaining peaks that may be differentially accessible.
Finally, any overlapping windows were filtered to obtain a set of non-overlapping windows. Overlapping windows were identified using bedtools cluster module. For each set of overlapping windows, only the one with the greatest number of normalized reads was kept. In the end, ~114,000 accessible regions (non-overlapping, uniform width) were obtained and used in subsequent analyses.
Normalizing read counts within accessible regions
The number of reads per peak was obtained to assess the differential chromatin landscape across samples. To estimate the effect of sequencing depth, a set of ‘housekeeping’ regions was obtained that was expected to have similar mean accessibility across samples. These ‘housekeeping’ regions were determined by finding the set of genes that were uniformly expressed across CMV tumors and metastases (as in Denny et al, 2016 (7)); peaks corresponding to the promoters of these genes were determined to be ‘housekeeping’ peaks (Supplementary Table S2). The inverse of the mean read count across all ‘housekeeping’ peaks was used as the sample-specific size factor for normalization in DESeq2 (57). These factors were divided by the geometric mean of all the size factors, as recommended in the DESeq2 vignette (https://bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html#sample-gene-dependent-normalization-factors). Size factors were converted to peak-specific normalization factors by using the sample-specific size factor for all peaks.
Number of reads expected from background fragmentation
The number of reads expected to come from background fragmentation during transposition was obtained by counting reads overlapping a set of background intervals (entire genome, excluding blacklist regions, and excluding any 10 Mb interval with evidence of copy number amplification in any sample), to form a total number of reads M for any sample. The number of reads within accessible regions for each sample (m) was subtracted from M, and divided by the number of base pairs in the background intervals (L), less the number of base pairs in accessible regions (l), such that the number of background reads per base pair per sample was b = (M − m)/(L − l). For accessible region k of size lk, the number of reads expected simply from background fragmentation was nk=lkb. In practice, because all of the accessible regions were of uniform size, all accessible regions had the same lk.
To obtain background-subtracted reads counts within accessible regions, nk was subtracted from the number of reads overlapping each accessible region for each sample. These subtracted values were rounded to the nearest integer, and any intervals with negative read counts were rounded up to zero. For all samples with enrichment score > 11, the number of background reads per each peak was less than ~5 (normalized for sequence depth).
Assessing differential accessibility
Through unsupervised clustering and principal component analysis, we determined that samples with low enrichment (calculated as the maximum number of reads around transcription start sites over the minimum number of reads, as in(7)) were associated with differences in chromatin accessibility. This effect was associated with batch and not with Nfib amplification (Supplementary Fig S5). To minimize this technical artifact, samples with low enrichment (<11) were filtered from the analysis.
Differential accessibility was determined using the package DESeq2 (57). Counts per peak were background subtracted to account for expected counts per peak due to background fragmentation (see above section). Each sample was annotated by the model (CGRP versus CMV), the metastatic state (either T for primary tumor or L for distal metastases (liver or lymph node), and the Nfib amplification state (1 if amplified, 0 if unamplified). This annotation can be found in the column “model_met_amp” in Supplementary Table S5. The model used in DESeq was then “~model_met_amp”. For the CGRP distal metastases versus primary tumor comparison, results were extracted using the command “res_G_LvsT <- results(dds, contrast=c(‘model_met_amp’, ‘G_L_0’, ‘G_T_0’))” (Supplementary Table S5).
Assessing copy number amplification from ATAC-seq data
Copy number amplification was estimated from ATAC-seq data by determining read counts in large (10 Mb) intervals across the genome (2 Mb step size) that have different read counts than expected, where the expectation is based on the average read count across 100 GC-matched intervals. Specifically, the GC content of each interval was found using bedtools nuc module. For each interval, the 100 other intervals with the closest GC-content to that region were found and used as the GC-matched intervals.
RNA-seq library preparation and RNA-seq analysis
RNA was extracted from FACS sorted cancer cells using Qiagen Allprep DNA/RNA kit. All the samples were checked by BioAnalyzer (Agilent) for RNA integrity (RIN>8). cDNA was prepared from 10 ng of RNA using the Nugen Ovation V2 kit, according to manufacturer’s instructions. RNA-seq libraries were generated using the Illumina Truseq kit.
We used Kallisto to quantify expression (58). We kept only transcripts with max TPM value greater than 1 in at least one sample and with standard deviation greater than 1. We performed the asinh() transformation to variance stabilize the data.
Because Illumina library preparation method had been slightly modified between the two library preparations, we observed a strong batch effect when comparing the two dataset. To correct for the batch effect, we included three RNA samples of our old CMV TKO samples while generating the CGRP TKO libraries.
We used SVA to remove batch effects from the data (59). To identify batch effects between the two categories of samples, we strategically incorporated three overlapping samples between the CGRP TKO and CMV TKO samples. These otherwise identical samples give an idea of what differences can arise purely from experimental procedure and other confounding effects. We used SVA to estimate confounding factors by protecting the sample name so that the algorithm only removed batch effects confounding the identical samples and otherwise knew nothing about the identity of the samples. We then removed the estimated confounding factors from the data. After batch correction, the batch difference of the three repeated samples was comparable to the difference between technical replicates generated in a single RNA-seq experiment.
Differential Expression and Gene Ontology analysis
To discover the genes driving transitions between different stages of metastasis we used the following procedure on data with the experimental batch corrected (see previous section): 1) fit a linear model (lmFit in R) modeling metastatic effect, 2) rank genes in order of evidence for differential expression using an empirical Bayes method (60). We only compared genes with non-zero variance. We compared CGRP primary tumors and CGRP metastases, CGRP primary tumors and CMV primary tumors, CMV primary tumors and CMV metastases, and CMV metastases versus CGRP metastases. Significantly differential genes were those that had an adjusted p-value below 0.05. Gene ontology analysis and neuroendocrine signature enrichment analysis were performed using gene set enrichment analysis (GSEA) (61).
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE116977 and the ATAC-seq data is GEO: GSE117177.
Comparison to Human Samples
We downloaded human SCLC RNA-Seq studies EGAD00001001431 and EGAD00001001244 from the European Genome-Phenome Archive. We quantified gene expression for all samples (n=74) from the two studies again using Kallisto to generate TPM for each sample.
In order to compare expression values for human samples and mouse samples, we subset all data sets to use only genes that shared the same gene names between the two species (n=14,813 genes). After merging human and mouse data sets on shared genes, we performed the asinh transformation for variance stabilization.
We then performed PCA on this merged data set. PCA allows us to inspect different axes of variation, such as those driven by species, cell type of origin, metastatic state, etc. without further transforming human and mouse data to make them comparable. We plotted PC1 versus PC2, PC1 versus PC3 and PC2 versus PC3. We found that PC1, as expected, is driven primarily by differences between mouse and human samples. We found that PC3 seems to be driven primarily by tumor type with a clear continuum among mouse samples from CGRP to CMV Met samples. We used this PC3 in order to understand how the distribution of human samples falls relative to the mouse distribution of samples.
Supplementary Material
SIGNIFICANCE.
We show that SCLC can arise from different cell-types of origin, which profoundly influences the eventual genetic and epigenetic changes that enable metastatic progression. Understanding inter-tumoral heterogeneity in SCLC, and across cancer types, may illuminate mechanisms of tumor progression and uncover how the cell-type of origin impacts tumor evolution.
ACKNOWLEDGMENTS
We thank Pauline Chu for technical assistance and the Stanford PAN Facility and Stanford Shared FACS Facility; Shin-Heng Chiou, Ian Winters, Barbara Grüner, Astrid Gillich, Christopher Murray, Sandra Cristea, Gokul Ramaswami and Rosanna Ma for sharing reagents and technical help; David Feldser, Laura Attardi, Mark Krasnow, Tushar Desai and members of the Winslow and Sage laboratories for helpful comments.
Financial Support: This work was supported by a Stanford Cancer Institute Cancer Biology Seed Grant (to M.M.W., W.J.G., and J.S.) and National Cancer Institute R01CA206540 (to J.S.) and R01CA194461 (to K.S.P.), the German-Israeli Foundation for Research and Development (I-65–412.20–2016 to H.C.R.), the Deutsche Krebshilfe (70113041, 1117240 and 70113041 to H.C.R.) and the German Ministry of Education and Research (BMBF e:Med 01ZX1303A to H.C.R.). D.Y. was supported by a Stanford Graduate Fellowship and by a TRDRP Dissertation Award (24DT-0001). S.K.D. was supported by the Stanford Biophysics training grant (T32 GM008294). A.C.C. was supported by the Stanford Cancer Biology training grant (T32 CA009302). A.K. was supported by NIH grant DP2OD022870. P.G.G. was supported by the Bio-X Stanford Interdisciplinary Graduate Fellowship (SIGF); P.G.G.). Y.O. was supported by a Stanford Graduate Fellowship. S.K.D., A.C.C., Y.O. were supported by the NSF GRFP. J.S.L. was supported by an A*STAR scholarship (Singapore). J.S. is the Harriet and Mary Zelencik Scientist in Children’s Cancer and Blood Diseases.
Footnotes
Disclosure of Potential Conflicts of Interests: W.J.G. is the scientific co-founder of Epinomics. A.K. is on the scientific advisory board of Epinomics. J.S. receives research funding from Abbvie. H.C.R. received consulting and lecture fees from Abbvie, Astra-Zeneca, Vertex and Merck and research funding from Gilead Pharmaceuticals.
REFERENCES
- 1.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell. 2017;168:670–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–52. [DOI] [PubMed] [Google Scholar]
- 3.Bunn PA, Minna JD Jr., Augustyn A, Gazdar AF, Ouadah Y, Krasnow MA, et al. Small Cell Lung Cancer: Can Recent Advances in Biology and Molecular Biology Be Translated into Improved Outcomes? J Thorac Oncol. 2016;11:453–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29:1447–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu N, Jia D, Ibrahim AH, Bachurski CJ, Gronostajski RM, MacPherson D. NFIB overexpression cooperates with Rb/p53 deletion to promote small cell lung cancer. Oncotarget. 2016;7:57514–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenova EA, Kwon MC, Monkhorst K, Song JY, Bhaskaran R, Krijgsman O, et al. Transcription Factor NFIB Is a Driver of Small Cell Lung Cancer Progression in Mice and Marks Metastatic Disease in Patients. Cell reports. 2016;16:631–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denny SK, Yang D, Chuang CH, Brady JJ, Lim JS, Gruner BM, et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell. 2016;166:328–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burger M, Glodek A, Hartmann T, Schmitt-Graff A, Silberstein LE, Fujii N, et al. Functional expression of CXCR4 (CD184) on small-cell lung cancer cells mediates migration, integrin activation, and adhesion to stromal cells. Oncogene. 2003;22:8093–101. [DOI] [PubMed] [Google Scholar]
- 9.Osborne JK, Larsen JE, Shields MD, Gonzales JX, Shames DS, Sato M, et al. NeuroD1 regulates survival and migration of neuroendocrine lung carcinomas via signaling molecules TrkB and NCAM. Proc Natl Acad Sci U S A. 2013;110:6524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirier JT, Dobromilskaya I, Moriarty WF, Peacock CD, Hann CL, Rudin CM. Selective tropism of Seneca Valley virus for variant subtype small cell lung cancer. J Natl Cancer Inst. 2013;105:1059–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirier JT, Gardner EE, Connis N, Moreira AL, de Stanchina E, Hann CL, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene. 2015;34:5869–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mollaoglu G, Guthrie MR, Bohm S, Bragelmann J, Can I, Ballieu PM, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell. 2017;31:270–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim JS, Ibaseta A, Fischer MM, Cancilla B, O’Young G, Cristea S, et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature. 2017;545:360–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jahchan NS, Lim JS, Bola B, Morris K, Seitz G, Tran KQ, et al. Identification and Targeting of Long-Term Tumor-Propagating Cells in Small Cell Lung Cancer. Cell reports. 2016;16:644–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson SC, Metcalf RL, Trapani F, Mohan S, Antonello J, Abbott B, et al. Vasculogenic mimicry in small cell lung cancer. Nat Commun. 2016;7:13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, et al. ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell reports. 2016;16:1259–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calbo J, van Montfort E, Proost N, van Drunen E, Beverloo HB, Meuwissen R, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell. 2011;19:244–56. [DOI] [PubMed] [Google Scholar]
- 19.Kwon MC, Proost N, Song JY, Sutherland KD, Zevenhoven J, Berns A. Paracrine signaling between tumor subclones of mouse SCLC: a critical role of ETS transcription factor Pea3 in facilitating metastasis. Genes Dev. 2015;29:1587–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park KS, Liang MC, Raiser DM, Zamponi R, Roach RR, Curtis SJ, et al. Characterization of the cell of origin for small cell lung cancer. Cell Cycle. 2011;10:2806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–64. [DOI] [PubMed] [Google Scholar]
- 22.Song H, Yao E, Lin C, Gacayan R, Chen MH, Chuang PT. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc Natl Acad Sci U S A. 2012;109:17531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517:621–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1:e90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 2010;70:3877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linnoila RI. Functional facets of the pulmonary neuroendocrine system. Lab Invest. 2006;86:425–44. [DOI] [PubMed] [Google Scholar]
- 28.Gazdar AF, Savage TK, Johnson JE, Berns A, Sage J, Linnoila RI, et al. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J Thorac Oncol. 2015;10:553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dooley AL, Winslow MM, Chiang DY, Banerji S, Stransky N, Dayton TL, et al. Nuclear factor I/B is an oncogene in small cell lung cancer. Genes Dev. 2011;25:1470–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20:897–903. [DOI] [PubMed] [Google Scholar]
- 31.Caswell DR, Chuang CH, Yang D, Chiou SH, Cheemalavagu S, Kim-Kiselak C, et al. Obligate progression precedes lung adenocarcinoma dissemination. Cancer Discov. 2014;4:781–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linnoila RI, Szabo E, DeMayo F, Witschi H, Sabourin C, Malkinson A. The role of CC10 in pulmonary carcinogenesis: from a marker to tumor suppression. Ann N Y Acad Sci. 2000;923:249–67. [DOI] [PubMed] [Google Scholar]
- 34.Zeng GQ, Yi H, Zhang PF, Li XH, Hu R, Li MY, et al. The function and significance of SELENBP1 downregulation in human bronchial epithelial carcinogenic process. PLoS One. 2013;8:e71865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cutz E Hyperplasia of pulmonary neuroendocrine cells in infancy and childhood. Semin Diagn Pathol. 2015;32:420–37. [DOI] [PubMed] [Google Scholar]
- 36.Kuo CS, Krasnow MA. Formation of a Neurosensory Organ by Epithelial Cell Slithering. Cell. 2015;163:394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156:1298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter BW, Glisson BS, Truong MT, Erasmus JJ. Small cell lung carcinoma: staging, imaging, and treatment considerations. Radiographics : a review publication of the Radiological Society of North America, Inc; 2014;34:1707–21. [DOI] [PubMed] [Google Scholar]
- 39.Lee D, Rho JY, Kang S, Yoo KJ, Choi HJ. CT findings of small cell lung carcinoma: Can recognizable features be found? Medicine (Baltimore). 2016;95:e5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yabuuchi H, Murayama S, Sakai S, Hashiguchi N, Murakami J, Muranaka T, et al. Resected peripheral small cell carcinoma of the lung: computed tomographic-histologic correlation. J Thorac Imaging. 1999;14:105–8. [DOI] [PubMed] [Google Scholar]
- 41.Hashimoto M, Miyauchi T, Heianna J, Sugawara M, Ishiyama K, Watarai J, et al. Accurate diagnosis of peripheral small cell lung cancer with computed tomography. Tohoku J Exp Med. 2009;217:217–21. [DOI] [PubMed] [Google Scholar]
- 42.Gephardt GN, Grady KJ, Ahmad M, Tubbs RR, Mehta AC, Shepard KV. Peripheral small cell undifferentiated carcinoma of the lung. Clinicopathologic features of 17 cases. Cancer. 1988;61:1002–8. [DOI] [PubMed] [Google Scholar]
- 43.Latil M, Nassar D, Beck B, Boumahdi S, Wang L, Brisebarre A, et al. Cell-Type-Specific Chromatin States Differentially Prime Squamous Cell Carcinoma Tumor-Initiating Cells for Epithelial to Mesenchymal Transition. Cell Stem Cell. 2017;20:191–204 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mainardi S, Mijimolle N, Francoz S, Vicente-Duenas C, Sanchez-Garcia I, Barbacid M. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014;111:255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagaraj AS, Lahtela J, Hemmes A, Pellinen T, Blom S, Devlin JR, et al. Cell of Origin Links Histotype Spectrum to Immune Microenvironment Diversity in Non-small-Cell Lung Cancer Driven by Mutant Kras and Loss of Lkb1. Cell reports. 2017;18:673–84. [DOI] [PubMed] [Google Scholar]
- 46.Sutherland KD, Song JY, Kwon MC, Proost N, Zevenhoven J, Berns A. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014;111:4952–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morel AP, Ginestier C, Pommier RM, Cabaud O, Ruiz E, Wicinski J, et al. A stemness-related ZEB1-MSRB3 axis governs cellular pliancy and breast cancer genome stability. Nat Med. 2017;23:568–78. [DOI] [PubMed] [Google Scholar]
- 48.Kareta MS, Gorges LL, Hafeez S, Benayoun BA, Marro S, Zmoos AF, et al. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell. 2015;16:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun. 2015;6:6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517:616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–9. [DOI] [PubMed] [Google Scholar]
- 54.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. [DOI] [PubMed] [Google Scholar]
- 55.Kuhn C Biotin stores in rodent lungs: localization to Clara and type II alveolar cells. Exp Lung Res. 1988;14:527–36. [DOI] [PubMed] [Google Scholar]
- 56.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–39. [DOI] [PubMed] [Google Scholar]
- 57.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 59.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 61.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.