Abstract
Gain-of-function mutations within the olfactomedin (OLF) domain of myocilin result in its toxic intracellular accumulation and hastens the onset of open-angle glaucoma. The absence of myocilin does not cause disease; therefore, strategies aimed at eliminating myocilin could lead to a successful glaucoma treatment. The endoplasmic reticulum Hsp90 paralog Grp94 accelerates OLF aggregation. Knockdown or pharmacological inhibition of Grp94 in cells facilitates clearance of mutant myocilin via a non-proteasomal pathway. Here we expand our support for targeting Grp94 over cytosolic paralogs Hsp90α and Hsp90β and then developed a high-throughput screening assay to identify new chemical matter capable of disrupting the Grp94/OLF interaction. When applied to a blind, focused library of 17 Hsp90 inhibitors, our miniaturized single-read in vitro thioflavin T -based kinetics aggregation assay exclusively identified compounds that target the chaperone N-terminal nucleotide binding site. In follow up studies, one compound (2) decreased the extent of co-aggregation of Grp94 with OLF in a dose-dependent manner in vitro. In cells stably expressing the full-length myocilin variant I477N, compound 2 enabled clearance of the aggregation-prone mutant protein without inducing the heat shock response or causing cytotoxicity. Comparison of the co-crystal structure of compound 2 and another non-selective hit in complex with the N-terminal domain of Grp94 reveals a docking mode tailored to Grp94 and explains its selectivity. A new lead compound has been identified, supporting a targeted chemical biology assay approach to develop a protein degradation-based therapy for myocilin-associated glaucoma by selectively inhibiting Grp94.
INTRODUCTION
Mutations in myocilin are causative for ~3–10% (~3 million patients) of the early-onset, heritable form of open angle glaucoma, a leading cause of blindness worldwide1–3. Myocilin is a protein secreted to the trabecular meshwork (TM)4–8, an extracellular matrix (ECM) located in the anterior eye segment. Though the specific function of myocilin in the ECM is not known3, the overall role of TM tissue is to regulate the outflow of nutrient-rich aqueous humor fluid4. Dysregulation of fluid outflow leads to ocular hypertension, currently the only known and clinically-addressable risk factor for glaucoma9, 10. Myocilin pathogenicity arises from coding mutations localized within the ~30 kDa myocilin C-terminal olfactomedin (OLF) domain11. OLF-directed mutations compromise protein stability12–14, resulting in its aggregation and accumulation in the endoplasmic reticulum (ER) of TM cells15, 16, which induces ER stress17–23 and ultimately causes cell death15, 19. Loss of TM cells is proposed to lead to loss of homeostatic control of intraocular pressure24, and initiates an accelerated path to vision loss and glaucoma.
The ER-associated degradation (ERAD) cellular system should efficiently respond to the insult of protein misfolding and aggregation25. In the case of mutant myocilin, however, glucose regulated protein 94 (Grp94)26, 27, the ER-resident heat shock protein 90 (Hsp90) molecular chaperone paralog, fails in its attempt to triage mutant myocilin for ERAD18. Grp94 and mutant myocilin co-aggregate and are retained in the ER18, 28 (Figure 1a). Cellular observations are recapitulated in vitro using an aggregation assay with purified proteins: Grp94 accelerates the rate at which the myocilin OLF domain forms thioflavin-T (ThT) positive amyloid aggregates, and Grp94 is recruited into the end-point aggregated material28. In cells, knockdown of Grp94 with siRNA or inhibition of Grp94 by pharmacologic intervention results in degradation of toxic mutant myocilin via autophagy, thus rescuing Grp94 from its co-aggregation fate18, 28. In vitro, pharmacologic inhibition of Grp94 rescues Grp94 from co-aggregation28. Grp94 has therefore been identified as a viable therapeutic target for the treatment of myocilin-associated glaucoma28–30, namely, as a means to enable mutant myocilin degradation and prevent TM cell death. Although Hsp90 isoforms share >85% sequence identity in nucleotide binding site region31, Grp94 is unique as a drug target because it is essential to cells only during early organismal development32, and its N-terminal domain nucleotide binding pocket includes a 5 amino acid insertion that has been exploited for selective inhibition33–35.
Figure 1.
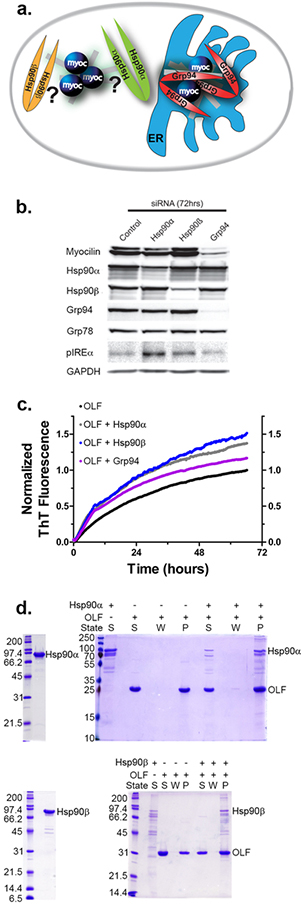
Effects of Hsp90 paralogs on mutant myocilin aggregation. a) Schematic of possible interactions between Hsp90 paralogs and aggregating mutant myocilin in a cellular context. b) Effects of Hsp90α, Hsp90β, and Grp94 siRNA knockdown on mutant myocilin levels, as well as ER stress markers Grp78 and pIRE1α. c) All Hsp90s enhance the rate of OLF aggregation in vitro. d) Post-assay analysis of aggregates from (c) by SDS-PAGE (right) reveals Hsp90α and Hsp90β co-aggregation with OLF. S = supernatant; W = wash; P = aggregate pellet. Left: Hsp90α or Hsp90β at the start of the aggregation experiment.
Here we first bolstered the premise of selective Grp94 inhibition to ameliorate mutant myocilin accumulation by comparing results of knockdown of cytosolic isoforms Hsp90α and Hsp90β on mutant myocilin degradation and their corresponding effects on OLF aggregation in vitro. We then developed a 384-well high-throughput screening (HTS) format single-readout biochemical aggregation assay with Grp94/OLF and used the assay to screen a focused library of Hsp90 family inhibitors to discover new chemical scaffolds capable of disrupting the Grp94/OLF interaction(s). Hits included only small molecules directed to the nucleotide-binding pocket. Secondary assays demonstrated dose-dependent rescue of Grp94 co-aggregation with OLF. Cellular studies confirmed one selective hit (compound 2) that enabled mutant myocilin degradation and did not induce the heat shock response, a critical issue for Hsp90 family drug development. The structural basis of selectivity of this compound was further revealed by comparing pan- and selective inhibitor-bound crystal structures. Taken together, our study has identified a promising chemical scaffold as well as supports a chemical biology approach for the discovery of other chemical matter with the potential to be developed into a disease-modifying therapeutic for myocilin-associated glaucoma.
RESULTS AND DISCUSSION
Effects of Cytosolic Hsp90s on Aggregation of OLF and Mutant Myocilin in Cells.
We first examined whether mutant myocilin aggregation is affected by levels of Hsp90α or Hsp90β, a scenario in which mutant myocilin is found in the cytosol36. Of particular interest was the result with Hsp90β, given analogous involvement with Alzheimer-associated tau37. Consistent with the observation that mutant myocilin aggregation predominantly occurs in the ER, knockdown of Hsp90α or Hsp90β does not promote mutant myocilin degradation nor its secretion in an inducible HEK293 cell model expressing I477N-mutant myocilin (iHEK) under a tetracycline promotor18, 38, 39 (Figure 1b, Supplementary Figure S1). Knockdown of Hsp90α or Hsp90β also increased levels of phosphorylated Ire1α (pIRE1α), an ER stress marker (Figure 1b, Supplementary Figure S1). By contrast, in vitro, OLF aggregation rates are enhanced in the presence of both Hsp90α and Hsp90β, and both proteins are observed in the end product (Figure 1c–d). The prominent enhancement observed with Hsp90β in vitro but not in cells suggests strict compartmentalization of aggregating mutant myocilin in the ER; otherwise, interactions between aggregating myocilin and Hsp90β would likely have led to significant toxicity37.
Design and Miniaturization of a Biochemical HTS Aggregation Assay.
After confirming the relevance of selective Grp94 inhibition to ameliorate myocilin aggregation, we developed a straightforward and statistically robust assay with a single end-point fluorescence readout compatible with HTS robotics. Our previously reported OLF aggregation assay28 takes advantage of wild-type OLF aggregation at slightly elevated temperatures (42°C) as a proxy for mutant OLF misfolding40. However, it requires mg-scale quantities of proteins and continuously sampling ThT fluorescence for up to 96 hours and is thus not practical for nor compatible with the instrumentation available at HTS screening facilities. To adapt the assay for a HTS setup, first the signal window (difference between OLF+Grp94 and OLF-only sample readings) and plate-to-plate reproducibility were iteratively optimized in 96-well format by testing numerous variables (final parameters in Supplementary Table S1). The Z-factor metric (Zʹ)41 was used to quantify improvements, with >0.5 an acceptable threshold. One key improvement was altering the ratio of OLF:Grp94 from 1:128 to 10:3, keeping OLF at 30 μM (~1 mg mL–1). Reproducibility was further enhanced when purified proteins (stock solutions prepared at > 100 μM for OLF and > 50 μM for Grp94) were pre-equilibrated at 4°C for 96 h prior to mixing and assay initiation. Finally, 18 hours at 42°C is sufficient for the optimal signal window (Supplementary Figure S2). The coefficient of variation (CV) for the signal window averaged ~10% under optimized conditions.
Next, the assay was converted to 384-well format (optimized parameters in Supplementary Table S2) for compatibility with Emory Chemical Biology Discovery Center (ECBDC) robotics and for reducing protein quantity (7.5-fold per well). Tissue-culture- (TC)- treated plates42 reproducibly yielded Zʹ scores of > 0.5 (average Zʹ = 0.65 over 2 plates, Figure 2a). Perhaps the oxidized plastic of TC plates stabilizes OLF because it mimics the extracellular milieu into which myocilin is secreted4–8. The CV for the 384-well plate setup averaged ~9%.
Figure 2.
Assay development. a) Plate-to-plate reproducibility of optimized, single-point readout, 384-well plate format miniaturized assay. A Zʹ score of 0.5 or greater was achieved per plate. Inhibitor 4-Br-BnIm was used as a positive control. b) Post-assay analysis of aggregates by SDS-PAGE reveals dose-responsive rescue of Grp94 from co-aggregation with OLF. S, W, P as in Figure 1.
Throughout assay optimization, the Grp94-selective inhibitor 4-Br-BnIm28, 29 was used as a positive control (Figure 2). This inhibitor attenuates the rate enhancement effect of Grp94 on OLF aggregation (Figure 2a) and it rescues Grp94 from co-aggregating with OLF (Figure 2b and reference28) in a dose-dependent fashion.
Hsp90 N-terminal Domain Inhibitors Identified in Focused Library Blind Screen.
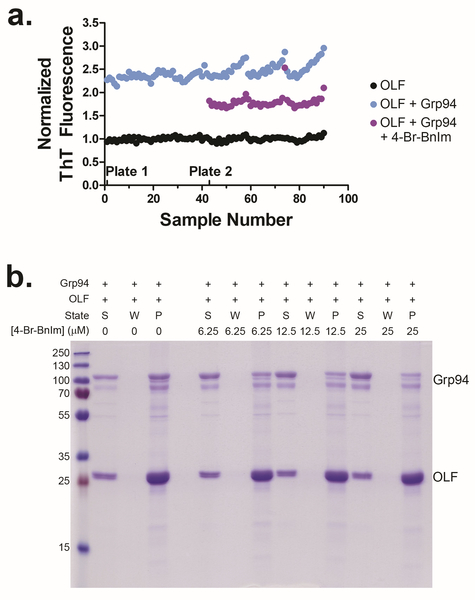
As a proof-of-concept test of the miniaturized HTS assay, we screened 17 Hsp90 inhibitors prepared by authors V.M.C. and B.S.J.B. This library contained both N- and C-terminal-directed Hsp90 inhibitors, and was blind to remaining authors involved in assay development and implementation (D.J.E.H., M.O.T., Y.D., H.F., R.L.L.) until after the screen was completed and results were ranked. An initial ThT fluorescence reading (t = 0 h) revealed that 4 of the small molecules (1, 5, 8, 17) were fluorescent under the assay conditions (25 μM compound), and were adjusted for in the final fluorescence reading (Figure 3a). A naïve readout of the screen results suggested 8 of the compounds (1–4, 6, 7, 9 and 11) exhibited the desired property of reverting the rate of OLF aggregation similar to samples where Grp94 is absent (Figure 3b–c). However, a heat gradient was apparent across the microplate row, complicating data interpretation. Specifically, the experimental controls, especially for the Grp94+OLF condition (Supplementary Figure S3a), each of which were dispensed in an individual row in the 384-well plate, exhibit an increase in ThT signal across-the-row from well positions 2–20. After correcting for this heat gradient and fluorescence (see Assay Data Analysis section of Supplemental Methods), the list of assay hits was expanded to 1–7 and 9–11 (Table 1, Supplementary Table S3). For future screening campaigns, minimizing this incubator-induced heat gradient will be necessary, by using a different incubator. After correcting for heat gradient effects, the average Zʹ score (0.57) and CV (12%) lent confidence to the assignment of compounds as preliminary hits.
Figure 3.
Blind screen of Blagg lab library. a) ThT fluorescence at time = 0 hours reveals intrinsic fluorescence of compounds 1, 5, 8, and 17 at 25 μM (purple line). b) Raw data from ThT fluorescence at 18 h suggests hit compounds are 1–7, 9–11. Average signals for OLF (solid black line) and OLF+Grp94 (solid blue line) controls are presented with ± 3 standard deviations (paired color bars). Data for odd-numbered compounds/replicates designated by black spheres; even-numbered compound data are shown by magenta spheres. c) Calculated percent inhibition scores using different methods. Heat gradient and compound intrinsic fluorescence adjustments classify compounds 5 and 10 as hits. d) Representative post-assay aggregate analysis SDS-PAGE gels. Grp94 co-aggregation with OLF seen in control (0, left) and non-hits (15, right). Hits (1, middle) resulted in partial rescue of Grp94. S, W, P as in Figure 1. For post-assay analysis of all compounds, see Supplementary Figure S3b–h.
Table 1.
Hit Ranking and Properties of Blagg Lab Blind Screen Compounds
| Compound # | Tier of Inhibitor |
Chemical Structure | Isoform Selectivity | Domain Targeted | % Inhibitiona |
|---|---|---|---|---|---|
| 5 | Top | 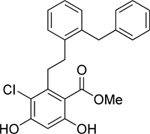 |
Grp94 | N-term | 159.6 |
| 1 | Top | 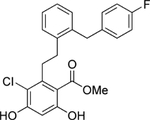 |
Grp94 | N-term | 148.4 |
| 3 | Mid | 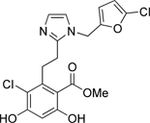 |
Grp94 | N-term | 54.3 |
| 7 | Mid | 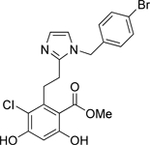 |
Grp94 | N-term | 52.8 |
| 2 | Mid | 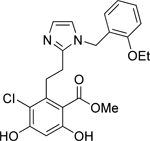 |
Grp94 | N-term | 52.3 |
| 11 | Mid | 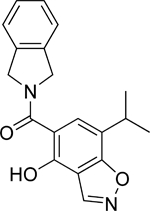 |
Hsp90β | N-term | 43.2 |
| 10 | Low | 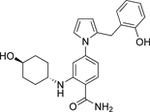 |
Grp94 | N-term | 31.4 |
| 4 | Low | 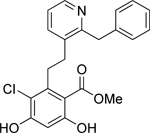 |
Non-selective | N-term | 28.4 |
| 6 | Low | Structure Not Disclosed | Hsp90α | N-term | 27.9 |
| 9 | Low | Structure Not Disclosed | Hsp90β | N-term | 27.4 |
| 13 | Non | 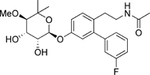 |
-- | C-term | −31.7 |
| 16 | Non | 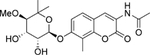 |
-- | C-term | −32.0 |
| 12 | Non | 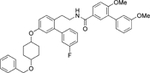 |
-- | C-term | −38.7 |
| 17 | Non | -- | C-term | −43.5 | |
| 14 | Non | 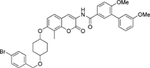 |
-- | C-term | −85.1 |
| 15 | Non | 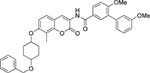 |
-- | C-term | −86.7 |
| 8 | Non | Structure Not Disclosed | Hsp90α/β | N-term | −98.3 |
See Supporting Information for calculation of % Inhibition after correction for the heat gradient experienced and intrinsic compound fluorescence. Tier ranking: >66% inhibition (top), >33% (mid), >0% (low), <0% (Non) non-hit).
To confirm the expected rescue of Grp94 from co-aggregation with OLF28, the end-point aggregate from the primary assay was visualized by SDS-PAGE (representative gel in Figure 3d, results for all compounds screened Supplementary Figure S3b–h, summary in Supplementary Table S3). Compounds 1–11 displayed the expected rescue of soluble Grp94. Compound 8, an Hsp90α/β inhibitor, was a false-negative in the assay, perhaps due to solubility or handling issues. Treatment with non-hits 12–17 failed to mitigate co-aggregation of Grp94 with OLF. Upon treatment with hit compounds (Figure 3d, Supplementary Figure S3b–h), there is an increase in soluble Grp94, and less co-aggregated Grp94, at the assay endpoint.
Secondary Dose-Response Assays Confirm Reduced Grp94/OLF Co-aggregation.
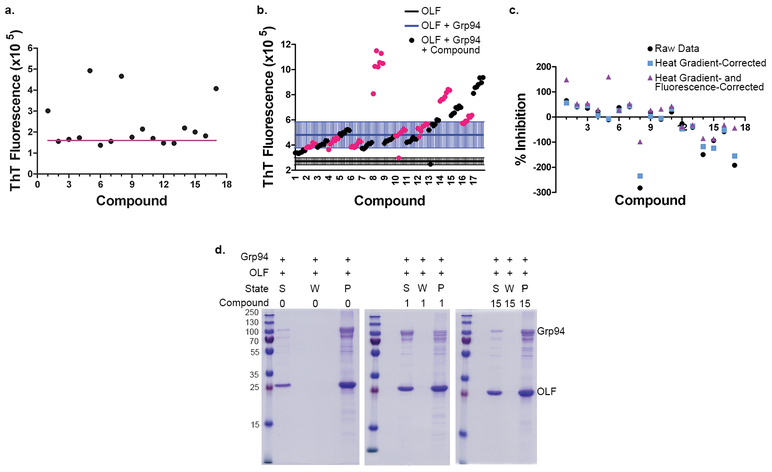
Representative hits with initial rank as top- (1), mid- (2, 7), or low-tier (4, 9) and negative control (15) were tested for dose-response rescue of soluble Grp94 in 96-well format using TC-treated plates. Post-assay products analyzed by SDS-PAGE (Figure 4 and Supplementary Figures S4 and S5) reveal dose-dependent rescue of Grp94 from co-aggregation with OLF in the presence of hit compounds (soluble Grp94 increased from ~33% to ~65%), irrespective of initial ranking. Compound 15 had no effect on Grp94 solubility (Figure 4b–c and Supplementary Figure S4) and OLF solubility was not impacted by the presence of hit compounds, either in the presence or absence of Grp94 (Supplementary Figure S4). This result reinforces the supposition that the HTS assay hits act via inhibition of Grp94.
Figure 4.
Dose-responsive rescue of Grp94 from co-aggregation with OLF. a-b) Representative post-assay SDS-PAGE analysis of hit compound (2, a) and negative control (15, b), with control conditions given at the left of each gel. S = supernatant; W = wash; P = pellet/aggregate. c) Densitometric analysis of post-dose-response aggregation assay bands on SDS-PAGE reveals that compounds identified as hits increase the percentage of soluble Grp94 by inhibiting chaperone activity. Data reflect at least 2 experiments and 2 gels per experiment. Error bars indicate standard deviation. Raw data appear in Supplementary Figures S4 and S5.
Cell-based Assays Reveal a Promising Grp94-Selective Inhibitor Scaffold.
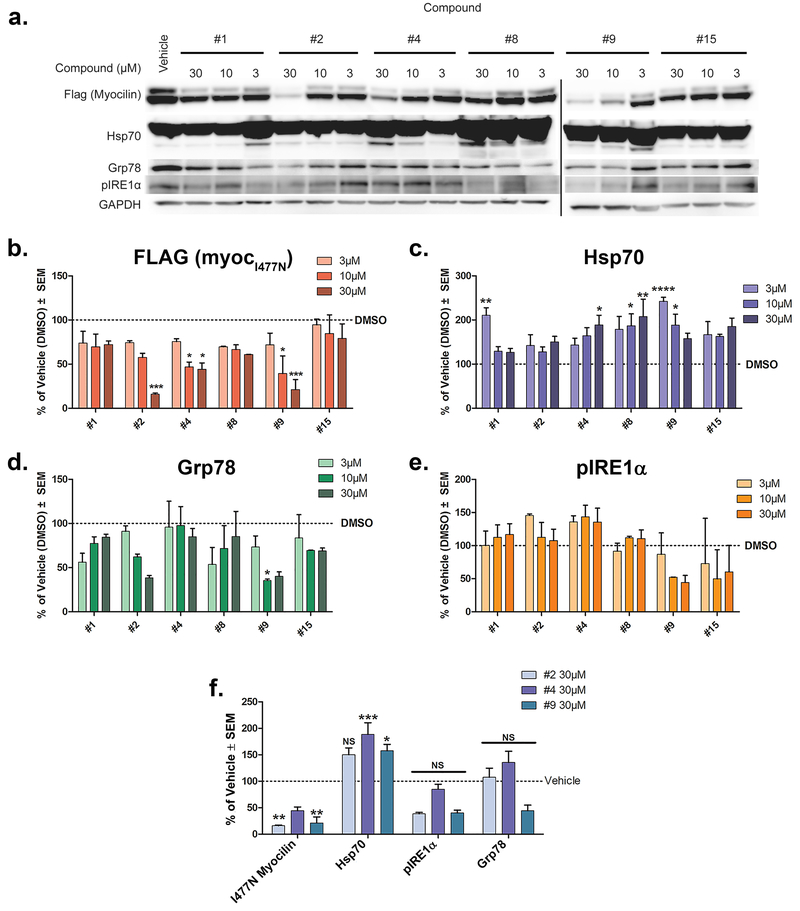
Inhibition of Grp94 with small molecules should enable the degradation of mutant myocilin18, 28 and thus reduce levels of accumulated intracellular mutant myocilin in cell culture, similar to the effect of 4-Br-BnIm. A selection of hit compounds (1, top-tier; 2 and 4, mid-level; 9, low-ranking; 8, false-negative, 15, negative control), were used to treat iHEK cells18, 38, 39. Only treatment with compound 2 or 9 resulted in a statistically significant clearance of I477N-mutant myocilin (Figure 5a–b). Treatment with 2 at up to 30 μM did not induce the pro-survival heat shock response as tracked by levels of Hsp70 (Figure 5c) and was not cytotoxic (Supplementary Figure S6). In addition, loss of Grp94 activity due to inhibition by 2 did not induce ER stress, as evidenced by reduced levels of Grp78 (Figure 5d), and no changes in pIRE1α (Figure 5e). By contrast, 9 induced the expression of Hsp70 (Figure 5c,f) suggesting undesirable non-selective Hsp90-family inhibition.
Figure 5.
Cellular profiling of hits. a) Western blot analysis of lysates from iHEK cells reveals that treatment with 2 results in dose-dependent clearance of mutant myocilin. Representative of two independent experiments is presented. b) Quantification of intracellular myocilin bands in (a). c-e) Compound 2 does not evoke the ER stress response (c, Hsp70; d, Grp78; e, pIRE1α) . f) Comparison of cellular responses to highest dose (30 μM) of compounds tested for compounds 2, 4, and 9; p <0.05, p<0.01, p<0.001, and p<0.0001 by Bonferoni Multiple Comparisons Post-hoc test. GAPDH serves as a load control and error bars are ± SEM.
After cell-based assays were concluded, the identities of the focused library molecules were disclosed. Compounds 1–11 are inhibitors of Hsp90 designed to target the N-terminal domain nucleotide-binding pocket. The top five ranked compounds based on the initial HTS assay are Grp94-selective. Compound 7 was revealed to be 4-Br-BnIm, the positive control used throughout assay development/optimization. Library members not positively identified as hits, 12–17, bind the C-terminal domain of Hsp90. Compounds 6, 8 and 9 are in early development and thus their chemical structures were not disclosed.
Proposed Structural Basis for Grp94 Selective Inhibition.
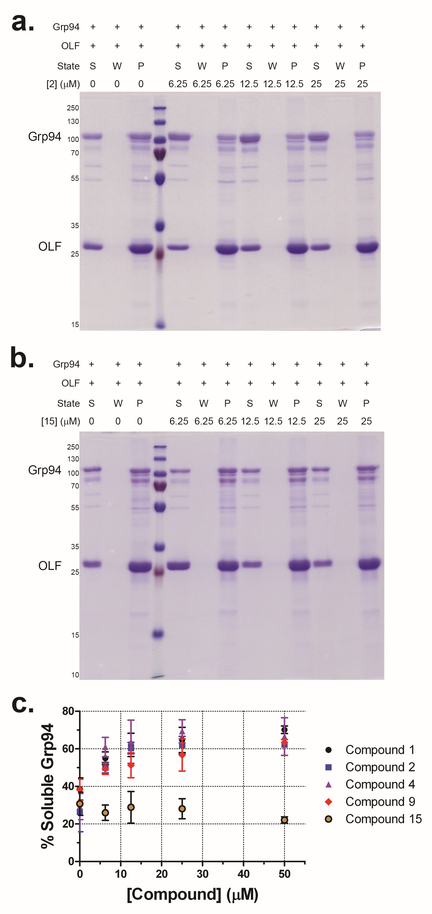
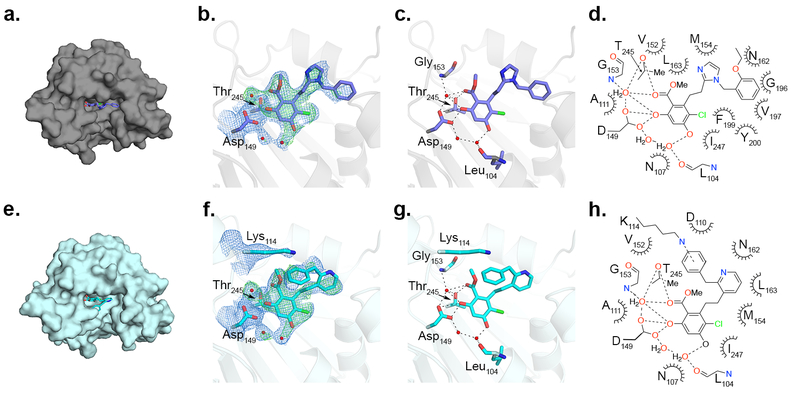
To gain molecular insight into the differing responses of compounds 2 and 4 in clearing mutant myocilin at 30 μM (Figure 5a,b,f), we solved co-crystal structures of Grp94 N-terminal domain construct NΔ41 (see Supporting Information and Supplementary Table S4). Compounds 2 and 4 feature the decorated resorcinol base of pan-Hsp90 inhibitor radamide43, plus either the BnIm-like44 imidazole linker (2) or a pyridine (4), resulting in two distinct poses in the nucleotide binding site of NΔ41 (Figure 6). The resorcinol anchors both molecules by direct interaction with Asp149 and Thr245, an interaction stabilized by a modest hydrogen bonding network (Figure 6). Compound 2 projects its benzyl side chain into the Grp94-specific hydrophobic pocket, and is stabilized by a number of nonpolar residues (Figure 6d); electron density coverage of the terminal ethyl ether substituent is absent, but the steric environment suggests a surface-directed orientation (Figure 6b–d). The benzyl moiety of 4, on the other hand, folds back away from the Grp94-exclusive pocket toward the solvent-exposed region of the nucleotide binding site and forms a cation-π interaction with Lys114. This residue is conserved among all Hsp90s (Figure 6f–h), suggesting that 4 is a pan-Hsp90 inhibitor and thus capable of inducing the heat shock response as indicated by Hsp70 induction (Figure 5c,f).
Figure 6.
Crystal structures of 2 (a-d) and 4 (e-h) bound to NΔ41. a, e) Surface representation of NΔ41 nucleotide binding pocket bound by inhibitors, shown as sticks. b, f) Zoomed view, with direct protein-inhibitor interactions highlighted. The final 2Fo-Fc map (blue mesh), contoured at 1.0 σ for ligands and 1.3 σ for amino acid residues, are superimposed with Fo-Fc densities (green mesh) contoured at 2.5 σ after initial molecular replacement (see Supporting Information). Inhibitors and residues are depicted as sticks, waters are red spheres. c, g) NΔ41-inhibitor interactions mediated by hydrogen bonds (dashes) in zoomed view. d, h) NΔ41-inhibitor interactions (dashed lines, hydrogen bonds or cation-π interactions; dashed crescents, hydrophobic interactions. Panels a-c, e-g were prepared with PyMOL (http://www.pymol.org). Panels d, h were generated with ChemDraw (version 16.0.1.4, Perkin Elmer).
In summary, our data support the strategy of specifically inhibiting Grp94 to abrogate co-aggregation of Grp94 and OLF. Our HTS ThT aggregation assay identified only molecules in the focused library that target the N-terminal nucleotide binding pocket, suggesting that the conformation of the N-terminal domain of Grp94 modulates its interaction with OLF. Thus, conformational dynamics localized to the C-terminal dimerization domain of Grp94, which are relevant to the overall conformational cycle of Hsp90s45, do not appear consequential with respect to the aberrant myocilin-Grp94 interaction. Compound 2, the most promising hit from secondary assays, is similar to 4-Br-BnIm, in vitro and in cells28. However, whereas the binding mode of 4-Br-BnIm in Grp94 is ambiguous beyond the resorcinol ring46, structural comparison of 2 versus 4 reveals that 2 interacts robustly with the unique secondary pocket of Grp94 furnishing selectivity; 4 forms a cation-π interaction with a conserved lysine residue that likely promotes non-selectivity among the Hsp90 isoforms.
Screening our assay against a chemically diverse compound library should be able to identify not just Hsp90-family N-terminal-directed small molecules, but also compounds that target the Grp94-OLF protein-protein interaction interface, as well as compounds that inhibit OLF aggregation by only binding to OLF. Compounds in the first two categories should result in mutant myocilin degradation by intracellular non-proteasomal degradation18; the third category may stabilize mutant myocilin and avoid detection by Grp94. In the latter scenario, mutant myocilin should be secreted where either it will be functional and thus the compound will serve as a pharmacological chaperone47 or myocilin will be removed by TM cell phagocytosis48, a major biological function of such cells in the TM ECM.
METHODS
Recombinant Protein Expression and Purification.
OLF was expressed and purified as described previously40. The truncated N-terminal domain of Canis lupus familiaris Grp94 (NΔ41, cloned by ATUM), which comprises amino acid residues 69–337 but 287–327 is replaced with Gly as in Dollins et al.34, was expressed and purified as before29. Procedures for expressing and purifying full-length Hsp90s (Grp94, Hsp90α, Hsp90β) are described in Supporting Information.
ThT Fluorescence Kinetics Aggregation Assay, 96-Well Format.
A working stock of 200 μM ThT (Sigma-Aldrich) was prepared in PBS, diluted from a 1 mg mL–1 master solution in water. A stock solution of 20 mM 4-Br-BnIm29 was prepared in DMSO and stored at –80°C prior to use. All aggregation assay samples contained 10 μM ThT, 0.5% (v/v) DMSO (absent in assay samples in Figure 1c), and PBS buffer. Protein-containing samples had either 30 μM OLF and/or 9 μM Grp94, diluted from concentrated stocks (>100 μM for OLF and >50 μM Grp94) that were equilibrated for at least 4 days at 4°C prior to assay setup. Hsp90 isoform experiments in Figure 1c were conducted similarly except 3 μM Grp94, Hsp90α, and Hsp90β were used without pre-equilibrarion. 4-Br-BnIm was used at 20 μM. Master mixes were prepared at room temperature, then dispensed in 150 μL aliquots into 96-well (black well, black bottom, medium binding) microplates (Grenier); each sample type was represented by at least 9–12 replicates per experiment (3 or greater replicates per sample type for the data in Figure 1c, representing 2 or more biological replicates). Plates were sealed with clear MicroAmp PCR film (Applied Biosystems) and loaded into a Biotek Synergy 2 microplate reader set to 42°C. ThT fluorescence (λex = 440 nm, λem = 485 nm) was measured every 10 minutes up to 66 hours (Figure 1c) or at 18 hours after incubation (Supplementary Figure S2). Dose-response assays were performed similarly (Supplementary Table S5), however TC-treated plates were used.
384-Well Format Aggregation Assay and Blind Library Screening.
To accommodate screening capabilities at ECBDC (Supplementary Table S5), 384-well low volume TC-treated plates with black wall and clear bottom from Corning (Cat#3542) were used. After dispensing samples (20 μL/well), plates were centrifuged (200 xg) for 5 min to remove air bubbles prior to reading ThT fluorescence (λex = 440 nm, λem = 485 nm) with an EnVision 2103 multilabel plate reader (Perkin Elmer). Plates were incubated at 42°C in a Cytomat incubator. Fluorescence measurements were taken with the plate-sealing film removed, and from top-down. The Blagg lab compound library was tested at 25 μM.
Assay Data Analysis.
Zʹ score, CV, as well as % inhibition are outlined in Supporting Information.
Post-assay Product Analysis by SDS-PAGE.
After ThT fluorescence measurements were acquired for aggregation assays (controls, blind screen, dose-response), the plates/samples were allowed to incubate for an additional 78 hours at 42°C; 30 additional hours of incubation for samples in Figure 1c. Wells containing identical samples were then harvested, pooled, and pelleted by centrifugation (10 minutes at 4°C and 17,000 xg), washed, and analyzed by SDS-PAGE as before28. After staining with Coomassie blue, densitometry was performed on the resulting bands with ImageJ software (http://imagej.nih.gov/ij/).
Cell Culture.
iHEK cells38 were grown and maintained in Dulbecco’s modified Eagle’s Medium (GE Healthcare) supplemented with 10% fetal bovine serum (VWR), 1% sodium pyruvate (Corning), and 1% GlutaMAX (Invitrogen) at 37°C under 5% CO2. Cells were plated and induced the next day with 5 μg mL–1 tetracycline. At 48 hours post induction, cells were treated with compounds (1, 2, 4, 8 or 9 at 3–30 μM) or DMSO vehicle and grown for an additional 24 hours. For Western blot experiments, cells were plated in a 6-well plate at a density of 3.0 ×105 cells/well. For cytotoxicity and cell viability assays, cells were plated 7.0 ×104 cells/well in 96-well plates (Corning, Cat#07-200-90). Experiments were conducted in duplicate.
siRNA Transfections.
siRNAs were transfected as reported previously18. Cells were harvested 48 hours post transfection with siRNA for Western blot analysis. Medium was collected for LDH analysis.
Western Blots.
Cell Viability Assays.
Lactose dehydrogenase (LDH) and sulforhodamine B (SRB) assays were performed as outlined in the Supporting Information.
Structure Determination of NΔ41 in Complex with Compounds 2 and 4.
Synthesis of Small Molecule Library.
Compounds 1, 4, and 5 were prepared as in reference49; 2, 3, and 7 were synthesized as in reference29; 10, 11 were synthesized as in reference50; 12, 14, 15 were synthesized as in reference51; 13 was synthesized as in reference52; 16 and 17 were synthesized as in reference53. The structures of compounds 6, 8, and 9, designed to target the nucleotide binding pocket of Grp94, are undisclosed; related manuscripts are in preparation.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01EY021205 to R. Lieberman, R01EY024232 to B. Blagg and C. Dickey, as well as NCATS award UL1TR000454 to R. Lieberman, H. Fu and Y. Du. V. Crowley is supported by F99CA212467 and M. Tomlin by the Petit Scholars Program (Georgia Tech).
Footnotes
Accession Codes.The structures of NΔ41 bound to compounds 2 and 4 have been deposited in the Protein Data Bank with PDB accession codes 6ASP and 6ASQ, respectively.
SUPPORTING INFORMATION. Figures S1–S6 and Tables S1–S5 appear in Supporting Information. Supporting Information Available: This material is available free of charge via the Internet.
REFERENCES
- [1].Quigley HA, and Broman AT (2006) The number of people with glaucoma worldwide in 2010 and 2020, Br. J. Ophthalmol 90, 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, and Sheffield VC (1997) Identification of a gene that causes primary open angle glaucoma, Science 275, 668–670. [DOI] [PubMed] [Google Scholar]
- [3].Resch ZT, and Fautsch MP (2009) Glaucoma-associated myocilin: a better understanding but much more to learn, Exp. Eye Res. 88, 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kwon YH, Fingert JH, Kuehn MH, and Alward WL (2009) Primary open-angle glaucoma, N. Engl. J. Med 360, 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hardy KM, Hoffman EA, Gonzalez P, McKay BS, and Stamer WD (2005) Extracellular trafficking of myocilin in human trabecular meshwork cells, J. Biol. Chem 280, 28917–28926. [DOI] [PubMed] [Google Scholar]
- [6].Filla MS, Liu X, Nguyen TD, Polansky JR, Brandt CR, Kaufman PL, and Peters DM (2002) In vitro localization of TIGR/MYOC in trabecular meshwork extracellular matrix and binding to fibronectin, Invest. Ophthalmol. Vis. Sci 43, 151–161. [PubMed] [Google Scholar]
- [7].Tawara A, Okada Y, Kubota T, Suzuki Y, Taniguchi F, Shirato S, Nguyen TD, and Ohnishi Y (2000) Immunohistochemical localization of MYOC/TIGR protein in the trabecular tissue of normal and glaucomatous eyes, Curr. Eye Res 21, 934–943. [DOI] [PubMed] [Google Scholar]
- [8].Wentz-Hunter K, Kubota R, Shen X, and Yue BY (2004) Extracellular myocilin affects activity of human trabecular meshwork cells, J. Cell Physiol 200, 45–52. [DOI] [PubMed] [Google Scholar]
- [9].Alward WL (1998) Medical management of glaucoma, N. Engl. J. Med 339, 1298–1307. [DOI] [PubMed] [Google Scholar]
- [10].Donegan RK, and Lieberman RL (2016) Discovery of molecular therapeutics for glaucoma: challenges, successes, and promising directions, J. Med. Chem 59, 788–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hewitt AW, Mackey DA, and Craig JE (2008) Myocilin allele-specific glaucoma phenotype database, Hum. Mutat 29, 207–211. [DOI] [PubMed] [Google Scholar]
- [12].Burns JN, Orwig SD, Harris JL, Watkins JD, Vollrath D, and Lieberman RL (2010) Rescue of glaucoma-causing mutant myocilin thermal stability by chemical chaperones, ACS Chem. Biol 5, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Burns JN, Turnage KC, Walker CA, and Lieberman RL (2011) The stability of myocilin olfactomedin domain variants provides new insight into glaucoma as a protein misfolding disorder, Biochemistry 50, 5824–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Donegan RK, Hill SE, Freeman DM, Nguyen E, Orwig SD, Turnage KC, and Lieberman RL (2015) Structural basis for misfolding in myocilin-associated glaucoma, Hum. Mol. Genet 24, 2111–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu Y, and Vollrath D (2004) Reversal of mutant myocilin non-secretion and cell killing: implications for glaucoma, Hum. Mol. Genet 13, 1193–1204. [DOI] [PubMed] [Google Scholar]
- [16].Jacobson N, Andrews M, Shepard AR, Nishimura D, Searby C, Fingert JH, Hageman G, Mullins R, Davidson BL, Kwon YH, Alward WL, Stone EM, Clark AF, and Sheffield VC (2001) Non-secretion of mutant proteins of the glaucoma gene myocilin in cultured trabecular meshwork cells and in aqueous humor, Hum. Mol. Genet 10, 117–125. [DOI] [PubMed] [Google Scholar]
- [17].Joe MK, Sohn S, Hur W, Moon Y, Choi YR, and Kee C (2003) Accumulation of mutant myocilins in ER leads to ER stress and potential cytotoxicity in human trabecular meshwork cells, Biochem. Biophys. Res. Commun 312, 592–600. [DOI] [PubMed] [Google Scholar]
- [18].Suntharalingam A, Abisambra JF, O’Leary JC 3rd, Koren J 3rd, Zhang B, Joe MK, Blair LJ, Hill SE, Jinwal UK, Cockman M, Duerfeldt AS, Tomarev S, Blagg BS, Lieberman RL, and Dickey CA (2012) Glucose-regulated protein 94 triage of mutant myocilin through endoplasmic reticulum-associated degradation subverts a more efficient autophagic clearance mechanism, J. Biol. Chem 287, 40661–40669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yam GH, Gaplovska-Kysela K, Zuber C, and Roth J (2007) Aggregated myocilin induces russell bodies and causes apoptosis: implications for the pathogenesis of myocilin-caused primary open-angle glaucoma, Am. J. Pathol 170, 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zode GS, Kuehn MH, Nishimura DY, Searby CC, Mohan K, Grozdanic SD, Bugge K, Anderson MG, Clark AF, Stone EM, and Sheffield VC (2011) Reduction of ER stress via a chemical chaperone prevents disease phenotypes in a mouse model of primary open angle glaucoma, J. Clin. Invest 121, 3542–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kanagavalli J, Pandaranayaka PJ, Krishnadas SR, Krishnaswamy S, and Sundaresan P (2007) In vitro and in vivo study on the secretion of the Gly367Arg mutant myocilin protein, Mol. Vis 13, 1161–1168. [PubMed] [Google Scholar]
- [22].Wang L, Zhuo Y, Liu B, Huang S, Hou F, and Ge J (2007) Pro370Leu mutant myocilin disturbs the endoplasm reticulum stress response and mitochondrial membrane potential in human trabecular meshwork cells, Mol. Vis 13, 618–625. [PMC free article] [PubMed] [Google Scholar]
- [23].Anholt RR, and Carbone MA (2013) A molecular mechanism for glaucoma: endoplasmic reticulum stress and the unfolded protein response, Trends Mol. Med 19, 586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Acott TS, Kelley MJ, Keller KE, Vranka JA, Abu-Hassan DW, Li X, Aga M, and Bradley JM (2014) Intraocular pressure homeostasis: maintaining balance in a high-pressure environment, J. Ocul. Pharmacol. Ther 30, 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Meusser B, Hirsch C, Jarosch E, and Sommer T (2005) ERAD: the long road to destruction, Nat. Cell Biol 7, 766–772. [DOI] [PubMed] [Google Scholar]
- [26].Marzec M, Eletto D, and Argon Y (2012) GRP94: an HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum, BBA-Mol. Cell Res 1823, 774–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Eletto D, Dersh D, and Argon Y (2010) GRP94 in ER quality control and stress responses, Semin. Cell Dev. Biol 21, 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stothert AR, Suntharalingam A, Huard DJ, Fontaine SN, Crowley VM, Mishra S, Blagg BS, Lieberman RL, and Dickey CA (2014) Exploiting the interaction between Grp94 and aggregated myocilin to treat glaucoma, Hum. Mol. Genet 23, 6470–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Crowley VM, Khandelwal A, Mishra S, Stothert AR, Huard DJE, Zhao JB, Muth A, Duerfeldt AS, Kizziah JL, Lieberman RL, Dickey CA, and Blagg BSJ (2016) Development of glucose regulated protein 94-selective inhibitors based on the BnIm and radamide scaffold, J. Med. Chem 59, 3471–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Stothert AR, Fontaine SN, Sabbagh JJ, and Dickey CA (2016) Targeting the ER-autophagy system in the trabecular meshwork to treat glaucoma, Exp. Eye Res 144, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen B, Piel WH, Gui L, Bruford E, and Monteiro A (2005) The HSP90 family of genes in the human genome: insights into their divergence and evolution, Genomics 86, 627–637. [DOI] [PubMed] [Google Scholar]
- [32].Maynard JC, Pham T, Zheng T, Jockheck-Clark A, Rankin HB, Newgard CB, Spana EP, and Nicchitta CV (2010) Gp93, the Drosophila GRP94 ortholog, is required for gut epithelial homeostasis and nutrient assimilation-coupled growth control, Dev. Biol 339, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gewirth DT (2016) Paralog specific Hsp90 inhibitors - a brief history and a bright future, Curr. Top. Med. Chem 16, 2779–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dollins DE, Warren JJ, Immormino RM, and Gewirth DT (2007) Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones, Mol. Cell 28, 41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Soldano KL, Jivan A, Nicchitta CV, and Gewirth DT (2003) Structure of the N-terminal domain of GRP94 - Basis for ligand specificity and regulation, J. Biol. Chem 278, 48330–48338. [DOI] [PubMed] [Google Scholar]
- [36].Joe MK, Kee C, and Tomarev SI (2012) Myocilin interacts with syntrophins and is member of dystrophin-associated protein complex, J. Biol. Chem 287, 13216–13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lackie RE, Maciejewski A, Ostapchenko VG, Marques-Lopes J, Choy WY, Duennwald ML, Prado VF, and Prado MAM (2017) The Hsp70/Hsp90 chaperone machinery in neurodegenerative diseases, Front. Neurosci 11, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Joe MK, and Tomarev SI (2010) Expression of myocilin mutants sensitizes cells to oxidative stress-induced apoptosis: implication for glaucoma pathogenesis, Am. J. Pathol 176, 2880–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Orwig SD, Chi PV, Du Y, Hill SE, Cavitt MA, Suntharalingam A, Turnage KC, Dickey CA, France S, Fu H, and Lieberman RL (2014) Ligands for glaucoma-associated myocilin discovered by a generic binding assay, ACS Chem. Biol 9, 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hill SE, Donegan RK, and Lieberman RL (2014) The glaucoma-associated olfactomedin domain of myocilin forms polymorphic fibrils that are constrained by partial unfolding and peptide sequence, J. Mol. Biol 426, 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang JH, Chung TDY, and Oldenburg KR (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays, J. Biomol. Screen 4, 67–73. [DOI] [PubMed] [Google Scholar]
- [42].Curtis ASG, Forrester JV, Mcinnes C, and Lawrie F (1983) Adhesion of cells to polystyrene surfaces, J. Cell Biol 97, 1500–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Clevenger RC, and Blagg BSJ (2004) Design, synthesis, and evaluation of a radicicol and geldanamycin chimera, radamide, Org. Lett 6, 4459–4462. [DOI] [PubMed] [Google Scholar]
- [44].Duerfeldt AS, Peterson LB, Maynard JC, Ng CL, Eletto D, Ostrovsky O, Shinogle HE, Moore DS, Argon Y, Nicchitta CV, and Blagg BSJ (2012) Development of a Grp94 inhibitor, J. Am. Chem. Soc 134, 9796–9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ratzke C, Mickler M, Hellenkamp B, Buchner J, and Hugel T (2010) Dynamics of heat shock protein 90 C-terminal dimerization is an important part of its conformational cycle, Proc. Natl. Acad. Sci. USA 107, 16101–16106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Stothert AR, Suntharalingam A, Tang X, Crowley VM, Mishra SJ, Webster JM, Nordues BA, Huard DJE, Passaglia C, Lieberman RL, Blagg BSJ, Blair LJ, Koren JI, and Dickey CA (2017) Isoform-selective Hsp90 inhibition rescues model of hereditary open-angle glaucoma Sci Rep. 7, 17951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cohen FE, and Kelly JW (2003) Therapeutic approaches to protein-misfolding diseases, Nature 426, 905–909. [DOI] [PubMed] [Google Scholar]
- [48].Buller C, Johnson DH, and Tschumper RC (1990) Human trabecular meshwork phagocytosis. Observations in an organ culture system, Invest. Ophthalmol. Vis. Sci 31, 2156–2163. [PubMed] [Google Scholar]
- [49].Crowley VM, Huard DJE, Lieberman RL, and Blagg BSJ (2017) Second generation Grp94-selective inhibitors provide opportunities for the inhibition of metastatic cancer, Chem. Eur. J 23, 15775–15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Khandelwal A, Kent CN, Balch M, Peng S, Mishra SJ, Deng J, Day VW, Liu W, Holzbeierlein JM, Matts R, and Blagg BSJ (2017) Structure-guided design of the first Hsp90b N-terminal isoform-selective inhibitor, Nat. Commun in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Anyika M, McMullen M, Forsberg LK, Dobrowsky RT, and Blagg BS (2016) Development of noviomimetics as C-terminal Hsp90 inhibitors, ACS Med. Chem. Lett 7, 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kusuma BR, Zhang L, Sundstrom T, Peterson LB, Dobrowsky RT, and Blagg BS (2012) Synthesis and evaluation of novologues as C-terminal Hsp90 inhibitors with cytoprotective activity against sensory neuron glucotoxicity, J. Med. Chem 55, 5797–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Burlison JA, Avila C, Vielhauer G, Lubbers DJ, Holzbeierlein J, and Blagg BSJ (2008) Development of novobiocin analogues that manifest anti-proliferative activity against several cancer cell lines, J. Org. Chem 73, 2130–2137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.