Abstract
Agent-based models (ABMs) integrate multiple scales of behavior and data to produce higher-order dynamic phenomena and are increasingly used in the study of important social complex systems in biomedicine, socio-economics and ecology/resource management. However, the development, validation and use of ABMs is hampered by the need to execute very large numbers of simulations in order to identify their behavioral properties, a challenge accentuated by the computational cost of running realistic, large-scale, potentially distributed ABM simulations. In this paper we describe the Extreme-scale Model Exploration with Swift (EMEWS) framework, which is capable of efficiently composing and executing large ensembles of simulations and other “black box” scientific applications while integrating model exploration (ME) algorithms developed with the use of widely available 3rd-party libraries written in popular languages such as R and Python. EMEWS combines novel stateful tasks with traditional run-to-completion many task computing (MTC) and solves many problems relevant to high-performance workflows, including scaling to very large numbers (millions) of tasks, maintaining state and locality information, and enabling effective multiple-language problem solving. We present the high-level programming model of the EMEWS framework and demonstrate how it is used to integrate an active learning ME algorithm to dynamically and efficiently characterize the parameter space of a large and complex, distributed Message Passing Interface (MPI) agent-based infectious disease model.
Keywords: Agent-based modeling, metamodeling, high performance computing, machine learning, parallel processing
I. INTRODUCTION
RECENT improvements in high-performance agent-based models (ABMs) have enabled the simulation of a variety of complex systems, including the spread of infectious diseases and community-based healthcare interventions [1],[2], critical materials supply chains [3], and land-use and resource management [4], [5]. As ABMs have become more complex, capturing more salient features of the systems under study, parameters that dictate the structural (e.g., social networks), behavioral, and other dynamical elements of the models have increased in number. Other complex systems modeling approaches (e.g., mathematical modeling, system dynamics) can rely on assumptions about model and parameter space structures to make use of relatively efficient methods for model calibration and optimization. However, the highly nonlinear relationship between ABM input parameters and model outputs, as well as feedback loops and emergent behaviors, require less efficient ensemble modeling approaches. These approaches execute large numbers of simulations, often in complex iterative workflows driven by sophisticated model exploration (ME) algorithms, such as active learning (AL), which adaptively refine model parameters through the analysis of recently generated simulation results and launch new simulations.
In order to facilitate these dynamic ME-based approaches, we have created the Extreme-scale Model Exploration with Swift (EMEWS) framework [6], [7]. EMEWS, which is built on Swift/T [8], offers the capability to run very large, highly concurrent ensembles of simulations of varying types while supporting a wide class of ME algorithms, including those increasingly available to the community via Python and R libraries. Furthermore, it offers a software sustainability solution, in that ME studies based around EMEWS can easily be compared and distributed. A central EMEWS design goal is to ease software integration while providing scalability to the largest scale (petascale plus) supercomputers, running millions of ABMs, thousands at a time. Initial scaling studies of EMEWS have shown robust scalability [9]. The tools are also easy to install and run on an ordinary laptop, requiring only an MPI (Message Passing Interface) implementation, which can be easily obtained from common OS package repositories. By combining novel stateful tasks with traditional run-to-completion many task computing, our framework solves many problems relevant to high-performance workflows, including scaling to very large numbers (millions) of tasks, maintaining state and locality information, and the multiple language problem.
EMEWS enables the user to plug in both ME algorithms and models (e.g., ABMs). Thus, researchers in various fields who may not be parallel programming experts can simply incorporate existing ME algorithms and run computational experiments on their scientific application without explicit parallel programming. A key feature of this approach is that the model is unmodified and the ME algorithm is only minimally aware of its existence within the EMEWS framework. EMEWS uses a novel form of inversion of control (IoC), where Swift/T instantiates the ME algorithm, that then provides model parameters back to Swift/T (over IPC, without returning). These parameters are distributed to worker processes for model execution. Swift/T provides a variety of methods for integrating models, including via built in interpreters, command line invocation, and as compiled libraries. Upon completion, the model outputs are registered back to the ME algorithm, which provides more parameters until a convergence criterion is satisfied or a computing budget is exhausted.
EMEWS also relies on new “many resident task computing” (MRTC) capabilities that extend the notion of many-task computing (MTC). This allows running tasks to effectively suspend, waiting for queries. We demonstrate that mixing resident tasks with traditional run-to-completion tasks is a powerful programming model that supports the development of calibrated and validated scientific applications, including realistic ABMs that can be used as electronic laboratories to answer important research and policy questions.
This paper offers the following contributions:
-
1)
It describes a software integration model for high-performance workflow-like applications, where advanced algorithms, such as AL, implemented in languages like R, can be integrated.
-
2)
It describes a compelling, real-world application infectious disease dynamics and presents results from running a large-scale AL workflow to characterize the parameter space of a distributed ABM.
-
3)
It proposed and investigates novel, flexible concurrency schemes for these workflows.
-
4)
It evaluates the performance and scalability of the application up to 10K cores on a Cray supercomputer.
The remainder of this paper is organized as follows. In §II we describe ABMs, ABM ensemble model exploration methods and our Susceptible Exposed Infected Recovered (SEIR) ABM. In §III we describe the EMEWS programming model and its implementation. In §IV we describe how the various components in our SEIR model and AL EMEWS workflow are connected. In §V we present the results from running a large-scale AL workflow to characterize the SEIR model parameter space. In §VI we report performance numbers for the workflow. In §VII we restate our contributions and offer conclusions.
II. ABM, ENSEMBLE MODEL EXPLORATION METHODS AND THE SEIR MODEL
Agent-based modeling and simulation (ABMS) is a method of computing the potential system-level consequences of the behaviors of sets of individuals [10]. ABMS allows modelers to specify the individual behavioral rules for each agent; to describe the circumstances in which the individuals reside; and then to execute the rules, via simulation, in order to determine possible system-level results. Agents themselves are individually identifiable components that usually represent decision makers at some level. Agents often are capable of some level of learning or adaptation ranging from simple parameter adjustment to the use of neural networks, evolutionary algorithms, and market models.
As larger and more complicated models of complex systems are developed, high-performance computing (HPC) resources are increasingly required to run the computational experiments needed for developing validated (i.e., trusted) models that can support decision-making. On the one hand, ABMS studies require the execution of many model runs to account for stochastic variation in model outputs and for the various ensemble model exploration (ME) methods that are required to calibrate and analyze them. These methods can be used to carry out:
adaptive parametric studies
large-scale sensitivity analyses and scaling studies
optimization and metaheuristics
inverse modeling
uncertainty quantification
data assimilation.
On the other hand, ABMs can also be distributed across processes to accommodate very large numbers of agents (e.g., > 109 [11]) or very complex agents1. These facts combine to make ABMs well suited for HPC resources and, through the EMEWS framework, they can easily and efficiently be run as part of large-scale scientific workflows.
A. Ensemble model exploration methods
Depending on the aims of a computational experiment, different dynamic ensemble ME methods are appropriate2. In the realm of stochastic optimization there are simulated annealing [15], adaptive mesh [16], genetic algorithms [17], approximate Bayesian computation [18], [19] and other techniques. Ensemble Kalman filtering [20] and particle filters [21], [22] are useful for combining ensembles of model outputs and empirical observations. AL [23] can be used to efficiently characterize large parameter spaces. These types of techniques are increasingly being used with ABMs [24]. Many of the methods are being actively developed and are implemented as free and open source libraries in popular data analysis programming languages (e.g., R) and general purpose languages (e.g., Python).
While sophisticated ME techniques have been a generally fruitful approach for combining ensemble mathematical (e.g., compartmental) models and empirical observations, for example in infectious disease modeling [25], [26], [27], we also see that such events as the 2013 West African Ebola outbreak have exposed some limits to the predictive power of these approaches [28]. The possible reasons for this are many, but some of the simplifying assumptions inherent in the compartmental models that are used for the infectious disease studies might be at issue. Compartmental models use differential equations relating aggregate variables (e.g., the fractions of the population that are susceptible, infected, or recovered/removed) to derive the dynamics of disease progression in a population. But such models are not able to capture “complex social networks and the direct contacts between individuals, who adapt their behaviors [29].” By developing more realistic models in the form of ABMs, the complexity, for example of the inter-agent and biological-social interactions inherent in many infectious diseases, can be encapsulated in the specification of processes such as agent activities and decision-making, agent interactions over social networks, demographic and geographic heterogeneity, and agent adaptation and learning.
With EMEWS, the ensemble ME techniques that have been applied to simpler modeling paradigms can be carried over to the ME of large, complex, parallel, and distributed ABMs. Furthermore, many of these techniques are being actively developed and are implemented as free and open source libraries in popular programming languages. As indicated earlier, rather than requiring the reimplementation of these algorithms in the Swift/T language, the goal of the EMEWS framework is to be able to have these libraries directly control large-scale HPC workflows, thereby making them more accessible to a wider range of researchers and, at the same time, enable them to run at HPC scales.
B. The SEIR model
Our SEIR model (Susceptible, Exposed, Infected, Recovered) is a distributed parallel agent-based model of the transmission of a flu-like disease using SEIR model dynamics [30]. The model represents each person in a selected geographical region (e.g., the City of Chicago) as an agent. Each person in the model is in one of four disease states: susceptible, exposed, infected, or recovered. Persons transition through states, moving from susceptible to exposed to infected and ending with recovered. While susceptible, a person can become exposed in the presence of infectious persons. Exposed persons are infectious but not yet infected, i.e., they can infect other persons but are not yet symptomatic. Infected persons are symptomatic and also infectious for at least part of the period of infection. Recovered persons are no longer infected or infectious and, being effectively immune to the disease, will not become susceptible again. The model begins with some specified number (parameter CI) of persons in the exposed state who subsequently transition through the infected and recovered state while, in turn, exposing other persons to the disease, triggering the transition of those persons through the disease states.
The transition and duration of each state is determined by model state, and user-specifiable input parameters. A susceptible person will transition to exposed in the presence of infectious persons with a base probability (PS→E) modified by the number of co-located infectious persons. The duration of a person’s stay in the exposed state is drawn from a triangular distribution specifed by a mode (Motinc), minimum (Mitinc) and maximum (Mxtinc) where the minimum (Mitinc) defaults to one day and the maximum (Mxtinc) to four [31]. After the exposed duration has elapsed, an exposed person enters the infected state. Exposed persons are infectious from one day prior to entering the infected state to seven days after entering it [31]. The length of the infected state is also drawn from a triangular distribution (MotI , MitI , MxtI ) with a default minimum of seven days and a default maximum of fourteen days [32]. While infected, a person will remain at home thus avoiding contact with anyone outside the household. With a user-specifiable probability (PhomeA) a person will remain at home as soon as they become infected, otherwise they will remain at home beginning one day after becoming infected. A person will remain at home for either five, six, or seven days depending on a user-specifiable probability (PhomeB,PhomeC ), after which they will resume their normal activities.
Once the infected period ends, a person transitions to the recovered state. The parameters of the triangular distributions, the “stay at home” probabilities, and the initial number of exposed persons are model parameters (see Table I) and thus can be altered to affect the number of persons in each state as the model progresses.
TABLE I:
SEIR model input parameters.
| Parameter | Description |
|---|---|
| CI | Initial number of infected individuals |
| PS→E | Hourly probability of going from susceptible to exposed per each collocated infectious agent |
| Motinc, Mitinc, Mxtinc | Mode, minimum, and maximum of the triangular distribution for exposed to infected incubation period |
| MotI, MitI, MxtI | Mode, minimum, and maximum of the triangular distribution for time in infected state |
| PhomeA, PhomeB, PhomeC | Probabilities for infected individual to remain home on first day, sixth day and seventh day of infection |
The SEIR model is implemented in C++ using the Repast for High Performance Computing (Repast HPC) [33] and the Chicago Social Interaction Model (chiSIM) [34] toolkits. Repast HPC is an agent-based model framework for implementing agent-based models in MPI and C++ on high performance distributed-memory computing platforms. chiSIM is a framework for implementing models that simulate the hourly mixing of a synthetic population, in this case the City of Chicago consisting of approximately 2.9 million individual agents and 1.2 million distinct places. Synthetic populations with baseline socio-demographic data, derived from combined U.S. Census files, are available from a growing number of sources. chiSIM uses baseline synthetic population data such as those developed through the NIH MIDAS network [35], [36]. The socio-demographic attributes of the synthetic population match that of the actual population for Chicago in the aggregate for the Census years of 2000 and 2010. Each agent has a baseline set of socio-demographic characteristics (e.g., race/ethnicity, age, gender, educational attainment, income). All places are characterized by place type, including households, schools, hospitals and workplaces, and have a geographic location. In the synthetic population agents are assigned to households, workplaces and schools (for those of school age). Places are categorized as having different types of activities that may occur there.
In a chiSIM based model, such as SEIR, each agent, that is, each person in the simulated population, resides in a place (a household, dormitory or retirement home/long term care facility, for example) and moves among other places such as schools, workplaces, hospitals, jails and sports facilities. Agents move between places according to their shared activity profiles. Each agent has a profile that determines at what times throughout the day they occupy a particular location [33]. Our Chicago agent activity profiles are empirically based on 24-hour time diaries collected as part of the U.S. Bureau of Labor Statistics annual American Time Use Survey (ATUS) for individuals aged 15 years and older and from the Panel Study of Income Dynamics (PSID) for children younger than 15 years. Both are nationally representative samples and collect diary data on randomly assigned days. In the SEIR model, two profiles (one weekday and one weekend) from ATUS /PSID respondents living in metropolitan areas are assigned to each agent in the model. This is done by stochastically matching each agent with an ATUS or PSID respondent who is either identical or similar with respect to socio-demographic characteristics. Agents move between places according to their activity profiles. Once in a place, an agent mixes with other agents in some model or domain-specific way. In the case of the SEIR model, infectious agents infect co-located susceptible agents, who having become infected can then in turn infect other agents as they move.
chiSIM itself is a generalization of a model of community associated methicillin-resistant Staphylococcus aureus (CAMRSA) [2]. The CA-MRSA model was a non-distributed model in which all the model components (all the agent, places, et cetera) run on a single process. chiSIM retains and generalizes the social interaction dynamics of the CA-MRSA model and allows models implemented using chiSIM to be distributed across multiple processes. Places are created on a process and remain there. Persons move among the processes according to their activity profiles. When a person agent selects a next place to move to, the person may stay on its current process or it may have to move to another process if its next place is not on the person’s current process. A load balancing algorithm has been applied to the synthetic Chicago population to create an efficient distribution of agents and places, minimizing this cross-process movement of persons and balancing the number of persons on each process [34]. In addition, chiSIM provides the ability to cache any constant agent state, given sufficient memory, lessening the amount of data transferred between processes.
The following sections describe how the EMEWS framework is used to perform an adaptive parametric study of the SEIR model by integrating it with a ME algorithm, in this case AL [23].
III. EMEWS PROGRAMMING MODEL
The EMEWS framework is designed to implement a high-level programming model that allows us to coordinate calls to scientific applications, such as large ABMs, as well as various control and analysis scripts over a scalable, MPI-based computing infrastructure. Specifically, EMEWS was implemented to meet the following requirements:
-
1)
The ability to construct a workflow of many (potentially millions of) calls to a scientific application (such as an ABM simulation) with different parameters;
-
2)
The ability to allow simulation results to feed forward into future application parameters;
-
3)
The ability to integrate a complex ME algorithm, like AL, into the parameter construction;
-
4)
The ability to call into the native code models and scientific applications (e.g., written in C++) and the 3rd-party implementation of a ME algorithm (written in R in the current application);
-
5)
The ability to maintain the state of the ME algorithm from call to call, and to programmatically access this state in the system.
We provide an overview of how EMEWS and Swift/T addresses these requirements in the following.
-
1)
The ability to manage an extreme quantity of tasks is a main design feature of the Swift/T implementation [8],[37], which essentially translates the Swift script into an MPI program for execution on the largest scale supercomputers. The Swift-Turbine Compiler (STC) [38] optimizes the script using multiple techniques, both conventional and oriented toward novel concurrency. In synthetic tests, Swift/T has been used to run trillions of tasks at over one billion tasks per second. It can also send very small tasks to GPUs at high rates [39], enabling powerful mixed programming models.
-
2)
Swift is a dataflow language. In this model, the user defines data items (numbers, strings, binary data, and various collections of these) and connects them with functional execution. Swift also offers conventional constructs such as if, for, foreach, and so on with their definition only slightly modified for automatic parallelism. Following dataflow (not control flow) functions execute when their inputs are available, possibly concurrently. Thus, typical Swift loops are automatically parallel loops. Dataflow analysis allows common expressions like g(f(1),f(2)); to expose available concurrency (2 simultaneous executions of f()).
-
3,4)Swift/T has rich support for integrating complex logic into workflows, including using scripting languages like Python and R. It enables this on HPC machines (where fork() may be undesirable or unavailable) by optionally bundling script interpreters for Python, R, Julia, Tcl, JVM languages, etc., into the Swift/T runtime [40]. These interpreters are called through their native code interfaces (thus reusing the Swift/T ability to call into native code libraries) but high-level interfaces are provided for Swift. For example, the Swift code:
string result = python(“a=2+2”,”str(a)”);
would store “4” in result. The python() function takes two string arguments, code and an expression. The code is executed and the string expression is returned as a Swift string. Users may set PYTHONPATH, load their own modules or 3rd-party modules such as Numpy, etc. They may also call through these scripting layers into native code.The Repast HPC code is called as an MPI library as described in §III-A.
-
5)
Various states may be maintained in the Swift/T implementation, while remaining outside the main dataflow model. This is typically done in the tasks, avoiding confusion with the dataflow. For example, a configuration file could be loaded from disk by a Python-based task, and cached in a global Python variable. This data would be available on the next invocation of a Python task on that process.
Developers can target different parts of the system by using the locality features in Swift/T. These were initially added to allow users to send tasks to data in a compute-node resident filesystem [41]. However, they can also be used to send tasks to state in a script interpreter. Tasks can be targeted at a rank or a node, and be strict or non-strict. We use strict rank targeting in this work, while non-strict, node-based targeting is used in (for example) cache storage systems.
The following subsections go into further details on the EMEWS and Swift/T features that address the programming model requirements.
A. Hierarchical concurrency
MPI enables the concurrent execution of multiple cooperating multiprocessing codes, each of which can have a separate communication context shared with only the MPI processes executing that code. MPI represents these contexts with communicators that typically form a tree hierarchy, starting from an initial world communicator, that encompasses all processes. Given a communicator, new child communicators can be created and passed to libraries for their exclusive use, allowing an application to be constructed through composition of existing parallel libraries and codes.
Our execution model has multiple levels of concurrency and a great deal of flexibility in how the workflow uses the available processing power of a supercomputer. Since the SEIR model itself uses MPI, it must be treated as an MPI library. Swift/T uses the MPI 3.0 MPI_Comm_create_group() feature to allocate a new communicator for each task [42]. These are handed to the application for each new task and deallocated (MPI_Comm_free()) at the end of the task. The user can specify the number of processes (e.g., p) for each task programmatically with:
@par=p f(…);
When launching a simulation task, Swift/T constructs the communicator and passes it to the SEIR model, which is a shared library loaded by Swift/T. The use of MPI in the SEIR model is completely independent of the use of MPI by Swift/T. There is no mixture of control flow from Swift to the SEIR model; once the SEIR task starts, it proceeds with normal MPI/C++ semantics, until returning control back to Swift/T.
B. Location-aware many-task scheduling
MTC workloads, on the one hand, generally allow the scheduler a great deal of leeway in determining where tasks will execute. Bag-of-tasks workloads, for example, are the most lenient, allowing tasks to execute anywhere in any order. Programming models such as MPI, on the other hand, give the programmer total control over execution locality.
Swift/T strikes a balance between these two extremes with its location annotation. By default, tasks can execute on any worker process, but the programmer has the option of specifying the annotation with @location=L f(), where f() is the task and L is a location value. A location value is constructed from an MPI rank r with optional accuracy and strictness qualifiers. (Swift/T features allow a hostname to be translated to one or more MPI ranks.) The accuracy may be RANK, specifying the process with rank r, or NODE, specifying any process that shares the same network host with r. The strictness may be SOFT, allowing the task to run anywhere in the system if there is nothing else to do at a given point in time, or HARD, specifying that the scheduler should wait until the location constraint can be satisfied (even at the expense of maintaining idle processors).
The location features in Swift/T were originally added for data-intensive workloads [41]. These provide a novel model for best effort, data-aware scheduling, when data is stored on the compute nodes. Compute node-resident storage systems that advertise data locations can be exploited by these programming features. In EMEWS, we extend the utility of this feature by using it to target program state instead of bulk data. By keeping program state resident, we avoid any cost associated with approaches that depend on data serialization. More importantly, we can more easily leverage third party libraries as resident programs without extensively modifying them to fit a data serialization based scheme.
C. Resident tasks for ensemble control
Previous uses of workflow languages to control ME typically take one of two approaches. In the first approach, the ME algorithm is encoded in the workflow language. While some workflow languages provide rich support for arithmetic operations (Swift/T is notable in this regard), many do not. Even so, this approach requires that such algorithms be coded from scratch in the workflow language, and makes it impossible to directly reuse code in other languages. In the second approach, the algorithm is provided as a built-in feature of the workflow system. This approach has been taken by Nimrod/O [43] and Dakota [44], among others. It does not allow the end users much control over the ME algorithm used, unless they can modify the source code of the workflow system itself.
EMEWS defines and uses resident tasks as a building block to implement user-defined ME workflows. The key technological feature is the ability to launch a task in a background process or thread. Background indicates that the foreground process or thread returns control to Swift/T after execution (as a normal task would), but the background task is still running. It retains state and potentially performs ongoing computation. For the current example, the background task maintains the state of an AL ME algorithm. The overarching workflow must simply query this task for instruction on what tasks to execute next. To do so, a task is issued to the same location as the resident task, which communicates with it over IPC.
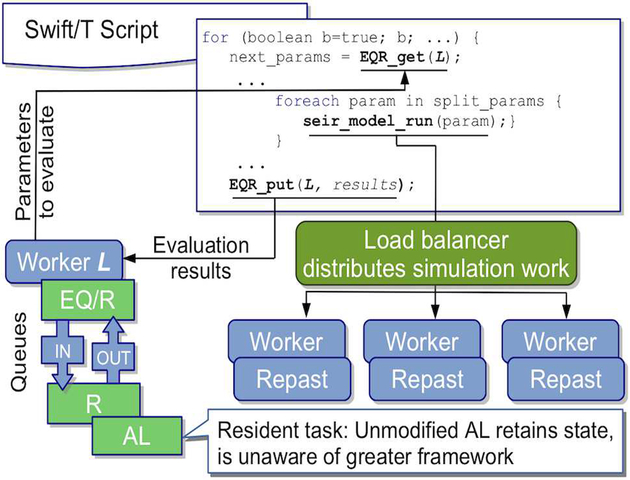
D. EMEWS Queues for R
To query the state of the AL algorithm, we designate one worker on location L for exclusive use by AL. Interaction with this worker via the EMEWS Queues for R (EQ/R) extension is shown in Figure 1. The EQ/R extension allows Swift/T workflows to communicate with a persistent embedded R interpreter on a worker at some location L via two blocking queues, IN and OUT. The extension provides C++ functions that allow string data to be pushed onto and retrieved from these queues. These functions are wrapped in an interface and are accessible to Swift/T and shared with the R environment. Upon initialization, EQ/R adds these functions to the R environment and spawns a thread in which the R script is run. Through these functions, the R script places string data in the OUT queue where the Swift/T parent thread can retrieve it with the EQR_get() function. Similar functionality exists for the IN queue, and in this way string data is passed back and forth from the R script to the Swift/T workflow. The queues themselves will block if the queue is empty, allowing the Swift/T workflow to pause and wait for data from the R script and vice versa. When the R script waits and control returns back to Swift/T, the R interpreter is not deallocated. When subsequent R tasks execute on location L, they have access to the IN and OUT queues via the same functions. Through blocking queues and resident tasks, EQ/R implements an inversion of control (IoC) pattern, where the logic embedded within the external ME algorithm, rather than in the Swift/T script, determines the progression of the workflow. As a note, a similar IoC pattern is employed with the EMEWS queues for Python (EQ/Py) extension, for Python-based ME algorithms.
Fig. 1:
EQ/R EMEWS workflow with an AL model exploration resident task.
E. Worker types
Swift/T offers worker types, a powerful, high-level way to map execution to various parts of the system. The user may specify any number of task types by simply providing a token. Then, functions that are defined with this token will execute only on workers (ranks) configured to accept these task types.
1 pragma worktypedef resident_work;
2 @dispatch=resident_work
3 register(string params) {
4 … // body
Similarly to the Swift/T locality features (§III-B), these offer a tradeoff between automated load balancing and full user control over execution location. They could be used to ensure that a small number of workers are allocated for performance-critical control tasks (e.g., tasks that produce input parameters for many other tasks), or to throttle the number of I/O-intensive tasks running at any point in time.
The EQ/R tasks have their own worker type resident work. This enables R based analysis code to be used for tasks, such as calculating complex objective functions from simulation outputs, without affecting the R interpreter used by the AL calculations.
F. Contiguous ranks
In previous work with Swift/T parallel tasks, worker ranks were assembled into per-task subcommunicators essentially randomly. This was the most flexible technique and was immune to fragmentation problems. For EMEWS, we extended the Swift/T parallel tasks feature to additionally support “parmod” (parallel-modulus) tasks. Communicators constructed to run tasks denoted with parmod = n have two additional constraints: 1) they must start on a rank r such that r 0 (mod n) and 2) the ranks in the new subcommunicator are contiguous in the parent communicator. For example, on a computer with 32 cores per node, the user could set parmod = 32, then a 32-process (@par=32) task would always consume exactly 1 node; when parmod = 64, a 64-process task would consume exactly 2 nodes, which are topologically neighbors (assuming the MPI implementation is configured to lay out ranks in such a manner).
In this work we use parmod tasks for two reasons. The first is simply to gain the benefit of achieving the intranode performance for parallel SEIR model tasks by ensuring that all the processes in that node are running the same model instance. We run each task, that is, each model instance on 256 processes with a per process node count of 8, and thus fully utilize 32 nodes. Second, it allows us to cache data more easily, since the task layout is always the same. If communicator layouts were more random, it would take a great deal more development time to correctly manage data cached in the SEIR model, in this case, the initial synthetic population, from one parallel task to the next.
IV. INTEGRATION
Our focus is on the identification of the viable regions within the parameter space of the SEIR model. These regions represent input parameters resulting in model outputs that fall within the range of plausible flu incidence trajectories. The SEIR model includes stochasticity in two of its key elements. First, the initially exposed population is randomly distributed across the synthetic population. Second, the collocation-based infection dynamics stochastically determines whether an infection has occurred. Thus, as modelers we are faced with the task of determining how to evaluate the “goodness” of a parameter set. We cannot simply look for time series fits to historical flu trends, since the empirical time series are individual trajectories of flu infection dynamics that have been observed. What the empirical data doesn’t show, for example, are all of the flu trajectories that did not occur (or possibly were not identified). Also, since the SEIR model distributes the initially exposed population randomly, it is highly unlikely that any actual distribution of initially exposed people would match this and, since the infections are not spread in aggregate but through contacts between collocated individuals, the initial spatial distribution has the potential to greatly affect the timing and size of the flu incidence peak. As such, as we describe below, we resolved to run twenty stochastic variations for each parameter combination and characterize the parameter set as viable or not based on two aggregate statistics.
In this current parameterization of the SEIR model, the inputs that are allowed to vary are the initial number of infected individuals (CI) and the hourly probability of going from susceptible to exposed per each collocated infectious agent (PS→E). CI ranged from 1 to 100 in increments of 1. PS→E ranged from 2e−5 to 4e−5 in increments of 0.02e−5. For each combination of these two parameters, the SEIR model outputs a table of newly infected agents for each week of a 35 week period. The objective function we use to characterize the model output calculates the mean and maximum values for each 35 week period. We define a threshold condition using the mean and maximum values within which the model outputs are deemed to adequately resemble empirically observed infection count trends for Chicago, obtained from [45]. The threshold condition used was less than 10,000 newly infected in any single week for the maximum and a mean across all 35 weeks of greater than 100 new infections per week. The computational challenge then becomes one of trying to characterize the SEIR model parameter space into viable and non-viable regions efficiently, that is, without having to run too many simulations to evaluate the viability of parameter combinations. While a number of different ensemble methods could potentially be used for this, the AL approach, described next, maps naturally to the problem.
A. The AL algorithm
AL [23] is a promising approach for characterizing large parameter spaces of computational models (see e.g., [46]) with less expensive reduced order models, or meta-models. AL combines concepts from adaptive design of experiments (e.g., [47]) and machine learning to iteratively and strategically sample from an unlabeled data set. AL works well in situations where, “… unlabeled data may be abundant or easily obtained, but labels are difficult, time-consuming, or expensive to obtain” [23]. The AL approach can be naturally mapped to the characterization of the parameter spaces of computer simulations when one considers the unlabeled data as points in a parameter space and the labelling activity as evaluating those points by running (possibly expensive) simulations.
In this paper we chose to implement an R-based AL algorithm in order to highlight the types of useful and sophisticated parameter search approaches that can be developed when leveraging existing functionality in widely used open source data analytics languages. Rather than requiring the time-intensive and error prone reimplementation of these algorithms in Swift/T for the sole purpose of running large ensembles of simulations, we are able to have these algorithms directly control large-scale HPC workflows.
AL is a general approach that can afford a fair amount of customization in its specific implementation. The overall goal is to iteratively pick points (individual or sets) to sample, where the sampled points are chosen through some query strategy. In our case, we choose an uncertainty sampling strategy, where we employ a machine learning classifier on the already collected data and then choose subsequent samples close to the classification boundary, i.e., where the uncertainty between classes is maximal. In this way we exploit the information that the classifier provides based on the existing data. To take advantage of the concurrency that we have available on HPC systems, the samples at each round of the AL procedure are batch collected (and evaluated) in parallel. In order to decrease the overlap in reducing classification uncertainty that nearby maximally uncertain sample points are likely to have, we cluster all the candidate points and choose an individual point within each cluster. This ensures a level of diversity in the sampled points and, therefore, a greater expected reduction of uncertainty [48]. We balance the exploitation of the classifier model with an exploratory component, where random points in the parameter space are sampled in order to investigate additional regions that may not have been sampled yet. This can prevent premature convergence to an incorrect or incomplete meta-model.
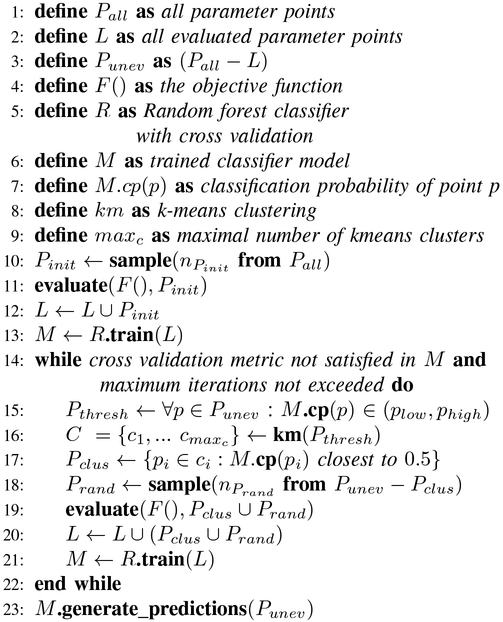
The pseudo-code for our AL algorithm is shown in Figure 2. The workflow proceeds until the cross validation metric, a proxy for out-of-sample model performance, is satisfied. Parallel evaluations of the objective function F () – the SEIR model simulation – are performed in lines 11 and 19 over some sample of parameters. At each iteration, the sampled results feed into the classifier R (lines 13 and 21). At the end of the workflow, the final meta-model predictions are generated for the remaining parameter space.
Fig. 2:
Pseudo-code for AL algorithm.
B. Inversion of Control Implementation
Our central EMEWS workflow pattern is shown in Figure 3. For our AL R algorithm, located at location L, the doAL function is called. The for loop continues to iterate while new sets of parameters are obtained from the AL algorithm via the EQ/R EQR_get call. The parameter sets are sent to run_model, where they are split up and evaluated concurrently via a Swift/T foreach loop (not shown). Objective function results results, indicating a viable parameter combination or not, are returned by run_model and passed back to the AL algorithm via the EQ/R EQR_put call. This loop continues until the EQR_get call obtains the special token “FINAL”. Note that the EQR_put and EQR_get calls take the location L as a parameter. The implementation of EQR_get and EQR_put use this location in a location-aware many-task scheduling annotation as described in §III-B.
Fig. 3:
Main Swift/T workflow loop.
Also as described earlier, this qualifies as an IoC pattern since rather than Swift/T, the R-based AL algorithm controls the overall workflow logic. The algorithm produces simulation parameters and consumes results, but instead of calling the model code directly, the parameters are intercepted and sent to Swift/T for distributed execution, with results seamlessly returned. This powerful pattern allows many 3rd-party algorithms to be easily dropped into our framework and coordinate large-scale ensemble ME workflows.
C. AL EQ/R communication interface
As described in §III-D, the interprocess communication is performed over queues. The queues are implemented in C++, but must also be accessible from Swift and R. The interface to these queues is shown in Figure 4. Their implementation uses a straightforward Standard Template Library (STL)-based locking scheme. This library is exposed to Swift/T by using its SWIG-based library calling technique [49]. It is exposed to R via RInside [50]. Thus, the C++ data structure is available to both the Swift/T workflow and the R-based algorithm, via Tcl and R wrapper interfaces, respectively.
Fig. 4:
Queue implementation header: Swift to C++ linkage.
D. SEIR model as parallel leaf function
Since the SEIR model is a MPI-application it must be compiled as a shared library and wrapped in a Swift/T Tcl interface [42]. Through this interface Swift/T passes a parameter string that contains all the parameters (i.e. the initial number of exposed persons, the various distribution values, and so forth) for the current model run to the SEIR model. In addition, the Tcl interface also passes the MPI communicator for the current run. When the model receives the first set of parameters, it fills a cache with the required input data from the files specified in the parameter string, virtually eliminating I/O overhead in subsequent model runs. The caches are per process and contain the data for that process rank. The input data consists of person, place and activity definitions. As part of the load balancing scheme mentioned above, places are assigned to particular process ranks and persons move among processes as they move to the next place in their activity schedules. Each cache then contains the data for its process rank. Consequently, the caching mechanism requires consistent contiguous process ranks such that the cache originally created on process n, remains on process n during subsequent runs. We make sure this is the case by setting the environment variable ADLB_PAR_MOD to the number of processes required to run the model (i.e., 256), enabling contiguous process ranks in communicators of that size.
For a more in-depth and technical description of the elements within an EMEWS workflow, including a complete AL workflow utilizing a distributed MPI-based model, the reader is referred to the EMEWS tutorial, accessible through the EWEWS site [7].
V. AL RESULTS
All experiments presented in the next two sections were performed on the Cray XE6 Beagle at the University of Chicago, hosted at Argonne National Laboratory. Beagle has 728 nodes, each with 2 AMD Operton 6300 processors, each having 16 cores, for a total of 32 cores per node; the system thus has 23,296 cores in all. Each node has 64 GB of RAM.
For the AL workflow run presented in this section, each SEIR model run was distributed over 256 processes (32 nodes) and we ran up to 6 models concurrently (192 nodes), demonstrating the hierarchical concurrency that EMEWS workflows can generate. Each model took approximately 7 seconds to run per simulated week, and we ran them for 35 weeks (≈245 seconds per model run). The initial cache loading of person, place and activity definitions occurred exactly once across each of the six sets of 32 nodes, and took a total of 2 minutes.
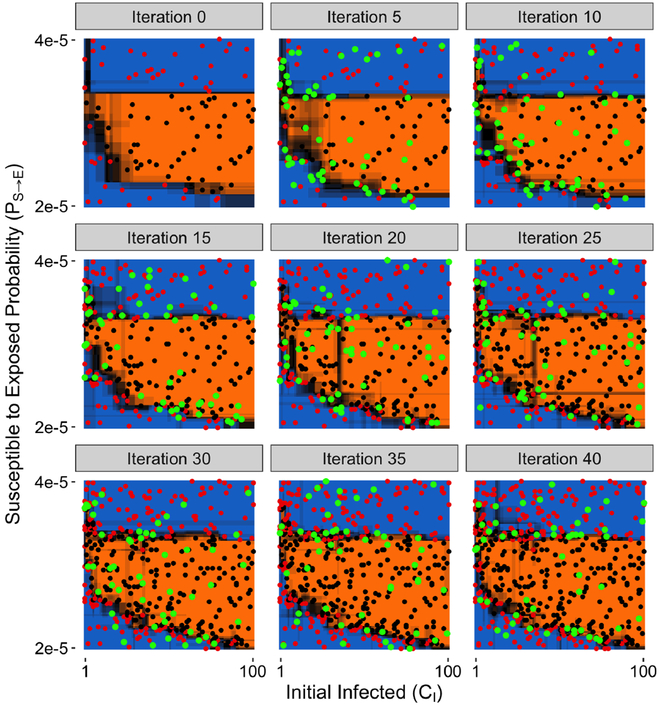
Figure 5 shows the progression of the AL algorithm evaluating parameter points, training the random forest model and generating predictions for the out-of-sample points in the two dimensional CI vs PS→E parameter space over 40 iterations, where the parameter space was gridded into 10100 discrete points (101 × 100). Each parameter point evaluation consists of twenty model runs of that parameter combination with the random seed varied for each of the runs, and with the viability of the parameter set determined as described in §IV. Iteration 0 shows the initial design, where 100 randomly chosen points were evaluated. The black and red dots signify parameter sets evaluated to be viable and nonviable, respectively. The orange and blue regions indicate the random forest meta-model out-of-sample prediction for viable and nonviable parameter space regions, respectively. The shading between the orange and blue regions represents the uncertainty in these predictions, where the darkest regions represent maximal uncertainty, i.e., equal probability of being viable or nonviable. As the iterations progress, points that were newly added since the last iteration panel are indicated by green dots. For this particular AL workflow, at each iteration we added 5 points close to the classification boundary (exploit) and 5 randomly sampled points (explore), for a total of 10 new points per iteration. Thus, at the end of iteration 40, about 5% of the parameter space was sampled. What can be observed is that as the AL progresses, the initial prediction boundary is gradually refined as additional points along it are evaluated, while the rest of the parameter space, where there is less uncertainty in the model prediction, e.g., the central part of the viable region, is not as densely explored. Importantly, regions of high uncertainty are seen to be reduced in width, sharpening the distinction between the two categories of interest. This pattern of parameter space evaluation is useful from the point of view of efficiently utilizing a computational budget, as the boundary points are the main drivers of an accurate meta-model. While the exploitation/exploration balance that we used appears to sufficiently cover and characterize our parameter space, other parameter spaces with, e.g., different dimensionality or granularity, may benefit from a different ratio.
Fig. 5:
Progression of the AL workflow, where the black/red dots indicate the evaluated (viable/nonviable) points, green points are newly added points since the previous panel, and orange/blue regions correspond to the out-of-sample predictions for (viable/nonviable) regions.
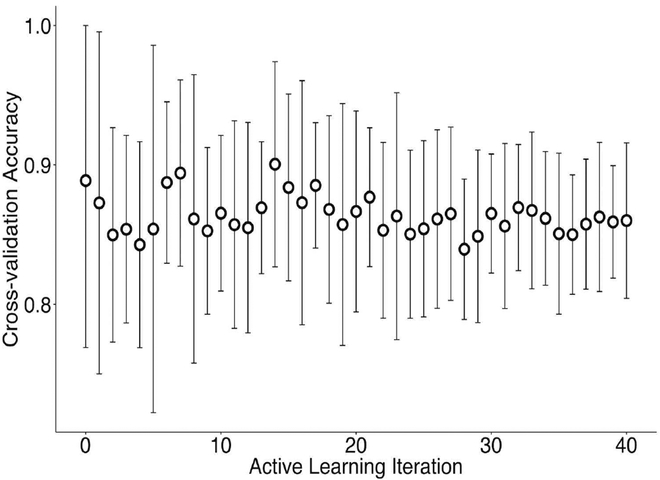
An iterative model exploration algorithm needs a termination condition. This can be based simply on a pre-determined computation budget or some expected performance metric. In this example we chose to monitor the cross-validation (CV) accuracy, both its sample mean and standard deviation. At each AL algorithm iteration the random forest model is trained and 10-fold cross validation is applied in order to get an estimate of the expected out-of-sample model performance. Figure 6 shows the progression of the CV accuracy and standard deviation. What is observed is that, while the CV accuracy is near constant, the standard deviation gradually decreases. This indicates that as the meta-model is being improved at each iteration with the addition of more data we are able to better trust its out-of-sample performance level. This also reflects the increased certainty of the meta-model as seen by the reduction of shaded regions in Figure 5. Finally, this also suggests additional AL experiments, such as varying the number of initial samples or the number of samples chosen at each iteration, to observe the effects on the trajectory of CV accuracy, or other CV metrics.
Fig. 6:
Cross-validation (CV) accuracy means and standard deviation based on 10-fold CV of the random forest meta-model at each AL iteration.
VI. PERFORMANCE RESULTS
A. Task parallelism
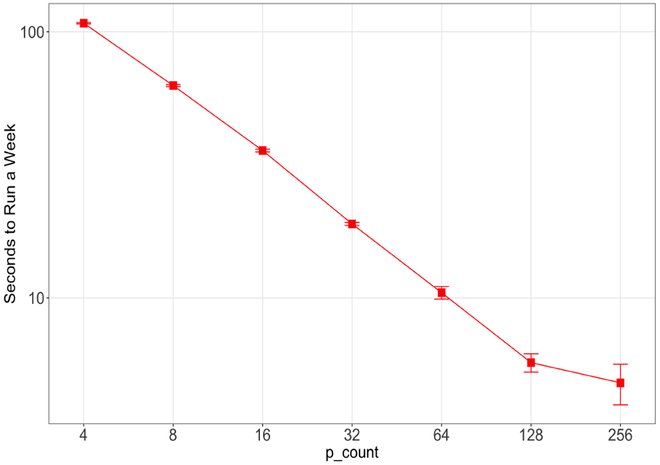
In our application model (cf. §III-A), there are multiple potential concurrency modes. Here we describe the task-specific parallelism. As described in § II-B, the SEIR model can be load balanced to run on any number of processes, parameterized by p_count. For these task parallelism experiments, we configured it to run on p_count=4,8,16,32,64,128,256 processes. We measured the average time it took for the SEIR model to simulate one week within the workflow and reported it in Figure 7.
Fig. 7:
Average time for the SEIR model to run a week as a function of p_count. Error bars are the sample standard deviation from 210 simulated weeks.
The results show that the SEIR model scales well to 128 and potentially to 256 processes. This scaling is important, as many of the ensemble methods of interest are iterative in nature, such that any performance increases that can be achieved for the simulation runs themselves are generally multiplied by the number of iterations required for the complete workflow, if the necessary concurrency is available. Thus, the simulation developer has the option to retain a model’s complexity rather than simplify it such that it “… be amenable to comprehensive and systematic analysis.” [24]
B. Total time to completion
For our SEIR/AL workflow performance evaluation, we constructed test AL workflows using a one ZIP code version (Ȉ 44k agents) of the SEIR model. The tests in this study exercised the full set of AL workflow components to observe their individual and collective performance characteristics. The cross validation metric condition was modified to run past satisfaction to produce a consistent number of tasks (and thus always ran to the provided maximum number of iterations).
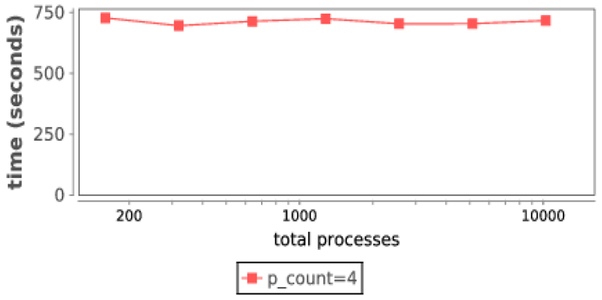
Our performance objective was to determine how workflow overheads might affect the total time to complete the AL workflow. For each number of total processes, we ran the workflow at p_count=4. This is the most challenging case for Swift, as higher p_count values reduces the number of tasks running at a time (as each task has more processes). For each increasing number of total processes, we increased the workload size (weak scaling). The total number of tasks in each workflow was hand-specified by selecting a maximum iterations number multiplied by the number of total processes; thus, the AL convergence criterion was disabled. The total number of tasks for each run was set to the number of total processes, and the maximal concurrency per round (Prand+Pclus) was equal to the number of total processes divided by 4, thus, there were 4 iterations. We recorded the total runtime reported by Swift and plotted it in Figure 8.
Fig. 8:
Total makespan times for the one ZIP code SEIR model.
As shown, the total workflow time is only minimally affected by scale. In our largest run, on 10,240 cores of Beagle, there is no utilization loss due to workflow overheads, demonstrating the robust scalability of EMEWS.
VII. CONCLUSION
In this paper we have presented EMEWS, a framework for running large ensembles of simulations in which sophisticated ME algorithms can iteratively and adaptively refine simulation parameters through the analysis of recently generated results and launch new scientific applications based on the refined parameters. The mechanism itself has been implemented by using the Swift/T dataflow language and exhibits a novel form of inversion of control using location-aware many task scheduling, resident tasks, and non-trivial IPC over HPC resources.
Using EMEWS, we developed an AL workflow through the selective reuse of 3rd-party R packages, highlighting the multiple parallel programming language and runtime innovations, including novel features for parallel tasks (§III-A, §III-F), task locality (§III-B), and stateful tasks (§III-C) that make such a workflow possible. We demonstrated how the AL workflow was able to efficiently characterize the parameter space of a stochastic, large-scale, distributed SEIR model into viable and non-viable regions while sampling only a small fraction of possible parameters.
Performance results illustrate the basic scalability of EMEWS on a typical supercomputer. We demonstrated that a flexible range of concurrency strategies are within the performance envelope of our tools, enabling anything from a massive battery of single-process simulations to a mix of varying multiprocess runs. Furthermore, while the focus here was the use of EMEWS for ABMs, EMEWS is being effectively applied to a variety of modeling methods (e.g., microsimulation [51], machine learning hyperparameter optimization [52], bio-physical modeling [53]) that require calibration, parameterization or optimization achieved through the iterative execution of large numbers of computations.
We believe that as application teams consider good uses of near-exascale resources, they will observe that defensible scientific investigations will have to be backed by large and novel many-task ensemble studies.
EMEWS has been released as an open source framework for the community [7], and we intend to continue to refine and improve it, while continuing to develop additional use case examples that exploit widely available model exploration libraries. Ultimately, the goal of EMEWS is to democratize the use of HPC resources by allowing non-expert researchers to tap into advanced 3rd party ensemble model exploration methods, such as optimization or AL algorithms, to take advantage of the extreme scale systems that will become available in coming years.
ACKNOWLEDGMENTS
This material is based upon work supported by the U.S. Department of Energy, Office of Science, under contract number DE-AC02–06CH11357, and by the NIH (awards R01GM115839 and R01GM121600). This work was completed in part with resources provided by the Beagle system and the Research Computing Center at the University of Chicago.
Biography
Jonathan Ozik, Ph.D., is Computational Scientist, in the Decision and Infrastructure Sciences Division at Argonne National Laboratory and Senior Scientist at the Consortium for Advanced Science and Engineering at the University of Chicago. He leads the Repast (https://repast.github.io) agent-based modeling toolkit and the EMEWS (http://emews.org) framework for large-scale model exploration.
Nicholson T Collier, Ph.D., is Software Engineer in the Decision and Infrastructure Sciences Division at Argonne National Laboratory and Research Staff at the Consortium for Advanced Science and Engineering at the University of Chicago. He is the lead developer of the Repast (https://repast.github.io) agent-based modeling toolkit and a core developer of the EMEWS framework.
Justin M Wozniak, Ph.D., is Computer Scientist in the Mathematics and Computer Science Division at Argonne National Laboratory and Scientist at the Consortium for Advanced Science and Engineering at the University of Chicago. He is the lead developer of the Swift/T (http://swift-lang.org/Swift-T/) parallel scripting language and a core developer of the EMEWS framework.
Charles M Macal, Ph.D., P.E., is Senior Systems Engineer, Argonne Distinguished Fellow, Group Leader of the Social, Behavioral and Decision Science Group within the Decision and Infrastructure Sciences Division of Argonne National Laboratory, and Senior Scientist at the Consortium for Advanced Science and Engineering at the University of Chicago. Dr. Macal is recognized globally as a leader in the field of agent-based modeling and simulation and has led interdisciplinary research teams in developing innovative computer simulation models in application areas including global and regional energy markets, critical materials, electric power, healthcare and infectious diseases, environment and sustainability, and technology adoption.
Gary An, M.D., is Associate Professor of Surgery in the Department of Surgery at the University of Chicago. His research involves the development of: mechanism-based computer simulations in conjunction with biomedical research labs, high-performance/parallel computing architectures for agent-based models, artificial intelligence systems for modular model construction, and community-wide meta-science environments, all with the goal of facilitating transformative scientific research. Towards this end he has developed agent-based models of sepsis, multiple organ failure, wound healing, surgical site infections, necrotizing enterocolitis, tumor metastasis, breast cancer, C. difficile colitis, and the link between oncogenesis and inflammation.
Footnotes
So called ‘thick’ agents may include sophisticated and computationally expensive cognitive abilities.
We note that there exist static parameter search techniques (e.g., full factorial design [12], Latin hypercube sampling [13], Morris method [14]) that a priori determine the sampling from a parameter space. While these can be useful for some purposes, they are not adaptive and do not require complex workflow logic and hence are not the focus of this paper.
Contributor Information
Jonathan Ozik, Argonne National Laboratory and The University of Chicago..
Nicholson T. Collier, Argonne National Laboratory and The University of Chicago.
Justin M. Wozniak, Argonne National Laboratory and The University of Chicago.
Charles Macal, Argonne National Laboratory and The University of Chicago..
Gary An, The University of Chicago..
REFERENCES
- [1].Germann TC, Kadau K, Longini IM, and Macken CA, “Mitigation strategies for pandemic influenza in the United States,” Proceedings of the National Academy of Sciences, vol. 103, no. 15, pp. 5935–5940, Apr. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Macal CM, North MJ, Collier N, Dukic VM, Wegener DT,David MZ, Daum RS, Schumm P, Evans JA, Wilder JR, Miller LG, Eells SJ, and Lauderdale DS, “Modeling the transmission of community-associated methicillin-resistant Staphylococcus aureus: a dynamic agent-based simulation,” Journal of Translational Medicine, vol. 12, no. 1, p. 124, May 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Riddle M, Macal CM, Conzelmann G, Combs TE, Bauer D, and Fields F, “Global critical materials markets: An agent-based modeling approach,” Resources Policy, vol. 45, pp. 307–321, Sep. 2015. [Google Scholar]
- [4].Ozik J, Collier N, Murphy JT, Altaweel M, Lammers RB, Prusevich AA, Kliskey A, and Alessa L, “Simulating Water, Individuals, and Management Using a Coupled and Distributed Approach,” in Proceedings of the 2014 Winter Simulation Conference, ser. WSC ‘14. Piscataway, NJ, USA: IEEE Press, 2014, pp. 1120–1131. [Google Scholar]
- [5].Bert F, North M, Rovere S, Tatara E, Macal C, and Podest G, “Simulating agricultural land rental markets by combining agent-based models with traditional economics concepts: The case of the Argentine Pampas,” Environmental Modelling & Software, vol. 71, pp. 97 – 110, 2015. [Google Scholar]
- [6].Ozik J, Collier NT, Wozniak JM, and Spagnuolo C, “From desktop to large-scale model exploration with swift/t,” in Proceedings of the 2016 Winter Simulation Conference, ser. WSC ‘16 Piscataway, NJ, USA: IEEE Press, 2016, pp. 206–220. [Online]. Available: http://dl.acm.org/citation.cfm?id=3042094.3042132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].“EMEWS: Extreme-scale Model Exploration with Swift.”. [Online]. Available: http://emews.org.
- [8].Wozniak JM, Armstrong TG, Wilde M, Katz DS, Lusk E, and Foster IT, “Swift/T: Scalable data flow programming for distributed-memory task-parallel applications,” in Proc. CCGrid, 2013. [Google Scholar]
- [9].Ozik J, Collier N, and Wozniak JM, “Many resident task computing in support of dynamic ensemble computations,” in Proc. MTAGS at SC, 2015. [Google Scholar]
- [10].North MJ and Macal CM, Managing Business Complexity: Discovering Strategic Solutions with Agent-Based Modeling and Simulation, 1st ed. Oxford University Press, USA, Mar. 2007. [Google Scholar]
- [11].Murphy JT, “Computational Social Science and High Performance Computing: A Case Study of a Simple Model at Large Scales,” in Computational Social Science Society of America Annual Conference Proceedings, Santa Fe, NM, USA, Oct. 2011. [Google Scholar]
- [12].Box GEP, Hunter JS, and Hunter WG, Statistics for Experimenters: Design, Innovation, and Discovery, 2nd Edition, 2nd ed. Hoboken NJ: Wiley-Interscience, May 2005. [Google Scholar]
- [13].McKay MD, Beckman RJ, and Conover WJ, “Comparison of Three Methods for Selecting Values of Input Variables in the Analysis of Output from a Computer Code,” Technometrics, vol. 21, no. 2, pp. 239–245, May 1979. [Google Scholar]
- [14].Morris MD, “Factorial Sampling Plans for Preliminary Computational Experiments,” Technometrics, vol. 33, no. 2, pp. 161–174, May 1991. [Google Scholar]
- [15].Kirkpatrick S, “Optimization by simulated annealing: Quantitative studies,” Journal of Statistical Physics, vol. 34, no. 5–6, pp. 975–986, Mar. 1984. [Google Scholar]
- [16].Verfrth R, “A posteriori error estimation and adaptive mesh-refinement techniques,” Journal of Computational and Applied Mathematics, vol. 50, no. 13, pp. 67 – 83, 1994. [Google Scholar]
- [17].Holland JH, Adaptation in Natural and Artificial Systems: An Introductory Analysis with Applications to Biology, Control, and Artificial Intelligence Cambridge, Mass: A Bradford Book, Apr. 1992. [Google Scholar]
- [18].Beaumont MA, “Approximate Bayesian Computation in Evolution and Ecology,” Annual Review of Ecology, Evolution, and Systematics, vol. 41, no. 1, pp. 379–406, 2010. [Google Scholar]
- [19].Hartig F, Calabrese JM, Reineking B, Wiegand T, and Huth A, “Statistical inference for stochastic simulation models theory and application,” Ecology Letters, vol. 14, no. 8, pp. 816–827, Aug. 2011. [DOI] [PubMed] [Google Scholar]
- [20].Evensen G, Data Assimilation - The Ensemble Kalman Filter, 2nd ed. Springer-Verlag Berlin Heidelberg, 2009. [Google Scholar]
- [21].Gordon N, Salmond D, and Smith A, “Novel approach to nonlinear/non-Gaussian Bayesian state estimation,” IEE Proceedings F Radar and Signal Processing, vol. 140, no. 2, p. 107, 1993. [Google Scholar]
- [22].Arulampalam M, Maskell S, Gordon N, and Clapp T, “A tutorial on particle filters for online nonlinear/non-Gaussian Bayesian tracking,” IEEE Transactions on Signal Processing, vol. 50, no. 2, pp. 174–188, Feb. 2002. [Google Scholar]
- [23].Settles B, “Active Learning,” Synthesis Lectures on Artificial Intelligence and Machine Learning, vol. 6, no. 1, pp. 1–114, Jun. 2012. [Google Scholar]
- [24].Thiele JC, Kurth W, and Grimm V, “Facilitating Parameter Estimation and Sensitivity Analysis of Agent-Based Models: A Cookbook Using NetLogo and R,” Journal of Artificial Societies and Social Simulation, vol. 17, no. 3, p. 11, 2014. [Google Scholar]
- [25].Shaman J and Karspeck A, “Forecasting seasonal outbreaks of influenza,” Proceedings of the National Academy of Sciences, vol. 109, no. 50, pp. 20 425–20 430, Dec. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shaman J, Karspeck A, Yang W, Tamerius J, and Lipsitch M, “Real-time influenza forecasts during the 20122013 season,” Nature Communications, vol. 4, Dec. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yang W, Karspeck A, and Shaman J, “Comparison of Filtering Methods for the Modeling and Retrospective Forecasting of Influenza Epidemics,” PLoS Comput Biol, vol. 10, no. 4, p. e1003583, Apr. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shaman J, Yang W, and Kandula S, “Inference and Forecast of the Current West African Ebola Outbreak in Guinea, Sierra Leone and Liberia,” PLoS Currents, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Epstein JM, “Modelling to contain pandemics,” Nature, vol. 460, no. 7256, p. 687, Aug. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brauer F, van den Driessche P, and Wu J, Eds., Compartmental Models in Epidemiology Berlin: Springer Berlin Heidelberg, 2008, ch. 2, pp. 19–79. [Google Scholar]
- [31].C. for Disease Control, “How flu spreads,” http://www.cdc.gov/flu/about/disease/spread.htm, 2016, [Online; accessed 25-March-2016]. [Online]. Available: http://www.cdc.gov/flu/about/disease/spread.htm
- [32].——, “Flu symptoms,” http://www.cdc.gov/flu/consumer/symptoms.htm, 2016, [Online; accessed 25-March-2016]. [Online]. Available: http://www.cdc.gov/flu/consumer/symptoms.htm
- [33].Collier N and North M, “Parallel agent-based simulation with Repast for High Performance Computing,” SIMULATION, Nov. 2012. [Google Scholar]
- [34].Collier NT, Ozik J, and Macal CM, “Large-scale agent-based modeling with repast HPC: A case study in parallelizing an agent-based model,” in Euro-Par 2015: Parallel Processing Workshops - Euro-Par 2015 International Workshops, Vienna, Austria, August 24–25, 2015, Revised Selected Papers, 2015, pp. 454–465. [Online]. Available: 10.1007/978-3-319-27308-2\37 [DOI] [Google Scholar]
- [35].Wheaton WD, Cajka JC, Chasteen BM, Wagener DK, Cooley PC, Ganapathi L, Roberts DJ, and Allpress JL, “Synthesized population databases: A us geospatial database for agent-based models,” Methods report (RTI Press), no. 10, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gallagher S, Richardson L, Ventura SL, and Eddy WF, “SPEW: Synthetic Populations and Ecosystems of the World,” arXiv:1701.02383 [physics, q-bio, stat], Jan. 2017, arXiv: 1701.02383 [Online]. Available: http://arxiv.org/abs/1701.02383 [Google Scholar]
- [37].Armstrong TG, Wozniak JM, Wilde M, and Foster IT, Programming Models for Parallel Computing, 2015, ch. Swift: Extreme-scale, implicitly parallel scripting, ed. Balaji P. [Google Scholar]
- [38].——, “Compiler techniques for massively scalable implicit task parallelism,” in Proc. SC, 2014. [Google Scholar]
- [39].Krieder SJ, Wozniak JM, Armstrong TG, Wilde M, Katz DS,Grimmer B, Foster IT, and Raicu I, “Design and evaluation of the GeMTC framework for GPU-enabled many task computing,” in Proc. HPDC, 2014. [Google Scholar]
- [40].Wozniak JM, Armstrong TG, Maheshwari KC, Katz DS, Wilde M, and Foster IT, “Interlanguage parallel scripting for distributed-memory scientific computing,” in Proc. WORKS @ SC, 2015. [Google Scholar]
- [41].Duro FR, Blas JG, Isaila F, Carretero J, Wozniak JM, and Ross R, “Experimental evaluation of a flexible I/O architecture for accelerating workflow engines in ultrascale environments,” Parallel Computing, vol. 61, 2017. [Google Scholar]
- [42].Wozniak JM, Peterka T, Armstrong TG, Dinan J, Lusk EL, Wilde M, and Foster IT, “Dataflow coordination of data-parallel tasks via MPI 3.0,” in Proc. EuroMPI, 2013. [Google Scholar]
- [43].Abramson D, Lewis A, Peachey T, and Fletcher C, “An automatic design optimization tool and its application to computational fluid dynamics,” in Proc. SuperComputing, 2001. [Google Scholar]
- [44].Adams B, Bauman L, Bohnhoff W, Dalbey K, Ebeida M, Eddy J, Eldred M, Hough P, Hu K, Jakeman J, Stephens J, Swiler L, Vigil D, and Wildey T, “Dakota, a multilevel parallel object-oriented framework for design optimization, parameter estimation, uncertainty quantification, and sensitivity analysis: Version 6.0 users manual,” July 2014, sandia Technical Report SAND2014–4633, Updated November 2015. (Version 6.3). [Google Scholar]
- [45].Shaman J, “Columbia Prediction of Infectious Diseases,” http://cpid.iri.columbia.edu, 2016.
- [46].Cevik M, Ergun MA, Stout NK, Trentham-Dietz A, Craven M, and Alagoz O, “Using Active Learning for Speeding up Calibration in Simulation Models,” Medical Decision Making, p. 0272989X15611359, Oct. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jin R, Chen W, and Sudjianto A, “On Sequential Sampling for Global Metamodeling in Engineering Design,” pp. 539–548, Jan. 2002. [Google Scholar]
- [48].Xu Z, Akella R, and Zhang Y, “Incorporating Diversity and Density in Active Learning for Relevance Feedback,” in Advances in Information Retrieval, ser. Lecture Notes in Computer Science, Amati G, Carpineto C, and Romano G, Eds. Springer Berlin; Heidelberg, Apr. 2007, no. 4425, pp. 246–257, dOI: 10.1007/978-3-540-71496-524. [DOI] [Google Scholar]
- [49].Wozniak JM, Armstrong TG, Maheshwari KC, Katz DS, Wilde M, and Foster IT, “Toward interlanguage parallel scripting for distributed-memory scientific computing,” in Proc. CLUSTER, 2015. [Google Scholar]
- [50].Eddelbuettel D and Francois R, “RInside CRAN package,” https://cran.r-project.org/web/packages/RInside.
- [51].Rutter C, Ozik J, DeYoreo M, and Collier N, “Microsimulation Model Calibration using Incremental Mixture Approximate Bayesian Computation,” arXiv:1804.02090 [stat], Apr. 2018, arXiv: 1804.02090 [Online]. Available: http://arxiv.org/abs/1804.02090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wozniak JM, Jain R, Balaprakash P, Ozik J, Collier N, Bauer J, Xia F, Brettin T, Stevens R, Mohd-Yusof J, Cardona CG, Van Essen B, and Baughman M, “Candle/supervisor: A workflow framework for machine learning applied to cancer research,” to appear in BMC Bioinformatics, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ozik J, Collier N, Wozniak J, Macal C, Cockrell C, Friedman S, Ghaffarizadeh A, Heiland R, An G, and Macklin P, “High-throughput cancer hypothesis testing with an integrated physicell-emews workflow,” to appear in BMC Bioinformatics, 2018. [Online]. Available: https://www.biorxiv.org/content/early/2018/02/12/196709 [DOI] [PMC free article] [PubMed] [Google Scholar]