Abstract
Nuclear factor of activated T cells (NFAT)c2 is important for the immune response and it compensates for NFATc1 for its effects on osteoclastogenesis, but its role in this process is not established. To study the function of NFATc2 in the skeleton, Nfatc2loxP/loxP mice, where the Nfact2 exon2 is flanked by loxP sequences, were created and mated with mice expressing the Cre recombinase under the control of the Lyz2 promoter. Bone marrow-derived macrophage (BMM) from Lyz2Cre/WT;Nfatc2Δ/Δ mice cultured in the presence of macrophage colony stimulating factor (M-CSF) and receptor activator of NF-κB ligand (RANKL) exhibited a decrease in the number and size of osteoclasts and a smaller sealing zone when compared to BMMs from Nfatc2loxP/loxP littermate controls. Bone resorption was decreased in osteoclasts from Lyz2Cre/WT;Nfatc2Δ/Δ mice. This demonstrates that NFATc2 is necessary for optimal osteoclast maturation and function in vitro. Male and female Lyz2Cre/WT;Nfatc2Δ/Δ mice did not exhibit an obvious skeletal phenotype by microcomputed tomography (μCT) at either 1 or 4 months of age when compared to Nfatc2loxP/loxP sex-matched littermates. Bone histomorphometry confirmed the μCT results, and conditional 4 month old Lyz2Cre/WT;Nfatc2Δ/Δ mice did not exhibit changes in parameters of bone histomorphometry. In conclusion, NFATc2 is necessary for optimal osteoclastogenesis in vitro, but its downregulation in the myeloid lineage has no consequences in skeletal remodeling in vivo.
Keywords: NFATc2, bone resorption, bone remodeling, osteoclasts
Nuclear factor of activated T cells (NFAT) are a family of five transcription factors (NFATc1 to c4 and NFAT5) that regulate the growth and differentiation of multiple cell lineages [Crabtree and Olson, 2002; Sitara and Aliprantis, 2010]. Initially studied for their role in the early immune response, NFATs were subsequently found to regulate cellular events in non-immune cells. Activation of NFATs requires dephosphorylation of specific serine residues in their regulatory domain by the phosphatase calcineurin. This results in the translocation of NFAT from the cytoplasm, where they reside in a phosphorylated state, to the nucleus where they regulate the transcription of target genes [Chow et al., 2008; Hogan et al., 2003; Okamura et al., 2000].
NFATc1 through c4 are expressed by skeletal cells, and NFATc1 plays an undisputable role in osteoclast differentiation [Aliprantis et al., 2008; Ikeda et al., 2006; Ikeda et al., 2004; Ishida et al., 2002; Matsuo et al., 2004; Takayanagi et al., 2002]. The functions of NFATc1 and c2 are not necessarily redundant, and the role of NFATc2 in osteoclast differentiation and function is less certain [Ikeda et al., 2006; Ranger et al., 1998]. This is in part because of limitations in the genetic manipulations used in former studies to explore the role of NFATs in skeletal cells [Hodge et al., 1996; Koga et al., 2005; Sun et al., 2005; Winslow et al., 2006; Yeo et al., 2007]. Whereas transgenic overexpression of constitutive active (ca)NFATc2 under the control of the Acp5 promoter results in increased osteoclastogenesis and bone resorption, the inactivation of Nfatc2 has led to conflicting results [Asagiri et al., 2005; Bauer et al., 2011; Ikeda et al., 2006; Koga et al., 2005]. Studies interrogating the function of NFATc2 in the skeleton have suffered from the use of a systemic gene inactivation strategy. Mice harboring the global inactivation of Nfatc2 were reported not to have a skeletal phenotype, to exhibit osteopenia due to reduced bone formation or osteopetrosis due to reduced osteoclast number [Asagiri et al., 2005; Bauer et al., 2011; Koga et al., 2005]. Nfatc2 null mutants display hyperproliferation of B, T and other immune cells and dysregulated expression of multiple cytokines, which may confound the interpretation of the skeletal phenotype [Hodge et al., 1996; Monticelli and Rao, 2002; Ranger et al., 2000]. Moreover, the Nfatc2 targeting vector used contains a Neo cassette, and retention of selectable marker cassettes, like phosphoglycerate kinase (PGK)-neo, can cause unexpected phenotypes by disrupting the expression of neighboring genes [Hodge et al., 1996; Olson et al., 1996; Pham et al., 1996]. Mouse models harboring either the activation or the downregulation of calcineurin also have generated conflicting skeletal phenotypes [Sun et al., 2005; Yeo et al., 2007], possibly because this approach does not discriminate between the independent effects of each NFAT isoform or other calcineurin-dependent signals.
The intent of the present study was to define the function of NFATc2 in osteoclast differentiation and function in vitro and in vivo. For this purpose, we created Nfatc2loxP/loxP conditional mice. Nfatc2 was inactivated by Cre recombination directed by the Lyz2 gene expressed in cells of the myeloid lineage including osteoclast precursors [Clausen et al., 1999; Takeda et al., 1999]. The skeletal phenotype of Nfatc2 conditional null mice was determined by microcomputed tomography (μCT) and by bone histomorphometry, and by the study of osteoclast differentiation and resorption activity in vitro.
MATERIALS AND METHODS
Generation of Nfatc2 Conditional Mice
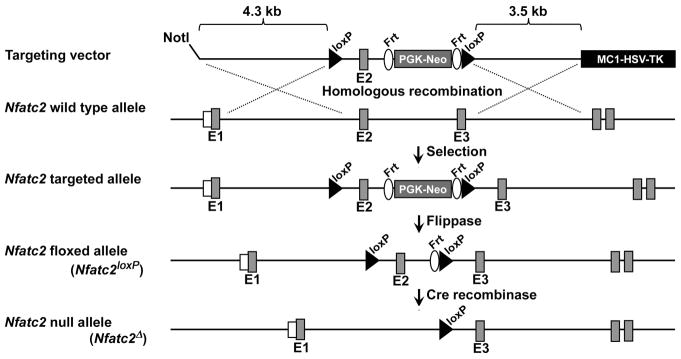
To create a conditional allele of Nfatc2, 9.7 kilobase pair (kb) of Nfatc2 sequence were selected from a bacterial artificial chromosome library of C57BL/6J mouse genomic DNA (id RP24-223B7) and retrieved into a PL253 vector. The vector contains an MC1 driven herpes simplex virus-thymidine kinase (MC1-HSV-TK) cassette for negative selection of embryonic stem (ES) cells [Liu et al., 2003; Mansour et al., 1988]. A 5′ loxP site was introduced approximately 0.4 kb 5′ of exon 2, followed by the insertion of a PGK promoter-driven neomycin selection cassette flanked by flippase recognition target (Frt) sequences and a 3′ loxP site (Frt-PGKneo-Frt-LoxP) approximately 0.4 kb 3′ of exon 2 (Figure 1) [Buchholz et al., 1996] (Figure 1). The targeting vector containing 4.3 kb of 5′ homology arm and 3.5 kb of 3′ homology arm was linearized and electroporated into ES cells derived from F1 (129Svj/C57BL/6J) embryos. G418 and gancyclovir resistant colonies were isolated and screened by long range nested polymerase chain reaction (PCR) [Lay et al., 1998]. Targeted ES clones were used for aggregations to generate chimeric mice at the Center for Mouse Genome Modification at UConn Health [Eakin and Hadjantonakis, 2006; Gertsenstein et al., 2010; Pettitt et al., 2009; Pluck and Klasen, 2009]. Chimeras that were transmitters of ES-derived sperm were bred with mice expressing the Flp recombinase under the control of the Rosa promoter (Gt(ROSA)26Sortm2(FLP+)Sor, Jackson Laboratory, Bar Harbor, ME, Stock 007844) for the removal of the PGK-neo selection cassette flanked by Frt [Buchholz et al., 1998; Canalis et al., 2010a; Canalis et al., 2010b; Farley et al., 2000; Mallo, 2006; Raymond and Soriano, 2007]. The excision of the cassette was confirmed by PCR (Table 1), and the Flp recombinase transgene was segregated by mating with C57BL/6J wild type mice. Mice with the Nfatc2 allele flanked by loxP sites were backcrossed into a C57BL/6J background for 7 generations. Cre recombination excises exon 2, encoding for the regulatory domain of NFATc2, and leads to a frame shift and the creation of a STOP codon in exon 3 so that only 42 amino acids of the mature protein are expressed, lacking both the regulatory and the DNA binding domain (aa 400–680) [Okamura et al., 2000].
Figure 1. Engineering of the Nfatc2 conditional allele showing the wild type and Nfatc2 targeted alleles.
In the conditional allele exon 2 is flanked by loxP sites upstream a PGK-neo selection cassette flanked by Frt sites. The Nfatc2 targeted allele is shown prior to and after Flp and Cre recombination for the removal of the neo cassette and excision of exon 2, respectively.
Table 1.
Primers used for genotyping by PCR.
| Allele | Strand | Sequence | Amplicon Size (bp) |
|---|---|---|---|
| Lyz2Cre | Forward1 Forward2 Reverse |
5′-TTACAGTCGGCCAGGCTGAC-3′ |
Lyz2WT = 350 Lyz2Cre = 700 |
| 5′-CCCAGAAATGCCAGATTACG-3′ 5′-CTTGGGCTGCCAGAATTTCTC-3′ | |||
| Nfatc2loxP | Forward Reverse |
5′-GGTGTCTCCCTCAAATGTTCA-3′ 5′-GCGAATATGAGGACCCAAGT-3′ |
WT = 160 Nfatc2loxP= 250 |
| Frt and LoxP recombination | Strand | Sequence | Amplicon Size (bp) |
| Frt recombination of Nfatc2 targeted allele | Forward Reverse |
5′-AGTATGTAACCTCTGCTTCC-3′ 5′-AGCACAATGCCCATGTTCAC-3′ |
WT = 250 Not recombined = no band Recombined = 351 (Nfatc2loxP) |
| LoxP recombination of Nfatc2loxP | Reverse Forward |
5′-GGTGTCTCCCTCAAATGTTCA-3′ 5′-AGCACAATGCCCATGTTCAC-3′ |
Not recombined = 2342 (Nfatc2loxP) Recombined = 358 (Nfatc2Δ) |
Deletion of Nfatc2 in the Myeloid Lineage
C57BL/6J mice where the Cre coding sequence was inserted into the endogenous Lyz2 locus (Lyz2Cre; Jackson Laboratory, Stock 004781) were used to express the Cre recombinase in cells of the myeloid lineage [Clausen et al., 1999; Takeda et al., 1999]. To induce the deletion of Nfatc2 in osteoclast precursors, homozygous Nfatc2loxP/loxP mice heterozygous for the Lyz2Cre allele (Lyz2Cre/WT;Nfatc2loxP/loxP) were bred with Nfatc2loxP/loxP mice to create Lyz2Cre/WT;Nfatc2Δ/Δ mice and Nfatc2loxP/loxP control littermates.
Culture of Bone Marrow-derived Macrophages (BMMs) and Osteoclast Formation
To obtain BMMs, the marrow from Lyz2Cre/WT;Nfatc2Δ/Δ and control Nfatc2loxP/loxP littermates was removed by flushing with a 26 gauge needle and erythrocytes were lysed in 150 mM NH4Cl, 10 mM KHCO3 and 0.1 mM EDTA (pH 7.4) as described previously [Canalis et al., 2017]. Cells were centrifuged and the sediment suspended in α-minimum essential medium (α-MEM) in the presence of 10% fetal bovine serum (FBS; both from Thermo Fisher Scientific, Waltham, MA) and recombinant human macrophage-colony stimulating factor (M-CSF) at 30 ng/ml. M-CSF cDNA and expression vector were obtained from D. Fremont (St. Louis, MO) and M-CSF was purified as previously reported [Lee et al., 2006]. Cells were seeded on plastic petri dishes at a density of 300,000 cells/cm2 and cultured for 3 to 4 days. Deletion of the Nfatc2 allele was documented by PCR of genomic DNA using primers specific for the Nfatc2Δ/Δ allele (Table 1). For osteoclast formation, cells were collected following treatment with 0.25% trypsin/EDTA for 5 min and seeded on tissue culture plates at a density of 47,000 cells/cm2 in α-MEM with 10% FBS, M-CSF at 30 ng/ml and recombinant murine receptor activator of NF-κB ligand (RANKL) at 10 ng/ml. Tnfsf11 encoding RANKL cDNA and expression vector were obtained from M. Glogauer (Toronto, Canada), and GST-tagged RANKL was expressed and purified as described [Wang et al., 2008]. Cultures were carried out until formation of multinucleated tartrate resistant acid phosphatase (TRAP)-positive cells. TRAP enzyme histochemistry was conducted using a commercial kit (Sigma-Aldrich, St. Louis, MO), in accordance with manufacturer’s instructions. TRAP-positive cells containing more than 3 nuclei were considered osteoclasts.
For Actin structure staining and bone resorption assay of osteoclasts in vitro, BMMs were seeded at a density of 4.7 × 104 cells/cm2 on bovine cortical bone slices and cultured in α-MEM with 10% FBS, M-CSF at 30 ng/ml and RANKL at 10 ng/ml. To visualize the sealing zone of osteoclasts on the bone slices, cells were fixed with 4% paraformaldehyde for 10 min and were permeabilized with 0.3% triton X-100 for 5 min. To block non-specific background staining, cells on bone discs were incubated with 2% BSA for 1 hour. Cells were stained with Alexa Fluor™ 594 Phalloidin (Thermo Fisher Scientific) at a 1:40 dilution for 20 min. The sealing zones were viewed on a Leica fluorescence microscope (Model DMI6000B), and collected images were processed using the Leica Application Suite × 1.5.1.1387 (Leica Microsystem, Buffalo Grove, IL). After visualizing the sealing zone, cells were stained for TRAP to assess their morphology. To visualize bone resorption pits, the bone slices were sonicated to remove osteoclasts and stained with 1% toluidine blue in 1% sodium borate. To evaluate the ability of osteoclasts to resorb bone, the total resorption area/total bone area was measured on images acquired with an Olympus DP72 camera using cellSens Dimension software v1.6 (Olympus Corporation, Center Valley, PA). The total resorption area/total bone area was corrected for the total number of TRAP-positive multinucleated cells [Canalis et al., 2017].
Microcomputed Tomography
Femoral microarchitecture was determined using a microcomputed tomography instrument (Scanco μCT 40; Scanco Medical AG, Bassersdorf, Switzerland), which was calibrated periodically using a phantom provided by the manufacturer [Bouxsein et al., 2010; Glatt et al., 2007]. Femurs were scanned in 70% ethanol at high resolution, energy level of 55 kVp, intensity of 145 μA, and integration time of 200 ms. A total of 100 slices at midshaft and 160 slices at the distal metaphysis were acquired at an isotropic voxel size of 216 μm3 and a slice thickness of 6 μm, and chosen for analysis. Trabecular bone volume fraction (bone volume/total volume) and microarchitecture were evaluated starting approximately 1.0 mm proximal from the femoral condyles. Contours were manually drawn every 10 slices, a few voxels away from the endocortical boundary, to define the region of interest for analysis, whereas the remaining slice contours were iterated automatically. Total volume, bone volume, bone volume fraction, trabecular thickness, trabecular number, connectivity density, structure model index (SMI) and material density were measured in trabecular regions using a Gaussian filter (σ = 0.8) and user defined thresholds [Bouxsein et al., 2010; Glatt et al., 2007]. For analysis of cortical bone, contours were iterated across 100 slices along the cortical shell of the femoral midshaft, excluding the marrow cavity. Analysis of bone volume/total volume, porosity, cortical thickness, total cross sectional and cortical bone area, periosteal and endosteal perimeter and material density were conducted using a Gaussian filter (σ = 0.8, support = 1) with operator-defined thresholds.
Bone Histomorphometry
Bone histomorphometry was carried out in 1 and 4 month old mice injected with calcein 20 mg/kg and demeclocycline 50 mg/kg at a 2 or 7 day interval, respectively, and sacrificed 2 days after demeclocycline administration. Femurs were dissected, fixed in 70% ethanol and embedded in methyl methacrylate. For cancellous bone analysis, bones were sectioned at a thickness of 5 μm along the sagittal plane on a Microm microtome (Richards-Allan Scientific, Kalamazoo, MI), and stained with 0.1% toluidine blue. Static and dynamic parameters of bone morphometry were measured in a defined area between 0.35 mm and 2.16 mm from the growth plate at a magnification of 10× using an OsteoMeasure morphometry system (Osteometrics, Atlanta, GA). Stained sections were used to draw bone tissue and to measure trabecular separation, number and thickness, osteoid and eroded surface, as well as to count osteoblast and osteoclast surface and number. Mineralizing surface per bone surface and mineral apposition rate were measured on unstained sections visualized under UV light and a triple diamidino-2-phenylindole/fluorescein/Texas red set long pass filter, and bone formation rate was calculated [Dempster et al., 2013].
RNA Integrity and Quantitative Reverse Transcription-PCR (qRT-PCR)
Total RNA was extracted from BMMs with the RNeasy kit (Qiagen, Valencia, CA) and from homogenized bones with the micro RNeasy kit (Qiagen), in accordance with manufacturer’s instructions. The integrity of the RNA extracted from bones was assessed by microfluidic electrophoresis on an Experion system (BioRad, Hercules, CA), and RNA with a quality indicator number equal to or higher than 7.0 was used for subsequent analysis. Equal amounts of RNA were reverse-transcribed using the iScript RT-PCR kit (BioRad) and amplified in the presence of specific primers (all primers from Integrated DNA Technologies, IDT, Coralville, IA; Table 2) with the SsoAdvanced™ Universal SYBR Green Supermix (BioRad) at 60°C for 40 cycles. Transcript copy number was estimated by comparison with a serial dilution of cDNA for Nfatc1 (Addgene plasmid 11793), Nfatc2 (Addgene plasmid 11791, both created by A. Rao, La Jolla, CA), Acp5, Ctsk and Calcr (Thermo Fisher Scientific). Tnfrsf11a copy number was estimated by comparison to a serial dilution of a 152 base pair (bp) synthetic DNA template (IDT) cloned into pcDNA3.1(−) (Thermo Fisher Scientific) by isothermal single reaction assembly using commercially available reagents (New England BioLabs, Ipswich, MA).
Table 2.
Primers used for qRT-PCR determinations. GenBank accession numbers identify transcript recognized by primer pairs.
| Gene | Strand | Sequence | GenBank Accession Number |
|---|---|---|---|
| Acp5 | Forward Reverse |
5′-GACAAGAGGTTCCAGGAGAC-3′ 5′-TTCCAGCCAGCACATACC-3′ |
NM_001102404; NM_001102405; NM_007388 |
| Calcr | Forward Reverse |
5′-TCTGAGAAACTGCAAAATGCGTAC-3′ 5′-AGCAACCAAAGCAGCAATCG-3′ |
NM_007588 |
| Ctsk | Forward Reverse |
5′-AGATATTGGTGGCTTTGGAA-3′ 5′-AACGAGAGGAGAAATGAAACA-3′ |
NM_007802 |
| Nfatc1 | Forward Reverse |
5′-GCGCAAGTACAGTCTCAATGGCC-3′ 5′-GGATGGTGTGGGTGAGTGGT-3′ |
NM_198429; NM_001164110; NM_001164111; NM_001164112; NM_00116641091; NM_016791 |
| Nfatc2 | Forward Reverse |
5′-AGAACAACATGAGAGCCACCATC-3′ 5′-AGCTCGATGTCAGCGTTTCG-3′ |
NM_010899 |
| Rpl38 | Forward Reverse |
5′-AGAACAAGGATAATGTGAAGTTCAAGGTTC-3′ 5′-CTGCTTCAGCTTCTCTGCCTTT-3′ |
NM_001048057; NM_001048058; NM_023372 |
| Tnfrsf11a | Forward Reverse |
5′-GATGCCTCACAGTTAAGAAGAAGA-3′ 5′-GAGCTGGAGTTACATGGAGTTG-3′ |
NM_009399.3 |
Amplification reactions were conducted in CFX96 qRT-PCR detection systems (BioRad), and fluorescence was monitored during every PCR cycle at the annealing step. Data are expressed as copy number corrected for Rpl38 expression estimated by comparison with a serial dilution of Rpl38 (from American Type Culture Collection, ATCC, Manassas, VA) [Kouadjo et al., 2007].
Immunoblotting
Osteoclasts from control or Lyz2Cre/WT;Nfatc2Δ/Δ mice were extracted in buffer containing 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5% glycerol, 1 mM EDTA, 0.5% Triton X-100, 1 mM sodium orthovanadate, 10 mM NaF, 1 mM phenyl methyl sulfonyl fluoride and a protease inhibitor cocktail (all from Sigma Aldrich, St. Louis, MO). Quantified total cell lysates (50 μg of total protein) were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) in 8% polyacrylamide gels and transferred to Immobilon-P membranes (Millipore, Billerica, MA). The blots were probed with anti-NFATc2 (4389) and anti-β-Actin (3700) antibodies, both from Cell Signaling Technology (Danvers, MA), and anti-NFATc1 (556602; BD Bioscience, San Jose, CA) antibodies. The blots were exposed to either anti-rabbit IgG or anti-mouse IgG conjugated to horseradish peroxidase (Sigma-Aldrich) and incubated with a chemiluminescence detection reagent (BioRad). The bands were detected by ChemiDoc™ XSR+ molecular imager (BioRad) with Image Lab™ software (version 5.2.1) [Zanotti et al., 2013].
Statistics
Data are expressed as means ± S.D. Statistical differences were determined by Student’s t test or two-way analysis of variance with Holm-Šídák post-hoc analysis for pairwise or multiple comparisons, respectively.
RESULTS
Nfatc2 mRNA and Protein Expression during Osteoclastogenesis
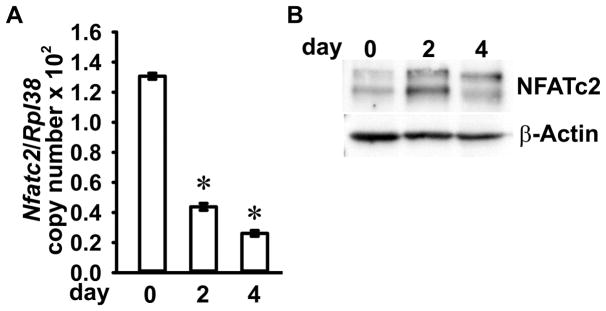
To verify Nfatc2 gene expression during osteoclast differentiation, we checked mRNA and protein levels of Nfatc2 in RANKL-stimulated BMMs undergoing differentiation into multinucleated osteoclasts. Cellular extracts were collected prior to, and after 2 and 4 days following RANKL stimulation for the determination of total RNA and protein. Nfatc2 mRNA levels were decreased, whereas NFATc2 protein levels were increased by RANKL during osteoclast differentiation (Figure 2).
Figure 2. NFATc2 expression during osteoclast differentiation.
BMMs were cultured for 4 days in the presence of M-CSF at 30ng/ml and RANKL at 10ng/ml. The cells were collected at the indicated times for extraction of total RNA and proteins. (A) Nfatc2 mRNA levels were measured by qRT-PCR. Transcript levels are reported as copy number corrected for Rpl38 mRNA levels. Values are means ± SD; n = 4 biological replicates. Two technical replicates were used for each qRT-PCR reaction. *Significantly different compared to day 0, p < 0.05. (B) 50 μg of total proteins were separated by SDS-PAGE and NFATc2 protein levels were detected by using anti-NFATc2 antibodies. β-Actin levels were detected using anti-β-Actin antibodies and served as a loading control in the same blot.
Conditional Inactivation of Nfatc2 in Cells of the Myeloid Lineage including Osteoclast Precursors
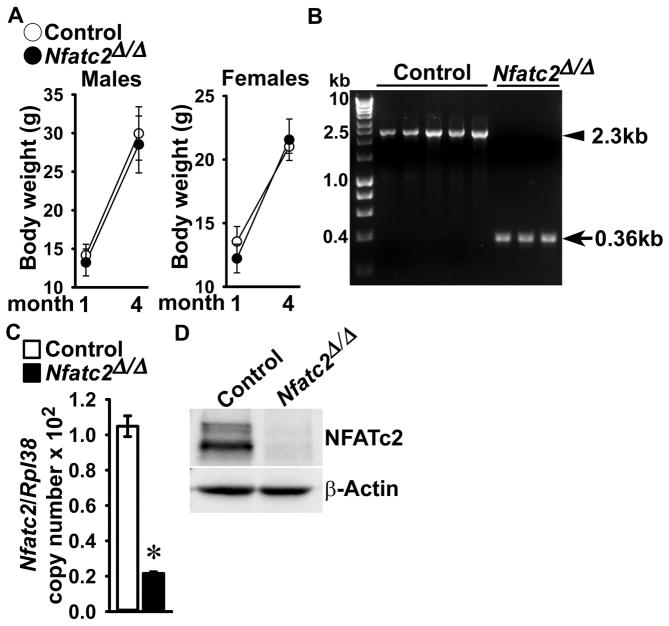
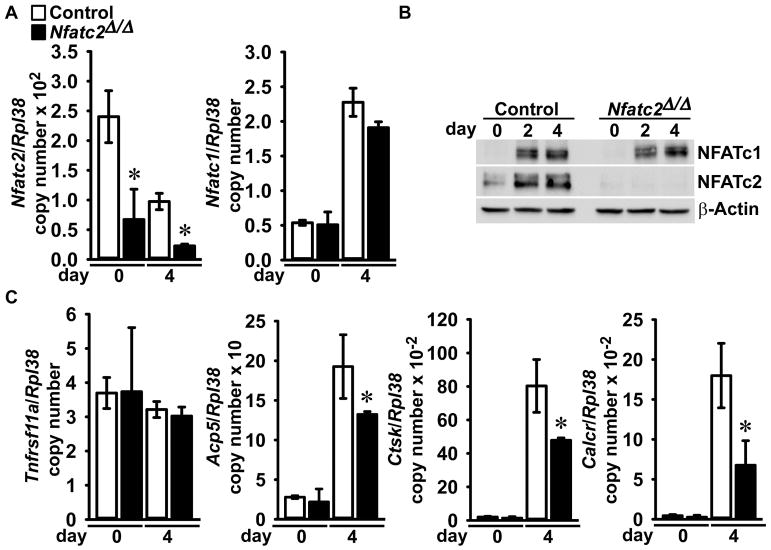
To establish the inactivation of Nfatc2 in cells of the myeloid lineage, the Nfatc2loxP allele (Figure 1) was introduced into Lyz2Cre/WT heterozygous mice. Subsequently, Lyz2Cre/WT;Nfatc2loxP/loxP mice were crossed with Nfatc2loxP/loxP mice for the creation of Lyz2Cre/WT;Nfatc2Δ/Δ experimental mice and Nfatc2loxP/loxP littermate controls. Lyz2Cre/WT;Nfatc2Δ/Δ mice appeared healthy and their weight was not different from that of Nfatc2loxP/loxP littermate controls (Figure 3). Lyz2Cre-mediated recombination was demonstrated in genomic DNA from Lyz2Cre/WT;Nfatc2Δ/Δ BMM cultures with the consequent decrease in Nfatc2 mRNA and NFATc2 protein levels. Nfatc2 transcripts were decreased by 80% in Lyz2Cre/WT;Nfatc2Δ/Δ BMM cultures when compared to control cultures, and NFATc2 protein levels were virtually undetectable in Lyz2Cre/WT;Nfatc2Δ/Δ cultures, documenting downregulation of Nfatc2 in osteoclast precursor BMMs (Figure 3).
Figure 3. Deletion of the Nfatc2 allele in Lyz2-expressing cells.
Weight, documentation of DNA recombination of the Nfatc2loxP allele by Cre and Nfatc2 deletion in BMMs. (A) Body weight of 1 and 4 month old Lyz2Cre/WT;Nfatc2Δ/Δ and sex-matched control Nfatc2loxP/loxP littermates. Values are means ± SD; n = 10 control and n = 7 Nfatc2Δ/Δ for 1 month old male mice; n = 8 control and n = 9 Nfatc2Δ/Δ for 4 month old male mice; n = 6 control and n = 6 Nfatc2Δ/Δ for 1month old female mice; n = 6 control and n = 9 Nfatc2Δ/Δ for 4 month old female mice. (B) Genomic DNA from BMMs obtained from 1 month old Lyz2Cre/WT;Nfatc2Δ/Δ mice and respective controls was isolated. Nfatc2 deletion by DNA recombination was demonstrated by gel electrophoresis of PCR amplification products obtained with primers for the Nfatc2loxP and Nfatc2Δ alleles. The arrow head indicates the position of the 2.3 kilobase pair (kb) amplicon verifying the Nfatc2loxP allele, and the arrow indicates a 0.36 kb amplicon verifying the Nfatc2Δ allele. n = 5 control and n = 3 Nfatc2Δ/Δ biological replicates. (C) Nfatc2 transcript levels were measured by qRT-PCR in total RNA from BMMs of Lyz2Cre/WT;Nfatc2Δ/Δ and respective controls. Transcript levels are reported as copy number corrected for Rpl38. Values are means ± SD; n = 4 biological replicates for control and Nfatc2Δ/Δ. Two technical replicates were used for each qRT-PCR reaction. *Significantly different between Nfatc2Δ/Δ and control, p < 0.05. (D) 50 μg of total proteins of BMMs from 1 month old Lyz2Cre/WT;Nfatc2Δ/Δ and respective controls were separated by SDS-PAGE and NFATc2 protein levels were detected by using anti-NFATc2 antibodies. β-Actin levels served as a loading control in the same blot.
NFATc2 Is Required for the Differentiation, Maturation and Resorption of Osteoclasts in vitro
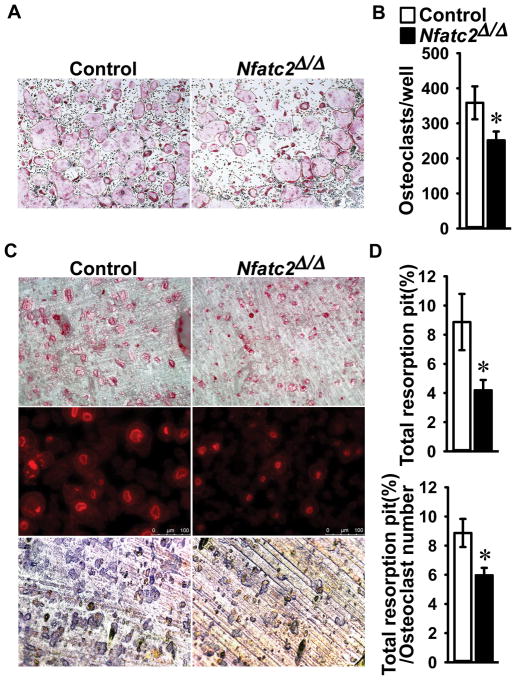
To determine the direct effects of NFATc2 on osteoclast differentiation and function, BMMs derived from Lyz2Cre/WT;Nfatc2Δ/Δ and control Nfatc2loxP/loxP littermates were cultured in the presence of M-CSF and RANKL. Cultured BMMs from Lyz2Cre/WT;Nfatc2Δ/Δ mice exhibited a 30% decrease in osteoclast number in comparison to cells from littermate controls (Figure 4). In addition, TRAP and Phalloidin staining of osteoclasts from Lyz2Cre/WT;Nfatc2Δ/Δ mice cultured on bone discs revealed smaller cells with smaller sealing zones than littermate controls. Accordingly, Lyz2Cre/WT;Nfatc2Δ/Δ osteoclasts exhibited a 50% decrease in total bone resorption area. When corrected for osteoclast number, bone resorption area was 30% lower in Lyz2Cre/WT;Nfatc2Δ/Δ than controls, indicating decreased osteoclast resorptive activity. These findings demonstrate that NFATc2 is necessary for optimal osteoclast maturation and function in vitro (Figure 4).
Figure 4. Number, size and resorption activity are decreased in Lyz2Cre;Nfatc2Δ/Δ osteoclasts.
BMMs derived from 1 month old Lyz2Cre/WT;Nfatc2Δ/Δ mice and Nfatc2loxP/loxP control littermates were cultured for 4 days in the presence of M-CSF at 30 ng/ml and of RANKL at 10 ng/ml in cell culture-coated plates (A and B) or bone discs (C and D). (A) After 4 days, cells were stained with TRAP and representative images of TRAP-stained multinucleated cells were shown (4× magnification). (B) TRAP-positive cells with more than 3 nuclei were counted as osteoclasts and values are means ± SD; n = 4 biological replicates for control and Nfatc2Δ/Δ. *Significantly different between Nfatc2Δ/Δ and control, p < 0.05. (C) Cultured osteoclasts on bone discs were stained by Alexa Fluor 594 Phalloidin (middle, 10X magnification) and TRAP (upper, 4× magnification). To stain for resorption pits, cells were removed by sonication and bone discs were stained by Toluidine blue (bottom, 10× magnification). n = 4 biological replicates for control and Nfatc2Δ/Δ and representative images for TRAP, Phalloidin and Toluidine blue are shown. (D) Data are expressed as total resorption pit area (%, upper) and total resorption pit area corrected for number of osteoclasts (%, lower). Values are means ± SD; n = 4 biological replicates for control and Nfatc2Δ/Δ. *Significantly different between Nfatc2Δ/Δ and control, p < 0.05.
Nfatc2 was downregulated specifically since neither Nfatc1 mRNA nor NFATc1 protein levels were affected in Lyz2Cre/WT;Nfatc2Δ/Δ osteoclasts compared to control (Figure 5). Tnfrsf11a encoding for RANK, the specific RANKL receptor was not affected in either BMMs or osteoclasts from Lyz2Cre/WT;Nfatc2Δ/Δ. Expression of gene markers representative of osteoclasts, such as Acp5 encoding for TRAP, Ctsk, encoding for Cathepsin K, and Calcr, encoding for the Calcitonin receptor, were all significantly decreased in Lyz2Cre/WT;Nfatc2Δ/Δ osteoclasts compared to cultures from control littermates (Figure 5).
Figure 5. Acp5, Ctsk and Calcr mRNA levels are decreased in Lyz2Cre;Nfatc2Δ/Δ Osteoclasts.
BMMs derived from 1 month old Lyz2Cre/WT;Nfatc2Δ/Δ mice and Nfatc2loxP/loxP control littermates were cultured for 4 days in the presence of M-CSF at 30 ng/ml and of RANKL at 10 ng/ml. The cells were collected at the indicated times for extraction of total RNA and proteins. (A) Nfatc2 and Nfatc1 mRNA levels were measured by qRT-PCR. Transcript levels are reported as copy number corrected for Rpl38 mRNA levels. Values are means ± SD; n = 5 control and n = 3 Nfatc2Δ/Δ biological replicates. Two technical replicates were used for each qRT-PCR reaction. *Significantly different between Nfatc2Δ/Δ and control, p < 0.05. (B) 50 μg of total protein were separated by SDS-PAGE and NFATc1 and NFATc2 levels were detected by using immunoblot anti-NFATc1 and anti-NFATc2 antibodies, respectively. β-Actin levels served as a loading control in the same blot. (C) Tnfrs11a, Acp5, Ctsk and Calcr mRNA levels in total RNA were measured by qRT-PCR. Transcript levels are reported as copy number corrected for Rpl38. Values are means ± SD; n = 5 control and n = 3 Nfatc2Δ/Δ biological replicates. Two technical replicates were used for each qRT-PCR reaction. *Significantly different between Nfatc2Δ/Δ and control, p < 0.05.
Conditional Inactivation of Nfatc2 in Lyz2-expressing Cells Does Not Cause a Skeletal Phenotype In Vivo
In preliminary experiments, we documented that 1 and 4 month old Lyz2Cre and 1 and 4 month old Nfatc2loxP/loxP mice do not have a skeletal phenotype as determined by μCT of distal femurs, when compared to wild type controls (data not shown). Femoral microarchitecture of 1 and 4 month old male and female Lyz2Cre/WT;Nfatc2Δ/Δ mice revealed no differences compared to Nfatc2loxP/loxP sex-matched littermate controls (Tables 3 and 4). Bone histomorphometry of 4 month old Lyz2Cre/WT;Nfatc2Δ/Δ male mice confirmed the absence of a skeletal phenotype when compared to Nfatc2loxP/loxP littermate sex-matched controls (Table 5).
Table 3.
Femoral microarchitecture assessed by μCT of 1 and 4 month old Lyz2Cre/WT;Nfatc2Δ/Δ male mice (Nfatc2Δ/Δ) and sex-matched littermate Nfatc2loxP/loxP controls.
| 1 Month | 4 Months | |||
|---|---|---|---|---|
| Control n = 10 |
Nfatc2Δ/Δ n = 7 |
Control n = 8 |
Nfatc2Δ/Δ n = 9 |
|
| Distal Femur Trabecular Bone | ||||
| Bone Volume/Total Volume (%) | 7.2 ± 2.5 | 8.6 ± 2.2 | 7.8 ± 3.0 | 6.8 ± 3.2 |
| Trabecular Separation (μm) | 226 ± 40 | 216 ± 21 | 257 ± 13 | 249 ± 23 |
| Trabecular Number (1/mm) | 4.6 ± 0.7 | 4.7 ± 0.5 | 3.9 ± 0.2 | 4.0 ± 0.4 |
| Trabecular Thickness (μm) | 28 ± 3 | 29 ± 2 | 38 ± 9 | 34 ± 6 |
| Connectivity Density (1/mm3) | 330 ± 190 | 426 ± 162 | 140 ± 45 | 135 ± 56 |
| Structure Model Index | 2.6 ± 0.3 | 2.4 ± 0.2 | 2.1 ± 0.4 | 2.3 ± 0.5 |
| Density of Material (mg HA/cm3) | 946 ± 27 | 942 ± 28 | 1012 ± 14 | 1004 ± 20 |
| Femoral Midshaft Cortical Bone | ||||
| Bone Volume/Total Volume (%) | 82.6 ± 4.2 | 82.6 ± 2.5 | 89.6 ± 0.6 | 89.4 ± 0.5 |
| Porosity (%) | 17.4 ± 4.2 | 17.4 ± 2.5 | 10.4 ± 0.6 | 10.6 ± 0.5 |
| Cortical Thickness (μm) | 95 ± 17 | 94 ± 7 | 171 ± 8 | 170 ± 9 |
| Total Area (mm2) | 1.6 ± 0.1 | 1.6 ± 0.1 | 2.3 ± 0.2 | 2.1 ± 0.2 |
| Bone Area (mm2) | 0.5 ± 0.1 | 0.5 ± 0.1 | 1.0 ± 0.1 | 0.9 ± 0.1 |
| Periosteal Perimeter (mm) | 4.4 ± 0.2 | 4.4 ± 0.1 | 5.3 ± 0.3 | 5.2 ± 0.2 |
| Endocortical Perimeter (mm) | 3.7 ± 0.1 | 0.6 ± 0.1 | 4.0 ± 0.2 | 3.9 ± 0.2 |
| Density of Material (mg HA/cm3) | 1049 ± 19 | 1051 ± 14 | 1235 ± 25 | 1224 ± 22 |
μCT was performed on distal femurs for trabecular bone and midshaft for cortical bone. Values are means ± SD.
Table 4.
Femoral microarchitecture assessed by μCT of 1 and 4 month old Lyz2Cre/WT;Nfatc2δ/δ female mice (Nfatc2δ/δ) and sex-matched littermate Nfatc2loxP/loxP controls.
| 1 Month | 4 Months | |||
|---|---|---|---|---|
| Control n = 6 |
Nfatc2Δ/Δ n = 6 |
Control n = 6 |
Nfatc2Δ/Δ n = 9 |
|
| Distal Femur Trabecular Bone | ||||
| Bone Volume/Total Volume (%) | 3.8 ± 0.5 | 3.8 ± 0.4 | 3.9 ± 1.2 | 3.1 ± 0.6 |
| Trabecular Separation (μm) | 285 ± 22 | 276 ± 20 | 314 ± 18 | 326 ± 18 |
| Trabecular Number (1/mm) | 3.6 ± 0.2 | 3.7 ± 0.2 | 3.2 ± 0.2 | 3.1 ± 0.1 |
| Trabecular Thickness (μm) | 26 ± 1 | 25 ± 1 | 37 ± 3 | 35 ± 3 |
| Connectivity Density (1/mm3) | 79 ± 36 | 89 ± 27 | 58 ± 27 | 52 ± 18 |
| Structure Model Index | 2.8 ± 0.2 | 2.8 ± 0.1 | 2.9 ± 0.3 | 3.0 ± 0.2 |
| Density of Material (mg HA/cm3) | 978 ± 10 | 973 ± 16 | 1014 ± 8 | 1009 ± 13 |
| Femoral Midshaft Cortical Bone | ||||
| Bone Volume/Total Volume (%) | 82.0 ± 3.6 | 81.5 ± 3.8 | 88.1 ± 0.6 | 87.6 ± 0.6 |
| Porosity (%) | 18.0 ± 3.6 | 18.5 ± 3.8 | 11.9 ± 0.6 | 12.4 ± 0.6 |
| Cortical Thickness (μm) | 91 ± 11 | 86 ± 13 | 171 ± 6 | 166 ± 4 |
| Total Area (mm2) | 1.6 ± 0.1 | 1.5 ± 0.2 | 1.8 ± 0.1 | 1.7 ± 0.1 |
| Bone Area (mm2) | 0.5 ± 0.1 | 0.5 ± 0.1 | 0.9 ± 0.1 | 0.8 ± 0.1 |
| Periosteal Perimeter (mm) | 4.4 ± 0.2 | 4.4 ± 0.2 | 4.7 ± 0.1 | 4.7 ± 0.1 |
| Endocortical Perimeter (mm) | 3.7 ± 0.2 | 3.6 ± 0.2 | 3.4 ± 0.1 | 3.4 ± 0.1 |
| Density of Material (mg HA/cm3) | 1060 ± 17 | 1058 ± 24 | 1269 ± 13 | 1262 ± 9 |
μCT was performed on distal femurs for trabecular bone and midshaft for cortical bone. Values are means ± SD.
Table 5.
Cancellous bone histomorphometry of 4 month old Lyz2Cre/WT;Nfatc2Δ/Δ male mice (Nfatc2Δ/Δ) and sex-matched littermate Nfatc2loxP/loxP controls.
| Control n = 6 |
Nfatc2Δ/Δ n = 6 |
|
|---|---|---|
| Distal Femur Trabecular Bone | ||
| Bone Volume/Tissue Volume (%) | 10.7 ± 3.2 | 9.7 ± 3.8 |
| Trabecular Number (1/mm) | 2.5 ± 0.3 | 2.4 ± 0.6 |
| Trabecular Thickness (μm) | 42.8 ± 11.7 | 40.5 ± 10.5 |
| Osteoblasts/Bone Perimeter (1/mm) | 7.2 ± 2.5 | 6.9 ± 3.5 |
| Osteoid Surface/Bone Surface (%) | 0.4 ± 0.3 | 0.6 ± 0.6 |
| Osteoclasts/Bone Perimeter (1/mm) | 2.1 ± 0.6 | 2.2 ± 0.6 |
| Eroded Surface/Bone Surface (%) | 1.6 ± 0.6 | 1.1 ± 0.6 |
| Mineral Apposition Rate (μm/day) | 1.0 ± 0.2 | 0.9 ± 0.1 |
| Mineralizing Surface/Bone Surface (%) | 1.7 ± 0.6 | 2.9 ± 1.8 |
| Bone Formation Rate (μm3/μm2/day) | 0.02 ± 0.01 | 0.03 ± 0.01 |
Bone histomorphometry was performed on saggital sections of distal femurs sagittal sections. Values are means ± SD.
DISCUSSION
In this study, the direct effects of NFATc2 on osteoclast differentiation and function were explored by the conditional inactivation of Nfatc2 in cells of the myeloid lineage. The present data revealed that Nfatc2 mRNA levels are decreased under the RANKL stimulation, but NFATc2 protein level is moderately increased during osteoclastogenesis, suggesting a possible post-translational regulation of NFATc2 expression during osteoclast differentiation. Previous studies reported that NFATc2 is recruited to the Nfatc1 promoter to promote amplification of NFATc1 by cooperating with NF-κB [Asagiri et al., 2005]. In the present study, Nfatc1 mRNA and NFATc1 protein levels were not affected by the downregulation of NFATc2 possibly because NFATc1 is sufficient for its own autoamplification suggesting that NFATc2 is dispensable for the induction of NFATc1. Although NFATc1 levels were not different between Nfatc2Δ/Δ and control osteoclasts, Nfatc2Δ/Δ osteoclasts exhibited a small sealing zone and decreased mRNA levels of osteoclast differentiation markers, such as Acp5, Ctsk and Calcr. Accordingly, bone resorptive activity of Nfatc2Δ/Δ osteoclasts was reduced suggesting a direct role of NFATc2 in osteoclast maturation and bone resorption. However, this effect was not translated in vivo and inactivation of Nfatc2 in Lyz2-expressing cells did not cause an obvious skeletal phenotype. This would indicate that NFATc2 is dispensable for the differentiation or function of cells of the osteoclast lineage in vivo. These findings are consistent with previous in vivo work demonstrating that the global inactivation of Nfatc2 does not alter osteoclast number or bone resorption in vivo [Asagiri et al., 2005]. It is likely that the absence of a phenotype following the deletion of Nfatc2 in vivo is because of genetic compensation by Nfatc1 which is considered essential for osteoclastogenesis [Asagiri et al., 2005].
Previous work has demonstrated that the delivery of caNFATc2 to cells of the osteoclast lineage can induce osteoclastogenesis and rescue the Nfatc1 null phenotype confirming that in the absence of NFATc1, NFATc2 has the capacity to induce osteoclast differentiation [Asagiri et al., 2005]. A similar role of NFATc2 in osteoclastogenesis and bone resorption was observed following the transgenic expression of caNFATc2 under the control of the Acp5 promoter. Acp5-Nfatc2 transgenic mice exhibit osteopenia due to increased osteoclast number and bone resorption [Ikeda et al., 2006]. The mechanism responsible for the increase in osteoclast maturation seemed to involve the induction of c-Src. It is possible that caNFATc2 targets signals similar to those targeted by NFATc1 to induce osteoclastogenesis, but its downregulation in the presence of NFATc1 results in a modest reduction in osteoclast differentiation.
Although NFATc1 and NFATc2 have important interactions with Notch signaling in osteoblasts, there is no evidence at present that these occur in the myeloid lineage [Zanotti et al., 2013]. Notch induces Nfatc2 expression in osteoblasts by post-transcriptional mechanisms, and NFATc2 competes for binding to DNA with the Notch transcriptional complex resulting in the downregulation of Notch signaling in what seems to be a negative feed-back regulatory loop. However, we have not detected consistent increases in Nfatc2 mRNA expression in the myeloid lineage in models of Notch2 gain-of-function (E. Canalis et al., unpublished observations).
Similar to the results observed with the delivery of caNFATc2 to cells of the myeloid lineage, the transgenic delivery of caNFATc2 under the control of the 3.6 kb Col1a1 promoter to cells of the osteoblast lineage results in a biological response. Col1a1-Nfatc2 transgenic mice exhibit osteopenia secondary to decreased bone formation [Zanotti and Canalis, 2015]. NFATc1 and NFATc2 also play an important role in chondrogenesis, and disruption of Nfatc1/c2 is associated with osteoarthritis, and alterations in NFAT signaling may explain bone and cartilage disorders associated with inflammation [Greenblatt et al., 2013; Zanotti and Canalis, 2013]. NFATc1 and NFATc2 oppose osteoarthritis progression, NFATc1 inhibit chondrogenesis, and the deletion of Nfatc1 and Nfatc2 result in entheseal osteochondromas reflecting a restrictive role of these transcription factors on osteochondral growth [Ge et al., 2016; Greenblatt et al., 2013; Zanotti and Canalis, 2013].
In conclusion, our studies reveal that the inactivation of Nfatc2 in osteoclast precursors decreases osteoclast differentiation and bone resorption in vitro but this effect is not translated in vivo possibly because of compensation by NFATc1.
Acknowledgments
Contract grant sponsor: National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS); Contract grant number: AR068160
The authors thank D. Fremont for M-CSF cDNA, M. Glogauer for Tnfsf11 cDNA, A. Rao for Nfatc1 and Nfatc2 cDNA, Tabitha Eller and David Bridgewater for technical assistance, and Mary Yurczak for secretarial support.
ABBREVIATIONS
The abbreviations used are
- α-MEM
α-minimum essential medium
- bp
base pair
- BMM
bone marrow-derived macrophage
- ca
constitutive active
- ES
embryonic stem
- FBS
fetal bovine serum
- Frt
flippase recognition target
- kb
kilobase pair
- MC1-HSV-TK
MC1 driven herpes simplex virus-thymidine kinase
- M-CSF
macrophage colony stimulating factor
- μCT
microcomputed tomography
- NFAT
Nuclear factor of activated T cells
- PGK
phosphoglycerate kinase
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- qRT-PCR
quantitative reverse transcription-PCR
- RANKL
receptor activator of NF-κB ligand
- SDS
sodium dodecyl sulfate
- SMI
structure model index
- TRAP
tartrate resistant acid phosphatase
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest with the contents of this article.
References
- Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM, Ostrowski MC, Olsen BR, Glimcher LH. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J Clin Invest. 2008;118:3775–3789. doi: 10.1172/JCI35711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asagiri M, Sato K, Usami T, Ochi S, Nishina H, Yoshida H, Morita I, Wagner EF, Mak TW, Serfling E, Takayanagi H. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med. 2005;202:1261–1269. doi: 10.1084/jem.20051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer W, Rauner M, Haase M, Kujawski S, Arabanian LS, Habermann I, Hofbauer L, Ehninger G, Kiani A. Osteomyelosclerosis, anemia and extramedullary hematopoiesis in mice lacking the transcription factor NFATc2. Haematologica. 2011;96:1580–1588. doi: 10.3324/haematol.2011.042515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 2010;25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- Buchholz F, Ringrose L, Angrand PO, Rossi F, Stewart AF. Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res. 1996;24:4256–4262. doi: 10.1093/nar/24.21.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Sanjay A, Yu J, Zanotti S. An Antibody to Notch2 Reverses the Osteopenic Phenotype of Hajdu-Cheney Mutant Male Mice. Endocrinology. 2017;158:730–742. doi: 10.1210/en.2016-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Smerdel-Ramoya A, Durant D, Economides AN, Beamer WG, Zanotti S. Nephroblastoma Overexpressed (NOV) Inactivation Sensitizes Osteoblasts To Bone Morphogenetic Protein-2 But NOV Is Dispensable For Skeletal Homeostasis. Endocrinology. 2010a;151:221–233. doi: 10.1210/en.2009-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Zanotti S, Beamer WG, Economides AN, Smerdel-Ramoya A. Connective Tissue Growth Factor Is Required for Skeletal Development and Postnatal Skeletal Homeostasis in Male Mice. Endocrinology. 2010b;151:3490–3501. doi: 10.1210/en.2010-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Hou G, Bendeck MP. Glycogen synthase kinase 3beta regulation of nuclear factor of activated T-cells isoform c1 in the vascular smooth muscle cell response to injury. Exp Cell Res. 2008;314:2919–2929. doi: 10.1016/j.yexcr.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109(Suppl):S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR, Parfitt AM. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28:2–17. doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakin GS, Hadjantonakis AK. Production of chimeras by aggregation of embryonic stem cells with diploid or tetraploid mouse embryos. Nat Protoc. 2006;1:1145–1153. doi: 10.1038/nprot.2006.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Ge X, Tsang K, He L, Garcia RA, Ermann J, Mizoguchi F, Zhang M, Zhou B, Zhou B, Aliprantis AO. NFAT restricts osteochondroma formation from entheseal progenitors. JCI Insight. 2016;1:e86254. doi: 10.1172/jci.insight.86254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertsenstein M, Nutter LM, Reid T, Pereira M, Stanford WL, Rossant J, Nagy A. Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLoS One. 2010;5:e11260. doi: 10.1371/journal.pone.0011260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt V, Canalis E, Stadmeyer L, Bouxsein ML. Age-Related Changes in Trabecular Architecture Differ in Female and Male C57BL/6J Mice. J Bone Miner Res. 2007;22:1197–1207. doi: 10.1359/jbmr.070507. [DOI] [PubMed] [Google Scholar]
- Greenblatt MB, Ritter SY, Wright J, Tsang K, Hu D, Glimcher LH, Aliprantis AO. NFATc1 and NFATc2 repress spontaneous osteoarthritis1. Proc Natl Acad Sci U S A. 2013;110:19914–19919. doi: 10.1073/pnas.1320036110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge MR, Ranger AM, Charles de la BF, Hoey T, Grusby MJ, Glimcher LH. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity. 1996;4:397–405. doi: 10.1016/s1074-7613(00)80253-8. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Nishimura R, Matsubara T, Hata K, Reddy SV, Yoneda T. Activation of NFAT signal in vivo leads to osteopenia associated with increased osteoclastogenesis and bone-resorbing activity. J Immunol. 2006;177:2384–2390. doi: 10.4049/jimmunol.177.4.2384. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Nishimura R, Matsubara T, Tanaka S, Inoue J, Reddy SV, Hata K, Yamashita K, Hiraga T, Watanabe T, Kukita T, Yoshioka K, Rao A, Yoneda T. Critical roles of c-Jun signaling in regulation of NFAT family and RANKL-regulated osteoclast differentiation. J Clin Invest. 2004;114:475–84. doi: 10.1172/JCI19657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida N, Hayashi K, Hoshijima M, Ogawa T, Koga S, Miyatake Y, Kumegawa M, Kimura T, Takeya T. Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J Biol Chem. 2002;277:41147–56. doi: 10.1074/jbc.M205063200. [DOI] [PubMed] [Google Scholar]
- Koga T, Matsui Y, Asagiri M, Kodama T, dC B, Nakashima K, Takayanagi H. NFAT and Osterix cooperatively regulate bone formation. Nat Med. 2005;11:880–885. doi: 10.1038/nm1270. [DOI] [PubMed] [Google Scholar]
- Kouadjo KE, Nishida Y, Cadrin-Girard JF, Yoshioka M, St-Amand J. Housekeeping and tissue-specific genes in mouse tissues. BMC Genomics. 2007;8:127. doi: 10.1186/1471-2164-8-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay JM, Friis-Hansen L, Gillespie PJ, Samuelson LC. Rapid confirmation of gene targeting in embryonic stem cells using two long-range PCR techniques. Transgenic Res. 1998;7:135–140. doi: 10.1023/a:1008876526826. [DOI] [PubMed] [Google Scholar]
- Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N, Kang JS, Miyamoto T, Suda T, Lee SK, Pignolo RJ, Koczon-Jaremko B, Lorenzo J, Choi Y. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med. 2006;12:1403–9. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M. Controlled gene activation and inactivation in the mouse. Front Biosci. 2006;11:313–327. doi: 10.2741/1799. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Galson DL, Zhao C, Peng L, Laplace C, Wang KZ, Bachler MA, Amano H, Aburatani H, Ishikawa H, Wagner EF. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J Biol Chem. 2004;279:26475–80. doi: 10.1074/jbc.M313973200. [DOI] [PubMed] [Google Scholar]
- Monticelli S, Rao A. NFAT1 and NFAT2 are positive regulators of IL-4 gene transcription. Eur J Immunol. 2002;32:2971–2978. doi: 10.1002/1521-4141(2002010)32:10<2971::AID-IMMU2971>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Okamura H, Aramburu J, Garcia-Rodriguez C, Viola JP, Raghavan A, Tahiliani M, Zhang X, Qin J, Hogan PG, Rao A. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6:539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- Olson EN, Arnold HH, Rigby PW, Wold BJ. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham CT, MacIvor DM, Hug BA, Heusel JW, Ley TJ. Long-range disruption of gene expression by a selectable marker cassette. Proc Natl Acad Sci U S A. 1996;93:13090–13095. doi: 10.1073/pnas.93.23.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluck A, Klasen C. Generation of chimeras by morula aggregation. Methods Mol Biol. 2009;561:219–229. doi: 10.1007/978-1-60327-019-9_14. [DOI] [PubMed] [Google Scholar]
- Ranger AM, Gerstenfeld LC, Wang J, Kon T, Bae H, Gravallese EM, Glimcher MJ, Glimcher LH. The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J Exp Med. 2000;191:9–22. doi: 10.1084/jem.191.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, de la Brousse FC, Hoey T, Mickanin C, Baldwin HS, Glimcher LH. The transcription factor NF-ATc is essential for cardiac valve formation. Nature. 1998;392:186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitara D, Aliprantis AO. Transcriptional regulation of bone and joint remodeling by NFAT. Immunol Rev. 2010;233:286–300. doi: 10.1111/j.0105-2896.2009.00849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Blair HC, Peng Y, Zaidi N, Adebanjo OA, Wu XB, Wu XY, Iqbal J, Epstein S, Abe E, Moonga BS, Zaidi M. Calcineurin regulates bone formation by the osteoblast. Proc Natl Acad Sci U S A. 2005;102:17130–17135. doi: 10.1073/pnas.0508480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, Taniguchi T. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, Akira S. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lebowitz D, Sun C, Thang H, Grynpas MD, Glogauer M. Identifying the relative contributions of Rac1 and Rac2 to osteoclastogenesis. J Bone Miner Res. 2008;23:260–70. doi: 10.1359/jbmr.071013. [DOI] [PubMed] [Google Scholar]
- Winslow MM, Pan M, Starbuck M, Gallo EM, Deng L, Karsenty G, Crabtree GR. Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Dev Cell. 2006;10:771–782. doi: 10.1016/j.devcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Yeo H, Beck LH, Thompson SR, Farach-Carson MC, McDonald JM, Clemens TL, Zayzafoon M. Conditional disruption of calcineurin B1 in osteoblasts increases bone formation and reduces bone resorption. J Biol Chem. 2007;282:35318–35327. doi: 10.1074/jbc.M702435200. [DOI] [PubMed] [Google Scholar]
- Zanotti S, Canalis E. Notch suppresses nuclear factor of activated T cells (Nfat) transactivation and Nfatc1 expression in chondrocytes. Endocrinology. 2013;154:762–772. doi: 10.1210/en.2012-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Canalis E. Activation of Nfatc2 in Osteoblasts Causes Osteopenia. J Cell Physiol. 2015;230:1689–1695. doi: 10.1002/jcp.24928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Smerdel-Ramoya A, Canalis E. Nuclear Factor of Activated T-cells (Nfat)c2 Inhibits Notch Signaling in Osteoblasts. J Biol Chem. 2013;288:624–632. doi: 10.1074/jbc.M112.340455. [DOI] [PMC free article] [PubMed] [Google Scholar]