Abstract
Colorectal cancer remains a leading malignancy in humans. The importance of epigenetic modification in the development of this disease is now being recognized. The reversible and dynamic nature of epigenetic modifications provides a promising strategy in colorectal cancer chemoprevention and treatment. Luteolin (LUT), a flavone dietary phytochemical, can modulate various signaling pathways involved in carcinogenesis. Many studies have demonstrated that LUT inhibits colorectal carcinogenesis by activating the Nrf2/ARE pathway. However, the potential epigenetic mechanism underlying Nrf2/ARE pathway activation remains unclear. In this study, we aimed to explore the anticancer potential of LUT in human colon cancer cells and the epigenetic regulation of the Nrf2/ARE pathway. Specifically, our data showed that LUT suppressed cell proliferation and cellular transformation of HCT116 and HT29 cells in a dose-dependent manner. Additionally, qPCR and western blotting were performed to determine the mRNA and protein expression of Nrf2 and its downstream genes after LUT treatment. Bisulfite genomic sequencing revealed that methylation of the Nrf2 promoter region was decreased by LUT, corresponding with the increased mRNA expression of Nrf2. Decreased protein levels and enzyme activities of epigenetic modifying enzymes, such as DNMTs and HDACs, were also observed in LUT-treated HCT116 cells. In summary, our findings suggest that LUT may exert its anti-tumor activity in part via epigenetic modifications of the Nrf2 gene with subsequent induction of its downstream antioxidative stress pathway.
Keywords: colorectal cancer, luteolin, Nrf2, DNA methylation
1. Introduction
Colorectal cancer (CRC) remains one of the leading contributors to cancer-related mortality and morbidity worldwide [Siegel et al., 2017]. Increasing evidence has revealed that accumulation of genetic and epigenetic alterations in colon epithelial cells transform them into adenocarcinomas [Guo et al., 2018; van Engeland et al., 2011]. Accumulating evidence has shown that high DNA methylation patterns within background mucosa might predispose patients to CRC [Ally et al., 2009; Figueiredo et al., 2009]. Emerging studies have indicated that aberrant DNA methylation serves as a crucial driving factor in CRC progression and metastasis. For example, the aberrant methylation of the CXCL12 gene can catalyze the metastatic behavior of colon cancer cells [Figueiredo et al., 2009], and multiple inactive genes, including CSLC12, TIMP3, ID4, and IRF8, with methylated promoter regions provide a clonal growth advantage, resulting in greater malignant phenotypes [Kim et al., 2010]. Epidemiological evidence indicates that the majority of CRC cases are sporadic, which may be greatly attributable to nutritional factors, and these dietary influences may account for oxidative metabolism by inducing a cascade of molecular alterations in cells and tissues [Pandurangan and Esa, 2013; Stigliano et al., 2014]. Increasing reports have demonstrated that oxidative stress can induce various types of cell dysregulation, such as DNA damage, mutagenesis and lipid peroxidation [Mathers et al., 2007]. Additionally, clinical studies have revealed a close link between oxidative stress biomarkers and the status and development of CRC [Aguirre-Portoles et al., 2017; Souglakos, 2007; Wu et al., 2017].
Nuclear factor erythroid 2-related factor 2 (Nrf2) has long been regarded as an important factor in protecting cells from oxidant stress, due to its regulation of various phase II detoxifying enzymes, such as hemeoxygenase-1 (HO-1), NADP(H):quinone oxidoreductase-1 (NQO1), glutathione S-transferase (GST), UDP-glucuronosyltransferase (UGT) and glutamate-cysteine ligase (GCL) [Leinonen et al., 2014; Na and Surh, 2014; Shelton and Jaiswal, 2013]. Ishaq et al. reported that Nrf2 plays a defensive role in HT29 cancer cells by inducing apoptosis via activating caspase-3/7 [Ishaq et al., 2014] and protects against oxidative stress-induced genotoxicity [Wondrak et al., 2010]. Furthermore, Nrf2-deficient (Nrf2−/−) mice are more susceptible to oxidative stress-induced diseases and chemical-induced DNA damage, which increases their risk for certain types of cancers (e.g., stomach, colorectal and skin) compared with that of wild-type mice [Khor et al., 2008; Saw et al., 2011; Xu et al., 2006]. For example, Khor et al. reported that compared with azoxymethane- and DSS-treated wild-type (WT) mice, azoxymethane- and DSS-treated Nrf2-knockout mice exhibited a higher tumor incidence (80% versus 29%, respectively) and the Nrf2-knockout mice had increased expression of inflammatory markers in tumor tissues (cyclooxygenase-2, 5-lipoxygenase, prostaglandin E2, and leukotriene B4 expression) and in inflamed colonic mucosa (nitrotyrosine expression). The underlying mechanism by which Nrf2 protects against chemical-induced carcinogenesis may be partly due to its ability to decrease cellular reactive oxygen species (ROS) levels and DNA damage [Morito et al., 2003], protecting cells against potentially harmful entities.
Luteolin (LUT), a common flavonoid derived from vegetables, fruits, and herbs, exerts various biological effects, including anti-inflammatory, anti-allergic, anticancer, antioxidant, and other beneficial activities. LUT inhibits critical events associated with carcinogenesis, including cell invasion, metastasis, transformation, and angiogenesis, by inhibiting transcription factors, kinase modification, and cell cycle arrest and inducing apoptosis [Birt et al., 2001; Lin et al., 2008]. Pandurangan AK et al. reported that LUT induced growth arrest in colon cancer HCT15 cells through the Wnt/β-catenin/GSK-3β signaling pathway. LUT was also found to effectively induce apoptosis through caspase-3/7-dependent pathways [Pandurangan et al., 2013; Wang et al., 2004].
Studies have also suggested that LUT could significantly decrease the number and size of colon polyps. When administered as a supplementation to aspirin in an animal study, LUT significantly decreased CEA, COX-2, and oxidative stress and increased antioxidant markers [Osman et al., 2015]. The anti-tumorigenesis effect was further demonstrated in a series of azoxymethane-induced mouse colon carcinogenesis models [Pandurangan et al., 2014b; Pandurangan et al., 2014c]. Interestingly, the anti-tumor activity of LUT is involved in oxidative stress homeostasis.
Recently, investigations have revealed that epigenetic modifications may be the underlying mechanism of the anti-neoplastic nature of LUT. For example, LUT has anticancer activity by up-regulating p16 expression through epigenetic alterations [Krifa et al., 2014]. In our previous study, LUT showed a dose-dependent activation of the Nrf2/ARE pathway in HepG2 cells [Paredes-Gonzalez et al., 2015]. However, it remains unclear how LUT modulates the CpG methylation of the Nrf2 gene and other epigenetic enzymes. This study aimed to investigate the effect of LUT in inhibiting the viability and colony formation of HCT116 cells and to determine the potential epigenetic mechanisms by which LUT activates the Nrf2/ARE pathway.
2. Materials and Methods
2.1. Materials and Chemicals
LUT (Figure 1A for structure; purity: > 98%) was obtained from Dalian Meilun Biotech Corp (Dalian, China). Dulbecco’s Modified Eagle’s Medium (DMEM), fetal bovine serum (FBS), penicillin-streptomycin (10,000 U/ml), and trypsin-EDTA were supplied by Gibco (Grand Island, NY, USA). A Cell-Titer 96 Aqueous One Solution Cell Proliferation (MTS) Assay Kit was obtained from Promega (Madison, WI, USA). Platinum Taq DNA polymerase was purchased from Takara (Mountain View, CA, USA). Power SYBR Green PCR Master Mix was purchased from Applied Biosystems (Carlsbad, CA, USA). Tris-HCl precast gels, turbo transfer buffer, and PVDF membranes were obtained from Bio-Rad (Hercules, CA, USA). Tris-Glycine-SDS running buffer and Super Signal enhanced chemiluminescent substrate were purchased from Boston BioProducts (Ashland, MA, USA) and Thermo Scientific (Rockford, IL, USA), respectively. An antibody against Nrf2 was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Protease inhibitor cocktail, radioimmunoprecipitation assay (RIPA) buffer, and antibodies against HO-1, NQO1, β-actin, HDACs (HDAC1-7) and DNMTs (DNMT1, DNMT3a/b) were supplied by Cell Signaling Technology (Beverly, MA, USA).
Figure 1.
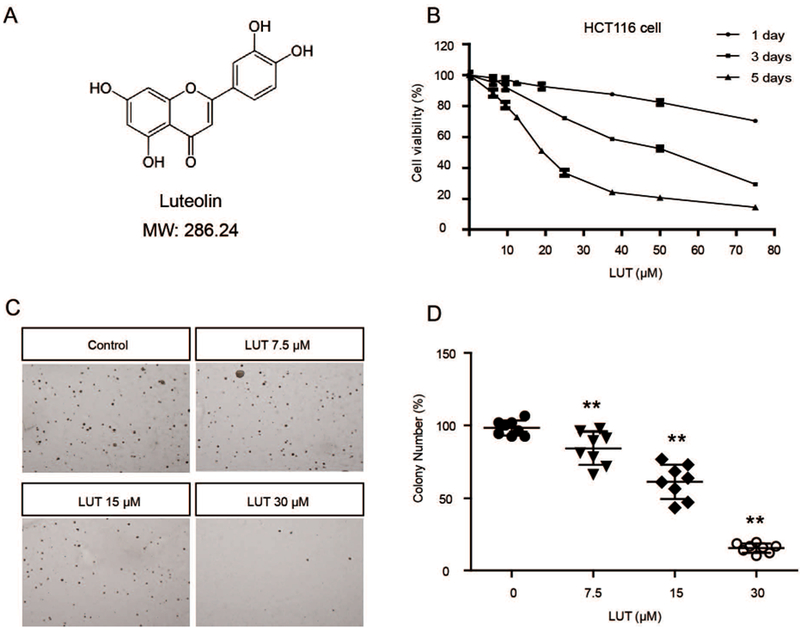
LUT inhibited the cell viability and anchorage-independent growth of HCT116 cells. (A) Structure and molecular weight of LUT. (B) LUT inhibited HCT116 cell viability in a dose-dependent manner. HCT116 cells were plated in 96-well plates at an initial density of 3000 cells/well then treated with various concentrations of LUT for 1, 3, or 5 days. Cell viability was detected using the MTS assay. (C, D) LUT inhibited the anchorage-independent growth of HCT116 cells. Eight thousand cells were seeded in soft agar containing 0.1% DMSO or various concentrations of LUT in 6-well plates for 14 days. Representative images of colonies are shown in C. The relative colony numbers from three independent experiments are shown in D. The data are presented as the mean ± SD. * P < 0.05 and ** P < 0.01 versus the control group.
All other chemicals, unless otherwise noted, were obtained from Sigma (St. Louis, MO, USA).
2.2. Cell Culture and Treatment
The human colorectal adenocarcinoma HCT116 cell line was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). HCT116 cells were routinely maintained in DMEM supplemented with 10% FBS at 37°C in a humidified 5% CO2 atmosphere.
HCT116 cells were seeded in 10-cm plates, incubated overnight to allow for adherence and then exposed to either 0.1% DMSO (vehicle control), 2.5 μM 5-Aza, or various concentrations of LUT in a 5% FBS-containing DMEM for 3 or 5 days. The medium was changed every other day. One day prior to cell harvesting, 100 nM TSA was added to the 5-Aza group. Then, all cells were collected for isolation of DNA, RNA, and protein.
2.3. Cell Viability Assay
HCT116 cells were seeded in 96-well plates at an initial density of 3000 cells/well for 1-, 3- or 5-day treatments. After a 24-h incubation, the cells were treated with either 0.1% DMSO (control) or LUT at various concentrations in DMEM with 5% FBS for 1, 3, or 5 days, and the medium was changed every other day. The cytotoxicity of LUT was determined using a Cell Titer 96 AQueous One Solution Cell Proliferation (MTS) assay kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
2.4. Anchorage-Independent Growth Assay
HCT116 cells (2 × 104 cells/ml) were suspended in 1 mL of basal medium Eagle (BME) containing 0.33% agar and plated over 3 mL of a solidified BME consisting of 0.5% agar and 10% FBS in 6-well plates in the presence of 7.5 μM, 15 μM or 30 μM LUT. Then, the cells were maintained at 37°C in a humidified 5% CO2 incubator for 2 weeks. Images of cell colonies were captured by the ACT-1 software (Version 2.20) using a Nikon microscope. Colonies were counted with the ImageJ program (Version 1.48d; NIH, Bethesda, MD, USA).
2.5. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qPCR)
HCT116 cells were seeded in 6-well plates at a density of 2 × 105 cells/well. After incubation for 24 h, the cells were treated with 0.1% DMSO (control), 2.5 μM 5-Aza and 100 nM TSA, or LUT at 15 and 30 μM for 3 days. The medium was changed every other day. Total RNA was extracted from the treated HCT116 cells using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and 1 μg of total RNA was reverse-transcribed to cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). The relative mRNA expression levels of Nrf2, HO-1, and NQO1 were determined by qPCR on an ABI7900HT system. The primer sequences for Nrf2, HO-1 and NQO1 are shown in Table 1.
Table 1.
qPCR primers used in the study
| Gene | Sequence (5’ - 3’) | |
|---|---|---|
| Nrf2 | sense | CAAAAGGAGCAAGAGAAAGCC |
| antisense | TCTGATTTGGGAATGTGGGC | |
| HO-1 | sense | GTGATGGAGCGTCCACAGC |
| antisense | TTGGTGGCCTCCTTCAAGG | |
| NQO1 | sense | TCACCGAGAGCCTAGTTCC |
| antisense | TCATGGCATAGTTGAAGGAACG | |
| Gapdh | sense | ACATCGCTCAGACACCATG |
| antisense | TGTAGTTGAGGTCAATGAAGGG |
2.6. Preparation of Protein Lysates and Western Blotting
Following treatment for 3 days, protein lysates of the treated cells were collected using RIPA buffer supplemented with a protein inhibitor cocktail (Sigma), and the concentration was measured using the BCA kit ((Pierce, Rockford, IL, USA). Identical concentrations of protein (20 μg) were separated by 4% to 15% SDS-polyacrylamide gel (Bio-Rad, Hercules, CA, USA) electrophoresis (SDS-PAGE) and then electro-transferred to PVDF membranes (Millipore, Billerica, MA, USA). After blocking with 5% BSA (Fisher Scientific, Pittsburgh, PA, USA) in Tris-buffered saline-0.1% Tween 20 (TBST) buffer (Boston BioProducts, Ashland, MA, USA), the PVDF membranes were incubated with a specific primary antibody and sequential horseradish peroxidase-conjugated secondary antibodies. The blots were visualized using Super Signal West Femto chemiluminescent substrate (Pierce, Rockford, IL, USA) and measured using a Gel Documentation 2000 system (Bio-Rad, Hercules, CA, USA).
2.7. DNA Isolation and Bisulfite Genomic Sequencing (BGS)
HCT116 cells were seeded in 10-cm plates for 24 h and then exposed to 0.1% DMSO, 2.5 μM 5-Aza and 100 nM TSA or LUT at 15 μM and 30 μM for 5 days. The medium was changed every 2 days. On day 5, the cells were collected to perform DNA isolation using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA). Then, 750 ng of genomic DNA was subjected to bisulfite conversion using EZ DNA Methylation Gold Kits (Zymo Research Corp., Orange, CA, USA) following the manufacturer’s instructions. To obtain products for sequencing, the converted DNA was amplified by PCR using Platinum PCR Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) and primers including the first three CpGs of the human Nrf2 gene spanning methylated CpG sites from −1576 to −1165, with the translational start sites (TSS) referenced as +1, as previously described [Khor et al., 2014; Yang et al., 2017b]. The forward and reverse primer sequences were 5′-TGT AAT TTA GAG AAA GTA AGT TTT GT-3′and 5′-ATT TCT CCT TAT TAC TTC CAA AT-3′. A TOPO TA Cloning kit (Invitrogen, Grand Island, NY, USA) was used to clone the PCR products into vector pCR4 TOPO. Plasmid DNA from at least 10 colonies per group from three independent repeats was amplified and purified with a QIAprep Spin Miniprep Kit (Qiagen) before sequencing (GeneWiz, South Plainfield, NJ, USA).
2.8. DNMT and HDAC Activity Assay
DNMT and HDAC activity/inhibition assay kits (EpiGentek, Farmingdale, NY, USA) were used to determine the activities of these enzymes. Nuclear protein extraction was conducted using the NEPER Nuclear and Cytoplasmic Protein Extraction Kit (Thermo Scientific, Pittsburgh, PA, USA), and the relative enzyme activity was estimated based on the ratio of the LUT treatment group to the control group with normalization to the protein amount.
2.9. Statistical Analyses
The data are presented as the mean ± SD. The statistical analyses were performed by Student’s t-test, and P values less than 0.05 were considered significant and are indicated with *; P values less than 0.01 are indicated with **.
3. Results
3.1. Inhibitory effect of LUT on cell viability and anchorage-independent growth capacity of human colon cancer cells
The LUT structure is shown in Figure 1A. To determine the cytotoxicity of LUT, an MTS assay was performed to analyze the viability of HCT116 cells after treatment with LUT for 1, 3, and 5 days. The results of the MTS assay showed that LUT reduced cell viability in a dose-dependent manner (Figure 1B). One-day and 3-day treatments with LUT were less toxic than the 5-day treatment. Anchorage-independent growth capacity is an indicator of tumorigenic and metastatic potential. A soft agar assay was employed to investigate the effect of LUT on inhibiting the colony formation of HCT116 cells. As shown in Figure 1C and 1D, LUT at 7.5, 15, and 30 μM significantly decreased the colony formation ability to 85.45%, 61.40%, and 15.41%, respectively. These results indicate that LUT plays an important role in inhibiting the cell viability and tumorigenicity of HCT116 cells. We also performed the same experiments in human colon adenocarcinoma HT29 cells and similar results were observed (Figure S1).
3.2. LUT activated Nrf2 and its target genes on both mRNA and protein levels
We previously demonstrated that both protein and mRNA expression of Nrf2 and Nrf2 downstream genes negatively correlate with the methylation ratio of the Nrf2 gene promoter region in colon cancer cells [Kang et al., 2016; Zhao et al., 2015]. In this study, the quantification of the mRNA level of Nrf2 and its target enzymes in HCT116 cells was performed by qPCR. As shown in Figure 2A-C, LUT increased the mRNA expression of Nrf2, HO-1, and NQO1 in a dose-dependent manner. Consistent with the qPCR results, LUT increased the protein levels of Nrf2 and NQO1 in a dose-dependent manner. However, no significant difference of HO-1 protein expression was observed between control and LUT-treated groups (Figure 2E). Representative images of western blots are shown in Figure 2D. These data suggest that LUT can increase mRNA and protein expression and thereby activate the expression of Nrf2-target antioxidant detoxifying enzymes in HCT116 cells.
Figure 2.
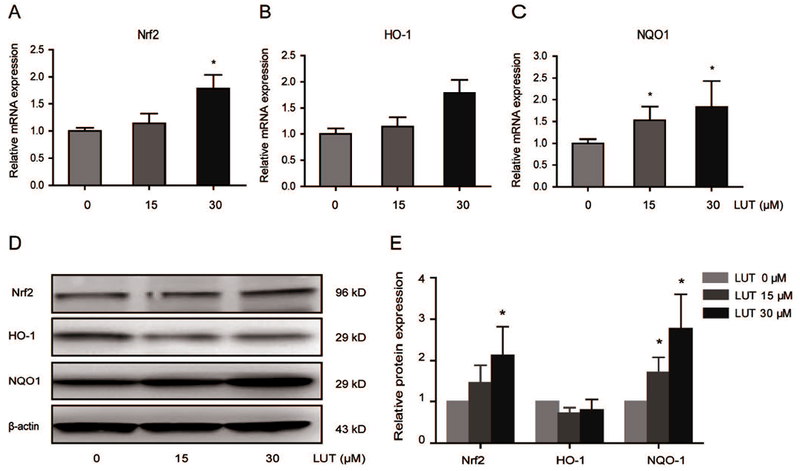
LUT activated Nrf2 and its downstream genes. (A-C) mRNA expression of Nrf2, HO-1, and NQO1 in HCT116 cells treated with LUT. HCT116 cells were treated with various concentrations of LUT for 3 days, and total RNA was extracted and converted to cDNA. Relative gene expression was assessed by qPCR. (D-E) Protein expression of Nrf2, HO-1, and NQO-1 in HCT116 cells treated with LUT. HCT116 cells were treated with various concentrations of LUT for 3 days, and then protein levels were measured by western blotting. Representative images of blots are shown in D. The relative densities of the blots were measured by ImageJ software. The data are presented as the mean ± SD from three independent experiments, as shown in E. * P < 0.05 and ** P < 0.01 versus the control group.
3.3. LUT reduced the CpG methylation of the Nrf2 gene promoter region
In this study, we performed bisulfite sequencing to investigate whether LUT could affect the methylation status of the Nrf2 promoter region. Previous reports have demonstrated that LUT can inhibit DNMT enzyme activity and reverse the CpG hypermethylation of DNA [Kanwal et al., 2016]. In our study, hypermethylation of the three CpGs (59.2% methylation) was observed in HCT116 cells without LUT treatment. Treatment with a combination of 5-Aza and TSA, a positive control of demethylation, significantly decreased the CpG methylation level to 41%. Treatment with LUT at 15 μM and 30 μM also showed a pronounced reduction in the methylation of these CpG sites from 59.2% to 55.5% and 48%, respectively (Figure 3A and 3B).
Figure 3.
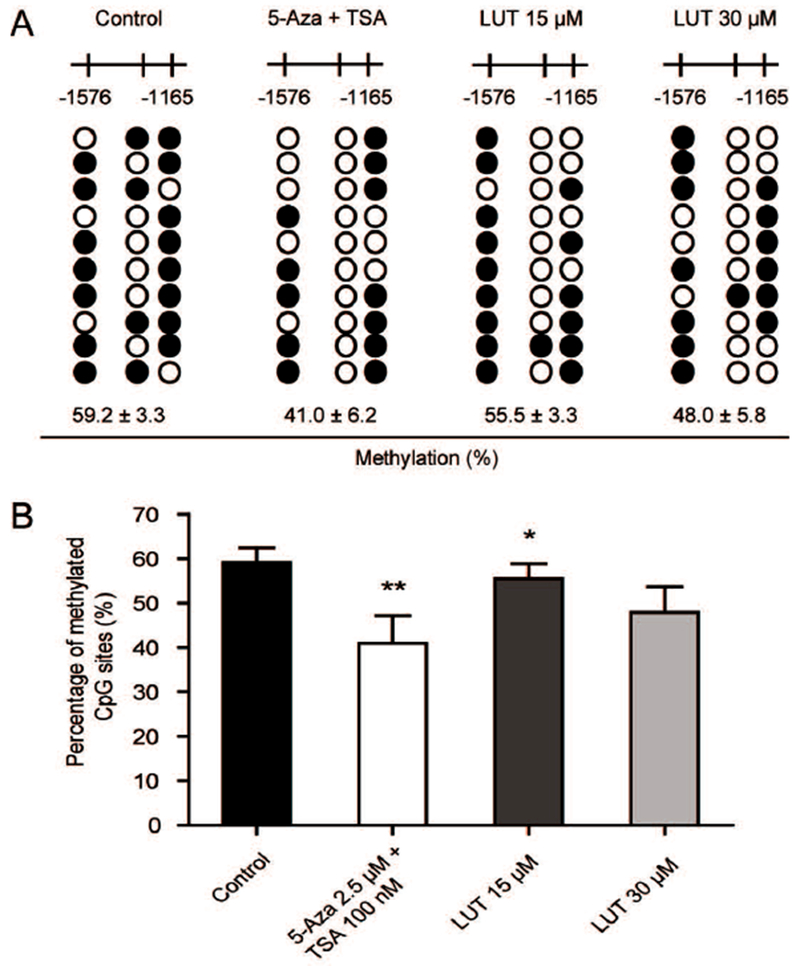
LUT reduced the CpG methylation ratio of the Nrf2 promoter region. (A-B) Detailed methylation patterns of 3 selected CpG sites (−1576 to −1165) in the promoter region of the Nrf2 gene in HCT116 cells were determined by bisulfite genomic sequencing. Filled dots indicate the methylated CpG sites, and open circles indicate the unmethylated CpG sites. HCT116 cells were treated with various concentrations of LUT or a positive control, 5-Aza and TSA, for 5 days, and genomic DNA was then bisulfite converted. Methylated cytosine was determined by Sanger sequencing. Ten clones were selected to represent three independent experiments. The methylation percentage was calculated from three independent experiments as the number of methylated CpG sites over the total number of CpG sites examined. The data are presented as the mean ± SD. * P < 0.05 and ** P < 0.01 versus the control group.
3.4. LUT altered the expression and activities of epigenetic modification enzymes in HCT116 cells
We performed western blotting to examine the effect of LUT on the protein expression and enzyme activity of epigenetic modifying enzymes to investigate the epigenetic mechanism by which LUT demethylated the Nrf2 promoter and increased Nrf2 transcription. DNMT1, DNMT3A, and DNMT3B are the most well-known DNA methyltransferases. As shown in Figure 4A and 4B, LUT at 15 μM and 30 μM significantly decreased the expression of DNMT1, DNMT3A and DNMT3B protein. Consistently, we found that the protein levels of HDAC1, HDAC2, HDAC3, HDAC6, and HDAC7 were significantly decreased by treatment with LUT for 5 days in HCT116 cells, while decreased expression of HDAC3, HDAC6, and HDAC7 was observed in a concentration-dependent manner (Figure 5A and 5B). In addition to the protein expression, the activities of these enzymes were also measured. As shown in Figure 4C and 5C, the combination of 5-Aza (2.5 μM) and TSA (100 nM) reduced the activities of DNMT and HDAC enzymes in HCT116 cells, and LUT also induced a comparable decrease in DNMT and HDAC activities. Interestingly, LUT at a low dose (15 μM) decreased HDAC activity more than LUT at a high dose (30 μM), while no such effect of higher inhibition was observed in DNMT activity.
Figure 4.
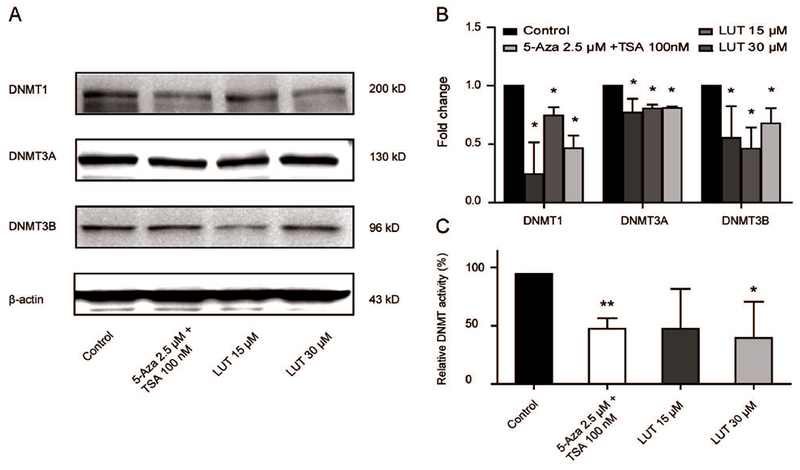
LUT decreased protein expression and enzyme activities of DNMTs in HCT116 cells. (A-B) Protein expression of DNMT1, DNMT3A, and DNMT3B in HCT116 cells treated with LUT. HCT116 cells were treated with various concentrations of LUT or 5-Aza and TSA for 3 days, and protein levels were measured by western blotting. Representative images of blots are shown in A. The relative densities of the blots were measured by the ImageJ software. The data are presented as the mean ± SD from three independent experiments. (C) Relative DNMT activities in HCT116 cells treated with various concentrations of LUT or 5-Aza and TSA for 3 days. The data are presented as the mean ± SD. * P < 0.05 and ** P < 0.01 versus the control group.
Figure 5.
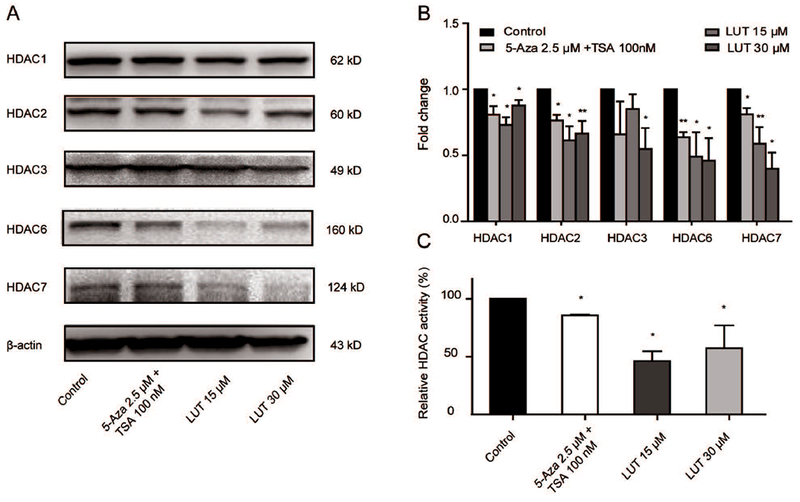
LUT decreased protein expression and enzyme activities of HDACs in HCT116 cells. (A-B) Protein expression of HDAC1/2/3/6/7 in HCT116 cells treated with LUT. HCT116 cells were treated with various concentrations of LUT or 5-Aza and TSA for 3 days, and protein levels were measured by western blotting. Representative images of blots are shown in A. The relative densities of the blots were measured by the ImageJ software. The data are presented as the mean ± SD from three independent experiments, as shown in B. (C) Relative HDAC activities in HCT116 cells treated with various concentrations of LUT or 5-Aza and TSA for 3 days. The data are presented as the mean ± SD. * P < 0.05 and ** P < 0.01 versus the control group.
4. Discussion
According to the latest report of Cancer Statistics in 2017, CRC remains among the top three types of tumors in all cancer incidence and deaths, in addition to nearly one million new cases of CRC diagnosed in 2017. Meanwhile, an estimated half of the new cases will turn into deaths and approximately 10% of cancer-caused deaths are due to CRC, suggesting that CRC is one of the most prevalent malignancies in humans [Siegel et al., 2017].
LUT is a bioflavonoid widely distributed in various fruits and vegetables. There are several reports suggesting the multiple actions of LUT in CRC [Osman et al., 2015; Pandurangan et al., 2014b; Xiao et al., 2017]. Previous reports have shown that LUT exerts dramatic inhibitory effects on colon carcinogenesis in an AOM-induced model, due to its strong antioxidant properties [Pandurangan et al., 2014a]. In our study, we employed an anchorage-independent colony assay to evaluate the tumorigenic and metastatic potential of HCT116 and HT29 cells in vitro. We found that the colony formation of HCT116 and HT29 was considerably attenuated by LUT in a dose-dependent manner (Figure 1C and S1), which is consistent with other published data and suggests, for the first time, that LUT has the potential to prevent colon carcinogenesis in vitro.
A large case-controlled study has revealed that a diet rich in fruit and vegetables has increased antioxidant potential, thus significantly decreasing the risk of CRC [La Vecchia et al., 2013]. Usually, the cellular levels of ROS are automatically altered by a natural antioxidant defense system to maintain redox homeostasis. Alternatively, ROS levels accumulate in breast, colon, pancreatic, prostate and other cancers [Afanas’ev, 2011; Kumar et al., 2008]. Nrf2, a redox-sensitive transcription factor, is stimulated by the presence of electrophonic chemicals and ROS, which contribute to the translocation of Nrf2 into the nucleus where it binds to antioxidant-responsive elements (AREs) and activates a series of cytoprotective genes, such as HO-1 and NQO1. The protective properties of Nrf2 have been supported by observations of decreased genotoxic damage, reduced cell proliferation and increased apoptosis of colon cancer cells [Tan et al., 2015; Trivedi et al., 2016; Xu et al., 2015], in addition to lower ROS levels in a carcinogen-induced CRC mouse model [Xi et al., 2013]. Recent studies have recognized the effect of LUT in modulating epigenetic mechanisms. For example, Kanwal1 et al. explored the dual interactions of three flavones with the methylation of genomic DNA, together with the trimethylation of lysine 27 at histone H3, and the results showed that dietary flavones could alter DNMT and HMT activities, which methylate DNA and histone proteins that regulate epigenetic modifications [Kanwal et al., 2016]. However, studies on the ability of LUT to epigenetically modify certain genes in CRC are relatively scarce. Thus, our present work provides evidence that LUT decreases the CpG methylation of the Nrf2 promoter and regulates an antioxidant gene potentially involved in the anchorage-independent growth of HCT116 cells. Furthermore, our data revealed that the demethylation phenomenon may be associated with LUT-mediated inhibition of DNMTs. DNMTs catalyze DNA methylation by transferring methyl groups to cytosine nucleotides. In mammals, DNMT1 is the most abundant enzyme and maintains methylation patterns following DNA replication and shows a preference for hemi-methylated DNA. The DNMT3a/b enzymes are responsible for de novo DNA methylation which is an essential mechanism of embryonic development and cell differentiation. DNA methylation in gene promoters correlates with low or no transcription by blocking transcription factors from binding.
In our previous study, curcumin, sulforaphane, and 3,3′-diindolylmethane were found to demethylate the Nrf2 promoter and re-stimulate Nrf2 signaling in the prostate of TRAMP mice and in TRAMP C1 cells, partly through the suppression of DNMTs and HDACs [Huang et al., 2017; Kong et al., 2013; Li et al., 2017; Su et al., 2013; Wu et al., 2013; Yang et al., 2017a; Zhang et al., 2016]. FN1, a curcumin analogue, restored Nrf2 expression and its downstream detoxification enzymes, which suppressed the colony formation of prostate cancer cells via epigenetic modification [Zhang et al., 2016]. Here, we showed that LUT suppressed either the protein expression or enzyme activity of DNMT1, DNMT3A, and DNMT3B in a dose-dependent manner in HCT116 cells, which in turn led to activation of Nrf2 and its downstream genes by demethylation of Nrf2 promoter.
Histone acetylation and deacetylation are the processes by which the lysine residues within the histone core of the nucleosome are acetylated and deacetylated as essential gene regulation. Acetylation removes the positive charge on the histones, thereby decreasing the interaction of the N termini of histones with the negatively charged phosphate groups of DNA. As a result, this relaxed chromatin structure facilitates gene transcription. The relaxation can be reversed by HDAC activity. HDAC inhibitors, such as TSA, suppress activity of HDACs and keep chromatin in a relaxed structure, which leads to increased transcription of the gene. In addition to DNA methylation, there is evidence that HDAC inhibitors activate the Nrf2/ARE signaling pathway and up-regulate Nrf2 downstream targets HO-1 and NQO1 in a cerebral ischemia mouse model [Wang et al., 2012]. Thus, histone deacetylation may also be involved in regulating the Nrf2/ARE pathway. Liu et al. reported the direct involvement of HDAC3 in the negative regulation of the Nrf2 pathway through NF-κB in response to inflammation-related stimuli [Liu et al., 2008]. Similarly, the impact of HDACs on the inhibition of Nrf2-mediated antioxidant defense in neuroinflammation has also been investigated [Correa et al., 2011]. In the present study, we found that LUT significantly down-regulated the enzyme expression and activity of HDACs in HCT116 cells (Figure 5A-C), possibly due to compromised HDAC activity after LUT treatment. Although the accurate mechanism underlying the interaction between LUT and Nrf2 activation requires further exploration, our results suggest that LUT may regulate the transcriptional activity of Nrf2 through epigenetic modifications.
In conclusion, our work confirmed the inhibitory role of LUT in the cell growth and colony formation of HCT116 cells. This work also confirmed the activation effect of LUT on the mRNA and protein expression of Nrf2 and its downstream phase II detoxifying and antioxidant enzymes HO-1 and NQO1. Most importantly, we demonstrated that LUT could epigenetically regulate Nrf2 expression through inhibiting DNMT and HDAC activities. These findings provide new insights into utilizing LUT for cancer prevention and treatment.
Supplementary Material
Acknowledgments
This work was supported in part by institutional funds and by R01 AT007065 from the National Center for Complementary and Integrative Health (NCCIH).
Footnotes
Conflicts of interest
None.
References
- Afanas’ev I. 2011. Reactive oxygen species signaling in cancer: comparison with aging. Aging Dis 2:219–30. [PMC free article] [PubMed] [Google Scholar]
- Aguirre-Portoles C, Fernandez LP, Ramirez de Molina A. 2017. Precision Nutrition for Targeting Lipid Metabolism in Colorectal Cancer. Nutrients 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ally MS, Al-Ghnaniem R, Pufulete M. 2009. The Relationship between Gene-Specific DNA Methylation in Leukocytes and Normal Colorectal Mucosa in Subjects with and without Colorectal Tumors. Cancer Epidemiology Biomarkers & Prevention 18:922–928. [DOI] [PubMed] [Google Scholar]
- Birt DF, Hendrich S, Wang W. 2001. Dietary agents in cancer prevention: flavonoids and isoflavonoids. Pharmacol Ther 90:157–77. [DOI] [PubMed] [Google Scholar]
- Correa F, Mallard C, Nilsson M, Sandberg M. 2011. Activated microglia decrease histone acetylation and Nrf2-inducible anti-oxidant defence in astrocytes: restoring effects of inhibitors of HDACs, p38 MAPK and GSK3beta. Neurobiol Dis 44:142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo JC, Grau MV, Wallace K, Levine AJ, Shen L, Hamdan R, Chen X, Bresalier RS, McKeown-Eyssen G, Haile RW, Baron JA, Issa JP. 2009. Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev 18:1041–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Wu R, Gaspar JM, Sargsyan D, Su ZY, Zhang C, Gao L, Cheng D, Li W, Wang C, Yin R, Fang M, Verzi MP, Hart RP, Kong AN. 2018. DNA Methylome and Transcriptome Alterations and Cancer Prevention by Curcumin in Colitis-accelerated Colon Cancer in Mice. Carcinogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wu R, Su ZY, Guo Y, Zheng X, Yang CS, Kong AN. 2017. A naturally occurring mixture of tocotrienols inhibits the growth of human prostate tumor, associated with epigenetic modifications of cyclin-dependent kinase inhibitors p21 and p27. J Nutr Biochem 40:155–163. [DOI] [PubMed] [Google Scholar]
- Ishaq M, Evans MD, Ostrikov KK. 2014. Atmospheric pressure gas plasma-induced colorectal cancer cell death is mediated by Nox2-ASK1 apoptosis pathways and oxidative stress is mitigated by Srx-Nrf2 anti-oxidant system. Biochim Biophys Acta 1843:2827–37. [DOI] [PubMed] [Google Scholar]
- Kang KA, Piao MJ, Ryu YS, Kang HK, Chang WY, Keum YS, Hyun JW. 2016. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget 7:40594–40620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwal R, Datt M, Liu X, Gupta S. 2016. Dietary Flavones as Dual Inhibitors of DNA Methyltransferases and Histone Methyltransferases. PLoS One 11:e0162956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor TO, Fuentes F, Shu L, Paredes-Gonzalez X, Yang AY, Liu Y, Smiraglia DJ, Yegnasubramanian S, Nelson WG, Kong AN. 2014. Epigenetic DNA methylation of antioxidative stress regulator NRF2 in human prostate cancer. Cancer Prev Res (Phila) 7:1186–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor TO, Huang MT, Prawan A, Liu Y, Hao XP, Yu SW, Cheung WKL, Chan JY, Reddy BS, Yang CS, Kong AN. 2008. Increased Susceptibility of Nrf2 Knockout Mice to Colitis-Associated Colorectal Cancer. Cancer Prevention Research 1:187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Lee J, Sidransky D. 2010. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev 29:181–206. [DOI] [PubMed] [Google Scholar]
- Kong AN, Zhang C, Su ZY. 2013. Targeting epigenetics for cancer prevention by dietary cancer preventive compounds--the case of miRNA. Cancer Prev Res (Phila) 6:622–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krifa M, Leloup L, Ghedira K, Mousli M, Chekir-Ghedira L. 2014. Luteolin Induces Apoptosis in BE Colorectal Cancer Cells by Downregulating Calpain, UHRF1, and DNMT1 Expressions. Nutrition and Cancer-an International Journal 66:1220–1227. [DOI] [PubMed] [Google Scholar]
- Kumar B, Koul S, Khandrika L, Meacham RB, Koul HK. 2008. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res 68:1777–85. [DOI] [PubMed] [Google Scholar]
- La Vecchia C, Decarli A, Serafini M, Parpinel M, Bellocco R, Galeone C, Bosetti C, Zucchetto A, Polesel J, Lagiou P, Negri E, Rossi M. 2013. Dietary total antioxidant capacity and colorectal cancer: a large case-control study in Italy. Int J Cancer 133:1447–51. [DOI] [PubMed] [Google Scholar]
- Leinonen HM, Kansanen E, Polonen P, Heinaniemi M, Levonen AL. 2014. Role of the Keap1-Nrf2 pathway in cancer. Adv Cancer Res 122:281–320. [DOI] [PubMed] [Google Scholar]
- Li W, Su ZY, Guo Y, Zhang C, Wu R, Gao L, Zheng X, Du ZY, Zhang K, Kong AN. 2017. Curcumin Derivative Epigenetically Reactivates Nrf2 Antioxidative Stress Signaling in Mouse Prostate Cancer TRAMP C1 Cells. Chem Res Toxicol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Shi RX, Wang X, Shen HM. 2008. Luteolin, a Flavonoid with Potential for Cancer Prevention and Therapy. Current Cancer Drug Targets 8:634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GH, Qu J, Shen X. 2008. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim Biophys Acta 1783:713–27. [DOI] [PubMed] [Google Scholar]
- Mathers JC, Coxhead JM, Tyson J. 2007. Nutrition and DNA repair--potential molecular mechanisms of action. Curr Cancer Drug Targets 7:425–31. [DOI] [PubMed] [Google Scholar]
- Morito N, Yoh K, Itoh K, Hirayama A, Koyama A, Yamamoto M, Takahashi S. 2003. Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene 22:9275–9281. [DOI] [PubMed] [Google Scholar]
- Na HK, Surh YJ. 2014. Oncogenic potential of Nrf2 and its principal target protein heme oxygenase-1. Free Radic Biol Med 67:353–65. [DOI] [PubMed] [Google Scholar]
- Osman NH, Said UZ, El-Waseef AM, Ahmed ES. 2015. Luteolin supplementation adjacent to aspirin treatment reduced dimethylhydrazine-induced experimental colon carcinogenesis in rats. Tumour Biol 36:1179–90. [DOI] [PubMed] [Google Scholar]
- Pandurangan AK, Ananda Sadagopan SK, Dharmalingam P, Ganapasam S. 2014a. Luteolin, a bioflavonoid inhibits Azoxymethane-induced colorectal cancer through activation of Nrf2 signaling. Toxicol Mech Methods 24:13–20. [DOI] [PubMed] [Google Scholar]
- Pandurangan AK, Ananda Sadagopan SK, Dharmalingam P, Ganapasam S. 2014b. Luteolin, a bioflavonoid, attenuates azoxymethane-induced effects on mitochondrial enzymes in BALB/c mice. Asian Pac J Cancer Prev 14:6669–72. [DOI] [PubMed] [Google Scholar]
- Pandurangan AK, Dharmalingam P, Sadagopan SK, Ramar M, Munusamy A, Ganapasam S. 2013. Luteolin induces growth arrest in colon cancer cells through involvement of Wnt/beta-catenin/GSK-3beta signaling. J Environ Pathol Toxicol Oncol 32:131–9. [DOI] [PubMed] [Google Scholar]
- Pandurangan AK, Esa NM. 2013. Dietary non-nutritive factors in targeting of regulatory molecules in colorectal cancer: an update. Asian Pac J Cancer Prev 14:5543–52. [DOI] [PubMed] [Google Scholar]
- Pandurangan AK, Sadagopan SKA, Dharmalingam P, Ganapasam S. 2014c. Luteolin, a bioflavonoid inhibits Azoxymethane-induced colorectal cancer through activation of Nrf2 signaling. Toxicology Mechanisms and Methods 24:13–20. [DOI] [PubMed] [Google Scholar]
- Paredes-Gonzalez X, Fuentes F, Jeffery S, Saw CLL, Shu LM, Su ZY, Kong ANT. 2015. Induction of NRF2-mediated gene expression by dietary phytochemical flavones apigenin and luteolin. Biopharmaceutics & Drug Disposition 36:440–451. [DOI] [PubMed] [Google Scholar]
- Saw CL, Huang MT, Liu Y, Khor TO, Conney AH, Kong AN. 2011. Impact of Nrf2 on UVB-Induced Skin Inflammation/Photoprotection and Photoprotective Effect of Sulforaphane. Molecular Carcinogenesis 50:479–486. [DOI] [PubMed] [Google Scholar]
- Shelton P, Jaiswal AK. 2013. The transcription factor NF-E2-related factor 2 (Nrf2): a protooncogene? FASEB J 27:414–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2017. Cancer Statistics, 2017. CA Cancer J Clin 67:7–30. [DOI] [PubMed] [Google Scholar]
- Souglakos J. 2007. Genetic alterations in sporadic and hereditary colorectal cancer: implementations for screening and follow-up. Dig Dis 25:9–19. [DOI] [PubMed] [Google Scholar]
- Stigliano V, Sanchez-Mete L, Martayan A, Anti M. 2014. Early-onset colorectal cancer: a sporadic or inherited disease? World J Gastroenterol 20:12420–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su ZY, Khor TO, Shu L, Lee JH, Saw CL, Wu TY, Huang Y, Suh N, Yang CS, Conney AH, Wu Q, Kong AN. 2013. Epigenetic reactivation of Nrf2 in murine prostate cancer TRAMP C1 cells by natural phytochemicals Z-ligustilide and Radix angelica sinensis via promoter CpG demethylation. Chem Res Toxicol 26:477–85. [DOI] [PubMed] [Google Scholar]
- Tan BL, Norhaizan ME, Huynh K, Yeap SK, Hazilawati H, Roselina K. 2015. Brewers’ rice modulates oxidative stress in azoxymethane-mediated colon carcinogenesis in rats. World J Gastroenterol 21:8826–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi PP, Jena GB, Tikoo KB, Kumar V. 2016. Melatonin modulated autophagy and Nrf2 signaling pathways in mice with colitis-associated colon carcinogenesis. Mol Carcinog 55:255–67. [DOI] [PubMed] [Google Scholar]
- van Engeland M, Derks S, Smits KM, Meijer GA, Herman JG. 2011. Colorectal cancer epigenetics: complex simplicity. J Clin Oncol 29:1382–91. [DOI] [PubMed] [Google Scholar]
- Wang B, Zhu X, Kim Y, Li J, Huang S, Saleem S, Li RC, Xu Y, Dore S, Cao W. 2012. Histone deacetylase inhibition activates transcription factor Nrf2 and protects against cerebral ischemic damage. Free Radic Biol Med 52:928–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, VanAlstyne PC, Irons KA, Chen S, Stewart JW, Birt DF. 2004. Individual and interactive effects of apigenin analogs on G2/M cell-cycle arrest in human colon carcinoma cell lines. Nutr Cancer 48:106–14. [DOI] [PubMed] [Google Scholar]
- Wondrak GT, Villeneuve NF, Lamore SD, Bause AS, Jiang T, Zhang DD. 2010. The cinnamon-derived dietary factor cinnamic aldehyde activates the Nrf2-dependent antioxidant response in human epithelial colon cells. Molecules 15:3338–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Feng J, Yang Y, Dai C, Lu A, Li J, Liao Y, Xiang M, Huang Q, Wang D, Du XB. 2017. Significance of Serum Total Oxidant/Antioxidant Status in Patients with Colorectal Cancer. PLoS One 12:e0170003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TY, Khor TO, Su ZY, Saw CL, Shu L, Cheung KL, Huang Y, Yu S, Kong AN. 2013. Epigenetic modifications of Nrf2 by 3,3′-diindolylmethane in vitro in TRAMP C1 cell line and in vivo TRAMP prostate tumors. AAPS J 15:864–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi MY, Jia JM, Sun HP, Sun ZY, Jiang JW, Wang YJ, Zhang MY, Zhu JF, Xu LL, Jiang ZY, Xue X, Ye M, Yang X, Gao Y, Tao L, Guo XK, Xu XL, Guo QL, Zhang XJ, Hu R, You QD. 2013. 3-aroylmethylene-2,3,6,7-tetrahydro-1H-pyrazino[2,1-a]isoquinolin-4(11bH)-ones as potent Nrf2/ARE inducers in human cancer cells and AOM-DSS treated mice. J Med Chem 56:7925–38. [DOI] [PubMed] [Google Scholar]
- Xiao B, Qin Y, Ying C, Ma B, Wang B, Long F, Wang R, Fang L, Wang Y. 2017. Combination of oncolytic adenovirus and luteolin exerts synergistic antitumor effects in colorectal cancer cells and a mouse model. Mol Med Rep 16:9375–9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CJ, Huang MT, Shen GX, Yuan XL, Lin W, Khor TO, Conney AH, Kong ANT. 2006. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Research 66:8293–8296. [DOI] [PubMed] [Google Scholar]
- Xu DG, Lv W, Dai CY, Zhu FF, Xu GH, Ma ZJ, Chen Z. 2015. 2-(Pro-1-ynyl)-5-(5,6-dihydroxypenta-1,3-diynyl) thiophene induces apoptosis through reactive oxygen species-mediated JNK activation in human colon cancer SW620 cells. Anat Rec (Hoboken) 298:376–85. [DOI] [PubMed] [Google Scholar]
- Yang J, Wu R, Li W, Gao L, Yang Y, Li P, Kong AN. 2017a. The triterpenoid corosolic acid blocks transformation and epigenetically reactivates Nrf2 in TRAMP-C1 prostate cells. Mol Carcinog. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fuentes F, Shu L, Wang C, Pung D, Li W, Zhang C, Guo Y, Kong AN. 2017b. Epigenetic CpG Methylation of the Promoter and Reactivation of the Expression of GSTP1 by Astaxanthin in Human Prostate LNCaP Cells. Aaps j 19:421–430. [DOI] [PubMed] [Google Scholar]
- Zhang C, Shu L, Kim H, Khor TO, Wu R, Li W, Kong AN. 2016. Phenethyl isothiocyanate (PEITC) suppresses prostate cancer cell invasion epigenetically through regulating microRNA-194. Mol Nutr Food Res 60:1427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XQ, Zhang YF, Xia YF, Zhou ZM, Cao YQ. 2015. Promoter demethylation of nuclear factor-erythroid 2-related factor 2 gene in drug-resistant colon cancer cells. Oncol Lett 10:1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


