Abstract
Human mesenchymal stem cells (MSCs) are a heterogeneous subset of non-hematopoietic multipotent stromal stem cells and can differentiate into mesodermal lineage, such as adipocytes, osteocytes and chondrocytes, as well as ectodermal and endodermal lineages. Human umbilical cord (UC) is one of the most promising sources of MSCs. However, the molecular and cellular characteristics of UC-MSCs require extensive investigations, which are hampered by the limited lifespan and the diminished potency over passages. Here, we employed the piggyBac transposon-based SV40T immortalization system and effectively immortalized UC-MSCs, yielding the iUC-MSCs. A vast majority of the immortalized lines are positive for MSC markers but not for hematopoietic markers. The immortalization phenotype of the iUC-MSCs can be effectively reversed by FLP recombinase-induced removal of SV40 T antigen. While possessing long-term proliferation capability, the iUC-MSCs are not tumorigenic in vivo. Upon BMP9 stimulation, the iUC-MSC cells effectively differentiate into osteogenic, chondrogenic and adipogenic lineages both in vitro and in vivo, which is indistinguishable from that of primary UC-MSCs, indicating the immortalized UC-MSCs possess the characteristics similar to that of their primary counterparts and retain tri-lineage differentiation potential upon BMP9 stimulation. Therefore, the engineered iUC-MSCs should be a valuable alternative cell source for studying UC-MSC biology and their potential utilities in immunotherapies and regenerative medicine.
Keywords: mesenchymal stem cells (MSCs), umbilical cord-derived MSCs (UC-MSCs), SV40 T antigen immortalization, BMP9-induced osteogenic differentiation, regenerative medicine, immunotherapy
1. INTRODUCTION
Human mesenchymal stem cells (MSCs) are a heterogeneous subset of non-hematopoietic multipotent stromal stem cells, which originate from human embryonic mesoderm and/or isolated from fetal and adult tissues, such as bone marrow, peripheral blood, cord blood, umbilical cord (UC), placenta, amniotic fluid, amniotic membrane, endometrium, menstrual blood, synovial fluid, skin and foreskin, adipose tissue, and dental pulp1–5. MSCs can differentiate into mesodermal lineage, such as osteocytes, chondrocytes, adipocytes, as well as ectodermal (e.g., neuronal cells) and endodermal lineages (e.g., hepatocytes). Human MSCs usually express cell surface markers, including CD29, CD44, CD73, CD90, CD105, but lack the expression of hematopoietic markers such as CD14, CD34, CD45 and HLA-DR2, 4. MSCs hold great promise as potential cell-based therapies in the treatment of immune disorders and in regenerative medicine1, 2, 4, 5.
Although MSCs were originally isolated from the bone marrow (BM), BM-derived MSCs have several limitations as a common source of MSCs, including MSC low frequency in BM, the painful collection procedure and the declined MSC potential in aged donors. Conversely, human umbilical cord (UC) is a rich and convenient source of progenitor cells, including MSCs derived either from the cord or from cord blood 6, 7. The human UC-derived MSCs (UC-MSCs) have several advantages as compared to other types and sources of stem cells as UC-MSCs can be easily isolated, without ethical concerns, from a tissue which is discarded after birth, and they represent a more primitive progenitor population than their adult counterparts, which may offer new perspectives for cell-based therapies8–10.
The human UC connects the fetus and mother and provides good blood circulation by preventing umbilical vessels from compression, torsion and bending during pregnancy. UC has two umbilical arteries and one umbilical vein, both of which are embedded within a specific mucous connective tissue, also known as Wharton’s jelly (WJ). The WJ is covered by amniotic epithelium6, 7, 10. With diverse protocols and culture methods, UC-MSCs can be isolated from several compartments of the umbilical cord, including umbilical cord blood, umbilical vein subendothelium, and the most commonly-used source, the WJ, within which MSCs can be isolated from three indistinct regions: the perivascular zone, the intervascular zone, and the subamnion6, 7, 10. Besides their clear advantages, such as an easy isolation and faster self-renewal, UC-MSCs show the ability to differentiate into three germ layers, to home in damaged tissue or inflamed regions, to promote tissue repair, and to modulate the immune response8–10. Thus, UC-MSCs are considered as a possible versatile tool for immunotherapy and regenerative medicine.
Although MSCs (including UC-MSCs) have great advantages over other types of stem cells, their clinical applications are hindered by many obstacles. One major challenge is to obtain adequate number of MSCs, as these cells often lose their potency during sub-culturing and at higher passages, most likely due to the shortening of the telomere length. For instance, early MSCs exhibit higher differentiation ability to chondrocytes, adipocytes and osteocytes whereas at higher passages and on long-term culture, this differentiation property declines. While UC-MSCs can be maintained in vitro longer than BM-MSCs or other sources of MSCs, the comparison of UC-MSC properties is challenging due to the variations in culture media and growth factors used.
To overcome the limited lifespan of in vitro culturing and provide a reliable cell source of UC-MSCs for basic and pre-clinical studies, here we sought to establish reversibly immortalized UC-MSCs (e.g., iUC-MSCs) using a piggyBac transposon-based SV40 T antigen (SV40T) immortalization system 11. We demonstrate that UC-MSCs are effectively immortalized, which can be reversed by FLP recombinase. The resulting iUC-MSCs express MSC markers and retain the ability to differentiate into osteogenic, chondrogenic and adipogenic lineages upon BMP9 stimulation. Therefore, the engineered iUC-MSCs should be a valuable cell source for studying UC-MSC biology and the potential utilities of their cells in immunotherapies and regenerative medicine.
2. MATERIALS AND METHODS
2.1. Cell Culture, Enzymes and Chemicals
HEK-293 cells were obtained from ATCC (Manassas, VA). The 293pTP and RAPA cells were derived from HEK-293 cells as described12, 13. All cells were maintained at 37°C with 5% CO2 in Dulbecco’s modified eagle medium (DMEM) containing 10% fetal bovine serum (Gemini Bio Products, West Sacramento, CA), 2mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin14, 15. All restriction enzymes used for cloning, the Phusion High-Fidelity PCR kit and the Gibson Assembly Master Mix were from New England Biolabs (Ipswich, MA, USA). Oligonucleotides were synthesized by IDT (Coralville, IA) or Sigma-Aldrich (St. Louis, MO). Unless indicated otherwise, all chemicals were purchased from Sigma-Aldrich or Thermo Fisher Scientific (Waltham, MA).
2.2. Generation and Amplification of Recombinant Adenoviruses Expressing BMP9, FLP Recombinase and GFP
Recombinant adenoviral vectors were constructed by using the AdEasy technology as described 16. Briefly, the coding regions of human BMP9 and bacteriophage flippase recombinase (FLP) were PCR amplified, subcloned into an adenoviral shuttle vector, and subsequently used to generate recombinant adenoviruses in HEK-293, 293pTP or RAPA cells as described12, 13. The resultant adenoviruses, designated as Ad-BMP9 and Ad-FLP, also express GFP as a marker for monitoring infection efficiency. An analogous adenovirus expressing GFP only (Ad-GFP) was used as a mock virus control. For all adenovirus infections, polybrene (8 μg/mL) was added to the culture medium to enhance adenoviral infection efficiency17.
2.3. Isolation and Culture of Primary Human Umbilical Cord-Derived-Mesenchymal Stem Cells (UC-MSCs)
The use of human UC-MSCs was approved by the Ethics and Research Conduct Committee of Chongqing Medical University. The primary human UC-MSCs were delinked from patients’ clinical data and obtained from the stem cell bank managed by Chongqing Engineering Research Center of Stem Cell Therapy (Chongqing, China). Briefly, fresh human umbilical cords were collected from healthy full-term and naturally- delivered newborns with written informed consents from the parents. After being dissected and washed with Hank’s buffer, the umbilical cord was cut into small pieces of 1 mm2 and placed in culture dishes in complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Hyclone, Logan, Utah, USA),100units/mL penicillin, 100 μg/mL streptomycin at 37 ℃ and 5 % CO2. The medium was exchanged every 2–3 days and the tissue was removed from culture after 7 days. At ~80% confluency, adherent cells were trypsinized and re-seeded into culture flasks. Primary UC-MSCs between passages #1 and #3 were used to establish immortalized UC-MSCs. Primary UC-MSCs derived from donors #79 (e.g., UC79) and #86 (e.g., UC86) were used in this study.
2.4. Establishment of Reversibly Immortalized UC-Mesenchymal Stem Cells (iUC-MSCs)
Primary human UC-MSCs were immortalized by using the piggyBac transposon-based immortalization vector MPH86 as described11, 18. Briefly, primary UC79 and UC86 cells were freshly seeded in T-25 cell culture flasks. The piggyBac vector MPH86 that expresses SV40 T antigen and hygromycin B gene flanked with FLP sites and a piggyBac transposase expression vector, pCMV-PBase, were co-transfected into the UC-MSCs as described11, 18. At 2 days after transfection, the transfected UC-MSCs were selected in the presence of 0.4 mg/ml hygromycin B (Invitrogen) for 7 days. The pooled, stably immortalized human umbilical cord-derived mesenchymal stem cells (iUC-MSCs) were designated as iUC79 and iUC86, respectively, and expanded and kept in liquid nitrogen tanks.
2.5. Western Blotting Analysis
The expression of immortalizing gene SV40 T antigen was examined by Western blotting as described19. Briefly, cells were lysed in 2x Laemmli sample buffer and subjected to 10% SDS-PAGE, followed by electrical transfer to Immobilon-P membranes. The membranes were blocked with 5% fat-free skimmed milk in TBST buffer at room temperature for 1h, followed by incubation with SV40 T (Santa Cruz Biotechnology) or β-actin (Santa Cruz Biotechnology) antibody at 4°C overnight. After being washed by TBST, the membranes were probed with a second antibody conjugated with horseradish peroxidase (Santa Cruz Biotechnology) at room temperature for 1hr. The presence of the protein of interest was visualized by using Enhanced Chemiluminescent Substrate (Kaiji, China) and exposed under the Syngene GBox Imaging System.
2.6. Mesenchymal Stem Cell Surface Marker Staining and Flow Cytometric Analysis
Exponentially growing iUC79 and iUC86 cells were trypsinized and resuspended in PBS containing 1% bovine serum albumin (BSA) at approximately 1×106 cells/ml. 100μl of the cell suspensions were incubated with CD73 (Santa Cruz), CD105 (Biolegend), CD90 (Biolegend), CD45 (BD Biosciences), HLA-DR (BD Biosciences) or CD34 (BD Biosciences) antibodies conjugated with phycoerythrin (PE) for 30 min at room temperature in the dark. After being washed with PBS, the labeled cells were resuspended in 0.2 ml of PBS and analyzed with the BD FACSCanto II System (BD Biosciences). The acquired data were analyzed by using CellQuest Pro software (BD Biosciences).
2.7. Crystal Violet Staining
Crystal violet assay was conducted as described20, 21. Briefly, exponentially growing iUC-MSC and/or primary cells were either directly seeded at low confluency, or infected with Ad-FLP or Ad-GFP and then seeded at low confluency. At the indicated time points, cells were carefully washed with PBS and stained with 0.5% crystal violet/formalin solution at room temperature for 20–30 min. The stained cells were washed with tap water and air dried for taking macrographic images22. For quantitative measurement, the stained cells were dissolved in 10% acetic acid at room temperature for 20 min with shaking. 500μl were taken and added to 2 ml ddH2O. Absorbance at 570–590nm was measured as described11, 22.
2.8. MTT Cell Proliferation Assay and Cell Viability Assay
MTT assay was carried out as described23. Briefly, approximately 5×103 cells were seeded into each well of 96-well plates and incubate overnight. At the indicate time points, 20 μl of freshly prepared 5 mg/ml MTT was added to each well. After 4h incubation, medium was carefully removed and 150ul of DMSO was added to dissolve MTT-formazan crystal. Plate was covered with tinfoil and agitated on an orbital shaker for 15 min, followed with reading absorbance at 590 nm with a reference filter of 620 nm.
Cell viability was measured using Trypan blue exclusion assay as described24. Briefly, cells were seeded in 24-well plates at a low density. At the indicated time points, both adherent and non-adherent cells were collected and mixed with 2x Trypan blue buffer. 10μl cell mixture (at approx. 105~106 cells per mL) were added to a hemocytometer and counted under a microscope. Viable cells were determined by the total cell count subtracted with nonviable or dead cells (cell stained as blue). Three independent experiments were performed in triplicate.
2.9. RNA Isolation, Semi-quantitative PCR (sqPCR) and Touchdown Quantitative Real-time PCR (TqPCR)
Total RNA was extracted from cells using the TRIZOL reagent according to the manufacturer’s instructions and reverse transcribed into cDNA with hexamer and superscript II reverse transcriptase RT (Thermo Fisher). Used as PCR templates, the first strand cDNA products were diluted 5- to 10-fold. PCR primers were designed with the Primer3.0 program to amplify the genes of interest (Supplemental Table. 1). PCR products were approximately 100~200 bps. The sqPCR reactions were carried out as follows: 94°C × 20’’, 68°C × 30’’, 70°C× 20’’ for 12 cycles, with 1°C decrease per cycle, followed by 25–30 cycles at 94°C× 20’’, 56°C × 30’’, 70°C × 20’’. PCR products were resolved on 1.5% agarose gels. All samples were normalized by the expression level of GAPDH.
TqPCR was carried out by using SYBR Green-based qPCR analysis on a CFX-Connect unit (Bio-Rad Laboratories, Hercules, CA) using our recently optimized TqPCR protocol25. Briefly, the PCR reactions were carried out by using a touchdown protocol: 95°C×3min for one cycle; 95℃×20 sec, 66℃×10 sec for 4 cycles, with 3℃ decrease per cycle; followed by 95℃×10 sec, 55℃×15 sec, 70℃×1 sec for 40 cycles, followed by plate read. All reactions were done in triplicate. TqPCR amplification was confirmed by performing the melting curve test and observing a single peak for each gene. GAPDH was used as a reference gene.
2.10. Alkaline Phosphatase (ALP) Activity Assay
ALP activity was assessed quantitatively with the modified Great Escape SEAP Chemiluminescence assay kit (BD Clontech, Mountain View, CA, USA) as described26. Each assay condition was performed in triplicate, and the results were derived from at least three independent experiments.
2.11. In vivo Tumorigenicity Assay and Xenogen Imaging
The use and care of animals was approved by the Ethics and Research Conduct Committee of Chongqing Medical University. All experimental procedures were in compliance with the NIH guidelines regarding the use and care of vertebrate animals. Experimentally, iUC86 and human osteosarcoma line 143B cells were retrovirally labeled with firefly luciferase and resulted in iUC86-FLuc and 143B-FLuc lines. Exponentially growing cells were collected and injected subcutaneously into the flanks of athymic nude mice (5×106 cells per injection, 4 injection sites/ animal; 4–6 wk old, female, 4 mice per group; from Nippon Crea Inc. Chongqing, China). At 1, 2, and 4 weeks after injection, animals were subjected to bioluminescence imaging by using Xenogen IVIS 200 system as described27. The pseudoimages were obtained by superimposing the emitted light over the gray-scale photographs of the animals. Xenogen’s Live Imaging V2.50.1 software was used to analyze the signaling of proliferating/surviving cells.
2.12. Subcutaneous Implantation of MSCs for Ectopic Bone Formation
Stem cell implantation and ectopic bone formation studies were carried out as described28, 29. Briefly, subconfluent iUC86 and primary UC86 cells were infected with Ad-BMP9 or Ad-GFP for 36h, collected and resuspended in 80μL of PBS for subcutaneous injection (5×106 cells/site) into the flanks of athymic nude mice (5 animals per group, 4–6 weeks old, female; Nippon Crea Inc. Chongqing, China). At 4 weeks after implantation, animals were sacrificed, and the implantation sites were retrieved for micro-computed tomography (μCT) imaging, histologic evaluation and other special staining (see below).
2.13. Micro-Computed Tomography (μCT) Analysis
The retrieved tissues were fixed in formalin and subjected to μCT imaging using the vivaCT 40 (SCANCO Medical AG, Switzerland). Serial images were acquired transverse to the longitudinal axis at 55 kVP (145 μA), employing 1000 conebeam projections per revolution at an integration time of 300 μs. The acquired imaging data were analyzed by using Micro-CT V6.1 software and Mimics 17.0 (Materialise NV, Leuven, Belgium) system.
2.14. Hematoxylin and Eosin, Trichrome, Alcian Blue, and Oil Red O Staining
After μCT imaging, the fixed tissues were decalcified and paraffin-embedded. Serial sections of the embedded specimens were mounted onto slides, deparaffinized and rehydrated in a graduated fashion. H & E, Masson’s Trichrome and Alcian Blue stains were carried out as previously described30. For Oil Red O staining, the decalcified specimens were subjected to frozen sectioning and stained with Oil Red O as described31.
2.15. Statistical Analysis
All quantitative assays were performed in triplicate and/or repeated in three independent batches. Data were expressed as mean ± SD. Statistical significances were determined by one-way analysis of variance and the student’s t test. A value of p<0.05 was considered statistically significant.
3. RESULTS
3.1. Primary human umbilical cord-derived mesenchymal stem cells (UC-MSCs) can be effectively immortalized by SV40 T antigen (SV40T) and express mesenchymal stem cell (MSC) markers
We obtained two primary UC-MSCs lines, which were derived from donors #79 (UC79) and #86 (UC86) and able to be maintained in culture for up to 10 passages with decreasing proliferation capability over passages (Figure 1A). To overcome the limited lifespan characteristic of primary cells, we sought to immortalize the primary UC-MSCs using our recently developed piggyBac transposon-based immortalization MPH86 vector11, 18 (Figure 1B). When the primary UC-MSCs were co-transfected with MPH86 and piggyBac transposase expression vectors, numerous stable clones were obtained, pooled and scaled up to establish two independently immortalized UC-MSC lines, iUC79 (Figure 1C-ac) and iUC86 (Figure 1C-bd). We conducted Western blotting analysis and confirmed the protein expression of SV40T in the immortalized cells (Figure 1D).
FIGURE 1. The establishment of reversibly immortalized umbilical cord-derived mesenchymal stem cell (UC-MSC) lines through piggyBac transposon-mediated expression of SV40 T antigen.

(A) Representative images of primary UM-MSCs derived from donors #79 (UC79) and #86 (UC86). Representative images of passage #3 cells are shown. (B) Schematic representation of the piggyBac transposon-based immortalization vector MPH86 as described in Refs. #11, 18. (C) Establishment of the immortalized UC-MSC lines iUC79 and iUC86. Primary UC-MSCs from the two donors were co-transfected with MPH86 and pCMV-PBase and selected in the presence of hygromycin. Clonal growth was visible after one week (a & b). The resultant stable pools, namely iUC79 and iUC86, can be passaged for long-term maintenance in culture. Representative images of passage #5 are shown (c & d). (D) Detection of SV40 T antigen expression in the immortalized lines. Subconfluent iUC79 and iUC86 cells and the respective primary cells were lysed in Laemmli sample buffer and subjected to Western blotting analysis with SV40 T antigen or β-actin (loading control) antibody. Representative results are shown.
We further analyzed whether the immortalized UC-MSCs express essential MSC markers. We found that the three consensus MSC markers CD73, CD105 and CD90 expressed in average 97.1%, 92.5% and 99.7% of the iUC79 cells, while hematopoietic cell markers CD45 (0.97%), CD34 (0.91%) and HLA-DR (1.33%) were lowly expressed (Figure 2A). Similarly, we found that vast majority of iUC86 cells expressed CD73 (99.7%), CD105 (92.5%) and CD90 (99.4%) while less 1% expressed CD45 (0.34%), CD34 (0.54%) and HLA-DR (0.45%) (Figure 2B). The positivity rates of MSC markers were similar to that detected in primary UC-MSCs (data not shown). Therefore, these results strongly suggest that the immortalized lines iUC79 and iUC86 may retain MSC-like characteristics.
FIGURE 2. The iUC-MSC lines express essential mesenchymal stem cell markers.

Subconfluent iUC79 (A) and iUC86 (B) cells were stained with antibodies against MSC markers CD73, CD105, and CD90 and hematopoietic progenitor cell markers CD45, CD34 and HLA-DR in triplicate. The stained cells were subjected to FACS analysis. Representative results are shown.
3.2. The immortalization phenotype of iUC-MSCs can be reversed by FLP recombinase-induced removal of SV40 T antigen
We compared the proliferative capability of the immortalized UC-MSCs with their primary counterparts. We found that the iUC86 cells qualitatively grew faster than UC86 cells after 3 days in culture (Figure 3Aa vs. b). Quantitative MTT assay revealed that iUC86 cells grew significantly faster than UC86 cells after day 4 than UC86 cells although there was a decrease in day 7, likely due to cell overgrowth (Figure 3B). Viable cell counting assay also indicated the iUC86 cells grew significantly faster than UC86 cells at days 4 and 5 (Figure 3C). Similarly, the iUC79 cells exhibited significant growth advantage over the early passage UC79 cells (data not shown).
FIGURE 3. The iUC-MSC lines exhibit higher proliferation capability, which can be effectively reversed by FLP recombinase.

(A) Cell proliferation assessed by crystal violet staining. Exponentially proliferating primary UC86 cells (a) and iUC86 cells (b) were seeded at the same density and subjected to crystal violet staining at the indicated time points. Representative results are shown. (B) & (C) Cell proliferation and viability assays. Exponentially proliferating primary UC86 cells (a) and iUC86 cells (b) were seeded at the same density and subjected to MTT assay (B) or counting for viable cells after Trypan blue staining (C). “*” p<0.05; “**” p<0.001, compared with that of the primary cell groups. (D) & (E) Decreased cell proliferation caused by FLP-mediated removal of SV40 T antigen. Subconfluent iUC86 and iUC79 (not shown in C) cells were infected with Ad-GFP (a) or Ad-FLP (b) and subjected to crystal violet staining at the indicated time points. Representative results are shown. The stained cells were dissolved and subjected to OD590nm reading. “*” p<0.05; “**” p<0.001, compared with that of the GFP groups. (F) FLP-mediated removal of SV40 T antigen in the iUC-MSC lines. Subconfluent iUC86 and iUC79 cells were infected with Ad-GFP or Ad-FLP and subjected to crystal violet staining at the indicated time points. Representative results are shown. The stained cells were dissolved and subjected to OD590nm reading. “*” p<0.05; “**” p<0.001, compared with that of the GFP groups. (F) FLP-mediated removal of SV40 T antigen in the iUC-MSC lines. Subconfluent iUC86 and iUC79 cells were infected with Ad-GFP or Ad-FLP for 72h. Total RNA was isolated and subjected to reverse transcription and semi-quantitative PCR using primers specific for SV40 T antigen. GAPDH was used as a reference control. PCR products were resolved on 1.5% agarose gels. PCR reactions were done in triplicate and representative results are shown.
We also analyzed whether FLP recombinase would effectively remove SV40T and hence reverse the immortalization phenotype. When the iUC86 and iUC79 cells were infected with Ad-FLP, cell proliferation was significantly slowed down, compared with those infected with Ad-GFP (Figure 3Da vs. b). Quantitative analysis indicated that cell proliferation of Ad-FLP infected cells decreased significantly at three days after infection, compared with that of the Ad-GFP infected cells (Figure 3E). Furthermore, using semi-quantitative PCR analysis we demonstrated that Ad-FLP infection effectively decreased the expression of SV40T in both iUC79 and iUC86 cells, compared with that of the Ad-GFP infected cells (Figure 3F). Collectively, these results strongly suggest that the immortalization phenotype of iUC86 and iUC79 cells may be effectively reversed by exogenous expression of FLP recombinase.
3.3. The iUC-MSCs are non-tumorigenic in subcutaneous cell implantation assays in immunodeficient mice
We further tested whether the immortalized cells were tumorigenic in vivo. When the iUC86 cells and a commonly-used human osteosarcoma line 143B cells were first labelled with firefly luciferase and injected subcutaneously into the flanks of athymic nude mice, we found that the bioluminescence signals were readily detectable at the iUC86 injection sites at the end of week 1, but the signals drastically decreased after two weeks and were almost undetectable after 4 weeks (Figure 4A). On the contrary, as expected the bioluminescence signals of the 143B injection sites increased significantly over time, which is consistent with our earlier reported results of the SV40 T antigen immortalized cells from different tissue origins21, 32–34. To ensure no significant tumor formation from any residually surviving iUC86 cells, we continued to monitor the mice for up to 8 weeks and found no palpable masses were detected in the iUC86 injection group, while the 143B injection group formed large tumor masses, some of which had to be sacrificed prior to the endpoint (data not shown). Similarly, we found that iUC79 cells were not tumorigenic under the same conditions (data not shown). The above in vivo tumorigenicity assay was carried out in two independent batches of experiments, and the results were highly reproducible. Thus, these in vivo results demonstrate that the immortalized cells exhibit long-term proliferation capability but are not tumorigenic.
FIGURE 4. The immortalized UC-MSC cells are not tumorigenic in vivo.
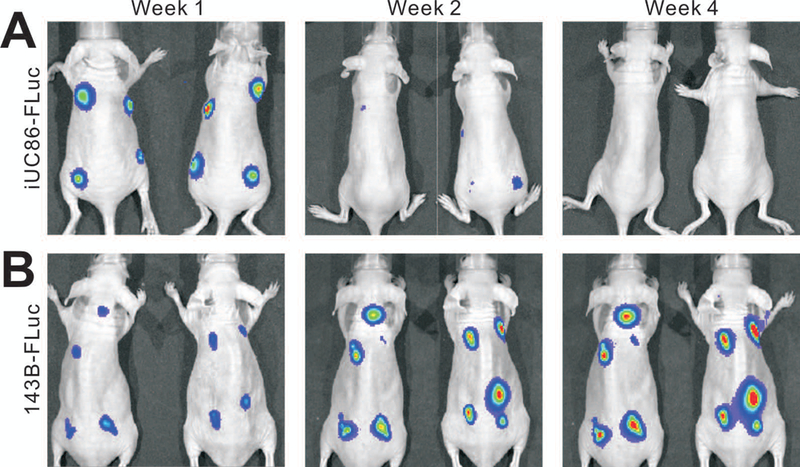
The firefly luciferase-tagged iUC86 (iUC86-FLuc) (A) and human osteosarcoma line 143B (143B-FLuc) (B) cells were injected subcutaneously into the flanks of athymic nude mice (n=4 per group). The animals were subjected to whole body bioluminescence imaging at the indicated time points. Representative images are shown.
3.4. The iUC-MSC cells can differentiate into osteogenic, chondrogenic and adipogenic lineages upon BMP9 stimulation
We next tested if the immortalized cells still possessed the multipotency of MSCs. We previously demonstrated that BMP9 can effectively induce osteogenic, chondrogenic and adipogenic lineages from MSCs31, 35. We analyzed the BMP9-induced expression of lineage-specific regulators RUNX2 (osteogenic), SOX9 (chondrogenic) and PPARG (adipogenic) in the iUC86 cells. Quantitative PCR analysis revealed that both RUNX2 and SOX9 were up-regulated as early as day 3 (p<0.05), and more drastically at days 5 and 7 (p<0.001) (Figure 5A). Interestingly, PPARG was drastically induced as early as day 3 by BMP9 in the iUC86 cells (p<0.001). Similar results were obtained in BMP9-stimulated iUC79 cells (data not shown). These results indicate that the immortalized UC-MSCs are responsive to BMP9-induced multi-lineage differentiation.
FIGURE 5. BMP9 effectively induces osteogenic, chondrogenic and adipogenic lineage regulators and osteogenic markers in the iUC-MSC cells.

(A) BMP9-induced expression of lineage-specific regulators in the iUC-MSCs. Subconfluent iUC86 cells were infected with Ad-GFP or Ad-BMP9. Total RNA was isolated at the indicated time points and subjected to TqPCR analysis using primers for human RUNX2, SOX9 and PPARG. GAPDH was used as a reference gene. The qPCR reactions were done in triplicate. “*” p<0.05; “**” p<0.001, compared with that of the GFP treated groups. (B) BMP9-induced alkaline phosphatase (ALP) activity in the iUC-MSCs. Subconfluent iUC86 and iUC79 cells were infected with Ad-GFP or Ad-BMP9. ALP activity was quantitatively assessed at the indicated time points in triplicate. “*” p<0.05; “**” p<0.001, compared with that of the GFP treated groups. (C) BMP9-induced the expression of late osteogenic marker osteopontin (OPN) in the iUC-MSCs. Subconfluent iUC86 cells were infected with Ad-GFP or Ad-BMP9. Total RNA was isolated at the indicated time points and subjected to TqPCR analysis using primers for human OPN at the indicated time points in triplicate. “**” p<0.001, compared with that of the GFP treated groups.
We further compared the osteogenic activity of the immortalized UC-MSCs with their primary counterparts. We found the alkaline phosphatase (ALP) activity was induced by BMP9 in UC79 cells as early as day 3 (p<0.05), and peaking at day 5, while BMP9-induced ALP activity peaked at day 7 in the iUC79 cells (Figure 5B). Similar patterns of BMP9-induced ALP activity were obtained in UC86 and iUC86 cells (Figure 5B). While it is currently unknown why BMP9-inudced ALP activities peaked at a later time point in the immortalized UC-MSCs than that in the primary UC-MSCs, it is conceivable that the iUC-MSCs have higher proliferative activities and thus may need longer time to differentiate towards osteogenic lineage. Nonetheless, we found the late osteogenic marker OPN was effectively induced by BMP9 after five days in iUC86 cells (Figure 5C). Similar results of BMP9-induced OPN expression were obtained in iUC79 cells (data not shown).
Lastly, we sought to determine whether the immortalized UC-MSCs can effectively differentiate into bone, cartilage and adipose in vivo. When the Ad-BMP9 or Ad-GFP transduced iUC86 and primary UC86 cells were implanted in the flanks of athymic nude mice for 4 weeks, apparent bony masses were formed in all BMP9 treatment groups in both iUC86 and UC86 cells, which did not show overall size differences between the two cell types (p>0.05) (Figure 6A-ab). No masses were found in the GFP groups, further confirming that the immortalized UC-MSCs are not tumorigenic. H & E staining revealed that the masses retrieved from both cell lines contained extensive trabecular bone and a high degree of mature osteoid matrix (Figure 6B). Trichrome staining further confirmed that there was the presence of well-mineralized osteoid matrix in both iUC86 and UC86 formed masses (Figure 6C). Nonetheless, Alcian Blue staining revealed the masses from both iUC86 and UC86 cells contained cartilage and chondroid matrix (Figure 6D). Lastly, Oil Red O staining indicated that the retrieved and decalcified masses contained numerous lipid droplets (Figure 6E). Taken these in vitro and in vivo results together, the immortalized UC-MSCs were shown to behave similar to their primary counterparts and retained tri-lineage differentiation potential upon BMP9 stimulation.
FIGURE 6. BMP9 effectively induces osteogenic, chondrogenic and adipogenic differentiation of the iUC-MSCs in vivo.
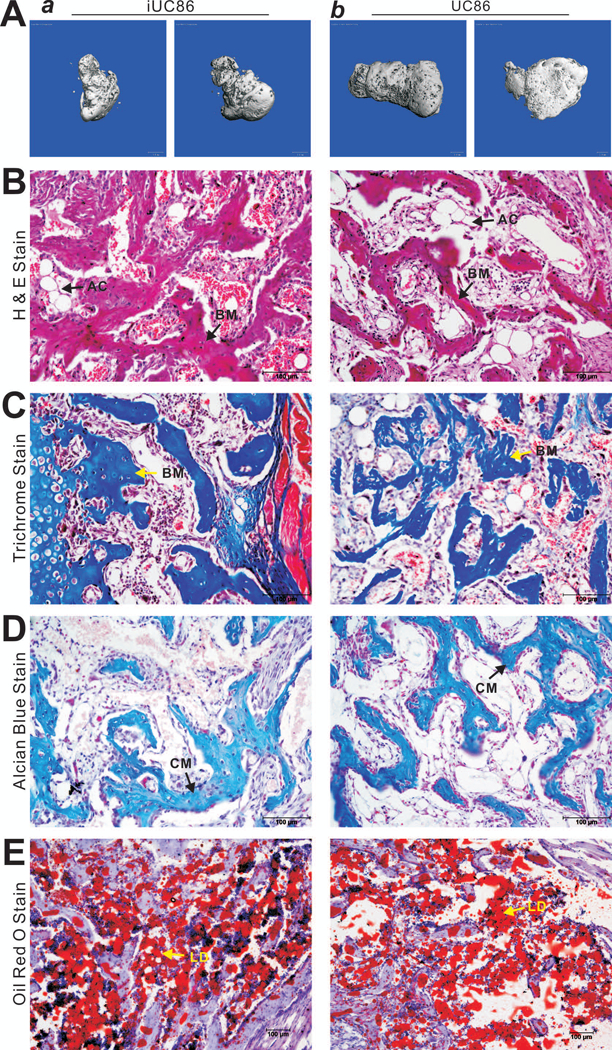
Subconfluent iUC86 and primary UC86 cells were infected with Ad-BMP9 or Ad-GFP for 24h, collected and injected subcutaneously into the flanks of athymic nude mice. The animals were sacrificed at 4 weeks after injection. Bony masses at the injection sites were retrieved, fixed and subjected to microCT imaging. Representative images are shown (A). No masses were retrievable from the sites injected with the cells infected with Ad-GFP. After microCT imaging, tissues were decalcified, and either paraffin-embedded for H & E staining (B), Trichrome staining (C), and Alcian Blue staining (D), or frozen-sectioned for Oil Red O staining (E). Representative images are shown. BM, osteoid matrix; CM, chondroid matrix; AC, adipocyte; LD, lipid droplet.
4. DISCUSSION
Human umbilical cord (UC) is one of the most promising sources of MSCs. However, the molecular and cellular characteristics of UC-MSCs require extensive investigation, which is hampered by the limited lifespan and the diminished potency over passages of the primary UC-MSCs. To that end, we employed the piggyBac transposon-based SV40T immortalization system and effectively immortalized UC-MSCs from two donors, yielding the iUC79 and iUC86 lines. A vast majority of the immortalized lines are positive for MSC markers but not for hematopoietic markers. We showed that the immortalization phenotype of the iUC-MSCs could be effectively reversed by FLP recombinase-induced removal of SV40 T antigen. Furthermore, the iUC-MSCs were not tumorigenic in vivo, while possessing long-term proliferation capability. Upon BMP9 stimulation, the iUC-MSC cells were shown to effectively differentiate into osteogenic, chondrogenic and adipogenic lineages both in vitro and in vivo, which was indistinguishable from primary UC-MSCs. Our in vitro and in vivo results demonstrate that the immortalized UC-MSCs possess the characteristics similar to that of their primary counterparts and retain tri-lineage differentiation potential upon BMP9 stimulation. Nonetheless, more detailed characterization of these immortalized UC-MSCs is needed.
The use of UC-MSCs has numerous advantages over other sources of MSCs6–10. First, human UC is considered as medical waste so the raw materials are readily available and ethically acceptable. Second, MSCs can be isolated from different compartments of the UC, which should increase the diversities of UC-MSCs and may have distinct features to preferentially differentiate into specific cell/tissue types. Third, UC-MSCs are more primitive and have higher proliferative capability, which allows longer culturing time in vitro while retaining multipotency. Fourth, UC-MSCs exhibit low immunogenicity with a good immunosuppressive ability. Compared with BM-MSCs and embryonic stem cells (ESCs), UC-MSCs exhibit a gene expression profile much more similar to that of ESCs’ and have faster self-renewal than BM-MSCs 10. Thus, it is easy to obtain a large number of UC-MSCs after several passages and ex vivo expansion.
Like other sources of MSCs such as BM-MSCs, UC-MSCs have autologous and allogeneic uses. Autologous UC-MSCs may be used for cell-based ex vivo gene therapy for genetic diseases and for regenerative or anti-inflammatory therapy for neonatal injury, such as cerebral palsy or hypoxic brain damage6–10. Conversely, allogeneic UC-MSCs can be expanded and cryopreserved in a cell bank for patients in need. For example, a recent study highlighted the therapeutic effects of human UC-MSCs in a mouse model of acute lung injury36. An early study compared the chondrogenic activity between BMSCs and UC-MSCs in a 3D scaffold model and concluded that UC-MSCs were a more desirable source for fibrocartilage tissue engineering37. Nonetheless, further investigation of the molecular and cellular characteristics between UC-MSCs and other sources of MSCs is highly warranted before UC-MSCs can be broadly used for immunotherapy and regenerative medicine.
Technically, we have employed both retroviral vector and/or piggyBac transposon system to stably expression SV40T and immortalize a panel of mouse progenitor cells, including human cranial suture progenitor cells38, mouse embryonic fibroblasts (MEFs)11, 39, mouse fetal hepatic progenitors15, 40–43, mouse cardiomyogenic progenitors44, mouse melanocyte progenitors45, mouse tendocytes46, mouse articular chondrocytes 22, mouse stem cells of dental apical papilla47, 48 and adipose-derived mesenchymal stem cells49. We found that the piggyBac transposon system is superior to retroviral vector system because the piggyBac transposon-mediated transgene integration occurs at AT-rich regions of the genome which are usually associated open regions of chromosomes11, 18. Furthermore, the piggyBac transposons can be excised from host genome by its transposase and hence achieve footprintless reversal of immortalization11, 18. Thus, our engineered iUC-MSCs should be a valuable alternative source of primary UC-MSCs for basic and translational studies.
In summary, we sought to establish reversibly immortalized UC-MSCs (e.g., iUC-MSCs) using a piggyBac transposon-based SV40 T antigen (SV40T) immortalization system. We showed that UC-MSCs were effectively immortalized, which could be reversed by FLP recombinase. A vast majority of the resulting iUC-MSCs expressed MSC markers and retained the ability to differentiate into osteogenic, chondrogenic and adipogenic lineages upon BMP9 stimulation. Therefore, the engineered iUC-MSCs should be a valuable cell source for studying UC-MSC biology and the potential utilities of their cells in immunotherapies and regenerative medicine.
Supplementary Material
List of PCR Primers
ACKNOWLEDGMENTS
The reported work was supported in part by research grants from the National Institutes of Health (CA226303, DE020140 to TCH and RRR), the U.S. Department of Defense (OR130096 to JMW), the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust (RRR, TCH), the Scoliosis Research Society (TCH and MJL), the National Key Research and Development Program of China (2016YFC1000803 and 2011CB707906 to TCH) and Chongqing Sciences and Technology Commission (Grants #CSTC2016JCYJA0445 to YL and #CSTC2013JCYJA10014 to XL). Funding sources were not involved in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol 2008;8:726–736. [DOI] [PubMed] [Google Scholar]
- 2.Nombela-Arrieta C, Ritz J, Silberstein LE. The elusive nature and function of mesenchymal stem cells. Nat Rev Mol Cell Biol 2011;12:126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianco P “Mesenchymal” Stem Cells. Annual Review of Cell and Developmental Biology 2014;30:677–704. [DOI] [PubMed] [Google Scholar]
- 4.Shenaq DS, Rastegar F, Petkovic D, et al. Mesenchymal Progenitor Cells and Their Orthopedic Applications: Forging a Path towards Clinical Trials. Stem Cells Int 2010;2010:519028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rastegar F, Shenaq D, Huang J, et al. Mesenchymal stem cells: Molecular characteristics and clinical applications. World J Stem Cells 2010;2:67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Troyer DL, Weiss ML. Wharton’s jelly-derived cells are a primitive stromal cell population. Stem Cells 2008;26:591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batsali AK, Kastrinaki MC, Papadaki HA, Pontikoglou C. Mesenchymal stem cells derived from Wharton’s Jelly of the umbilical cord: biological properties and emerging clinical applications. Curr Stem Cell Res Ther 2013;8:144–155. [DOI] [PubMed] [Google Scholar]
- 8.Fan CG, Zhang QJ, Zhou JR. Therapeutic potentials of mesenchymal stem cells derived from human umbilical cord. Stem Cell Rev 2011;7:195–207. [DOI] [PubMed] [Google Scholar]
- 9.Bongso A, Fong CY. The therapeutic potential, challenges and future clinical directions of stem cells from the Wharton’s jelly of the human umbilical cord. Stem Cell Rev 2013;9:226–240. [DOI] [PubMed] [Google Scholar]
- 10.Nagamura-Inoue T, He H. Umbilical cord-derived mesenchymal stem cells: Their advantages and potential clinical utility. World J Stem Cells 2014;6:195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang N, Zhang W, Cui J, et al. The piggyBac transposon-mediated expression of SV40 T antigen efficiently immortalizes mouse embryonic fibroblasts (MEFs). PLoS One 2014;9:e97316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu N, Zhang H, Deng F, et al. Overexpression of Ad5 precursor terminal protein accelerates recombinant adenovirus packaging and amplification in HEK-293 packaging cells. Gene Ther 2014;21:629–637. [DOI] [PubMed] [Google Scholar]
- 13.Wei Q, Fan J, Liao J, et al. Engineering the Rapid Adenovirus Production and Amplification (RAPA) Cell Line to Expedite the Generation of Recombinant Adenoviruses. Cell Physiol Biochem 2017;41:2383–2398. [DOI] [PubMed] [Google Scholar]
- 14.Liao J, Wei Q, Fan J, et al. Characterization of retroviral infectivity and superinfection resistance during retrovirus-mediated transduction of mammalian cells. Gene Ther 2017;24:333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan J, Wei Q, Liao J, et al. Noncanonical Wnt signaling plays an important role in modulating canonical Wnt-regulated stemness, proliferation and terminal differentiation of hepatic progenitors. Oncotarget 2017;8:27105–27119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo J, Deng ZL, Luo X, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc 2007;2:1236–1247. [DOI] [PubMed] [Google Scholar]
- 17.Zhao C, Wu N, Deng F, et al. Adenovirus-mediated gene transfer in mesenchymal stem cells can be significantly enhanced by the cationic polymer polybrene. PLoS One 2014;9:e92908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Cui J, Yan Z, et al. Sustained high level transgene expression in mammalian cells mediated by the optimized piggyBac transposon system. Genes Dis 2015;2:96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo Q, Kang Q, Si W, et al. Connective Tissue Growth Factor (CTGF) Is Regulated by Wnt and Bone Morphogenetic Proteins Signaling in Osteoblast Differentiation of Mesenchymal Stem Cells. J Biol Chem 2004;279:55958–55968. [DOI] [PubMed] [Google Scholar]
- 20.Deng Y, Wang Z, Zhang F, et al. A Blockade of IGF Signaling Sensitizes Human Ovarian Cancer Cells to the Anthelmintic Niclosamide-Induced Anti-Proliferative and Anticancer Activities. Cell Physiol Biochem 2016;39:871–888. [DOI] [PubMed] [Google Scholar]
- 21.Luther GA, Lamplot J, Chen X, et al. IGFBP5 Domains Exert Distinct Inhibitory Effects on the Tumorigenicity and Metastasis of Human Osteosarcoma. Cancer Lett 2013;336:222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamplot JD, Liu B, Yin L, et al. Reversibly Immortalized Mouse Articular Chondrocytes Acquire Long-Term Proliferative Capability while Retaining Chondrogenic Phenotype. Cell Transplant 2015;24:1053–1066. [DOI] [PubMed] [Google Scholar]
- 23.Si W, Kang Q, Luu HH, et al. CCN1/Cyr61 Is Regulated by the Canonical Wnt Signal and Plays an Important Role in Wnt3A-Induced Osteoblast Differentiation of Mesenchymal Stem Cells. Mol Cell Biol 2006;26:2955–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo X, Chen J, Song WX, et al. Osteogenic BMPs promote tumor growth of human osteosarcomas that harbor differentiation defects. Lab Invest 2008;88:1264–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Wang J, Deng F, et al. TqPCR: A Touchdown qPCR Assay with Significantly Improved Detection Sensitivity and Amplification Efficiency of SYBR Green qPCR. PLoS One 2015;10:e0132666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Qin J, Luo Q, et al. Cross-talk between EGF and BMP9 signalling pathways regulates the osteogenic differentiation of mesenchymal stem cells. J Cell Mol Med 2013;17:1160–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang E, Zhu G, Jiang W, et al. Growth hormone synergizes with BMP9 in osteogenic differentiation by activating the JAK/STAT/IGF1 pathway in murine multilineage cells. J Bone Miner Res 2012;27:1566–1575. [DOI] [PubMed] [Google Scholar]
- 28.Liao J, Wei Q, Zou Y, et al. Notch Signaling Augments BMP9-Induced Bone Formation by Promoting the Osteogenesis-Angiogenesis Coupling Process in Mesenchymal Stem Cells (MSCs). Cell Physiol Biochem 2017;41:1905–1923. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Liao J, Zhang F, et al. NEL-Like Molecule-1 (Nell1) Is Regulated by Bone Morphogenetic Protein 9 (BMP9) and Potentiates BMP9-Induced Osteogenic Differentiation at the Expense of Adipogenesis in Mesenchymal Stem Cells. Cell Physiol Biochem 2017;41:484–500. [DOI] [PubMed] [Google Scholar]
- 30.Ye J, Wang J, Zhu Y, et al. A thermoresponsive polydiolcitrate-gelatin scaffold and delivery system mediates effective bone formation from BMP9-transduced mesenchymal stem cells. Biomed Mater 2016;11:025021. [DOI] [PubMed] [Google Scholar]
- 31.Kang Q, Song WX, Luo Q, et al. A comprehensive analysis of the dual roles of BMPs in regulating adipogenic and osteogenic differentiation of mesenchymal progenitor cells. Stem Cells Dev 2009;18:545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Y, Luo X, He BC, et al. Establishment and characterization of a new highly metastatic human osteosarcoma cell line. Clin Exp Metastasis 2009;26:599–610. [DOI] [PubMed] [Google Scholar]
- 33.Su Y, Wagner ER, Luo Q, et al. Insulin-like growth factor binding protein 5 suppresses tumor growth and metastasis of human osteosarcoma. Oncogene 2011;30:3907–3917. [DOI] [PubMed] [Google Scholar]
- 34.Liao Z, Nan G, Yan Z, et al. The Anthelmintic Drug Niclosamide Inhibits the Proliferative Activity of Human Osteosarcoma Cells by Targeting Multiple Signal Pathways. Curr Cancer Drug Targets 2015;15:726–738. [DOI] [PubMed] [Google Scholar]
- 35.Lamplot JD, Qin J, Nan G, et al. BMP9 signaling in stem cell differentiation and osteogenesis. Am J Stem Cells 2013;2:1–21. [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu H, Xiong Y, Xia Y, et al. Therapeutic Effects of Human Umbilical Cord-Derived Mesenchymal Stem Cells in Acute Lung Injury Mice. Sci Rep 2017;7:39889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Tran I, Seshareddy K, Weiss ML, Detamore MS. A comparison of human bone marrow-derived mesenchymal stem cells and human umbilical cord-derived mesenchymal stromal cells for cartilage tissue engineering. Tissue Eng Part A 2009;15:2259–2266. [DOI] [PubMed] [Google Scholar]
- 38.Song D, Zhang F, Reid RR, et al. BMP9 induces osteogenesis and adipogenesis in the immortalized human cranial suture progenitors from the patent sutures of craniosynostosis patients. J Cell Mol Med 2017;21:2782–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang E, Bi Y, Jiang W, et al. Conditionally Immortalized Mouse Embryonic Fibroblasts Retain Proliferative Activity without Compromising Multipotent Differentiation Potential. PLoS One 2012;7:e32428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bi Y, Gong M, Zhang X, et al. Pre-activation of retinoid signaling facilitates neuronal differentiation of mesenchymal stem cells. Dev Growth Differ 2010;52:419–431. [DOI] [PubMed] [Google Scholar]
- 41.Bi Y, He Y, Huang J, et al. Functional characteristics of reversibly immortalized hepatic progenitor cells derived from mouse embryonic liver. Cell Physiol Biochem 2014;34:1318–1338. [DOI] [PubMed] [Google Scholar]
- 42.Bi Y, Huang J, He Y, et al. Wnt antagonist SFRP3 inhibits the differentiation of mouse hepatic progenitor cells. J Cell Biochem 2009;108:295–303. [DOI] [PubMed] [Google Scholar]
- 43.Huang J, Bi Y, Zhu GH, et al. Retinoic acid signalling induces the differentiation of mouse fetal liver-derived hepatic progenitor cells. Liver Int 2009;29:1569–1581. [DOI] [PubMed] [Google Scholar]
- 44.Li M, Chen Y, Bi Y, et al. Establishment and characterization of the reversibly immortalized mouse fetal heart progenitors. Int J Med Sci 2013;10:1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang K, Chen J, Jiang W, et al. Conditional Immortalization Establishes a Repertoire of Mouse Melanocyte Progenitors with Distinct Melanogenic Differentiation Potential. J Invest Dermatol 2012;132:2479–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denduluri SK, Scott B, Lamplot JD, et al. Immortalized Mouse Achilles Tenocytes Demonstrate Long-Term Proliferative Capacity While Retaining Tenogenic Properties. Tissue Eng Part C Methods 2016;22:280–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Zhang H, Zhang W, et al. Bone Morphogenetic Protein-9 (BMP9) Effectively Induces Osteo/Odontoblastic Differentiation of the Reversibly Immortalized Stem Cells of Dental Apical Papilla. Stem Cells Dev 2014;23:1405–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Wang J, Deng F, et al. Canonical Wnt signaling acts synergistically on BMP9-induced osteo/odontoblastic differentiation of stem cells of dental apical papilla (SCAPs). Biomaterials 2015;39:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu S, Wang J, Ye J, et al. Bone morphogenetic protein 9 (BMP9) induces effective bone formation from reversibly immortalized multipotent adipose-derived (iMAD) mesenchymal stem cells. Am J Transl Res 2016;8:3710–3730. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of PCR Primers


