Abstract
As transcription of the human genome is quite pervasive it is possible that many novel functions of the noncoding genome have yet to be identified. Often the noncoding genome’s functions are carried out by their RNA transcripts which may rely on their structures and/or extensive interactions with other molecules. Recent technology developments are transforming the fields of RNA biology from studying one-RNA-at-a-time to transcriptome-wide mapping of structures and interactions. Here, we highlight the recent advances in transcriptome-wide RNA interaction analysis. These technologies revealed surprising versatility of RNA to participate in diverse molecular systems. For example, tens of thousands of RNA-RNA interactions have been revealed in cultured cells as well as in mouse brain, including interactions between transposon-produced transcripts and mRNAs. Additionally, most transcription start sites in the human genome are associated with noncoding RNA transcribed from other genomic loci. These recent discoveries expanded our understanding of RNAs’ roles in chromatin organization, gene regulation, and intracellular signaling.
RNA interactions regulate diverse molecular functions
RNA is produced from most human genomic sequences, although only a relatively small portion of these transcripts are translated and/or have known associated functions. The vast amounts of transcripts with unknown functions may not be translated and present an opportunity to investigate the functions of the noncoding genome. Previous studies of noncoding RNA interactions have led to major discoveries, including RNA interference [1, 2], essential steps of RNA splicing through snRNA binding to intronic splice sites [3, 4], and site-specific rRNA pseudouridination through snoRNA-rRNA interactions [5]. On the other hand, RNA-chromatin interactions constitute essential steps in X chromosome silencing [6, 7], RNAi mediated epigenetic inheritance [8], telomere replication [9], de novo DNA methylation on an imprinting locus [10], transcriptional activation [11,12], and a negative feedback between paralogous genes [13]. We therefore anticipate novel functions to be revealed by identifying novel classes of RNA-participating interactions.
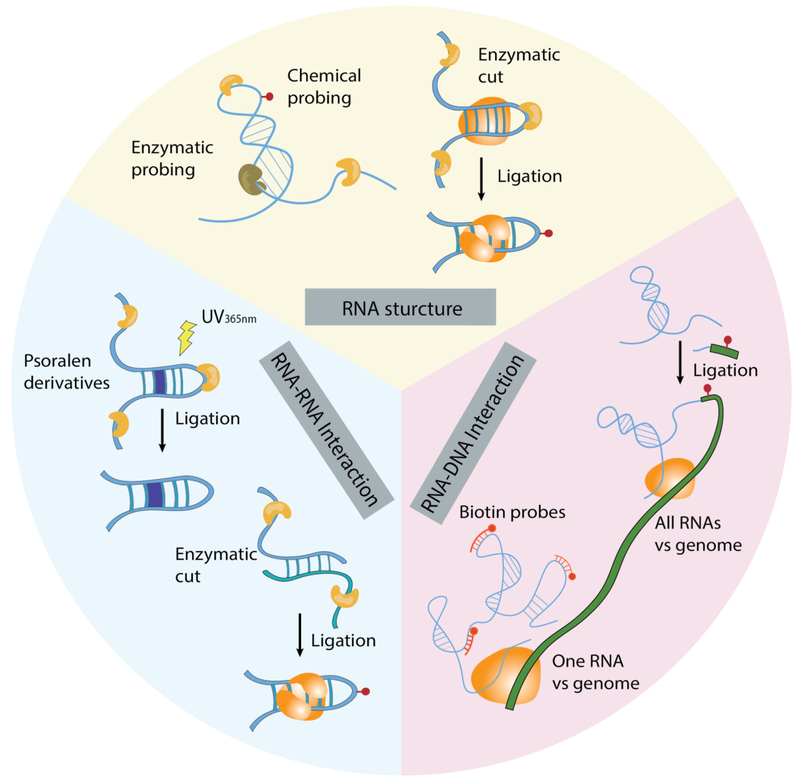
Recent technology developments are transforming the way RNA structure and interactions are analyzed. Instead of studying one RNA or one interaction at-a-time, recent technologies have enabled transcriptome-wide analysis of RNA structures, RNA-RNA interactions, and RNA-DNA interactions (Figure 1). These developments were achieved by combining biochemical reactions with next-generation sequencing. The general strategy of investigating RNA-RNA and RNA-DNA interactions is to convert interacting sequence (RNA-RNA or RNA-DNA) pairs into chimeric DNA, and leverage DNA sequencing as a high-throughput readout of the underlying interactions. Many of these technologies can be applied to analyze intact cells and primary tissues without requiring genetic perturbation or ectopic expression. In this article, we review sequencing-based approaches for mapping RNA structures, RNA-RNA and RNA-DNA interactions, summarize the major findings, and point out the new hypotheses derived from these findings.
Figure 1.
Overview of sequencing-based technologies for mapping RNA structures, RNA-RNA interactions, and RNA-DNA interactions.
RNA STRUCTURE
Sequencing-based methods for mapping RNA structures
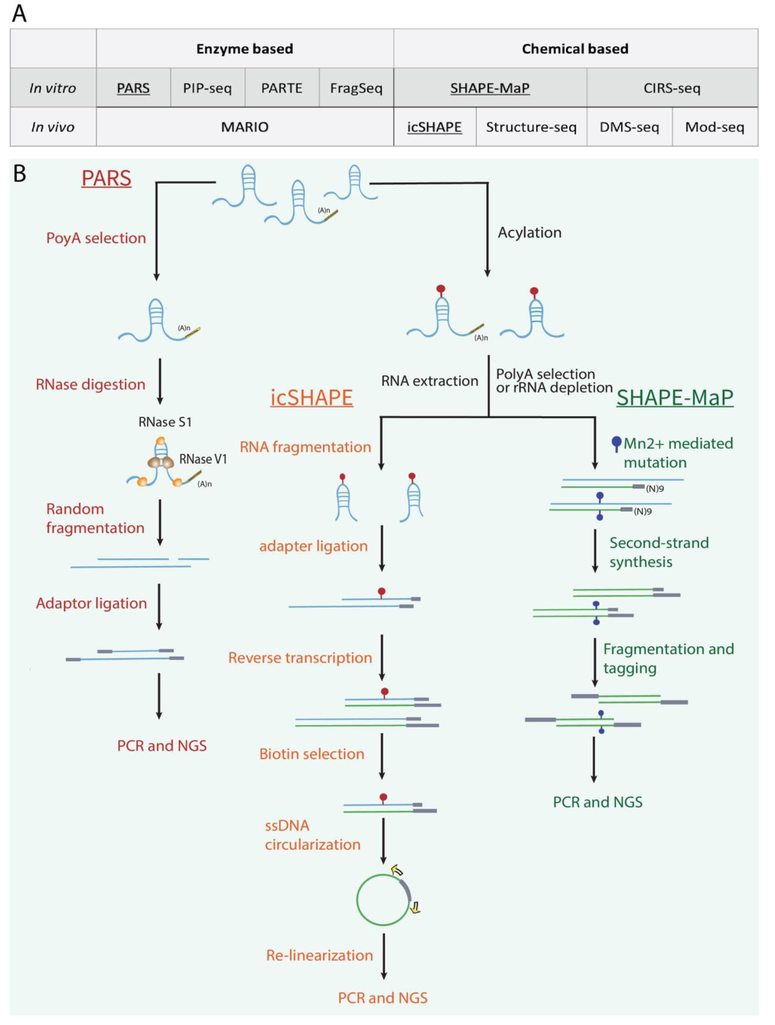
The flexibility of RNA provides a physical basis for forming a diverse array of secondary and tertiary structures. The structures of RNA and their interactions with other molecules are modulated by physiochemical environment [14-18], RNA sequences, and posttranscriptional modifications [19-22]. A general strategy employed in systematic mapping of RNA structures is to leverage enzymes or chemicals that specifically react with certain local structures [23]. These reactions include RNA cleavage or modification. The cleaved or modified sites could then be systematically revealed by sequencing. We have classified the sequencing-based RNA structure analysis methods: 1) by reagents, into enzyme-based and chemical-based approaches (columns, Figure 2, Panel A), and 2) by application scenarios, into in vitro and in vivo approaches (rows, Figure 2, Panel A). Briefly, enzyme-based in vitro RNA structure analysis methods include Protein Interaction Profile Sequencing (PIP-seq) [24, 25], Parallel Analysis of RNA Structure (PARS) [26], Parallel Analysis of RNA structures with Temperature Elevation (PARTE) [27], Fragmentation Sequencing (FragSeq) [28]. Chemical-based in vitro methods include Dimethyl Sulfate Sequencing (DMS-seq) [29], in vivo Click Selective 2-hydroxyl Acylation And Profiling Experiment (icSHAPE) [30], Structure-seq [31], and Mod-seq [32]. Chemical-based in vivo methods include Chemical Inference of RNA Structures (CIRS-seq) [33] and Selective 2'-hydroxyl Acylation analyzed by Primer Extension and Mutational Profiling (SHAPE-MaP) [34]. Finally, Mapping RNA Interactome and Structure in vivo (MARIO) is an enzyme-based analysis method that in theory captures in vivo structures [35]. In addition to revealing the single-stranded regions, MARIO also identifies all the spatially proximal regions of an RNA molecule, thus providing unique information about the secondary and tertiary structures. Figure 2B shows selected examples of these methods to illustrate their major experimental steps.
Figure 2. Sequencing-based technologies for mapping RNA structures.
(A) Summary of enzyme-based and chemical-based RNA structure technologies (columns) and their application domains (rows). Selected technologies (underscored) are expanded in detail in panels B. (B) Major steps of selected technologies. In PARS, polyA-tailed RNA is selected and divided into two pools. One pool is treated with RNase S1 that cleaves single-stranded sequence, and the other pool is treated with RNase V1 that cuts at double-stranded regions. The produced RNA segments are subjected to random fragmentation and converted into a sequencing library. In icSHAPE, cells are treated with NAI-azide, allowing for attaching a biotin moiety through copper-free CLICK reactions. SHAPE-reacted RNA segments are enriched by streptavidin-biotin interaction, and are subsequently converted into a sequencing library. In SHAPE-MaP, RNA is treated with 1M7 and is reverse transcribed in a reaction mixture that induces mutation at SHAPE-reacted sites.
The enzyme-based approaches leverage different ribonucleases (RNases) based on their selectivity in cutting either single-stranded or double-stranded regions. The resulting mixture of RNA fragments when analyzed by sequencing, allows for assessment of nucleotide accessibility and base-pairing regions, and thereby inference of secondary structures. The commonly used dsRNA-specific RNase with predefined structural preferences is RNase V1, which is, but the specificity is not absolute [36]. RNase S1 (process all four nucleotides), RNase P1 (process all four nucleotides), RNase A (ssC/U-specific) and RNase T1 (ssG-specific) are ssRNA-specific, but these enzymes may miss small bulges, loops, or mismatches [28]. Thus, integration of the sequencing data obtained from treatments with different RNases may generate more complete mapping of single- and double-stranded regions. A limitation of enzyme-based methods is that the applications are often limited to in vitro structural analysis. This is in part due to the large sizes of RNases (>10 kDa) and hence the difficulty of crossing cell membranes and susceptibility to steric hindrance in the presence of bound proteins or other RNA-associated macromolecules.
Chemical-based methods utilize small molecules to probe RNA structure. These membrane permeable molecules are utilized for in vivo analyses of RNA structures, which often achieve single nucleotide resolution. Frequently used chemicals include nucleobase-specific chemicals, carbodiimide modifying reagents, and ribose-specific probes. Nucleobase-specific chemicals including dimethyl sulfate (DMS) can modify the functional groups on the Watson-Crick (WC) face of the base. DMS alkylates the unprotected N1 position of adenine (N1A), unprotected N3 position of cytosine (N3C), and unprotected N7 position of guanine (N7G) [37]. Carbodiimide modifying reagents react with guanosine and uridine. These chemicals detect the presence of base-paired regions, allowing for mapping of the secondary structures and protein binding sites. Ribose-specific probes acylate the flexible C2’-hydroxyl group of the ribose (C2’-OH). Using such a probe, Selective 2’-Hydroxyl acylation Analyzed by Primer Extension (SHAPE) resolves the local structural environment at nucleotide resolution [38]. Flexible bases exhibit a higher tendency to adapt to specific local structural environments, which facilitates acetylation, resulting in higher SHAPE activity [39]. An advantage of SHAPE reagents over nucleobase-specific probes lies in their capability of targeting the ribose of all four nucleotides (Table 1). Ideally, combining the sequencing data generated from treatments of multiple chemicals and enzymes may release the complimentary advantages of these methods and potentially reveal more comprehensive structural information.
Table 1.
Comparison of high-throughput methods for mapping RNA structure and interactions.
| Sequencing-based for mapping RNA structures | ||
|---|---|---|
| Method | Advantages | Limitations |
| Enzyme based methods | ||
| PIP-Seq | ● Reveals both protein-bound RNA regions and RNA secondary structure. ● Provides strand-specific information. |
● Limited resolution at small nucleotide bulges and loops. |
| PARS | ● Increased sensitivity by sequencing both single-stranded and double-stranded regions. | ● RNA was folded in vitro. |
| PARTE | ● Measures melting temperature. ● Single nucleotide resolution. ● Preserves in vivo RNA modifications. ● Can infer RNA regulatory motifs. |
|
| FragSeq | ● Simple and fast protocol. ● Accompanied with modifiable software. |
|
| Chemical based methods | ||
| DMS-Seq | ● Identifies RNA structure in native conditions. ● Single nucleotide resolution. |
● Limited to the analysis of two bases (As and Cs). ● RNA binding proteins can block DMS activity. |
| icSHAPE | ● Measures base flexibility. ● Single nucleotide resolution. |
● Limited to the analysis of relatively short (~300 nt) in vitro transcribed RNAs. |
| Structure-seq | ● Single nucleotide resolution ● Applicable to both in vitro and in vivo analyses. |
● Limited to the analysis of two bases (As and Cs). ● RNA binding proteins can block DMS activity. |
| Mod-Seq | ● Can probe structures of long RNAs in vivo. ●Single nucleotide resolution. |
● Limited to the analysis of two bases (As and Cs). |
| CIRS-Seq | ● Single nucleotide resolution. ● Can identify structural requirements for RNA binding proteins. |
|
| SHAPE-MaP | ● Can be customized for different applications. ● Applicable to analysis of long RNAs. ● Can infer structural changes of single-nucleotide and other allelic polymorphisms |
● Length of the RNA must be at least ~150 nt for the randomer and native workflow, and at least ~40 nt for the small-RNA workflow. |
| Sequencing-based for mapping RNA-RNA Interactions | ||
| Method | Advantages | Limitations |
| CLASH | ● Stringent purification conditions remove non-physiological interactions. | ● Require prior knowledge of an RNA binding protein. ● Requires a good antibody. |
| hiCLIP | ● Incorporation of an adaptor between two RNA molecules increases ligation efficiency and improves accuracy in sequence mapping. | ● Require prior knowledge of an RNA binding protein. Requires a good antibody. ● No in vivo crosslinking step may incur challenges in differentiating bona fide and spurious RNA attachments. |
| PARIS | ● Many to many mapping. | ● AMT preferentially crosslinks pyrimidine bases and may introduce bias. |
| SPLASH | ● Improves signal to noise ratio by leveraging biotinylated psoralen. ● Many to many mapping. |
● Psoralen preferentially crosslinks pyrimidine bases and may introduce bias. |
| LIGR-Seq | ● Many to many mapping. | ● AMT preferentially crosslinks pyrimidine bases and may introduce bias. |
| MARIO | ● Many to many mapping. ● Incorporation of an adaptor between two RNA molecules increases ligation efficiency and improves accuracy in sequence mapping. ● Reports both between- and within-molecule interactions. ● Captures proximal regions of an RNA molecule in 3D. ● Reveals single-stranded regions of each RNA. |
● Loses RNA duplexes that are not associated with any proteins. |
| Sequencing-based for mapping RNA-DNA Interactions | ||
| Method | Advantages | Limitations |
| ChIRP | ● Tilling the entire transcript with antisense DNA. | ● Limited to analyzing RNA at a time. |
| CHART | ● Tilling the RNase H accessible region by antisense DNA. | ● Limited to analyzing RNA at a time. |
| RAP | ● Tilling the entire transcript with complimentary RNA. | ● Limited to analyzing RNA at a time. ● Limited to analysis of long RNA. |
| MARGI | ● Many to many mapping. ● Captures interaction at native conditions. |
● Require a large number (107) of cells. |
| ChAR-seq | ● Many to many mapping. ● Proximity ligation is performed in nuclei, which reduces nonspecific interactions. |
● Only sequencing reads that cover the entire bridge sequence are informative, reducing the number of informative reads. |
| GRID-seq | ● Many to many mapping. ● Proximity ligation is performed in nuclei, which reduces nonspecific interactions. |
● The informative sequence lengths on the RNA side and the DNA side are both limited to ~20 bases, resulting in challenges in unambiguous sequence mapping. |
Toward understanding sequence and environmental determinants of RNA structures
High-throughput mapping of RNA structures has been completed for the HIV-1 RNA genome [38], bacteria [40] [41], yeast [26], Arabidopsis [31], Drosophila [42], Caenorhabditis elegans [42], and selected cell types in mouse [30, 33] and human [25, 29, 43]. These high-throughput analyses offered insights to sequence and environmental determinants of RNA structures.
Sequence determinants of RNA structures have been identified using the above based methods. In addition to base pairing, a triplet repeat pattern emerged from both in vitro [41] and in vivo [31] experiments. This repeat pattern in chemical/enzyme reactivity is indicative of existence of combinatorial rules of sequence motif arrangement for determining RNA structure [26, 31, 33, 41, 43-47]. Furthermore, single-nucleotide polymorphisms (SNP) were found to correlate with variations in RNA structures [43]. Thousands of riboSNitches (SNP-mediated RNA structure switch) were discovered in healthy human parent-offspring trios. Approximately 200 riboSNitches overlapped with expression quantitative trait loci (eQTL), and 22 riboSNitches overlapped with disease-associated SNPs. These data suggested that personal genomic difference could result in variation of personal traits through altering RNA structure [43]. However, it is likely that only a small portion of sequence determinants of mRNA structures have been identified. Taking E. coli for example, “operonic mRNAs are comprised of ORF-wide units of secondary structure that vary across ORF boundaries such that adjacent ORFs on the same mRNA molecule are structurally distinct’ [40]. It is difficult to conceive a model with currently available sequence-to-structure information to completely explain how sequence could orchestrate such a structural arrangement.
Diverse environmental factors can modulate RNA structures [14-18]. For example, cold shock induces a global decrease of mRNA secondary structures in E. coli, which correlates with increased translation [48]. In vitro, RNA generally appears more structured than in vivo [29-31], which is partially attributable to different Mg2+ concentrations [29] and accessibility to RNA-binding proteins. Melting and refolding of an mRNA results in different structures as revealed by SHAPE-MaP [49]. By quantifying the reactivity differences between in vivo and in vitro conditions [30] and between in cellulo and ex vivo conditions [50], two teams were able to reveal protein-bound RNA regions. By adding cross-linking and proximity ligation steps, the MARIO team identified a case of protein-assisted RNA folding [35]. It remains a challenge to integrate RNA sequence and cellular context for deriving the most compatible structure from diverse types of structure probing assays.
RNA-RNA INTERACTIONS
Sequencing-based methods for mapping RNA-RNA interactions
Methods for analysis of intermolecular RNA-RNA interactions were restricted to targeting a specific RNA that participates in RNA-RNA interactions, until it was discovered that chimeric RNAs can be extracted from RNA sequencing data [51]. Although these chimeric RNAs are present in low frequencies, they could represent pairs of interacting RNAs [52]. Two subsequent methods, Crosslinking, Ligation, And Sequencing of Hybrids (CLASH) [53] and RNA Hybrid and Individual-Nucleotide Resolution UV Cross-Linking and Immunoprecipitation (hiCLIP) [54] enriched for the interacting RNAs by purifying a specific protein that is required for such interactions. The major difference between these two methods lies in utility of ectopic expression of a tagged protein (CLASH) versus antibody-based isolation of the protein of interest from unperturbed cells (hiCLIP). CLASH and hiCLIP broke the barrier of having to target a specific RNA in identifying RNA-RNA interactions. These technologies enabled identification of RNA interactions mediated by a specific protein.
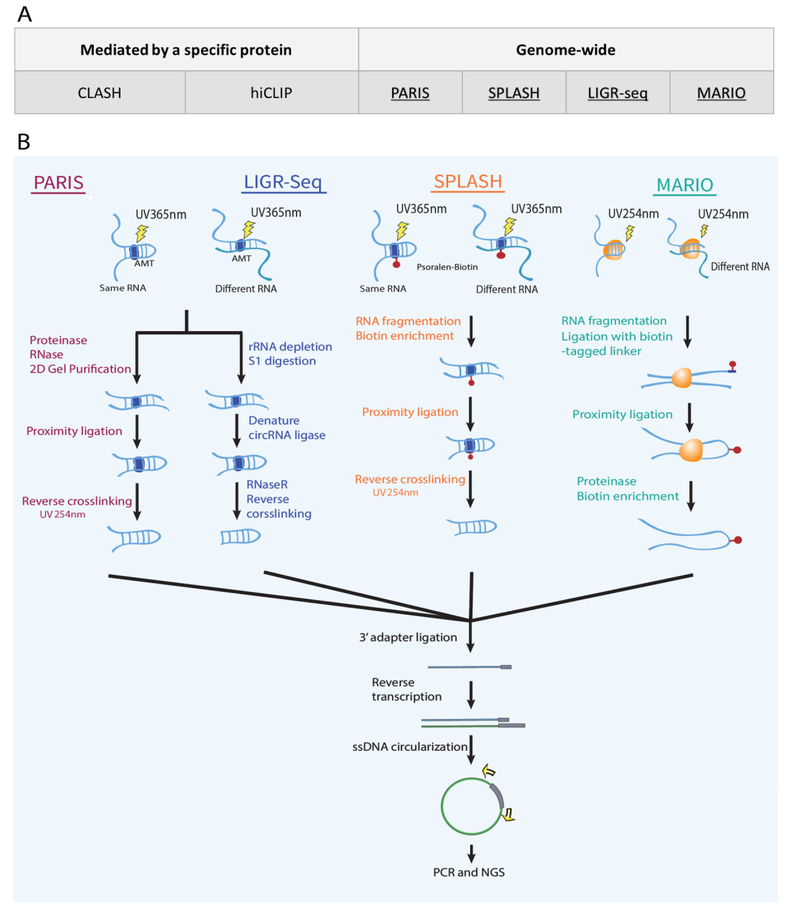
High-throughput RNA interactome analysis was enabled by a cohort of four methods, including Psoralen Analysis Of RNA Interactions And Structures (PARIS) [55], Sequencing Of Psoralen-Crosslinked, Ligated, And Selected Hybrids (SPLASH) [56], Ligation of Interacting RNA Followed By High-Throughput Sequencing (LIGR-seq) [57], and Mapping RNA Interactome and Structure in vivo (MARIO) [35] (Figure 3). The central idea of these technologies is to leverage proximity ligation to produce chimeric sequences. All methods used in vivo crosslinking of RNA, either by UV-mediated RNA-protein crosslinking (MARIO), or RNA duplex crosslinking enabled by psoralen derivatives (PARIS, SPLASH, LIGR-seq), followed by RNA fragmentation to produce single-stranded RNA ends, which were subjected to intramolecular ligation and reverse-crosslinking to convert into a sequencing library. The different choice of crosslinking reagents led to revelation of several types of RNA interactions. PARIS, SPLASH and LIGR-Seq used psoralen or its derivatives including 4’-aminomethyltrioxsalen (AMT) and biotinylated psoralen, which intercalate in RNA helices and undergo interstranded cross-link upon 365 nm UV irradiation. MARIO crosslinked RNAs with proteins and ligated the RNAs bound by the same protein molecules. PARIS, SPLASH, LIGR-seq were designed for identifying hybridized RNA pairs, whereas MARIO was designed for identifying all RNA pairs brought together by any protein without requiring RNA-RNA hybridization. A bias of psoralen-based crosslinking methods is introduced by psoralen’s preferential activities with pyrimidines [58]. Combining MARIO with one of the psoralen-based methods (PARIS, SPLASH, and LIGR-seq) may lead to a more comprehensive view of the RNA interactome.
Figure 3. Sequencing-based technologies for mapping RNA-RNA interactions.
(A) Summary of antibody-based methods that analyze interactions mediated by a specific protein (left column) and genome-wide methods without targeting any specific proteins (right column). Selected technologies (underscored) are expanded in panels B. (B) Major steps of selected technologies. In PARIS, double-stranded RNA regions are crosslinked by AMT and UV. RNA is purified and subjected to proximity ligation. The resulting RNA is ligated with a 3’ adaptor and converted into a sequencing library. SPLASH procedure is similar to PARIS, except that instead of AMT, biotinylated psoralen is used as the crosslinking reagent, which allows for enrichment of double-stranded regions. LIGR-Seq used a similar experimental strategy, with different choices of RNA purification, treatment and ligation steps. In MARIO, RNA-protein complexes are crosslinked by UV. RNA is randomly fragmented and ligated with a biotinylated linker sequence and then subjected to proximity ligation. The resulting RNA-linker-RNA chimeric sequences are purified by streptavidin-biotin interaction and converted into a sequencing library.
RNA interactome as a scale-free network
Lack of specificity was once considered a theme in miRNA interaction with its target mRNAs. This phenomenon was also referred to as promiscuity in miRNA targeting. The promiscuity was supported by many complementary sequences in the transcriptome, as well as changes in transcript abundances when the endogenous concentration of a miRNA was perturbed [59-61]. However, when applied to unperturbed cells, none of the four high-throughput assays (PARIS, SPLASHs, LIGR-seq, MARIO) reported many targets for most of the miRNAs. Instead, in embryonic stem cells and in mouse brain, most of the miRNAs exhibited only 1 to 3 mRNA targets [35]. Only a handful of miRNAs exhibited more than 10 mRNA targets. In addition, most lincRNAs also appeared to each target only one or a few mRNAs. More generally, the MARIO authors found that the RNA interactome follows the power-law and is a scale-free network [35]. Nearly all other molecular networks being studied were reported to be scale-free [62, 63], whereas the promiscuity of miRNA-involved interactions would argue against the scale-free property in an RNA interactome. However, the in vivo high-throughput data suggested that in endogenous cellular conditions, the RNA interactome remains scale-free and therefore does not present an exception to the power-law, a physics rule of biological networks [62].
Sno-miR: A new gene repertoire of regulatory RNAs
Abundant interactions between snoRNAs and mRNAs were reported from all four assays (PARIS, SPLASH, LIGR-seq, MARIO). The identified snoRNA interaction sites on mRNAs were enriched with pseudouridylation sites [35], consistent with the contribution of snoRNAs to the pseudouridylation process. However, many identified interactions involved truncated forms of snoRNAs rather than the entire snoRNAs [35]. These truncation forms were present in the cells, as revealed by small RNA sequencing, and were bound by AGO2 as revealed by High-Throughput Sequencing of RNA Isolated By Crosslinking Immunoprecipitation (CLIP-seq) [35]. Knocking down the SNORD83B snoRNA resulted in increased abundances of three out of four of LIGR-seq identified SNORD83B targeting mRNAs [57]. Taken together, more than 170 snoRNA genes appeared to produce miRNA-like RNAs, which interact with mRNAs. The snoRNA-originated miRNA-like RNAs (sno-miR) could be a new repertoire of regulatory RNAs. Indeed, one of the human snoRNAs is processed by DICER and mediate mRNA silencing through AGO1 and AGO2 [64].
Connections between mRNA-mRNA interaction and coordinated regulation
Many mRNA-mRNA interactions were identified by all four assays (PARIS, SPLASH, LIGR-seq, and MARIO) [35, 55-57]. In humans and mice, approximately 1,000 mRNA pairs interact by base pairing [56], and more than 5,000 mRNA pairs were brought together by the same protein [35]. Base complementation was significant even in MARIO identified mRNA-mRNA interactions, where the experimental procedure did not select for base paired RNA pairs [35]. Interactions at sites near start codons negatively correlated with translation efficiency, whereas intra-molecular interactions of the two ends of mRNA molecules positively correlated translation efficiency, suggesting a link between RNA interaction and translational control [56]. Furthermore, interacting mRNA pairs tended to encode for proteins that co-localize to the same subcellular compartments and sometimes exhibited similar translation efficiencies or RNA decay rates, suggesting mRNA-mRNA interaction as a means of co-regulation of gene expression [56]. Although the mechanisms underscoring the similar kinetic rates remain largely unknown, mRNA duplexes with complementary Alu sequences in their 3’ untranslated regions (UTRs) could trigger Staufen1-mediated RNA decay [65]. In addition, intermolecular hybridization of RNA molecules could promote phase transition [49], a process underscoring the formation of liquid droplets including stress granules, P-bodies, and nuclear speckles [66]. The created liquid droplets may provide physical substrates for segregating mRNAs into different pools for coordinated RNA metabolism in each pool [67, 68].
Pseudogenes and transposons produce RNAs that interact with mRNAs
Large numbers of transcripts produced from pseudogenes and transposons were reported to interact with mRNAs [35]. Pseudogene RNAs interacted with both exonic and intronic regions of mRNAs. Both pseudogene-exon and pseudogene-intron interactions exhibited significant base pairing [35]. Significant base pairing was also observed in interacting Long Interspersed Nuclear Element (LINE)_RNA-mRNA pairs and Long Terminal Repeat (LTR)_RNA-mRNA pairs [35]. The interaction sites on pseudogene RNAs and mRNAs exhibited increased interspecies conservation levels than other parts of the pseudogenes and mRNAs, suggesting the pseudogene_RNA-mRNA interactions were evolutionarily selected for [35]. These novel interactions indicate a subset of pseudogenes and transposons may function by providing mRNA-interacting transcripts. In line with this conjecture, Alu-repeat-containing polyadenylated long noncoding RNA (lncRNA) when duplexed with mRNAs’ 3’UTR could trigger Staufen1-mediated RNA decay [69]. However, the in vivo identified RNA duplexes contained imperfect base-pairing of many other types of repeat sequences, which may serve as starting points for searches of Staufen-independent regulatory pathways.
RNA-DNA INTERACTIONS
Sequencing-based methods for mapping RNA-DNA interactions
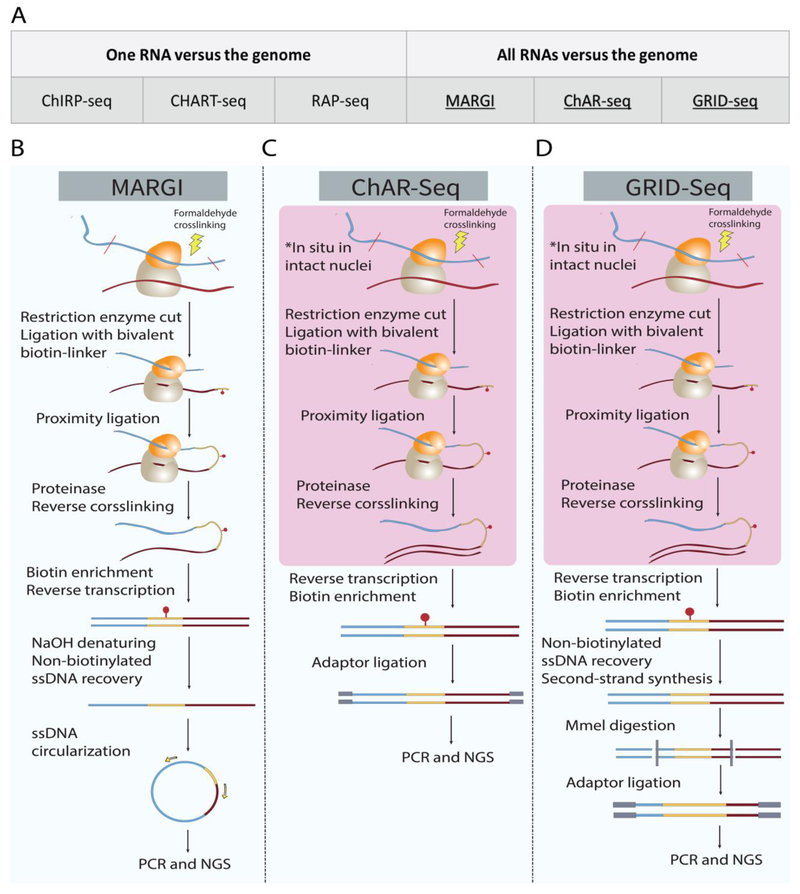
Earlier technology developments focused on mapping genome-wide locations of a specific RNA (one RNA versus the genome, Figure 4A), including Chromatin Isolation by RNA Purification (ChIRP) [70], Capture Hybridization Analysis of RNA Targets (CHART) [71] and RNA Antisense Purification (RAP) [72]. These technologies utilize biotinylated complementary oligonucleotides to pull down a specific target RNA together with its binding partners. The identities of its DNA- or protein-binding partners are subsequently revealed by sequencing or mass spectrometry. A more recent cohort of technologies enabled mapping (possibly) all chromatin-interacting RNAs together with each RNA’s genomic interacting regions (all RNAs versus the genome, Figure 4A), including Mapping RNA-Genome Interactions (MARGI) [73], Chromatin-Associated RNA Sequencing (ChAR-seq) [74] and Mapping Global RNA Interactions With DNA by Deep Sequencing (GRID-seq) [75]. These methods leverage proximity ligation to convert RNA and its proximal DNA sequence into a chimeric sequence that can be read out by sequencing. A major advantage of these ligation-based methods is their capability of discovering de novo chromatin-associated RNAs.
Figure 4. Sequencing-based technologies for mapping RNA-DNA interactions.
(A) Summary of technologies for RNA-DNA interactions based on a specific RNA (left column) or any RNA (right column). Selected technologies (underscored) are expanded in detail in panels B-D. (B-D) Major steps of selected technologies. (B) In MARGI, protein-RNA-DNA complexes are crosslinked by formaldehyde. DNA is fragmented. RNA is ligated with the RNA-end of a biotinylated half-RNA-half-DNA linker, and the DNA-end of this linker is subsequently ligated to DNA through proximity ligation. The resulting chimeric RNA-DNA sequences are selected by streptavidin-biotin interactions and converted into a sequencing library. (C-D) The ChAR-Seq and GRID-seq procedures are similar to MARGI. The major difference is that many steps are conducted in intact nuclei, including restriction enzyme digestion, RNA-linker ligation, and proximity ligation.
In all these techniques, cells are first subjected to cross-linking reagents to preserve protein-nucleic acid interactions. ChIRP, CHART, and RAP focus on capturing chromatin interactions of individual RNAs. All use synthetic biotinylated antisense DNA oligonucleotides designed specifically to capture and purify lncRNA-chromatin complexes from the cells. Due to the inherent stickiness of RNA and propelled by the need to maximize both specificity and recovery of the RNA of interest, chemical crosslinking is used to allow stringent manipulations of pulldown experiments. Crosslinking coupled with sonication as well as denaturing washing conditions are to ensure that non-physiological bindings formed in vitro upon cell lysis are removed. ChIRP, CHART, and RAP are very similar in overall approach with differences in specific crosslinking reagents, strength of chromatin shearing, strength of washing buffer, density and length of antisense probes. Without much prior knowledge about the local structures of RNA such as folding, and interacting proteins, it is difficult to design only a few probes that ensure consistent performance of every pulldown experiments. Taking this consideration into account, ChIRP and RAP do not rely on any knowledge of the RNA of target. Instead, tiling probes that are spaced across the entire RNA are used. This design maximized the chances of capturing the entire length of fragmented RNAs, which is usually sheared in advance into smaller species. Chromatin shearing by sonication, however brief, is almost always required to efficiently lyse chemically crosslinked cells. ChIRP uses substantial sonication to fragment RNA into hundreds of nucleotides length. On the other hand, RAP only employs brief sonication for solubilizing the chromatin to minimize the chances of damaging the target RNA. ChIRP uses 20-mer probes that can be easily synthesized while RAP uses 120-mer probes which would require in vitro transcription to prepare [76]. A variation of ChIRP, domain ChIRP (dChIRP) [77] designs probe sets by iteratively finding the minimal set of probes targeting the chromatin-interacting region of a RNA, which can result in higher signal-to-noise ratio.
In all-RNA-versus-genome approaches (MARGI [73], ChAR-Seq [74], GRID-seq [75]), a bivalent, and biotinylated linker comprising both single-stranded RNA at one end and double-stranded DNA at another end is used to link RNA to DNA by proximity ligation. The RNA end of the linker is first ligated to RNA molecule. Next, the DNA end of the linker is proximity-ligated to the DNA molecule. The biotinylated linker enables enrichment of desirable chimeric RNA-DNA segments. The procedure is followed by amplification and deep sequencing to detect the RNA-DNA interactions. In MARGI, the linker ligation and proximity ligation were performed on RNA-DNA protein complexes that are tethered to the solid surface of streptavidin beads [73]. In contrast, ChAR-Seq and GRID-seq performed those steps in intact nuclei [74, 75].
Diverse modes of RNA-chromatin interactions
Several modes of RNA-chromatin interactions have been revealed. Some lincRNAs bind to chromatin in a localized fashion reminiscent of transcription factor binding while others spread wider in binding locations [70-72, 78]. lincRNAs can interact in cis near their sites of transcription or in trans to regions on different chromosomes [11, 70, 79]. Some lncRNAs only interact with a few genomic loci while others interact promiscuously with multiple genomic regions. Some lncRNAs or 5’UTRs [79] act as repressors, whereas others function as activators [11] of gene expression. Some lncRNAs, Xist for example, spread their binding regions by using the three-dimensional organization of the chromatin to spread to spatially proximal genomic loci [72]. It remains an open question whether there are principles that can explain the diverse types and functions of observed RNA-chromatin interactions.
RNA-DNA interaction on transcription start sites: a genome-wide phenomenon
An overriding theme emerged from the genome perspective. That is nearly all promoters [75] or more specifically nearly all transcription start sites (TSS) [73] are associated with trans-interacting RNAs. Even the TSSs of silent genes are associated with RNAs. However, the amount of TSS-associated RNAs exhibited weak correlation with the expression level of TSS-specified genes [73], suggesting RNA attachment on TSS may promote transcription. Consistent with this idea, a case study of TSS-associated antisense RNA suggested such an interaction promotes the transcription of the TSS-specified gene [80].
Enhancer-promoter interaction offers a possible explanation for the large amounts of TSS-RNA interactions. In this model, transcripts produced from enhancers can associate with promoters either as a result of or as a contributing cause to enhancer-promoter interactions. Conversely, many enhancers were found associated with the transcripts of their supposedly regulating genes [75]. However, enhancer-promoter interactions cannot completely explain the symmetric pattern of RNA attachment, centering at each TSS [73]. It remains to be tested if RNA attachment on TSSs is a molecular mechanism for specifying the start sites of transcription. After all, unlike yeasts that have characteristic binding sites including TATA and CAAT boxes located at defined distances which could help the transcription machinery to pinpoint the locations to initiate transcription [81], the vertebrate promoters do not necessarily contain a predefined set of transcription factor binding sites at fixed distances to TSS [82].
RNA decoration on chromatin as a new layer of epigenome
RNA attachment was found to positively correlate with histone modifications H3K27ac and H3K4me3, both of which are associated with more open chromatin regions and active transcription [73]. The genomic regions enriched with trans- interacting RNAs (RNA attachment hotspots) were clearly correlated with H3K9me3 depleted regions [73]. These general observations, however, do not seem to apply to every type of RNAs. An exception lies in snoRNAs, of which both MARGI [73] and CHAR-seq [74] reported extensive interactions with chromatin, but primarily with heterochromatin [74]. More intriguingly, ChAR-Seq analysis revealed that TSS-associated RNAs are enriched at topologically associated domain (TAD) boundaries, corroborating with the potential role of active transcription in shaping the topological organization of the genome [74]. Although the above-mentioned associations await further validations, these RNA attachments may act inter-dependently or coordinately to the hitherto better characterized DNA-and histone- modifications, thus constitute a novel layer of chromatin modifications that contribute to gene regulation.
Concluding remarks
The recent eruption of technologies propelled RNA biology to new frontiers. High-throughput methods derived rich information on RNA-participating interactions. These interactions are indicative of a wide range of new functions. Coupling the identified interactions with genome editing or RNA perturbation experiments may lead to new insights on the functions of the RNA-producing genomic sequences (See Outstanding Questions).
RNA could exhibit surprising functions in different subcellular compartments, as exemplified by the miR-1 miRNA in enhancing translation in mitochondria [83] and the LINK-A lncRNA in binding with inner membrane lipids and transducing PI3K-Akt signaling [84, 85]. It would be exciting to reveal all RNA-participating interactions in each type of membrane- and membraneless- organelles including mitochondria, exosomes, stress granules, and P-bodies. A potential approach is to combine the recently developed P-body purification method [67] or its variation with a high-throughput RNA-interaction detection method. The resulting organelle-level RNA interactomes could illuminate intricate organelle-specific molecular machinery, thus revealing the context-specificity of the multiple functions of each (class of) RNA. Organelle-specific RNA interactomes would facilitate studies on liquid-liquid phase separations in nucleus and cytoplasm [86], thus contributing to addressing fundamental questions including how the 3 dimensional (3D) organization of the nucleus is coordinated [87].
Revealing the principles underlying 3D organization of nuclear components has become a focal international endeavor [87]. A bottleneck is the shortage of high-throughput methods capable of identifying spatially proximal molecular units that do not physically interact [88] [89]. Most methods require introduction of some forms of engineered Ascorbate Peroxidase (APEX) as proximity labeling reagents [88, 89]. It turns out that endogenous RNAs could serve as proximity labeling media for identifying proximal regions of different membraneless organelles, due to the nature of in vivo proximity ligation in MARGI [90]. Combining in vivo proximity-ligation methods with innovative use of endogenous RNA distribution could transform the means of studying spatial organization of membraneless organelles, both in nucleus and in cytoplasm. Future work on between- and within- organelle RNA interactions may connect the dots from “structure” to function, where the structure is the 3D organization of subcellular organelles.
Outstanding questions.
What are the sequence motifs and grammar that determine RNA structures? How do the structures change in response to defined changes of environmental factors?
What are the functions of pseudogene_RNA-mRNA, transposon_RNA-mRNA, lincRNA-mRNA, and mRNA-mRNA interactions?
SnoRNA genes produce miRNA-like small RNAs (sno-miR). What are the regulatory functions of these sno-miRs?
Do RNA-RNA interactions promote protein-protein interactions or facilitate signal transduction?
Why are most transcription start sites attached with RNAs that are transcribed from other genomic regions?
How does personal genomic difference affect personal variations in RNA-RNA interactions and RNA-DNA interactions?
RNA attachment to chromatin could be regarded as a type of epigenomic modifications. Could RNA-DNA interaction be passed on across generations?
Highlights.
New technologies have enabled systematic mapping of RNA-RNA interactions, revealing tens of thousands of such interactions.
In endogenous cellular conditions, miRNAs and lincRNAs tend to specifically target one or a few mRNAs, indicating that the entire RNA interactome is a scale-free network.
Hundreds of snoRNA genes produce miRNA-like short RNAs which interact with mRNAs, thus providing a gene repertoire of new regulatory RNAs.
Pseudogenes and transposons produce RNAs that interact with mRNAs, through base pairing. The interaction regions exhibit increased inter-species conservation levels.
New technologies enabled systematic mapping of RNA-DNA interactions, revealing hundreds of chromatin-interacting RNAs.
Acknowledgements
We thank Dr. Zhen Chen for critical comments to this work. We apologize for not able to cite all relevant work due to limited space. This work is supported by NIH DP1HD087990. S.Z. is a co-founder and a board member of Genemo Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Lee RC et al. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 (5), 843–54. [DOI] [PubMed] [Google Scholar]
- 2.Fire A et al. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 (6669), 806–811. [DOI] [PubMed] [Google Scholar]
- 3.Burge C et al. (1999) Splicing of Precursors to mRNAs by the Spliceosomes. In The RNA World, CSH Monographs. [Google Scholar]
- 4.Valadkhan S and Manley JL (2001) Splicing-related catalysis by protein-free snRNAs. Nature 413 (6857), 701–7. [DOI] [PubMed] [Google Scholar]
- 5.Ni J et al. (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89 (4), 565–73. [DOI] [PubMed] [Google Scholar]
- 6.Penny GD et al. (1996) Requirement for Xist in X chromosome inactivation. Nature 379 (6561), 131–7. [DOI] [PubMed] [Google Scholar]
- 7.Yang F et al. (2015) The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol 16, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu R et al. (2018) Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation. Nature 558 (7711), 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singer MS and Gottschling DE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266 (5184), 404–9. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe T et al. (2011) Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332 (6031), 848–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao Y et al. (2018) Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nature communications 9 (1), 292–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Place RF et al. (2008) MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A 105 (5), 1608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinn JL et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129 (7), 1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgs PG (2000) RNA secondary structure: physical and computational aspects. Quarterly reviews of biophysics 33 (3), 199–253. [DOI] [PubMed] [Google Scholar]
- 15.Kolkenbeck K and Zundel G (1975) The significance of the 2' OH group and the influence of cations on the secondary structure of the RNA backbone. Biophysics of structure and mechanism 1 (3), 203–219. [DOI] [PubMed] [Google Scholar]
- 16.Egli M et al. (2005) Probing the influence of stereoelectronic effects on the biophysical properties of oligonucleotides: comprehensive analysis of the RNA affinity, nuclease resistance, and crystal structure of ten 2'-O-ribonucleic acid modifications. Biochemistry 44 (25), 9045–9057. [DOI] [PubMed] [Google Scholar]
- 17.Downey CD et al. (2007) Influence of Hydrostatic Pressure and Cosolutes on RNA Tertiary Structure. Journal of the American Chemical Society 129 (30), 9290–9291. [DOI] [PubMed] [Google Scholar]
- 18.Ribitsch G et al. (1985) Small-angle X-ray and light scattering studies on the influence of Mg2+ ions on the structure of the RNA from bacteriophage MS2. Zeitschrift fur Naturforschung. Section C, Biosciences 40 (3–4), 234–241. [DOI] [PubMed] [Google Scholar]
- 19.Lewis CJT et al. (2017) RNA modifications and structures cooperate to guide RNA-protein interactions. Nature Reviews Molecular Cell Biology 18, 202–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu N et al. (2015) N-6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. NATURE 518 (7540), 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y et al. (2014) N-6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. NATURE CELL BIOLOGY 16 (2), 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng G et al. (2013) ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Molecular Cell 49 (1), 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson JF and Hearst JE (1983) Structure of E. coli 16S RNA elucidated by psoralen crosslinking. Cell 32 (4), 1355–65. [DOI] [PubMed] [Google Scholar]
- 24.Silverman IM et al. (2014) RNase-mediated protein footprint sequencing reveals protein-binding sites throughout the human transcriptome. Genome Biology 15 (1, SI). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silverman IM and Gregory BD (2015) Transcriptome-wide ribonuclease-mediated protein footprinting to identify RNA-protein interaction sites. Methods 72, 76–85. [DOI] [PubMed] [Google Scholar]
- 26.Kertesz M et al. (2010) Genome-wide measurement of RNA secondary structure in yeast. NATURE 467 (7311), 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan Y et al. (2012) Genome-wide Measurement of RNA Folding Energies. MOLECULAR CELL 48 (2), 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Underwood JG et al. (2010) FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. NATURE METHODS 7 (12), 995–U81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouskin S et al. (2014) Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. NATURE 505 (7485), 701+–701+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spitale RC et al. (2015) Structural imprints in vivo decode RNA regulatory mechanisms. NATURE 519 (7544), 486+–486+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y et al. (2014) In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. NATURE 505 (7485), 696+–696+. [DOI] [PubMed] [Google Scholar]
- 32.Talkish J et al. (2014) Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA 20 (5), 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Incarnato D et al. (2014) Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. GENOME BIOLOGY 15 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siegfried NA et al. (2014) RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). NATURE METHODS 11 (9), 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen TC et al. (2016) Mapping RNA-RNA interactome and RNA structure in vivo by MARIO. Nature Communications 7, 12023–12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowman HB and Draper DE (1986) On the recognition of helical RNA by cobra venom V1 nuclease. JOURNAL OF BIOLOGICAL CHEMISTRY 261 (12), 5396–5403. [PubMed] [Google Scholar]
- 37.Kwok CK (2016) Dawn of the in vivo RNA structurome and interactome. Biochemical Society Transactions 44 (5), 1395–1410. [DOI] [PubMed] [Google Scholar]
- 38.Watts JM et al. (2009) Architecture and secondary structure of an entire HIV-1 RNA genome. NATURE 460 (7256), 711–U87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGinnis JL et al. (2012) The Mechanisms of RNA SHAPE Chemistry. Journal of the American Chemical Society 134 (15), 6617–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burkhardt DH et al. (2017) Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Del Campo C et al. (2015) Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function. PLOS GENETICS 11 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li F et al. (2012) Global Analysis of RNA Secondary Structure in Two Metazoans. CELL REPORTS 1 (1), 69–82. [DOI] [PubMed] [Google Scholar]
- 43.Wan Y et al. (2014) Landscape and variation of RNA secondary structure across the human transcriptome. NATURE 505 (7485), 706+–706+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Editorial (2002) Folding as grammar. NATURE STRUCTURAL & MOLECULAR BIOLOGY 9 (10), 713. [DOI] [PubMed] [Google Scholar]
- 45.Wj Anderson J et al. (2012) Evolving stochastic context--free grammars for RNA secondary structure prediction. BMC Bioinformatics 13, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bevilacqua PC et al. (2016) Genome-Wide Analysis of RNA Secondary Structure. Annual Review of Genetics 50 (1), 235–266. [DOI] [PubMed] [Google Scholar]
- 47.Shabalina SA et al. (2006) A periodic pattern of mRNA secondary structure created by the genetic code. NUCLEIC ACIDS RESEARCH 34 (8), 2428–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y et al. (2018) A Stress Response that Monitors and Regulates mRNA Structure Is Central to Cold Shock Adaptation. Mol Cell 70 (2), 274–286 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langdon EM et al. (2018) mRNA structure determines specificity of a polyQ-driven phase separation. Science 360 (6391), 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smola MJ et al. (2015) Detection of RNA-Protein Interactions in Living Cells with SHAPE. BIOCHEMISTRY 54 (46), 6867–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kudla G et al. (2011) Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA 108 (24), 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bohnsack MT et al. (2012) Identification of RNA helicase target sites by UV cross-linking and analysis of cDNA. Methods in enzymology 511, 275–288. [DOI] [PubMed] [Google Scholar]
- 53.Helwak A et al. (2018) Mapping the Human miRNA Interactome by CLASH Reveals Frequent Noncanonical Binding. Cell 153 (3), 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugimoto Y et al. (2015) hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. NATURE 519 (7544), 491+–491+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu Z et al. (2016) RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. CELL 165 (5), 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aw JGA et al. (2016) In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. MOLECULAR CELL 62 (4), 603–617. [DOI] [PubMed] [Google Scholar]
- 57.Sharma E et al. (2016) Global Mapping of Human RNA-RNA Interactions. MOLECULAR CELL 62 (4), 618–626. [DOI] [PubMed] [Google Scholar]
- 58.Nawy T (2016) Topographical transcriptomes. NATURE METHODS 13 (7), 544–545. [Google Scholar]
- 59.Du T and Zamore PD (2007) Beginning to understand microRNA function. Cell Research 17, 661–661. [DOI] [PubMed] [Google Scholar]
- 60.Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 (2), 281–297. [DOI] [PubMed] [Google Scholar]
- 61.Chi SW et al. (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460 (7254), 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barabasi A-L and Oltvai ZN (2004) Network biology: understanding the cell's functional organization. Nature reviews. Genetics 5 (2), 101–113. [DOI] [PubMed] [Google Scholar]
- 63.Oltvai ZN and Barabasi A-L (2002) Systems biology. Life's complexity pyramid. Science (New York, N.Y.) 298 (5594), 763–764. [DOI] [PubMed] [Google Scholar]
- 64.Ender C et al. (2008) A human snoRNA with microRNA-like functions. Molecular cell 32 (4), 519–528. [DOI] [PubMed] [Google Scholar]
- 65.Gong C et al. (2013) mRNA-mRNA duplexes that autoelicit Staufen1-mediated mRNA decay. Nat Struct Mol Biol 20 (10), 1214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polymenidou M (2018) The RNA face of phase separation. Science 360 (6391), 859–860. [DOI] [PubMed] [Google Scholar]
- 67.Hubstenberger A et al. (2017) P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol Cell 68 (1), 144–157 e5. [DOI] [PubMed] [Google Scholar]
- 68.Spector DL and Lamond AI (2011) Nuclear speckles. Cold Spring Harb Perspect Biol 3 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gong C and Maquat LE (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature 470 (7333), 284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chu C et al. (2011) Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. MOLECULAR CELL 44 (4), 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simon MD et al. (2011) The genomic binding sites of a noncoding RNA. PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA 108 (51), 20497–20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Engreitz JM et al. (2013) The Xist lncRNA Exploits Three-Dimensional Genome Architecture to Spread Across the X Chromosome. SCIENCE 341 (6147), 767+–767+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sridhar B et al. (2017) Systematic Mapping of RNA-Chromatin Interactions In Vivo. Current Biology 27 (4), 602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bell JC et al. (2018) Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li X et al. (2017) GRID-seq reveals the global RNA-chromatin interactome. Nature Biotechnology 35, 940–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chu C et al. (2015) Technologies to probe functions and mechanisms of long noncoding RNAs. Nature structural & molecular biology 22 (1), 29–35. [DOI] [PubMed] [Google Scholar]
- 77.Quinn JJ et al. (2014) Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. NATURE BIOTECHNOLOGY 32 (9), 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simon MD et al. (2013) High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. NATURE 504 (7480), 465+–465+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Colak D et al. (2014) Promoter-Bound Trinucleotide Repeat mRNA Drives Epigenetic Silencing in Fragile X Syndrome. SCIENCE 343 (6174), 1002–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo S et al. (2016) Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell stem cell 18 (5), 637–652. [DOI] [PubMed] [Google Scholar]
- 81.Pugh BF and Tjian R (1991) Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev 5 (11), 1935–45. [DOI] [PubMed] [Google Scholar]
- 82.Smale ST and Kadonaga JT (2003) The RNA polymerase II core promoter. Annu Rev Biochem 72, 449–79. [DOI] [PubMed] [Google Scholar]
- 83.Zhang X et al. (2014) MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158 (3), 607–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin A et al. (2017) The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol 19 (3), 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krause HM (2018) New and Prospective Roles for lncRNAs in Organelle Formation and Function. Trends Genet. [DOI] [PubMed] [Google Scholar]
- 86.Maharana S et al. (2018) RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360 (6391), 918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dekker J et al. (2017) The 4D nucleome project. Nature 549 (7671), 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han S et al. (2017) Proximity Biotinylation as a Method for Mapping Proteins Associated with mtDNA in Living Cells. Cell Chem Biol 24 (3), 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Myers SA et al. (2018) Discovery of proteins associated with a predefined genomic locus via dCas9-APEX-mediated proximity labeling. Nat Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen W et al. (2018) RNAs as Proximity-Labeling Media for Identifying Nuclear Speckle Positions Relative to the Genome. iScience 4, 204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]