Abstract
Purpose
BRAF inhibitors are clinically active in patients with advanced BRAFV600-mutant melanoma, although acquired resistance remains common. Preclinical studies demonstrated that resistance could be overcome using concurrent treatment with the HSP90 inhibitor XL888.
Methods
Vemurafenib (960 mg PO BID) combined with escalating doses of XL888 (30, 45, 90 or 135 mg PO twice weekly) was investigated in 21 patients with advanced BRAFV600-mutant melanoma. Primary endpoints were safety and determination of a maximum tolerated dose. Correlative proteomic studies were performed to confirm HSP inhibitor activity.
Results
Objective responses were observed in 15/20 evaluable patients (75%; 95% CI: 51–91%), with 3 complete and 12 partial responses. Median progression-free and overall survival were 9.2 months (95% CI: 3.8–not reached) and 34.6 months (6.2–not reached), respectively. The most common grade 3/4 toxicities were skin toxicities such as rash (n=4, 19%) and cutaneous squamous cell carcinomas (n=3, 14%), along with diarrhea (n=3, 14%). Pharmacodynamic analysis of patients’ PBMCs showed increased day 8 HSP70 expression compared to baseline in the three cohorts with XL888 doses ≥45 mg. Diverse effects of vemurafenib-XL888 upon intratumoral HSP-client protein expression were noted, with the expression of multiple proteins (including ERBB3 and BAD) modulated on therapy.
Conclusion
XL888 in combination with vemurafenib has clinical activity in patients with advanced BRAFV600-mutant melanoma, with a tolerable side-effect profile. HSP90 inhibitors warrant further evaluation in combination with current standard-of-care BRAF plus MEK inhibitors in BRAFV600-mutant melanoma.
Keywords: melanoma, BRAF, HSP90, phase I
Introduction
The discovery of activating mutations in the serine/threonine kinase BRAF and the appreciation of its role as a driver of melanoma growth and progression has transformed the treatment of disseminated BRAF-mutant melanoma (1, 2). Although the use of small molecule BRAF inhibitors, such as vemurafenib and dabrafenib, frequently leads to rapid and impressive responses in patients with metastatic melanoma, resistance resulting in therapeutic escape is common, with median progression-free survivals of 5.3 and 5.1 months demonstrated for vemurafenib and dabrafenib, respectively (1, 2). This therapeutic escape is frequently characterized by the recovery of signaling through the mitogen-activated protein kinase (MAPK) pathway, an effect that is mediated through multiple and diverse mechanisms including mutations in NRAS and MEK and BRAF splice mutations, and increased receptor tyrosine kinase (RTK) signaling (3–6). The continued dependence of resistant tumors upon MAPK signaling led to the development of strategies designed to vertically inhibit the pathway. Preclinical studies showed that dual BRAF-MEK inhibition abrogated therapeutic escape in vitro and delayed treatment failure in human melanoma mouse xenograft models (7, 8). In randomized clinical trials, the combination of a MEK inhibitor and a BRAF inhibitor (including vemurafenib and cobimetinib, dabrafenib and trametinib, and encorafenib and binimetinib) was associated with improved progression-free and overall survival compared to BRAF inhibitor therapy alone (9–11).
Despite these improvements in efficacy, most patients eventually fail to respond to therapy, with similar mechanisms of resistance being reported for both single agent BRAF inhibitor and BRAF-MEK inhibitor combination therapy (12). Clinical strategies to delay or prevent acquired BRAF and BRAF-MEK inhibitor resistance are complicated by the diversity of resistance mechanisms. Recent work from our group suggested that many of the proteins involved in developing resistance to BRAF and BRAF-MEK inhibitor therapy are dependent upon heat shock protein (HSP)-90 for their stabilities (13). Proteins stabilized by HSP90 (HSP90 client proteins) implicated in BRAF inhibitor resistance include CRAF, ARAF, cyclin D1, AKT and CDK4, as well as multiple receptor tyrosine kinases (RTKs) including c-MET, PDGFR, IGF1R and ERBB3 (4, 14, 15). There are also preclinical data supporting the notion that HSP90 inhibitors can reverse BRAF and BRAF-MEK inhibitor resistance and delay the onset of BRAF inhibitor resistance in vivo (13, 16). XL888 (Exelixis, South San Francisco, CA) is a potent, orally-administered small molecular inhibitor of HSP90. In a phase I study of 33 patients with refractory solid tumors, the maximum tolerated dose (MTD) of XL888 was determined to be 135 mg twice weekly (BIW) with diarrhea as a DLT (dose-limiting toxicity); the study did not include any patients with melanoma. In the current study, we performed a phase I dose escalation clinical trial of vemurafenib in combination with XL888 to determine an MTD and to evaluate the safety and potential efficacy of the combination in patients with advanced BRAFV600-mutant melanoma.
Methods
Study design and treatments
This was an open-label, single-center phase I trial of escalating doses of XL888 in combination with vemurafenib in patients with unresectable or metastatic melanoma. Patients were enrolled using a modified Ji design (17) with the primary objective to determine the MTD of XL888 in combination with vemurafenib. There were four dose-level cohorts of XL888: 30 mg PO BIW, 45 mg BIW, 90 mg BIW and 135 mg BIW, each given together with standard doses of vemurafenib (960 mg BID, see Supplemental Figure 1.) Secondary objectives were to assess overall response rate (ORR), progression-free survival (PFS) and overall survival (OS), in addition to assessing the biological activity and pharmacodynamics of XL888 utilizing proteomics-based biomarkers. Tumor responses were assessed using Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1, with radiologic assessments performed every 8 weeks. The study was approved by the institutional review board at the University of South Florida.
Inclusion and exclusion criteria
Patients were ≥18 years of age, with cytologically or histologically-confirmed unresectable (Stage IIIC or IV) melanoma harboring a BRAFV600E/K mutation determined by a CLIA-certified assay. Patients had adequate hepatic, renal and bone marrow function, along with an ECOG performance status of 0 or 1. Treatment-naïve and previously treated patients were included, but patients could not have received prior BRAF inhibitors. Measurable disease defined by RECIST 1.1 was required. Patients were excluded if they had received systemic therapy or radiotherapy within 4 weeks of enrollment or if they had not recovered from adverse events caused by prior therapy. Untreated or uncontrolled central nervous system (CNS) metastases or evidence of leptomeningeal disease were also exclusions, although brain metastases treated with radiation and/or surgery were allowable if they had been stable for ≥4 weeks.
Pharmacodynamic assessments
HSP70 levels in peripheral blood mononuclear cells (PBMC) were analyzed as a marker of HSP90 inhibition (18,19) since its inhibition leads to compensatory increases in the expression of the related chaperone protein HSP70 subtype 1. Serum samples were collected at treatment days 1 and 8 to evaluate PBMC HSP70 levels. Tumor biopsies on treatment days 1 and 8 were also obtained from consenting patients for analysis of HSP client protein levels. We utilized discovery proteomics and developed liquid chromatography-multiple reaction monitoring (LC-MRM) mass spectrometry assay panels for proteomics analysis of PBMC and biopsy specimens to validate increased HSP70 protein expression as a pharmacodynamic marker of HSP90 inhibition, and to determine the role of HSP90 inhibition in blocking the signaling pathways implicated in therapeutic escape to BRAF-inhibitors (13, 18). Frozen PBMC samples were thawed using a water bath at 37°C and spun at 500xg for 5 minutes at 4°C to pellet the cells. Pellets were washed with ice-cold PBS and then resuspended in aqueous 50% tetrafluoroethanol and sonicated (Branson 150). The lysates were reduced with tris (2-carboxyethyl)phosphine followed by alkylation with iodoacetamide. Protein samples were diluted 10-fold with 30 mM ammonium bicarbonate and digested with trypsin prior to LC-MRM data acquisition. High abundance proteins, including HSPs, were quantified from ~2,000 cells (~200 ng of total protein digest).
Tumor homogenates were resuspended in denaturing buffer (100 mM ammonium bicarbonate with 8 M urea), sonicated to maximize protein recovery (Branson 150), and clarified by centrifugation at 21,000xg for 10 minutes. Protein concentrations were determined by Bradford assay and 50 μg of sample was fractionated by SDS-PAGE into five regions of 4–12% BisTris gels (Criterion XT, Bio-Rad, Berkeley, CA), visualized with Coomassie Brilliant Blue G-250 (Sigma-Aldrich, St. Louis, MO) (see Supplemental Figure 2) and excised as previously published. Gel regions were diced to ~1 mm3 for processing. After destaining, disulfides were reduced with 2 mM tris(carboxyethyl)phosphine and then cysteines were alkylated with 20 mM iodoacetamide prior to overnight digestion with sequencing grade trypsin (Promega, Madison, WI). The resulting proteolytic peptides were extracted with aqueous 50% acetonitrile, 0.01% trifluoroacetic acid and concentrated by vacuum centrifugation (SC210A, Speedvac, Thermo, San Jose, CA). Peptides were resuspended in 2% acetonitrile with 0.1% formic acid (loading solvent), containing the stable isotope-labeled internal standards (at 10 fmol/μl, so 50 fmol of each standard is injected for LC-MRM). The equivalent of 8.3 micrograms of total protein digest was analyzed in each LC-MRM experiment.
LC-MS/MS discovery proteomics was performed as described previously (19). LC-MRM analysis was performed in triplicate on a nanoLC interfaced with an electrospray triple quadrupole mass spectrometer (RSLCnano and Quantiva, Thermo). The following solvent system is used for LC-MRM analysis: solvent A is aqueous 5% acetonitrile with 0.1% formic acid, and solvent B is aqueous 90% acetonitrile with 0.1% formic acid. For each sample, an aliquot of the peptide mixture (5 μl, ~1/6 of the sample) was loaded onto the trap column at 6 μl/min and washed with loading solvent for 5 minutes. Then, a gradient of 5% B to 50% B was applied over 35 min prior to washing the column at 90% B and re-equilibrating over 10 minutes, for a total of 55 minutes for the LC experiment. Mass spectrometry instrument parameters included the following: 2200V spray voltage; 250°C transfer tube temperature; Q1 resolution 0.4 when transitions were monitored for the entire LC separation (HSPs and band 1) and 0.7 when scheduled methods were used (bands 2–5); 2 milliTorr collision gas pressure; and Q3 resolution 0.7. Time per transition (peptide precursor and fragment ion pair) is optimized using a 2.5 second cycle time, so it is based on the total number of transitions measured that that time. Collision energy values were optimized for this instrument by infusion of the standard peptides (See Supplemental Table 1).
Data analysis for protein quantification
Skyline version 3.7 was used for data evaluation (20). Peaks were evaluated by comparison of their elution time and fragment ion signal ratios to their matched internal standards. All transitions above 10% of the base peak were used for quantification. Data were exported to Excel for calculations of protein quantity, standard deviation and coefficient of variation (%).
Statistical and analytical methods
Sample size, safety and dose-limiting toxicity definitions
The trial was designed to enroll up to 36 patients but would be successfully concluded earlier if at least 15 patients were accrued at the dose determined to be the MTD, with no more than 4 dose-limiting toxicities (DLTs) using the modified Ji Design. Adverse events were graded according to NCI CTCAE v4. Cohorts of 3 patients with possible expansions to 6, 9, 12 or 15 patients were treated at each dose level until the MTD was defined. The cohort size at each dose depended on the observed toxicity, as the design allowed sufficient number of patients to be explored with targeting the toxicity at 18%. The modifications to the Ji design were to escalate the dose whenever the observed toxicity rate was below 17% and de-escalate the dose when the toxicity rate exceeded 25%, and N ≥9 to account for the fact that the design was being used in cohorts of 3 rather than continuously as designed by Ji et al (see Supplemental Figure 3.). Once the MTD of XL888 with vemurafenib was determined, six additional patients were to be treated at this dose to further define the safety and efficacy profile of the combination. If there was de-escalation to a dose that had already accrued 15 patients, that dose was declared the MTD.
DLT was defined as any of the following occurring during cycle 1 (the first 8 weeks) of treatment: 1) non-hematologic Grade 3/4 adverse event not easily managed or corrected by medical intervention; 2) Grade 4 neutropenia that did not resolve within 7 days, or any grade 3/4 febrile neutropenia, or any grade 4 thrombocytopenia, 3) any treatment-related AE that in the investigator’s opinion warranted a dose reduction or where any further dose-escalation would expose patients to unacceptable risk, 4) inability to take 75% or more of the planned XL888 doses in cycle 1 due to study-related AEs, 5) failure to recover from any study-related AE within 14 days.
Efficacy analysis
The ORR and its 95% confidence interval was estimated using the exact binominal method. For PFS and OS, medians and 95% confidence intervals were estimated using the Kaplan-Meier method. One-year PFS and OS rates were also estimated using the Kaplan-Meier method. PFS was defined from start of treatment to progression or death from any cause. OS was defined from start of treatment to death from any cause; patients still alive were censored at date of last follow-up.
Results
Safety and dose-limiting toxicity
Twenty-one patients with advanced BRAFV600E/K mutant melanoma were enrolled from July 2012 to May 2016. Fourteen (67%) patients had stage IV M1c disease, with 12 (57%) having an elevated baseline LDH; 39% of all patients had prior systemic therapy (most commonly immunotherapy, Table 1). There were three patients each enrolled in cohorts 1 and 2, nine patients in cohort 3, and six patients in cohort 4. The most common grade 3 adverse events were skin toxicities such as rash (n=4, 19%), cutaneous squamous cell carcinomas (n=3, 14%) and new primary melanomas (n=2, 10%), along with diarrhea (n=3, 14%), headache (n=2, 10%), fatigue (n=2, 14%). (Table 2) There were three grade 4 adverse events noted: asymptomatic lipase elevation, AST elevation and arthralgia. Skin events, including squamous cell carcinomas (SCC) and keratoacanthomas (KA), were reduced in cohorts 3 and 4 compared to cohorts 1 and 2, as previously reported (21). In the phase I study of XL888, blurred vision and visual impairment were reported in two of 33 patients. In this trial, six patients experienced grade 2 visual disturbances possibly or probably related to treatment, most commonly blurry vision, and one patient had macular edema; no retinopathy was observed. Two patients had treatment interruption and subsequently resumed treatment with no recurrence of symptoms, while two patients discontinued study treatment. All visual symptoms resolved.
Table 1.
Patient characteristics
| Characteristic | N=21 | % |
|---|---|---|
| Median age (range) | 60 years (19–75) | |
|
| ||
| Gender = male | 13/21 | 62% |
|
| ||
| ECOG | ||
| PS = 0 | 8/21 | 38% |
| PS = 1 | 13/21 | 62% |
|
| ||
| Staging | ||
| IIIC | 2/21 | 10% |
| M1A | 2/21 | 10% |
| M1B | 3/21 | 14% |
| M1C | 14/21 | 67% |
|
| ||
| BRAF V600E | 20/21 | 95% |
| BRAF V600K | 1/21 | 5% |
|
| ||
| Elevated LDH | 12/21 | 57% |
|
| ||
| Prior therapy | ||
| Surgery | 21/21 | 100% |
| Radiation | 6/21 | 29% |
| Systemic therapy | 8/21 | 39% |
| Anti-CTLA-4 therapy | 5/21 | 24% |
| Anti-PD-1 therapy | 1/21 | 5% |
Table 2.
Summary of maximum grade 3 or 4 toxicity at patient level -Attributions are definitely/probably/possibly related to study drugs
| Toxicity description | Grade 3 n (%) |
Grade 4 n (%) |
N |
|---|---|---|---|
| Diarrhea | 3(14) | - | 21 |
| Nausea | 1(5) | - | 21 |
| Pancreatitis | 1(5) | - | 21 |
| Fatigue | 2(10) | - | 21 |
| Alanine aminotransferase increased | 1(5) | - | 21 |
| Aspartate aminotransferase increased | - | 1(4.8) | 21 |
| Electrocardiogram QT corrected interval prolonged | 1(5) | - | 21 |
| Lipase increased | - | 1(4.8) | 21 |
| Serum amylase increased | 1(5) | - | 21 |
| Hyponatremia | 1(5) | - | 21 |
| Arthralgia | 1(5) | 1(4.8) | 21 |
| Generalized muscle weakness | 1(5) | - | 21 |
| Cutaneous squamous cell carcinoma | 3(14) | - | 21 |
| Basal cell carcinoma | 1(5) | - | 21 |
| New primary melanoma | 2(10) | ||
| Headache | 2(10) | - | 21 |
| Rash - maculo-papular | 3(14) | - | 21 |
| Rash - morbiliform | 1(5) | - | 21 |
| Overall | 11(52.4) | 3(14.3) | 21 |
With enrollment of 3 patients to each of the first three cohorts without DLTs, six patients were enrolled onto the highest dose cohort of XL888 at 135 mg BIW. However, two DLTs (grade 3 diarrhea and pancreatitis) were observed, along with one additional patient receiving <75% of the planned XL888 dose in the first cycle of treatment. Therefore, with 3 DLTs observed in this dose cohort, the decision was made to de-escalate to cohort 3 (90 mg BIW of XL888 with vemurafenib), which was declared as the MTD. Six additional patients were enrolled at this dose level; in total, two patients out of nine in this cohort experienced DLTs (one with grade 3 diarrhea and arthalgia, one patient had a grade 3 rash.)
Efficacy
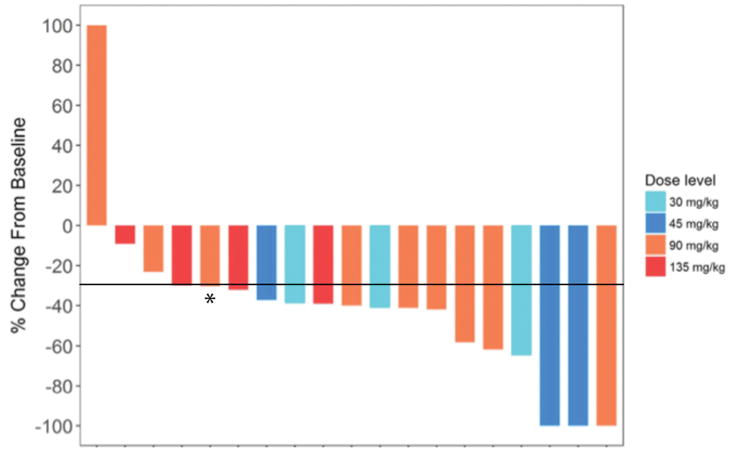
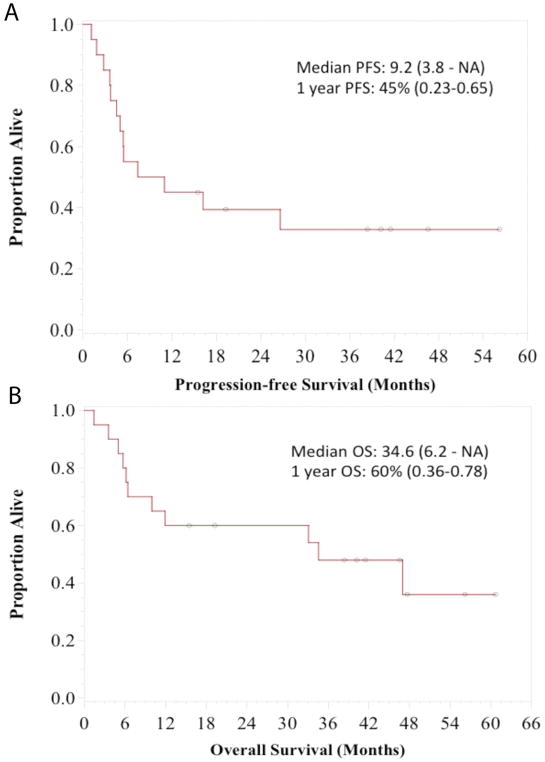
One patient on the trial switched to dabrafenib/trametinib after only 6 doses of XL888 prior to the first restaging scans (unrelated to toxicity) with a subsequent partial response. As this patient received a MEK inhibitor they were excluded from the subsequent efficacy analysis. We observed objective responses to the vemurafenib-XL888 combination in 15 of 20 evaluable patients (ORR 75%; 95% CI 51%–91%) with three complete (15%) and 12 partial (60%) responses (see waterfall plot, Figure 1). There was no clear correlation between level of response and XL888 dose cohort. Two patients with a partial response underwent resection of all residual disease and both were found to have a pathological complete response. At a median follow-up of 26.4 months, the median PFS was 9.2 months (95% CI 3.8 months–not reached), with one-year PFS of 45% (95% CI 23%–65%, Figure 2A). Among patients surviving beyond 2 years without progression, all of them had a normal LDH at baseline, as an elevated baseline LDH was significantly associated with worse PFS (HR=1.969, p = 0.042). Median OS was 34.6 months (6.2 months – not reached), with one-year OS of 60% (95% CI 36%–78%, Figure 2B). At the time of data cut-off, nine out of 21 patients were still alive. Of these nine patients, 7 were without disease progression with a median follow-up of 35 months.
Figure 1.
Waterfall plot of best overall response (ORR) on therapy for 20 evaluable patients treated with vemurafenib/XL888, ORR of 75%; 95% CI: 51–91%). Line delineates −30% decreases in tumor size. *Patient who had regression in target lesions, but new subcm brain mets, therefore counted as PD. One patient who switched to another treatment prior to tumor assessment for non-toxicity related reasons was considered unevaluable.
Figure 2. Survival data.
(A): Kaplan-Meier plot of progression free survival, n=20, median 9.2 months (95% CI: 3.8–not reached) (B): Kaplan-Meier plot of overall survival, n=20, median 34.6 months (6.2–not reached)
Pharmacodynamics
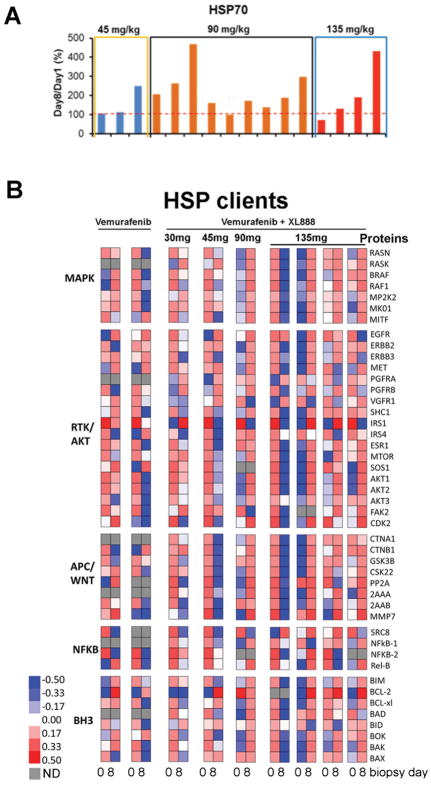
Our LC-MRM assay was used to quantify levels of the HSP70 chaperone protein in the blood of 10 patients from cohorts 2–4, and the expansion cohort (90 mg XL888), at baseline and following 8 days of therapy. These analyses showed increased HSP70 expression above baseline indicating that XL888 was inhibiting HSP90 for most, but not all, patients (Figure 3A).
Figure 3. Multiplexed quantification of heat shock and cancer signaling proteins as candidate biomarkers for drug response.
Patients are organized in cohorts by the dose of XL888. Bar graphs of relative HSP70 expression in PBMCs from Pre-Treatment Baseline to Day 8 Post-Treatment present that average data and their standard deviation (A); data were normalized to GAPDH expression to control for sample cellularity. Heat maps (B) of changes in the expression of cancer signaling proteins in tumor tissues from biopsies pre-treatment and day 8 post-treatment. Protein measurements are clustered by similarity across the dataset. Highest protein expression is shown in red; lowest in blue. Gray indicates that the protein was not observed in that sample (ND: not determined).
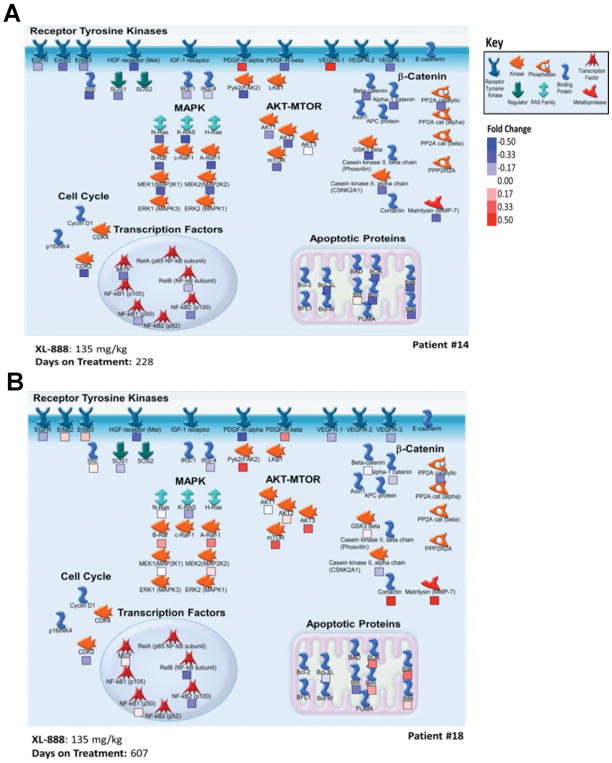
We next analyzed the expression of 45 HSP client proteins across seven paired set of pre- (day 0) and on-therapy (day 8) tumor biopsies from patients receiving vemurafenib-XL888 (30, 45, 90 and 135mg of XL888) and from two other (non-protocol) patients on vemurafenib alone (960mg BID). Overall, the patterns of protein expression observed on vemurafenib and vemurafenib-XL888 were heterogeneous, with large patient-to-patient variability seen (Figure 3B). Some trends were, however, noted. Expression of ERBB3, a known mediator of BRAF inhibitor resistance, increased in the day 8 samples of both patients receiving vemurafenib alone (Figure 3B), but decreased in the day 8 samples of 4 out of 7 patients receiving the vemurafenib-XL888 combination. The decreased ERBB3 expression was seen at all dose levels of XL888 evaluated in the proteomic studies. Decreased expression of the anti-apoptotic protein BAD was also observed in 5 out of 7 of the day 8 vemurafenib-XL888 treated samples, but not in those receiving vemurafenib alone (Figure 3B). In general, however, similar trends in protein expression were noted for both the vemurafenib alone and vemurafenib-XL888 treated specimens with regards to multiple proteins including Bcl-2, BIM, Bok, c-MET and IRS1. To better understand some of the changes in HSP client protein expression at a systems level, we produced signaling maps of one patient (#14) who stayed on vemurafenib and 135 mg XL888 for 228 days, and one (#18) who was treated at the same dose of both drugs for 607 days (Figures 4A and 4B, respectively). The best response for both patients was partial response. These high-level overviews showed patient #14 to have decreased expression of multiple proteins involved in RTK signaling, the MAPK pathway, AKT signaling, components of the β-catenin pathway and negative apoptosis regulators (Figure 4A). Increases in some RTKs, such as PDGFR-α and VEGFR1 were also noted. Patient #18 showed decreases in multiple RTKs including EGFR, c-MET, PDGFR-α, VEGFR1 and increases in components of the MAPK and AKT pathways. In a similar vein to patient #14, patient #18 also demonstrated decreases in NFκB family transcription factors and the β-catenin signaling pathway (Figure 4B). Together these data illustrate that proteome level responses to the BRAF-HSP90 inhibitor are diverse and heterogeneous, even among patients who respond to therapy. The relatively small cohort of patients being analyzed limited our ability to perform detailed statistical analyses of these trends in HSP90 client expression.
Figure 4. Annotation of patient HSP client measurements on a simplified pathway diagram.
Protein targets are grouped by pathway on a map created using GeneGO Metacore; icons in the key are derived from the GeneGO functions associated with each protein (see key). Each protein is labeled with a box showing the increase or decrease of protein expression between samples captured pre-treatment and those collected after 8 days of treatment on a red to blue scale (white indicates that little change occurred and the absence of the box indicates that the ratio could not be measured). Data are provided for patient 14 and patient 18 (both patients received 135 mg/kg XL888 and 960 mg of vemurafenib twice daily and had a partial response on treatment). Days on therapy are the total days that the patient remained on BRAF-HSP90 inhibitor therapy, although the on-treatment biopsies were done on day 8 of treatment.
Discussion
Although initial responses of melanoma patients to BRAF inhibitors are typically impressive, development of resistance is commonplace. Relapse is associated with recovery of signaling in the MAPK pathway in 52% of cases, PI3K/AKT signaling in 4% of individuals, and both the MAPK and PI3K/AKT pathways in 18% of cases (3). The mechanism of resistance in the remaining 26% patients is not well understood, but may result from non-genomic drug tolerance mechanisms that are associated with novel epigenetic states or immune-mediated effects, or other mechanisms of resistance that are as yet undefined (22, 23). Effective strategies to prevent the onset of BRAF-MEK inhibitor resistance in the clinic are currently lacking, with the diversity of resistance mechanisms posing a formidable problem. The HSP90 family of chaperone proteins play a critical role in regulating the stability of a great number of receptor tyrosine kinases, serine/threonine kinases and other signaling molecules required to maintain the transformed state of cancer cells (24). Many of the key drivers of melanoma progression, including CRAF, CDK4, EGFR, IGF1R, mTOR, COT and AKT, are known to be clients of HSP90, and are degraded following treatment with small molecule HSP90 inhibitors (13, 16). We reasoned that HSP90 inhibition, which has a broad “network level” of activity, was a promising strategy to overcome multiple mechanisms of BRAF inhibitor resistance. In preclinical studies, we and others demonstrated that HSP90 inhibition could overcome acquired BRAF and BRAF-MEK inhibitor resistance mediated through multiple mechanisms and, more importantly, that the BRAF-HSP90 inhibitor combination, when used upfront, prevented treatment failure in melanoma xenograft models (13, 16, 25). HSP90 inhibitors have also been demonstrated to reverse resistance to EGFR, KIT and ALK inhibitors in triple negative breast cancer, gastrointestinal stromal tumors and ALK-mutant lung cancers, respectively (26–29). The purpose of the current study was to determine the safety and tolerability of the HSP90 inhibitor XL888 in combination with the FDA-approved BRAF inhibitor vemurafenib. The most thoroughly investigated HSP90 inhibitor in melanoma thus far in the single-agent setting is 17-allylamino, 17 demethoxygeldanamycin (17-AAG, tanespimycin), a benzoquinone ansamycin with good preclinical activity against BRAF-mutant melanoma cell lines and xenografts (30). In both phase I and phase II clinical trials of patients with advanced melanoma, single-agent 17-AAG activity was modest (30–32). Some evidence for target engagement was observed in one phase I study, with decreased expression of CRAF and CDK4 and increased HSP70 expression reported (30). In a subsequent phase II clinical trial, 17-AAG was not found to alter CRAF expression or to inhibit phospho-ERK in melanoma biopsies, despite increases in HSP70 expression and decreased levels of intratumoral cyclin D1 being observed (32).
In the current study of the BRAF inhibitor vemurafenib in combination with escalating doses of the HSP90 inhibitor XL888, PFS and OS rates compared favorably to previously published data on single agent vemurafenib with a median PFS of 5.3 to 6.2 months, with results similar to the median PFS observed with vemurafenib/cobimetinib of 9.9 months. (9) The response rate of 75% experienced by the vemurafenib-XL888 treated patients in our study also compares favorably to the 48% response rate seen to single agent vemurafenib, and is similar to the 68% response rate reported with vemurafenib/cobimetinib (1, 33). There were also differences in the side effect profile seen when compared to single agent vemurafenib. The most frequent off-target effect seen with the vemurafenib-XL888 combination (when all cohorts were combined) was the development of proliferative skin lesions including SCCs, KAs, melanomas and verruca vulgaris (VV). These are known side effects of treatment with vemurafenib monotherapy, where up to 26% of patients developed either SCC or KA (34). In the case of vemurafenib monotherapy, secondary skin lesions are known to arise following the paradoxical activation of MAPK signaling in clones of skin keratinocytes that harbor pre-existing HRAS mutations (35, 36). Of interest, no SCCs or KAs were observed in patients at the higher XL888 doses (cohorts 3–4). Although it is tempting to speculate that this resulted from XL888 blocking the paradoxical activation of MAPK (as previously shown by our group in preclinical studies) (21), the limited number of patients treated in each cohort of the present study makes this difficult to determine. The addition of higher dose (90–135mg) XL888 to vemurafenib, as experienced by patients in cohorts 3 and 4, did not reduce the incidence of VV, suggesting these may have a different etiology than that underlying SCC or KA development (21). Two patients developed new primary stage 1 melanomas that were resected in entirety; the association of new primary melanomas with single-agent vemurafenib has been well documented.(37)
Measuring HSP90 inhibitor activity in a clinical setting has proven to be challenging. The most commonly used method has been the quantification of co-chaperone HSP70 expression in PBMCs. Approaches used to quantify HSP70 expression clinically have included ELISA assays and semi-quantitative assessment of limited numbers of client proteins (typically 2–4) using immunohistochemistry (IHC) or Western blot (30, 38, 39). HSP chaperone proteins are known to regulate the stability of >200 clients, making these limited pharmacodynamic assessments insufficient to determine the breadth of target engagement. To address these issues, and to provide mechanistic insights into the effects of HSP90 inhibition, we developed a mass spectrometry-based proteomic assay for the quantification of HSP70 expression in PBMCs and HSP90 client proteins in tumor samples that allows the changes in the expression of multiple HSP client proteins to be determined in pre- and post-treatment melanoma biopsies (18). These assays showed HSP70 expression to be robustly increased in PBMC from most patients on vemurafenib-XL888 therapy.
At day 8, decreased client protein expression was frequently seen in pathways implicated in melanoma progression and adaptation to BRAF inhibitor therapy (3, 7, 40, 41). Increases in some client proteins were also observed. Proteomic responses were heterogeneous between patients, with different client proteins being degraded or increasing in expression in different pairs of matched tumor specimens. This variation in response likely reflects the high level of genomic and phenotypic diversity and heterogeneity of melanoma (42). Although the limited numbers patients enrolled on this trial, and the small numbers of paired samples available for analysis, made it difficult to ascertain the link between HSP client degradation and clinical response, possible indicators of HSP90 inhibitor activity were observed. ERBB3 is an RTK implicated in BRAF inhibitor resistance, whose expression increases as an adaptation to BRAF inhibitor therapy (15). ERBB3 is known to be an HSP client protein, with multiple studies showing that HSP90 inhibitors lead to its degradation and a decrease in its signaling activity (43, 44). In agreement with published data, we noted that ERBB3 expression was increased in the day 8 biopsies from two patients receiving vemurafenib alone. In contrast, 4/7 patients receiving vemurafenib-XL888 experienced a decrease in ERBB3 expression at day 8. Heterogeneous responses were also seen with regards to c-MET (another RTK implicated in BRAF inhibitor resistance) (45, 46), with 4/7 patients showing increased expression at day 8 and 3/7 showing decreased expression. CRAF (or RAF1) is another HSP90 client implicated in BRAF inhibitor failure (47, 48). Analysis of CRAF expression levels showed decreased expression in 3/7 day 8 samples from patients receiving vemurafenib-XL888. Our results herein agree with previous Western blot studies in which patients responding to the HSP90 inhibitor 17-AAG frequently demonstrated decreased expression of the HSP90 client RAF-1 after drug dosing (30). Unexpectedly, increased KRAS expression was seen in 6/7 post-treatment samples from patients on the BRAF-HSP90 inhibitor combination, which was not observed in two patients on BRAF inhibitor alone. It was the evident that XL888 was impacting pathways other than the MAPK pathway, supporting our preclinical findings that HSP90 inhibitors also impact RTK signaling, AKT, mTOR etc. (13, 16). At the same time, there was also significant overlap between the proteins impacted by vemurafenib and vemurafenib-XL888; these included robust induction/degradation of both anti-apoptotic (Bcl-2, Bak) and pro-apoptotic proteins (BIM, Bad, Bok). The complexity of these results illustrates the difficulties associated with predicting responses from the measurement of individual proteins. We therefore believe that a systems-level analysis of the HSP clientome performed on greater numbers of patients will be required to determine which combination of clients must be degraded for robust clinical responses to be observed and/or maintained. Recent preclinical studies have demonstrated that HSP90 inhibitors also improve anti-melanoma immunity through upregulation of interferon response genes, and that this can potentiate responses to anti-CTLA-4 and PD-1 therapy (49). This raises the possibility that the vemurafenib-XL888 combination could also positively impact the tumor microenvironment.
Besides XL888, other HSP90 inhibitors such as AT13387 and ganetespib are also in clinical testing; trials studying the combination of BRAF and MEK inhibitors with both XL888 and AT13387 are ongoing in patients with BRAFV600-mutated advanced melanoma (NCT02721459, NCT02097225). Results of these trials will provide further insight into the role of HSP90 inhibitors in treatment of these patients. BRAF-MEK inhibitor combinations have now replaced single agent BRAF inhibitor therapy as a standard of care targeted therapy for advanced melanoma. As with single agent BRAF inhibition, patients on BRAF-MEK inhibitor therapy also show signs of resistance; there is evidence that escape from BRAF-MEK inhibitor therapy is also dependent upon HSP client proteins and that HSP90 inhibitors can reverse this (16). We are currently investigating a triplet combination of vemurafenib-cobimetinib-XL888, with the goal of further improving the PFS and OS seen with vemurafenib-cobimetinib. However, given the toxicity profile seen with vemurafenib-XL888, it is possible that alternative doses of vemurafenib and cobimetinib may need to be explored when combined with XL888.
Supplementary Material
Statement of translational relevance.
BRAF inhibitor monotherapy is frequently associated with the development of acquired resistance. Preclinical work from our group and others has demonstrated that clients of heat shock protein (HSP)-90 are involved in multiple mechanisms of BRAF inhibitor resistance and that concurrent HSP90 inhibition abrogates or reverses therapeutic escape. In the present study we performed a phase I clinical trial of the BRAF inhibitor vemurafenib in combination with the HSP90 inhibitor XL888 in patients with advanced BRAFV600-mutant melanoma; we found that the combination had clinical activity and a tolerable side-effect profile. Pharmacodynamic analyses, using a novel proteomic assay, demonstrated the combination of vemurafenib and XL888 to reduce the expression of multiple HSP client proteins implicated in resistance. These studies suggest XL888 could be a valuable addition to the BRAF-MEK inhibitor combination, a current standard-of-care treatment for advanced BRAFV600 mutant melanoma.
Acknowledgments
Supported by SPORE grant P50CA168536-01A1 from the National Cancer Institute to K Smalley and V Sondak
Footnotes
Conflicts of interest
Zeynep Eroglu served on an advisory board for Compugen and has received travel support from Genentech/Roche and Tesaro. Jane Messina is a consultant for Castle Biosciences. Jeffrey S. Weber has acted as a consultant and member of the advisory boards of Bristol-Myer Squibb, GlaxoSmithKline, Genentech, Novartis, Merck, AstraZeneca, Nektar, Medivation, Roche, Celldex, Incyte, and EMD Serono; has participated in advisory boards for Lion Bioscience, Celldex, CytomX Therapeutics, and cCAM Biotherapeutics; received research support from Mirati Therapeutics and Acetylon Pharmaceuticals; holds equity in Celldex, CytomX Therapeutics, and Altor; and has a patent pending for an ipilimumab biomarker by Moffitt Cancer Center and a patent pending by Biodesix for a programmed cell death protein 1 (PD-1) biomarker. Geoffrey T. Gibney has served as a consultant for Novartis and Genentech/Roche, and as a member of the Speakers’ Bureau for Merck and Genentech for work performed outside of the current study. Joseph Markowitz has served as a consultant for Newlink Genetics and Idera. Vernon K. Sondak has participated in advisory boards for Merck, Genentech/Roche, Bristol-Myers Squibb, and Novartis, and has served on data safety monitoring boards for Bristol-Myers Squibb, Array Biopharma, Novartis, Oncolys, Polynoma, and Pfizer for work performed outside of the current study. Nikhil Khushlani has served as a consultant for Bristol-Myers Squibb, Astra-Zeneca, EMD Serono and Regeneron. Ragini Kudchadkar serves as a consultant for Bristol-Myers Squibb and Array Biopharma and receives research funding from Merck. All other authors declare no conflicts of interest.
References
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–65. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 3.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A. 2009;106:20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poulikakos PI, Persaud Y, Janakiraman M, Kong XJ, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–U144. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paraiso KH, Fedorenko IV, Cantini LP, Munko AC, Hall M, Sondak VK, et al. Recovery of phospho-ERK activity allows melanoma cells to escape from BRAF inhibitor therapy. Br J Cancer. 2010;102:1724–30. doi: 10.1038/sj.bjc.6605714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, et al. Disruption of CRAF-Mediated MEK Activation Is Required for Effective MEK Inhibition in KRAS Mutant Tumors. Cancer Cell. 2014 doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371:1867–76. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 10.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved Overall Survival in Melanoma with Combined Dabrafenib and Trametinib. New Engl J Med. 2015;372:30–9. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 11.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Overall survival for dabrafenib and trametinib versus dabrafenib and placebo in V600 BRAF-mutant melanoma: a multi-center, double-blind, phase 3 randomised controlled trial. Lancet. 2015 doi: 10.1016/S0140-6736(15)60898-4. [DOI] [PubMed] [Google Scholar]
- 12.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–8. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paraiso KHT, Haarberg HE, Wood E, Rebecca VW, Chen YA, Xiang Y, et al. The HSP90 Inhibitor XL888 Overcomes BRAF Inhibitor Resistance Mediated through Diverse Mechanisms. Clinical Cancer Research. 2012;18:2502–14. doi: 10.1158/1078-0432.CCR-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abel EV, Basile KJ, Kugel CH, 3rd, Witkiewicz AK, Le K, Amaravadi RK, et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. The Journal of clinical investigation. 2013;123:2155–68. doi: 10.1172/JCI65780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smyth T, Paraiso KH, Hearn K, Rodriguez-Lopez AM, Munck JM, Haarberg HE, et al. Inhibition of HSP90 by AT13387 Delays the Emergence of Resistance to BRAF Inhibitors and Overcomes Resistance to Dual BRAF and MEK Inhibition in Melanoma Models. Mol Cancer Ther. 2014 doi: 10.1158/1535-7163.MCT-14-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji Y, Liu P, Li Y, Bekele BN. A modified toxicity probability interval method for dose-finding trials. Clinical trials (London, England) 2010;7:653–63. doi: 10.1177/1740774510382799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rebecca VW, Wood ER, Fedorenko IV, Paraiso KH, Haarberg HE, Chen Y, et al. Evaluating Melanoma Drug Response and Therapeutic Escape with Quantitative Proteomics. Molecular & cellular proteomics: MCP. 2014 doi: 10.1074/mcp.M113.037424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma R, Fedorenko I, Spence PT, Sondak VK, Smalley KS, Koomen JM. Activity-Based Protein Profiling Shows Heterogeneous Signaling Adaptations to BRAF Inhibition. J Proteome Res. 2016;15:4476–89. doi: 10.1021/acs.jproteome.6b00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phadke M, Gibney GT, Rich CJ, Fedorenko IV, Chen YA, Kudchadkar RR, et al. XL888 Limits Vemurafenib-Induced Proliferative Skin Events by Suppressing Paradoxical MAPK Activation. J Invest Dermatol. 2015;135:2542–4. doi: 10.1038/jid.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith MP, Brunton H, Rowling EJ, Ferguson J, Arozarena I, Miskolczi Z, et al. Inhibiting Drivers of Non-mutational Drug Tolerance Is a Salvage Strategy for Targeted Melanoma Therapy. Cancer Cell. 2016;29:270–84. doi: 10.1016/j.ccell.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell. 2015;162:1271–85. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nature reviews Cancer. 2010;10:537–49. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acquaviva J, Smith DL, Jimenez JP, Zhang C, Sequeira M, He S, et al. Overcoming acquired BRAF inhibitor resistance in melanoma via targeted inhibition of Hsp90 with ganetespib. Molecular Cancer Therapeutics. 2014;13:353–63. doi: 10.1158/1535-7163.MCT-13-0481. [DOI] [PubMed] [Google Scholar]
- 26.Chandarlapaty S, Scaltriti M, Angelini P, Ye Q, Guzman M, Hudis CA, et al. Inhibitors of HSP90 block p95-HER2 signaling in Trastuzumab-resistant tumors and suppress their growth. Oncogene. 2010;29:325–34. doi: 10.1038/onc.2009.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Modi S, Stopeck A, Linden H, Solit D, Chandarlapaty S, Rosen N, et al. HSP90 Inhibition Is Effective in Breast Cancer: A Phase II Trial of Tanespimycin (17-AAG) Plus Trastuzumab in Patients with HER2-Positive Metastatic Breast Cancer Progressing on Trastuzumab. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:5132–9. doi: 10.1158/1078-0432.CCR-11-0072. [DOI] [PubMed] [Google Scholar]
- 28.Bauer S, Yu LK, Demetri GD, Fletcher JA. Heat shock protein 90 inhibition in imatinib-resistant gastrointestinal stromal tumor. Cancer Res. 2006;66:9153–61. doi: 10.1158/0008-5472.CAN-06-0165. [DOI] [PubMed] [Google Scholar]
- 29.Sang J, Acquaviva J, Friedland JC, Smith DL, Sequeira M, Zhang C, et al. Targeted inhibition of the molecular chaperone Hsp90 overcomes ALK inhibitor resistance in non-small cell lung cancer. Cancer Discov. 2013;3:430–43. doi: 10.1158/2159-8290.CD-12-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerji U, O’Donnell A, Scurr M, Pacey S, Stapleton S, Asad Y, et al. Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. J Clin Oncol. 2005;23:4152–61. doi: 10.1200/JCO.2005.00.612. [DOI] [PubMed] [Google Scholar]
- 31.Pacey S, Gore M, Chao D, Banerji U, Larkin J, Sarker S, et al. A Phase II trial of 17-allylamino, 17-demethoxygeldanamycin (17-AAG, tanespimycin) in patients with metastatic melanoma. Invest New Drugs. 2012;30:341–9. doi: 10.1007/s10637-010-9493-4. [DOI] [PubMed] [Google Scholar]
- 32.Solit DB, Osman I, Polsky D, Panageas KS, Daud A, Goydos JS, et al. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:8302–7. doi: 10.1158/1078-0432.CCR-08-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ascierto PA, McArthur GA, Dreno B, Atkinson V, Liszkay G, Di Giacomo AM, et al. Cobimetinib combined with vemurafenib in advanced BRAF(V600)-mutant melanoma (coBRIM): updated efficacy results from a randomised, double-blind, phase 3 trial. The Lancet Oncology. 2016;17:1248–60. doi: 10.1016/S1470-2045(16)30122-X. [DOI] [PubMed] [Google Scholar]
- 34.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. The New England journal of medicine. 2012;366:707–14. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, et al. RAS Mutations in Cutaneous Squamous-Cell Carcinomas in Patients Treated with BRAF Inhibitors. New Engl J Med. 2012;366:207–15. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibney GT, Messina JL, Fedorenko IV, Sondak VK, Smalley KS. Paradoxical oncogenesis--the long-term effects of BRAF inhibition in melanoma. Nat Rev Clin Oncol. 2013;10:390–9. doi: 10.1038/nrclinonc.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmer L, Hillen U, Livingstone E, Lacouture ME, Busam K, Carvajal RD, et al. Atypical Melanocytic Proliferations and New Primary Melanomas in Patients With Advanced Melanoma Undergoing Selective BRAF Inhibition. Journal of Clinical Oncology. 2012;30:2375–83. doi: 10.1200/JCO.2011.41.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro GI, Kwak E, Dezube BJ, Yule M, Ayrton J, Lyons J, et al. First-in-human phase I dose escalation study of a second-generation non-ansamycin HSP90 inhibitor, AT13387, in patients with advanced solid tumors. Clin Cancer Res. 2015;21:87–97. doi: 10.1158/1078-0432.CCR-14-0979. [DOI] [PubMed] [Google Scholar]
- 39.Pacey S, Wilson RH, Walton M, Eatock MM, Hardcastle A, Zetterlund A, et al. A phase I study of the heat shock protein 90 inhibitor alvespimycin (17-DMAG) given intravenously to patients with advanced solid tumors. Clin Cancer Res. 2011;17:1561–70. doi: 10.1158/1078-0432.CCR-10-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frederick DT, Salas Fragomeni RA, Schalck A, Ferreiro-Neira I, Hoff T, Cooper ZA, et al. Clinical profiling of BCL-2 family members in the setting of BRAF inhibition offers a rationale for targeting de novo resistance using BH3 mimetics. PLoS One. 2014;9:e101286. doi: 10.1371/journal.pone.0101286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi H, Hong A, Kong X, Koya RC, Song C, Moriceau G, et al. A Novel AKT1 Mutant Amplifies an Adaptive Melanoma Response to BRAF Inhibition. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A Landscape of Driver Mutations in Melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safavi S, Jarnum S, Vannas C, Udhane S, Jonasson E, Tomic TT, et al. HSP90 inhibition blocks ERBB3 and RET phosphorylation in myxoid/round cell liposarcoma and causes massive cell death in vitro and in vivo. Oncotarget. 2016;7:433–45. doi: 10.18632/oncotarget.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerbin CS, Landgraf R. Geldanamycin selectively targets the nascent form of ERBB3 for degradation. Cell Stress Chaperones. 2010;15:529–44. doi: 10.1007/s12192-009-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du JY, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–U118. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fedorenko IV, Wargo JA, Flaherty KT, Messina JL, Smalley KS. BRAF Inhibition Generates a Host-Tumor Niche that Mediates Therapeutic Escape. J Invest Dermatol. 2015;135:3115–24. doi: 10.1038/jid.2015.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, et al. Relief of Profound Feedback Inhibition of Mitogenic Signaling by RAF Inhibitors Attenuates Their Activity in BRAFV600E Melanomas. Cancer Cell. 2012;22:668–82. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–61. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mbofung RM, McKenzie JA, Malu S, Zhang M, Peng W, Liu C, et al. HSP90 inhibition enhances cancer immunotherapy by upregulating interferon response genes. Nat Commun. 2017;8:451. doi: 10.1038/s41467-017-00449-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.