Abstract
Dendritic cells (DCs) play an integral role in regulating mucosal immunity and homeostasis, but the signaling network mediating this function of DCs is poorly defined. We identified the noncanonical NF-κB inducing kinase (NIK) as a crucial mediator of mucosal DC function. DC-specific NIK deletion impaired intestinal IgA secretion and microbiota homeostasis, rendering mice sensitive to an intestinal pathogen, Citrobacter rodentium. DC-specific NIK was required for expression of the IgA transporter poly Ig receptor (pIgR) in intestinal epithelial cells, which in turn relied on the cytokine IL-17 produced by TH17 cells and innate lymphoid cells (ILCs). NIK-activated noncanonical NF-κB induced expression of IL-23 in DCs, contributing to the maintenance of TH17 cells and type 3 ILCs. Consistent with the duel functions of IL-23 and IL-17 in mucosal immunity and inflammation, NIK deficiency also ameliorated colitis induction. Thus, our data suggest a pivotal role for the NIK signaling axis in regulating DC functions in intestinal immunity and homeostasis.
Keywords: Dendritic cells, NIK, IgA, TH17 cells, ILCs, mucosal immunity
Immunoglobulin A (IgA) is a major immune component in the mucosa that plays an important role in mediating host defenses against pathogen infections, shaping the commensal communities, and maintaining intestinal homeostasis1. Upon production by B cells in gut-associated lymphoid tissues, IgA is secreted into the intestinal lumen via transcytosis that involves binding of dimeric IgA to polymeric Ig receptor (pIgR) on intestinal epithelial cells (IECs)2. Secreted IgA interacts with surface antigens of bacteria in the intestinal lumen, which serve as a mechanism to prevent invasion by pathogenic species and maintain a healthy composition of microbiota2, 3.
Being the major type of antigen-presenting cells, dendritic cells (DCs) play a crucial role in regulating mucosal immunity and homeostasis4. DCs uptake and present gut microbial antigens to promote the differentiation and expansion of effector T cells, particularly T helper (TH) 17 and TH1 cells, as well as regulatory T (Treg) cells4, 5. Intestinal DCs also serve as an important regulator of type 3 innate lymphoid cells (ILC3s), which resemble TH17 cells in expressing the transcription factor RORγt and producing the pro-inflammatory cytokines IL-17A and IL-225, 6, 7. The DC-derived cytokine IL-23 promotes expansion and activation of TH17 cells and ILC3s, which in turn are involved in mucosal immunity and inflammation5. In addition, lamina propria (LP) DCs sense bacterial components via Toll-like receptors (TLRs) and other pattern-recognition receptors (PRRs) and produce various immunoregulatory factors, such as TGF-β and the B cell survival factors B cell-activating factor belonging to TNF superfamily (BAFF) and a proliferation inducing ligand (APRIL), which are involved in induction of IgA class switching in B cells8, 9, 10. Despite these important findings, the signaling network that mediates the functions of DCs in intestinal immunity is poorly defined.
The NF-κB family of transcription factors regulates diverse aspects of immune functions11. Two different signaling pathways, the canonical and noncanonical pathways, mediate activation of distinct NF-κB members12. The noncanonical NF-κB pathway is based on inducible processing of p100, a protein that serves as both the precursor of NF-κB2 p52 and an IκB-like molecule that predominantly regulates the NF-κB member RelB. Proteasome-mediated p100 processing not only generates p52 but also disrupts the IκB-like function of p100, leading to nuclear translocation of the noncanonical NF-κB members, p52 and RelB13. A central signaling component of the noncanonical NF-κB pathway is NF-κB inducing kinase (NIK), which responds to signals from specific immune receptors, predominantly members of the TNF receptor superfamily. Upon activation, NIK stimulates its downstream kinase IKKα, which in turn phosphorylates specific C-terminal serines of p100 to trigger its ubiquitin-dependent processing14, 15. The steady amount of NIK is extremely low due to its constant degradation mediated via ubiquitination by an E3 ubiquitin ligase complex composed of TRAF2, TRAF3, and cIAP, and noncanonical NF-κB activation involves TRAF3 degradation and NIK accumulation13.
The noncanonical NF-κB pathway is known to regulate lymphoid organ development and B cell maturation12. However, the role of this NF-κB pathway in the regulation of mucosal immunity and homeostasis is largely elusive. In this study, we demonstrated a DC-intrinsic role for NIK in regulating IgA secretion and microbiota regulation.
Results
NIK is dispensable for DC development and homeostasis
To examine the role of NIK in DCs, we specifically deleted the NIK-encoding gene Map3k14 by crossing Map3k14-flox mice with Cd11c-Cre mice to generate Map3k14 DC-conditional knockout (Map3k14-cKO) and wild-type mice (Supplementary Fig. 1a). RT-PCR detecting the deleted exon of Map3k14 confirmed the Map3k14 gene deletion in DCs, but not in T cells (Supplementary Fig. 1b). We further confirmed the NIK protein deficiency in Map3k14-cKO DCs by immunoblot using cells treated with a CD40 agonistic antibody together with the proteasome inhibitor MG132 known to inhibit NIK degradation16 (Supplementary Fig. 1c). The Map3k14-cKO and wild-type mice had similar frequencies of CD11c+MHCII+ DCs in different lymphoid tissues (Supplementary Fig. 1d). Consistent with a prior study17, the NIK deficiency also did not influence the expression of DC maturation markers CD80 and CD86 (Supplementary Fig. 1e). The wild-type and Map3k14-cKO mice also did not display significant differences in LP DC subpopulation frequencies or surface marker expression (Supplementary Fig. 1f–h). These results suggest that NIK deletion in DCs had no obvious effect on the development or maturation of DCs.
NIK deletion in DCs alters intestinal microbiota
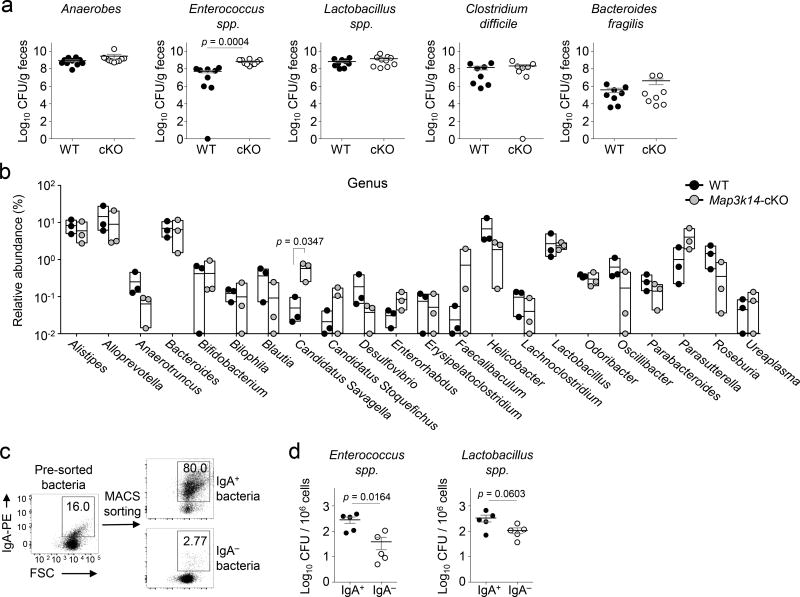
To examine the role of DC-specific NIK in regulating microbiota, we analyzed the profile of commensal bacteria in the Map3k14-cKO and wild-type mice housed under SPF conditions. Selective-medium cultivation analyses revealed that most of the commensal species had similar abundance in the Map3k14-cKO and wild-type mice (Fig. 1a). However, the number of Enterococcus spp. was significantly higher in the Map3k14-cKO mice than in the control mice (Fig. 1a). Enterococci are low abundant gut commensals of human and mice but can cause opportunistic infections, and increased Enterococcus infections have been associated with intestinal inflammation and other disorders18, 19, 20. To further examine the role of DC-specific NIK in regulating microbiota, we performed 16S rRNA sequencing analyses. While the overall microbiota profile of the wild-type and Map3k14-cKO mice was similar, the Map3k14-cKO mice had significantly higher abundance of Candidatus (C.) Savagella (Fig. 1b), segmented filamentous bacteria (SFB) from the gut known to be involved in dynamic interaction between mucosal immunity and microbiota21, 22. Consistent with its low abundance20, Enterococcus spp. was not detected in the 16S rRNA sequencing. Nevertheless, increased CFUs of Enterococcus spp. was reproducibly detected in the Map3k14-cKO mice using the cultivation-based approach.
Figure 1.
Decreased levels of secretory IgA accompanied with increased fecal Enterococci spp. and Candidatus Savagella in Map3k14-cKO (cKO) mice. (a,b) Analysis of commensal microbiota in the fecal extracts of age-matched wild-type (WT) and Map3k14-cKO mice using selective agar plates (a; n=8–10 mice/group) and 16S rRNA sequencing strategies (b; n = 3 mice/group). (c,d) Flow cytometry (c) and selective agar plate quantification (d) of the indicated MACS-sorted IgA+ and IgA− fecal bacteria (d, n=5 mice/group). Each symbol (a, b, d) represents an individual mouse; small horizontal lines (a, d) indicate the mean (± s.e.m.). Panel b is presented as box plots (middle line indicates mean value, and all individual plots are shown). Data are analyzed by unpaired two-tailed Student’s t-test (a, d) or Holm-Sidak method (b) with P values shown above plots. Data are representative of three independent experiments.
Flow cytometric analyses revealed a large proportion of IgA-coated fecal bacteria (Fig. 1c). Importantly, the majority of Enterococci were IgA positive (Fig. 1d). Parallel analysis also revealed a larger proportion of Lactobacillus spp. in the IgA-coated fraction, although this result was less striking than that of Enterococcus spp. (Fig. 1d). 16S rRNA sequencing also revealed that IgA+ and IgA− bacteria were clearly separated and that the SFB C. Savagella was one of the IgA+ genera (Supplementary Fig. 2). These results suggest that DC-specific NIK regulates the homeostasis of both SFB and Enterococcus spp. in the gut.
DC NIK facilitates pIgR expression and IgA secretion
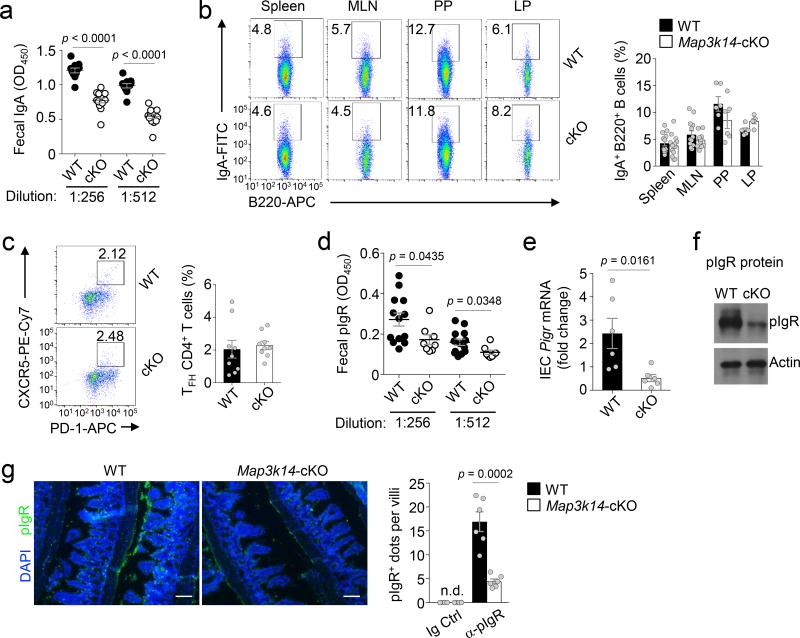
The results described above raised the question of whether the DC-specific NIK deficiency had an effect on the concentration of intestinal secretary IgA. Indeed, the Map3k14-cKO mice had reduced concentration of fecal IgA compared to the wild-type mice (Fig. 2a). Interestingly, Map3k14-cKO and wild-type mice had comparable concentrations of serum IgA as well as similar frequencies of total B cells and IgA+ B cells in LP and other lymphoid tissues (Fig. 2b, Supplementary Fig. 3a–c). Furthermore, DC-specific NIK deletion did not significantly alter the size, number, cellularity, or follicular T (TFH) cell frequency of Peyer’s patches (Supplementary Fig. 3d–g, Fig. 2c).
Figure 2.
DC-specific NIK deficiency impairs pIgR expression in IECs and IgA secretion. (a) ELISA of IgA concentration in fecal extracts of age-matched wild-type (WT) (n=11) and Map3k14-cKO (cKO) mice (n=14). (b) Flow cytometric analysis of IgA+ cells in the B cell population (gated on CD45+B220+ cells) of the spleen, mesenteric lymph nodes (MLN), Peyer’s patches (PP), and lamina propria (LP) of wild-type and Map3k14-cKO mice. Data are presented as a representative FACS plot (left) and a summary graph (right) based on multiple mice (spleen and MLN: n=13 mice/group; PP: n=6 mice/group; LP: n=5 mice/group). (c) Flow cytometric analysis of follicular T helper cells (TFH) in Peyer’s patch CD4+ T cells of wild-type and Map3k14-cKO mice, presented as a representative plot (left) and a summary graph (right) based on multiple mice (n=9 mice/group). (d) ELISA of pIgR concentration in the feces of wild-type (n=13) and Map3k14-cKO (n=8) mice. (e,f) qRT-PCR (e, n=6 mice/group) and immunoblot (f) analysis of pIgR expression in IECs of wild-type and Map3k14-cKO mice. (g) Confocal immunofluorescence analysis of pIgR expression in IECs of colon tissue derived from wild-type and Map3k14-cKO mice (n=6 mice/group). Scale bar represents 50 µm. Each symbol (a–e, g) represents an individual mouse; small horizontal lines (a–e and g) indicate the mean (± s.e.m.). Data are analyzed by unpaired two-tailed Student’s t-test (a–e, g) and representative of two (b, c) or three (a, d–g) independent experiments. P values are shown above plots.
ELISA detected significantly reduced fecal pIgR concentration in Map3k14-cKO mice compared to wild-type mice (Fig. 2d). Consistently, IECs from Map3k14-cKO mice expressed much lower amounts of Pigr mRNA and pIgR protein than IECs from wild-type mice (Fig. 2e,f). The reduced pIgR expression in IECs of Map3k14-cKO mice was also detected by an immunofluorescence assay using intestinal tissues (Fig. 2g). Thus, DC-specific NIK facilitates IgA secretion by maintaining the expression of the IgA transporter pIgR in IECs.
Because microbiota, particularly SFB, promote secretary IgA production in the intestine23, we examined whether DC-specific NIK directly regulated IgA secretion or acted indirectly through controlling microbiota homeostasis. As expected, co-housed wild-type and Map3k14-cKO mice no longer displayed significant microbiota differences (Supplementary Fig. 4a,b). In contrast, the fecal IgA and pIgR defects of Map3k14-cKO were not rescued by their co-housing with wild-type mice (Supplementary Fig. 4c,d), thus suggesting a role for NIK in regulating IgA secretion independently of microbiota.
DC NIK promotes IgA secretion via TH17 cell induction
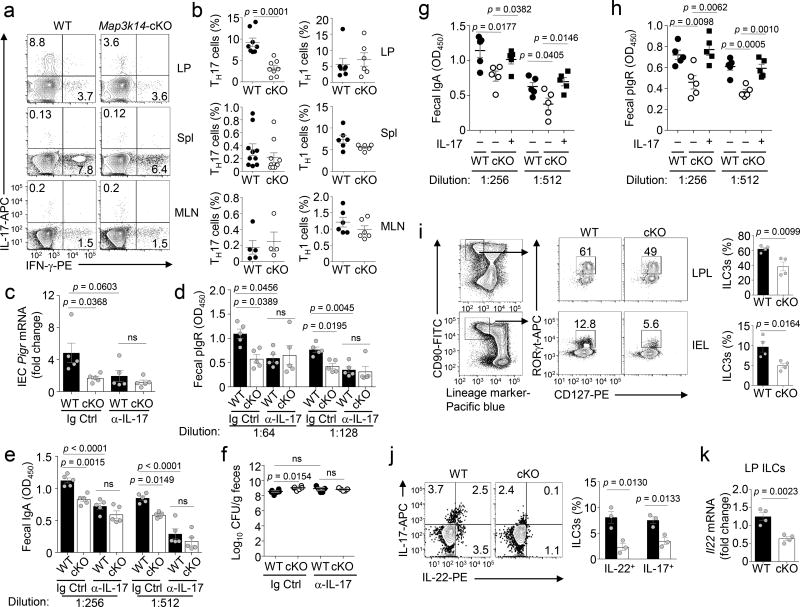
Our finding that DC-specific NIK regulates pIgR expression in IECs raised the question regarding the functional link between DCs and IECs. In this regard, DCs regulate mucosal T cell subsets, particularly the CD4+ TH1 and TH17 cells5. Flow cytometric analysis revealed that the IL-17-producing TH17 cells were abundant in the LP, but scarce in the spleen and mesenteric lymph nodes (MLNs), whereas the IFN-γ-producing TH1 cells were readily detected in all three types of lymphoid tissues (Fig. 3a,b). Importantly, DC-specific NIK deficiency greatly reduced the frequency of TH17, but not TH1, cells in LP, and this phenotype was not seen in the spleen or MLNs (Fig. 3a,b). This finding provided important insight into the mechanism by which NIK regulates intestinal immunity and homeostasis. Specifically, the cytokine IL-17 has been shown to stimulate IECs for pIgR expression and IgA secretion24, 25.
Figure 3.
DC-specific deletion of NIK reduces the frequency of intestinal TH17 cells and ILC3s. (a,b) Flow cytometric and ICS analysis of IFNγ-producing TH1 cells and IL-17-producing TH17 cells in the CD3+CD4+ lymphocyte population of lamina propria (LP), spleen (Spl), and mesenteric lymph nodes (MLN) of wild-type (WT) or Map3k14-cKO mice. Data are presented as a representative plot (a) and summary graph (b) based on multiple mice (TH17: LP, n=8 mice/group; spleen, n=10 mice/group; MLN, wild-type, n=5; cKO, n=4. TH1: LP, wild-type, n=7; cKO, n=6; spleen and MLN, n=6 mice/group). (c–f) qRT-PCR analysis of pIgR mRNA in IECs (c, n=5 mice/group), ELISA of fecal pIgR (d, n=5 mice/group) and IgA concentration (e, n=5 mice/group), and selective agar analysis of fecal Enterococci spp. colony forming units (CFU) (f, n=5–6 mice/group) in wild-type and Map3k14-cKO mice injected i.p. with anti-IL-17 neutralization antibody (10 µg/g bodyweight) or an IgG isotype control every other day for 2 weeks. (g,h) ELISA analysis of IgA (g) and pIgR (h) concentrations in fecal extracts of age-matched wild-type (n=5) and Map3k14-cKO (n=5) mice that had been injected i.p. with recombinant IL-17 (+; 2µg in 100 µl of PBS) or PBS control (−) every other day for two weeks (n=5 mice/group). (i) Flow cytometric analysis of RORγt+CD127+ ILC3s in LP lymphocytes (LPL) and intraepithelial lymphocytes (IEL) of wild-type (n=4) and Map3k14-cKO (n=4) mice, gated on CD90+ and lineage marker-negative cells. (j) Flow cytometry and ICS analysis of IL-22- and IL-17-expressing ILCs (gated on CD90+Lineage− cells) in LP lymphocytes of wild-type and Map3k14-cKO mice (n=3 mice/group). (k) qRT-PCR analysis of Il22 mRNA expression in sorted LP ILCs (n=4 mice/group). Each symbol represents an individual mouse, and small horizontal lines indicate the mean ± s.e.m. (b–k). Data are analyzed by one-way ANOVA followed by Tukey’s multiple-comparisons test (c–e) or unpaired two-tailed Student’s t-test (b, f–k), and P values are shown above plots. Data are representative of three independent experiments.
To assess the functional connection between NIK-mediated TH17 cell regulation and IgA secretion, we injected the wild-type and Map3k14-cKO mice i.p. with an IL-17-specific neutralizing antibody and analyzed pIgR expression. Administration of anti-IL-17 efficiently inhibited the expression of pIgR in IECs of wild-type mice to a level similar to that detected in the IECs of Map3k14-cKO mice (Fig. 3c). Consistent with fewer TH17 cells in the Map3k14-cKO LPs, injection of the anti-IL-17 to these mutant mice only moderately reduced pIgR expression (Fig. 3c). Injection with anti-IL-17 also profoundly reduced the concentration of pIgR and IgA in the feces of wild-type mice (Fig. 3d,e). Furthermore, upon injection with anti-IL-17, the number of fecal Enterococci in wild-type and Map3k14-cKO mice became comparable, as a result of substantial increase in the wild-type mice (Fig. 3f). In line with these findings, injection of Map3k14-cKO mice with a recombinant IL-17 rescued the abundance of fecal IgA and pIgR (Fig. 3g,h). These results suggest that DC-specific NIK maintains the homeostasis of gut TH17 cells, which in turn promotes pIgR expression and IgA secretion via IL-17 secretion.
DC NIK regulates intestinal ILC3s
Like TH17 cells, ILC3s express the signature cytokines IL-17 and IL-22 and play a role in regulating intestinal immunity and homeostasis7. ILC3-derived IL-22 has been shown to mediate mucosal immunity against intestinal pathogens, such as Citrobacter (C.) rodentium, as well as opportunistic pathogens from the microbiota, including Enterococci26, 27. Compared to wild-type control mice, the Map3k14-cKO mice had a significantly reduced frequency of ILC3s in both the LP cells and intraepithelial lymphocytes (IELs) (Fig. 3i). Furthermore, the ILCs from Map3k14-cKO mice expressed less IL-22 and IL-17 (Fig. 3j,k). Thus, NIK is crucial for DC functions in regulating the homeostasis of both TH17 cells and ILC3s in the intestine.
The transcription factor RORγt is required for the generation of both TH17 cells and ILC3s as well as several other subsets of T cells28. To further emphasize the role of IL-17-producing cells in IgA regulation, we examined IgA secretion in Rorc−/− and wild-type control mice. The Rorc−/− mice had a drastic reduction in fecal IgA and pIgR concentrations and moderately increased CFUs of fecal Enterococcus spp.compared to their wild-type control mice (Supplementary Fig. 5a–c). Furthermore, like the Map3k14-cKO mice, the Rorc−/− mice had significantly increased abundance of the SFB C. Savagella (Supplementary Fig. 5d). Consistent with the involvement of RORγt in regulating various other types of mucosal immune cells, such as lymphoid tissue inducer cells, natural killer T cells, γδ T cells, and Treg cells28, the Rorc−/− mice also had additional microbiota alterations (Supplementary Fig. 5d). Nevertheless, these results reveal that the Rorc−/− and Map3k14-cKO mice share major phenotypes in mucosal immunity, particularly reduced fecal IgA and pIgR.
NIK mediates IL-23 induction by TLRs in DCs
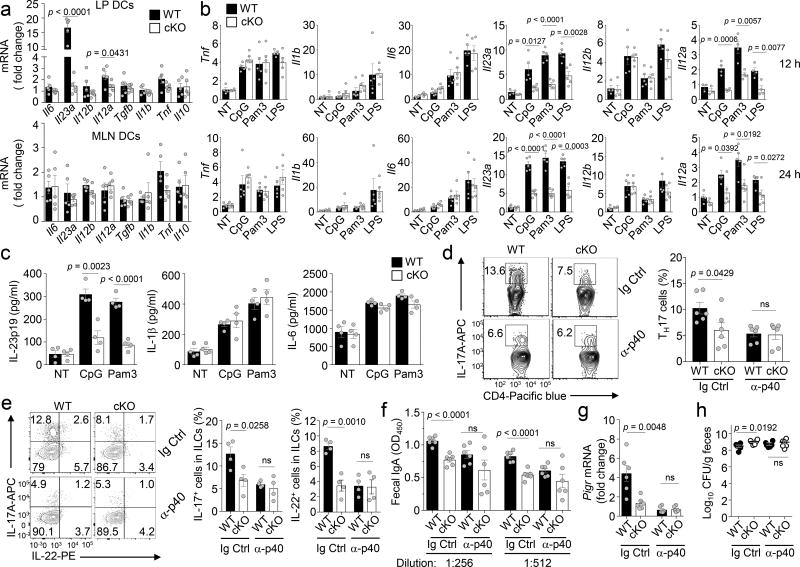
To understand the molecular mechanism by which DC-specific NIK regulates the abundance of TH17 cells and ILC3s in the intestine, we analyzed the cytokine gene expression profile of LP DCs freshly isolated from wild-type or Map3k14-cKO mice. The expression of Il23a, encoding the IL-23p19 subunit, was drastically reduced in the NIK-deficient LP DCs (Fig. 4a). The Il12a gene, encoding the IL-12p35 subunit, was also down regulated, albeit less profoundly than the downregulation of Il23a (Fig. 4a). This phenotype was not detected in DCs derived from the MLNs (Fig. 4a). Since LP DCs are involved in dynamic interactions with commensal bacteria, this result suggested an important role for NIK in mediating induction of Il23a and Il12a genes by bacterial products.
Figure 4.
NIK regulates intestinal homeostasis by mediating TLR-stimulated IL-23 expression in DCs. (a) qRT-PCR analysis of the indicated mRNAs in untreated DCs (CD45+CD11c+ cells) freshly sorted from the lamina propria (LP DCs) or mesenteric lymph nodes (MLN DCs) of wild-type (WT, n=5) and Map3k14-cKO (n=5) mice. (b) qRT-PCR analysis of the indicated mRNAs in wild-type (n=5) or Map3k14-cKO (n=5) bone marrow derived DCs (BMDCs) that were either not treated (NT) or stimulated with the indicated TLR ligands for 12 h (upper) or 24 h (lower). (c) ELISA of the indicated cytokines in the supernatant of wild-type or Map3k14-cKO BMDCs that were either not treated (NT) or stimulated for 48 h with CpG or Pam3CSK4 (n=4 mice/group). (d–h) Flow cytometric and ICS analysis of IL-17-expressing TH17 cells in the LP CD3+CD4+ T cell population (d, n=6 mice/group) and IL-17-expressing ILC3s in the LP Lin−CD90+ cell population (e, n= 4 mice/group), ELISA of fecal IgA (f, n=6 mice/group), qRT-PCR analysis of pIgR mRNA expression in IECs (g, n=6 mice/group), and selective agar analysis of fecal Enterococci spp. colony forming units (CFUs) (h, n=6 mice/group) in wild-type and Map3k14-cKO mice injected i.p. with anti-IL-12/IL-23 p40 (α-p40) neutralization antibody (100 µg/mouse) or an IgG isotype control every other day for 2 weeks. Each symbol represents an individual mouse, and small horizontal lines indicate the mean (± s.e.m.) (a–h). Data are analyzed by unpaired two-tailed Student’s t-test with P values shown above the plots (a–h). Data are representative of three independent experiments.
In vitro studies revealed that TLR ligands stimulated the expression of several pro-inflammatory cytokine genes in bone marrow DCs (BMDCs) (Fig. 4b). In agreement with the results obtained from freshly isolated LP DCs, NIK deficiency attenuated TLR-mediated induction of Il23a and Il12a without significantly affecting the induction of the other cytokine genes analyzed (Fig. 4b). Parallel ELISA further confirmed the crucial role for NIK in mediating TLR-stimulated production of IL-23p19 (Fig. 4c). To examine TLR-stimulated cytokine gene expression in LP DCs, we rested the LP DCs to reduce their spontaneous cytokine gene expression and then stimulated them with TLR ligands. NIK deficiency also attenuated TLR-stimulated expression of Il23a and Il12a in the LP DCs (Supplementary Fig. 6). Collectively, these data demonstrated a pivotal function of NIK in regulating TLR-stimulated expression of IL-23 and, to a lesser extent, IL-12.
To examine the contribution of IL-23 and IL-12 to the impaired intestinal immune homeostasis of the Map3k14-cKO mice, we employed an antibody neutralization approach using the commercially available IL-12/IL-23 p40 neutralizing antibody. Injection of anti-p40, but not an isotype control, every other day for 2 weeks profoundly reduced the frequency of LP TH17 cells and ILC3s in the wild-type mice, making the frequencies of these immune cells comparable between the wild-type and Map3k14-cKO mice (Fig. 4d,e). The IL-12/IL-23 p40 neutralization also erased the differences between wild-type and Map3k14-cKO mice in the fecal IgA concentration, pIgR expression in IECs, and Enterococci CFUs (Fig. 4f–h). Given the previous finding that IL-23, but not IL-12, is required for maintenance of intestinal TH17 cells29, 30, 31, 32, these studies emphasize an important role for IL-23 in mediating the mucosal function of NIK.
Noncanonical NF-κB mediates IL-23 induction in DCs
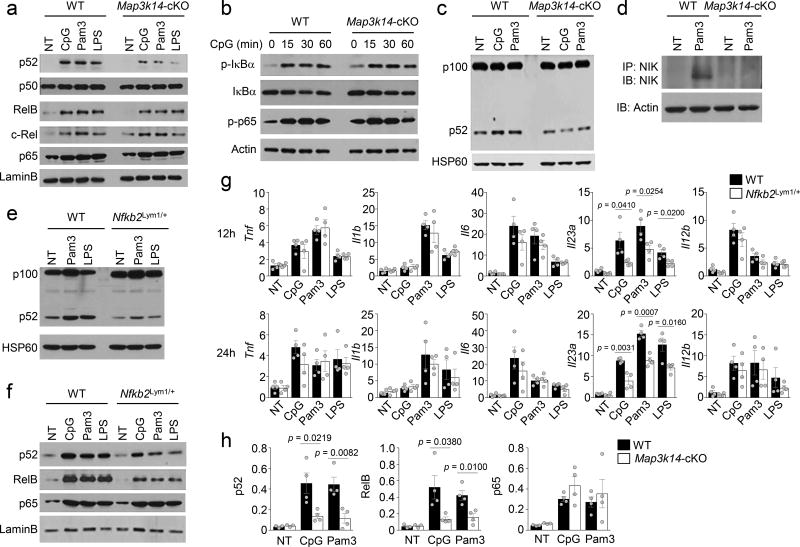
NIK is a central component in the noncanonical NF-κB signaling pathway, which typically responds to signals from the TNF receptor superfamily12, although the role of noncanonical NF-κB in mediating TLR signaling is poorly defined. We found that stimulation of BMDCs with several TLR ligands resulted in the induction of nuclear p52, which was largely, although not completely, dependent on NIK (Fig. 5a). The TLR ligands also induced the nuclear translocation of RelB, an NF-κB member activated by both the canonical and noncanonical NF-κB pathways12, 33, 34. Consistently, the TLR-stimulated RelB nuclear expression was partially inhibited in the NIK-deficient DCs (Fig. 5a). NIK deficiency did not affect the activation of canonical NF-κB members, RelA, c-Rel, and p50 (Fig. 5a). TLR-stimulated early phase activation of canonical NF-κB signaling events, including phosphorylation of IκBα and RelA, was also not affected by the NIK deficiency (Fig. 5b).
Figure 5.
NIK-dependent noncanonical NF-κB activation contributes to TLR-stimulated IL-23 expression. (a–c) Immunoblot analysis of the indicated proteins and phosphorylated (p-) proteins in the nuclear (a) or whole-cell (b, c) extracts of wild-type (WT) or Map3k14-cKO BMDCs stimulated with the indicated TLR ligands for 24 h (a, c) or as indicated (b). (d) immunoblot analysis of IP-concentrated NIK (upper) and loading control Actin (lower) in whole-cell extracts of wild-type and Map3k14-cKO BMDCs that were either not treated (NT) or stimulated for 24 h with Pam3CSK4. (e,f) Immunoblot analysis of the indicated proteins in whole-cell lysates (e) and nuclear extracts (f) of wild-type or Nfkb2Lym1 BMDCs stimulated for 24 h with the indicated TLR ligands. (g) qRT-PCR analysis of the indicated mRNAs in wild-type or Nfkb2Lym1/+ BMDCs stimulated for 12 h (upper) or 24 h (lower) with the indicated TLR ligands (n=4 mice/group). (h) Chip assays, quantified by QPCR, to detect the binding of NF-κB members to a region of Il23a promoter (−121 to −15) covering a κB enhancer (located at −105 to −95) using wild-type or Map3k14-cKO BMDCs that were either not treated (NT) or stimulated for 12 h with CpG and Pam3CSK4. Data are presented as percentage of the total input DNA (NT groups: n=3 mice/group; CpG or Pam3CSK4 stimulated groups: n=4 mice/group). Each symbol represents an individual mouse, and small horizontal lines indicate the mean (± s.e.m.) (g, h). Data are analyzed by unpaired two-tailed Student’s t-test with P values shown above the plots (g, h). Data are representative of two (h) or three (a–g) independent experiments.
NIK-mediated activation of noncanonical NF-κB members is based on the induction of p100 processing12. TLR ligands induced p100 processing in wild-type DCs, as revealed by enhanced generation of p52 (Fig. 5c). Although NIK deficiency did not block the basal p52 expression, TLR-induced production of p52 was inhibited in the NIK-deficient DCs (Fig. 5c). Furthermore, the induction of p100 processing by TLR ligands was coupled with robust accumulation of NIK in wild-type DCs (Fig. 5d). These results suggest that TLR signals activate NIK, thereby inducing noncanonical NF-κB activation in DCs.
The induction of p100 processing requires phosphorylation of 2 specific serine residues (S866 and S870)15, 35. A mutant mouse strain, Nfkb2lym1, carries a point mutation in the Nfkb2 gene that results in generation of a p100 mutant lacking its C-terminal phosphorylation residues and, thus, defective in processing36. As seen previously in B cells36, the BMDCs derived from Nfkb2lym1/+ heterozygous mice predominantly expressed the mutated p100, which had smaller molecular size than wild-type p100 due to the loss of its C-terminal region (Fig. 5e). Therefore, for the convenience of breeding, we employed the Nfkb2lym1/+ heterozygous mice. TLR ligands stimulated p100 processing to generate p52 in wild-type DCs but not in the Nfkb2lym1/+ DCs (Fig. 5e). Furthermore, the TLR-stimulated nuclear translocation of p52 and RelB was also diminished in the Nfkb2lym1/+ DCs (Fig. 5f).
Consistent with the results obtained with NIK-deficient DCs, Il23a gene expression was significantly attenuated in TLR-stimulated Nfkb2lym1/+ BMDCs as well as freshly isolated Nfkb2lym1/+ LP DCs (Fig. 5g and Supplementary Fig. 7a). Furthermore, as seen with the Map3k14-cKO mice, the Nfkb2lym1/+ mice had significantly reduced concentration of fecal IgA and pIgR compared to the wild-type control mice (Supplementary Fig. 7b,c). Similarly, the Nfkb2lym1/+ mice also had reduced frequencies of LP TH17 cells and ILCs (Supplementary Fig. 7d,e), associated with significantly increased Enterococcus spp. and C. Savagella (Supplementary Fig. 7f,g).
To examine whether Il23a was a direct target gene of noncanonical NF-κB, we performed chromatin immunoprecipitation (ChIP) assays. The TLR ligands CpG and Pam3CSK4 stimulated the association of several NF-κB members to the Il23a promoter in wild-type DCs (Fig. 5h). Consistent with its NIK-independent activation, RelA bound to the Il23a promoter to a similar level in wild-type and NIK-deficient DCs in response to TLR stimulation (Fig. 5h). In contrast, the association of p52 and RelB with Il23a promoter was significantly reduced in NIK-deficient DCs (Fig. 5h), which was in line with the defect of these cells in TLR-stimulated p52/RelB nuclear translocation (Fig. 5a). Together, these results suggest that TLR stimulates NIK-dependent activation of noncanonical NF-κB, which in turn is required for the induction of Il23a gene expression and mucosal immune homeostasis.
TLR activation of noncanonical NF-κB requires TNFRII
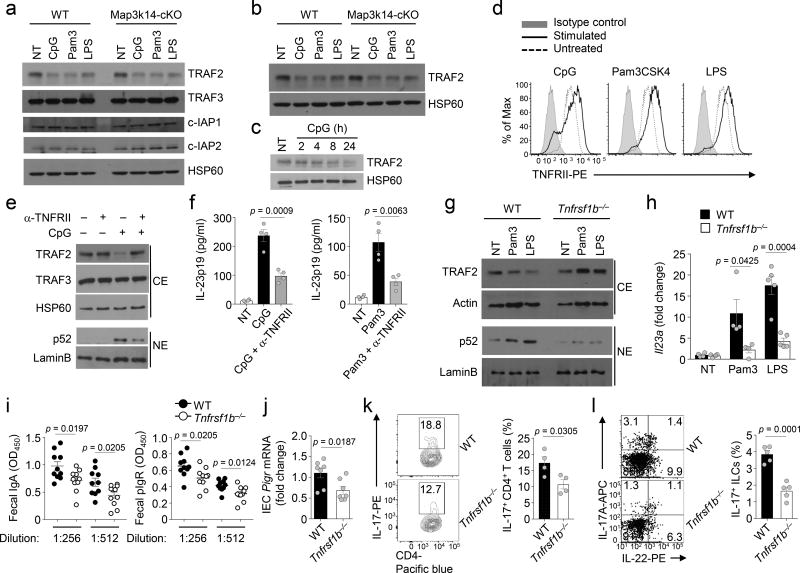
NIK is constantly degraded through its ubiqutination by an E3 ubiquitin ligase complex composed of cIAP (cIAP1 or cIAP2), TRAF2, and TRAF3. Induction of noncanonical NF-κB signaling by TNFR superfamily members involves disruption of the cIAP-TRAF2-TRAF3 ubiquitin ligase complex, typically mediated through degradation of TRAF313. Stimulation of DCs with TLR ligands did not induce appreciable loss of TRAF3, cIAP1, or cIAP2, but triggered a dramatic loss of TRAF2 (Fig. 6a,b). This result was unexpected, since TRAF2 typically functions in the TNFR signaling pathways, instead of the TLR pathways. We noticed that the TLR-stimulated TRAF2 degradation involved delayed kinetics (Fig. 6c). Thus, we surmised that TLR-stimulated TRAF2 degradation and noncanonical NF-κB activation might be mediated by an indirect mechanism, likely involving the autocrine action of a TNFR superfamily member. One TNFR family member known to stimulate TRAF2 degradation is TNFRII (also called CD120b)37. We found that TNFRII was abundantly expressed on DCs and further induced by TLR ligands (Fig. 6d). Because TLR ligands are also strong inducers of the TNFRII ligand TNF in DCs, we examined the role of TNFRII in TLR-stimulated noncanonical NF-κB signaling and Il23a gene expression. A TNFRII neutralizing antibody profoundly inhibited CpG-stimulated TRAF2 degradation and p52 nuclear translocation as well as induction of IL-23 p19 by CpG and Pam3CSK4 (Fig. 6e,f). Parallel studies using BMDCs prepared from mice deficient in the TNFRII-encoding gene Tnfrsf1b (Tnfrsf1b−/−) revealed a complete blockade of TLR-stimulated TRAF2 degradation, associated with impaired p52 nuclear expression and Il23a gene induction (Fig. 6g,h). Consistently, Tnfrsf1b−/− mice had a significant reduction in the concentration of fecal IgA and pIgR, pIgR expression in IECs, and the frequency of TH17 cells and ILCs in LP as compared to wild-type controls (Fig. 6i–l). These data suggest that induction of noncanonical NF-κB activation and Il23a gene expression involves TNFRII, highlighting a dynamic mechanism triggering noncanonical NF-κB signaling in DCs.
Figure 6.
TLR stimulates TRAF2 degradation in a TNFRII-dependent manner. (a–c) Immunoblot analysis of the indicated proteins in cytoplasmic (a) and whole-cell (b,c) extracts of wild-type (WT) or Map3k14-cKO BMDCs that were either not treated (NT) or stimulated for 24 h (a,b) or the indicated times (c). (d) Flow cytometric analysis of TNFRII (CD120b) in wild-type BMDCs stimulated for 24 h with the indicated TLR ligands. (e) Immunoblot analysis of the indicated proteins in cytoplasmic (CE) and nuclear (NE) extracts of wild-type BMDCs stimulated for 24 h with CpG either in the absence (−) or presence (+) of an anti-TNFRII blocking antibody. (f) ELISA of IL-23p19 in the culture supernatant of wild-type BMDCs that were not treated (NT) or stimulated with the indicated TLR ligands either alone or together with anti-TNFRII (n=4 mice/group). (g) Immunoblot analysis of the indicated proteins in cytoplasmic (CE) and nuclear (NE) extracts of wild-type or Tnfrsf1b−/− BMDCs stimulated with Pam3CSK4 (Pam3) or LPS for 24 h. (h) qRT-PCR analysis of Il23a mRNA in wild-type or Tnfrsf1b−/− BMDCs that were either not treated (NT) or stimulated with Pam3CSK4 (Pam3) or LPS for 24 h (NT and Pam3 stimulated groups: n=4 mice/group; LPS stimulated groups: n=5 mice/group). (i,j) ELISA of fecal IgA and pIgR concentration (i, mice: wild-type, n=10; Tnfrsf1b−/−, n=14) and qRT-PCR of pIgR mRNA level in IECs (j, n=7 mice/group) of wild-type and Tnfrsf1b−/− mice. (k,l) Flow cytometric analysis of IL-17-producing Th17 cells (k, n=4 mice/group) and ILC3s (l, n=5 mice/group) in lamina propria of wild-type and Tnfrsf1b−/− mice. Each symbol represents an individual mouse, and small horizontal lines indicate the mean (± s.e.m.) (f, h, l). Data are analyzed by unpaired student t test with P values shown above the plots (f, h, l). Data were representative of two independent experiments (h, l) or three independent experiments (a–g).
NIK regulates host defense against pathogen infections
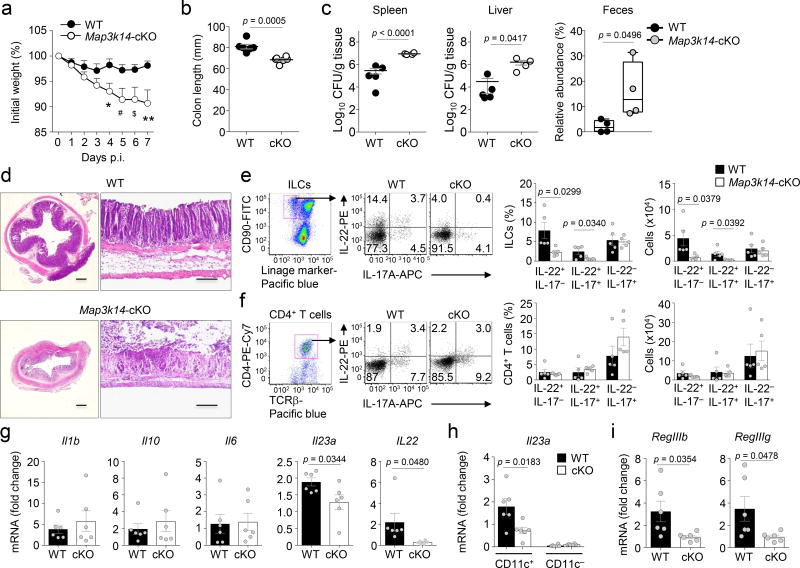
In addition to regulating intestinal homeostasis, IL-23 and IL-22 play an important role in promoting mucosal immunity against pathogens, particularly the mouse intestinal bacterial pathogen Citrobacter (C.) rodentium26, 38. Early phase of host defense against C. rodentium infection requires IL-23-dependent induction of IL-22 in ILC3s26, 39, although the late phase induction of IL-22-producing TH22 and TH17 subsets of CD4+ T cells is largely independent of IL-23 and dependent on IL-626, 38, 39. We infected the wild-type and Map3k14-cKO mice with C. rodentium and monitored inflammation caused during the early phase of infection. With the dose used in our experiment, the wild-type mice were largely resistant to C. rodentium infection and did not display substantial bodyweight loss (Fig. 7a). In contrast, the Map3k14-cKO mice were much more sensitive to C. rodentium infection, showing a severe bodyweight loss (Fig. 7a), significant colon shortening characteristics of colon inflammation (Fig. 7b), and higher C. rodentium titer in the spleen, liver, and feces (Fig. 7c). Consistently, histological analysis revealed much more severe damage in the colon tissue of the Map3k14-cKO mice compared to the wild-type mice (Fig. 7d). Studies using the Nfkb2Lym1/+ mice revealed similar results (Supplementary Fig. 8), emphasizing the function of NIK-dependent noncanonical NF-κB pathway in the regulation of mucosal immunity against C. rodentium infection.
Figure 7.
DC-specific NIK deletion impairs host defenses against C. rodentium infection. (a–c) Bodyweight change of wild-type (WT) and Map3k14-cKO (cKO) mice infected orally with 4×109 CFUs of C. rodentium (mice: wild-type, n=16; cKO, n=12). (b) Colon length of C. rodentium-infected (day 7) wild-type and Map3k14-cKO mice (mice: wild-type, n=6; cKO, n=6). (c) C. rodentium titer in spleen, liver and feces of wild-type and Map3k14-cKO mice 7 days after infection (mice: spleen and liver, wild-type, n=5; cKO, n=4; feces, wild-type, n=4; cKO, n=4). (d) Histological analysis of H&E stained colon tissue in vertical (left) and parallel (right) sections. Scale bar represents 200 µm. (e,f) Flow cytometric analysis of ILC (e) and CD4+ T cell (f) populations producing IL-17A and IL-22. Data are presented as a representative FACS plot (left) and summary graphs based on multiple mice (wild-type, n=5; cKO, n=5). (g) qRT-PCR analysis of the indicated cytokine mRNAs in colonic tissues of wild-type or Map3k14-cKO mice (wild-type, n=6; cKO, n=6). (h) qRT-PCR analysis of Il23a mRNA in sorted LP DCs of wild-type or Map3k14-cKO mice (wild-type, n=6; cKO, n=6). (i) qRT-PCR analysis of RegIIIb and RegIIIg mRNAs in purified IECs of wild-type or Map3k14-cKO mice (wild-type, n=6; cKO, n=6). Each symbol represents an individual mouse, and small horizontal lines indicate the mean (± s.e.m.) (b, c, e–i). Data are analyzed by two-way ANOVA followed by Sidak’s multiple comparisons test (a) or unpaired two-tailed Student’s t-test (b, c and e–i), with P values shown above the plots (b, c, e–i). For panel a, *P=0.0139, #P=0.0177, $P=0.0201 and **P=0.0061. Data were representative of two independent experiments.
In response to C. rodentium challenge, the Map3k14-cKO mice had a significantly lower level of IL-22-producing ILCs than wild-type control mice (Fig. 7e), which was in line with the critical role of ILC-derived IL-22 in mediating the early phase of host defense against C. rodentium infections26, 27. On the other hand, the Map3k14-cKO and wild-type mice had comparable frequencies of CD4+ T cells expressing IL-22 (TH22 cells), IL-17 or both IL-17 and IL-22 (TH17 cells) upon C. rodentium infection (Fig. 7f). Parallel qRT-PCR analysis revealed reduced expression of Il23a and Il22 in the colonic tissue of Map3k14-cKO mice (Fig. 7g). The phenotype of Il23a expression became more striking when we used purified LP DCs (Fig. 7h). DC-specific NIK deficiency also reduced the expression of anti-bacterial genes, RegIIIb and RegIIIg, known to be induced by IL-22 (Fig. 7i). These results demonstrate an important role for the DC-specific NIK in regulating mucosal immunity against C. rodentium infection and further emphasized the role of NIK in mediating IL-23 induction in DCs and generation of IL-22-producing ILCs.
DC-specific NIK deletion ameliorates colonic inflammation
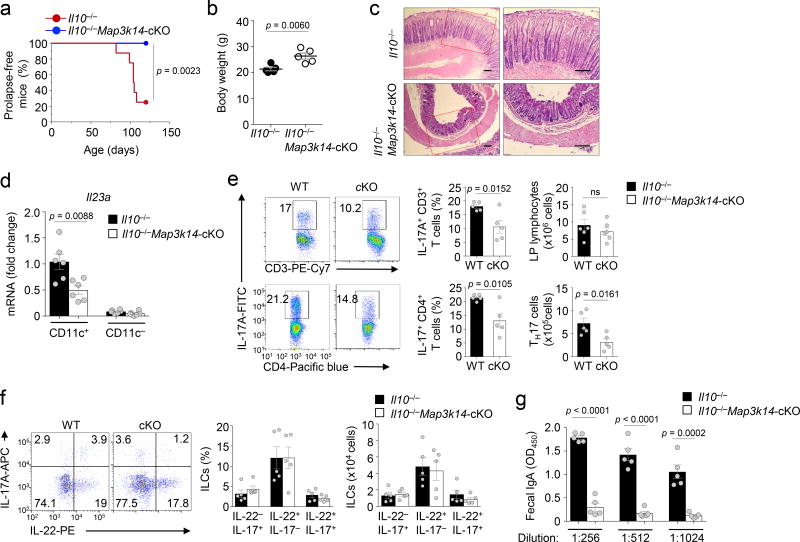
While IL-23 and IL-17 are important for host defense against infections, these cytokines also contribute to colon inflammation under conditions that disrupt intestinal homeostasis and have been associated with human inflammatory bowel disease (IBD)29, 31, 40, 41. A well-defined animal model of chronic intestinal inflammation is the Il10−/− mouse, which spontaneously develops colitis associated with bodyweight loss, rectal prolapse, and increased mortality42, 43, 44. The chronic colon inflammation of Il10−/− mice is dependent on IL-23, which appears to act through promoting TH17 cell differentiation and expansion31. As expected, the Il10−/− mice spontaneously developed signs of colitis by 3 months of age, including rectal prolapse, body weight loss, and histological features of inflammation characterized by the thickening of mucosa (Fig. 8a–c). Remarkably, DC-specific deletion of NIK in the Il10−/− mice largely prevented the development of these symptoms of inflammation (Fig. 8a–c). Consistently, DC-specific deletion of NIK in Il10−/− mice inhibited the expression of IL-23 in LP DCs (Fig. 8d), coupled with reduced frequencies of IL-17-producing TH17 cells (Fig. 8e). Interestingly, however, the NIK deficiency did not significantly alter the frequency of ILC3s (Fig. 8f). The spontaneous colitis of Il10−/− mice is also associated with an increase in the level of secreted IgA45. We found that DC-specific NIK deletion drastically reduced the level of fecal IgA in Il10−/− mice (Fig. 8g). Thus, NIK has a crucial role in the regulation of colonic inflammation in the Il10−/− mouse model.
Figure 8.
DC-specific NIK deletion ameliorates colon inflammation in IL-10-deficient mice. (a) Summary graph of rectal prolapse in age- and sex-matched IL10−/− and Il10−/− Map3k14-cKO mice (n=8 mice/group). (b) Summary graph of bodyweight of 3-month old IL10−/− and Il10−/− Map3k14-cKO mice (n=5 mice/group). (c) Histological analysis by H&E staining of distal colon from IL10−/− and Il10−/− Map3k14-cKO mice (3-month old). Scale bar represents 200 µm, left panels (×100) and right panels (×200). (d) qRT-PCR analysis of Il23a mRNA in LP DCs of 3-month old IL10−/− and Il10−/− Map3k14-cKO mice (n=6 mice/group). (e,f) Flow cytometric analysis of IL-17-producing CD4+ T cells (e, n=5 mice/group) and IL-22/IL-17-producing ILCs (f, n=5 mice/group), presented as representative FACS plots (left) and summary graphs based on multiple mice. (g) ELISA of fecal IgA of IL10−/− and Il10−/− Map3k14-cKO mice (n=5 mice/group). Each symbol represents an individual mouse, and small horizontal lines indicate the mean ± s.e.m. (b, d–g). Data are compared using the Kaplan-Meier method, with differences determined by log-rank test (a) or analyzed by unpaired two-tailed Student’s t-test (b, d–g), with P values shown above the plots.
Discussion
In this study, we identified the noncanonical NF-κB kinase NIK as a pivotal mediator of DC functions in the regulation of IgA secretion and microbiota homeostasis. Our data suggest that DCs control pIgR expression and IgA secretion through a signaling mechanism that requires NIK and its downstream noncanonical NF-κB pathway. The NIK signaling axis facilitates TLR-stimulated IL-23 production and, thereby, maintains the abundance of Th17 cells and ILC3s, major sources of IL-17. IL-17 in turn induces the expression of pIgR in IECS to support IgA secretion.
Consistent with its involvement in mucosal immunity, the NIK pathway played a role in regulating the homeostasis of intestinal microbiota. DC-specific NIK deletion or Nfkb2lym1 mutation increased the abundance of Enterococcus spp. and the SFB C. Savagella. SFB induces mucosal immunity, including IgA production, whereas the secretary IgA is required for controlling SFB expansion in the intestine22, 46. Enterococci are known as low abundance gut commensals that are induced to cause opportunistic infections under pathogenic conditions and are associated with intestinal inflammation18, 19, 20. A recent report suggests that ILC-derived IL-22 is crucial for controlling the expansion and invasion by Enterococci27. Interestingly, we found that the Enterococcus expansion in Map3k14-cKO mice was associated with a severe reduction in IL-22-producing ILCs, supporting the involvement of IL-22 in Enterococcus spp control. However, we believe that the defective IgA secretion may also contribute to the microbiota deregulation in Map3k14-cKO mice. We found that Enterococcus spp., like C. Savagella, are IgA-coated bacteria. Interference of IgA secretion by antibody-mediated IL-17 neutralization causes an increase in Enterococcus numbers. Although the relative contribution of IgA and IL-22 to the regulation of Enterococci is unclear, it is apparent that IL-23 plays an important role in controlling these enteric bacteria, since antibody-mediated IL-23 neutralization also causes Enterococcus upregulation.
Noncanonical NF-κB activation is typically mediated by signals from the TNFR superfamily members13. Our current study suggests that the noncanonical NF-κB pathway may be stimulated by TLR ligands in intestinal DCs. However, it appeared that this novel function of TLRs is coupled with specific members of the TNFR superfamily, particularly TNFRII. TNFRII was highly expressed on DCs and further upregulated upon TLR stimulation. We obtained strong evidence that TNFRII is involved in TLR-stimulated TRAF2 degradation and noncanonical NF-κB activation. Consistently, the Tnfrsf1b−/− mice had reduced frequency of TH17 cells and ILCs and impaired IgA secretion. It is possible that TLRs stimulate TNFRII-mediated noncanonical NF-κB activation through upregulation of TNFRII and induction of its ligand, TNF. On the other hand, our data do not exclude the involvement of additional TNFR members. In fact, another TNFR member, CD40, was also strongly induced in DCs by LPS, although not by another TLR ligand, Pam3CSK4. Unlike the ligand of TNFRII, which is expressed in DCs, the CD40 ligand is expressed on T cells and is involved in T cell-dependent CD40 stimulation. Another TNFR member that may be involved in noncanonical NF-κB activation in DCs is lymphotoxin beta receptor (LTβR), since it has been implicated in the regulation of intestinal DCs47.
The function of noncanonical NF-κB in DCs has been poorly studied, since the DC-conditional NIK KO mice were not available until recently17. Our finding that DC-specific NIK is crucial for intestinal homeostasis is novel and unexpected. We identified Il23a as a major target gene of the noncanonical NF-κB pathway in DCs stimulated with different TLR ligands. The requirement of noncanonical NF-κB for IL-23 expression was also demonstrated in freshly isolated LP DCs, thus emphasizing the physiological relevance of this finding. IL-23 and its downstream cytokine IL-17 have paradoxical roles in the regulation of intestinal homeostasis. While these cytokines promote host defenses against infections and shape the microbiota, they are also important players in colon inflammation and have been associated with human inflammatory bowel disease (IBD)29, 31, 40, 41. In agreement with its role in mediating Il23a gene induction, NIK plays a crucial role in the Il10−/− animal model of IBD.
In summary, our data provide genetic evidence that NIK signaling axis is crucial for the functions of DCs in regulating intestinal immunity and homeostasis. The DC-specific NIK is required for maintaining pIgR expression in IECs and, thus, IgA secretion as well as involved in modulation of microbiota composition. Our work identifies Il23a as a novel target gene of the NIK-regulated noncanonical NF-κB pathway in DCs. Given the crucial role of IL-23 in regulating mucosal immune homeostasis and inflammation, this finding provides important insight into the molecular mechanism that mediates the mucosal function of DCs. Our work also has important implications for preventive and therapeutic approaches for colon inflammation.
ONLINE METHODS
Mice
Map3k14-flox mice (on C57BL/6 background), provided by Genentech, were generated using loxP system targeting exon 2 of the Map3k14 gene48. The Map3k14-flox mice were crossed with Cd11c-Cre transgenic mice (B6 genetic background, Jackson Laboratories) to create Map3k14 DC-conditional KO or Map3k14-cKO (Map3k14 fl/flCd11c-Cre) and wild-type (Map3k14+/+Cd11c-Cre) mice. Il10−/− mice were purchased from Jackson Lab and crossed with Map3k14-cKO mice to produce Il10−/−Map3k14-cKO and control Il10−/− mice. Rorc−/− and Tnfrsf1b−/− mice were purchased from Jackson Lab. Nfkb2lym1 mice were provided by R. Starr (Walter and Eliza Hall Institute of Medical Research)36. Nfkb2lym1/+ heterozygous mice were used in experiments since they display strong phenotype in impaired noncanonical NF-κB activation and function36. All KO and conditional KO mice were crossed using heterozygous breeders to generate littermate KO and wild-type control mice for experiments. Mice were maintained in a specific pathogen-free facility, and all animal experiments were in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of Texas MD Anderson Cancer Center.
Antibodies and reagents
The following fluorochrome-labeled antibodies specific to mouse (m) proteins and their corresponding isotype controls were purchased from eBioscience: APC- or PE-conjugated anti-mIL-17A (eBio17B7), FITC- or APC-conjugated anti-mIFN-γ (XMG1.2), PE-conjugated anti-mIL-22 (1H8PWSR), PE-Cy7-conjugated anti-mCD3 (17A2), Pacific blue-conjugated anti-mCD4 (GK1.5), PerCp-Cy5.5-conjugated anti-mCD8 (53-6.7), PE-Cy7-conjugated anti-mNK1.1 (PK136), PE-Cy7-conjugated anti-mB220 (RA3-6B2), PE-Cy7-conjugated anti-mCD11b (M1/70), PE-Cy7-conjugated anti-mCD11c (N418), PE-Cy7-conjugated anti-mGr-1 (RB6-8C5), PE-Cy7-conjugated anti-mTer-119 (TER-119), FITC-conjugated anti-mCD45 (30-F11), APC-conjugated anti-mRORγt (B2D), APC- or PE-Cy7-conjugated anti-mMHCII (M5/114.15.2), PE- or FITC-conjugated anti-mCD80 (16-10A1), PE- or FITC- conjugated anti-mCD86 (GL1), PE- or Pacific blue-conjugated anti-mCD127 (A7R34), PE-conjugated anti-mCD103 (2E7), APC-conjugated anti-mCD62L (MEL-14), PE-conjugated anti-mCD44 (IM7), APC-conjugated anti-mEpCAME (G8.8), APC-conjugated anti-mCD317 (BST2), PerCp-Cy5.5-conjugated anti-mCCR7 (4B12) and eFluor 506 conjugated fixable viability dye. Purified anti-mCD16/32 (2.4G2), PE conjugated anti-mCD120b (TR75-89), FITC-conjugated anti-mIgA (c10-3), FITC-conjugated anti-mCD90.2 (30-H12), PE-Cy7 conjugated anti-mCXCR5 (2G8), and APC-conjugated anti-mPD-1 (J43) were purchased from BD Pharmingen. PE-conjugated anti-mAPRIL (A3D8), and Alexa Fluor647-conjugated anti-mTLR5 (ACT5) were purchased from BioLegend. PE-conjugated anti-mLTβR (5G11b) was purchased from Bio-Rad. PE-conjugated anti-mBAFF (121808) was purchased from R&D Systems.
Anti-Actin (C-4,1:10,000) was from Sigma, and anti-p100/p52 (TB4, 1:8,000) was from National Cancer Institute Preclinical Repository. Antibodies for RelB (C-19, 1:1000), Lamin B (C-20, 1:1000), NIK (H248, 1:1000), TRAF2 (C-20, 1:1000), TRAF3 (H-122, 1:1000), cIAP2 (H85, 1:1000), phospho-p65 (Ser529, 1:1000), and c-Rel (sc71, 1:3000) as well as a control rabbit IgG (sc-2027, 1:1000) were from Santa Cruz Biotechnology. Antibodies for phospho-IκBα (Ser32, 1:1000), p65 were purchased from Cell Signaling Technology Inc. MG132 was from Cayman Chemical.
LPS (derived from Escherichia coli strain 0127:B8) and CpG (2216) were purchased from Sigma-Aldrich, Pam3CSK4 was from Alexis, recombinant murine GM-CSF was from Peprotech, and recombinant murine IL-17 was from R&D Systems.
Preparation of bone marrow-derived DCs (BMDCs)
BMDCs were generated by cultivating bone marrow cells of indicated wild-type and KO mice in growth medium supplemented with recombinant GM-CSF (20 ng/ml), as described previously49. Fresh GM-CSF-containing medium was added on day 3 and day 6, and the fully differentiated DCs were harvested on day 8 for function assay. The generated DC population was analyzed by flow cytometry based on CD11c expression and further enriched using CD11c microbeads (Miltenyi Biotec).
Microbiome analysis
Littermates of the indicated mutant and wild-type mice were used for microbiome analysis. The mutant and wild-type mice were weaned at 3 weeks after birth and co-caged until 4 weeks after birth. The mutant and wild-type mice were then either separately housed or, as control, co-housed for the experiments, and all mice were housed under pathogen-free conditions in ventilated cages located on the same rack to minimize environmental influences. Fresh feces were harvested from individual mice and placed into sterilized PBS (10 µl/mg). The samples were vigorously vortexed for 1 min and then thoroughly mashed on 100-µm cell strainers. Each sample was diluted and placed on universal medium for the growth of anaerobic (Luria-Bertani broth) and aerobic (Bacto agar) bacteria. The commensal bacteria were grown in an anaerobic chamber, and colonies were counted after incubation at 37 °C for 72 h. Colonies were further classified using selective media, including MacConkey agar for E. coli, Bacteroides Bile Esculin agar for Bacteroides fragilis, Enterococcosel agar for Enterococci and group D streptococci, and lactobacilli MRS agar for Lactobacillus species. All agar plates were made according to the manufacturer's protocol (BBL).
16S rRNA gene sequencing was performed at the Alkek Center for Metagenomics and Microbiome Research (CMMR), Baylor College of Medicine, as previously reported50. Briefly, bacterial genomic DNA was extracted from feces using MO BIO PowerSoil DNA Isolation Kit (MO BIO Laboratories). The 16S rDNA V4 region was amplified by PCR and sequenced in the MiSeq platform (Illumina) using the 2 × 250 bp paired-end protocol yielding pair-end reads that overlap almost completely. The primers used for amplification contain adapters for MiSeq sequencing and single-index barcodes so that the PCR products may be pooled and sequenced directly51, targeting at least 10,000 reads per sample. 16S rRNA sequencing data analysis was performed by ATIMA software developed at the Alkek Center for Metagenomics and Microbiome Research (CMMR), Baylor College of Medicine50.
Analysis of IgA-coated bacteria
Fresh feces collected directly from individual mice were placed on ice and incubated in 1 ml PBS per 100 mg feces for 1 h, and then thoroughly mashed on 100-µm cell strainers. After passing through strainers, the resulting suspension was briefly centrifuged (50g, 15 min, 4 °C) to remove large aggregates. 100 µl of supernatants containing fecal bacteria were transferred to new tubes, washed with 1% BSA in PBS, and centrifuged (8,000g, 5 min, 4 °C). After wash, bacterial pellets were resuspended in a blocking buffer (100 µl) containing 20% fetal bovine serum (FBS), incubated for 20 min on ice, and then stained in a buffer (100 µl) containing PE-conjugated anti-Mouse IgA (1:12.5; eBioscience clone 11-44-2) for 30–40 min on ice. Samples were then washed 3 times with 1 ml staining buffer before cell sorting. Anti-IgA stained fecal bacteria were incubated in 1 ml staining buffer containing 50 µl anti-PE Magnetic Activated Cell Sorting (MACS) beads for 15 min at 4 °C, washed twice with 1ml staining buffer (10,000g, 5 min, 4 °C), and then sorted by MACS. After MACS separation, each 50 µl of the positive and negative fraction was collected for confirmation of cell separation and for cell counting. Each sample was diluted and smeared on several selective agar media to detect IgA-positive commensal bacteria. The sorted IgA-positive and negative fractions were further analyzed by 16S rRNA gene sequencing.
Histology analysis
The entire colon was removed, opened longitudinally, rinsed to clean internal contents, and rolled over the full length to obtain a ‘Swiss roll’, which was then fixed in 4% formaldehyde in PBS for 1 h at 25 °C. Fixed tissues were dehydrated by gradually soaking in alcohol and xylene and then embedded in paraffin. The paraffin-embedded specimens were cut into 5 µm sections, stained with hematoxylin-eosin (H&E), and viewed with a digital inverted light microscope (EVOS, Thermo Fisher Scientific,).
Confocal analysis
For pIgR staining, small intestinal tissue was cut longitudinally and rolled over a ‘Swiss roll’, which was then fixed in 4% formaldehyde in PBS for 1 h at 25 °C. Fixed tissues were dehydrated by gradually soaking in sucrose (10 % ~ 30 %) and then embedded in cryomatrix (Thermo scientific). The frozen sections were sequentially reacted with goat anti-pIgR IgG Antibody (R&D Systems) at 4 °C overnight, and then incubated with FITC-conjugated donkey anti-goat antibody (Jackson ImmunoResearch) at room temperature for 2 h and viewed under a confocal scanning laser microscope (Zeiss). The pIgR+ dots were counted on the intestinal villi that were randomly selected in 5 regions per slide. Data shown are the mean values ±s.e.m. of individual mice from 2 independent experiments with 3 mice per experiment.
C. rodentium infection
C. rodentium preparation and infection were as previously described52. Briefly, C. rodentium strain DBS100 (ATCC 51459) was grown in LB broth at 37 °C overnight with shaking. The number of bacteria was measured by checking optical density at 600nm and further confirmed as colony-forming units (CFUs) by smearing to LB agar plates after serial dilution. For oral infection, each mouse was administered 4 × 109 CFUs of bacteria in a total volume of 200 µl.
Cell preparation from intestine
For isolation of lymphocytes from small intestinal LP (SI-LP), tissue pieces were treated with RPMI 1640 containing 0.5 mM EDTA for 20 min at 37 °C to remove epithelial cells. This washing step was repeated twice. Tissues were minced and then treated with 500 µg/ml collagenase D (Sigma-Aldrich) and 100 µg/ml DNase I (Roche Applied Science,) in RPMI 1640 containing 10% FBS and digested for 30 min at 37 °C with shaking. This enzyme reaction was also repeated twice. The lymphocytes of LP were enriched by using 40% and 75% Percoll gradients (Amersham Biosciences,). For isolation of intestinal epithelial cells (IECs) and intestinal epithelial lymphocytes (IELs) from SI-LP, the filtered liquid in the previous washing step was centrifuged (700g, 20 min, 25°C) in 3 different Percoll gradients (25% / 40% / 75%) with no brake. IECs and IELs were enriched in the boundary between 25% and 40% and between 40% and 75% Percoll, respectively.
For sorting LP-DCs and intestinal epithelial cells (IECs), LP cells were isolated and stained with FITC-conjugated anti-mouse CD45, Pacific Blue-conjugated anti-mouse CD11c and APC-conjugated anti-mouse MHCII. LP-DCs were sorted as CD45+CD11c+MHCII+ cells using FACS Aria. IECs were isolated as described previously53, and cells were further sorted as CD45−EpCAM+ population using FACS Aria.
Flow cytometry and intracellular cytokine staining (ICS)
Single-cell suspensions of spleen, mesenteric lymph nodes, Peyer’s patches, lamina propria, bone marrow and thymus were subjected to flow cytometry using an LSRFortessa (Becton Dickinson). The cells were incubated with anti-CD16/CD32 (eBioscience) to block nonspecific antibody binding via Fc receptors and then stained with fluorochrome-labeled antibodies, and subsequently processed on an LSRFortessa. The data were analyzed using FlowJo software (Tree Star). Gating strategies are summarized in Supplementary Fig. 9. For ICS detection of IFN-γ, IL-17 and IL-22, cells were stimulated for 4–5 h with PMA (50 ng/ml) and ionomycin (750 ng/ml) in the presence of GolgiStop (BD Bioscience). After incubation, cells were stained with fixable viability dye, blocked with FcγR blocker (CD16/32), and stained for specific surface molecules. After surface staining, cells were fixed, permeabilized and stained for intracellular cytokines by using a fixation/permeabilization kit (BD Biosciences). Samples were processed on an LSRFortessa (Becton Dickinson) and analyzed by using FlowJo software (TreeStar).
IL-17 and IL-12/IL-23 p40 neutralization
For IL-17 neutralization, mice were i.p. injected with anti-mouse IL-17 antibody (Clone: 17F3) or isotype control IgG1 (Clone: MOPC-21) (10 µg/g mouse weight) every other day for 2 weeks. For IL-12/IL-23 p40 neutralization, mice were i.p. injected with anti-mouse IL-12/IL-23 p40 antibody (Clone: C17.8) or isotype control IgG2a (Clone: 2A3) (100 µg per mice) every other day for 2 weeks. Anti-mouse IL-17 (Clone: 17F3), anti-mouse IL-12/IL-23 p40 (Clone: C17.8), mouse IgG1 isotype control (Clone: MOPC-21), and mouse IgG2a isotype control (Clone: 2A3) were bought from BioXcell company.
Real-time quantitative RT-PCR
Total RNA was isolated from DC cells or colon tissue using TRI reagent (Invitrogen) and subjected to cDNA synthesis using MMLV reverse transcriptase (Invitrogen) and oligo (dT) primers. Real-time quantitative PCR (qRT-PCR) was performed using iCycler Sequence Detection System (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad). The expression of individual genes was calculated by a standard curve method and was normalized to the expression of Actb. The primers used in qRT-PCR assays are shown in Supplementary Table 1.
Immunoblot assays
Whole-cell and subcellular extracts were prepared and subjected to IB essentially as described49. In brief, protein samples were separated by SDS-PAGE, transferred to PVDF membranes (0.45 µm, Millipore), and blocked for 1 h with 5% bovine serum albumin (BSA), 0.1 % Tween-20 in TBS. The membranes were then washed and incubated with primary antibodies overnight. The goat anti-rabbit or goat anti-mouse IgG secondary antibodies (1:5000 dilution) were used to incubate for 1 h. After washing, immunoreactive bands were visualized by chemiluminescent detection (ECL, Roche Diagnostics,) and exposure to X-ray film (Thermo Fisher Scientific).
Chromatin immunoprecipitation (ChIP) assays
BMDCs were stimulated for 12 h with CpG, Pam3CSK4 and LPS and then fixed with 1% formaldehyde and sonicated as previously described54. Lysates (from 2×107 cells in 3 ml) were subjected to IP with the indicated antibodies, and the precipitated DNA was then purified by Qiaquick columns (Qiagen) and quantified by qPCR using a pair of primers that amplify the Il23a promoter (Supplementary Table 1). The y axis is the percentage (%) of target transcript factor-binding DNA normalization for total input DNA.
ELISA
For IgA and pIgR ELISA analysis, fresh feces were collected from individual mice, homogenized in sterile PBS, and centrifuged to remove bacteria and insoluble debris. Fecal samples were diluted in sterile PBS as a 2-fold serial dilution and added to 96-well plates pre-coated overnight (at 4 °C) with 1 µg/ml anti-mouse IgA (Southern Biotechnology) or 0.5 µg/ml anti-mouse pIgR (R&D Systems). Samples were incubated at room temperature for 2 h. For IgA analysis, HRP conjugated anti-IgA (Southern Biotechnology) was added for 1 h. For pIgR ELISA analysis, biotinylated mouse pIgR antibody was added for 1 h, followed by incubation with avidin-HRP for 30 min. Plates were developed using dTMB substrate according to the manufacturer’s instructions (Thermo Fisher Scientific) and analyzed at 450nm using a microplate reader.
For ELISA analysis of cytokines (IL-23p19, IL-1β and IL-6), equal numbers of wild-type and Map3k14-cKO BMDCs were stimulated with CpG or Pam3CSK4 for 48 h, and supernatants were collected for ELISA analysis using an ELISA kits (Thermo Fisher Scientific) according to the manufacturer’s instructions. Detection limits were 16 pg/ml for IL-23p19, 1.2 pg/ml for IL-1β, and 4 pg/ml for IL-6.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (GraphPad,). Significant differences between two groups were analyzed with two-tailed unpaired t-test. Multiple groups were analyzed by one-way ANOVA, where applicable, was performed to determine whether an overall statistically significant change existed before the multiple Student’s t test to analyze the difference between any two groups. The body weight loss were analyzed by multiple t tests, and in turn statistical significance were determined by using Holm-Sidak method. The prolapse-free rate was compared using the Kaplan-Meier method, with differences determined by log-rank test.
Reporting summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank Genentech Inc for providing the Map3k14-flox mice, Walter and Eliza Hall Institute of Medical Research for Nfkb2Lym1 mice, and National Cancer Institute Preclinical Repository for NF-κB p100/p52 antibody. This work was supported by grants from the National Institutes of Health (GM84459, AI057555, AI104519, and AI64639). This study also used the NIH/NCI-supported resources under award number P30CA016672 at The MD Anderson Cancer Center.
Footnotes
AUTHOR CONTRIBUTIONS
Z.J. and J.-Y.Y. designed and performed the research, prepared the experiments, and wrote part of the manuscript; M.G., H.W., X.X., Y.L., T.L., L.Zhu, J.S., L.Zha., X.Z., D.J., D.L., X.C. contributed experiments; H.D.B. contributed critical reagents; Y.C. made supervising contributions to intestinal immune cells analyses, and S.-C.S. supervised the work and wrote the manuscript.
Competing financial interests
H.D.B. is an employee of Genentech, Inc., and the other authors declare no competing financial interests. Genentech, Inc. provided the Map3k14-flox mice.
References
- 1.Gutzeit C, Magri G, Cerutti A. Intestinal IgA production and its role in host-microbe interaction. Immunol Rev. 2014;260:76–85. doi: 10.1111/imr.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandtzaeg P. Secretory IgA: Designed for Anti-Microbial Defense. Front Immunol. 2013;4:222. doi: 10.3389/fimmu.2013.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nat Rev Immunol. 2008;8:435–446. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briseno CG, Murphy TL, Murphy KM. Complementary diversification of dendritic cells and innate lymphoid cells. Curr Opin Immunol. 2014;29:69–78. doi: 10.1016/j.coi.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker JA, Barlow JL, McKenzie AN. Innate lymphoid cells--how did we miss them? Nat Rev Immunol. 2013;13:75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 7.Buela KA, Omenetti S, Pizarro TT. Cross-talk between type 3 innate lymphoid cells and the gut microbiota in inflammatory bowel disease. Curr Opin Gastroenterol. 2015;31:449–455. doi: 10.1097/MOG.0000000000000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerutti A. The regulation of IgA class switching. Nat. Rev. Immunol. 2008;8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massacand JC, et al. Intestinal bacteria condition dendritic cells to promote IgA production. PLoS One. 2008;3:e2588. doi: 10.1371/journal.pone.0002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruane D, et al. Microbiota regulate the ability of lung dendritic cells to induce IgA class-switch recombination and generate protective gastrointestinal immune responses. J Exp Med. 2016;213:53–73. doi: 10.1084/jem.20150567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh H, Ghosh S. NF-kappaB: roles and regulation in different CD4(+) T-cell subsets. Immunol. Rev. 2013;252:41–51. doi: 10.1111/imr.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun SC. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat Rev Immunol. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun SC. The noncanonical NF-kappaB pathway. Immunol. Rev. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senftleben U, et al. Activation of IKKa of a second, evolutionary conserved, NF-kB signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 15.Xiao G, Harhaj EW, Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol. Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 16.Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 17.Katakam AK, et al. Dendritic cells require NIK for CD40-dependent cross-priming of CD8+ T cells. Proc Natl Acad Sci U S A. 2015;112:14664–14669. doi: 10.1073/pnas.1520627112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vedantam G, Viswanathan VK. Unlocking the gates to inflammatory bowel disease: the role of Enterococcus faecalis gelatinase. Gastroenterology. 2011;141:795–798. doi: 10.1053/j.gastro.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y, et al. Increased Enterococcus faecalis infection is associated with clinically active Crohn disease. Medicine (Baltimore) 2016;95:e5019. doi: 10.1097/MD.0000000000005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jett BD, Huycke MM, Gilmore MS. Virulence of enterococci. Clin Microbiol Rev. 1994;7:462–478. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson CL, Vier R, Mikaelyan A, Wienemann T, Brune A. 'Candidatus Arthromitus' revised: segmented filamentous bacteria in arthropod guts are members of Lachnospiraceae. Environ Microbiol. 2012;14:1454–1465. doi: 10.1111/j.1462-2920.2012.02731.x. [DOI] [PubMed] [Google Scholar]
- 22.Ericsson AC, Hagan CE, Davis DJ, Franklin CL. Segmented filamentous bacteria: commensal microbes with potential effects on research. Comp Med. 2014;64:90–98. [PMC free article] [PubMed] [Google Scholar]
- 23.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaffar Z, Ferrini ME, Herritt LA, Roberts K. Cutting edge: lung mucosal Th17-mediated responses induce polymeric Ig receptor expression by the airway epithelium and elevate secretory IgA levels. J Immunol. 2009;182:4507–4511. doi: 10.4049/jimmunol.0900237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao AT, Yao S, Gong B, Elson CO, Cong Y. Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J Immunol. 2012;189:4666–4673. doi: 10.4049/jimmunol.1200955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 27.Pham TA, et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 2014;16:504–516. doi: 10.1016/j.chom.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberl G. RORgammat, a multitask nuclear receptor at mucosal surfaces. Mucosal Immunol. 2017;10:27–34. doi: 10.1038/mi.2016.86. [DOI] [PubMed] [Google Scholar]
- 29.Hue S, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J. Exp. Med. 2006;203:2473–2483. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kullberg MC, et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J. Exp. Med. 2006;203:2485–2494. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yen D, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clinic. Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maloy KJ, Kullberg MC. IL-23 and Th17 cytokines in intestinal homeostasis. Mucosal Immunol. 2008;1:339–349. doi: 10.1038/mi.2008.28. [DOI] [PubMed] [Google Scholar]
- 33.Shih VF, et al. Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-kappaB pathways. Nat. Immunol. 2012;13:1162–1170. doi: 10.1038/ni.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J, et al. T Cell-Intrinsic Function of the Noncanonical NF-kappaB Pathway in the Regulation of GM-CSF Expression and Experimental Autoimmune Encephalomyelitis. Pathogenesis. J. Immunol. 2014;193:422–430. doi: 10.4049/jimmunol.1303237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang C, Zhang M, Sun SC. beta-TrCP binding and processing of NF-kappaB2/p100 involve its phosphorylation at serines 866 and 870. Cell Signal. 2006;18:1309–1317. doi: 10.1016/j.cellsig.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 36.Tucker E, et al. A novel mutation in the Nfkb2 gene generates an NF-kappa B2 "super repressor". J. Immunol. 2007;179:7514–7522. doi: 10.4049/jimmunol.179.11.7514. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Yang Y, Ashwell JD. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature. 2002;416:345–347. doi: 10.1038/416345a. [DOI] [PubMed] [Google Scholar]
- 38.Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 39.Basu R, et al. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity. 2012;37:1061–1075. doi: 10.1016/j.immuni.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Catana CS, et al. Contribution of the IL-17/IL-23 axis to the pathogenesis of inflammatory bowel disease. World J Gastroenterol. 2015;21:5823–5830. doi: 10.3748/wjg.v21.i19.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neurath MF. Current and emerging therapeutic targets for IBD. Nat Rev Gastroenterol Hepatol. 2017;14:269–278. doi: 10.1038/nrgastro.2016.208. [DOI] [PubMed] [Google Scholar]
- 42.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 43.Specht S, Arriens S, Hoerauf A. Induction of chronic colitis in IL-10 deficient mice requires IL-4. Microbes Infect. 2006;8:694–703. doi: 10.1016/j.micinf.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Davidson NJ, Fort MM, Muller W, Leach MW, Rennick DM. Chronic colitis in IL-10−/−mice: insufficient counter regulation of a Th1 response. Int Rev Immunol. 2000;19:91–121. doi: 10.3109/08830180009048392. [DOI] [PubMed] [Google Scholar]
- 45.Gomes-Santos AC, et al. New insights into the immunological changes in IL-10-deficient mice during the course of spontaneous inflammation in the gut mucosa. Clin Dev Immunol. 2012;2012:560817. doi: 10.1155/2012/560817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki K, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tumanov AV, et al. Lymphotoxin controls the IL-22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host Microbe. 2011;10:44–53. doi: 10.1016/j.chom.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brightbill HD, et al. Conditional Deletion of NF-kappaB-Inducing Kinase (NIK) in Adult Mice Disrupts Mature B Cell Survival and Activation. J. Immunol. 2015;195:953–964. doi: 10.4049/jimmunol.1401514. [DOI] [PubMed] [Google Scholar]
- 49.Jin J, et al. Epigenetic regulation of the expression of Il12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid. Nat Immunol. 2016;17:259–268. doi: 10.1038/ni.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ajami NJ, Cope JL, Wong MC, Petrosino JF, Chesnel L. Impact of Oral Fidaxomicin Administration on the Intestinal Microbiota and Susceptibility to Clostridium difficile Colonization in Mice. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.02112-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu H, et al. OTUD7B controls non-canonical NF-kappaB activation through deubiquitination of TRAF3. Nature. 2013;494:371–374. doi: 10.1038/nature11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu J, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.