Abstract
With the continuous increase in the population of developing countries and decline of natural resources, there is an urgent need to qualitatively and quantitatively augment crop productivity by using new tools and technologies for improvement of agriculturally important traits. The new scientific and technological omics-based approaches have enabled us to deal with several issues and challenges faced by modern agricultural system and provided us novel opportunities for ensuring food and nutritional security. Recent developments in sequencing techniques have made available huge amount of genomic and transcriptomic data on model and cultivated crop plants including Arabidopsis thaliana, Oryza sativa, Triticum aestivum etc. The sequencing data along with other data generated through several omics platforms have significantly influenced the disciplines of crop sciences. Gene discovery and expression profiling-based technologies are offering enormous opportunities to the scientific community which can now apply marker-assisted selection technology to assess and enhance diversity in their collected germplasm, introgress essential traits from new sources and investigate genes that control key traits of crop plants. Utilization of omics science and technologies for crop productivity, protection and management has recently been receiving a lot of attention; the majority of the efforts have been put into signifying the possible applications of various omics technologies in crop plant sciences. This article highlights the background of challenges and opportunities for augmentation of crop productivity through interventions of omics technologies in India.
Keywords: Agriculture, Omics, High-throughput omics data, Crop plants, Crop productivity
Introduction
Agriculture is the pride of India. Indian agriculture has witnessed various revolutions (green revolution, white revolution, blue revolution and now brown revolution), which have made significant strides in terms of production and productivity, availability of food grains, horticultural produce, milk, meat and fish products in India. A critical role was played by government bodies such as the Indian Council of Agricultural Research (ICAR), Department of Biotechnology (DBT), Council of Scientific and Industrial Research (CSIR) and other government-funded as well as private organizations in ushering these revolutions. Although the total area under cultivation has remained the same, 40 ± 2 million hectares for the last 40 years, production has increased apparently. Production of food crops, horticultural crops, fisheries and eggs has increased by 4.5, 8, 9 and 27 times since 1950–1951, respectively. India has become a leading producer of many crops, which were previously unknown and unrecognized, but have now emerged as economically important. Although Indian agriculture has faced numerous challenges and shortcomings, yet it has exhibited impressive growth (Singh et al. 2013a; Ma et al. 2013). If Indian agriculture has to achieve the broad goal of sustainable growth, it is important that the omics-based approaches are extended to the total agricultural production consumption system, that is, across the complete agricultural value chain. This would need the use of omics technologies for further increase in agricultural productivity, product quality and resource use efficiencies that reduce farm costs, increase the rate of production and raise incomes besides conserving and enhancing the quality of natural resources (Narsaiah et al. 2012; Ma et al. 2013; Hollister 2014; Jung and Main 2014).
As per recent FAO report, India is the leading producer of eight commodities with higher production and productivity than the rest of the world. For example, two to three times increase in banana production has been possible due to adoption of in vitro quality planting material, fertigation, efficient mat and bunch management. Some vegetables such as tomato, cauliflower and potato, which were earlier temperate, can now be grown under tropical environmental conditions (Singh et al. 2013a; Mohanty et al. 2017). At the global level, fulfilling the food requirements of a population which is expected to be about 9.0 billion in 2050 is a cause of concern which has been discussed across the world. This situation is further complicated by climate change that results in an increase in abiotic and biotic stresses in crop plants, which further impose constraints on food production. The increasing temperature along with shifting pattern of rainfall and rising incidence of extreme weather events such as floods, droughts and frost further add to the complexity of the problem (Narsaiah et al. 2012; Ma et al. 2013; Singh et al. 2013a; Hollister 2014; Jung and Main 2014). Consequently, the challenge before the agriculture sector is to produce more food with declining arable land and water. Addressing the growing needs of the people and reducing unequal social satisfaction, research focus, therefore, has to be reoriented to utilize the emerging knowledge (Shekoofa et al. 2014). Omics-based approaches are now being utilized by agriculturists for improving crop productivity (Weckwerth 2011; Grover et al. 2013; Gupta et al. 2017). For example, a hybrid pigeonpea variety (GTH-1) has been developed by the International Crops Research Institute for the Semi-arid Tropics (ICRISAT) and Indian Council of Agriculture Research (ICAR) using genomics-assisted breeding technology. This variety has produced 32% more yield as compared to the best local variety (Saxena and Nadarajan 2010). Several high-yielding Basmati rice varieties including Pusa-1509 have also been developed by the Indian Agricultural Research Institute (IARI), New Delhi, through marker-assisted selection (MAS) and molecular breeding approaches. These varieties offer several benefits over other Basmati varieties in terms of less number of days required for maturity, better cooking quality and higher productivity (Singh et al. 2014). Previous research has demonstrated the impacts of Bt cotton in India; its cultivation reduced application of pesticides by 40% and provided yield advantages of 30–40% over conventional varieties (Sadashivappa and Qaim 2009). Furthermore, DuPont Pioneer (https://www.pioneer.com) and Monsanto (https://monsanto.com/) have, respectively, developed T series soybean cultivars and sunflower, soybean and maize cultivars, respectively, for commercial production using genomics-assisted breeding approach (Sam et al. 2007; Scheben et al. 2016). Besides, some mustard varieties, i.e., Coral-432, NRCHB 5-6, NRCDR 601, NPJ 112 (Pusa Mustard 25), RYSKS-2 and DMH-I, recently released by the Indian Council of Agriculture Research (ICAR), have at least 10% higher yield than other local varieties along with better tolerance to several abiotic and biotic stresses (https://timesofindia.indiatimes.com/city/nagpur/5-varieties-of-rapeseed-mustard-released-for-entire-country/articleshow/4872591.cm).
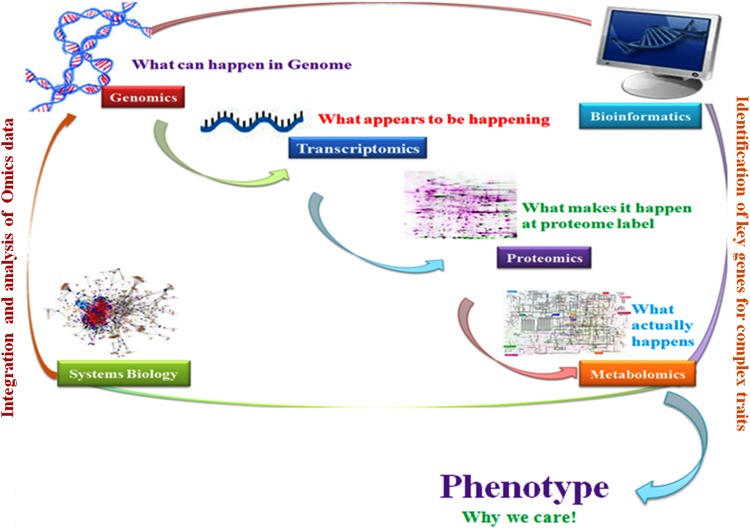
During the past few decades, there have been tremendous progresses in genetic and genomic technologies, assisting us to understand the complex biological systems in an integrative and predictive manner (Filipp 2013; Mewalal et al. 2014). Since the sequencing of the human and several plant genomes, efforts were made to elucidate the data to discover a number of agriculturally important traits/genes and their roles (Kumar et al. 2015a). Several tools for disclosing the functionality of the genes/traits have recently emerged. Initially, functional genomics tools evolved based on comparative studies in relation to model organisms. Accumulation of massive uncharacterized data demanded the requirements to develop novel approaches and technologies that may be useful in assigning functions of the sequenced genes. Advances in computational biology and software packages have opened the gates for further evolution of novel technologies to be capable of taking an applied approach by studying the components of biological systems that are required for proper functioning of the cell and acquire a holistic systems biology-based approach in agricultural research. Besides genomics, certain other tools and technologies have been developed in the last few decades which take an ‘omics’ approach, i.e., an integrated systems-based approach for the study of cellular structure, function and its dynamics. It is expected that the applied integrative omics-based approaches are helpful in understanding the cause and effect relations between genotype and phenotype (Cowie et al. 2013; Kumar et al. 2015a). These ‘omics’-based approaches include the incorporation of genomics, transcriptomics, proteomics, metabolomics and other omics science and technologies to perform the non-targeted studies of biological molecules involved in the appropriate working of the cells and their responses with respect to environmental changes (Fig. 1) (Jung and Main 2014).
Fig. 1.
The omics workflow for revealing the complexity of the crop plant systems and its key components linked to productivity via integration and analysis of big molecular data obtained through omics-based experimentation technology
Agricultural constraints and their mitigation through omics technologies
The world’s population may exceed 10 billion by the end of the century. Besides, the increased energy demand, water scarcity, climate change and environmental destruction, which have been brought about by the increasing population, would all pose a tremendous challenge to future agricultural production (Lamberth et al. 2013; Agrawal et al. 2013; Wortman and Lovell 2013; Nelson et al. 2014; Haddeland et al. 2014). The available statistical data with respect to climate change, groundwater depletion and its effect on crop productivity and malnutrition along with their mitigation through omics technologies are discussed in following sections.
Food security
Food availability is a prerequisite for food security, which is an important element to alleviate poverty. India is more or less self-sufficient in cereals; however, the productivity of pulses and oilseeds is limited. Due to modifications in consumption patterns, the demand for fruits, vegetables and dairy products has been increasing. Recently, a QTL that influenced grain yield in wheat was identified in double haploid lines which were obtained from a cross between two Chinese varieties, Hanxuan 10 and Lumai 14 (Wu et al. 2012). Though Li et al (2012) observed QTL controlling grain weight on chromosomes 1B, 2A, 2D, 4A, 4B and 7B, yet no single QTL was seen to be effective at all phases of grain development. Utilization of omics to identify QTLs and other key genes controlling grain yield and their development in different crop plants is necessary to increase crop productivity for meeting the growing demands for food via adoption of omics-based technology in future (Curtis and Halford 2014; Kumar et al. 2015a; Nadolska-Orczyk et al. 2017; Varshney et al. 2018; Wambugu et al. 2018; Xie and Wang 2018).
Nutritional security
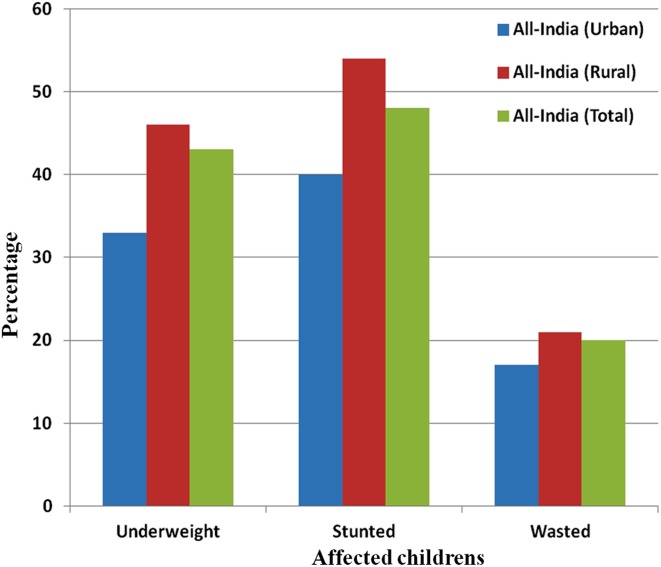
Generally, nutritional security is referred to as the measurement and availability of nutrient in food for development of society. Nutritional security is very important for the individual, home and community as well as for nation. In developing countries like India, large sections of the population are under-nourished. Due to malnutrition, 46% of the rural and 33% of the urban population with a total of 43% of children are underweight; 51% of the rural and 40% of the urban population with a total of 48% of children are stunted, whereas 21% of the rural and 17% of the urban population with a total of 20% children are wasted throughout the India (Fig. 2) (Gupta 2015). Since it is a major barrier to economic and social upliftment, there is a need of developing functional food with improved nutritional quality and quantity affordable to the masses (Nelson et al. 2014). Advanced efforts have been attempted by scientists to improve nitrogen use efficiency in plants such as Arabidopsis, rice, wheat and maize through omics approaches for improving nutritional security, because N is a key source for the growth and development of crop plants that ultimately leads to accumulation of nutrients, but excessive external use of N can result in environmental damage and high production costs (Tiwari et al. 2018). Omics-based sciences have the ability to overcome malnutrition and hunger through development of superior environmentally friendly crops for society through genomics-assisted crop breeding approaches and genome engineering (Kumar et al. 2015a).
Fig. 2.
The number of stunted, wasted and underweight children due to malnutrition, unavailability of food and insufficient amount of nutrients in both rural and urban areas of India in terms of percentage
Food safety
Crop productivity is far from satisfying the requirement of the people for food under current conditions and there is a question confronting agri-food researcher whether the emphasis should be on quantity or quality (Kumar et al. 2015a). Food safety is receiving heightened attention worldwide, as the key links between food and health are increasingly recognized. Improving food safety is a vital constituent of providing food security, which is possible when people have access to enough healthy food. In this age of globalization, as trade in crop produce expands all over the world, food safety is a common concern among both developed and developing countries (Kulasekaran 2012). Efforts have been made by researchers to reduce allergenicity in soybean via construction of a gene silencing vector to suppress the expression of P34 protein responsible for allergenic effects, in transgenic soybean lines (Herman et al. 2003). Besides, most food safety-related traits have been also discovered which can be utilized to develop soybean varieties with hypoallergenic properties (Joseph et al. 2006; Watanabe et al. 2018). Recent transcriptomics study has uncovered an indispensable and profitable knowledge into pathogenicity, survival and adjustment of pathogens in food (Joseph et al. 2006). It has been utilized to examine intracellular expression of genes in Listeria monocytogenes amid contamination and host adaptation, and those genes initiated by the transcriptional regulars PrfA, sigma B regulon and VirR (Hain et al. 2008) The omics-based innovation specifically for RNA profiling will prompt revelation of more novel tags that are differentially expressed. While pragmatic uses and utilization of metagenomics, transcriptomics and proteomics information and related apparatuses are less conspicuous, these instruments have likewise begun to yield practice food safety solution (Chawla et al. 2018; Ojiewo et al. 2018). Therefore, the problem related to food safety can be resolved by the use of omics for obtaining higher productivity.
Energy security
With the decrease in fossil fuel deposits, there is increase in energy prices, because of which progressively developed countries are vigorously researching to develop viable bioenergy sources, the generation of which would require large amounts of crop plants products (biomass). Energy demand is still on the rise now, which means that huge quantity of crop products will be used to produce more fuel (Nelson et al. 2014; Haddeland et al. 2014). Recently, novel GM loblolly pine plants with altered cell wall and lignin composition are being optimized, in a xylem particular way, by means of control of glucommann (LpGluM) genes and three genes encoding NAC (NAM, ATAF1/2 and CUC2) transcription factor and domain proteins, i.e., LpNAC1, LpNAC2, and LpNAC3 that are involved in the plant polysaccharides regulation. A definitive objective is to enhance the physical and chemical properties of wood for proficient deconstruction and sugar discharge for large-scale production of cellulosic ethanol. Besides, Populus and Eucalyptus plants are also utilized for the nproduction of bioenergy through biotechnological approaches (Al-Ahmad 2018). Therefore, collaborative approaches of genetics, advanced breeding and omics technologies, such as those carried out on Pinus and other plants for characterization of novel genes and transcription factors, are expected to open up new avenues for increasing biomass which can be harvested for generation of fuels (Al-Ahmad 2018; Jouzani and Sharafi 2018; Wambugu et al. 2018).
Climate change
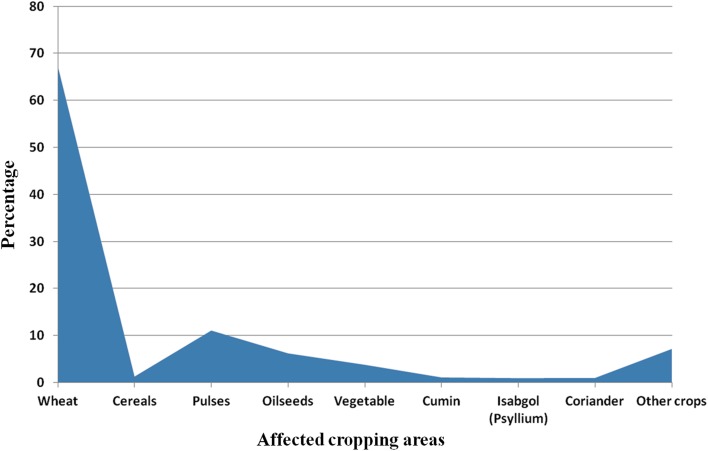
Climate is one of the most important factors influencing agricultural practices and crop productivity. Climate change exerts harmful influence on productivity of the crop plants, which decreases the per unit yield of main crops such as wheat, pulses and oilseeds. Influence of this kind is more conspicuous in low-latitude area of India. There has been increasing concern regarding the changes associated with climate becoming warmer, occurrence of new crop diseases and pests, expanded area, increased hazard and difficulty in control (Wortman and Lovell 2013; Nelson et al. 2014; Haddeland et al. 2014). In this way, agricultural production management would become extremely challenging. Wheat was a highly affected crop, followed by pulses and oilseeds. 67.22% of the cultivated area of wheat, 1.32% of cereals, 11.09% of pulses, 6.24% of oilseeds, 3.82% of vegetable, 1.13% of cumin, 1.02% of coriander, 0.98% of isabgol and 7.18% area of other crops are affected due to climate change and unusual rainfall in the country (Fig. 3) (http://businesseconomics.in/?p=2439). Transcriptomic and proteomics studies conducted in different plants have determined several putative genes. i.e.. Apx1, Mbf1c, At1g26580 associated with multiple abiotic stress regulation in plants, which could be potential targets for the development of stress tolerance. Besides, some achievement has just been accomplished in the development of crops tolerant to a specific abiotic stress condition by genetic manipulation of key genes (Zandalinas et al. 2018). Therefore, omics-based approaches have potential to produce climate-resilient crops through genetic improvement.
Fig. 3.
The affected cropping area in India due to climate change in terms of percentage. Wheat is an extremely affected crop followed by pulses and oilseeds
Shortage of water
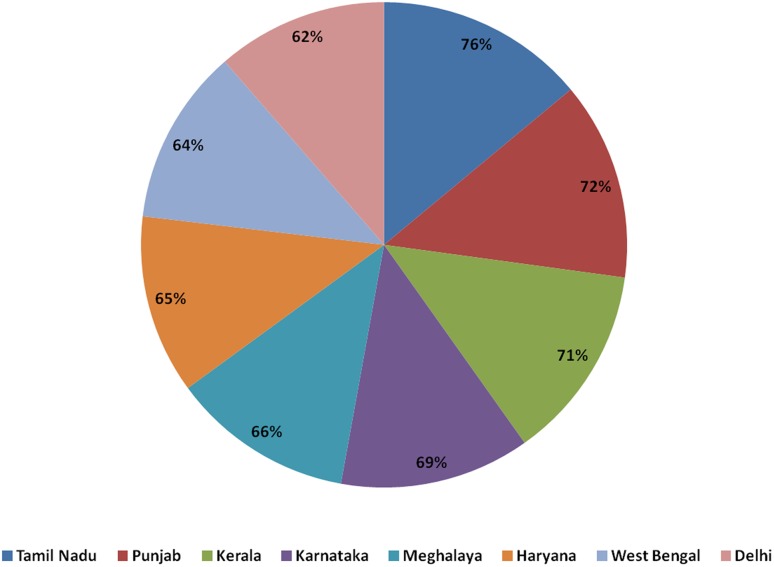
Freshwater for irrigation is a backbone of agriculture and directly linked to crop productivity. At present, demands for agriculture water accounts for 69% of the world total water consumption and surpasses more than 89% in some arid countries. As population growth and urbanization continues, the proportion of domestic water consumption has also increased. The Tamil Nadu state is dealing with the maximum groundwater depletion at 76%, whereas Delhi has its share of 62%. The top Indian states in terms of percentage of groundwater depletion of are shown in Fig. 4 (http://scroll.in/article/676038/imagine-the-qutub-minar-underground-thats-how-low-delhis-water-table-has-fallen). The alternation of physiological activities in response to drought and heat stress and their combination have been determined in several plant species. It was investigated that the photosynthetic machinery is repressed in responses to several abiotic stresses. However, the impact of drought, heat and their mix on photosynthetic activity could be distinctive depending upon plant species. Transcriptomic studies conducted on Arabidopsis thaliana in response to drought, heat and their combination clearly demonstrated the expression of many genes including different heat shock proteins (HSPs), MYB transcription factor and several protein kinases (Zandalinas et al. 2018). Besides, Apx1 gene was investigated in A. thaliana that is specifically required for the tolerance to the combination of drought and heat stress (Koussevitzky et al. 2008) Therefore, omics-based approaches have potential to decode the molecular mechanism of drought for identification of key genes which can further be exploited for developing drought-resistant crop plants through genomics-assisted plant breeding and genetic engineering to combat the problem of water shortage.
Fig. 4.
Pie chart depicting the percentage of groundwater depletion in different Indian states
Loss of soil fertility and degradation
The severity of soil degradation and loss of soil fertility have been growing in many places of the world. Land degradation and desertification, which are caused by unauthorized cultivation of marginal land, deforestation for farmland expansion with bad farming practices and soil degradation, have all degraded soil to a certain extent. The harmful effects of land degradation include decreased crop productivity, population migration, food insecurity, destruction of basic resources and ecological system, and loss of biological diversity due to habitat changes (Montgomery 2007). It was already determined that the legumes plants fix soil nitrogen and enhance soil health (Ojiewo et al. 2018). Besides, the establishment of artificial symbioses among nitrogen-fixing microbes and non-legume plants is the major breakthrough in the field of agricultural sciences. Ongoing advances in understanding endosymbiotic and endophytic N fixation with non-legume crop plants have demonstrated a unique opportunity for designing non-legume N-fixing crops. This has become possible with the availability of several genomes of N-fixing microbes, i.e., Azoarcus sp. BH72, Herbaspirillum seropedicae SmR1, Gluconacetobacter diazotrophicus Pal5, Pseudomonas stutzeri A1501, Azospirillum sp. B510 and Klebsiella pneumoniae 342 along with genomes of crop plants decoded by omics technologies (Santi et al. 2013). Therefore, artificial symbiosis in many crop plants with N-fixing microbes will ultimately increase soil fertility and decrease the risk of their degradation, population migration and food insecurity as well as increase crop productivity.
Human activities influencing ecosystems
To protect our ecosystem from extensive use of fertilizers by growers is the major challenge. To improve agricultural sustainability together with long-term income, special attention has to be given for newer strategies for crop protection such as integrated practices, to eradicate or control pests (Wortman and Lovel 2013; Haddeland et al. 2014; Diepens et al. 2014). Some newly designed crop plant varieties of rice and wheat have potential to protect themselves from several abiotic and biotic stresses and providing higher yield with less use of fertilizer (Lesser and Kolady 2011; Singh et al. 2011; Huang et al. 2017). Therefore, advances in omics are very useful in designing of smart crops for sustainable agriculture to protect the ecosystem (Kumar et al. 2015a).
Roles of omics technologies in agricultural production
In India, agriculture has always been regarded as the backbone of economy. Therefore, it is important that scientists, extension workers and farmers are aware of the current agriculture practices, technologies and research for better and sustainable development (Grover et al. 2014; Kumar et al. 2015a; Kumar 2015). Large-scale genome sequencing projects are giving biological and agricultural scientists access to an exponentially growing lists of genomes, from organisms covering all three forms of life. This has brought about a paradigm shift in the way scientists address the problem of food and nutritional security. Exciting molecular technologies to elucidate genomic data have taken the investigation of gene structure and function to an unprecedented level and more comprehensive understanding of cellular processes. Conventional biological research has changed fundamentally with the advent of the science of ‘omics’. Although a majority of the omics approaches are still in the developmental phase, tremendous effects have been shown on world agriculture in the form of “GENE REVOLUTION” (Kumar 2015). Omics technologies have substantially changed both the approach and the design of research experiments. These technologies allow the generation of large-scale data at each level of information from gene sequence, expression to protein as well as metabolite patterns determining variability in cellular networks and function at a systems level. This has already signaled a new era of approaching scientific queries, that is, the advent of ‘big biology’ and a system based approach to scientific practice with global measurements of metabolic pathways in agricultural sciences (Koyuturk 2010; Kumar et al. 2015a).
In view of the amplified throughput data availability, the process of research has fundamentally been altered in ‘omics science’. Consequently, a scientist addresses a scientific problem by postulating a hypothesis and working through the experimentation to prove or disprove the hypothesis. With the omics-based approach, inquiring an initial research question is not always compulsory or a pre-requisite (Weisenburger 1993; Sachs et al. 2005; Ozdemir et al. 2009; Koyuturk 2010; An and Lin 2011; Martinez-Gomez et al. 2012). Genomics or proteomics data can be collected in an omics-based experimentation without an existing hypothesis. The data in turn can be analyzed for deriving useful information that can lead to inventions and testing of novel hypotheses. ‘First hypothesize-then-experiment’ tradition has changed to ‘first experiment-then-hypothesize’ approach which offers the promise of discovering unprecedented patho-physiological mechanisms of stress tolerance and other complex traits in the case of crop plants (Nadella et al. 2012; Martinez-Gomez et al. 2012; Kujur et al. 2013; Henry et al. 2014; Mano et al. 2014; Rahman et al. 2014; Tohge et al. 2014).
Unraveling the genome sequences is only the beginning of the era of genome biology and genomics. This is followed by the study of the function of numerous genes, comparison of genes in one crop with another and to generate the holistic structures of proteins from several protein families. Thus, such approaches can help in deriving clues for their functional analysis (Henry et al. 2014; Rahman et al. 2014). In agriculture, the major applications of genomics is to gain a bigger understanding of plants so that agronomically important genes can be discovered and targeted to develop better, healthy and safe food. The attention is also on the protection of the environment by not adopting a sustainable agricultural practice (Martinez-Gomez et al. 2012).
Unlike the genome, the proteome is highly dynamic and changes in response to different environmental stimuli from time to time. The goal of proteomics research is to understand how the structure and functional dynamics of proteins allow them to function, work together and determine individual roles that can contribute to crop productivity (Mano et al. 2014; Tohge et al. 2014). Similarly, metabolomics can be used to identify and characterize differences among the quantity of thousands of biological molecules between a healthy and unhealthy or diseased plant to decipher plant defense metabolites. The technology can also be used to investigate the nutritional difference between traditional, commercial and GM crops. Contiguous to this interpretation and integration of the experimental data within the context of the whole cell as well as cell physiology, development of bioinformatics tools and databases to store and integrate different types of data sets is still a major task ahead (Kumar et al. 2015a; Martinez-Gomez et al. 2012).
Opportunities in omics technologies
To make modern agriculture sustainable, it is necessary that scientists adopt innovative technologies for crop improvement. Molecular approaches including genomics and genetic engineering have emerged as powerful tools to assure rapid and precise selection and implementation of the trait(s) of interest for crop improvements (Kumar et al. 2015a).
Next generation sequencing and high-throughput genotyping have helped immensely in understanding the functions and regulation of genes in crop plants. Plant genomics and bioinformatics are rapidly developing and deeply improving understanding of plant biology through development of novel tools and techniques to understand plant systems which will ultimately lead to enhanced and sustained agriculture production (Wei et al. 2013).
The ever-increasing availability of genome sequences in crop plants has facilitated the development of genetic and genomic resources that will allow us to address biological functions and a number of basic processes relevant to crop production (Tohge et al. 2014). The major key areas of omics sciences are genomics, transcriptomics, proteomics, metabolomics, bioinformatics and systems biology, and its associated techniques as well as technologies such as nutrigenomics for designing of functional foods and nutraceuticals, other omics for microalgal-based biofuel production, transgenic plant analysis, computational regulomics and network biology, and analysis of RNA splicing, with new fields, including glycomics, miRNA and metagenomics (Fig. 1) having incredible prospects for robust crop productivity and sustainability (Martinez-Gomez et al. 2012; Tohge et al. 2014; Kumar et al. 2015a).
This article provides a comprehensive overview of ‘omics’ technologies and application in the area of agriculture to improve crop productivity. The approximate experimental cost of sequencing technologies in India as per NxGenBio Life Sciences, New Delhi, for sequencing of one sample with single library is provided in Table 1. Some of the opportunities of these technologies given in Fig. 5 are summarized.
Table 1.
Intervention of sequencing technology and its approximate experimental cost in India; the mentioned prices are for sequencing of one sample with a single library
| S. No. | Technology/methods | Read length | Output per lane | Output per run | Per lane cost (INR) | Per run cost (INR) |
|---|---|---|---|---|---|---|
| Second-generation sequencing technology | ||||||
| 1. | 454 Sequencing | Up to 1 kb | 500 MB | – | – | 500,000 Outdated/discontinued |
| 2. | Illumina HiSeq1000 | 100 bp | 15 GB | – | 300,000 | 2,100,000 Outdated/discontinued |
| 3. | ABI SOLiD system | 50–100 bp | – | – | – | Outdated/discontinued |
| 4. | Polonator G.007 | 26 bases per amplicon | – | – | – | Outdated/discontinued |
| 5. | Ion Torrent Sequencing | Up to 200 bp | 1 GB | 1 GB | 100,000 | |
| Third-generation sequencing technology | ||||||
| 6. | PacBio | 800–1000 bp | – | – | – | 100,000 |
| 7. | HeliscopeTM Sequencer | 33-nt read length | – | – | – | Outdated/discontinued |
| Fourth-generation sequencing technology | ||||||
| 8. | Oxford Nanopore | 62–70 bases sequences per DNA nanoball (DNB) | – | – | – | 110,000 |
| 9. | Illumina HiSeq3000 | 150 bp | 50 GB | 400 GB | 275,000 | 2,000,000 |
| 10. | ABI SOLID 550XL system | 75 bp | 120 GB | 120 GB | – | 200,000 Outdated/discontinued |
| 11. | Ion Proton | 200 bp | 6 GB | 6 GB | – | 150,000 |
| 12. | Pac-Bio RSII | 1–30 kb | 750 MB | 750 MB | – | 125,000 |
Fig. 5.
Science of omics to increasing crop productivity: an overview
Genomics and bio-resource
Bio-resource refers to the total biological variation manifested as microbes, plants, animals from which certain product could be derived and utilized as drugs, food and feed. They could also be a source for the development of improved crops and animals for higher yield and tolerance to biotic and abiotic stresses. There is dependence on these bio-resources for the continued existence of mankind. Therefore, human beings have to bear the responsibility of using and preserving them for future generations (Sayers et al. 2009). India harbors two hot spots of biodiversity in the world. The Eastern Himalayan region and the Western Ghats are the abode of numerous plants, animals and microbial species. Since utilization of the available bio-resources is an inevitable part of human existence, it is to be ensured that a balance exists between the use of resources and their conservation for their sustainable exploitation (Sayers et al. 2009; Jung and Main 2014; Haddeland et al. 2014).
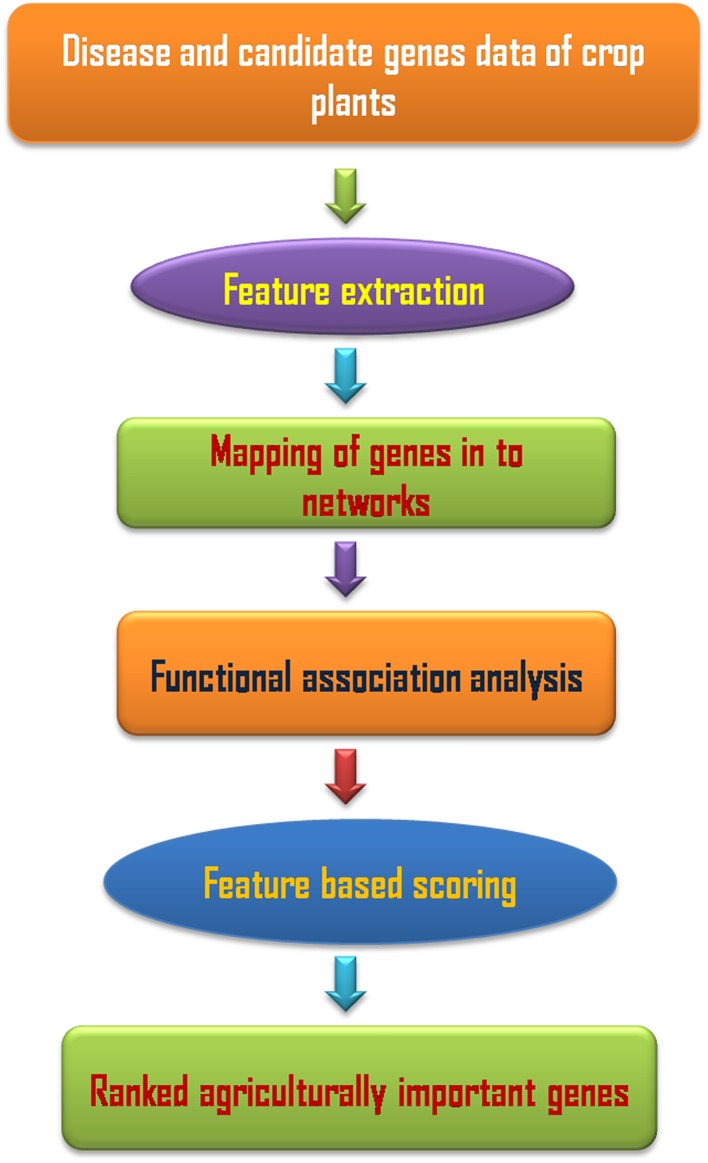
At the level of DNA, genomic data reveal the information that is stored in the genomes of organisms and passed along generations (Liu et al. 2014). These include sequences of genes coding for functional proteins, regulatory motifs that serve as markers for the regulation of the expression of specific genes, as well as individual differences in the genetic composition of populations, such as single nucleotide polymorphisms (SNPs) and copy number variants (CNVs—multiplicity or lack of certain DNA segments in genomic sequences), inversions or transpositions. Sequence data drives translational research through genome-wide association studies (GWAS), discovery of driver mutations and structural variants in progressive diseases of plants, and transfer of biological knowledge across model organisms via comparative genomics (Ning and Lo 2010; Mochida and Shinozaki 2011; Ma et al. 2013; Martinez 2013).Genome sequencing is traditionally achieved by exploiting the natural process of DNA replication. On the other hand, identification of SNPs and CNVs has been usually carried out using DNA microarrays, which exploit the natural process of hybridization. Besides, sequencing technology and associated computational techniques are now being transformed by the appearance of short read sequence data, also known as next-generation sequencing (NGS) (Martinez 2013; Kumar et al. 2015a). NGS has broadly been used for the de novo sequencing, whole genome sequencing (WGS), whole genome resequencing (WGRS) and genotyping by sequencing (GBS). It would serve as a powerful tool for decoding the genetic basis of agriculturally important traits in crop plants (Muthamilarasan et al. 2016) (Fig. 6).
Fig. 6.
Schematic diagram of the network-based disease gene prioritization at the crop improvement level
The whole sequencing of the Arabidopsis thaliana genome has been regarded as a landmark in plant biology. After that, more than 60 plant genomes have been sequenced so far and sequencing of several other genomes is underway. In India, more than seven genomes including rice, tomato, pigeonpea, wheat, Puccinia triticina, Tilletia indica and tulsi have been successfully completed with the effort of national and international group of scientists (Sasaki 2005; Consortium 2012, 2014; Singh et al. 2012, 2014a, b; Upadhyay et al. 2015a; Kiran et al. 2016; Sharma et al. 2016). The sequencing of mango genome and its analysis is ongoing (Singh et al. 2014a, b). Over the last two decades, comparative genomics has revealed that the organization of genes within crop plant genomes has remained conserved over the evolutionary periods. Advances in the field of structural and functional genomics and equitational bio-resource utilization will encompass a wider range of technologies and disciplines such as biocomputational engineering, bioenergies and genome systems analysis so as to help achieve the goal of world food and nutritional security (Vij and Tyagi 2007; Hamblin et al. 2011; Anderson et al. 2014; Kumar et al. 2015a; Thao and Tran 2016). The scientists of GBPUAT Pantnagar, India, are currently working on deciphering the complete genome sequence of an economically important fungal pathogen Karnal bunt (Tillitia indica) of wheat, for identification of genes/proteins involved in pathogenesis (Kumar et al. 2017). This study will enable to boost scientific efforts in molecular breeding and disease management (Kumar et al. 2015a).
Transcriptome
NGS has revolutionized transcriptome sequencing as compared to genome sequencing. Due to the profit of cost-effectiveness and high coverage, transcriptome sequencing of the majority of the crop plants has been performed to find out genes and gene families involved in various biological processes. In addition, RNA-sequencing (RNA-seq) reveals the relative abundance of transcripts in a given sample (tissue and/or condition specific) and also allows high-throughput development of molecular markers (Muthamilarasan et al. 2016). In the past years, transcriptomics has been executed by a comparative approach using cDNA-AFLP, suppression subtraction hybridization (SSH) and microarray analysis. Whole transcriptome sequencing in millets was first decoded in foxtail millet, in which RNA-seq was performed in root, stem, leaf and spica samples using the Illumina GA II platform. The average size of transcripts was predicted to be 2522 bp with an intron length of 442 bp, average exon length 256 bp and average exons per gene 4.3. This analysis showed that ∼ 81.7% of genes were expressed, whereas 1367 were pseudogenes (Muthamilarasan et al. 2016). For the first time, the transcriptome of developing spikes of finger millet (Eleusine coracana) was sequenced in India for understanding the molecular basis of three complex agriculturally important traits, viz., nutritional quality (micronutrient Ca2+, Fe2+, Zn2+ and protein quality), nitrogen use efficiency and stress responsiveness (Kumar et al. 2015b). In addition, several other efforts in transcriptome sequencing were made by Indian scientists in different crop plants and many more are underway to decode the complexity of important agricultural traits for improving crop productivity (Dutta et al. 2011; Thakur et al. 2013; Prasath et al. 2014; Paritosh et al. 2014; Garg et al. 2015, 2016; Shukla et al. 2015; Reddy et al. 2015; Mahato et al. 2016; Meena et al. 2016; Devi et al. 2016).
Functional genomics aims at developing and synthesizing genomic and proteomic knowledge into recognizable, sensible information, which would unravel the dynamic properties of plant systems at the molecular and cellular level (Vij and Tyagi 2007; Hamblin et al. 2011; Kumar et al. 2015a, b, 2017). This could offer a complete picture of biological functioning that is taking place in the plant cell. Further, it will also provide an opportunity to explore genomics for identification of novel genes from bio-resources having hidden value for engineering superior traits. This could also help in superior trait identification and their introgression into plants (Sayers et al. 2009; Rahman et al. 2014). In the twenty-first century, optimal utilization of available bio-resources in sync with the genomics approaches is poised to become a major platform for driving significant progress over the next 20–50 years. The knowledge and understanding of genome sequences and their relation with metabolic control mechanisms will allow a sound scientific basis for a healthier and more reliable food supply (Kumar et al. 2015b). The partial list of genomes and transcriptomes sequencing done by Indian scientists are given in Table 2 (Sasaki 2005; Dutta et al. 2011; Consortium 2012, 2014; Singh et al. 2012, 2014; Thakur et al. 2013; Prasath et al. 2014; Paritosh et al. 2014; Kumar et al. 2015b, 2017; Upadhyay et al. 2015a; Garg et al. 2015, 2016; Shukla et al. 2015; Reddy et al. 2015; Kiran et al. 2016; Sharma et al. 2016; Mahato et al. 2016; Meena et al. 2016; Devi et al. 2016; Shankar et al. 2016; Hittalmani et al. 2017; Varshney et al. 2017).
Table 2.
Partial list of sequencing of the genome(s) and transcriptome(s) by Indian scientists
| S. no. | Name of crops | Genomes/or transcriptomes | Research institutes/university | References |
|---|---|---|---|---|
| 1. | Rice | Genome | Indian Initiative for Rice Genome Sequencing (IIRGS), University of Delhi and Indian Agriculture Research Institute | Project IRGS (2005) |
| 2. | Tomato | Genome | Indian Initiative on Tomato Genome Sequencing, University of Delhi, National Research Centre on Plant Biotechnology and National Plant Genome Research Institute | Tomato Genome Consortium (2012) |
| 3. | Pigeonpea | Genome | National Research Centre on Plant Biotechnology | Singh et al. (2012) |
| 4. | Wheat | Genome | Punjab Agricultural University, National Research Centre on Plant Biotechnology and University of Delhi | International Wheat Genome Sequencing Consortium (2014) |
| 5. | Mango | Genome | National Research Centre on Plant Biotechnology | Singh et al. (2014) |
| 6. |
Puccinia triticina
Wheat rust pathogen |
Genome | National Research Centre on Plant Biotechnology | Kiran et al. (2016) |
| 7. | Tilletia indica; Karnal bunt Wheat pathogen | Genome | G. B. Pant University of Agriculture & Technology; Indian Institute of Wheat and Barley Research and Indian Agricultural Statistics Research Institute | Sharma et al. (2016), Kumar et al. (2017) |
| 8. | Tulsi | Genome | National Centre for Biological Sciences (TIFR); Manipal University; Centre for Cellular and Molecular Platforms and SASTRA University | Upadhyay et al. (2015a, b) |
| 9. | Finger millet | Genome | University of Agricultural Sciences, Bengaluru | Hittalmani et al. (2017) |
| 10. | Pearl millet | Genome | International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Hyderabad with International collaboration of the several other institutions | Varshney et al. (2017) |
| 11. | Pigeonpea | Transcriptome | National Research Centre on Plant Biotechnology | Dutta et al. (2011) |
| 12. |
Venturia inaequalis
Apple scab pathogen |
Transcriptome | Institute of Himalayan Bioresource Technology | Thakur et al. (2013) |
| 13. | Ginger | Transcriptome | Indian Institute of Spice Research | Prasath et al. (2014) |
| 14. | Mustard | Transcriptome | University of Delhi | Paritosh et al. (2014) |
| 15. | Finger millet | Developing spikes transcriptome | G. B. Pant University of Agriculture & Technology | Kumar et al. (2015b) |
| 16. | Kalmegh | Transcriptome | Central Institute of Medicinal and Aromatic Plants, National Botanical Research Institute | Garg et al. (2015) |
| 17. | Bitter gourd | Transcriptome | Indian Institute of Vegetable Research, Banaras Hindu University and Genotypic Technology (P) Ltd | Shukla et al. (2015) |
| 18. | Senna | Transcriptome | Directorate of Medicinal and Aromatic Plants Research | Reddy et al. (2015) |
| 19. | Chickpea | Transcriptome | National Institute of Plant Genome Research, International Crops Research Institute for the Semi-Arid Tropics , RMIT University, Jawaharlal Nehru University | Garg et al. (2016) |
| 20. | Mango | Leaf transcriptome | National Research Centre on Plant Biotechnology | Mahato et al. (2016) |
| 21. | Lemongrass | Transcriptome | Central Institute of Medicinal and Aromatic Plants | Meena et al. (2016) |
| 22. | Cymbopogon winterianus | Transcriptome | Assam Agricultural University | Devi et al. (2016) |
| 23. | Rice | Transcriptome | National Institute of Plant Genome Research and Jawaharlal Nehru University | Shankar et al. (2016) |
Big data in genomics: challenges and solutions
These revolutionary modifications in big data generation and gaining are profound challenges for storage, transfer and security (Sayers et al. 2009). Indeed, it may now be less costly to generate the data than to store it. One model of this issue is the National Centre for Biotechnology Information (NCBI). The NCBI has been leading big data efforts in plants and other organisms since 1988, but neither the NCBI nor anyone in the private sector has a complete, low-cost, and secure solution to the problem related to data storage and security (Sayers et al. 2009; Lal et al. 2013; Gour et al. 2013). Since these potentials are beyond the reach of small laboratories and small institutions, therefore it imposes various challenges for future agricultural research. Another challenge is to transfer data from one site to another; it is mainly done by shipping external hard disks through mail (Marx 2013).
Proteomics and biofortification
One of the greatest problems for low income developing country including India is undernourishment and malnutrition among the children and women. According to the estimates of World Bank, India holds second rank in the world, after Bangladesh, in terms of the number of children suffering from malnutrition (Kulasekaran 2012; Gupta 2015). In India, 47% of the children exhibit a degree of malnutrition. 50% of poor women of reproductive age are anemic in India. Protein energy malnutrition and micronutrient deficiencies of vitamin A, iron and zinc contribute to high mortality rate in children and women due to onset of various illnesses and diseases. Biofortification is seen as an effective strategy for coping with the problems of protein energy malnutrition and micronutrient deficiencies, prevalent among children and women in the developing world including India (Kulasekaran 2012; Lee et al. 2012; Kumar et al. 2015a).
The objective of biofortification is to enrich plant foods in essential micronutrients and proteins as plants grow naturally (Lee et al. 2012; Kumar et al. 2015b). It has been realized that biofortification of staple food crops such as rice and wheat would solve the malnutrition problem associated with the rural poor, as these staple foods predominate the diet of poor people (Blancquaert et al. 2014). In this regard, genetic engineering methods hold a great promise for development of biofortified foods. The genetic engineering-based strategies include insertion of novel genes from heterologous system, overexpression of already existing genes, disruption of pathways involved in synthesis of inhibitors of trace elements absorption and synthesis of the enhancers of trace element absorption to increase the nutrient contents in staple food crops. Its application has already been seen in the form of golden rice developed to solve the problem of vitamin A deficiency (Lee et al. 2012; Blancquaert et al. 2014; Kumar et al. 2015b). However, implementation of such methods requires identification of nutritionally important genes and promoters which are associated with accumulation of high quality of proteins and micronutrients in seeds of staple crops (Lee et al. 2012; Kadiyala et al. 2012; Ramachandran 2013; Blancquaert et al. 2014).
Proteomics technologies aim at studying the global patterns of protein content, protein activities, modifications and localization, and interactions of proteins with other proteins or molecules of the cell (Deracinois et al. 2013). The proteomics techniques such as 2D gel electrophoresis and MALDI-TOF have potential for the detection and identification of proteins expressed in a particular tissue, organ or organelle which in turn facilitates identification of relevant genes through reverse genetics approach. Such approaches, therefore, can be utilized to identify key candidate genes involved in uptake, transport, accumulation and availability of a potentially useful protein or micronutrient in the seeds (Deracinois et al. 2013; Vanderschuren et al. 2013; Kumar et al. 2015a). These studies also facilitate the understanding of the molecular mechanism of remobilization of nutrient from leaves to grain seeds and its subsequent compartmentalization within the seed. The unraveling of such molecular mechanism involved in synthesis and accumulation of seed storage proteins and other nutritionally important nutrients would help in the identification of key genes/transcription factors/promoters which could subsequently be transferred into staple food crops for biofortification (Lee et al. 2012; Kadiyala et al. 2012; Ramachandran 2013; Deracinois et al. 2013; Vanderschuren et al. 2013; Blancquaert et al. 2014). Some examples of such biofortification strategies may include the manipulation of enzyme levels in biosynthetic pathways to increase the levels of accumulated vitamins, higher expression of the molecular “pumps” that allow plants to take up more minerals from the soil or deposit a specific nutrient in seed grain and elevation of protein content by integration of genes from protein-rich crops into staple food crops (Goel et al. 2012; Blancquaert et al. 2014). A recent example of the latter is the development of genetically engineered potato expressing the gene of an Amaranthus protein so as to enhance the protein content of potato (Chakraborty et al. 2010). Another approach is to transfer the genes for good-quality proteins rich in essential amino acids into staple crops. Ethnic cereal crops, which unfortunately are neglected crops, are bestowed with good-quality proteins such as prolamins and mineral sequestering proteins (calcium-binding protein) and micronutrients such as calcium, iron and zinc (Kumar et al. 2015b; Chinchole et al. 2017). In these crops, the seed storage protein genes are regulated by the concerted or combinatorial action of seed-specific transcription factors which bind to their respective DNA binding sites on the promoter region. Advances in proteomics can be utilized to investigate the role of these components in the accumulation of seed storage proteins in seeds by performing proteomics-based co-expression analysis. Besides, isolation, identification and expression analysis of plant-specific Dof transcription factors genes were done to understand their role in the plant developmental process (Kadiyala et al. 2012; Ramachandran 2013; Deracinois et al. 2013; Vanderschuren et al. 2013; Gupta et al. 2014a, b, 2018; Gujjar et al. 2018). The transporter proteins involved in the accumulation of micronutrient in the seeds can be identified and their genes can subsequently be overexpressed in staple crops so as to increase their bioavailability in the seeds. It will certainly help in opening new vistas for biofortification research programs (Lee et al. 2012; Blancquaert et al. 2014; Kokane et al. 2018).
Proteome
Although the transcriptomics-based analysis of gene expression gives an approximate idea about the relative abundance of the related protein in a sample, it cannot necessarily be related to proteome and its function. The latter is because the expression of gene, after transcription, is regulated through different mechanisms including mRNA degradation, alternative splicing and post-translational modification (Ma et al. 2013; Filipp 2013; Kumar et al. 2015a, b). Proteomic screening identifies molecular abundance and activity at the functional stage. A universal method, 2-D polyacrylamide gel electrophoresis, facilitates the partition or separation of proteins in a sample of interest based on their isoelectric point and other electrochemical properties. Separated proteins can then be determined using mass spectrometry (MS) (Tsugita and Kamo 1999; Weiss and Gorg 2007). Although proteomic screening methods are helpful in quantifying the expression and variation or modification of proteins at the functional stage, setup of proteomic screening techniques can only control and manage the expression of a partial or limited subset of proteins in the particular cell at a time. Furthermore, advanced techniques such as flow cytometry facilitate the activity of screening of the protein at the resolution of thousands of cells; but this comes at the price of a limited coverage of the proteome (Galbraith 2014). Proteomic data enable detection of functional proteins and their modifications that have a role in the development and progression of disease and development of causal models for cellular signaling, driving translational science at the functional level in crop plants (Quirino et al. 2010; Lodha et al. 2013; Gong et al. 2015).
Interactome
High-throughput screening techniques have made it possible to identify the physical interactions among proteins. A commonly used technique, yeast two-hybrid (Y2H) identifies interactions involving a couple of proteins by using the modularity of the activating and binding domains of eukaryotic transcription factors. In yeast two-hybrid, the activating and binding domains of a particular transcription factor are separated, and each domain is merged to one of the two (prey and bait) proteins (Damon et al. 2012). Then, the interaction between the two proteins is identified by reporter gene expression that is the target of the transcription factor (Cao and Yan 2013). On the other hand, tandem affinity purification (TAP) identifies interactions between an individual bait protein and several other proteins (Van Leene et al. 2015). This is achieved by tagging the target protein of interest and inserting it into the host. When the bait protein is retrieved with other proteins associated with it, these interacting proteins are investigated using MS (Weiss and Gorg 2007).
The identified protein–protein interactions (PPIs) using wet laboratory experimentation are known as protein–protein interaction networks, which provide a high-level as well as static understanding of cellular organization, commonly called as the interactome (Boruc et al. 2010; Zhang et al. 2010). Recently, established protein–protein interaction network models imagine binary interactions between pairs of proteins, which describe the outcome of Y2H experiments (Cao and Yan 2013). On the other hand, multiple protein–protein interactions identified by TAP are shown by either a star network at around the bait protein (spoke model) or a cluster of all proteins obtained by the bait protein, within itself (matrix model) (Zhang et al. 2010; Cao and Yan 2013). A key limitation of high-throughput protein–protein interactions, however, is their unfinished and noisy nature. In addition, these interactions just signify a picture of the complex and dynamical organization of the proteins in the cell (Zhang et al. 2010; Pathak et al. 2013a; Cao and Yan 2013; Kumar et al. 2015a). In recent years, PPI networks have been extremely useful in understanding the systems biology of complex diseases, through identification of protein complexes, functional modules and signaling pathways network-based functional annotation, network-based disease gene prioritization, and identification of dysregulated pathways in plant systems (Pathak et al. 2013a, b; Kumar et al. 2015a).
Bioinformatics and systems biology
The life sciences are now in the midst of a truthful biological revolution similar to what biophysics experienced after the turn of the last century (Mount 2001). Nowadays, researchers are making constant effort to gain insight into biological systems through their molecular function. Subsequent coordinated activities are just in a stage of unparalleled growth and development that is reflected by the quantity of data generated from every experiment (Kumar et al. 2015a). This huge information from seemingly disparate data sets is converted into useful knowledge through bioinformatics (Mount 2001; Wang et al. 2005). Bioinformatics is the application of information technology to facilitate gathering, linking and manipulating diverse types of biological data to understand new biological insights.
In other words, bioinformatics is an information management and manipulation system for molecular biology, biochemistry, health sector, environmental biology and agriculture by addressing biological data collection, data mining, data analyses, interpretation and finding of genes and protein, modeling and product design especially drug/agrochemicals discovery (Jayaram and Priyanka 2010; Verma et al. 2017). It is used to predict the structure as well as function of newly investigated proteins and protein sequences for making a cluster of associated sequences into families and constructing phylogenetic trees for evolutionary relationships analysis (Mount 2001; Wang et al. 2005; Jayaram and Priyanka 2010).
In an agriculture-dependent country like India, bioinformatics has a very important role to play where it can be used for improving the nutritional content, increasing the yield of the agricultural produce and implanting resistance to several biotic and abiotic stresses (Jayaram and Priyanka 2010). Recently, the defense inducer(s) and antifungal molecule(s) against Alternaria blight disease of oilseed Brassica were investigated through bioinformatics approaches (Upadhyay et al. 2015a, b, 2016; Pathak et al. 2016, 2017a, b). In India, the national level high performance computing for agricultural research “Advanced Supercomputing Hub for Omics Knowledge in Agriculture (ASHOKA)” has been established at the Indian Agricultural Statistics Research Institute as a program “Centre for Agricultural Bioinformatics” (Rai 2014) for providing computational support to ICAR as well as other scientific and educational institutions in India, working in the area of computational biology and bioinformatics with respect to agriculture and crop productivity (http://cabgrid.res.in/cabin/). Department of Information Technology, Government of India, is starting an “Agri Bio-informatics Promotion Research Program” for crop improvement, disease detection and pest control (http://meity.gov.in/content/bio-informatics). The BTISNet program of DBT for bioinformatics research and education has also been adopted by various institutions across the country (http://btisnet.gov.in/).
Plants and animal genome sequencing projects hold enormous contribution for agricultural scientific community. Bioinformatics plays a vital role in the integration as well as analysis of genomics, transcriptomics and other high-throughput sequencing data, which hold great potential to redesign crops for improved productivity (Consortium 2012, 2014; Singh et al. 2012, 2014a, b; Kiran et al. 2016).
There has been a paradigm shift from single gene approach (i.e., gene by gene approach) in understanding the mechanism of biological phenomenon to working with several genes at a time (Kumar et al. 2015a). This shift has resulted from the observation that any biological response results from cross talk of many biomolecules which act in an inter-dependent manner. As a result, many high-throughput technologies have been evolved which provide a glimpse of all the molecules involved in the process (Kumar et al. 2015a; Gupta et al. 2015). The process has been accelerated by research in the field of genomics. However, there exists a wide gap between genotype and phenotype in manifestation of a trait (Kumar et al. 2015a). To fill this gap, research is conducted at different levels—whole plant, cellular, biochemical, gene and protein levels. All these areas of unparalleled scientific efforts have led to the accumulation of large quantities of biological information. Bioinformatics which is culmination of biology and computational technology attempts to use these data to design novel strategies to get a holistic view of biological system and to design novel strategies to address the problems of agricultural, medicinal and industrial importance (Gupta et al. 2015; Kumar et al. 2016). Systems biology aims to develop and use well-organized and efficient algorithms, data structure, visualization and communication tools for the integration of these biological data with the goal of computational modeling and simulation. It studies crop plant systems by systematically perturbing them, checking the gene, protein and informational pathway responses; integrating these data; and finally, formulating mathematical models that described the structure of system and its response to individual perturbations. Consequently, systems biology approaches such as integrative and predictive ones hold immense potential in understanding the molecular mechanism of agriculturally important complex traits linked to crop productivity (Kitano 2002; Kumar et al. 2015a; Pathak et al. 2017b).
Bioinformatics and systems biology tools including simulation, docking and protein–protein interaction can be employed to search the important genes for specific function or manipulate the sequence for better fit and study the function of these genes or protein at the system level (Mount 2001; Kitano 2002; Wang et al. 2005; Jayaram and Priyanka 2010; Rai 2014; Avashthi et al. 2014; Gupta et al. 2015; Pathak et al. 2016, 2017a). This specified genetic and genomic knowledge could then be utilized to produce resistant, nutritionally improved and productive crops (Kumar et al. 2015a). Several bioinformatics tools and databases, viz., RetroPred (Naik et al. 2008), pTAREF (Jha and Shankar 2011), QlicRice (Smita et al. 2011), MirtronPred (Joshi et al. 2012), STIFDB2 (Naika et al. 2013) and RiceSRTFDB (Priya and Jain 2013). Rice blast infection prediction (Kaundal et al. 2006), MCDRP (Gaur et al. 2014), Bhageerath (Jayaram et al. 2006), etc., have been developed by Indian scientists and many more are underway for conducting research in area of agricultural sciences with respect to plant molecular biology and biotechnology.
Expression data analysis for diagnosis and prediction
Systems-based approaches have proven of big utility for the management of plant diseases, with growing power expected to continue to appear in the future (Dodds and Rathjen 2010; Kumar et al. 2015a). Although notable and major challenges remain, one of the areas that has revealed significant promise is the mining of data sets related to gene expression for investigation of molecular signatures that can be utilized for disease management. These types of studies normally involve the collection of data samples from two or more sources with respect to infected and normal plant, by using these data sets to train a molecular classifier and another set on which to test (Furey et al. 2000). In the absence of the true test set data, resampling techniques or methods such as cross validation are commonly used to determine the probable performance of the classifier on analysis of future data (Furey et al. 2000; Yue et al. 2003). Challenges often occur in these studies when several measurement platforms are utilized in training and test data sets (Wing 2006). The capability of creating a precise classifier is a function of factors, e.g., (i) the mass of the training set relative to the number of characteristic features, (ii) the computational method used, and (iii) the natural distinctness of the chosen phenotypes. Usually, the number of samples is far fewer than the number of transcripts, leading to overfitting being a major problem. This leads to the requirement for bioinformatics methods that aid in evading overfitting after selecting a classifier (Mount 2001; Wing 2006).
Bio-prospecting and metabolomics
Bioprospecting is the science of investigation, screening and extraction of biological diversity for genetic, genomic and biochemical resources. The indigenous knowledge is also applied to develop a commercially valuable product (Purkayastha 2016). In other words, it can be said that it is a scientific research that looks for a useful application, process or product in nature. A common perception is that bioprospecting is a new science linked to modern biotechnology (Purkayastha 2016; Talukdar et al. 2012; Zotchev et al. 2012). However, the fact is that humankind has been studying, manipulating and exploiting natural diversity ever since the emergence of Homo sapiens over 150,000 years ago. Early ancestors explored biodiversity and learned how to derive benefits from nature. Early bioprospecting led to the improvement of methods for growing food, building shelters and maintaining health. In the genomics era, scientists continue to find useful applications for compounds from nature. Bioprospecting is simply an extension of a long history of exploring nature to improve the quality of life. Bioprospecting should bolster economic and conservation goals underpinning bio-medical, agricultural and veterinary advances required to combat disease and sustain growing populations (Wing 2006; Beattie et al. 2011; Talukdar et al. 2012; Zotchev et al. 2012; Saslis-Lagoudakis et al. 2012; Silva et al. 2012; Sprocati et al. 2014; Purkayastha 2016).
The term metabolomics is the newly emerging field of the omics research and an integral part of bioprospecting (Joshi et al. 2002). It is a broad and simultaneous methodical investigation of metabolite levels in the metabolome and their changes over time as a consequence of stimuli. Metabolome is the qualitative and quantitative assessment of all low molecular weight compounds present in cells and is required for maintenance, growth and cellular normal function. Metabolomics facilitate the identification and quantification of economical valuable products from natural resources (Joshi et al. 2002; Seifert et al. 2013). Several techniques have been applied for metabolome analysis according to the type of sample to be analyzed such as NMR, HPLC, GC-MS and CE-MS (Seifert et al. 2013; Gong et al. 2013; Wen et al. 2014; Caspi et al. 2013). Bioprospecting has frequently been cited as a sustainable use of biodiversity. But there is a growing concern that many pharmaceutical firms and biotechnology companies explore natural resources to develop profitable and patented product without giving any recognition and money to the people who maintained and improved the traditional plant varieties for medicines. It is termed as biopiracy and there are no effective guidelines and conditions to stop it. To overcome the problem of biopiracy, it is necessary that the terms and conditions applying for exploring biodiversity to investigate useful and novel information should be equally shared with pharmaceutical companies and local people (Joshi et al. 2002; Silva et al. 2012; Seifert et al. 2013; Gong et al. 2013; Wen et al. 2014; Caspi et al. 2013). Besides, it will provide an opportunity to learn how to convert bio-resource into wealth and also rejuvenate old principles of Ayurvedic biology into newer scientific perspectives for exploring optimum use of huge bio-resources for biomedical and agricultural applications (An and Lin 2011). The scientists of GBPUAT, Pantnagar, are currently working on genetic enhancement of aromatic rice landraces for blast resistance and quality traits through molecular breeding to develop high-yielding ideal plant type, blast-resistant and good-quality aromatic rice diversity for wider adaptability by farmers.
Metabolism, i.e., chains of chemical reactions that change various forms of matter and energy into one another, is one of the fundamental processes in living systems (Kumar et al. 2015a). The organization of metabolic reactions is generally abstracted using metabolic network models, which represent the complex web of relationships between metabolites (compounds consumed and/or produced by reactions) and enzymes (gene products that catalyze reactions) (Filipp 2013; Babu et al. 2012). Today, several well-characterized metabolic pathways for diverse species, including humans and other model organisms, are available in public databases (Caspi et al. 2013). However, large-scale analyses of the kinetics of metabolic networks are bound by data availability and computational complexity. Nevertheless, flux balance analyses that rely on steady-state assumption provide significant insights into the dynamics of metabolism (Filipp 2013; Kumar et al. 2015a). These analyses are enabled by monitoring of the abundance of metabolites via NMR and MS, as well as monitoring of the abundance and functional activity of enzymes through transcriptomic and proteomic screening for decoding of complex agricultural traits (Joshi et al. 2002; Filipp 2013; Babu et al. 2012; Gong et al. 2013; Agrawal et al. 2013; Seifert et al. 2013; Wen et al. 2014; Caspi et al. 2013; Schweiger et al. 2014; Bueno 2015).
Advances in metabolic engineering have enabled to overcome the challenges of heterologous production of the plant-based natural products in microorganisms. Omics-based approaches are being used to identify all the genes responsible for biosynthesis of the desired product, followed by their expression in microbes to synthesize the desired product (Pickens et al. 2011). In E. coli and yeast, several metabolites such as terpenoids, paclitaxel (Huang et al. 2001; Engels et al. 2008), artemisinin (Martin et al. 2003; Ro et al. 2006) and alkaloid intermediate reticuline have been successfully produced by following the above approach (Minami et al. 2008; Hawkins and Smolke 2008). Besides, metabolic pathway was also engineered to successfully produce flavonoids and stilbenes: a plant aromatic polyketide in E. coli (Horinouchi 2009). Moreover, some progress has been made to disclose the expression problem of the plant-derived cytochrome P450 (Gillam 2007). Recent progress in omics science and technology is therefore being used to produce the desired product with high yield through metabolic engineering approaches (Pickens et al. 2011).
Plant stress and molecular cell biology
The world today faces societal challenges in the areas of agri-food nutrition, energy, environment and health sector. Although, the “green revolution” had an immense impact on agriculture in India since the 1960s, its profits were limited (Kesavan and Swaminathan 2008). For the farmers in stress-prone agriculture, the “green revolution” had made only a moderate impact. A major challenge in modern agriculture is to cope with biotic and abiotic stresses in an economical and environmentally sustainable way. Additionally, the global population is likely to cross 7 billion by 2025 and 10 billion by 2050, which will put extra burden on the already saturated crop yields (Singh et al. 2013a; Kumar et al. 2015a).
The role of mitogen activated protein kinases (MAPKs) cascades in plants has been well documented during various biotic and abiotic stresses (Pathak et al. 2013a, b). A recent study suggested the role of a least studied C group of rice MAPK during biotic stress response. The study revealed the role of OsMPK7 in imparting disease resistance against Xanthomonas oryzae the major pathogen of disease (Jalmi and Sinha 2016). Comparative microarray studies have indicated that the genes, i.e., serine carboxypeptidase S28, SCPL20, RD20, LCAT3 and NHX2 are up-regulated in response to B. cinerea and abiotic stresses such as heat, salinity and osmotic stress in Arabidopsis thaliana (Gujjar et al. 2014; Sham et al. 2015). Previous studies have also demonstrated that various transcription factor genes, i.e., WRKY17, WRKY22, WRKY44, WRKY51, WRKY57, WRKY58, WRKY65, WRKY70, WRKY72, WRKY98, WRKY104, CBF/DREB1 and NAC are differentially expressed in response to many abiotic stresses (Kayum et al. 2016; Gujjar et al. 2018; Karkute et al. 2018).
Tackling recalcitrant diseases like Karnal bunt caused by Tilletia indica in wheat and Alternaria blight incited by Alternaria brassicae in Brassica is a major problem and several approaches have been followed by Indian scientists to address the complexity of these agriculturally and economically important diseases. These approaches include molecular and immune diagnostics for plant disease surveillance (Singh et al. 2013a, b; Pathak et al. 2016), molecular pathotyping for pathogen indexing program and molecular signaling for characterization of resistance (Kumar et al. 2015a).
Environmental stresses, including biotic as well as abiotic stresses, present some of the most enduring factors which limit crop productivity (Weckwerth 2011; Filipp 2013; Mewalal et al. 2014; Kumar et al. 2015a). Apart from various biotic stresses caused by a diversity of plant pathogens and several abiotic stresses such as extreme temperatures, drought, salinity and radiation have detrimental effects on plant growth and productivity, especially when several of them occur simultaneously. In the world, only 9% of the area is conducive for crop production, while 91% is under one or the other kind of environmental stress. Ironically, the area under stress is likely to increase further due to land degradation and urbanization (Nelson et al. 2014; Haddeland et al. 2014; Gupta 2015). There is also impending concern about the impacts of climate change and its variability on agricultural production. Research proves that while plants would react positively to elevated CO2 in the absence of environment change, the impacts of high temperatures, changed patterns of precipitation and likely increased frequency of disastrous events, such as flood and droughts, will possibly combine to decrease yields and increase production risks worldwide (Soon et al. 1999).
In a few years, the effect of ‘global warming’ on the production of crop will become an extremely ‘contentious’ problem (Sultan 2012). Resolving this issue at the plant systems level is approximate exclusively to a question of coping with stress on plant. It is to be noted that global environmental concerns are born out of the recognition that ecological processes do not always respect national boundaries and that environmental problems often have impacts beyond borders; sometimes globally (Sultan 2012; Nelson et al. 2014; Haddeland et al. 2014). Viruses, bacteria, weeds, fungi, insects and other pests and pathogens are a key constraint to crop productivity from fields to markets, in the developing world (Thakur et al. 2013; Kumar et al. 2015a; Sharma et al. 2016). With little resources to fight or stop infection and infestation, humans farming small tracts of land are most susceptible to these stresses and can have devastating crop losses. Presently, most crop plant protection strategies and management involve genetic improvement to develop resistance against pests and pathogens as well as develop agrochemicals with desired affinity, efficacy and potency (Lamberth et al. 2013; Rai 2014; Gupta et al. 2015; Kumar et al. 2015a; Pathak et al. 2016, 2017a, b; Mamgain et al. 2018).
International agricultural and environmental research institutions are now re-discovering plant stress as a key constituent of global warming effect on local as well as global food production. Advances in plant molecular cell biology have offered new information and technologies required to address these issues and challenges (Filipp 2013; Kumar et al. 2015a). A promising source of excitement are the powerful tools of modern plant molecular cell biology in functional genomics and gene expression profiling in stress response that seems to provide a ray of new hope (Toenniessen et al. 2003). These include the use of microarrays and genome mapping to detect and precisely manipulate stress-response genes in breeding programs, and in the growing ability to engineer new or ‘foreign’ genes into plant genomes (Datta 2013). Great advances have been made in recent years in understanding the molecular basis of plant response and plant tolerance to various biotic and abiotic stresses (Pathak et al. 2013b). Despite this, it will be prudent to admit that a huge gap persists between the findings at the molecular level and the application of this knowledge at the whole plant level for improved yield on-farm. Nevertheless, plant genomics remains a major component of food security, at the international level, and prosperity for the foreseeable future (Filipp 2013; Kumar et al. 2015a).
Network-based disease gene prioritization
Characterization of disease-associated difference in plant genome is an important step toward enhancing understanding of the cellular mechanisms that drive complex diseases, with profound applications in modeling, diagnosis and management intervention (Kumar et al. 2015a; Pathak et al. 2016). Genome-wide linkage and association studies in healthy and affected plants provide chromosomal regions containing hundreds of polymorphisms that are potentially associated with certain diseases (Gupta et al. 2014a, b; Li et al. 2016). These polymorphisms often span up to the 300 genes, only a few of which probably have a role in the manifestation of disease (Koyuturk et al. 2011; Sheikh et al. 2013). Investigation of that many candidates via sequencing is clearly an expensive task, thus not always a feasible option. Consequently, computational methods are primarily used to integrate omics data sets to prioritize and identify the most likely disease-associated gene and proteins (Kumar et al. 2015a). Protein–protein interactions provide an invaluable resource in this regard, since they provide functional information in a network context and can be obtained at a large scale via high-throughput screening (Sheikh et al. 2013). Network-based analyses of diverse phenotypes demonstrate that products of genes that are implicated in similar diseases are clustered together into “hot spots” in PPI networks (Ma’ayan 2011; Kanaya et al. 2014).
Diversity and molecular breeding
Diversity is one of today’s buzzwords and concept of diversity can be examined as a positive aspect of globalized society. Diversity is in several ways reflective of current world order, but there are methods of taking this further without compulsorily endangering its alternatives (Arif et al. 2010). There is a lot of scope to discover the series of what diversity means and investigate modes of diversity in real-life conditions. Genetic and genomic diversity offers opportunities to utilize various genomic resources and technologies such as molecular marker technology and molecular breeding in an effort to breed new varieties of desirable traits. Improved varieties, landraces and wild species can be explored for their desirable traits and this knoweldge can be further utilized to develop new varieties through marker-assisted selection (Arif et al. 2010; Singh et al. 2011; Govindaraj et al. 2015).
Molecular markers are DNA-based markers which are either coding or noncoding sequence in nature and that assist in crop breeding programs (Singh and Singh 2015). The restriction fragment length polymorphism (RFLP) was the first DNA-based marker to be used successfully in crop improvement, followed by others such as AFLP, SSR, DarT and SNPs, for significant use in plant breeding programs, and molecular markers essentially need to be present in abundance in the genome and be polymorphic in nature, among the various other salient features required. Continuous progress in development of such new and specific types of markers play crucial role in identifying the genomic variability and diversity present within the same and across the species of the crop plants (Gupta et al. 2014c; Kumar et al. 2016). These markers help in effective utilization of germplasm and improve the genetic gains more precisely and efficiently by using marker-assisted selection (Arif et al. 2010; Singh et al. 2011; Fahad et al. 2014; Broughton et al. 2014; Govindaraj et al. 2015; Singh and Singh 2015).
In a study, 113 finger millet genotypes were collected from different parts of Uttarakhand and analyzed by genotype by sequencing (GBS) method using the Illumina platform. The GBS data were used for the analysis, identification, association mapping and functional validation of important agricultural traits of finger millet through experimental and computational approaches (Kumar et al. 2016).
Plant transformation and developmental biology
In the last 25 years, various key components associated with the development of plants and their physiological and biochemical mechanisms were identified (Schmid et al. 2005). Arabidopsis thaliana was considered as model organisms for developmental biology that facilitated researchers in the improvement of other economic and agriculturally important crops plants through omics sciences and technology (Schmid et al. 2005; Van Norman and Benfey 2009). The practice of plant tissue culture has changed the approach of some nurserymen for performing plant propagation. Plant tissue culture consists of a set of in vitro techniques, methodology and strategies that are a branch of plant biology and biotechnology (De Filippis 2014). Advances in omics dramatically changed the outcome of tissue culture through genetic transformation technology by using key genes investigated via omics-based experimentation for development of the desired product. It has been playing a significant role in basic and applied research, including crop improvement (De Filippis 2014). In modern agriculture systems, only about 150 species of plants are extensively cultivated. a large number of these have reached the limit of their improvement by traditional methods (Johnson 2008). The utilization of tissue culture technology, as a key tool or attached to other methods, including rDNA technology, is at the vanguard in the modification and improvement of crop plants for agriculture and related discipline such as horticulture and forestry (Schmid et al. 2005; Johnson 2008; Van Norman and Benfey 2009; Singh et al. 2011; Naik and Chand 2011; Rathore et al. 2011; Wang et al. 2012; Brookes and Barfoot 2013; Fahad et al. 2014; Broughton et al. 2014; De Filippis 2014; Valkov et al. 2014; Okay et al. 2014; Pant 2014; Motte et al. 2014; Singh and Singh 2015; Kumar et al. 2016). Tissue culture techniques, in association with molecular biology techniques, have been successfully applied to incorporate specific traits by gene transfer technology. Commercially glyphosate-tolerant soybeans, lepidoptera-tolerant corn, glyphosate-tolerant cotton, Bt cotton and Bt maize are commercially released varieties. Since their availabilities, these technologies have been adopted at an effective rate (Naik and Chand 2011; Wang et al. 2012; Brookes and Barfoot 2013; Okay et al. 2014). The major reason for the quick acceptance is that the new technologies provide an excellent deal for farmers. Recent estimates place cost or price reductions in the case of soybeans in the USA at about US$20 per hectare, mainly because of decrease in energy costs (weed management cost) (McFarlane et al. 2010).
In vitro techniques for the protoplast, anther, microspore, ovule and embryo culture have been utilized to create novel genetic variant in the breeding lines (Chawla 2002). Cell culture technology has also made somaclonal and gametoclonal variants with the potential of crop improvement. The single cell and meristem culture can be successfully used to eradicate plant pathogens from planting material and improve the growth as well as yield of established cultivars (Chawla 2002; De Filippis 2014). Large-scale micropropagation and tissue culture laboratories are offering millions of plants for commercial purposes (Giles and Morgan 1987). With respect to medicinal plants, useful metabolite production by plant cell and tissues culture has been performed on a growing scale since the end of the 1950s (Filova 2014). Their outcomes stimulated recent studies on the commercial application of this technology in various countries including India. However, there are still small barriers that must be defeated before commercialization of several other products (Giles and Morgan 1987; Chawla 2002; Filova 2014). To overcome the barriers hindering commercial and industrial applications of plant tissue cultures, however, it is essential to conduct more basic and fundamental research, including deciphering of the biosynthetic pathways of many useful bioactive compounds and secondary metabolites in plants and the mechanisms for their biosynthesis. Association with a number of researchers working in other scientific fields is also very useful.
Similarly, foreign proteins can be synthesized using plant tissue culture and transgenic plants. For choosing the technique for commercial production, a case-by-case analysis is needed. A broad range of issues must be considered, including production cost, market volume, efficacy, safety as well as product stability, and whether the foreign protein has various biochemical or pharmacological features compared with material from available sources (Ferrante and Simpson 2001). Plant cell and root cultures have been verified for recombinant protein production. The role of plant cell culture as a way for commercial production of protein is not clear at the time, but is likely to depend on the capability to express proteins of high levels in vitro by changing the culture conditions (Gharelo et al. 2016). The stability and extracellular localization of secreted proteins in the medium of the plant cell cultures may be significant keys to the economic feasibility, permitting easier product recovery without destroying the biomass, for the reason that the controlled environmental condition in plant cell cultures facilitates advantages for regulatory compliance and GMP. A potential niche for reactor-scale plant cell and tissue culture is the fast production of low-to-medium volume of therapeutic proteins (Ferrante and Simpson 2001; Gharelo et al. 2016). Technologies for changing the glycans of plant glycoproteins may be needed before plant systems can be completely exploited for the production of vaccines and injectable drugs (Doran 2000). Considering the recent trends in plant tissue culture and transformation, there is need to explore “what is being completed” and “what can be completed” in case of tissue culture of commercially valuable plants.
Plant cell physiology and input use efficiency
Molecular biology and cell physiology mostly concern themselves with recognizing the interactions between the various cell systems, including the interactions between nucleic acid (DNA, RNA) and protein biosynthesis and studying how these interactions are regulated (Alberts et al. 1995). This discipline of science overlaps with biochemistry which is the learning of the chemical substances and fundamental processes happening in living organisms. Biochemists focus heavily on the role, structure and function of biomolecules. Rapid advances in the field of plant cell physiology and input use efficiency have enabled identification of molecular principles which underlie plant behavior (Buchanan et al. 2015). The identification of various factors and their cognate target sites involved in the regulation of plant development and adaption processes has led to distinct molecular models which explain how plants cope with different environmental stresses of light, water, nutrient conditions and various biotic stresses. It can be used to model and analyze important and complex traits linked with crop productivity such as water use efficiency, nitrogen use efficiency, photosynthetic efficiency and other essential traits associated with plant architecture and plant cell physiology for future Omics-based applications in crop plants (Kumar et al. 2015a).
This in turn has facilitated formulation of effective strategies to develop stress-tolerant crop varieties through molecular manipulation of key genes or proteins (Bita and Gerats 2013). The recently developed skill to recognize entire genomic sequences has provided the data required to accomplish massive assessments and comparisons of derived protein sequences, the outcome of which may be utilized to formulate and test hypotheses regarding biochemical function of a specific plant protein (Collins et al. 2003). With the advent of omics technologies it has become possible to understand the behavior of whole genome and gene networks involved in regulation of complex agronomic traits. This has been possible due to availability of molecular maps, quantitative trait loci (QTLs), genomic and expressed tag sequences from model plant Arabidopsis thaliana and various crop plants such as rice and sorghum (Li et al. 2014). Comparative and functional genomics approaches are being utilized for understanding gene functions and for studying simultaneous interaction of various genes governing a complex agronomic trait (Kumar et al. 2015a). With the inception of genomic research, coordination of regulation of a set of genes is possible by transfer of key regulatory master switch gene.
Nanobiotechnology and diagnostics
Molecular diagnostics technologies will also play an important role in practice of agriculture, food and biological warfare in the twenty-first century (Dawidziuk et al. 2014). Nanobiotechnology broadens the limitation of molecular diagnostics to the nanoscale level. Recent advances in genomics and proteomics have provided us a number of different molecular markers, which are specific for various crop plant diseases. Nanotechnology can utilize high-throughput data obtained through omics-based experimentations, which are able to allow us for more specific and efficient diagnosis of diseases linked to crop productivity (Hood et al. 2012; Lee 2014).
In general, it has been most significant innovations of recent years and has quickly become the basis of various products that are currently in use, especially in crop plants (Rai and Ingle 2012; Grobe and Rissanen 2012). The success of this advanced technology has been in its power to present smarter solutions that have broad applications in agriculture (Grobe and Rissanen 2012). The potential of nanodiagnostics arises from the fact that most biological molecules and cell organelles fall within the nanometer scale. Conventional methods of pathogen detection have often depended on recognition of disease symptoms, isolation, culturing and identification of organisms by morphology as well as biochemical tests. The major limitations of these culture-based morphological approaches, however, are the dependence on the ability of organism to be cultured, time-consuming nature and the need of a wide taxonomic expertise (Perez-Fernandez 2012; Kole et al. 2013; Husen and Siddiqi 2014; Kumari and Yadav 2014). Laboratory diagnostics for plant pathogens have traditionally relied on methods of detecting the pathogen by antibodies or culture using a variety of techniques such as neutralization, enzyme-linked immunosorbent assay, complement fixation and agar gel immunodiffusion. The field of nanobiotechnology and molecular diagnostics has mainly increased sensitivity and faster detection (Singh et al. 2013a, b).
Nanobiotechnology can be exploited to develop biosensors and other efficient diagnostic tools for crop diagnostics (Moreau et al. 2012). Nanobarcodes and submicrometer metallic barcodes along with striping patterns made by sequential electrochemical depositon of the metal demonstrate differential reflectivity of the adjacent stripes facilitating investigation of the striping patterns by using conventional light microscopy (Kawadkar et al. 2011). The processes and products that are precise and impossible to achieve through conventional systems can be achieved by atom by atom manipulation using nanobiotechnology (Singh et al. 2013a). Nanoparticles are able to control crop diseases. Nutrient deficiency and food contaminants can also be detected using nanosensors (Manjunatha et al. 2016). “Smart delivery systems” for agriculture can have timely controlled, spatially targeted, self-regulated and remotely regulated, pre-programmed as well as multi-functional features to avoid biological difficulties to successful targeting. It can examine and monitor the effects of delivery of bioactive molecules or nutrients or any important molecules such as pesticide. A small sealed package brings the drug which opens up only when the required location or site of the infection of the human and animal system is reached in the case of smart delivery system. The nano food packaging sector has experienced some of the most significant developments in terms of commercialization (Scrinis and Lyons 2007). Manufacturers are using nano techniques with the aim of improving the quality, durability and shelf life of packaged foods. Nanobiotechnology can be employed to synthesize smart nano-devices that release active herbicide and insecticide molecules only when moisture is available in rain-fed system besides targeting both leaves and tubers (Grobe and Rissanen 2012; Singh et al. 2013a). Nanoinsecticides are better than the conventional insecticides due to their small size, reduced run off, higher water suspension/solubility, less damage to the environment. Nanofertilizers may be applied as a strategy to regulate and control smart release of the nutrients that commensurate with crop requirement (Kah 2015).
In a study conducted at GBPUAT, Pantnagar, antisera were successfully produced in albino white New Zealand rabbits against teliospores, soluble cytoplasmic and the insoluble cell wall antigens of vegetative mycelium of fungal pathogen. The various immunological laboratory-scale formats such as dot immunoblot binding assay (DIBA), seed immunoblot binding assay (SIBA), indirect immunofluorescence test (IIFT) and indirect microtiter ELISA have been developed for quick detection of infection and infestations of KB teliospores in wheat samples for declaring it pathogen free and will strengthen the quality and regulatory aspects of seed certification and quarantine regulation. The ELISA-based immuno-dipstick assay and gold nanoparticles-based lateral flow immunodiffusion tests have been also developed and are quite sensitive to detect only few teliospores (as low as five) and apply for on-site diagnostics at field level. The ultimate goals of nanodiagnostics, nano delivery and nano safety are revolutionizing crop plant biology (Gupta et al. 2001; Kumar et al. 2004).
Bio-Inoculant, bioprocessing and metagenomics
Bioprocessing is associated with the utilization of biological resources such as microorganisms, plant, animal cells and enzymes in converting raw materials into different new value-added food as well as non-food products (Beattie et al. 2011; Saslis-Lagoudakis et al. 2012; Talukdar et al. 2012; Zotchev et al. 2012; Sprocati et al. 2014; Purkayastha 2016). Research on bioprocessing focuses on three key thrust areas, namely enzyme technology, fermentation and microbial technology, plant metabolites and cell culture technology (Costa et al. 2013). There is need for extensive research which has the potential for facilitate the agri-food industry to enhance process efficiency, improve product quality and extend shelf life of processed as well as fresh agri-food products. In the genomic era, vast genome sequence information can be utilized to manipulate the metabolism of the organism resulting in more efficient strains for bioprocessing purposes. The science of omics can be used to produce more efficient enzymes, optimize fermentation conditions and identify new gene targets for enhancing the bioprocessing of crop produce (Beattie et al. 2011; Talukdar et al. 2012; Zotchev et al. 2012; Sprocati et al. 2014). Functional genomics approaches provide insight into cell’s metabolism which can be exploited for metabolic engineering to develop value-added products (Georgiev et al. 2009).
Bioinoculants have revealed big potential as a renewable, supplementary and environmental-friendly resource of plant nutrients, and are a key element of integrated nutrient management (INM) (Jat et al. 2015). Bioinoculants are the live microbial cells of single species or consortium of different microbial isolates that are applied to the soil or treated with plant/seed material to improve plant health. Broadly, bioinoculants include biofertilizers and biopesticides. Omics technologies can be utilized to authenticate different strains of bioinoculants to maintain quality standards and to identify different genes which could be used to develop improved strains of bioinoculants (Balachandar 2012; Malusá et al. 2012).
Metagenomics is the study of metagenomes, genetic material recovered directly from environmental samples. It holds immense potential to study useful microbial diversity present in the environment, which is non-culturable and cannot be studied by traditional microbiological methods (Singh et al. 2009). Science of “Omics” has revolutionized the study of microbes present in the environment and enhanced understanding of the composition, physiology and phylogeny of microorganism communities. At present, there is a major emphasis on the use of “omics”-based approaches such as genomics, proteomics and transcriptomics to determine the uniqueness and functional features of microbes inhabiting various environments (Singh et al. 2009, 2010; Suyal et al. 2014). Needless to say, such recent developments will be of significant value in discovering new microbes and microbial genes and to exploit them in solving the urgent challenges facing the environment, agriculture and human health.
Challenges in omics
We have entered into the era of omics from the past decades that may lead to holistic approach for interpreting the complex nature of biological systems at the molecular to cellular level. The completion of several genome projects will allow us to define the functional relationship among one or more particular genes involved in the productivity, disease resistance and physiological processes of crop plants. Additionally, the expression of these gene, mRNA stability and final protein product can be altered by a wide range of physiological processes, alternative splicing, post-translational modification, epigenetic modifications and single nucleotide polymorphisms, etc., but they present a complex challenge to the scientific community. With the beginning of newer techniques and technologies, the omics science and its applications in diverse areas are quickly increasing. Such types of emerging fields including next generation sequencing, regulomics, spliceomics, metagenomics, environomics and bioinformatics present great solutions to fight global challenges to crop productivity and the environment (Ning and Lo 2010). Currently, the potential challenges of omics are availability of computational resources. Big amount of data have been and are being generated. This has imposed certain challenges including storage and analysis of the data: lack of reference database, and the reference phytochemicals for metabolomics, where some metabolites with unknown structure might be produced; reproducibility: inability of one group’s findings to be replicated by another group or person; noise: omics-based science can generate huge data that noise overwhelms indication. Why is it that researchers get fewer genes as apparatus and methodologies advance?; fishing: the look for novel genes in complex samples, i.e., discovery-mode “omics”, has been frequently criticized as a “fishing expedition” in which one blindly expects to find something attractive and often falls short (Ning and Lo 2010; Costa 2012; Gomez-Cabrero et al. 2014; Cambiaghi et al. 2017).
Future perspectives
The growing demand for improved crop varieties, effectiveness and quality of crop product and major challenges faced by modern agriculture are the key factors that force the scientific community to adopt innovative omics-based technology for enhancing crop productivity in India. Omics science and technology, viz., genomics, transcriptomics, proteomics, metabolomics, bioinformatics, systems biology and other omics-based approaches having tremendous potential to decipher the complexity of important and complex traits linked to productivity of the crops such as nitrogen use efficiency, photosynthetic efficiency, water use efficiency, plant architecture, biotic and abiotic stress tolerance and nutritional quality. Currently, the cost of omics-based applications for crop plants is still extremely high, but it is expected to decrease with technological advancements in the future. Advances in these approaches can lead to bridging the gap between genotype to phenotype by utilizing quantitative and automated screening and selection methods that target whole crop plants system and quality traits, which enhance the release of new high yielding varieties with rich amount of nutrients to the farmers, and reduces large-scale field studies and developmental process. Besides, the omics-based technology will also enable bridging the gap between gene–environment interactions, because many key traits depend upon the interaction with environment for a particular biological function. It will help in the creation of ‘smart crops’ for higher yield and sustainability that are capable of fulfilling the need of rapidly growing population for securing food and nutritional security in future.
Conclusion
This article highlighted the importance of new omics-based tools and technologies in the improvement of crop productivity. A thorough understanding of genomics, transcriptomics, metagenomics, proteomics, metabolomics, bioinformatics and other related areas of omics have opened up new perspectives for investigating functions and interactions of plants with abiotic and biotic stress agents. The omics-based researches have become very helpful for solving various agricultural challenges. Therefore, additional resources should be allocated for improvement of omics-based technologies to effectively target plant systems for crop improvement programs. A public conversation is necessary in addition to research and development for promoting the application of omics science and technology in India for upliftment of the agri-food nutrition sector in the future.
Acknowledgements
The authors gratefully acknowledge the Department of Biotechnology (DBT), Government of India, for providing financial assistance by the grant of program mode support for research and development in Agricultural Biotechnology (Grant no. BT/PR7849/AGR/02/374/2006-Part II). Author R.K.P. is supported by fellowship from CSIR, India. NxGenBio Life Sciences, New Delhi is gratefully acknowledged for providing the cost of several sequencing technologies.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Agrawal GK, Sarkar A, Righetti PG, Pedreschi R, Carpentier S, Wang T, Barkla BJ, Kohli A, Ndimba BK, Bykova NV. A decade of plant proteomics and mass spectrometry: translation of technical advancements to food security and safety issues. Mass Spectrom Rev. 2013;32:335–365. doi: 10.1002/mas.21365. [DOI] [PubMed] [Google Scholar]
- Al-Ahmad H. Biotechnology for bioenergy dedicated trees: meeting future energy demands. Zeitschrift für Naturforschung C. 2018;73:15–32. doi: 10.1515/znc-2016-0185. [DOI] [PubMed] [Google Scholar]
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD, Grimstone A. Molecular biology of the Cell. Trends Biochem Sci. 1995;20:210–210. [Google Scholar]
- An Y-Q, Lin L. Transcriptional regulatory programs underlying barley germination and regulatory functions of gibberellin and abscisic acid. BMC Plant Biol. 2011;11:105. doi: 10.1186/1471-2229-11-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JE, Kantar MB, Kono TY, Fu F, Stec AO, Song Q, Cregan PB, Specht JE, Diers BW, Cannon SB (2014) A roadmap for functional structural variants in the soybean genome. G3 114.011551 [DOI] [PMC free article] [PubMed]
- Arif IA, Bakir MA, Khan HA, Al Farhan AH, Al Homaidan AA, Bahkali AH, Al Sadoon M, Shobrak M. A brief review of molecular techniques to assess plant diversity. Int J Mol Sci. 2010;11:2079–2096. doi: 10.3390/ijms11052079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avashthi H, Gautam B, Jain PA, Tiwari A, Pathak RK, Srivastava A, Taj G, Kumar A. In silico identification of MAPK3/6 substrates in WRKY, bZIP, MYB, MYB-related, NAC and AP-2 transcription factor family in Arabidopsis thaliana. Int J Comput Bioinf In Silico Model. 2014;3:454–459. [Google Scholar]
- Babu BK, Agrawal PK, Gupta HS, Kumar A, Bhatt JC. Identification of candidate gene–based SSR markers for lysine and tryptophan metabolic pathways in maize (Zea mays) Plant Breed. 2012;131:20–27. [Google Scholar]
- Balachandar D. Biofertilizers–what next. J Biofertil Biopestici. 2012;3:e108. [Google Scholar]
- Beattie AJ, Hay M, Magnusson B, de Nys R, Smeathers J, Vincent JF. Ecology and bioprospecting. Aust Ecol. 2011;36:341–356. doi: 10.1111/j.1442-9993.2010.02170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bita C, Gerats T. Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci. 2013;4:273. doi: 10.3389/fpls.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancquaert D, De Steur H, Gellynck X, Van Der Straeten D. Present and future of folate biofortification of crop plants. J Exp Bot. 2014;65:895–906. doi: 10.1093/jxb/ert483. [DOI] [PubMed] [Google Scholar]
- Boruc J, Inzé D, Russinova E. A high-throughput bimolecular fluorescence complementation protein–protein interaction screen identifies functional Arabidopsis CDKA/B-CYCD4/5 complexes. Plant Signal Behav. 2010;5:1276–1281. doi: 10.4161/psb.5.10.13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes G, Barfoot P. Key environmental impacts of global genetically modified (GM) crop use 1996–2011. GM Crops Food. 2013;4:109–119. doi: 10.4161/gmcr.24459. [DOI] [PubMed] [Google Scholar]
- Broughton Sue, Sidhu Parminder K., Davies Philip A. Methods in Molecular Biology. New York, NY: Springer New York; 2014. In vitro Culture for Doubled Haploids: Tools for Molecular Breeding; pp. 167–189. [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL. Biochemistry and molecular biology of plants. Amsterdam: Wiley; 2015. [Google Scholar]
- Bueno J. Metabolomics in antimicrobial drug discovery: the success of the chemical diversity. J Microb Biochem Technol. 2015;7:380–383. [Google Scholar]
- Cambiaghi A, Ferrario M, Masseroli M. Analysis of metabolomic data: tools, current strategies and future challenges for omics data integration. Brief Bioinform. 2017;18:498–510. doi: 10.1093/bib/bbw031. [DOI] [PubMed] [Google Scholar]
- Cao S, Yan L. Construction of a high-quality yeast two-hybrid (Y2H) library and its application in identification of interacting proteins with key vernalization regulator Ta VRN-A1 in wheat. BMC Res Notes. 2013;6:81. doi: 10.1186/1756-0500-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2013;42(D1):D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Chakraborty N, Agrawal L, Ghosh S, Narula K, Shekhar S, Naik PS, Pande P, Chakrborti SK, Datta A. Next-generation protein-rich potato expressing the seed protein gene AmA1 is a result of proteome rebalancing in transgenic tuber. Proc Natl Acad Sci USA. 2010;14:201006265. doi: 10.1073/pnas.1006265107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla H. Introduction to plant biotechnology. New York: Science Publishers; 2002. [Google Scholar]
- Chawla R, Arora JS, Dubey RK, Mukhopadhyay CS (2018) Omics approaches and applications in dairy and food processing technology. In: Omics Technologies and Bio-Engineering. Elsevier, pp 271–295
- Chinchole M, Pathak RK, Singh UM, Kumar A. Molecular characterization of EcCIPK24 gene of finger millet (Eleusine coracana) for investigating its regulatory role in calcium transport. 3 Biotech. 2017;7:267. doi: 10.1007/s13205-017-0874-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422:835. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- Consortium TG. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium IWGS. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- Costa FF. Big data in genomics: challenges and solutions. GIT Lab J. 2012;11:1–4. [Google Scholar]
- Costa C, Antonucci F, Pallottino F, Aguzzi J, Sarriá D, Menesatti P. A review on agri-food supply chain traceability by means of RFID technology. Food Bioprocess Tech. 2013;6:353–366. [Google Scholar]
- Cowie P, Ross R, MacKenzie A. Understanding the dynamics of gene regulatory systems; characterisation and clinical relevance of cis-regulatory polymorphisms. Biology. 2013;2:64–84. doi: 10.3390/biology2010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis T, Halford NG. Food security: the challenge of increasing wheat yield and the importance of not compromising food safety. Ann A Biol. 2014;164:354–372. doi: 10.1111/aab.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon C, Dmitrieva J, Muhovski Y, Francis F, Lins L, Ledoux Q, Luwaert W, Markó IE, Mauro S, Ongena M. Interaction network of antimicrobial peptides of Arabidopsis thaliana, based on high-throughput yeast two-hybrid screening. Plant Physio Biochem. 2012;58:245–252. doi: 10.1016/j.plaphy.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Datta A. Genetic engineering for improving quality and productivity of crops. Agricult Food Secur. 2013;2:15. [Google Scholar]
- Dawidziuk A, Koczyk G, Popiel D, Kaczmarek J, Buśko M. Molecular diagnostics on the toxigenic potential of Fusarium spp. plant pathogens. J Appl Micro. 2014;116:1607–1620. doi: 10.1111/jam.12488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippis L (2014) Crop improvement through tissue culture. In: Ahmad P, Wani MR, Azooz MM, Phan Tran L-S (eds) Improvement of crops in the era of climatic changes. Springer, pp 289–346
- Deracinois B, Flahaut C, Duban-Deweer S, Karamanos Y. Comparative and quantitative global proteomics approaches: an overview. Proteomes. 2013;1:180–218. doi: 10.3390/proteomes1030180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi K, Mishra SK, Sahu J, Panda D, Modi MK, Sen P. Genome wide transcriptome profiling reveals differential gene expression in secondary metabolite pathway of Cymbopogon winterianus. Sci Rep. 2016;6:21026. doi: 10.1038/srep21026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepens NJ, Pfennig S, Van den Brink PJ, Gunnarsson JS, Ruepert C, Castillo L. Effect of pesticides used in banana and pineapple plantations on aquatic ecosystems in Costa rica. J Environ Biol. 2014;35:73–84. [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet. 2010;11:539. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Doran PM. Foreign protein production in plant tissue cultures. Curr Opin Biotechnol. 2000;11:199–204. doi: 10.1016/s0958-1669(00)00086-0. [DOI] [PubMed] [Google Scholar]
- Dutta S, Kumawat G, Singh BP, Gupta DK, Singh S, Dogra V, Gaikwad K, Sharma TR, Raje RS, Bandhopadhya TK. Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh] BMC Plant Biol. 2011;11:17. doi: 10.1186/1471-2229-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels B, Dahm P, Jennewein S. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab Eng. 2008;10:201–206. doi: 10.1016/j.ymben.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Fahad S, Nie L, Khan FA, Chen Y, Hussain S, Wu C, Xiong D, Jing W, Saud S, Khan FA. Disease resistance in rice and the role of molecular breeding in protecting rice crops against diseases. Biotechnol Lett. 2014;36:1407–1420. doi: 10.1007/s10529-014-1510-9. [DOI] [PubMed] [Google Scholar]
- Ferrante E, Simpson D. A review of the progression of transgenic plants used to produce plantibodies for human usage. J Young Investig. 2001;4:11. [Google Scholar]
- Filipp FV. A Gateway between omics data and systems biology. J Metab Syst Biol. 2013;1:1–1. [PMC free article] [PubMed] [Google Scholar]
- Filova A (2014) Production of secondary metabolities in plant tissue cultures. Res J Agri Sci 46
- Furey TS, Cristianini N, Duffy N, Bednarski DW, Schummer M, Haussler D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000;16:906–914. doi: 10.1093/bioinformatics/16.10.906. [DOI] [PubMed] [Google Scholar]
- Galbraith David W. Methods in Molecular Biology. Totowa, NJ: Humana Press; 2013. Flow Cytometry and Sorting in Arabidopsis; pp. 509–537. [DOI] [PubMed] [Google Scholar]
- Garg A, Agrawal L, Misra RC, Sharma S, Ghosh S. Andrographis paniculata transcriptome provides molecular insights into tissue-specific accumulation of medicinal diterpenes. BMC Genet. 2015;16:659. doi: 10.1186/s12864-015-1864-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Shankar R, Thakkar B, Kudapa H, Krishnamurthy L, Mantri N, Varshney RK, Bhatia S, Jain M. Transcriptome analyses reveal genotype-and developmental stage-specific molecular responses to drought and salinity stresses in chickpea. Sci Rep. 2016;6:19228. doi: 10.1038/srep19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev MI, Weber J, Maciuk A. Bioprocessing of plant cell cultures for mass production of targeted compounds. Appl Microbiol Biotechnol. 2009;83:809–823. doi: 10.1007/s00253-009-2049-x. [DOI] [PubMed] [Google Scholar]
- Gharelo RS, Oliaei ED, Bandehagh A, Khodadadi E, Noparvar PM. Production of therapeutic proteins through plant tissue and cell culture. J BioSci Biotechnol. 2016;5:1. [Google Scholar]
- Giles KL, Morgan WM. Industrial-scale plant micropropagation. Trends Biotechnol. 1987;5:35–39. [Google Scholar]
- Gillam EM. Engineering cytochrome P450 enzymes. Chem Res Toxicol. 2007;21:220–231. doi: 10.1021/tx7002849. [DOI] [PubMed] [Google Scholar]
- Goel A, Gaur VS, Arora S, Gupta S, Kumar A. In silico analysis of expression data for identification of genes involved in spatial accumulation of calcium in developing seeds of rice. OMICS. 2012;16:402–413. doi: 10.1089/omi.2012.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cabrero D, Abugessaisa I, Maier D, Maier D, Teschendorff A, Merkenschlager M, Gisel A, Ballestar E, Bongcam-Rudloff E, Conesa A, Tegnér J. Data integration in the era of omics: current and future challenges. Bio Med Central. 2014 doi: 10.1186/1752-0509-8-S2-I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Chen W, Gao Y, Liu X, Zhang H, Xu C, Yu S, Zhang Q, Luo J. Genetic analysis of the metabolome exemplified using a rice population. Proc Natl Acad Sci USA. 2013;110:20320–20325. doi: 10.1073/pnas.1319681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Hu X, Wang W. Proteomic analysis of crop plants under abiotic stress conditions: where to focus our research? Front Plant Sci. 2015;6:418. doi: 10.3389/fpls.2015.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gour P, Garg P, Jain R, Joseph SV, Tyagi AK, Raghuvanshi S. Manually curated database of rice proteins. Nucleic Acids Res. 2013;42(D1):D1214–D1221. doi: 10.1093/nar/gkt1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindaraj M, Vetriventhan M, Srinivasan M. Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genetics research international. 2015;2015:431487. doi: 10.1155/2015/431487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobe A, E Rissanen M. Nanotechnologies in agriculture and food-An overview of different fields of application, risk assessment and public perception. Recent Pat Food Nutr Agric. 2012;4:176–186. doi: 10.2174/2212798411204030176. [DOI] [PubMed] [Google Scholar]
- Grover A, Patade V, Kumari M, Gupta SM, Arif M, Ahmed Z. Omics approaches in biofuel production for a green environment. OMICS: applications in biomedical, agricultural, and environmental sciences. Boca Raton: CRC Press, Taylor and Francis Group; 2013. pp. 623–636. [Google Scholar]
- Grover A, Patade V, Kumari M, Gupta SM, Arif M, Ahmed Z. Bio-energy crops enter the omics era. OMICS applications in crop science. Taylor: CRC Press; 2014. pp. 549–562. [Google Scholar]
- Gujjar RS, Akhtar M, Rai A, Singh M. Expression analysis of drought-induced genes in wild tomato line (Solanum habrochaites) Curr Sci. 2014;00113891:107. [Google Scholar]
- Gujjar RS, Karkute SG, Rai A, Singh M, Singh B. Proline-rich proteins may regulate free cellular proline levels during drought stress in tomato. Curr Sci. 2018;00113891:114. [Google Scholar]
- Gupta A (2015) State-wise malnutrition analysis in India: findings from NFHS III. IJSR 482–483. https://www.ijsr.net/archive/v5i4/NOV162624.pdf. Accessed 12 Dec 2016
- Gupta V, Kumar A, Lakhchaura B, Garg G. Generation of anti-teliospores antibodies for immunolocalization and characterization of antigenic epitopes of teliospores of Karnal bunt (Tilletia indica) of wheat. Indian J Exp Biol. 2001;39(7):686–90. [PubMed] [Google Scholar]
- Gupta AK, Gaur VS, Pathak RK, Gupta S, Kumar A. Dof1 transcription factor interacts with only specific regions of the promoters driving the expression of genes involved in carbon and nitrogen metabolism. Int J Comput Bioinf In Silico Model. 2014;3:412–422. [Google Scholar]
- Gupta S, Gupta SM, Gupta AK, Gaur VS, Kumar A. Fluctuation of Dof1/Dof2 expression ratio under the influence of varying nitrogen and light conditions: involvement in differential regulation of nitrogen metabolism in two genotypes of finger millet (Eleusine coracana L.) Gene. 2014;546:327–335. doi: 10.1016/j.gene.2014.05.057. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Kulwal PL, Jaiswal V. Association mapping in crop plants: opportunities and challenges. Adv Genet. 2014;85:109–147. doi: 10.1016/B978-0-12-800271-1.00002-0. [DOI] [PubMed] [Google Scholar]
- Gupta S, Chavan S, Deobagkar DN, Deobagkar DD. Bio/chemoinformatics in India: an outlook. Brief Bioinf. 2014;16:710–731. doi: 10.1093/bib/bbu028. [DOI] [PubMed] [Google Scholar]
- Gupta R, Lee SE, Agrawal GK, Rakwal R, Park S, Wang Y, Kim ST. Understanding the plant-pathogen interactions in the context of proteomics-generated apoplastic proteins inventory. Front Plant Sci. 2015;6:352. doi: 10.3389/fpls.2015.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SM, Arora S, Mirza N, Pande A, Lata C, Puranik S, Kumar J, Kumar A. Finger millet: a “certain” crop for an “uncertain” future and a solution to food insecurity and hidden hunger under stressful environments. Front Plant Sci. 2017;8:643. doi: 10.3389/fpls.2017.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Pathak RK, Gupta SM, Gaur VS, Singh N, Kumar A. Identification and molecular characterization of Dof transcription factor gene family preferentially expressed in developing spikes of Eleusine coracana L. Biotechnology. 2018;8(3):82. doi: 10.1007/s13205-017-1068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddeland I, Heinke J, Biemans H, Eisner S, Flörke M, Hanasaki N, Konzmann M, Ludwig F, Masaki Y, Schewe J. Global water resources affected by human interventions and climate change. Proc Natl Acad Sci USA. 2014;111:3251–3256. doi: 10.1073/pnas.1222475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain T, Hossain H, Chatterjee SS, Machata S, Volk U, Wagner S, Brors B, Haas S, Kuenne CT, Billion A, Otten S. Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e σ B regulon. BMC Microbiol. 2008;8:20. doi: 10.1186/1471-2180-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin MT, Buckler ES, Jannink J-L. Population genetics of genomics-based crop improvement methods. Trends Genet. 2011;27:98–106. doi: 10.1016/j.tig.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Hawkins KM, Smolke CD. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat Chem Biol. 2008;4:564. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Nagalakshmi U, Lieberman MC, Ngo KJ, Krasileva KV, Vasquez-Gross H, Akhunova A, Akhunov E, Dubcovsky J, Tai TH. Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant Cell. 2014;113:121590. doi: 10.1105/tpc.113.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman EM, Helm RM, Jung R, Kinney AJ. Genetic modification removes an immunodominant allergen from soybean. Plant Physiol. 2003;132:36–43. doi: 10.1104/pp.103.021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittalmani S, Mahesh H, Shirke MD, Biradar H, Uday G, Aruna Y, Lohithaswa H, Mohanrao A. Genome and Transcriptome sequence of Finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genet. 2017;18:465. doi: 10.1186/s12864-017-3850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD. Genomic variation in Arabidopsis: tools and insights from next-generation sequencing. Chromosome Res. 2014;22:103–115. doi: 10.1007/s10577-014-9420-1. [DOI] [PubMed] [Google Scholar]
- Hood LE, Omenn GS, Moritz RL, Aebersold R, Yamamoto KR, Amos M, Hunter-Cevera J, Locascio L, Participants W. New and improved proteomics technologies for understanding complex biological systems: addressing a grand challenge in the life sciences. Proteomics. 2012;12:2773–2783. doi: 10.1002/pmic.201270086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S. Combinatorial biosynthesis of plant medicinal polyketides by microorganisms. Curr Opin Chem Biol. 2009;13:197–204. doi: 10.1016/j.cbpa.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Huang Q, Roessner CA, Croteau R, Scott AI. Engineering Escherichia coli for the synthesis of taxadiene, a key intermediate in the biosynthesis of taxol. Bioorg Med Chem. 2001;9:2237–2242. doi: 10.1016/s0968-0896(01)00072-4. [DOI] [PubMed] [Google Scholar]
- Huang M, Jiang P, Shan S, Gao W, Ma G, Zou Y, Uphoff N, Yuan L. Higher yields of hybrid rice do not depend on nitrogen fertilization under moderate to high soil fertility conditions. Rice. 2017;10:43. doi: 10.1186/s12284-017-0182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husen A, Siddiqi KS. Carbon and fullerene nanomaterials in plant system. J Nanobiotechnol. 2014;12:16. doi: 10.1186/1477-3155-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalmi SK, Sinha AK. Functional involvement of a mitogen activated protein kinase module, OsMKK3–OsMPK7–OsWRK30 in mediating resistance against Xanthomonas oryzae in rice. Sci Rep. 2016;6:37974. doi: 10.1038/srep37974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jat LK, Singh Y, Meena SK, Meena SK, Parihar M, Jatav H, Meena RK, Meena VS. Does integrated nutrient management enhance agricultural productivity. J Pure Appl Microbiol. 2015;9:1211–1221. [Google Scholar]
- Jayaram B, Priyanka D (2010) Bioinformatics for better tomorrow. Supercomputing facility for bioinformatics and computational biology. Indian Institute of Technology, New Delhi, http://www.scfbio-iitd.org. Accessed 21 Dec 2016
- Jayaram B, Bhushan K, Shenoy SR, Narang P, Bose S, Agrawal P, Sahu D, Pandey V. Bhageerath: an energy based web enabled computer software suite for limiting the search space of tertiary structures of small globular proteins. Nucleic Acids Res. 2006;34:6195–6204. doi: 10.1093/nar/gkl789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A, Shankar R. Employing machine learning for reliable miRNA target identification in plants. BMC Genet. 2011;12:1. doi: 10.1186/1471-2164-12-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. Gene banks pay big dividends to agriculture, the environment, and human welfare. PLoS Biol. 2008;6:e148. doi: 10.1371/journal.pbio.0060148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph LM, Hymowitz T, Schmidt MA, Herman EM. Evaluation of Glycine germplasm for nulls of the immunodominant allergen P34/Gly m Bd30 k. Crop Sci. 2006;46:1755–1763. [Google Scholar]
- Joshi L, Van Eck JM, Mayo K, Di Silvestro R, Blake ME, Ganapathi T, Haridas V, Gutterman JU, Arntzen CJ. Metabolomics of plant saponins: bioprospecting triterpene glycoside diversity with respect to mammalian cell targets. OMICS. 2002;6:235–246. doi: 10.1089/15362310260256891. [DOI] [PubMed] [Google Scholar]
- Joshi PK, Gupta D, Nandal UK, Khan Y, Mukherjee SK, Sanan-Mishra N. Identification of mirtrons in rice using MirtronPred: a tool for predicting plant mirtrons. Genomics. 2012;99:370–375. doi: 10.1016/j.ygeno.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Jouzani GS, Sharafi R (2018) New “Omics” Technologies and Biogas Production. In: Tabatabaei M, Ghanavati H (eds) Biogas. Biofuel and Biorefinery Technologies, vol 6. Springer, Cham
- Jung S, Main D. Genomics and bioinformatics resources for translational science in Rosaceae. Plant Biotechnol Rep. 2014;8:49–64. doi: 10.1007/s11816-013-0282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadiyala S, Joshi P, Dev SM, Kumar TN, Vyas V (2012) A nutrition secure India: role of agriculture. Econ Polit Weekly 21–25
- Kah M. Nanopesticides and nanofertilizers: emerging contaminants or opportunities for risk mitigation? Front Chem. 2015;3:64. doi: 10.3389/fchem.2015.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S, Altaf-Ul-Amin M, Kiboi SK, Mochamad Afendi F. Big data and network biology. BioMed Res Int. 2014;2014:836708. doi: 10.1155/2014/836708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkute SG, Gujjar RS, Rai A, Akhtar M, Singh M, Singh B. Genome wide expression analysis of WRKY genes in tomato (Solanum lycopersicum) under drought stress. Plant Gene. 2018;13:8–17. [Google Scholar]
- Kaundal R, Kapoor AS, Raghava GP. Machine learning techniques in disease forecasting: a case study on rice blast prediction. BMC Bioinf. 2006;7:485. doi: 10.1186/1471-2105-7-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawadkar J, Chauhan MK, Maharana M. Nanobiotechnology: application of nanotechnology in diagnosis, drug discovery and drug development. Asian J Pharm Clin Res. 2011;4:23–25. [Google Scholar]
- Kayum MA, Kim H-T, Nath UK, Park J-I, Kho KH, Cho Y-G, Nou I-S. Research on biotic and abiotic stress related genes exploration and prediction in Brassica rapa and B. oleracea: a review. Plant Breed Biotechnol. 2016;4:135–144. [Google Scholar]
- Kesavan P, Swaminathan M. Strategies and models for agricultural sustainability in developing Asian countries. Philos Trans R Soc B. 2008;363:877–891. doi: 10.1098/rstb.2007.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran K, Rawal HC, Dubey H, Jaswal R, Devanna B, Gupta DK, Bhardwaj SC, Prasad P, Pal D, Chhuneja P. Draft genome of the wheat rust pathogen (Puccinia triticina) unravels genome-wide structural variations during evolution. Genome Biol Evol. 2016;8:2702–2721. doi: 10.1093/gbe/evw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Kokane S, Pathak R, Singh M, Kumar A. The role of tripartite interaction of calcium sensors and transporters in the accumulation of calcium in finger millet grain. Biol Plant. 2018 doi: 10.3835/plantgenome2015.07.0058. [DOI] [Google Scholar]
- Kole C, Kole P, Randunu KM, Choudhary P, Podila R, Ke PC, Rao AM, Marcus RK. Nanobiotechnology can boost crop production and quality: first evidence from increased plant biomass, fruit yield and phytomedicine content in bitter melon (Momordica charantia) BMC Biotechnol. 2013;13:37. doi: 10.1186/1472-6750-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Suzuki N, Huntington S, Armijo L, Sha W, Cortes D, Shulaev V, Mittler R. Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J Biol Chem. 2008;283:34197–34203. doi: 10.1074/jbc.M806337200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyutürk M. Algorithmic and analytical methods in network biology. Wiley Interdiscip Rev Syst Biol Med. 2010;2:277–292. doi: 10.1002/wsbm.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyutürk M, Subramaniam S, Grama A. Functional coherence of molecular networks in bioinformatics. New York: Springer; 2011. [DOI] [PubMed] [Google Scholar]
- Kujur A, Saxena MS, Bajaj D, Parida SK. Integrated genomics and molecular breeding approaches for dissecting the complex quantitative traits in crop plants. J Biosci. 2013;38:971–987. doi: 10.1007/s12038-013-9388-6. [DOI] [PubMed] [Google Scholar]
- Kulasekaran RA. Influence of mothers’ chronic energy deficiency on the nutritional status of preschool children in Empowered Action Group states in India. Int J Nutr Pharmacol Neurol Dis. 2012;2:198. [Google Scholar]
- Kumar A. Science of omics for agricultural productivity: future perspective-A national conference report. Int J Comp Biol In Silico Model. 2015;4:607610. [Google Scholar]
- Kumar A, Tripathi K, Rana M, Purwar S, Garg G. Dibutyryl c-AMP as an inducer of sporidia formation: biochemical and antigenic changes during morphological differentiation of Karnal bunt (Tilletia indica) pathogen in axenic culture. J Biosci. 2004;29:23–31. doi: 10.1007/BF02702558. [DOI] [PubMed] [Google Scholar]
- Kumar A, Pathak RK, Gupta SM, Gaur VS, Pandey D. Systems biology for smart crops and agricultural innovation: filling the gaps between genotype and phenotype for complex traits linked with robust agricultural productivity and sustainability. OMICS. 2015;19:581–601. doi: 10.1089/omi.2015.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Gaur VS, Goel A, Gupta AK. De novo assembly and characterization of developing spikes transcriptome of finger millet (Eleusine coracana): a minor crop having nutraceutical properties. Plant Mol Biol Rep. 2015;33:905–922. [Google Scholar]
- Kumar Anil, Sharma Divya, Tiwari Apoorv, Jaiswal J.P., Singh N.K., Sood Salej. Genotyping-by-Sequencing Analysis for Determining Population Structure of Finger Millet Germplasm of Diverse Origins. The Plant Genome. 2016;9(2):0. doi: 10.3835/plantgenome2015.07.0058. [DOI] [PubMed] [Google Scholar]
- Kumar A, Pandey V, Singh M, Pandey D, Saharan M, Marla SS. Draft genome sequence of Karnal bunt pathogen (Tilletia indica) of wheat provides insights into the pathogenic mechanisms of quarantined fungus. PLoS One. 2017;12:e0171323. doi: 10.1371/journal.pone.0171323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A, Yadav SK. Nanotechnology in agri-food sector. Crit Rev Food Sci Nutr. 2014;54:975–984. doi: 10.1080/10408398.2011.621095. [DOI] [PubMed] [Google Scholar]
- Lal SB, Pandey PK, Rai PK, Rai A, Sharma A, Chaturvedi KK. Design and development of portal for biological database in agriculture. Bioinformation. 2013;9:588. doi: 10.6026/97320630009588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberth C, Jeanmart S, Luksch T, Plant A. Current challenges and trends in the discovery of agrochemicals. Science. 2013;341:742–746. doi: 10.1126/science.1237227. [DOI] [PubMed] [Google Scholar]
- Lee BH. Fundamentals of food biotechnology. Amsterdam: Wiley; 2014. [Google Scholar]
- Lee S, Kim Y-S, Jeon US, Kim Y-K, Schjoerring JK, An G. Activation of rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol Cells. 2012;33:269–275. doi: 10.1007/s10059-012-2231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser W, Kolady DE. Disease resistance of wheat varieties: can private varieties withstand the pressure? Economics Research International. 2011;2011:575192. [Google Scholar]
- Li S, Wang C, Chang X, Jing R. Genetic dissection of developmental behavior of grain weight in wheat under diverse temperature and water regimes. Genetica. 2012;140:393–405. doi: 10.1007/s10709-012-9688-z. [DOI] [PubMed] [Google Scholar]
- Li R, Han Y, Lv P, Du R, Liu G. Molecular mapping of the brace root traits in sorghum (Sorghum bicolor L. Moench) Breed Sci. 2014;64:193–198. doi: 10.1270/jsbbs.64.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou Z, Ding J, Wu Y, Zhou B, Wang R, Ma J, Wang S, Zhang X, Xia Z. Combined linkage and association mapping reveals QTL and candidate genes for plant and ear height in maize. Front Plant Sci. 2016;7:833. doi: 10.3389/fpls.2016.00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA, Zhao M, Ma J, Yu J, Huang S. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodha TD, Hembram P, Nitile Tep JB. Proteomics: a successful approach to understand the molecular mechanism of plant-pathogen interaction. Am J Plant Sci. 2013;4:1212. [Google Scholar]
- Ma NL, Rahmat Z, Lam SS. A review of the “omics” approach to biomarkers of oxidative stress in Oryza sativa. Int J Mol Sci. 2013;14:7515–7541. doi: 10.3390/ijms14047515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma’ayan A. Introduction to network analysis in systems biology. Sci Signal. 2011;4:tr5–tr5. doi: 10.1126/scisignal.2001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahato AK, Sharma N, Singh A, Srivastav M, Singh SK, Singh AK, Sharma TR, Singh NK. Leaf transcriptome sequencing for identifying genic-SSR markers and SNP heterozygosity in crossbred mango variety ‘Amrapali’ (Mangifera indica L.) PLoS One. 2016;11:e0164325. doi: 10.1371/journal.pone.0164325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malusá E, Sas-Paszt L, Ciesielska J (2012) Technologies for beneficial microorganisms inocula used as biofertilizers. Sci World J 2012 [DOI] [PMC free article] [PubMed]
- Mamgain S, Dhiman S, Pathak RK, Baunthiyal M. In silico identification of agriculturally important molecule (s) for defense induction against bacterial blight disease in Soybean (Glycine max) Plant Omics. 2018;11:98–105. [Google Scholar]
- Manjunatha S, Biradar D, Aladakatti Y. Nanotechnology and its applications in agriculture: a review. J Farm Sci. 2016;29:1–13. [Google Scholar]
- Mano J, Nagata M, Okamura S, Shiraya T, Mitsui T. Identification of oxidatively modified proteins in salt-stressed Arabidopsis: a carbonyl-targeted proteomics approach. Plant Cell Physiol. 2014;55:1233–1244. doi: 10.1093/pcp/pcu072. [DOI] [PubMed] [Google Scholar]
- Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Martinez M. From plant genomes to protein families: computational tools. Comput Struct Biotechnol J. 2013;8:e201307001. doi: 10.5936/csbj.201307001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gomez P, Sánchez-Pérez R, Rubio M. Clarifying omics concepts, challenges, and opportunities for Prunus breeding in the postgenomic era. OMICS. 2012;16:268–283. doi: 10.1089/omi.2011.0133. [DOI] [PubMed] [Google Scholar]
- Marx V. Biology: the big challenges of big data. Nature. 2013;498:255–260. doi: 10.1038/498255a. [DOI] [PubMed] [Google Scholar]
- McFarlane I, Park J, Ceddia G, Phipps R (2010) Economics of transgenic soyabean production: implications for Eu. In: 14th ICABR Conference, June 16–18, 2010, Ravello, Italy, 2010. vol 188103. International Consortium on Applied Bioeconomy Research (ICABR)
- Meena S, Kumar SR, Venkata Rao D, Dwivedi V, Shilpashree H, Rastogi S, Shasany AK, Nagegowda DA. De novo sequencing and analysis of lemongrass transcriptome provide first insights into the essential oil biosynthesis of aromatic grasses. Front Plant Sci. 2016;7:1129. doi: 10.3389/fpls.2016.01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewalal R, Mizrachi E, Mansfield SD, Myburg AA. Cell wall-related proteins of unknown function: missing links in plant cell wall development. Plant Cell Physiol. 2014;55:1031–1043. doi: 10.1093/pcp/pcu050. [DOI] [PubMed] [Google Scholar]
- Minami H, Kim J-S, Ikezawa N, Takemura T, Katayama T, Kumagai H, Sato F. Microbial production of plant benzylisoquinoline alkaloids. Proc Natl Acad Sci USA. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Shinozaki K. Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant Cell Physiol. 2011;52:2017–2038. doi: 10.1093/pcp/pcr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty S, Das MP, Surabhi GK. ‘OMICS’-approach to regulate ripening and enhance fruit shelf-life in banana: an important fruit crop for food security. Can J Biotechnol. 2017;1:289. [Google Scholar]
- Montgomery DR. Soil erosion and agricultural sustainability. Proc Natl Acad Sci USA. 2007;104:13268–13272. doi: 10.1073/pnas.0611508104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau AL, Janissen R, Santos CA, Peroni LA, Stach-Machado DR, de Souza AA, de Souza AP, Cotta MA. Highly-sensitive and label-free indium phosphide biosensor for early phytopathogen diagnosis. Biosens Bioelectron. 2012;36:62–68. doi: 10.1016/j.bios.2012.03.038. [DOI] [PubMed] [Google Scholar]
- Motte H, Vereecke D, Geelen D, Werbrouck S. The molecular path to in vitro shoot regeneration. Biotechnol Adv. 2014;32:107–121. doi: 10.1016/j.biotechadv.2013.12.002. [DOI] [PubMed] [Google Scholar]
- Mount David W. Bioinformatics–sequence and genome analysis. New York: CSHL; 2001. pp. 75–85. [Google Scholar]
- Muthamilarasan M, Dhaka A, Yadav R, Prasad M. Exploration of millet models for developing nutrient rich graminaceous crops. Plant Sci. 2016;242:89–97. doi: 10.1016/j.plantsci.2015.08.023. [DOI] [PubMed] [Google Scholar]
- Nadella K, Marla SS, Kumar PA. Metabolomics in agriculture. OMICS. 2012;16:149–159. doi: 10.1089/omi.2011.0067. [DOI] [PubMed] [Google Scholar]
- Nadolska-Orczyk A, Rajchel IK, Orczyk W, Gasparis S. Major genes determining yield-related traits in wheat and barley. Theor Appl Genet. 2017;130:1081–1098. doi: 10.1007/s00122-017-2880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik SK, Chand PK. Tissue culture-mediated biotechnological intervention in pomegranate: a review. Plant Cell Rep. 2011;30:707–721. doi: 10.1007/s00299-010-0969-7. [DOI] [PubMed] [Google Scholar]
- Naik PK, Mittal VK, Gupta S. RetroPred: a tool for prediction, classification and extraction of non-LTR retrotransposons (LINEs and SINEs) from the genome by integrating PALS, PILER, MEME and ANN. Bioinformation. 2008;2:263. doi: 10.6026/97320630002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naika M, Shameer K, Mathew OK, Gowda R, Sowdhamini R. STIFDB2: an updated version of plant stress-responsive transcription factor database with additional stress signals, stress-responsive transcription factor binding sites and stress-responsive genes in Arabidopsis and rice. Plant Cell Physiol. 2013;54:e8–e8. doi: 10.1093/pcp/pcs185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsaiah K, Jha SN, Bhardwaj R, Sharma R, Kumar R. Optical biosensors for food quality and safety assurance—a review. J Food Sci Technol. 2012;49:383–406. doi: 10.1007/s13197-011-0437-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson GC, Valin H, Sands RD, Havlík P, Ahammad H, Deryng D, Elliott J, Fujimori S, Hasegawa T, Heyhoe E. Climate change effects on agriculture: economic responses to biophysical shocks. Proc Natl Acad Sci USA. 2014;111:3274–3279. doi: 10.1073/pnas.1222465110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning M, Lo EH. Opportunities and challenges in omics. Transl Stroke Res. 2010;1:233–237. doi: 10.1007/s12975-010-0048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojiewo C, Monyo E, Desmae H, Boukar O, Mukankusi-Mugisha C, Thudi M, Pandey MK, Saxena RK, Gaur PM, Chaturvedi SK. Genomics, genetics and breeding of tropical legumes for better livelihoods of smallholder farmers. Plant Breed. 2018 doi: 10.1111/pbr.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okay S, Derelli E, Unver T. Transcriptome-wide identification of bread wheat WRKY transcription factors in response to drought stress. Mol Genet Genom. 2014;289:765–781. doi: 10.1007/s00438-014-0849-x. [DOI] [PubMed] [Google Scholar]
- Ozdemir V, Suarez-Kurtz G, Stenne R, Somogyi AA, Someya T, Kayaalp SO, Kolker E. Risk assessment and communication tools for genotype associations with multifactorial phenotypes: the concept of “edge effect” and cultivating an ethical bridge between omics innovations and society. OMICS. 2009;13:43–61. doi: 10.1089/omi.2009.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant Bijaya. Advances in Experimental Medicine and Biology. New Delhi: Springer India; 2014. Application of Plant Cell and Tissue Culture for the Production of Phytochemicals in Medicinal Plants; pp. 25–39. [DOI] [PubMed] [Google Scholar]
- Paritosh K, Gupta V, Yadava SK, Singh P, Pradhan AK, Pental D. RNA-seq based SNPs for mapping in Brassica juncea (AABB): synteny analysis between the two constituent genomes A (from B. rapa) and B (from B. nigra) shows highly divergent gene block arrangement and unique block fragmentation patterns. BMC Genet. 2014;15:396. doi: 10.1186/1471-2164-15-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak RK, Giri P, Taj G, Kumar A. Molecular modeling and docking approach to predict the potential interacting partners involved in various biological processes of MAPK with downstream WRKY transcription factor family in Arabidopsis thaliana. Int J Comput Bioinf In Silico Model. 2013;2:262–268. [Google Scholar]
- Pathak RK, Taj G, Pandey D, Arora S, Kumar A. Modeling of the MAPK machinery activation in response to various abiotic and biotic stresses in plants by a system biology approach. Bioinformation. 2013;9:443. doi: 10.6026/97320630009443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak RK, Taj G, Pandey D, Kasana VK, Baunthiyal M, Kumar A. Molecular modeling and docking studies of phytoalexin (s) with pathogenic protein (s) as molecular targets for designing the derivatives with anti-fungal action on Alternaria spp. of Brassica. Plant Omics. 2016;9:172. [Google Scholar]
- Pathak RK, Baunthiyal M, Shukla R, Pandey D, Taj G, Kumar A. In silico identification of mimicking molecules as defense inducers triggering jasmonic acid mediated immunity against alternaria blight disease in brassica species. Front Plant Sci. 2017;8:609. doi: 10.3389/fpls.2017.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak RK, Baunthiyal M, Pandey N, Pandey D, Kumar A. Modeling of the jasmonate signaling pathway in Arabidopsis thaliana with respect to pathophysiology of Alternaria blight in Brassica. Sci Rep. 2017;7:16790. doi: 10.1038/s41598-017-16884-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Fernández V, Dominguez-Vega E, Chankvetadze B, Crego AL, García M, Marina ML. Evaluation of new cellulose-based chiral stationary phases Sepapak-2 and Sepapak-4 for the enantiomeric separation of pesticides by nano liquid chromatography and capillary electrochromatography. J Chromatogr. 2012;1234:22–31. doi: 10.1016/j.chroma.2012.01.035. [DOI] [PubMed] [Google Scholar]
- Pickens LB, Tang Y, Chooi YH. Metabolic engineering for the production of natural products. Annu Rev Chem Biomol Eng. 2011;2:211–236. doi: 10.1146/annurev-chembioeng-061010-114209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasath D, Karthika R, Habeeba NT, Suraby EJ, Rosana OB, Shaji A, Eapen SJ, Deshpande U, Anandaraj M. Comparison of the transcriptomes of ginger (Zingiber officinale Rosc.) and mango ginger (Curcuma amada Roxb.) in response to the bacterial wilt infection. PLoS One. 2014;9:e99731. doi: 10.1371/journal.pone.0099731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priya P, Jain M. RiceSRTFDB: a database of rice transcription factors containing comprehensive expression, cis-regulatory element and mutant information to facilitate gene function analysis. Database. 2013 doi: 10.1093/database/bat027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha J. Bioprospecting of indigenous bioresources of North-East India. New York: Springer; 2016. [Google Scholar]
- Quirino B, Candido E, Campos P, Franco O, Krüger R. Proteomic approaches to study plant–pathogen interactions. Phytochemistry. 2010;71:351–362. doi: 10.1016/j.phytochem.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Rahman H, Jagadeeshselvam N, Valarmathi R, Sachin B, Sasikala R, Senthil N, Sudhakar D, Robin S, Muthurajan R. Transcriptome analysis of salinity responsiveness in contrasting genotypes of finger millet (Eleusine coracana L.) through RNA-sequencing. Annu Rev Plant Biol. 2014;85:485–503. doi: 10.1007/s11103-014-0199-4. [DOI] [PubMed] [Google Scholar]
- Rai A. Establishment of national agricultural bioinformatics grid in ICAR. NAIP project report. New Delhi: Indian Agricultural Statistics Research Institute; 2014. [Google Scholar]
- Rai M, Ingle A. Role of nanotechnology in agriculture with special reference to management of insect pests. Appl Microbiol Biotechnol. 2012;94:287–293. doi: 10.1007/s00253-012-3969-4. [DOI] [PubMed] [Google Scholar]
- Ramachandran P. Food and nutrition security: challenges in the new millennium. Indian J Med Res. 2013;138:373. [PMC free article] [PubMed] [Google Scholar]
- Rathore MS, Chikara J, Shekhawat N. Plantlet regeneration from callus cultures of selected genotype of Aloe vera L.—an ancient plant for modern herbal industries. Appl Biochem Biotechnol. 2011;163:860–868. doi: 10.1007/s12010-010-9090-1. [DOI] [PubMed] [Google Scholar]
- Reddy NRR, Mehta RH, Soni PH, Makasana J, Gajbhiye NA, Ponnuchamy M, Kumar J. Next generation sequencing and transcriptome analysis predicts biosynthetic pathway of sennosides from Senna (Cassia angustifolia Vahl.), a non-model plant with potent laxative properties. PLoS One. 2015;10:e0129422. doi: 10.1371/journal.pone.0129422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro D-K, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- Sadashivappa P, Qaim M. Bt cotton in India: Development of benefits and the role of government seed price interventions. Agric Biol Forum. 2009;12:172–183. [Google Scholar]
- Sam RE, Theodore MC, Marlin DE, Robert SR, Jason K. Molecular markers in a commercial breeding program. Crop Sci. 2007;47:154–163. [Google Scholar]
- Santi C, Bogusz D, Franche C. Biological nitrogen fixation in non-legume plants. Ann Bot. 2013;111:743–767. doi: 10.1093/aob/mct048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T. The map-based sequence of the rice genome. Nature. 2005;436:793. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Saslis-Lagoudakis CH, Savolainen V, Williamson EM, Forest F, Wagstaff SJ, Baral SR, Watson MF, Pendry CA, Hawkins JA. Phylogenies reveal predictive power of traditional medicine in bioprospecting. Proc Natl Acad Sci USA. 2012;109:15835–15840. doi: 10.1073/pnas.1202242109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena K, Nadarajan N. Prospects of pigeonpea hybrids in Indian agriculture. Electron J Plant Breed. 2010;1:1107–1117. [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2009;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheben A, Yuan Y, Edwards D. Advances in genomics for adapting crops to climate change. Curr Plant Biol. 2016;6:2–10. [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schweiger R, Baier MC, Persicke M, Müller C. High specificity in plant leaf metabolic responses to arbuscular mycorrhiza. Nat Commun. 2014;5:3886. doi: 10.1038/ncomms4886. [DOI] [PubMed] [Google Scholar]
- Scrinis G, Lyons K. The emerging nano-corporate paradigm: nanotechnology and the transformation of nature, food and agri-food systems. Int J Soc Agric Food. 2007;15:22–44. [Google Scholar]
- Seifert J, Herbst FA, Halkjær Nielsen P, Planes FJ, Jehmlich N, Ferrer M, Von Bergen M. Bioinformatic progress and applications in metaproteogenomics for bridging the gap between genomic sequences and metabolic functions in microbial communities. Proteomics. 2013;13:2786–2804. doi: 10.1002/pmic.201200566. [DOI] [PubMed] [Google Scholar]
- Sham A, Moustafa K, Al-Ameri S, Al-Azzawi A, Iratni R, AbuQamar S. Identification of Arabidopsis candidate genes in response to biotic and abiotic stresses using comparative microarrays. PLoS One. 2015;10:e0125666. doi: 10.1371/journal.pone.0125666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar R, Bhattacharjee A, Jain M. Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci Rep. 2016;6:23719. doi: 10.1038/srep23719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Tiwari R, Saharan M, Sharma I, Kumar J, Mishra S, Muthusamy SK, Gupta R, Jaiswal S, Iquebal M. Draft genome sequence of two monosporidial lines of the Karnal bunt fungus Tilletia indica Mitra (PSWKBGH-1 and PSWKBGH-2) Genome Ann. 2016;4:e00928–e00916. doi: 10.1128/genomeA.00928-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh AH, Raghuram B, Jalmi SK, Wankhede DP, Singh P, Sinha AK. Interaction between two rice mitogen activated protein kinases and its possible role in plant defense. BMC Plant Biol. 2013;13:121. doi: 10.1186/1471-2229-13-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekoofa A, Emam Y, Shekoufa N, Ebrahimi M, Ebrahimie E. Determining the most important physiological and agronomic traits contributing to maize grain yield through machine learning algorithms: a new avenue in intelligent agriculture. PLoS One. 2014;9:e97288. doi: 10.1371/journal.pone.0097288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Singh V, Bharadwaj D, Kumar R, Rai A, Rai A, Mugasimangalam R, Parameswaran S, Singh M, Naik P. De novo assembly of bitter gourd transcriptomes: gene expression and sequence variations in gynoecious and monoecious lines. PLoS One. 2015;10:e0128331. doi: 10.1371/journal.pone.0128331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MSP, Brandao DO, Chaves TP, Formiga Filho AL, Costa EMMdB, Santos VL, Medeiros ACD. Study bioprospecting of medicinal plant extracts of the semiarid northeast: contribution to the control of oral microorganisms. Evid Based Complement Alternat Med. 2012 doi: 10.1155/2012/681207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Singh AK. Marker-assisted plant breeding: principles and practices. New Delhi: Springer; 2015. [Google Scholar]
- Singh J, Behal A, Singla N, Joshi A, Birbian N, Singh S, Bali V, Batra N. Metagenomics: concept, methodology, ecological inference and recent advances. Biotechnol J Health Nutr Technol. 2009;4:480–494. doi: 10.1002/biot.200800201. [DOI] [PubMed] [Google Scholar]
- Singh C, Soni R, Jain S, Roy S, Goel R. Diversification of nitrogen fixing bacterial community using nifH gene as a biomarker in different geographical soils of Western Indian Himalayas. J Environ Biol. 2010;31:553–556. [PubMed] [Google Scholar]
- Singh A, Gopalakrishnan S, Singh V, Prabhu K, Mohapatra T, Singh N, Sharma T, Nagarajan M, Vinod K, Singh D. Marker assisted selection: a paradigm shift in Basmati breeding. Indian J Genet Plant Breed. 2011;71:120. [Google Scholar]
- Singh N, Gupta D, Jayaswal P, Mahato M, Dutta S, Singh S, Bhutani S, Dogra V, Singh B, Kumawat G. The first draft of the pigeonpea genome sequence. J Plant Biochem Biot. 2012;20:15. doi: 10.1007/s13562-011-0088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh HP, Kumar A, Parthasarathy VA, Singh B. Advances in horticulture biotechnology nanotechnology in agriculture. Westv Publ House. 2013;6:367. [Google Scholar]
- Singh S, Gupta S, Gupta AK, Singh M, Kumar A. Surface plasmon resonance sensogram based characterization of antibodies raised against intact teliospores and purified diagnostic antigen for development of nano-immunosensor for fungal spore antigen of karnal bunt (Tilletia indica) of wheat. Proc Natl Acad Sci India Sect B Biol Sci. 2013;83:551–560. [Google Scholar]
- Singh N, Mahato A, Sharma N, Gaikwad K, Srivastava M, Tiwari K, Dogra V, Rawal S, Rajan S, Singh A (2014a) A draft genome of the king of fruit, mango (Mangifera indica L.). In: Plant and Animal Genome XXII Conference
- Singh A, Krishnan SG, Nagarajan M, Vinod K, Bhowmick P, Atwal S, Seth R, Chopra N, Chander S, Singh V. Variety Pusa Basmati 1509. Indian J Genet Pl Br. 2014;74:123–123. [Google Scholar]
- Smita S, Lenka SK, Katiyar A, Jaiswal P, Preece J, Bansal KC. QlicRice: a web interface for abiotic stress responsive QTL and loci interaction channels in rice. Database. 2011 doi: 10.1093/database/bar037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon W, Baliunas SL, Robinson AB, Robinson ZW. Environmental effects of increased atmospheric carbon dioxide. Climate Res. 1999;13:149–164. [Google Scholar]
- Sprocati AR, Alisi C, Tasso F, Fiore A, Marconi P, Langella F, Haferburg G, Nicoara A, Neagoe A, Kothe E. Bioprospecting at former mining sites across Europe: microbial and functional diversity in soils. Environ Sci Pollut Res. 2014;21:6824–6835. doi: 10.1007/s11356-013-1907-3. [DOI] [PubMed] [Google Scholar]
- Sultan B. Global warming threatens agricultural productivity in Africa and South Asia. Environ Res Lett. 2012;7:041001. [Google Scholar]
- Suyal DC, Yadav A, Shouche Y, Goel R. Differential proteomics in response to low temperature Diazotrophy of Himalayan psychrophilic nitrogen fixing Pseudomonasmigulae S10724 strain. Curr Microbiol. 2014;68:543–550. doi: 10.1007/s00284-013-0508-1. [DOI] [PubMed] [Google Scholar]
- Talukdar M, Duarah A, Talukdar S, Gohain MB, Debnath R, Yadav A, Jha DK, Bora TC. Bioprospecting Micromonospora from Kaziranga National Park of India and their anti-infective potential. World J Microbiol Biotechnol. 2012;28:2703–2712. doi: 10.1007/s11274-012-1080-8. [DOI] [PubMed] [Google Scholar]
- Thakur K, Chawla V, Bhatti S, Swarnkar MK, Kaur J, Shankar R, Jha G. De novo transcriptome sequencing and analysis for Venturia inaequalis, the devastating apple scab pathogen. PLoS One. 2013;8:e53937. doi: 10.1371/journal.pone.0053937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao NP, Tran VL-SP. Enhancement of plant productivity in the post-genomics era. Curr Genom. 2016;17:295. doi: 10.2174/138920291704160607182507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari JK, Plett D, Garnett T, Chakrabarti SK, Singh RK. Integrated genomics, physiology and breeding approaches for improving nitrogen use efficiency in potato: translating knowledge from other crops. Funct Plant Biol. 2018;45:587–605. doi: 10.1071/FP17303. [DOI] [PubMed] [Google Scholar]
- Toenniessen GH, O’Toole JC, DeVries J. Advances in plant biotechnology and its adoption in developing countries. Curr Opin Plant Biol. 2003;6:191–198. doi: 10.1016/s1369-5266(03)00002-5. [DOI] [PubMed] [Google Scholar]
- Tohge T, de Souza LP, Fernie AR. Genome-enabled plant metabolomics. J Chromatogr B. 2014;966:7–20. doi: 10.1016/j.jchromb.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Tsugita A, Kamo M (1999) 2-D electrophoresis of plant proteins. In: Andrew J (ed) 2-D Proteome Analysis Protocols. Springer, pp 95–97
- Upadhyay AK, Chacko AR, Gandhimathi A, Ghosh P, Harini K, Joseph AP, Joshi AG, Karpe SD, Kaushik S, Kuravadi N. Genome sequencing of herb Tulsi (Ocimum tenuiflorum) unravels key genes behind its strong medicinal properties. BMC Plant Biol. 2015;15:212. doi: 10.1186/s12870-015-0562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay P, Rai A, Kumar R, Singh M, Sinha B. Microarray analyses during early stage of the tomato/Alternaria solani interaction. Genom Data. 2015;6:170–172. doi: 10.1016/j.gdata.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay P, Ganie SH, Rai A, Singh M, Sinha B. Identification of transcription factors in tomato, potentially related to early blight resistance at invasion in host tissue, using microarray expression profiling. S Afr J Bot. 2016;106:165–173. [Google Scholar]
- Valkov Vladimir T., Gargano Daniela, Scotti Nunzia, Cardi Teodoro. Methods in Molecular Biology. Totowa, NJ: Humana Press; 2014. Plastid Transformation in Potato: Solanum tuberosum; pp. 295–303. [DOI] [PubMed] [Google Scholar]
- Van Norman JM, Benfey PN. Arabidopsis thaliana as a model organism in systems biology. Wiley Interdiscip Rev Syst Biol Med. 2009;1:372–379. doi: 10.1002/wsbm.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Eeckhout D, Cannoot B, De Winne N, Persiau G, Van De Slijke E, Vercruysse L, Dedecker M, Verkest A, Vandepoele K. An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat Protoc. 2015;10:169. doi: 10.1038/nprot.2014.199. [DOI] [PubMed] [Google Scholar]
- Vanderschuren H, Lentz E, Zainuddin I, Gruissem W. Proteomics of model and crop plant species: status, current limitations and strategic advances for crop improvement. J Proteomics. 2013;93:5–19. doi: 10.1016/j.jprot.2013.05.036. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Shi C, Thudi M, Mariac C, Wallace J, Qi P, Zhang H, Zhao Y, Wang X, Rathore A. Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat Biotechnol. 2017;35:969. doi: 10.1038/nbt.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney Rajeev K., Pandey Manish K., Chitikineni Annapurna. Plant Genetics and Molecular Biology. Cham: Springer International Publishing; 2018. Plant Genetics and Molecular Biology: An Introduction; pp. 1–9. [DOI] [PubMed] [Google Scholar]
- Verma S, Pathak RK, Kasana V, Kumar A. Binding affinity analysis of cinnamanilide and alpha-aminophosphonic acid derivatives for acetohydroxyacid synthase through molecular docking. Int J Agric Environ Biotechnol. 2017;10:271. [Google Scholar]
- Vij S, Tyagi AK. Emerging trends in the functional genomics of the abiotic stress response in crop plants. Plant Biotechnol J. 2007;5:361–380. doi: 10.1111/j.1467-7652.2007.00239.x. [DOI] [PubMed] [Google Scholar]
- Wambugu PW, Ndjiondjop M-N, Henry RJ. Role of genomics in promoting the utilization of plant genetic resources in genebanks. Brief Funct Genomics. 2018;17:198–206. doi: 10.1093/bfgp/ely014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Zaki MJ, Toivonen HT, Shasha D (2005) Introduction to data mining in bioinformatics. In: Data Mining in Bioinformatics. Springer, New York, pp 3–8
- Wang B, Zhang Z, Yin Z, Feng C, Wang Q. Novel and potential application of cryopreservation to plant genetic transformation. Biotechnol Adv. 2012;30:604–612. doi: 10.1016/j.biotechadv.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Losak T, Vollmann J. From proteomics to ionomics: soybean genetic improvement for better food safety. Genetika-belgrade. 2018;50:333–350. [Google Scholar]
- Weckwerth W. Green systems biology—from single genomes, proteomes and metabolomes to ecosystems research and biotechnology. J Proteom. 2011;75:284–305. doi: 10.1016/j.jprot.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Wei L, Xiao M, Hayward A, Fu D. Applications and challenges of next-generation sequencing in Brassica species. Planta. 2013;238:1005–1024. doi: 10.1007/s00425-013-1961-6. [DOI] [PubMed] [Google Scholar]
- Weisenburger DD. Human health effects of agrichemical use. Hum Pathol. 1993;24:571–576. doi: 10.1016/0046-8177(93)90234-8. [DOI] [PubMed] [Google Scholar]
- Weiss W, Görg A (2007) Two-dimensional electrophoresis for plant proteomics. In: Plant proteomics, Springer, pp 121–143 [DOI] [PubMed]
- Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun. 2014;5:3438. doi: 10.1038/ncomms4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing JM. Computational thinking. Commun ACM. 2006;49:33–35. [Google Scholar]
- Wortman SE, Lovell ST. Environmental challenges threatening the growth of urban agriculture in the United States. J Environ Qual. 2013;42:1283–1294. doi: 10.2134/jeq2013.01.0031. [DOI] [PubMed] [Google Scholar]
- Wu X, Chang X, Jing R. Genetic insight into yield-associated traits of wheat grown in multiple rain-fed environments. PLoS One. 2012;7:e31249. doi: 10.1371/journal.pone.0031249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Wang X (2018) Comparative transcriptomic analysis identifies genes responsible for fruit count and oil yield in the oil tea plant Camellia chekiangoleosa. Sci Rep 8 [DOI] [PMC free article] [PubMed]
- Yue S, Li P, Hao P. Svm classification: its contents and challenges. Appl Math J Chin Univ. 2003;18:332–342. [Google Scholar]
- Zandalinas SI, Mittler R, Balfagón D, Arbona V, Gómez-Cadenas A. Plant adaptations to the combination of drought and high temperatures. Physiol Plant. 2018;162:2–12. doi: 10.1111/ppl.12540. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao P, Yuan JS. Plant protein-protein interaction network and interactome. Curr Genom. 2010;11:40–46. doi: 10.2174/138920210790218016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotchev SB, Sekurova ON, Katz L. Genome-based bioprospecting of microbes for new therapeutics. Curr Opin Biotechnol. 2012;23:941–947. doi: 10.1016/j.copbio.2012.04.002. [DOI] [PubMed] [Google Scholar]