Abstract
Vancomycin is one of the most commonly used glycopeptide antiobiotics, and as such is an important emerging environmental contaminant. Pharmaceuticals and personal care products (PPCPs), such as antibiotics, are problematic since wastewater treatment processes are not completely effective at removing these chemical compounds. Since wastewater treatment processes are not completely effective, vancomycin occurs in surface water. Vancomycin and its metabolites in vivo and degradation products in aquatic environment may lead to undesirable ecological effects that threaten the environment or cause undesirable reactions that affect human health. We aimed to study vancomycin metabolism in vitro and its natural degradation in aquatic environment, as well as explore for related metabolites and degradation products. Accordingly, we established four systems, using a constant temperature oscillator at 37 °C for 10 days for vancomycin in activated rat liver microsomes (experimental system), inactivated rat liver microsomes (control system), phosphate buffer saline (PBS system) and pure water (pure water system), as well as an additional system of activated rat liver microsomes without vancomycin (blank system). The metabolism and degradation of vancomycin were studied using a high resolution and high sensitivity ultra-high performance liquid chromatography (UHPLC)-Triple-time of flight (TOF)-mass spectrometry (MS) method in positive ion mode. The compared result of activated rat liver microsomes system and inactivated rat liver microsomes system confirms that vancomycin is not metabolized in the liver. Vancomycin was degraded in the four non-blank incubation systems. The MetabolitePilot 2.0 software was used for screening the probable degradation products, as well as for establishing its associated degradation pathways. Eventually, four degradation products were identified and their chemical structures were deduced. The results of this study provide a foundation for evaluation of the effects of vancomycin and its degradation products on environmental safety and human health in the future.
Introduction
Pharmaceuticals and personal care products (PPCPs) are widely concerning as a group of emerging environmental contaminants due to their possible threats to aquatic environment and human health1,2. PPCPs comprises numerous diverse products, including pharmaceuticals (human clinical medicine and veterinary drugs) and active ingredients in personal care products (PCPs)3. The deposited PPCPs may either retain their original concentrations and structures or be metabolized and converted into other active (or inactive) compounds during their lifespan in aquatic matrices4. Indeed, not only PPCPs itself, but also their metabolites and degradation products in the environment can have a potential negative impact on the environment, living organisms and human health by means of migration or accumulation5,6.
As a commonly used clinical drug, vancomycin is one of the PPCPs that is of significant concern. Among all the glycopeptide antibiotics in clinical use, vancomycin is the most commonly used to treat serious infections like endocarditis, pneumonia and meningitis, caused by vancomycin-susceptible Gram-positive organisms, such as methicillin-resistant Staphylococcus aureus (MRSA)7,8. In Italy and Germany, treated urban discharges can be an entry route of vancomycin into surface water, because wastewater treatment processes are not completely effective in removing vancomycin9. In French Rivers, vancomycin has been detected at concentrations reaching 90 ng/L10. Vancomycin is ingested by humans and animals through drinking water and may be biotransformed into related metabolites in vivo. Vancomycin may be degraded in an aquatic environment to produce related degradation products. Vancomycin and its metabolites and degradation products in aquatic environment may lead to detrimental ecological effects that threaten the environment or produce adverse reactions that affect human health. The ecological risk of antibiotics in the aquatic environment is a growing concern. Vancomycin, as an antibiotic, may have acute or chronic ecotoxicity on bacteria, algae, invertebrates, and fish in the aquatic environment9. The increased risk of renal toxicity due to vancomycin was found to be likely associated with its higher daily dose and longer duration of ingestion by eating, drinking and treating11,12. Drug toxicity may result from the parent compound or from its metabolites13–15. Nevertheless, no data on the metabolism or degradation of vancomycin are currently available. Therefore, we aim to find related metabolites or degradation products by studying vancomycin metabolism in vitro and natural degradation in aquatic environment. This would provide a foundation for the study of the effects of vancomycin and its degradation products on environmental safety and human health in the future.
Drug metabolism research can be divided into in vivo and in vitro metabolism studies16–18. The liver is the main organ for biotransformation, with cytochrome P450 enzyme system playing an important role in drug metabolism19,20. As drug metabolism may be altered by NADPH concentrations in vivo and in vitro21, appropriate concentrations of NADPH should be used. The in vitro metabolism model based on liver microsomes has been widely used in drug metabolism research and has the advantages of being fast and accurate, and having a high throughput22–24. In vitro, liver microsomal incubation simulates the physiological environment of metabolism in the liver.
To the best of our knowledge, there are only a few studies about the metabolism of vancomycin. Aboleneen et al., used high-performance liquid chromatography (HPLC) to determine the serum concentration of vancomycin and the major and minor isomers of its degradation product, CDP-1, in patients with renal impairment25. White et al., used a microbiological assay, HPLC and a polarisation fluoroimmunoassay to measure the vancomycin concentration in phosphate-buffered saline, peritoneal dialysis effluent fluid and human serum after daily sampling for 10 days of incubation at 37 °C, but vancomycin metabolites were not determined26. The aquatic ecological environment is complex and diverse, containing organics and mineral salt, etc27,28. Accordingly, the existing rat liver microsomes system and phosphate buffer saline (PBS) system may simulate the aquatic environment containing organics and mineral salt respectively. Besides being a simulation of the basic aquatic environment, the pure water system is also a contrasting environment to other types of aquatic environments. Therefore, we elected to establish five systems including activated rat liver microsomes (experimental system), inactivated rat liver microsomes (control system), activated rat liver microsomes without vancomycin (blank system), phosphate buffer saline (PBS system) and pure water (pure water system) to study vancomycin metabolism in vitro and natural degradation in an aquatic environment.
For drug metabolism or degradation studies, liquid chromatography mass spectrometry (LC-MS) has become the most commonly used technique to discover previously unreported metabolites and degradation products. This is especially true for time-of-flight (TOF) mass spectrometry (MS) instruments, which have the advantages of excellent sensitivity, high resolution, accurate mass detection, fast analysis, and comprehensive detection of the nature of the metabolite29,30. Widespread use of the AB SCIEX triple TOF 5600 MS/MS system is attributed to its online data acquisition, which is followed by comprehensive data processing techniques including extracted ion chromatography (EIC), mass defect filtering (MDF), product ion filtering (PIF), neutral loss filtering (NLF) and isotope pattern filtering. The information dependent acquisition (IDA) criteria are provided to the ions that match the mass defect window to obtain the MS/MS spectra. This enables the chemical structure of the compound to be deduced31. Thus, in this study, samples were analyzed by ultra-high performance liquid chromatography connected to an AB SCIEX triple TOF 5600 MS/MS system to identify related metabolites and degradation products. This study has an important hygienic significance for evaluation of the effects of vancomycin on environmental safety and human health in the future.
Materials and Methods
Chemicals and reagents
Vancomycin hydrochloride was purchased from Dr. Ehrenstorfer GmbH (Augsburg,Germany). Nicotinamide adenine dinucleotide phosphate (NADPH) and phosphate buffer saline (PBS) were purchased from Shanghai Yisheng Biotechnology Co., Ltd (Shanghai, China) and Sangon Biotech Co., Ltd (Shanghai, China), respectively. Analytical grade magnesium chloride (MgCl2) was obtained from Yongda Chemical Reagent Co., Ltd (Tianjin, China). Acetonitrile and formic acid of HPLC grade were purchased from Thermo Fisher Scientific Co., Ltd (Shanghai, China) and DikmaPure Technologies Company (Beijing, China), respectively. HPLC grade methanol was purchased from Sunrise Chem, INC (New York, USA). Pure water was prepared by the Milli-Q system made in Millipore Corporation (Billerica, USA).
Standard solutions preparation
Vancomycin and NADPH were dissolved in pure water at the exact concentrations of 6 g/L and 10 mmol/L when they were used, respectively. MgCl2 was previously dissolved in PBS at a concentration of 13.3 mmol/L.
Chromatographic and mass spectrometric conditions
UHPLC-Triple-TOF-MS/MS analysis was performed on a Shimadzu LC-30A UHPLC system (Kyoto, Japan) consisting of an autosampler (SIL-30AC), a binary pump (LC-30AD) and a column oven (CTO-30A) connected to an AB SCIEX triple TOF 5600 MS/MS system (Redwood, USA) equipped with Duo-Spray ion source. The compound separation was conducted with a Poroshell 120 EC-C18 (2.1 × 150 mm, 2.7 μm, Agilent InfinityLab, USA) chromatographic column maintained at 40 °C. The mobile phases A (0.1% aqueous formic acid) and B (acetonitrile) were used to optimized the gradient elution program, which was operated at follows: 0–1 min, 5% B; 1–9 min, 5–10% B; 9–14 min, 10–45% B; 14–20 min, 45–90% B; 20–25 min, 90% B for washing the column; 25–25.1 min, 90–5% B; 25.1–30 min, 5% B for equilibrating the column. The constant flow rate was 0.4 mL/min, and the injection volume was 5 µL.
The full-scan MS spectra and MS/MS spectra data were obtained in positive electrospray ionization (ESI) mode by Analyst TF 1.6 software (AB Sciex, USA). The optimized parameters of the high resolution mass spectrometer are shown in Table 1. Full scan mode was applied, the parent ions scan ranged from m/z 100 to 2000 Da with a 200 ms accumulation time and the product ions scan range from 100 to 1500 Da with a 70 ms accumulation time. Simultaneously, the calibration delivery system calibrated mass numbers by every five samples to acquire exact mass of ions. The running time of data independent acquisition (DIA) was 20 min.
Table 1.
The optimized parameters of the high resolution mass spectrometer.
| Parameters | Value |
|---|---|
| Ion source temperature (°C) | 550 |
| IonSpray voltage floating (V) | 5500 |
| Ion source gas 1 (psi) | 50 |
| Ion source gas 2 (psi) | 50 |
| Curtain gas (psi) | 35 |
| Collision energy (eV) | 46 |
| Declustering potential (eV) | 80 |
Preparation of rat liver microsomes
Ten male Sprague-Dawley (SD) rats (250 ± 20 g) were provided by the Experimental Animal Research Center at Hebei Medical University. The animal experimentation were approved by the Ethics Committee of Hebei Medical University (Approval Number 2018012). Rat liver was removed immediately after decapitation and then weighed blotting the excess blood with a filter paper. Liver microsomes were prepared by differential centrifugation after the liver was repeatedly washed with a cold solution (10 mmol/L Tris-HCl, 250 mmol/L sucrose and 1 mmol/L ethylene diamine tetraacetic acid (EDTA) at pH 7.4), shredded and homogenized on ice. A full description of the procedures can be obtained elsewhere32. Eventually, the Lowery protein assay was used to measure protein concentrations in the rat liver microsome suspension33.
Experimental design
In this study, five systems (experimental, control, blank, PBS and pure water systems) were established. The five systems each included 11 samples, in a final volume of 200 µL. All systems, except the blank system, contained a final vancomycin concentration of 60 mg/L26. The experimental, control and blank systems contained PBS and MgCl2 at a final concentration of 3.3 mmol/L. The experimental and blank systems contained 1 mg/mL of activated rat liver microsome protein, and the control system contained 1 mg/ml of inactivated rat liver microsome protein. After preheating the incubation system at 37 °C for 5 min, NADPH at 1 mmol/L was added to the experimental and blank systems, while the same volume of pure water was added to the control system. Vancomycin was diluted in PBS and pure water in the PBS system and pure water system, respectively (details in Table 2). One sample from each system was placed directly in the freezer at −20 °C (0 h), and the remaining samples of the incubation systems were placed in a constant temperature oscillator at 37 °C for 1–10 days. One sample from each system was removed every 24 hours for ten consecutive days, adding 1 mL pre-cooled methanol immediatel yafter removal to the experimental, control and blank systems. All samples were stored at −20 °C until further analysis.
Table 2.
Five systems of vancomycin.
| 200 µL System | MgCl2 (µL) | NADPH (µL) | Active rat liver microsomes(µL) | Inactiverat liver microsomes(µL) | Vancomycin (µL) | PBS (µL) | Pure water (µL) |
|---|---|---|---|---|---|---|---|
| experimental | 50 | 20 | 20 | — | 2 | 108 | — |
| control | 50 | — | — | 20 | 2 | 108 | 20 |
| blank | 50 | 20 | 20 | — | — | 108 | 2 |
| PBS | — | — | — | — | 2 | 198 | — |
| pure water | — | — | — | — | 2 | — | 198 |
Pretreatment of samples
All the samples were centrifuged (10,625 g for 10 min) including experimental, control and blank systems. The supernatant was completely transferred to a clean glass tube and dried with a slow stream of nitrogen at 37 °C. The dried substances were dissolved with 200 µL 30% methanol before determining.
Quality control (QC) sample
A QC sample of vancomycin in pure water was prepared with a concentration of 60 mg/L. The same QC samples were injected at the start of the run and then again every eleven samples.
Data analysis
Comparing of the raw data in five systems by MetabolitePilot 2.0 software (AB Sciex, USA), we can acquired the information on probable metabolites or degradation products, such as biotransformation, molecular formula, retention time (RT), m/z values, relative error (ER) and score. According to the full-scan MS spectra and MS/MS spectra of the probable metabolites or degradation products provided by PeakView 2.2 software (AB Sciex, USA) and the structure characterization of vancomycin analysed by Mass Frontier 5.0 software (Thermo Fisher Scientific, USA), the probable chemical structures of the metabolites or degradation products could be calculated.
Results and Discussion
Degradation of vancomycin
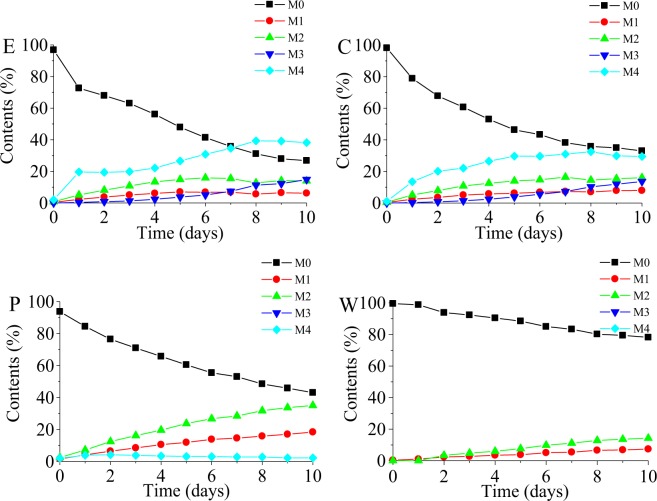
During the 1–10-day incubation period, the concentration of vancomycin (M0) decreased in the experimental, control, PBS and pure water systems (Fig. 1). A similar degradation rate in the experimental and control systems over the early time period of the incubation, when the microsomes are viable, may be attributed to the weak effect of activated rat liver microsomes cytochrome P450 enzymes on vancomycin. The results confirmed that vancomycin is not metabolized in the liver and most of the vancomycin ingested in the body is excreted in the urine. The most marked loss of vancomycin was in rat liver microsomes, which was degraded 50% over approximately 6 days. The time to 50% loss of vancomycin in PBS or in water was approximately 9 days and over 10 days, respectively, which showed that vancomycin can be degraded in an aquatic environment. Organics and mineral salt may be favorable to the degradation of vancomycin.
Figure 1.
The degradation of vancomycin and the generated trends of four degradation products in experimental system (E), control system (C), PBS system (P) and pure water system (W) at 37 °C for 10 days.
Mass spectrometry fragmentation patterns of vancomycin
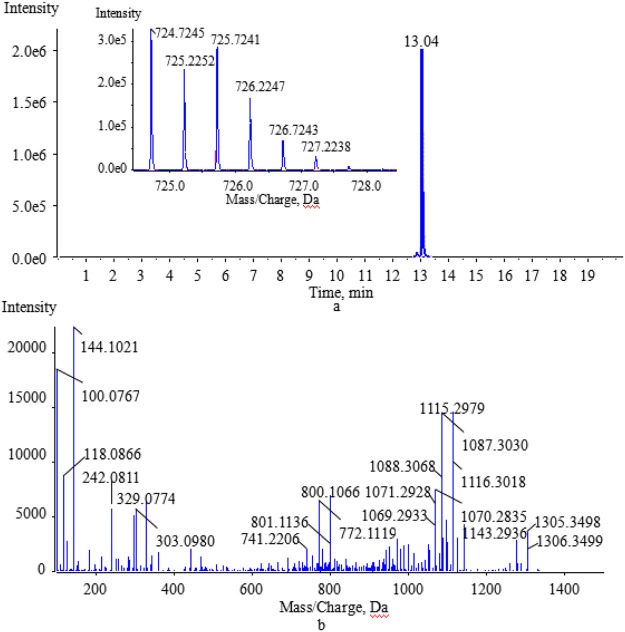
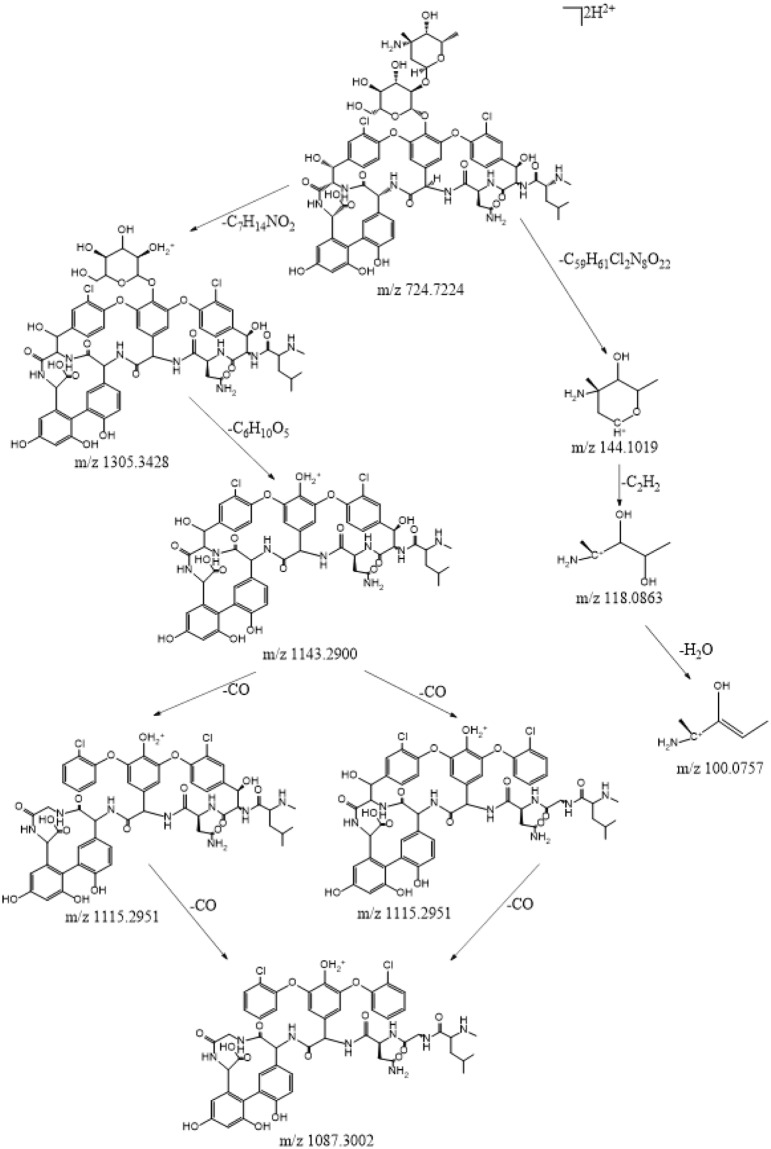
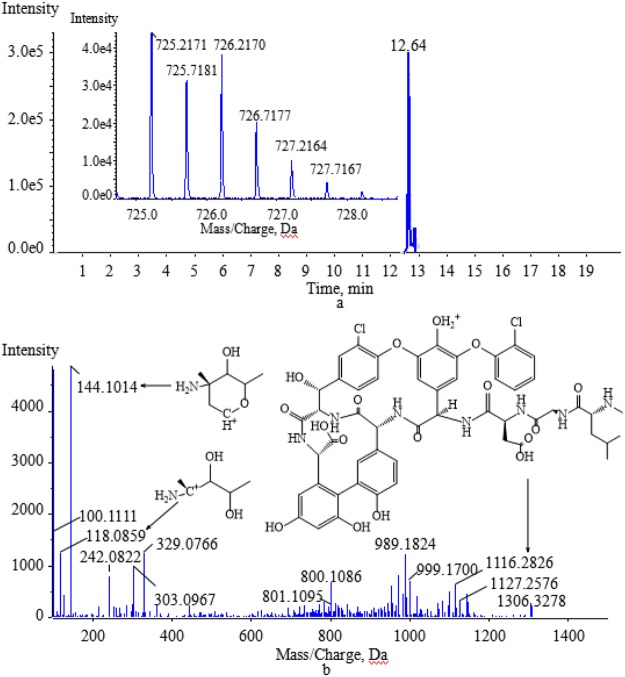
The extracted ion chromatogram and the MS fragmentation patterns of vancomycin, reveal that vancomycin eluted at 13.04 min with a parent ion at m/z 724.7246 with two charges (Fig. 2a). The chemical structure of vancomycin is very complex, and includes aminoglycoside and polypeptide. Combined with the MS/MS spectra of vancomycin (Fig. 2b), the MS/MS fragmentation behaviors were analyzed by inference and the ‘Fragmentation Library function’ in the Mass Frontier 5.0 software. The results of the analysis suggested that vancomycin was initially fragmented into the fragment ions m/z 144.1019 (F1) and 1305.3428 (F2). The two fragment ions then produced a series of characteristic product ions at m/z 118.0863 [F1-(-C2H2)]+ (F3), 100.0757 [F3-(-H2O)]+ (F4), 1143.2900 [F2-(-C6H10O5)]+ (F5), 1115.2951 [F5-(-CO)]+ (F6,F7) and 1087.3002 [F6,F7-(-CO)]+ (F8). The main ions were at m/z 100.0767, 144.1021, 1115.2979, 1087.3030 in the MS/MS spectrum (Fig. 2b)34–36. The proposed fragmentation pathways for vancomycin are shown in Fig. 3. The fragment ions at m/z 100.0767, 118.0866 and 144.1021 were produced from fragmentation of the aminoglycoside of vancomycin. Fragmentation of the polypeptide occurred with the loss of two CO to produce characteristic product ions at m/z 1115.2979 and 1087.3030. According to the characteristic fragmentation patterns of vancomycin, the probable chemical structures of the degradation products could be further deduced37.
Figure 2.
The extract ion chromatogram, MS(a) and MS/MS(b) spectra of vancomycin.
Figure 3.
The proposed fragmentation pathways of vancomycin.
Identification of the degradation products
The procedure to identify likely degradation products
The Triple TOF MS/MS instrument, data independent acquisition (DIA) and various techniques to process the raw data were used to identify likely degradation products.
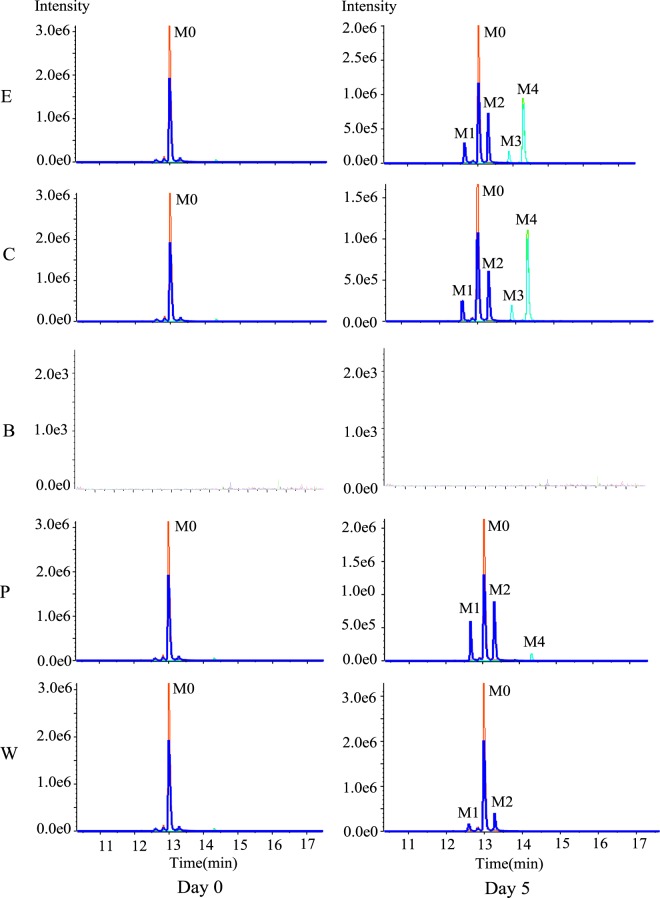
First, the raw data from the experimental, control, PBS and pure water systems were compared with the blank system and a prepared standard solution of vancomycin using the MetabolitePilot 2.0 software, and the likely degradation products were acquired. Subsequently, the data for likely degradation products were acquired and confirmed using many data-mining functions in the PeakView software, such as extracted ion chromatogram (XIC), information dependent acquisition (IDA) explorer, mass calculator and formula finder. Ultimately, four likely degradation products (M1, M2, M3 and M4) of vancomycin (M0) were identified. The biotransformation pathway, molecular formula, retention time, theoretical and observed mass, mass error and score for vancomycin and each of its four likely degradation products are outlined in Table 3. For high quality and accurate mass spectra, the relative error (RE) between the theoretical and observed values for the ion mass were less than 5 ppm38,39. Accordingly, the inferred element composition of the four likely degradation products was acceptable, as the RE for vancomycin and the four likely degradation products was less than 5 ppm. Using the XIC function in the PeakView software, the extracted ion chromatograms of the experimental, control, blank, PBS and pure water systems were obtained by applying the characteristic ion at m/z 724.7245, 725.2171, 725.2167, 731.2164, 730.7239 of vancomycin and the four likely degradation products. The comparison between day 0 and day 5 of incubation is shown in Fig. 4. The chromatograms reveal that M1, M2, M3 and M4 were present in the experimental and control systems. Additionally, no degradation products were detected in the blank system, but M1, M2 and M4 were detected in the PBS system, and only M1 and M2 were detected in the pure water system.
Table 3.
The information of vancomycin and four degradation products.
| Peak ID | Metabolite discription | Molecular Formula | R.T. (min) | Theoretical Mass | Observed Mass | RE (ppm) | MS m/z [M + 2H]2+ | MS/MS Fragments | Score (%) |
|---|---|---|---|---|---|---|---|---|---|
| M0 | Parent | C66H75Cl2N9O24 | 13.04 | 1447.4302 | 1447.4344 | 2.9 | 724.7245 | 100.0767 | 97.7 |
| 118.0866 | |||||||||
| 144.1021 | |||||||||
| 1087.303 | |||||||||
| 1115.2979 | |||||||||
| 1043.2936 | |||||||||
| 1305.3498 | |||||||||
| M1 | Oxidative Deamination to Alcohol | C66H74Cl2N8O25 | 12.64 | 1448.4142 | 1448.4197 | 3.8 | 725.2171 | 118.0859 | 86.6 |
| 144.1014 | |||||||||
| 1116.2826 | |||||||||
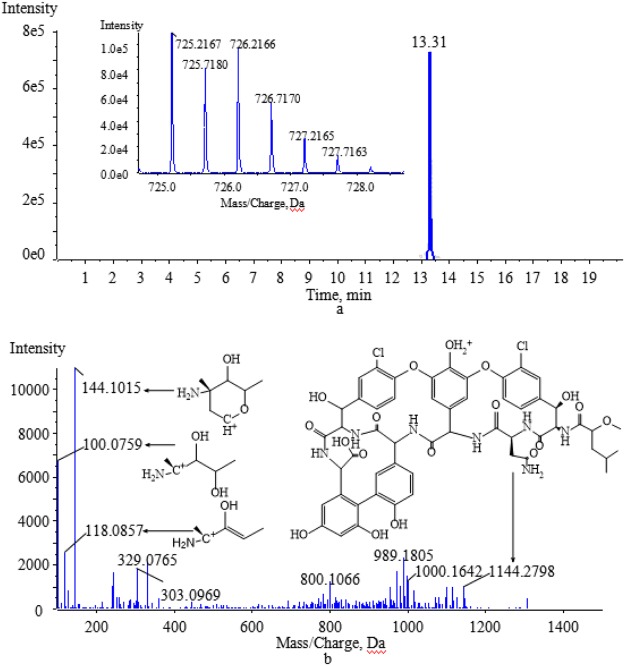
| M2 | Loss of CH3N + Oxidation and Methylation | C66H74Cl2N8O25 | 13.31 | 1448.4142 | 1448.4189 | 3.2 | 725.2167 | 100.0759 | 82.4 |
| 118.0857 | |||||||||
| 144.1015 | |||||||||
| 1144.2798 | |||||||||
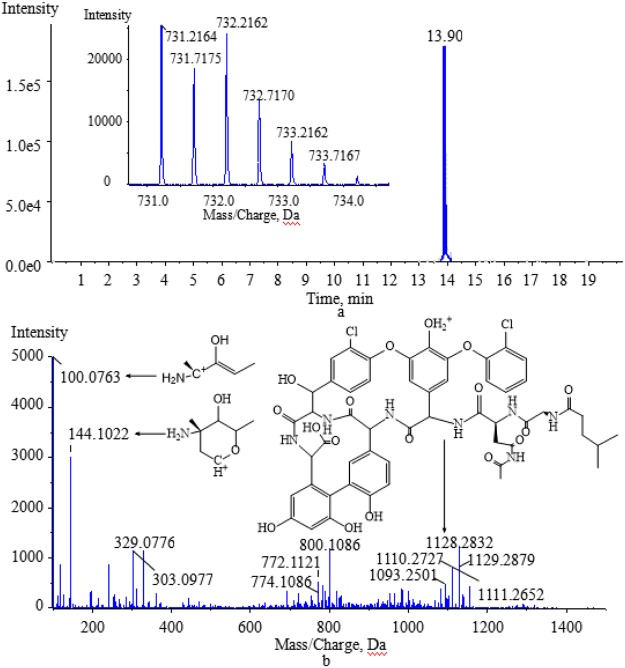
| M3 | Loss of CH3N + N-Acetylation | C67H74Cl2N8O25 | 13.9 | 1460.4142 | 1460.4185 | 2.9 | 731.2164 | 100.0763 | 83.8 |
| 144.1022 | |||||||||
| 1128.2832 | |||||||||
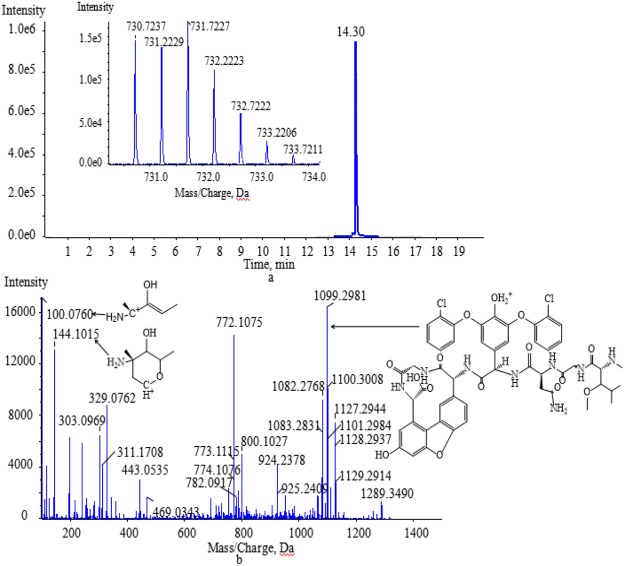
| M4 | Loss of water + Oxidation and Methylation | C67H75Cl2N9O24 | 14.3 | 1459.4302 | 1459.4344 | 2.9 | 730.7239 | 100.076 | 82.9 |
| 144.1015 | |||||||||
| 1099.2981 |
Figure 4.
The XIC of vancomycin and the four vancomycin degradation products. M0, M1, M2, M3, M4 in experimental system (E) and control system (C); no metabolites in blank system (B); M0, M1, M2, M4 in PBS system (P); M0, M1, M2 in pure water system (W).
The contents of the degradation products
The relative standard deviation (RSD) of the vancomycin intensity in the QC samples was 14.41%, which was less than 15%, thus indicating that the repeatability and stability were acceptable40,41. Based on the definition that the total content of vancomycin and its four degradation products is 100%, the related content of vancomycin and its four degradation products were calculated using the normalization method. The trends for vancomycin and the four degradation products during the 1–10-day incubation period in the experimental, control, PBS and pure water systems are shown in Fig. 1. In the experimental and control systems, M1 and M2 increased with time over the first seven days and then decreased slightly from day 8. Compared with the other three systems, M1 and M2 had the fastest increase in the PBS system, with M2 increasing faster than M1. This may be due to the stimulation and inhibition of the formation of M1 and M2 by the mineral salt and organics, respectively. M3 was only produced in the experimental and control systems. Compared with the other three degradation products, M3 was the lowest produced in the first seven days and then exceeded M1 after seven days. M4 showed an obvious upward trend with the longer time in both the experimental and control systems, and increased at the fastest rate of the four degradation products. M4 showed a trend to increase in the first two days in the PBS system and then decreased slightly from day 3. The M4 content was the lowest in the PBS system, and was not found in the pure water system. A possible explanation for this is that organics were favorable to M3 and M4 production.
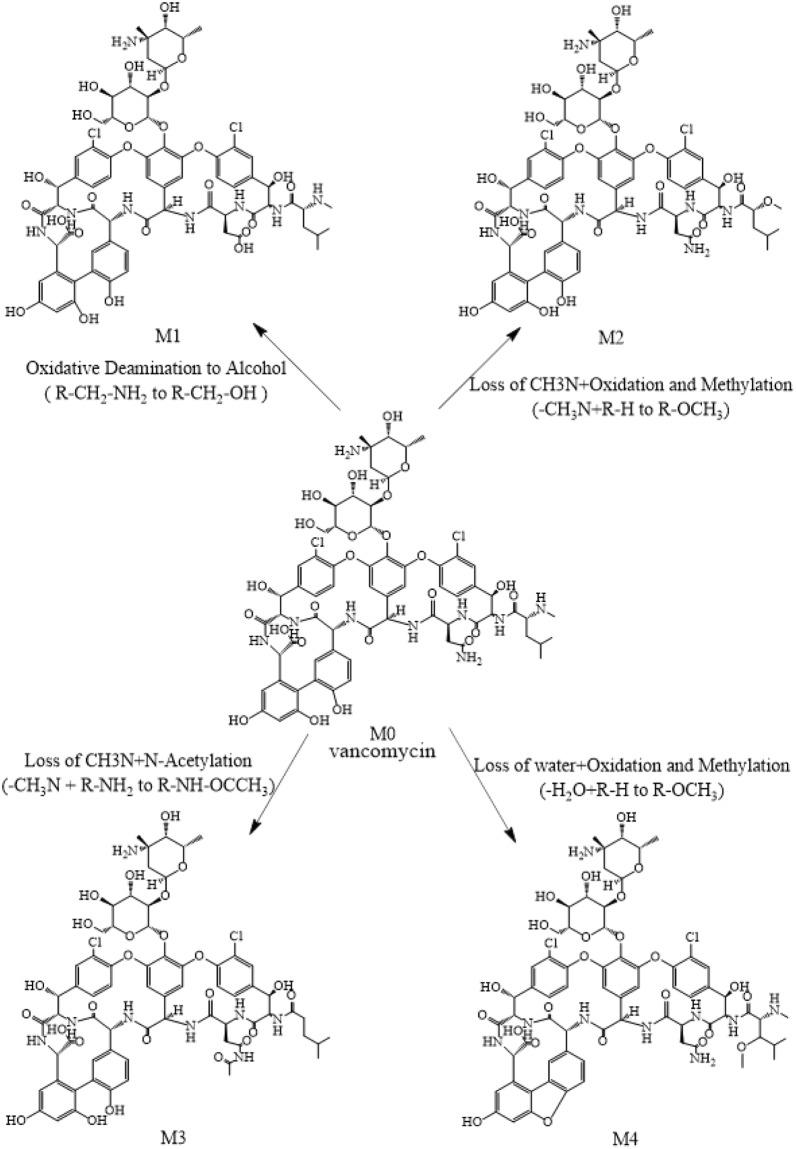
Structural deduction of the degradation products
The degradation products formed by biotransformation generally retain the basic skeleton and some substructures of the original parent molecule. This means that the degradation products may have similar cleavage laws to the raw drugs in HPLC-MS/MS analysis. Vancomycin contains two amino groups (one on the aminoglycoside moiety and the other on the polypeptide component). The MS/MS spectra of vancomycin and its four degradation products (Figs 2b, 5b, 6b, 7b and 8b) reveal that all the compounds contained the same fragment ions at m/z 100.07, 118.08 and 144.10. These fragment ions were produced from the fragmentation of the aminoglycoside fragmentation of vancomycin, thus the amino moiety in the aminoglycoside structure is consistent across all four degradation products. Based on the finding that vancomycin easily loses the CO part from the polypeptide under collision energy, we reasoned that the degradation products had similar principles and this provided the basis for deducing their chemical structures.
Figure 5.
The extract ion chromatogram, MS(a) and MS/MS(b) spectra of M1.
Figure 6.
The extract ion chromatogram, MS(a) and MS/MS(b) spectra of M2.
Figure 7.
The extract ion chromatogram, MS(a) and MS/MS(b) spectra of M3.
Figure 8.
The extract ion chromatogram, MS(a) and MS/MS(b) spectra of M4.
The retention time for M1 ([M + 2H]2+ at m/z 725.2171) was 12.64 min (Fig. 5a). M1 was acquired by the biotransformation process of oxidative deamination to alcohol, which suggested that M1 was obtained by changing the amino group in the polypeptide structure of vancomycin. M1 lost part of the aminoglycoside and one CO to produce the characteristic product ion at m/z 1116.2826. The likely structure of the characteristic product ion at m/z 1116.2826 is shown in Fig. 5b.
M2 eluted at 13.31 min and had the same protonated ion ([M + 2H]2+ at m/z 725.2167) as M1 (Fig. 6a). The biotransformation process for M2 involved the loss of CH3N, and oxidation and methylation. Vancomycin contains only one CH3N group within the polypeptide structure. M2 lost the part of aminoglycoside to produce the characteristic product ion at m/z 1144.2798 (Fig. 6b). M1 and M2 are isomers, and the polarity of M1 is greater than that of M2 according to the value of the CLogP. As a result, M1 eluted before M2.
Vancomycin lost a CH3N group and was N-acetylated to produce M3 ([M + 2H]2+ at m/z 731.2164), which eluted at 13.90 min (Fig. 7a). The loss of CH3N was the same as for M2, and the position of the change in the amino moiety was the same as for M1. The likely characteristic product ion at m/z 1128.2832 was acquired by M3, which lost the aminoglycoside component and a CO (Fig. 7b).
M4 ([M + 2H]2+ at m/z 730.7237) was obtained by the biotransformation process involving the loss of water and oxidation and methylation and eluted at 14.30 min (Fig. 8a). Vancomycin contains many hydroxyl groups, and two adjacent hydroxyl groups could be dehydrated to form ethers. The likely characteristic product ion at m/z 1099.2981 was acquired by M4, which lost the aminoglycoside and two CO moieties (Fig. 8b).
The Fragmentation Library Function in the Mass Frontier 5.0 software played a guiding role in the process to deduce the chemical structure of the four degradation products. The chemical structures of the four degradation products are illustrated in Fig. 9. The theoretical and observed MS and MS/MS mass of the four degradation products are listed in Table 4. All relative errors of the ion mass were less than 10 ppm.
Figure 9.
Probable degradation pathways of vancomycin in vitro and deductive chemical structure of four degradation products.
Table 4.
The theoretical and observed MS and MS/MS mass of four probable degradation products of vancomycin.
| Peak ID | Theoretical MS m/z [M + 2H]2+ | Observed MS m/z [M+2H]2+ | RE (ppm) | Theoretical MS/MS Fragments | Observed MS/MS Fragments | RE (ppm) |
|---|---|---|---|---|---|---|
| M0 | 724.7224 | 724.7245 | 2.9 | 100.0757 | 100.0767 | 10.0 |
| 118.0863 | 118.0866 | 2.5 | ||||
| 144.1019 | 144.1021 | 1.4 | ||||
| 1087.3002 | 1087.303 | 2.6 | ||||
| 1115.2951 | 1115.2979 | 2.5 | ||||
| 1043.29 | 1043.2936 | 3.4 | ||||
| 1305.3428 | 1305.3498 | 5.4 | ||||
| M1 | 725.2144 | 725.2171 | 3.7 | 118.0863 | 118.0859 | −3.4 |
| 144.1019 | 144.1014 | 3.5 | ||||
| 1116.2791 | 1116.2826 | 3.1 | ||||
| M2 | 725.2144 | 725.2167 | 3.2 | 100.0757 | 100.0759 | 2.0 |
| 118.0863 | 118.0857 | −5.1 | ||||
| 144.1019 | 144.1015 | −2.8 | ||||
| 1144.274 | 1144.2798 | 5.1 | ||||
| M3 | 731.2144 | 731.2164 | 2.7 | 100.0757 | 100.0763 | 6.0 |
| 144.1019 | 144.1022 | 2.1 | ||||
| 1128.2791 | 1128.2832 | 3.6 | ||||
| M4 | 730.7224 | 730.7239 | 2.0 | 100.0757 | 100.076 | 3.0 |
| 144.1019 | 144.1015 | −2.8 | ||||
| 1099.3002 | 1099.2981 | 1.9 |
The degradation pathway of vancomycin
In this study, a total of 4 metabolites were identified. The plausible degradation pathways of vancomycin are shown in Fig. 9. Additionally, the principle of the degradation of vancomycin into four metabolites was described in detail in the “Structural deduction of the degradation products” section above. The results showed that the amino group, methylamino group, adjacent hydroxyl group and hydrogens on the alkyl group in the polypeptide were the main degradation sites.
Conclusions
We used a high resolution and high sensitivity UHPLC-Triple-TOF-MS/MS method in positive ion mode to investigate the degradation of vancomycin in activated and inactivated rat liver microsomes, PBS and pure water systems. The compared result for activated rat liver microsomes system and inactivated rat liver microsomes system confirmed that the effect of activated rat liver microsomes cytochrome P450 enzymes on vancomycin is so weak that vancomycin is essentially not metabolized in the liver. A comparison of the degradation trends of vancomycin in the four systems, reveals that organics and mineral salt may be favorable to the degradation of vancomycin in aquatic environment. Ultimately, four likely novel degradation products were identified. The structural characteristics and cleavage of the four novel degradation products were deduced based on the mass spectra analysis of vancomycin. These finding could be a foundation for future research on the effects of vancomycin and its degradation products on environmental safety and human health.
Acknowledgements
This work was supported by the Natural Science Foundation of China (No. 81872628) and the Natural Science Foundation of Hebei Province (Nos H2018206122 and Nos C2015321005).
Author Contributions
L.A. and K.L. designed research; M.C. and Y.F. performed experiment and analyzed data. M.C. edit the manuscript; K.L. wrote the paper. Y.Z. and W.K. provided the experimental materials. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kaoqi Lian, Email: liankq@hebmu.edu.cn.
Lianfeng Ai, Email: ai_lianfeng@126.com.
References
- 1.Liu JL, Wong MH. Pharmaceuticals and personal care products (PPCPs): A review on environmental contamination in China. Environ. Int. 2013;59:208–224. doi: 10.1016/j.envint.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Ali AM, et al. Detection of PPCPs in marine organisms from contaminated coastal waters of the Saudi Red Sea. Sci. Total Environ. 2018;621:654–662. doi: 10.1016/j.scitotenv.2017.11.298. [DOI] [PubMed] [Google Scholar]
- 3.Subedi B, Mottaleb MA, Chambliss CK, Usenko S. Simultaneous analysis of select pharmaceuticals and personal care products in fish tissue using pressurized liquid extraction combined with silica gel cleanup. J. Chromatogr. A. 2011;1218:6278–6284. doi: 10.1016/j.chroma.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y, Ok YS, Kim KH, Kwon EE, Tsang YF. Occurrences and removal of pharmaceuticals and personal care products(PPCPs) in drinking water and water/sewage treatment plants: A review. Sci. Total Environ. 2017;596-597:303–320. doi: 10.1016/j.scitotenv.2017.04.102. [DOI] [PubMed] [Google Scholar]
- 5.Gracia-Lor E, Martínez M, Sancho JV, Peñuela G, Hernández F. Multi-class determination of personal care products and pharmaceuticals in environmental and wastewater samples by ultra-high performance liquid-chromatography-tandem mass spectrometry. Talanta. 2012;99:1011–1023. doi: 10.1016/j.talanta.2012.07.091. [DOI] [PubMed] [Google Scholar]
- 6.Sun CL, Dudley S, Trumble J, Gan J. Pharmaceutical and personal care products-induced stress symptoms and detoxification mechanisms in cucumber plants. Environ. Pollut. 2018;234:39–47. doi: 10.1016/j.envpol.2017.11.041. [DOI] [PubMed] [Google Scholar]
- 7.Liu C, et al. Clinical Practice Guidelines by the Infectious Diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections in Adults and Children: Executive Summary. Clin. Infect. Dis. 2011;52:285–292. doi: 10.1093/cid/cir034. [DOI] [PubMed] [Google Scholar]
- 8.Mei SH, et al. A UPLC-MS/MS method for analysis of vancomycin in human cerebrospinal fluid and comparison with the chemiluminescence immunoassay. Biomed. Chromatogr. 2017;31:1–7. doi: 10.1002/bmc.3939. [DOI] [PubMed] [Google Scholar]
- 9.Isabel TC, Lúcia S. Antibiotics in the aquatic environments: A review of the European scenario. Environ. Int. 2016;94:736–757. doi: 10.1016/j.envint.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 10.Dinh QT, et al. Measurement of trace levels of antibiotics in river water using on-line enrichment and triple-quadrupole LC-MS/MS. Talanta. 2011;85:1238–1245. doi: 10.1016/j.talanta.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 11.van Hal SJ, Paterson DL, Lodise TP. Systematic Review and Meta-Analysis of Vancomycin-Induced Nephrotoxicity Associated with Dosing Schedules That Maintain Troughs between 15 and 20 Milligrams per Liter. Antimicrob. Agents Ch. 2013;57:734–744. doi: 10.1128/AAC.01568-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rostas SE, Kubiak DW, Calderwood MS. High-Dose Intravenous Vancomycin Therapy and the Risk of Nephrotoxicity. Clin. Ther. 2014;36:1098–1101. doi: 10.1016/j.clinthera.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 13.Matthewsa AM, Hinson JA, Robertsa DW, Pumford NR. Comparison of covalent binding of acetaminophen and the regioisomer 3’-hydroxyacetanilide to mouse liver protein. Toxicol Lett. 1997;90:77–82. doi: 10.1016/S0378-4274(96)03831-3. [DOI] [PubMed] [Google Scholar]
- 14.Cabrera-Pérez, L. C., et al. Evaluation of a new benzothiazole derivative with antioxidant activity in the initial phase of acetaminophen toxicity. Arab. J. Chem., 10.1016/j.arabjc.2016.02.004 (2016).
- 15.Hassanzadeh K, et al. Riluzole prevents morphine-induced apoptosis in rat cerebral cortex. Pharmacol. Rep. 2011;63:697–707. doi: 10.1016/S1734-1140(11)70581-3. [DOI] [PubMed] [Google Scholar]
- 16.Yao DG, et al. Study on the metabolites of isoalantolactone in vivo and in vitro by ultra performance liquid chromatography combined with Triple TOF mass spectrometry. Food Chem. 2017;214:328–338. doi: 10.1016/j.foodchem.2016.07.100. [DOI] [PubMed] [Google Scholar]
- 17.Xie WW, et al. A practical strategy for the characterization of ponicidin metabolites in vivo and in vitro by UHPLC-Q-TOF-MS based on nontargeted SWATH data acquisition. J. Pharmaceut. Biomed. 2017;145:865–878. doi: 10.1016/j.jpba.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Boevre MD, Graniczkowska K. Metabolism of modified mycotoxins studied through in vitro and in vivo models: An overview. Toxicol. Lett. 2015;223:24–28. doi: 10.1016/j.toxlet.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Yuan L, et al. Study of in vitro metabolism of m-nisoldipine in human liver microsomes and recombinant cytochrome P450 enzymes by liquid chromatography-mass spectrometry. J. Pharmaceut. Biomed. 2014;97:65–71. doi: 10.1016/j.jpba.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 20.Hua XJ, et al. Metabolic detoxification of bakuchiol is mediated by oxidation of CYP 450s in liver microsomes. Food Chem. Toxicol. 2018;111:385–392. doi: 10.1016/j.fct.2017.11.048. [DOI] [PubMed] [Google Scholar]
- 21.Paolini M, et al. NADPH as rate-limiting factor for microsomal metabolism An alternative and economic NADPH-generating system for microsomal mono-oxygenase in in vitro genotoxicity studies. Muta. Res. 1987;178:11–20. doi: 10.1016/0027-5107(87)90080-7. [DOI] [PubMed] [Google Scholar]
- 22.Uutela P, et al. Identification of metabolites of fosinopril produced by human and rat liver microsomes with liquid chromatography-mass spectrometry. Eur. J. Pharm. Sci. 2014;53:86–94. doi: 10.1016/j.ejps.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Wang GJ, et al. Comprehensive characterization of the in vitro and in vivo metabolites of ziyuglycoside I in rat microsome, intestinal flora, excretion specimen and fresh tissues based on LC–Q-TOF/MS. J. Pharmaceut. Biomed. 2016;128:191–200. doi: 10.1016/j.jpba.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 24.Tang XG, et al. In vitro metabolism of L-corydalmine, a potent analgesic drug, in human, cynomolgus monkey, beagle dog, rat and mouse liver microsomes. J. Pharmaceut. Biomed. 2016;128:98–105. doi: 10.1016/j.jpba.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 25.Backes DW, Aboleneen HI, Simpson JA. Quantitation of vancomycin and its crystalline degradation product (CDP-1) in human serum by high performance liquid chromatography. J. Pharmaceut. Biomed. 1998;16:1281–1287. doi: 10.1016/S0731-7085(97)00140-4. [DOI] [PubMed] [Google Scholar]
- 26.White LO, et al. The in-vitro degradation at 37 °C of vancomycin in serum, CAPD fluid and phosphate-buffered saline. J. Antimicrob. Chemoth. 1988;22:739–745. doi: 10.1093/jac/22.5.739. [DOI] [PubMed] [Google Scholar]
- 27.Jaffé R. Fate of Hydrophobic Organic Pollutants in the Aquatic Environment: A Review. Environ. Pollut. 1991;69:237–257. doi: 10.1016/0269-7491(91)90147-O. [DOI] [PubMed] [Google Scholar]
- 28.Fernández-López C, Cano ÁF, Arocena JM, Alcolea A. Elemental and mineral composition of salts from selected natural and mine-affected areas in the Poopó and Uru-Uru lakes (Bolivia) Carmen. J. Great. Lakes. Res. 2014;40:841–850. doi: 10.1016/j.jglr.2014.08.003. [DOI] [Google Scholar]
- 29.Tian TT, et al. Identification of metabolites of oridonin in rats with a single run on UPLC-Triple-TOF-MS/MS system based on multiple mass defect filter data acquisition and multiple data processing techniques. J. Chromatogr. B. 2015;1006:80–92. doi: 10.1016/j.jchromb.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Hu K, et al. Screening and identification of the main metabolites of 2-amino-9H-pyrido[2,3-b]indole (AαC) in liver microsomes and rat urine by using UPLC-Q-TOF-MS/MS. J. Chromatogr. B. 2017;1046:110–121. doi: 10.1016/j.jchromb.2017.01.038. [DOI] [PubMed] [Google Scholar]
- 31.Liao M, Cheng XY, Diao XP, Sun YP, Zhang LT. Metabolites identificaion of two bioactive constituents in Trollius ledebourii in rats using ultra-high-performance liquid chromatography coupled to quadrupole time-of-flight mass spectrometry. J. Chromatogr. B. 2017;1068-1069:297–312. doi: 10.1016/j.jchromb.2017.10.061. [DOI] [PubMed] [Google Scholar]
- 32.Zhang. X, Yin JT, Liang CJ, Sun YP, Zhang LT. A simple and sensitive UHPLC-Q-TOF-MS/MS method for sophoricoside metabolism study in vitro and in vivo. J. Chromatogr. B. 2017;1061-1062:193–208. doi: 10.1016/j.jchromb.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 34.Javorska L, Krcmova LK, Solich P, Kaska M. Simple and rapid quantification of vancomycin in serum, urine and peritoneal/pleural effusion via UHPLC–MS/MS applicable to personalized antibiotic dosing research. J. Pharmaceut. Biomed. 2017;142:59–65. doi: 10.1016/j.jpba.2017.04.029. [DOI] [PubMed] [Google Scholar]
- 35.Brozmanová H, Kacířová I, Uřinovskáb R, Šištíkb P, Grundmann M. New liquid chromatography-tandem mass spectrometry method for routine TDM of vancomycin in patients with both normal and impaired renal functions and comparison with results of polarization fluoroimmunoassay in light of varying creatinine concentrations. Clin. Chim. Acta. 2017;469:136–143. doi: 10.1016/j.cca.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Oyaert M, et al. Novel LC-MS/MS method for plasma vancomycin: Comparison with immunoassays and clinical impact. Clin. Chim. Acta. 2015;441:63–70. doi: 10.1016/j.cca.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Li H, et al. Identification of rotundic acid metabolites after oral administration to rats and comparison with the biotransformation by Syncephalastrum racemosum AS 3.264. J. Pharmaceut. Biomed. 2018;150:406–412. doi: 10.1016/j.jpba.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 38.Yang SP, et al. Comparative metabolism of Lappaconitine in rat and human liver microsomes and in vivo of rat using ultra high-performance liquid chromatography–quadrupole/time-of-flight mass spectrometry. J. Pharmaceut. Biomed. 2015;110:1–11. doi: 10.1016/j.jpba.2015.02.048. [DOI] [PubMed] [Google Scholar]
- 39.Tang YN, et al. UPLC-QTOF-MS identification of metabolites in rat biosamples after oral administration of Dioscorea saponins: A comparative study. J. Ethnopharmacol. 2015;165:127–140. doi: 10.1016/j.jep.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 40.Want EJ, et al. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013;8:17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
- 41.Sun JH, et al. A high fat, high cholesterol diet leads to changes in metabolite patterns in pigs – A metabolomic study. Food Chem. 2015;173:171–178. doi: 10.1016/j.foodchem.2014.09.161. [DOI] [PMC free article] [PubMed] [Google Scholar]