Abstract
Genetic alterations in the transcriptional repressor ETV6 are associated with hematological malignancies. Notably, the t(12;21) translocation leading to an ETV6-AML1 fusion gene is the most common genetic alteration found in childhood acute lymphoblastic leukemia. Moreover, most of these patients also lack ETV6 expression, suggesting a tumor suppressor function. To gain insights on ETV6 DNA-binding specificity and genome wide transcriptional regulation capacities, we performed chromatin immunoprecipitation experiments coupled to deep sequencing in a t(12;21)-positive pre-B leukemic cell line. This strategy led to the identification of ETV6-bound regions that were further associated to gene expression. ETV6 binding is mostly cell type-specific as only few regions are shared with other blood cell subtypes. Peaks localization and motif enrichment analyses revealed that this unique binding profile could be associated with the ETV6-AML1 fusion protein specific to the t(12;21) background. This study underscores the complexity of ETV6 binding and uncovers ETV6 transcriptional network in pre-B leukemia cells bearing the recurrent t(12;21) translocation.
Introduction
ETV6 is a member of the ETS superfamily of transcription factors that are critical modulators of cellular homeostasis in several tissues. The normal function of ETS factors is mandatory for appropriate cell fate as dysregulation or deleterious events affecting these factors are frequently observed in a variety of cancers1. ETV6 is essential in the establishment and maintenance of hematopoiesis within the bone marrow compartment2,3. ETV6 translocations are frequently observed in various hematological disorders4 and germline mutations have been associated to predispositions for such diseases5–10.
The most common ETV6 aberration is the t(12;21)(p13;q22) translocation which fuses ETV6 to the AML1 gene (or RUNX1) and generates an in-frame ETV6-AML1 chimeric protein11. This is the most frequent chromosomal abnormality in childhood pre-B cell acute lymphoblastic leukemia (pre-B ALL), occurring in 20% of cases12. However, the ETV6-AML1 fusion protein seems insufficient to induce leukemia by itself13–15, suggesting that additional events are required to fully develop pre-B ALL16. Interestingly, the complete inactivation of ETV6 in t(12;21)-positive pre-B ALL cases was underscored by several studies17–21 and indicates that ETV6 depletion could lead to pre-B ALL initiation.
Unlike the majority of ETS members, ETV6 acts as a transcriptional repressor22,23. ETV6 has a N-terminal pointed (PNT) helix-loop-helix domain required for protein-protein interactions and homodimerization24. Its central repressive domain is also implicated in protein-protein interactions with members of the SMRT/N-CoR/mSin3A/HDAC co-repressor complexes25–27. The C-terminal part of ETV6 contains an ETS DNA-binding domain that recognizes a consensus ETS-binding site consisting of a core GGAA/T sequence with adjacent purine-rich sequences24. Interestingly, the ETV6-AML1 fusion protein combines both PNT and central repressive domains of ETV6 with Runt DNA binding and transactivation domains of AML1, thus converting AML1 from a transcriptional activator to a putative repressor28.
Although the molecular functions of ETV6-AML1 have been studied29–31, the exact role of ETV6 remains poorly understood. To gain insights into ETV6 function in t(12;21)-positive pre-B leukemia cells, we sought to identify ETV6 binding sites using chromatin immunoprecipitation coupled to high throughput sequencing (ChIP-seq). By including expression data32, we extensively described ETV6 binding properties and transcriptional activity in this particular context.
Methods
Constructs
The complete wild-type coding sequence of ETV6 was subcloned into pcDNA3.1 (pcDNA3.1 ETV6). The C-terminal HA-tagged ETV6 construct was generated by restriction enzyme digestion as described previously32 using an oligomer containing 3 tandem HA tag repeats. Both ETV6 and ETV6-HA were subcloned into pCCL lentiviral vector (kindly provided by Dr. Christian Beauséjour) through enzymatic digestion and ligation.
Cell culture
Reh (ATCC ® CRL-8286™), a t(12;21)-positive pre-B ALL cell line, was maintained in RPMI 1640 (Wisent) 10% Fetal Bovine Serum (FBS; Wisent) in a 5% CO2 incubator at 37 °C.
Lentiviral production
1.5 × 107 HEK293T cells were seeded into 15 cm petri dishes in DMEM (Wisent) 10% FBS. The next day, cells were transfected with 9 μg pCCL plasmids together with 6 μg pRSV-Rev, 7.8 μg pMD2.VSVG and 15 μg pMDL third generation encapsidation plasmids (kindly provided by Dr. Christian Beauséjour) in fresh RPMI 1640 10% FBS medium using polyethylenimine (Polysciences) at a final concentration of 6.5 μg/mL. Media was removed 16 h post-transfection and replaced by fresh DMEM 10% FBS. After 30 h, viral particles were retrieved from media by ultracentrifugation (50 000 g) and quantified by p24 antigen ELISA (Advanced Bioscience Laboratories).
Lentiviral infection
2 × 106 Reh cells and two different Reh clones (generated in methylcellulose media) were seeded in 2 mL of RPMI 1640 10% FBS medium. 200 ng of concentrated virus were added to cells with polybrene (Sigma) to a final concentration of 8 μg/mL. 24 h post-infection, medium was changed with fresh RPMI 1640 10% FBS. These cells were maintained 2 weeks in culture before carrying out further experiments.
Western blotting
20 μg of nuclear protein extracts were diluted in Laemmli buffer and migrated on SDS-denaturating 10% polyacrylamide gels. Transfer on polyvinylidene difluoride membranes was performed at 4 °C overnight. Membranes were blocked in Blotto A solution (1X TBS, 5% milk and 0.05% Tween-20) prior to immunoblotting using the primary antibodies against ETV6 (1:1000; ab54705; Abcam) or GAPDH (1:1000; sc-31915; Santa Cruz) and HRP-coupled secondary antibodies anti- mouse (1:5000; sc-358914; Santa Cruz) and anti-goat (1:5000; sc-2961; Santa Cruz) IgG, respectively. Membranes were then assayed by enhanced chemiluminescence detection with Western Lightning Plus-ECL (PerkinElmer) according to the manufacturer protocol.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) has been performed as previously described32. Briefly, cross-linked chromatin isolated from 1.0 × 107 transduced Reh cells was used for immunoprecipitation with anti-HA magnetic beads (Thermo Fisher Scientific). DNA-protein complexes were eluted from the beads by competition with HA peptides prior to reverse-crosslinking and standard purification using phenol/chloroform/isoamyl alcohol (Sigma). The purified ChIP DNA was processed through TruSeq ChIP Sample Preparation Kit (Illumina) according to the manufacturer protocol. As positive control, a fraction of the amplified ChIP material was used to assess the ETV6 binding enrichment at the CLIC5A promoter32 by quantitative PCR (qPCR) using primers listed in Supplementary Table S1. Libraries were sequenced on the HiSeq 2500 system (Illumina) in paired-end mode (2 × 100 bp).
ChIP-seq data analysis
Raw reads were aligned on the Hg19 reference genome using bowtie v2.2.333 and filtered using a mapping quality threshold of 20. Reads were merged for both ETV6 and ETV6-HA populations (n = 3). Peaks were called in the ETV6-HA condition over the background obtained in the ETV6 condition (negative IP control) using MACS v2.1.1.2016030934 with a q-value threshold of 0.1. Peaks overlapping blacklisted regions (ENCODE’s EncodeDacMapabilityConsensus35, svelter’s exclude file36 and canva’s filter file37) or having a fold enrichment above 50 were discarded. Peaks were associated to ensembl genes using the closest gene transcription start site (TSS) annotations from HOMER and genomic coordinates from bedops’ closest-features v2.4.1938. To define the minimal peak fold enrichment threshold (≥4.5), a Fisher’s exact test p-value was calculated for the overlaps between ETV6 peaks and ETV6-modulated genes32 against the proportion of ETV6 peaks in the complete ensembl genes repertoire. Genomic annotations of peaks and motif enrichment analyses were performed using HOMER v4.839.
ETV6 ChIP-seq data in GM12878 and K562 cells are publically available as part of the ENCODE project.
GM12878:https://www.encodeproject.org/files/ENCFF272DJU/@@download/ENCFF272DJU.bed.gz
K562:https://www.encodeproject.org/files/ENCFF514SLV/@@download/ENCFF514SLV.bed.gz
ETV6 peaks in GM12878 and K562 cells were thus obtained directly from the ENCODE platform as conservative IDR bed files (narrowPeak). The intersection of ETV6 peaks between cell types was obtained using pybedtools v.0.7.7 venn_mpl with default parameters40.
Chromatin state data of GM12878 cells were obtained from the NIH Roadmap Epigenomics Project Portal (E116, 15 coreMarks)41.
Results
Identification of genome wide ETV6-bound regions
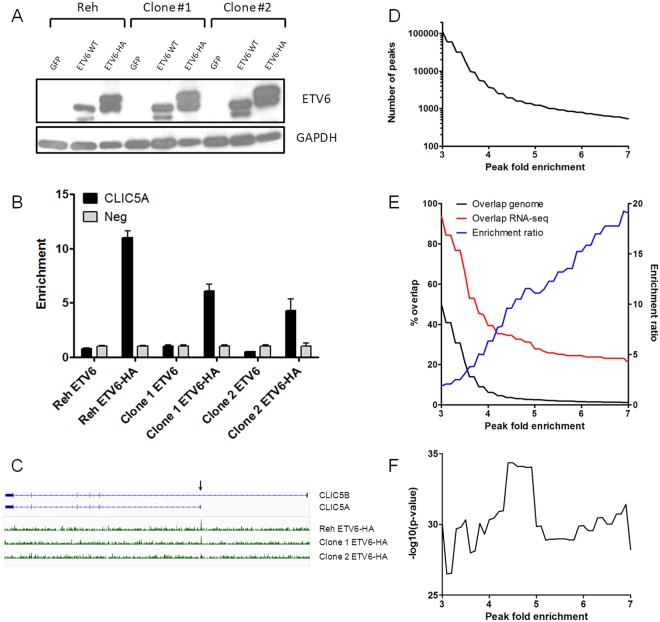
To determine the genomic regions bound by ETV6, we expressed ETV6 and HA-tagged ETV6 in 3 biological replicates of Reh pre-B leukemic cells (Fig. 1A). Of note, Reh cells lack endogenous wild-type ETV6 expression as a result of a t(12;21) translocation and a 12p13 locus deletion. These cells were then used for ChIP experiments using the HA epitope as a bait. We confirmed ETV6 binding to the CLIC5A promoter region, a validated ETV6 target gene32, in the 3 replicates, both by qRT-PCR (Fig. 1B) and high throughput sequencing (Fig. 1C), indicating that our ChIP-seq data are suitable for the identification of ETV6-bound regions. Note that the mapping and the peak calling procedures were performed individually for each sample to confirm the reproducibility of the biological replicates (Supplementary Fig S1). To retrieve the maximum of significant and robust ETV6 peaks, the reads from the replicates were pooled.
Figure 1.
ChIP-seq analysis of ETV6 binding sites in pre-B leukemia cells. (A) The Reh cell line, a pre-B leukemia cell line which is t(12;21)-positive and ETV6 negative, was transduced with either pCCL-GFP, ETV6 or ETV6-HA. The expression of ETV6 and ETV6-HA was confirmed by western blot. Results from two derived clones are shown. This blot was cropped and adjusted for brightness and contrast. The original blot is shown in the supplementary information. (This figure is the same control Western Blot as the one we published as Supplementary Figure S2 in Haematologica. 2016 Dec;101(12):1534 – 1543 PMID:2754013632; with permission of the Ferrata Storti Foundation©. The exact same cells were used in both publications). (B) ChIP-qPCR analysis of the CLIC5 locus from the HA-immunoprecipitated DNA. The CLIC5A promoter, a known bound region of ETV632, was successfully enriched in ETV6-HA populations compared to a negative control region (neg), but not in the untagged ETV6 populations. Error bars represent the standard deviation (n = 4) (C). Distribution of the reads mapped to the CLIC5 gene after sequencing of the HA-immunoprecipitated DNA. A ChIP signal at the CLIC5A promoter region (arrow) is seen in all three ETV6-HA populations. (D) The number of ETV6 ChIP peak signals using variable peak fold enrichment thresholds (0.1 increments in a 3 to 7 range). The number of peaks drastically increases with peak fold enrichment thresholds <4. (E) Overlap percentage (left y axis) between the genes associated with putative ETV6 binding sites and known ETV6-modulated genes32 (red line) or total ensembl genes (black line). The ratio between the percentages of overlap was calculated (blue line; right y axis) and increases with the stringency of the peak fold enrichment thresholds. (F) A Fisher’s exact test p-value was calculated for the overlaps between ETV6 peaks and ETV6-modulated genes against the proportion of ETV6 peaks in the complete ensembl genes repertoire. The most significant enrichment is calculated at a peak fold enrichment of ≥4.5.
The number of ETV6 peaks called varies markedly with the fold enrichment threshold (Fig. 1D; Supplementary Table S2). Effective ETV6 binding is expected to impact the expression of nearby genes. Accordingly, ETV6 peaks were associated to genes and compared to known ETV6-modulated genes (88 downregulated and 59 upregulated genes following ETV6-His expression in Reh cells) obtained by RNA-seq32. This strategy was performed through the entire peak fold enrichment range (Fig. 1E; Supplementary Table S2). The overlap between ETV6 peaks and ETV6-modulated genes is systematically higher than the random probability given by the proportion of ETV6 peaks in the complete ensembl genes repertoire. The ratio calculated between these overlaps is directly correlated to the peak fold enrichment (Fig. 1E, enrichment ratio; Supplementary Table S2), indicating that stronger ETV6 peaks are more likely associated with ETV6-mediated transcriptional regulation.
A peak fold enrichment ≥4.5 shows the strongest peak-to-gene expression association (Fisher’s exact test p-value = 4.46E−35; Fig. 1F; Supplementary Table S2). Using 4.5 as a specified threshold, 1,931 peaks were associated to 2,223 genes (Supplementary Table S3), representing 3.6% of the total ensembl genes set (n = 60,235). However, 51 of the 147 (34.6%) known ETV6-modulated genes in Reh cells32 have at least one associated peak called in this condition (74 expression-correlated peaks; Supplementary Table S4). By including expression data, we thus selected 1,931 high priority ETV6-bound regions that were used in the subsequent analyses.
ETV6 distribution across hematopoietic cell lines genomes
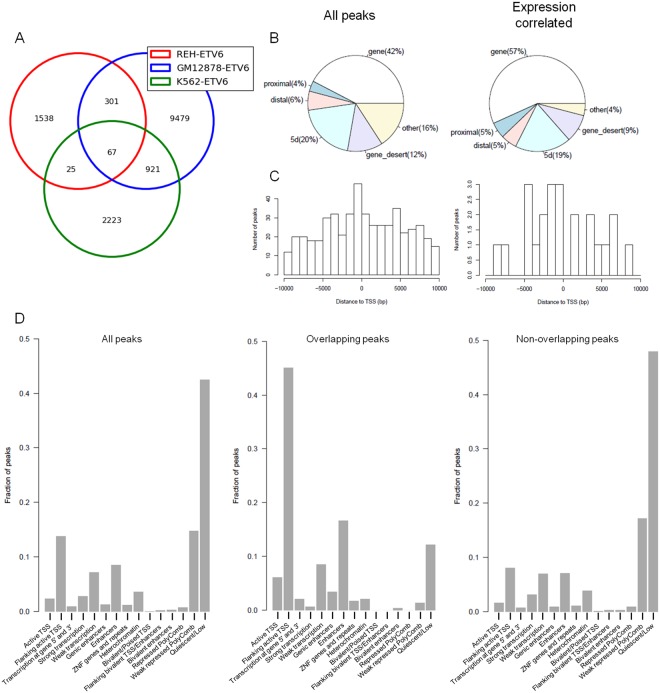
We next compared ETV6-bound regions identified in pre-B leukemia cells (Reh) to those of normal lymphoblastoid cells (GM12878) and myelogenous leukemia cells (K562) obtained from the ENCODE project (Fig. 2A). The overlap between Reh and GM12878 cells (368 of the 1,931 Reh peaks; 19.05%) was greater than with K562 cells (92 of the 1,931 Reh peaks; 4.76%), suggesting that the lineage (lymphoid vs. myeloid) is more likely to shape ETV6 binding compared to the differentiation stage (pre-B vs. B). The strongest overlap was however observed between K562 and GM12878 cells (988 of the 3,236 K562 peaks; 30.53%). Of note, the same antibody and immunoprecipitation protocol were used to generate these ENCODE datasets and might favor this overlap. Only 67 regions were bound by ETV6 in all 3 cell lines (3.47% of Reh peaks). Peaks shared across Reh and GM12878 cells were however not more associated to ETV6-modulated genes in Reh32 (Supplementary Table S5).
Figure 2.
Genomic annotations of ETV6-bound regions. (A) Comparison of ETV6 binding sites in Reh-ETV6 cells and two other hematopoietic cell lines, GM12878 and K562. (B) Genomic distribution of all Reh-derived ETV6 peaks (left panel; n = 1,931) and expression-correlated peaks (right panel; n = 74). Proximal: ≤2 kb upstream of TSS; Distal: 2 kb to 10 kb upstream of TSS; 5d: 10 kb to 100 kb upstream of TSS. ETV6 binding occurs mostly in genes. (C) Distribution of all Reh-derived ETV6 peaks (left panel) and expression-correlated peaks (right panel) in a 20 kb region across the TSS (+/−10 kb). Expression-correlated peaks are not enriched for proximal TSS binding. (D) Left panel: Reh-derived ETV6-bound regions were associated to GM12878 chromatin states (x axis) based on epigenetic profiles. Center panel: The same analysis was restricted to Reh and GM12878 overlapping ETV6 peaks. Those shared ETV6-bound regions are mainly flanking active TSS regions and enhancers. Right panel: Reh-specific ETV6 peaks are mostly associated to quiescent regions in GM12878 cells.
Genomic features of ETV6-bound regions
We next interrogated the genomic localization of ETV6 peaks. Figure 2B (“All peaks”; left panel) shows the percentage of peaks located within distinct genomic regions. The peaks are more often found (42%) within genes bodies (exons and introns). This distribution was similar in both GM12878 and K562 cells (Supplementary Fig S2). When we assessed the localization of expression-correlated ETV6 peaks, a stronger association (57%) was observed within gene (Fig. 2B, right panel). ETV6 binding within genes seems to induce a change in gene expression.
The number of peaks located in the vicinity of the transcription start site (TSS) was assessed (Fig. 2C). Of the 1,931 ETV6 peaks, 502 (26%) were in the flanking region (+/− 10 kb) of a TSS. The highest number of peaks within this range is observed in the first 1,000 bp upstream of the TSS (Fig. 2C, left panel). The distribution of ETV6 peaks observed in Reh cells is less TSS centered compared to GM12878 and K562 cells (Supplementary Fig S2). Although the relatively small sample size of expression-correlated ETV6 peaks, they did not display a tighter distribution around the TSS. (Fig. 2C, right panel), suggesting that ETV6-mediated transcription can be induced by distant TSS binding in Reh cells.
Epigenetic modifications in GM12878 cells were extensively investigated41 compared to Reh. Considering the number of shared ETV6 peaks between Reh and GM12878 cell lines (368 peaks, Fig. 2A), we assigned Reh-derived ETV6 peaks to specific chromatin states based on epigenetic data from the corresponding regions in GM12878 cells. Most of ETV6 peaks were associated to a quiescent state (Fig. 2D, left panel). However, when focusing on shared Reh and GM12878 ETV6 peaks, 45% of these were categorized as flanking active TSS (Fig. 2D, center panel; enriched for H3K4me3 and H3K4me1). Enhancers were also significantly enriched (Fig. 2D, center panel; enriched for H3K4me1 and high CpG methylation). In contrast, Reh-specific peaks are depleted for both flanking active TSS and enhancers chromatin states and are instead further associated with quiescent regions in GM12878 cells (Fig. 2D, right panel), indicating that ETV6 binding to a given region appears to depend on a prerequisite chromatin environment.
Motifs enrichment of ETV6-bound regions
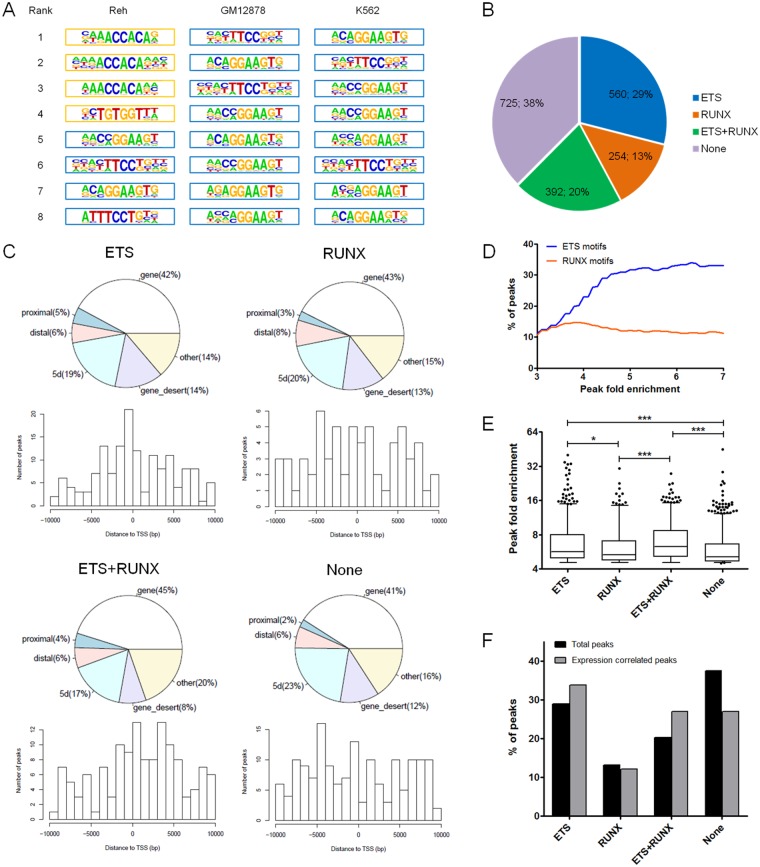
ETS transcription factors such as ETV6 have well-known binding motifs24. As expected, motif enrichment analyses revealed significant over-representation of ETS motifs within ETV6 peaks in Reh cells as well as in GM12878 and K562 cells (Fig. 3A,B; Supplementary Table S6). Interestingly, based on the ranking of the enriched motifs, RUNX motifs were significantly more over-represented in Reh cells compared to GM12878 and K562 cells (Fig. 3A,B; Supplementary Table S6). RUNX motifs are found centered on the peak summits similarly to ETS motifs (Supplementary Fig S3), indicating that RUNX-containing peaks are also properly bound. Unlike GM12878 and K562 cells, Reh cells express the ETV6-AML1 fusion protein that can interact with the bait protein ETV6-HA through their PNT domain and bind RUNX motifs42. Therefore, ETV6 ChIP-seq in this particular t(12;21)-positive background may have retrieved putative ETV6-AML1-bound regions in addition to ETV6-bound regions.
Figure 3.
Consensus binding motifs enrichment analyses. (A) Consensus binding motifs associated with ETV6-bound regions are shown for Reh, GM12878 and K562 cell lines. ETS motifs (blue border) were significantly enriched in all three cell lines. RUNX motifs (orange border) were also significantly enriched in Reh cells. (B) Reh-derived ETV6-bound regions were classified based on the presence or absence of ETS or RUNX motifs (identified in (A) (C) Genomic annotations of peaks for each motif groups. Proximal: ≤2 kb upstream of TSS; Distal: 2 kb to 10 kb upstream of TSS; 5d: 10 kb to 100 kb upstream of TSS. Notably, ETS-unique peaks shows a tighter distribution across the TSS region. (D) Percentages of ETV6-bound regions containing either ETS or RUNX using variable peak fold enrichment thresholds. ETS but not RUNX motifs become increasingly more frequent with stronger peak fold enrichments. (E) Fold enrichment of all ETV6 peaks in Reh cells according to each motif groups. ETS containing peaks (ETS and the ETS/RUNX compound) have significantly greater fold enrichments. Whiskers represent the 5–95 percentiles. Statistical significance is calculated by two-tailed Student’s t test. (F) Proportion of each motif groups in expression-correlated ETV6 peaks (n = 74) compared to all ETV6 peaks (n = 1,931). ETS containing peaks (ETS and the ETS/RUNX compound) are even more over-represented in expression-correlated peaks.
Only minor differences were observed when we assessed the global genomic localization pattern of ETV6 peaks according to the motif groups (refer to Fig. 3B) in Reh cells (Fig. 3C). However, ETV6 binding occurs predominantly in the first 1,000 bp upstream of TSS only in ETS-unique peaks (Fig. 3C). With ETS motifs peaks being directly bound by ETV6-HA, the intensity (or peak fold enrichment) of these peaks is expected to be higher than RUNX motifs peaks which are presumably bound through ETV6-AML1. A striking difference is indeed observed when we compared the percentage of peaks containing only ETS (direct binding) or RUNX (indirect binding) motifs according to different peak fold enrichment (Fig. 3D). ETS motifs become increasingly predominant among ETV6-bound regions with higher peak fold enrichment thresholds. Inversely, the fraction of ETV6-bound regions containing RUNX-unique motifs remains constant through the peak fold enrichment range.
ETV6 peaks containing ETS motifs (unique or with RUNX) are therefore significantly stronger (i.e. higher fold enrichment) than RUNX-unique peaks and ETS/RUNX depleted peaks (Fig. 3E). Interestingly, peaks correlated with gene expression modulation have significantly higher fold enrichments (Supplementary Table S7) without significant differences between motif groups (Supplementary Fig S4). We then assessed the relationship between motifs and expression. As shown in Fig. 3F, expression-correlated peaks are slightly more enriched for ETS motifs (unique or with RUNX) compared to all the peaks. Inversely, peaks lacking both ETS and RUNX motifs are less likely to induce a change in gene expression. All together, these data indicate that ETV6 predominantly binds ETS-containing sequences in vivo. ETS-driven binding of ETV6 is generally stronger than other binding events and are even more associated with a change in gene expression.
Although most ETV6 binding sites contain either ETS or RUNX motifs, a non-negligible fraction does not (38%, Fig. 3B). Additional motif enrichment analyses revealed that only 10 motifs were significantly over-represented among this subset of ETS/RUNX free peaks (q-value < 0.1; Supplementary Table S8). IRF consensus sequences were present in 3 of these enriched motifs with one of them in conjunction with an ETS motif (Supplementary Table S8, see motif #4). Interestingly, this ETS-IRF compound motif was also strongly associated to expression regulation (Supplementary Fig S5). In fact, 10 of the 20 expression-correlated peaks lacking ETS or RUNX motifs instead contain this ETS-IRF compound motif. It is known that ETV6 can interact with IRF843, suggesting a potential regulation through this interaction similarly to what was observed with RUNX motifs and ETV6-AML1. These results suggest a role for ETV6 as a cofactor at these loci.
Discussion
In this study, we provided the first in-depth mapping of ETV6 binding sites in t(12;21)-positive pre-B leukemia cells. We further associated ETV6 binding to transcriptional regulation by integrating expression data32. Despite the clear correlation between ETV6 binding and gene expression in the Reh cell line, ETV6 binding profile in these cells was markedly different of those obtained in other hematopoietic cell types. By including epigenetic data, we were able to unveil the importance of the cell type-specific chromatin environment on ETV6 binding. In this regard, roughly half of Reh-specific ETV6 peaks maps in quiescent regions of the lymphoblastoid cell line GM12878. The cell type-specific transcriptional programs and related active chromatin states thus modulate ETV6 binding. Although ubiquitously expressed, ETV6 impact on expression profiles might be different across tissues and cell types as its binding depends on higher order chromatin conformations. It is noteworthy that ETV6 silencing or overexpression in purified CD34+ cells from cord blood led to differentially expressed genes with however no recurrence with genes obtained in Reh cells (unpublished observations32;), further demonstrating the cell-specific role of ETV6 in transcriptional regulation.
ETV6 binding might as well rely on other proteins43,44 whose availability would lead to additional complexity and divergences across cell types. For instance, ETV6-AML1 seems to shape ETV6 binding profile in Reh cells. Although additional experiments could further strengthen our observations, the over-representation of RUNX motifs is likely a consequence of their interaction42 as these sequences are expectedly targeted by the Runt domain of the fusion protein29. The contrasting distribution across TSS of ETV6 peaks in Reh cells compared to other cell types may be attributed to this interaction as the dissection per motif groups revealed a tighter distribution of ETS-unique peaks. Furthermore, not only ETV6 is recruited to RUNX-containing sequences but also induced changes in gene expression at these loci. RUNX-unique motifs were found in similar proportions within ETV6 peaks and expression-correlated peaks. More importantly, ETV6 peaks containing both ETS and RUNX consensus binding sequences were even more associated with differentially expressed genes. These regions may be bound synergistically by the complex formed by ETV6-HA and endogenous ETV6-AML1 through their ETS and Runt domains, respectively. Interestingly, the expression of ETV632 or the knock-down of ETV6-AML131 in Reh cells lead to common differentially expressed genes (37 of the 147 ETV6-modulated genes are also differentially expressed upon ETV6-AML1 knock-down), further suggesting that ETV6 and ETV6-AML1 cooperate in transcriptional regulation. Additionally, it remains possible that a fraction of RUNX-containing peaks could have been retrieved through the recruitment of ETV6 to AML1 itself as several reports described an interaction between AML1 and other ETS transcription factors45–48.
In addition to its canonical function to directly bind ETS sequences, ETV6 seems to be recruited to additional sites, indicating that ETV6 can act as a cofactor without direct DNA-binding. The most striking example is the high occurrence of RUNX motifs that could be explained by the ETV6-AML1 fusion protein in Reh cells. However this is unique to the specific t(12;21)-positive background of these cells and might not reflect the general impact of ETV6 in normal cells. Nonetheless, a significant proportion of peaks contained neither ETS nor RUNX motifs but were still associated to a change in gene expression. These peaks were enriched for IRF binding sites in combination or not with ETS sites. This result is supported by the known interaction between ETV6 and IRF843 and further suggests that the interaction between ETV6 (and potentially other ETS factors) and IRF proteins is a rather common mechanism of transcriptional regulation. The function of ETV6 as a cofactor is however poorly understood and additional efforts are required to uncover putative partners and characterize their functions.
All together, this report indicates that ETV6 binding is highly flexible and therefore ETV6 inactivation is expected to induce unique transcriptional modifications in a given cellular context. The wide spectrum of hematological diseases associated with ETV6 alterations5–10 could be explained, at least partially, by the distinct ETV6 regulatory network of the cell originally affected by the mutation. Accordingly, the complete disruption of ETV6 observed in most t(12;21)-positive childhood pre-B ALL cases17–21 may induce specific transcriptional changes required for complete leukemic transformation. Although this study dissected ETV6 binding and clarified its transcriptional network in t(12;21)-positive pre-B leukemia cells32, it remains challenging to associate ETV6 target genes to leukemia-related phenotypes as their functions are, for most of them, still unclear.
Conclusions
Molecular characterization of ETV6 function is mandatory to fully understand its role in leukemogenesis. Towards this goal we built the first genome wide map of ETV6 binding sites in pre-B leukemia cells bearing the recurrent t(12;21) translocation. By including expression data, we obtained the detailed transcriptional network of ETV6 in these cells. This comprehensive analysis exposed the binding properties of ETV6 and suggests that ETV6 could also act as a cofactor to regulate gene expression. With the recent reports connecting germline ETV6 mutations to a panel of familial hematological diseases and given the complexity of ETV6-mediated transcription, further characterization of ETV6 remains valuable.
Electronic supplementary material
Author Contributions
D.S. is the principal investigator and takes primary responsibility for the paper. B.N., M.C. and D.S. contributed to the conception and design of the study. K.L. performed ChIP experiments. C.R. was involved in sample and library preparation. M.C. performed bioinformatics analyses and data processing. B.N. contributed to data integration and analyses. B.N. drafted the paper and D.S. contributed to interpretation of the data and was involved in critical revision of the manuscript. All authors approved the final version.
Availability of Materials and Data
The datasets generated and/or analysed during the current study are available in the GEO repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102785.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33947-1.
References
- 1.Kar A, Gutierrez-Hartmann A. Molecular mechanisms of ETS transcription factor-mediated tumorigenesis. Critical reviews in biochemistry and molecular biology. 2013;48:522–543. doi: 10.3109/10409238.2013.838202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang LC, et al. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes & development. 1998;12:2392–2402. doi: 10.1101/gad.12.15.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang LC, et al. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. The EMBO journal. 1997;16:4374–4383. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Braekeleer E, et al. ETV6 fusion genes in hematological malignancies: a review. Leukemia research. 2012;36:945–961. doi: 10.1016/j.leukres.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Zhang MY, et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nature genetics. 2015;47:180–185. doi: 10.1038/ng.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noetzli L, et al. Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia. Nature genetics. 2015;47:535–538. doi: 10.1038/ng.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topka S, et al. Germline ETV6 Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia. PLoS genetics. 2015;11:e1005262. doi: 10.1371/journal.pgen.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moriyama T, et al. Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukaemia: a systematic genetic study. The Lancet. Oncology. 2015;16:1659–1666. doi: 10.1016/S1470-2045(15)00369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkpatrick G, Noetzli L, Di Paola J, Porter CC. ETV6 mutations define a new cancer predisposition syndrome. Oncotarget. 2015;6:16830–16831. doi: 10.18632/oncotarget.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romero D. Haematological cancer: ETV6 germline mutation - a risk for ALL. Nature reviews. Clinical oncology. 2016;13:4. doi: 10.1038/nrclinonc.2015.211. [DOI] [PubMed] [Google Scholar]
- 11.Golub TR, et al. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tasian SK, Loh ML, Hunger SP. Childhood acute lymphoblastic leukemia: Integrating genomics into therapy. Cancer. 2015;121:3577–3590. doi: 10.1002/cncr.29573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreasson P, Schwaller J, Anastasiadou E, Aster J, Gilliland DG. The expression of ETV6/CBFA2 (TEL/AML1) is not sufficient for the transformation of hematopoietic cell lines in vitro or the induction of hematologic disease in vivo. Cancer genetics and cytogenetics. 2001;130:93–104. doi: 10.1016/S0165-4608(01)00518-0. [DOI] [PubMed] [Google Scholar]
- 14.van der Weyden L, et al. Modeling the evolution of ETV6-RUNX1-induced B-cell precursor acute lymphoblastic leukemia in mice. Blood. 2011;118:1041–1051. doi: 10.1182/blood-2011-02-338848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori H, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greaves M. Childhood leukaemia. Bmj. 2002;324:283–287. doi: 10.1136/bmj.324.7332.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel H, et al. Analysis of TEL proteins in human leukemias. Oncogene. 1998;16:2895–2903. doi: 10.1038/sj.onc.1201817. [DOI] [PubMed] [Google Scholar]
- 18.Patel N, et al. Expression profile of wild-type ETV6 in childhood acute leukaemia. British journal of haematology. 2003;122:94–98. doi: 10.1046/j.1365-2141.2003.04399.x. [DOI] [PubMed] [Google Scholar]
- 19.Lilljebjorn H, et al. The correlation pattern of acquired copy number changes in 164 ETV6/RUNX1-positive childhood acute lymphoblastic leukemias. Human molecular genetics. 2010;19:3150–3158. doi: 10.1093/hmg/ddq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montpetit A, Boily G, Sinnett D. A detailed transcriptional map of the chromosome 12p12 tumour suppressor locus. European journal of human genetics: EJHG. 2002;10:62–71. doi: 10.1038/sj.ejhg.5200766. [DOI] [PubMed] [Google Scholar]
- 21.Montpetit A, et al. Mutational and expression analysis of the chromosome 12p candidate tumor suppressor genes in pre-B acute lymphoblastic leukemia. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2004;18:1499–1504. doi: 10.1038/sj.leu.2403441. [DOI] [PubMed] [Google Scholar]
- 22.Lopez RG, et al. TEL is a sequence-specific transcriptional repressor. The Journal of biological chemistry. 1999;274:30132–30138. doi: 10.1074/jbc.274.42.30132. [DOI] [PubMed] [Google Scholar]
- 23.Mavrothalassitis G, Ghysdael J. Proteins of the ETS family with transcriptional repressor activity. Oncogene. 2000;19:6524–6532. doi: 10.1038/sj.onc.1204045. [DOI] [PubMed] [Google Scholar]
- 24.Bohlander SK. ETV6: a versatile player in leukemogenesis. Seminars in cancer biology. 2005;15:162–174. doi: 10.1016/j.semcancer.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Chakrabarti SR, Nucifora G. The leukemia-associated gene TEL encodes a transcription repressor which associates with SMRT and mSin3A. Biochemical and biophysical research communications. 1999;264:871–877. doi: 10.1006/bbrc.1999.1605. [DOI] [PubMed] [Google Scholar]
- 26.Guidez F, et al. Recruitment of the nuclear receptor corepressor N-CoR by the TEL moiety of the childhood leukemia-associated TEL-AML1 oncoprotein. Blood. 2000;96:2557–2561. [PubMed] [Google Scholar]
- 27.Wang L, Hiebert SW. TEL contacts multiple co-repressors and specifically associates with histone deacetylase-3. Oncogene. 2001;20:3716–3725. doi: 10.1038/sj.onc.1204479. [DOI] [PubMed] [Google Scholar]
- 28.Zelent A, Greaves M, Enver T. Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene. 2004;23:4275–4283. doi: 10.1038/sj.onc.1207672. [DOI] [PubMed] [Google Scholar]
- 29.Linka Y, et al. The impact of TEL-AML1 (ETV6-RUNX1) expression in precursor B cells and implications for leukaemia using three different genome-wide screening methods. Blood cancer journal. 2013;3:e151. doi: 10.1038/bcj.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linka Y, Ginzel S, Borkhardt A, Landgraf P. Identification of TEL-AML1 (ETV6-RUNX1) associated DNA and its impact on mRNA and protein output using ChIP, mRNA expression arrays and SILAC. Genomics data. 2014;2:85–88. doi: 10.1016/j.gdata.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghazavi, F. et al. Unique long non-coding RNA expression signature in ETV6/RUNX1-driven B-cell precursor acute lymphoblastic leukemia. Oncotarget, 10.18632/oncotarget.12063 (2016). [DOI] [PMC free article] [PubMed]
- 32.Neveu B, et al. CLIC5: a novel ETV6 target gene in childhood acute lymphoblastic leukemia. Haematologica. 2016;101:1534–1543. doi: 10.3324/haematol.2016.149740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X, Emery SB, Myers B, Kidd JM, Mills RE. Resolving complex structural genomic rearrangements using a randomized approach. Genome biology. 2016;17:126. doi: 10.1186/s13059-016-0993-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roller E, Ivakhno S, Lee S, Royce T, Tanner S. Canvas: versatile and scalable detection of copy number variants. Bioinformatics. 2016;32:2375–2377. doi: 10.1093/bioinformatics/btw163. [DOI] [PubMed] [Google Scholar]
- 38.Neph S, et al. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012;28:1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dale RK, Pedersen BS, Quinlan AR. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–3424. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roadmap Epigenomics C, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gunji H, et al. TEL/AML1 shows dominant-negative effects over TEL as well as AML1. Biochemical and biophysical research communications. 2004;322:623–630. doi: 10.1016/j.bbrc.2004.07.169. [DOI] [PubMed] [Google Scholar]
- 43.Kuwata T, et al. Gamma interferon triggers interaction between ICSBP (IRF-8) and TEL, recruiting the histone deacetylase HDAC3 to the interferon-responsive element. Molecular and cellular biology. 2002;22:7439–7448. doi: 10.1128/MCB.22.21.7439-7448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schick N, Oakeley EJ, Hynes NE, Badache A. TEL/ETV6 is a signal transducer and activator of transcription 3 (Stat3)-induced repressor of Stat3 activity. The Journal of biological chemistry. 2004;279:38787–38796. doi: 10.1074/jbc.M312581200. [DOI] [PubMed] [Google Scholar]
- 45.Petrovick MS, et al. Multiple functional domains of AML1: PU.1 and C/EBPalpha synergize with different regions of AML1. Molecular and cellular biology. 1998;18:3915–3925. doi: 10.1128/MCB.18.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao S, Frank RC, Zhang J, Miyazaki Y, Nimer SD. Functional and physical interactions between AML1 proteins and an ETS protein, MEF: implications for the pathogenesis of t(8;21)-positive leukemias. Molecular and cellular biology. 1999;19:3635–3644. doi: 10.1128/MCB.19.5.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goetz TL, Gu TL, Speck NA, Graves BJ. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Molecular and cellular biology. 2000;20:81–90. doi: 10.1128/MCB.20.1.81-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang H, et al. Differentiation-dependent interactions between RUNX-1 and FLI-1 during megakaryocyte development. Molecular and cellular biology. 2009;29:4103–4115. doi: 10.1128/MCB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the GEO repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102785.