Abstract
A gasotransmitter is defined as a small, generally reactive, gaseous molecule that, in solution, is generated endogenously in an organism and exerts important signalling roles. It is noteworthy that these molecules are also toxic and antimicrobial. We ask: is this definition of a gasotransmitter appropriate in the cases of nitric oxide, carbon monoxide and hydrogen sulfide (H2S) in microbes? Recent advances show that, not only do bacteria synthesise each of these gases, but the molecules also have important signalling or messenger roles in addition to their toxic effects. However, strict application of the criteria proposed for a gasotransmitter leads us to conclude that the term ‘small molecule signalling agent’, as proposed by Fukuto and others, is preferable terminology.
Keywords: carbon monoxide, cellular signalling, gasotransmitters, hydrogen sulfide, nitric oxide
Introduction: Don't shoot the messenger!
The term ‘gasotransmitter’ or ‘gaseous transmitter’ appears to have been coined ca. 2002 [1] to describe endogenously generated gaseous molecules that play important physiological roles. Specifically, they were defined [1,2] by the following six criteria:
(i) They are small (molecular mass 28–34) molecules of gas [such as nitric oxide (NO), carbon monoxide (CO) and hydrogen sulfide (H2S)].
(ii) They are freely permeable through membranes and therefore act without cognate membrane receptors. In this respect, they differ from better-established transmitters such as hormones, neurotransmitters and drugs.
(iii) Their endogenous generation is enzymatic and highly regulated, a characteristic that chimes with the potentially toxic nature of the best established of the gasotransmitters — currently NO, CO and H2S.
(iv) They have well-defined, specific functions at physiologically relevant, generally exceptionally low, concentrations. Manipulating endogenous levels evokes specific physiological changes.
(v) Their functions can be mimicked by exogenous counterparts (for example, NO-releasing compounds or CO-releasing molecules, CORMs)
(vi) Their cellular effects may or may not be mediated by secondary messengers, but specific final targets are implicated. For example, NO may result in vasodilation by prior interaction with soluble guanylate cyclase (GC, formerly sGC), or may bind directly to haem proteins and a multitude of other targets.
However, as observed by Samuel Butler, ‘Definitions are a kind of scratching and generally leave a sore place more sore than it was before’. In this short review, we do not wish to scratch too vigorously, but feel that, in microbial science in particular, it is useful to question the early definitions. The term has been widely adopted: a recent Web of Science search reveals almost 1200 hits. A major criticism of the term is that each of these three molecules is completely soluble at physiologically relevant concentrations: the term ‘gas’ does not accurately describe their physical state or chemical reactivity [3]. Nevertheless, one uniting feature lies in their biochemical targets, i.e. the reactive centres with which they interact (redox-active metals or amino acids, notably cysteine thiols or tyrosine phenols). The commonality of targets disguises, however, the distinctions in reaction mechanisms that underpin the ability of ‘gasotransmitters’ to achieve exquisite control.
Here, we consider how relevant and precise the term ‘gasotransmitter’ is when applied to bacteria [4,5]. To what extent are the above criteria met? We must question, for example, the nature of the signal (if any) being transmitted by the molecule and its targets. Is the term ‘small molecule signalling agent’ proposed before [3] more apt in microbes? Note that we do not detail those chemical species that are chemically related to the gases NO, CO and H2S. For example, the oxidation/reduction products of NO, carbonylation reactions that introduce CO onto organic and inorganic substrates and the multiple low-molecular-mass persulfides are discussed only en passant. The topic has been reviewed on numerous occasions before, including in this journal [6] and The Biochemist (e.g. [7–9]). New members of the gasotransmitter family are proposed, such as sulfur dioxide [10].
The eukaryotic exemplars: NO, CO and H2S
This trinity comprises the first ‘gasotransmitters’ to be identified and they remain the best studied. Since its discovery as a signal mediator in the early 1980s, NO has been implicated in a myriad of physiological processes with roles in the immune, cardiovascular and nervous systems [11]. Nitric oxide synthase (NOS) is the enzyme responsible for endogenous NO production and three isoforms exist in mammals: the constitutive forms: neuronal NOS1 (nNOS) and endothelial NOS3 (eNOS), and the inducible NOS2 (iNOS). NOS enzymes were initially classified by their localisation and expression patterns, but this is misleading: eNOS is found in cells and tissues other than the endothelium, nNOS is found in cells other than neurons and iNOS is found constitutively in some tissues [12]. Nevertheless, the diverse expression and subcellular locations of NOS leads to an array of NO-mediated physiological effects.
Whereas NO was discovered as the chemical mediator of an important biological function (most famously vasodilation), the production of CO in mammalian physiology was recognised long before much attention had been devoted to its biological function [13]. Its presence in biology was thought to be merely as a waste product of the degradation of haem, a reaction catalysed by the enzyme haem oxygenase (HO), also liberating Fe and biliverdin. However, CO has latterly been recognised as a potent biological messenger in mammalian systems. Haem oxygenases exist in constitutive (HO-2) and inducible (HO-1) isoforms [14]. While HO-2 is predominantly expressed in testes, brain and the endothelium, HO-1 is up-regulated in tissues following various stress stimuli, including oxidative stress, which is an underlying factor in different pathological states [15]. At physiological levels, CO has been shown to mediate a broad array of signalling processes, including the production of inflammatory mediators [16], cell survival and apoptosis [17], signalling in the central nervous system [18] and in bacterial infection [19].
By comparison, the acceptance of H2S as a gasotransmitter is more recent. Although the presence of H2S in tissue had been noted for decades, and its potential signalling capacity was first noted in the late 1990s [20], the argument for gasotransmitter status, founded on its endogenous metabolism and physiological functions, was made much later [1]. H2S is endogenously generated via both non-enzymatic and enzymatic pathways. In mammalian tissues, the enzymatic production of H2S from l-cysteine is catalysed by cystathionine-β-synthase (CBS), cystathionine-γ-synthase (CSE) and the 3-mercaptopyruvate sulfurtransferase/cysteine aminotransferase (3-MST/CAT) pathways (e.g. [21,22]). Like NO and CO, H2S at very low levels affects human physiology at molecular, cellular, tissue and system levels [22,23].
Thus, gasotransmitters in mammalian systems are endogenous signalling molecules; cellular production leads to their binding to cell surface receptors, intracellular receptors and proteins, triggering a cascade of events that lead to multiple physiological outcomes. Are these effects mirrored in microbes?
NO in microbes: gasotransmitter or not?
The products of NO oxidation by oxygen (NO2), the nitrosonium cation (NO+), the nitroxyl anion (NO−) and peroxynitrite (ONOO−; the product of the reaction of NO with superoxide) are outside the scope of this short review. As we have argued before [24,25], although these congeners of NO are of immense biological importance, and are ‘close cousins’, they are not NO. That NO satisfies criteria (i) and (ii), namely in being a small molecule that permeates membranes, is evident, but what is the origin of NO in microbes?
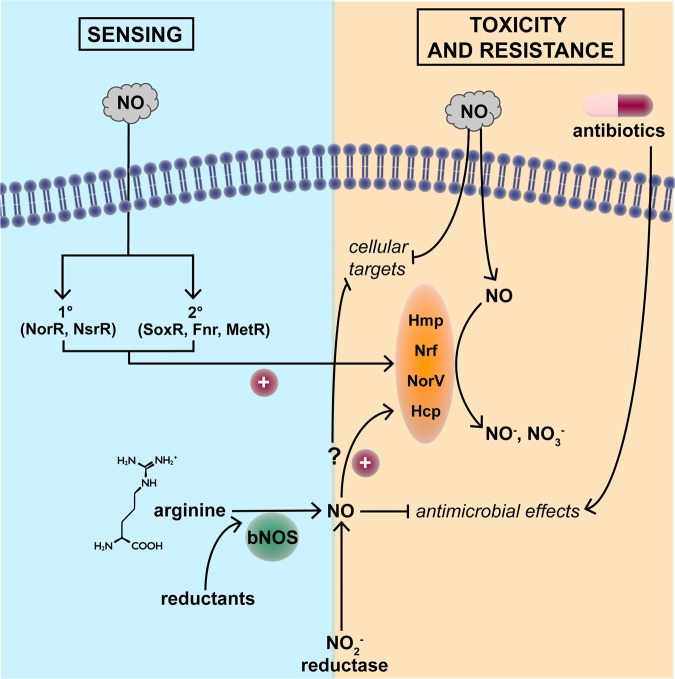
One criterion for a gasotransmitter is that it should be endogenously generated. The most significant sources of NO in a bacterial context are (a) synthesis by NOS in the gut epithelium, (b) release by denitrifying bacteria and nitrite reduction [26] and (c) synthesis by iNOS [27] as a component of the antibacterial weaponry in phagocytic cells (macrophages and monocytes) [28] on stimulation by cytokines. Minor sources include anaerobic reduction of nitrite to ammonia or the chemical reduction of nitrite by FeS proteins or haemproteins [29]. However, bacteria also possess NOS enzymes (bacterial NOS, bNOS; Figure 1) that share similarities with the mammalian forms [27], but lack the reductase domain of eukaryotes. Electron donation is achieved from specialist or promiscuous cellular reductases [30,31]. Specialised roles for bacterial NOS have been proposed in nitration reactions [32], resistance to oxidative stress and antibiotics [33,34], and host colonisation [35].
Figure 1. NO sensing and mechanisms of resistance to toxicity.
NO readily enters microbial cells from diverse exogenous sources, but may also be synthesised from arginine by bacterial NO synthase (bNOS). Primary and secondary sensors detect NO, which exerts numerous antimicrobial effects, but paradoxically may protect bacteria from antibiotic action. The toxicity of NO is minimised by NO-detoxifying proteins (Hmp, Nrf, NorV and Hcp). However, it is unclear whether endogenously generated NO elicits equivalent defence responses. For details, see the text.
Further criteria (iv–vi) are that the small molecule should have well-defined and specific functions at physiologically relevant concentrations and that its levels should evoke specific physiological changes. A clear example of a function for bNOS-generated NO is its role as a virulence factor in Bacillus anthracis [36], in which NO-mediated activation of bacterial catalase and suppression of the damaging Fenton reaction protects the bacterium from the immune oxidative burst. More striking is the role of NO from bNOS in increasing the resistance to antibiotics [34]. This appears to be achieved in two ways: chemical modification of the drugs (e.g. nitrosation reactions with arylamino moiety of acriflavin) and alleviating the impact of oxidative stress that is proposed to accompany antibiotic action. Certain bacteria exploit bNOS-derived NO in competition with others: Bacillus sp. use NO to reduce the oxidative stress associated with pyocyanin production by Pseudomonas [34]. However, many aspects of their functions, particularly in the NO-producing cell, remain obscure.
With such exceptions, it is unclear what function NO fulfils in bacteria; this is for semantic debate, perhaps. However, if we accept that the function of intracellular NO may be to signal the presence of exogenous NO as a potentially toxic agent, then the sensing mechanism(s) are relevant.
NO sensing and signalling
An interesting example of NO acting as a signal between bacterium and host is that of Moraxella catarrhalis [37]. The nitrite reductase NirK (also known as AniA) generates NO and it is thought that this diffuses into the proximate environment of the bacterium where it increases the secretion of tumour necrosis factor alpha and modulates the expression of apoptotic proteins, therefore triggering host cell programmed death, thus dysregulating host cell gene expression. Conversely, nitrite-derived NO is toxic towards M. catarrhalis in maturing biofilms [37,38]. An opposite case might be the consumption of NO by meningococci [39]. We demonstrated that expression of meningococcal NorB increased the rate at which low-molecular-mass S-nitrosothiols (SNO) decompose in vitro. We then induced SNO formation in murine macrophages by activation with lipopolysaccharide and γ-interferon, resulting in a reduced abundance of SNO during co-incubation with Neisseria meningitidis, Salmonella enterica or Escherichia coli. In each case, this was dependent on bacterial NO detoxification genes, which prevented SNO formation through the removal of NO. This may represent a novel mechanism of NO-mediated host cell injury by bacteria.
NO sensors (Figure 1) may be subdivided into those that are specialised (or dedicated), and those that are secondary, in the sense that the sensor responds to another, primary signal. The prototypical secondary sensor is SoxR, its primary function being sensing of superoxide anion by virtue of a reactive [2Fe-2S] cluster that becomes oxidised, presumably by superoxide itself [40]. However, NO also activates SoxR and induces expression of genes in the SoxRS regulon. A further example is the bacterial redox sensor Fnr: its primary signal is O2, which reacts with the [4Fe-4S], via a mechanism that has been the subject of intensive studies [41]. However, Fnr also senses NO [42]. The Fnr homologue in Azotobacter vinelandii, CydR, is also both oxygen- and NO-reactive [43]. Subsequently, many other bacterial FeS proteins were reported to sense NO, e.g. the NO-responsive regulator of flavohaemoglobin gene expression in Streptomyces coelicolor[44]. Other secondary sensors include OxyR, MetR, DosS and Fur (for a review, see ref. [45]).
A well-established example of a dedicated NO sensor is that of NorR. In E. coli, NorR activates the divergently transcribed norVW genes in response to NO (or nitroprusside, GSNO or acidified nitrite, which we do not discuss here; see ref. [25]). These NorR-activated genes encode a flavorubredoxin and its redox partner that reduce NO to nitrous oxide at the expense of NADH [46–48]. NorR is activated by the formation of a mono-nitrosyl complex at a mononuclear iron centre (not an [FeS] cluster or haem) in the GAF domain [49,50]. Ferrous NorR reacts with NO in vitro, suggesting that NorR is a sensor of NO per se. However, in other bacteria, as in Pseudomonas aeruginosa, NorR activates transcription of the fhp gene (encoding flavohaemoglobin) in response to NO [51]. Thus, NorR is a true NO sensor and turns on genes exclusively involved in NO metabolism, but the physiological cue signalled by this mechanism in nature is unclear.
NsrR is also an NO-responsive regulator in many bacteria including E. coli [52,53], Bacillus subtilis [54], Neisseria gonorrhoae[55] and S. coelicolor [56]. Numerous genes may be so regulated but in S. coelicolor only two flavohaemoglobin genes (hmpA1 and hmpA2) are targets. These genes encode the NO-detoxifying flavohaemoglobin, rationalising the control exerted. The S. coelicolor NsrR contains an NO-reactive [4Fe-4S] cluster and exhibits high-affinity DNA binding to the operators of the NsrR-regulated genes. The reactions with NO are complex with up to 8–10 NO molecules reacting per cluster, leading to the formation of several iron-nitrosyl species [44].
Distinct classes of NO sensors are represented by the H-NOX proteins (haem-NO or oxygen-binding domain) [57,58] and the NosP protein discovered by Boon and colleagues. These haem proteins are widely distributed in bacterial genera and encoded in operons with genes for kinases or cyclic di-AMP synthesis/phosphodiesterase enzymes. In P. aeruginosa, it is well recognised that NO ‘directs’ the bacterium to disperse biofilms at picomolar concentrations and several proteins have been implicated, namely BdlA, DipA and others (see [58]). NosP is in the same operon as a gene for kinase previously implicated in biofilm formation. Biofilms of a mutant in the NosP signalling pathway lose the ability to disperse in response to NO [57], so NosP may be a global biofilm regulator. In Vibrio cholerae, NO binding to NosP inhibits the autophosphorylation activity of a histidine kinase implicated in quorum sensing. In other words, NO seems to regulate quorum sensing and biofilm formation, and may be a clear case of NO acting as a signal that alters bacterial behaviour. Beneficial effects of NO signalling are also reported in yeast [59]. However, why such physiological responses should be controlled by NO or what the source of that NO might be in natural environments is less clear.
CO sensing in microbes?
Unlike NO, CO has no obvious function in the vast majority of bacteria, with a few notable exceptions. Some prokaryotes that survive in hostile conditions sense and metabolise CO [60,61]. Often, these sensing proteins bear a haem cofactor that provides an allosteric binding site for CO due to the high affinity of the iron (witness the binding of CO to globin proteins). The most well-studied haem-dependent gas-sensing transcription factor is CooA (CO oxidation activator) from Rhodospirillum rubrum [62,63]. CooA, a member of the FNR/CRP superfamily of transcriptional regulators, is a homodimeric protein comprising an N-terminal haem-binding domain and a C-terminal helix–turn–helix DNA-binding domain. The haem iron undergoes a redox-mediated ligand switch as the mechanism for activation. A further haem-containing transcription factor associated with CO metabolism occurs in Burkholderia xenovorans (LB400) [64], which catalyses dissimilatory nitrate reduction and aerobic CO oxidation [65]. The cox genes that encode the molybdo-flavoprotein complex required for CO oxidation are regulated by the transcription factors RcoM-1 and RcoM-2 (regulator of CO metabolism). In this sense, then, CO may be considered as a ‘transmitter’, the signal being the availability of a useable, if unlikely, carbon source. It has been suggested that CO, occurring at a concentration of ∼700 ppm on Mars, could support near-surface bacterial activity; an abundant potential oxidant is perchlorate [66].
A more familiar part played by CO in biology is that of poison. CO is a notoriously toxic gas, binding primarily to ferrous oxygen-reactive haem proteins [67] in competition with oxygen. CO is generally considered to be toxic to microorganisms. It is used to preserve meat [68], but many bacteria are relatively resistant, in part, because they possess CO-insensitive oxidases, such as cytochrome bd [69]. Indeed, airborne bacteria survive high urban CO concentrations [70], and bacterial cultures may be bubbled with CO with little toxicity [71,72].
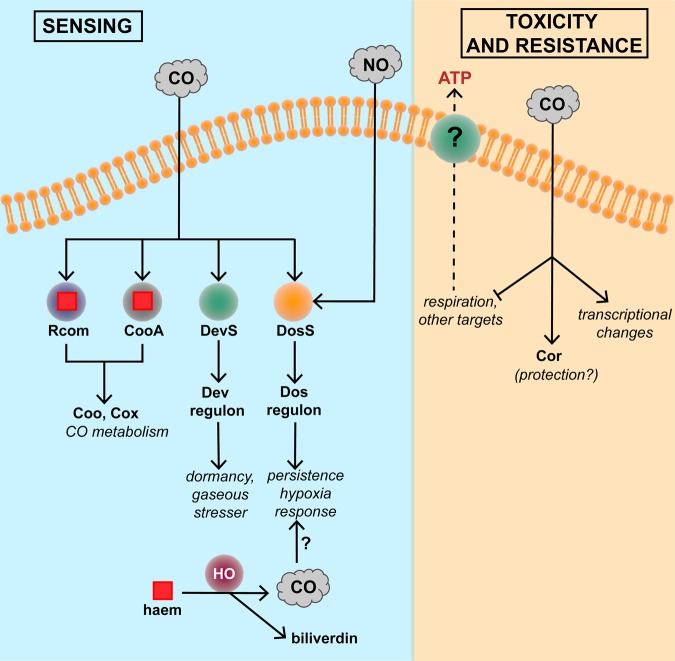
As described above, CO is generated endogenously in mammals by HO-catalysed breakdown of haem and, although relatively inert from a chemical standpoint, CO is an established gasotransmitter. Certain bacteria (e.g. Mycobacterium tuberculosis) sense host-derived CO (Figure 2). The DevRS/DosT (also called DosRS/DosT) system regulates an ∼50-member regulon in response to hypoxia, NO and CO [73–75]. More recently, the DevR regulon has been implicated in the transition of M. tuberculosis from an actively respiring to a latent non-replicating persistent state [76].
Figure 2. Carbon monoxide sensing and mechanisms of resistance to toxicity.
CO readily enters microbial cells from diverse exogenous sources, but may also be synthesised by endogenous haem oxygenase activity; the role of endogenously derived CO is unclear. Numerous sensors detect CO, leading to CO metabolism or downstream gene regulation. CO exerts numerous antimicrobial effects, but Cor is reported to afford protection. The reported effects of CO on ATP release from bacteria are unexplained. For details, see the text.
Curiously, infection with M. tuberculosis induces (and co-localises with) HO1 in both mouse and human tuberculosis lesions and primary human macrophages. This induction itself appears to be a virulence mechanism since experimental inhibition of HO1 reduces inflammatory cytokine production and restricts mycobacterial growth [77]. It is unclear whether HO1 is beneficial or detrimental during human infections by mycobacteria.
Wegiel et al. [71] recently hypothesised that bacteria exposed to CO release ATP (Figure 2), which activates inflammatory pathways. This appears to be a clear example of a gasotransmitter role, albeit between species: it is proposed that HO-1 is induced by endotoxin (LPS) (from bacteria) and generates CO. CO emitted by the host macrophage ‘compels the bacteria to generate ATP’, which acts as a danger-associated molecular pattern for the macrophage. The bacteria-derived ATP binds to a purinergic receptor on the macrophage, leading to activation of the inflammasome to drive pathogen clearance. There are several puzzling aspects of this claim. First, we are unaware of any extracellular or periplasmic location of ATP in bacteria, implying that the ATP ‘leaked’ from damaged cells, although the authors state that bacteria were viable. Second, the ATP release was attributed to a CO-dependent increase in bacterial ATP levels, a finding hard to reconcile with inhibition of respiration by CO.
If CO is toxic, it might be expected that bacteria could possess genes that confer CO resistance. One such gene, cor, has been described in M. tuberculosis (Figure 2). Zacharia et al. [78] screened an M. tuberculosis transposon library for mutants susceptible to an atmosphere of 2% CO and found that disruption of Rv1829 (carbon monoxide resistance, Cor) led to marked CO sensitivity. Heterologous expression of Cor in E. coli rescued it from CO toxicity, although this assay used CORM-2 [tricarbonyldichlororuthenium(II) dimer] as a CO source, not CO itself. The impact of the redox-active ruthenium ion [79] should be assessed in further studies. Importantly, the virulence of the cor mutant was attenuated in a mouse model of tuberculosis. Thus, Cor appears to protect bacteria from host-derived CO. Conceivably, this example could serve as an illustration of a bacterium receiving a signal on the presence of CO, but the molecular mechanisms of the CO sensing and its detoxification are unknown.
H2S sensing and signalling in microbes
H2S is emerging as an important mediator or gasotransmitter, promoting the resolution of inflammation and injury repair [22,80]. A recent bibliometric analysis of publications in biology and medicine shows almost 6000 papers between 1990 and 2016 [81]. Although the term ‘hydrogen sulfide’ is most commonly used [20], H2S is a weak acid in aqueous solutions — dissociating into its anions HS− and S2−. As the dissociation equilibrium between H2S and HS− is at pH ∼7, in physiological conditions ‘H2S’ exists predominantly as its anion HS− (∼80%), H2S (∼20%) and negligible amounts of S2−, which are collectively termed the ‘sulfide pool’ [82]. Thus, although it has not been possible to determine which form of H2S (H2S, HS− or S2−) is active, the term ‘hydrogen sulfide’ has been used in toxicity studies [20], and it is customarily regarded as a gasotransmitter [23].
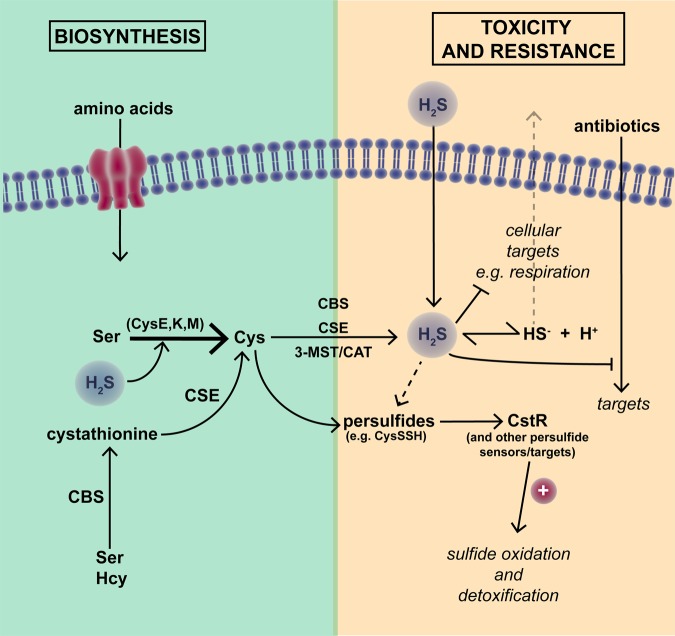
The principal pathways that mediate H2S production in mammalian systems (e.g. [21,22]) are shown in Figure 3. A more recently discovered route involves 3-MST and d-amino acid oxidase [83]. H2S in higher organisms also arises in part from bacterial sulfur metabolism. Homologues of CBS, CSE and 3-MST/CAT also appear to be ubiquitous across bacterial genomes [84], although in most bacteria l-cysteine is produced enzymatically (via CysE, CysK and CysM) from l-serine. For a review, see [85]. H2S may also be generated in microbes by other physiological processes, i.e. via degradation of cysteine and other sulfur-containing amino acids/peptides, or non-enzymatically, by dissimilatory reduction of inorganic sulfur compounds [84,86] (Figure 3). Non-enzymatic generation of H2S by obligate or facultative anaerobic sulfate-reducing bacteria occurs via the utilisation of inorganic sulfur compounds such as sulfates, sulfites or polysulfides as alternative terminal electron acceptors for reduced organic substrates [86].
Figure 3. Hydrogen sulfide biosynthesis, signalling and regulation.
As described in the text, H2S is synthesised by diverse pathways from cysteine and cystathionine, but also readily permeates membranes in the unionised form. Specific H2S synthesis enzymes include cystathionine-β-synthase (CBS), cystathionine-γ-synthase (CSE) and the 3-mercaptopyruvate sulfurtransferase/cysteine aminotransferase (3-MST/CAT) pathway. Persulfides (e.g. CysSSR) are key to H2S signalling pathways. CstR is a persulfide-sensing sulfide stress-response regulator in S. aureus.
H2S, like NO but unlike CO, is reactive. Its signalling effects are complex since it interacts with oxygen and NO. These reaction products are outside the scope of this review. At elevated concentrations, extracellular H2S is toxic to bacteria, complexing with transition metals, leading to respiratory inhibition and covalent modification of haem-containing and Fe–S containing enzymes [82]. Recently, a specific terminal oxidase, cytochrome bd, has been shown to be required for sulfide tolerance [87]; the same oxidase also confers tolerance to NO [88] and CORM-2 [69]. Although H2S-induced toxicity is linked to oxidative damage via inhibition of antioxidant enzymes [89], low levels of intracellular H2S protect against metal ion toxicity, antibiotics and oxidative damage. H2S directly scavenges metals ions and reactive oxygen species (ROS) and prevents Fenton chemistry and related antibiotic-induced oxidative damage [82,84,90]. H2S-mediated signalling processes appear to play an important role in microbial resistance to oxidative stress [82,91].
Low-molecular-mass persulfides such as cysteine hydropersulfide (CysSSH) and glutathione (GSSG) have long been recognised as important metabolites, and recent advances in analytical tools have identified many such species in prokaryotes and eukaryotes [85,92], but these are outside the scope of this review. H2S signalling occurs via post-translational modification of protein cysteine residues (–SH groups) to form the persulfide moiety (RSSH), which in turn modulates protein activity [91]. This process (persulfidation or ‘S-sulfhydration’) is the major mechanism of H2S signalling in mammalian systems leading to the classification of H2S as the third gasotransmitter. In evolutionarily diverse bacteria, sensing of persulfides typically involves transcriptional repressors that contain conserved cysteine residues that regulate DNA-binding activity. Persulfidation or oxidation of these residues induces structural changes leading to derepression of sulfide oxidation genes when reactive sulfide species (RSS) are present [93]. Proteomic profiling of Staphylococcus aureus in response to exogenous sulfide revealed that persulfidation regulates many key metabolic enzymes and ROS and RNS stress-response systems and other virulence-related genes, including the persulfide stress-response regulator CstR [91]. In addition, there appears to be a dedicated thioredoxin-mediated hydrosulfide reduction system that functions to reverse persulfidation in S. aureus [91,94].
To what extent is H2S a true signalling effector molecule in microbes? Though it is clear that bacteria both produce and sense H2S, albeit indirectly via related RSS, it is currently unclear whether endogenously produced H2S is an effector molecule that induces a signalling response in the same bacterial cell or if H2S-mediated responses are more a response to environmental and/or host-derived H2S or RSS.
Conclusions
There can be no doubt that in microbes, as in higher organisms, small molecules that can exist as gases (‘gasotransmitters’) are both generated and sensed. In the bacterial world, an extraordinarily sophisticated body of knowledge has been acquired on the generation of these signals/molecules and on their perception. However, in very few cases, do we understand what the signals mean. Many bacteria respond, for example, to NO, but why? The clearest rationalisation may be the protective response of bacteria that have been engulfed by phagocytic cells. Strictly adopting the criteria laid down in eukaryotic cells leads us to the view that, in such a case, the gas molecule is not a transmitter, being not generated endogenously (i.e. within the bacterium). Conversely, why do bacteria make NO? Similar doubts are raised regarding CO and H2S: to be regarded as a true ‘gasotransmitter’, the small signalling molecule should derive from, and be active in, the same cell/organism. Because many aspects of the microbial effects of these gases remain poorly resolved, it is perhaps dangerous to adopt the term ‘gasotransmitter’, which has been so precisely defined (Table 1). This leads us to conclude that the term ‘small molecule signalling agent’, as proposed by others, is preferable terminology in the case of bacteria.
Table 1. Do NO, CO and H2S qualify as gasotransmitters?
For criteria (iv) to (vi), examples are given.
| Criterion | NO | CO | H2S |
|---|---|---|---|
| (i) Small gas | ✓ | ✓ | ✓ |
| (ii) Membrane-permeable | ✓ | ✓ | ✓ |
| (iii) Endogenously synthesised | ✓ | ✓ | ✓ |
| (iv) Specific functions; manipulating endogenous levels evokes changes | ✓ antibiotic resistance, virulence, quorum sensing, biofilms, etc. | unclear, except in CO metabolism1 | ✓ antibiotic resistance, oxidative stress resistance |
| (v) Effects mimicked by exogenous counterparts | ✓ | Only toxicity can be mimicked by CO or CORMs? | ✓ |
| (vi) Specific cellular/molecular targets2 | ✓ | unclear | Only via persulfide chemistry |
We do not regard use of CO as a metabolic substrate as signal transduction or gasotransmitter functions; endogenous CO is not used in this way.
We exclude targets such as metal centres that may lead to toxicity.
Conductors must give unmistakable and suggestive signals to the orchestra — not choreography to the audience
(George Szell, conductor, 1897–1970)
Abbreviations
- 3-MST
3-mercaptopyruvate sulfurtransferase
- CAT
cysteine aminotransferase
- CBS
cystathionine-β-synthase
- CO
carbon monoxide
- CooA
CO oxidation activator
- Cor
carbon monoxide resistance
- CORMs
CO-releasing molecules
- CSE
cystathionine-γ-synthase
- eNOS
endothelial NOS
- H2S
hydrogen sulfide
- Hcy
homocysteine
- HO
haem oxygenase
- iNOS
inducible NOS
- nNOS
neuronal NOS
- NO
nitric oxide
- NOS
Nitric oxide synthase
- ROS
reactive oxygen species
- SNO
S-nitrosothiols
Funding
The UK Biotechnology and Biological Sciences Research Council (BBSRC) most recently [BB/M022579/1] funded work leading to this manuscript.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Wang R. (2002) Two's company, three's a crowd: can H2S be the third endogenous gaseous transmitter? FASEB J. 16, 1792–1798 10.1096/fj.02-0211hyp [DOI] [PubMed] [Google Scholar]
- 2.Wang R. (2004) The evolution of gasotransmitter biology and medicine. From atmospheric toxic gases to endogenous gaseous signaling molecules In Signal Transduction and the Gasotransmitters. NO, CO and H2S in Biology and Medicine (Wang R., ed.), pp. 3–31, Humana Press, Totowa, New Jersey [Google Scholar]
- 3.Fukuto J.M., Carrington S.J., Tantillo D.J., Harrison J.G., Ignarro L.J., Freeman B.A. et al. (2012) Small molecule signaling agents: the integrated chemistry and biochemistry of nitrogen oxides, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species. Chem. Res. Toxicol. 25, 769–793 10.1021/tx2005234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luhachack L. and Nudler E. (2014) Bacterial gasotransmitters: an innate defense against antibiotics. Curr. Opin. Microbiol. 21, 13–17 10.1016/j.mib.2014.06.017 [DOI] [PubMed] [Google Scholar]
- 5.Chinta K.C., Saini V., Glasgow J.N., Mazorodze J.H., Rahman M.A., Reddy D. et al. (2016) The emerging role of gasotransmitters in the pathogenesis of tuberculosis. Nitric Oxide Biol. Chem. 59, 28–41 10.1016/j.niox.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L. and Moore P.K. (2007) An overview of the biological significance of endogenous gases: new roles for old molecules. Biochem. Soc. Trans. 35, 1138–1141 10.1042/BST0351138 [DOI] [PubMed] [Google Scholar]
- 7.Forte E. and Giuffre A. (2016) How bacteria breathe in hydrogen sulfide-rich environments. Biochemist 38, 8–11 [Google Scholar]
- 8.Zivanovic J. and Filipovic M.R. (2016) Hydrogen sulfide: stench from the past as a mediator of the future. Biochemist 38, 12–17 [Google Scholar]
- 9.Nichols A., Ahmed K.A. and Patel R.P. (2016) Yes to ‘NO’ host flora symbiosis. Biochemist 38, 18–21 [Google Scholar]
- 10.Huang Y.Q., Tang C.S., Du J.B. and Jin H.F. (2016) Endogenous sulfur dioxide: a new member of gasotransmitter family in the cardiovascular system. Oxid. Med. Cell Longev. 2016, 8961951 10.1155/2016/8961951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryan N.S. and Loscalzo J. (2017) Nitrite and Nitrate in Human Health and Disease, Springer [Google Scholar]
- 12.Mattila J.T. and Thomas A.C. (2014) Nitric oxide synthase: non-canonical expression patterns. Front. Immunol. 5, 1–5 10.3389/fimmu.2014.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maines M.D. and Gibbs P.E.M. (2005) 30 some years of heme oxygenase: from a ‘molecular wrecking ball’ to a ‘mesmerizing’ trigger of cellular events. Biochem. Biophys. Res. Commun. 338, 568–577 10.1016/j.bbrc.2005.08.121 [DOI] [PubMed] [Google Scholar]
- 14.Maines M.D. (1997) The heme oxygenase system: a regulator of second messenger gases. Annu. Rev. Pharmacol. Toxicol. 37, 517–554 10.1146/annurev.pharmtox.37.1.517 [DOI] [PubMed] [Google Scholar]
- 15.Motterlini R. and Foresti R. (2017) Biological signaling by carbon monoxide and carbon monoxide-releasing molecules. Am. J. Physiol. Cell Physiol. 312, C302–C313 10.1152/ajpcell.00360.2016 [DOI] [PubMed] [Google Scholar]
- 16.Otterbein L.E. (2002) Carbon monoxide: innovative anti-inflammatory properties of an age-old gas molecule. Antiox. Redox Signal. 4, 309–319 10.1089/152308602753666361 [DOI] [PubMed] [Google Scholar]
- 17.Ryter S.W. and Choi A.M.K. (2013) Carbon monoxide: present and future indications for a medical gas. Korean J. Intern. Med. 28, 123–140 10.3904/kjim.2013.28.2.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Queiroga C.S.F., Vercelli A. and Vieira H.L.A. (2015) Carbon monoxide and the CNS: challenges and achievements. Br. J. Pharmacol. 172, 1533–1545 10.1111/bph.12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wegiel B., Hauser C.J. and Otterbein L.E. (2015) Heme as a danger molecule in pathogen recognition. Free Radic. Biol. Med. 89, 651–661 10.1016/j.freeradbiomed.2015.08.020 [DOI] [PubMed] [Google Scholar]
- 20.Abe K. and Kimura H. (1996) The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 16, 1066–1071 10.1523/JNEUROSCI.16-03-01066.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shibuya N., Tanaka M., Yoshida M., Ogasawara Y., Togawa T., Ishii K. et al. (2009) 3-Mercaptopyruvate sulfurtransferase produces hydrogen sulfide and bound sulfane sulfur in the brain. Antiox. Redox Signal. 11, 703–714 10.1089/ars.2008.2253 [DOI] [PubMed] [Google Scholar]
- 22.Rose P., Moore P.K. and Zhu Y.Z. (2017) H2s biosynthesis and catabolism: new insights from molecular studies. Cell. Mol. Life Sci. 74, 1391–1412 10.1007/s00018-016-2406-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang R. (2003) The gasotransmitter role of hydrogen sulfide. Antiox. Redox Signal. 5, 493–501 10.1089/152308603768295249 [DOI] [PubMed] [Google Scholar]
- 24.Poole R.K. and Hughes M.N. (2000) New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36, 775–783 10.1046/j.1365-2958.2000.01889.x [DOI] [PubMed] [Google Scholar]
- 25.Bowman L.A.H., McLean S., Poole R.K. and Fukuto J. (2011) The diversity of microbial responses to nitric oxide and agents of nitrosative stress: close cousins but not identical twins. Adv. Microb. Physiol. 59, 135–219 10.1016/B978-0-12-387661-4.00006-9 [DOI] [PubMed] [Google Scholar]
- 26.Lundberg J.O., Weitzberg E., Cole J.A. and Benjamin N. (2004) Nitrate, bacteria and human health. Nat. Rev. Microbiol. 2, 593–602 10.1038/nrmicro929 [DOI] [PubMed] [Google Scholar]
- 27.Stuehr D.J. and Vasquez-Vivar J. (2017) Nitric oxide synthases—from genes to function. Nitric Oxide Biol. Chem. 63, 29–29 10.1016/j.niox.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevanin T.M., Poole R.K., Demoncheaux E.A.G. and Read R.C. (2002) Flavohemoglobin Hmp protects Salmonella enterica serovar Typhimurium from nitric oxide-related killing by human macrophages. Infect. Immun. 70, 4399–4405 10.1128/IAI.70.8.4399-4405.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole J.A. (2018) Anaerobic bacterial response to nitrosative stress. Adv. Microb. Physiol. 72, 193–237 10.1016/bs.ampbs.2018.01.001 [DOI] [PubMed] [Google Scholar]
- 30.Gusarov I., Starodubtseva M., Wang Z.Q., McQuade L., Lippard S.J., Stuehr D.J. et al. (2008) Bacterial nitric-oxide synthases operate without a dedicated redox partner. J. Biol. Chem. 283, 13140–13147 10.1074/jbc.M710178200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crane B.R., Sudhamsu J. and Patel B.A. (2010) Bacterial nitric oxide synthases. Annu. Rev. Biochem. 79, 445–470 10.1146/annurev-biochem-062608-103436 [DOI] [PubMed] [Google Scholar]
- 32.Kers J.A., Wach M.J., Krasnoff S.B., Widom J., Cameron K.D., Bukhalid R.A. et al. (2004) Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature 429, 79–82 10.1038/nature02504 [DOI] [PubMed] [Google Scholar]
- 33.Gusarov I. and Nudler E. (2005) NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc. Natl Acad. Sci. U.S.A. 102, 13855–13860 10.1073/pnas.0504307102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gusarov I., Shatalin K., Starodubtseva M. and Nudler E. (2009) Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325, 1380–1384 10.1126/science.1175439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinkel T.L., Ramos-Montanez S., Pando J.M., Tadeo D.V., Strom E.N., Libby S.J. et al. (2017) An essential role for bacterial nitric oxide synthase in Staphylococcus aureus electron transfer and colonization. Nat. Microbiol. 2, 16224 10.1038/nmicrobiol.2016.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shatalin K., Gusarov I., Avetissova E., Shatalina Y., McQuade L.E., Lippard S.J. et al. (2008) Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc. Natl Acad. Sci. U.S.A. 105, 1009–1013 10.1073/pnas.0710950105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mocca B., Yin D.D., Gao Y.M. and Wang W. (2015) Moraxella catarrhalis-produced nitric oxide has dual roles in pathogenicity and clearance of infection in bacterial-host cell co-cultures. Nitric Oxide Biol. Chem. 51, 52–62 10.1016/j.niox.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 38.Mocca B. and Wang W. (2012) Bacterium-generated nitric oxide hijacks host tumor necrosis factor alpha signaling and modulates the host cell cycle in vitro. J. Bacteriol. 194, 4059–4068 10.1128/JB.00476-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laver J.R., Stevanin T.M., Messenger S.L., Lunn A.D., Lee M.E., Moir J.W. et al. (2010) Bacterial nitric oxide detoxification prevents host cell S-nitrosothiol formation: a novel mechanism of bacterial pathogenesis. FASEB J. 24, 286–295 10.1096/fj.08-128330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiro S. (2007) Regulators of bacterial responses to nitric oxide. FEMS Microbiol. Rev. 31, 193–211 10.1111/j.1574-6976.2006.00061.x [DOI] [PubMed] [Google Scholar]
- 41.Green J. and Paget M.S. (2004) Bacterial redox sensors. Nat. Rev. Microbiol. 2, 954–966 10.1038/nrmicro1022 [DOI] [PubMed] [Google Scholar]
- 42.Cruz-Ramos H., Crack J., Wu G., Hughes M.N., Scott C., Thomson A.J. et al. (2002) NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 21, 3235–3244 10.1093/emboj/cdf339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu G., Cruz-Ramos H., Hill S., Green J., Sawers G. and Poole R.K. (2000) Regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii by CydR (Fnr). Sensitivity to oxygen, reactive oxygen species, and nitric oxide. J. Biol. Chem. 275, 4679–4686 10.1074/jbc.275.7.4679 [DOI] [PubMed] [Google Scholar]
- 44.Crack J.C., Svistunenko D.A., Munnoch J., Thomson A.J., Hutchings M.I. and Le Brun N.E. (2016) Differentiated, promoter-specific response of 4Fe-4S NsrR DNA binding to reaction with nitric oxide. J. Biol. Chem. 291, 8663–8672 10.1074/jbc.M115.693192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spiro S. (2006) Nitric oxide-sensing mechanisms in Escherichia coli. Biochem. Soc. Trans. 34, 200–202 10.1042/BST0340200 [DOI] [PubMed] [Google Scholar]
- 46.Gardner A.M., Gessner C.R. and Gardner P.R. (2003) Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. Role of NorR and s54 in the nitric oxide stress response. J. Biol. Chem. 278, 10081–10086 10.1074/jbc.M212462200 [DOI] [PubMed] [Google Scholar]
- 47.Gomes C.M., Giuffre A., Forte E., Vicente J.B., Saraiva L.M., Brunori M. et al. (2002) A novel type of nitric-oxide reductase — Escherichia coli flavorubredoxin. J. Biol. Chem. 277, 25273–25276 10.1074/jbc.M203886200 [DOI] [PubMed] [Google Scholar]
- 48.Hutchings M.I., Mandhana N. and Spiro S. (2002) The NorR protein of Escherichia coli activates expression of the flavorubredoxin gene norV in response to reactive nitrogen species. J. Bacteriol. 184, 4640–4643 10.1128/JB.184.16.4640-4643.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D'Autreaux B., Tucker N.P., Dixon R. and Spiro S. (2005) A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature 437, 769–772 10.1038/nature03953 [DOI] [PubMed] [Google Scholar]
- 50.Tucker N., D'Autreaux B., Spiro S. and Dixon R. (2005) DNA binding properties of the Escherichia coli nitric oxide sensor NorR: towards an understanding of the regulation of flavorubredoxin expression. Biochem. Soc. Trans. 33, 181–183 10.1042/BST0330181 [DOI] [PubMed] [Google Scholar]
- 51.Arai H., Hayashi M., Kuroi A., Ishii M. and Igarashi Y. (2005) Transcriptional regulation of the flavohemoglobin gene for aerobic nitric oxide detoxification by the second nitric oxide-responsive regulator of Pseudomonas aeruginosa. J. Bacteriol. 187, 3960–3968 10.1128/JB.187.12.3960-3968.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bodenmiller D.M. and Spiro S. (2006) The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188, 874–881 10.1128/JB.188.3.874-881.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Partridge J.D., Bodenmiller D.M., Humphrys M.S. and Spiro S. (2009) Nsrr targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol. Microbiol. 73, 680–694 10.1111/j.1365-2958.2009.06799.x [DOI] [PubMed] [Google Scholar]
- 54.Kommineni S., Lama A., Popescu B. and Nakano M.M. (2012) Global transcriptional control by NsrR in Bacillus subtilis. J. Bacteriol. 194, 1679–1688 10.1128/JB.06486-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heurlier K., Thomson M.J., Aziz N. and Moir J.W.B. (2008) The nitric oxide (NO)-sensing repressor NsrR of Neisseria meningitidis has a compact regulon of genes involved in NO synthesis and detoxification. J. Bacteriol. 190, 2488–2495 10.1128/JB.01869-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crack J.C., Munnoch J., Dodd E.L., Knowles F., Al Bassam M.M., Kamali S. et al. (2015) Nsrr from Streptomyces coelicolor is a nitric oxide-sensing 4Fe-4S cluster protein with a specialized regulatory function. J. Biol. Chem. 290, 12689–12704 10.1074/jbc.M115.643072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hossain S. and Boon E.M. (2017) Discovery of a novel nitric oxide binding protein and nitric-oxide-responsive signaling pathway in Pseudomonas aeruginosa. ACS Infect. Dis. 3, 454–461 10.1021/acsinfecdis.7b00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hossain S., Nisbett L.M. and Boon E.M. (2017) Discovery of two bacterial nitric oxide-responsive proteins and their roles in bacterial biofilm regulation. Acc. Chem. Res. 50, 1633–1639 10.1021/acs.accounts.7b00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Astuti R.I., Nasuno R. and Takagi H. (2018) Nitric oxide signalling in yeast. Adv. Microb. Physiol. 72, 29–63 10.1016/bs.ampbs.2018.01.003 [DOI] [PubMed] [Google Scholar]
- 60.McDuff S., King G.M., Neupane S. and Myers M.R. (2016) Isolation and characterization of extremely halophilic CO-oxidizing Euryarchaeota from hypersaline cinders, sediments and soils and description of a novel CO oxidizer, Haloferax namakaokahaiae Mke2.3(T), sp. nov. FEMS Microbiol. Ecol. 92, fiw028 10.1093/femsec/fiw028 [DOI] [PubMed] [Google Scholar]
- 61.Weber C.F. and King G.M. (2017) Volcanic soils as sources of novel CO-oxidizing Paraburkholderia and Burkholderia: Paraburkholderia hiiakae sp nov., Paraburkholderia metrosideri sp nov., Paraburkholderia paradisi sp nov., Paraburkholderia peleae sp nov., and Burkholderia alpina sp nov. a member of the Burkholderia cepacia complex. Front. Microbiol. 8, 207 10.3389/fmicb.2017.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shelver D., Kerby R.L., He Y.P. and Roberts G.P. (1997) Cooa, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc. Natl Acad. Sci. U.S.A. 94, 11216–11220 10.1073/pnas.94.21.11216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shelver D., Thorsteinsson M.V., Kerby R.L., Chung S.Y., Roberts G.P., Reynolds M.F. et al. (1999) Identification of two important heme site residues (cysteine 75 and histidine 77) in CooA, the CO-sensing transcription factor of Rhodospirillum rubrum. Biochemistry 38, 2669–2678 10.1021/bi982658j [DOI] [PubMed] [Google Scholar]
- 64.Kerby R.L., Youn H. and Roberts G.P. (2008) Rcom: a new single-component transcriptional regulator of CO metabolism in bacteria. J. Bacteriol. 190, 3336–3343 10.1128/JB.00033-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.King G.M. (2006) Nitrate-dependent anaerobic carbon monoxide oxidation by aerobic CO-oxidizing bacteria. FEMS Microbiol. Ecol. 56, 1–7 10.1111/j.1574-6941.2006.00065.x [DOI] [PubMed] [Google Scholar]
- 66.Myers M.R. and King G.M. (2017) Perchlorate-coupled carbon monoxide (CO) oxidation: evidence for a plausible microbe-mediated reaction in Martian brines. Front. Microbiol. 8, 2571 10.3389/fmicb.2017.02571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davidge K.S., Sanguinetti G., Yee C.H., Cox A.G., McLeod C.W., Monk C.E. et al. (2009) Carbon monoxide-releasing antibacterial molecules target respiration and global transcriptional regulators. J. Biol. Chem. 284, 4516–4524 10.1074/jbc.M808210200 [DOI] [PubMed] [Google Scholar]
- 68.Ramamoorthi L., Toshkov S. and Brewer M.S. (2009) Effects of carbon monoxide-modified atmosphere packaging and irradiation on E. coli K12 survival and raw beef quality. Meat Sci. 83, 358–365 10.1016/j.meatsci.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 69.Jesse H.E., Nye T.L., McLean S., Green J., Mann B.E. and Poole R.K. (2013) The terminal oxidase cytochrome bd-I in Escherichia coli has lower susceptibility than cytochromes bd-II or bo′ to inhibition by the carbon monoxide-releasing molecule, CORM-3: N-acetylcysteine reduces CO-RM uptake and inhibition of respiration. Biochim. Biophys. Acta 1834, 1693–1703 10.1016/j.bbapap.2013.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lighthart B. (1973) Survival of airborne bacteria in a high urban concentration of carbon monoxide. Appl. Microbiol. 25, 86–91 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wegiel B., Larsen R., Gallo D., Chin B.Y., Harris C., Mannam P. et al. (2014) Macrophages sense and kill bacteria through carbon monoxide-dependent inflammasome activation. J. Clin. Invest. 124, 4926–4940 10.1172/JCI72853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wareham L.K., Begg R., Jesse H.E., van Beilen J.W.A., Ali S., Svistunenko D. et al. (2016) Carbon monoxide gas is not inert, but global, in its consequences for bacterial gene expression, iron acquisition, and antibiotic resistance. Antiox. Redox Signal. 24, 1013–1028 10.1089/ars.2015.6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voskuil M.I., Schnappinger D., Visconti K.C., Harrell M.I., Dolganov G.M., Sherman D.R. et al. (2003) Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198, 705–713 10.1084/jem.20030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar A., Deshane J.S., Crossman D.K., Bolisetty S., Yan B.S., Kramnik I. et al. (2008) Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. 283, 18032–18039 10.1074/jbc.M802274200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shiloh M.U., Manzanillo P. and Cox J.S. (2008) Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microb. 3, 323–330 10.1016/j.chom.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leistikow R.L., Morton R.A., Bartek I.L., Frimpong I., Wagner K. and Voskuil M.I. (2010) The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J. Bacteriol. 192, 1662–1670 10.1128/JB.00926-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scharn C.R., Collins A.C., Nair V.R., Stamm C.E., Marciano D.K., Graviss E.A. et al. (2016) Heme oxygenase-1 regulates inflammation and mycobacterial survival in human macrophages during Mycobacterium tuberculosis infection. J. Immunol. 196, 4641–4649 10.4049/jimmunol.1500434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zacharia V.M., Manzanillo P.S., Nair V.R., Marciano D.K., Kinch L.N., Grishin N.V. et al. (2013) Cor, a Novel carbon monoxide resistance gene, is essential for Mycobacterium tuberculosis pathogenesis. mBio 4, e00721-00713 10.1128/mBio.00721-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Southam H.M., Butler J.A., Chapman J.A. and Poole R.K. (2017) The microbiology of ruthenium complexes. Adv. Microb. Physiol. 71, 1–96 10.1016/bs.ampbs.2017.03.001 [DOI] [PubMed] [Google Scholar]
- 80.Wallace J.L., Ferraz J.G. and Muscara M.N. (2012) Hydrogen sulfide: an endogenous mediator of resolution of inflammation and injury. Antioxid. Redox Signal. 17, 58–67 10.1089/ars.2011.4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang G.D. and Wu L.Y. (2017) Trend in H2S biology and medicine research — a bibliometric analysis. Molecules 22, E2087 10.3390/molecules22122087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pal V.K., Bandyopadhyay P. and Singh A. (2018) Hydrogen sulfide in physiology and pathogenesis of bacteria and viruses. IUBMB Life 70, 393–410 10.1002/iub.1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shibuya N., Koike S., Tanaka M., Ishigami-Yuasa M., Kimura Y., Ogasawara Y. et al. (2013) A novel pathway for the production of hydrogen sulfide from d-cysteine in mammalian cells. Nat. Commun. 4, 1366 10.1038/ncomms2371 [DOI] [PubMed] [Google Scholar]
- 84.Shatalin K., Shatalina E., Mironov A. and Nudler E. (2011) H2s: a universal defense against antibiotics in bacteria. Science 334, 986–990 10.1126/science.1209855 [DOI] [PubMed] [Google Scholar]
- 85.Sawa T., Ono K., Tsutsuki H., Zhang T., Ida T., Nishida M. et al. (2018) Reactive cysteine persulphides: occurrence, biosynthesis, antioxidant activity, methodologies, and bacterial persulphide signalling. Adv. Microb. Physiol. 72, 1–28 10.1016/bs.ampbs.2018.01.002 [DOI] [PubMed] [Google Scholar]
- 86.Tomasova L., Konopelski P. and Ufnal M. (2016) Gut bacteria and hydrogen sulfide: the new old players in circulatory system homeostasis. Molecules 21, E1558 10.3390/molecules21111558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Forte E., Borisov V.B., Falabella M., Colaco H.G., Tinajero-Trejo M., Poole R.K. et al. (2016) The terminal oxidase cytochrome bd promotes sulfide-resistant bacterial respiration and growth. Sci. Rep. 6, 23788 10.1038/srep23788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mason M.G., Shepherd M., Nicholls P., Dobbin P.S., Dodsworth K.S., Poole R.K. et al. (2009) Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat. Chem. Biol. 5, 94–96 10.1038/nchembio.135 [DOI] [PubMed] [Google Scholar]
- 89.Fu L.H., Wei Z.Z., Hu K.D., Hu L.Y., Li Y.H., Chen X.Y. et al. (2018) Hydrogen sulfide inhibits the growth of Escherichia coli through oxidative damage. J. Microbiol. 56, 238–245 10.1007/s12275-018-7537-1 [DOI] [PubMed] [Google Scholar]
- 90.Mironov A., Seregina T., Nagornykh M., Luhachack L.G., Korolkova N., Lopes L.E. et al. (2017) Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc. Natl Acad. Sci. U.S.A. 114, 6022–6027 10.1073/pnas.1703576114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peng H., Zhang Y., Palmer L.D., Kehl-Fie T.E., Skaar E.P., Trinidad J.C. et al. (2017) Hydrogen sulfide and reactive sulfur species impact proteome S-sulfhydration and global virulence regulation in Staphylococcus aureus. ACS Infect. Dis. 3, 744–755 10.1021/acsinfecdis.7b00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kimura Y., Toyofuku Y., Koike S., Shibuya N., Nagahara N., Lefer D. et al. (2015) Identification of H2S3 and H2S produced by 3-mercaptopyruvate sulfurtransferase in the brain. Sci. Rep. 5, 14774 10.1038/srep14774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shimizu T., Shen J., Fang M., Zhang Y., Hori K., Trinidad J.C. et al. (2017) Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc. Natl Acad. Sci. U.S.A. 114, 2355–2360 10.1073/pnas.1614133114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luebke J.L., Shen J., Bruce K.E., Kehl-Fie T.E., Peng H., Skaar E.P. et al. (2014) The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol. Microbiol. 94, 1343–1360 10.1111/mmi.12835 [DOI] [PMC free article] [PubMed] [Google Scholar]