Abstract
Background
Bacterial meningitis remains a big threat to the integrity of the central nervous system (CNS), despite the advancements in antimicrobial reagents. Escherichia coli is a bacterial pathogen that can disrupt the CNS function, especially in neonates. E. coli meningitis occurs after bacteria invade the brain microvascular endothelial cells (BMECs) that form a direct and essential barrier restricting the entry of circulating microbes and toxins to the brain. Previous studies have reported on several cellular proteins that function during meningitic E. coli infections; however, more comprehensive investigations to elucidate the potential targets involved in E. coli meningitis are essential to better understand this disease and discover new treatments for it.
Methods
The isobaric tags for relative and absolute quantification (iTRAQ) approach coupled with LC-MS/MS were applied to compare and characterize the different proteomic profiles of BMECs in response to meningitic or non-meningitic E. coli strains. KEGG and gene ontology annotations, ingenuity pathways analysis, and functional experiments were combined to identify the key host molecules involved in the meningitic E. coli-induced tight junction breakdown and neuroinflammatory responses.
Results
A total of 13 cellular proteins were found to be differentially expressed by meningitic E. coli strains PCN033 and RS218, including one that was also affected by HB101, a non-meningitic E. coli strain. Through bioinformatics analysis, we identified the macrophage migration inhibitory factor (MIF), granzyme A, NF-κB signaling, and mitogen-activated protein kinase (MAPK) pathways as being biologically involved in the meningitic E. coli-induced tight junction breakdown and neuroinflammation. Functionally, we showed that MIF facilitated meningitic E. coli-induced production of cytokines and chemokines and also helped to disrupt the blood-brain barrier by decreasing the expression of tight junction proteins like ZO-1, occludin. Moreover, we demonstrated the significant activation of NF-κB and MAPK signaling in BMECs in response to meningitic E. coli strains, which dominantly determined the generation of the proinflammatory cytokines including IL-6, IL-8, TNF-α, and IL-1β.
Conclusions
Our work identified 12 host cellular targets that are affected by meningitic E. coli strains and revealed MIF to be an important contributor to meningitic E. coli-induced cytokine production and tight junction disruption, and also the NF-κB and MAPK signaling pathways that are mainly involved in the infection-induced cytokines production. Characterization of these distinct proteins and pathways in BMECs will facilitate further elucidation of meningitis-causing mechanisms in humans and animals, thereby enabling the development of novel preventative and therapeutic strategies against infection with meningitic E. coli.
Electronic supplementary material
The online version of this article (10.1186/s12974-018-1325-z) contains supplementary material, which is available to authorized users.
Keywords: iTRAQ, Proteomics, Blood-brain barrier, BMECs, Meningitic E. coli
Background
Bacterial meningitis is a severe, life-threatening infection of the central nervous system (CNS) with high morbidity and mortality. It is currently recognized as one of the top ten killers in infection-related deaths worldwide, with almost half of the survivors suffering from diverse neurological sequelae (e.g., mental retardation, hearing impairment and blindness), despite the advancements made in the field of antimicrobial treatment [1–3]. Most bacterial meningitis cases are initiated by hematogenous spread and develop when the circulating bacteria penetrate the blood-brain barrier (BBB), destroy brain parenchyma, and finally cause CNS disorders [1]. Among the meningitis-causing microbes, extraintestinal pathogenic Escherichia coli (ExPEC) has recently emerged as an important zoonotic bacterial pathogen with the potential to colonize multiple tissues outside the intestine and cause severe infections, with one typical outcome being meningitis. The evidence from recent in vivo and in vitro studies indicates that meningitic E. coli strains possess the ability to invade the brain, and the infection-induced BBB disruption that occurs is the hallmark event in the development of E. coli meningitis [4, 5].
The availability of in vitro and in vivo BBB infection models has made the study of meningitic E. coli penetration of the brain possible [6–9]. The in vitro BBB model uses brain microvascular endothelial cells (BMECs) that form distinctive tight junctions and exhibit high trans-endothelial electrical resistance, thereby mimicking the features of the natural in vivo barrier that protects the brain from circulating microorganisms and toxins [10–13]. The in vivo model is established by inducing experimental hematogenous meningitis in newborn rats and mice [9, 14, 15]. With these models, it is now well-established that successful traversal of the BBB by circulating E. coli strains requires the following prerequisites: a high bacteremia, binding to and invasion of BMECs, rearrangement of actin cytoskeleton, and crossing the BBB as live bacteria [1, 2]. These require a series of complicated interactions between meningitic E. coli and the host. So far, several host targets have been found to be associated with this invasion process, including certain intracellular signaling molecules like focal adhesion kinase, phosphatidylinositol 3-kinase (PI3K), Rho GTPases, cytosolic phospholipase A2, nuclear factor-κB (NF-κB), inducible nitric oxide synthase (NOS), and several cellular surface molecules/receptors such as caveolin-1, Toll-like receptors, the intercellular adhesion molecule (ICAM-1), and some actin-binding molecules like ERM family proteins (ezrin, radixin, and moesin), most likely through their influences on the aforementioned prerequisites [8, 16–19]. We have previously identified and characterized two essential cellular targets, S1P and EGFR, which are exploited by meningitic E. coli for successful invasion of the BBB [20]. In other work, we have also found that vascular endothelial growth factor A (VEGFA) and Snail-1, which are inducible by meningitic E. coli, can mediate the BBB disruption [5]. Despite these advances, the mechanisms involved in CNS infection by meningitic E. coli are still poorly understood, and a more comprehensive investigation to elucidate the cellular targets in infected BMECs is now required.
In the current study, we compared the different proteomic profiles of BMECs in response to meningitic and non-meningitic E. coli strains via the isobaric tags for relative and absolute quantification (iTRAQ) approach and investigated the potential host factors and mechanisms that were hijacked by meningitic E. coli to penetrate the BBB. Characterization of these potential host targets will expand our current knowledge on meningitic E. coli-induced CNS infections and provide new strategies to prevent this infection and develop novel therapeutic reagents against it.
Methods
Bacterial strains, cell culture, and infection
The E. coli K1 strain RS218 (O18:K1:H7) [GenBank: CP007149.1], whose genomic sequencing has been finalized and annotated, is a well-characterized cerebrospinal fluid (CSF) isolate from a neonatal meningitis case [21]. The porcine-originated ExPEC strain PCN033 (O11: K2) [GenBank: CP006632.1], which was isolated from swine CSF in China [22, 23], is evidenced to be highly virulent and capable of invading and disrupting the BBB, thereby causing CNS dysfunction [5, 24]. E. coli K12 strain HB101 is an avirulent and non-meningitic strain normally used as a negative control strain [25, 26]. All E. coli strains were grown aerobically at 37 °C in Luria–Bertani medium unless otherwise specified.
The immortalized human BMECs (hereafter called hBMECs) were kindly provided by Prof. Kwang Sik Kim in Johns Hopkins University School of Medicine and routinely cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 1 mM sodium pyruvate, essential amino acids, nonessential amino acids, vitamins, and penicillin and streptomycin (100 U/mL) in a 37 °C incubator under 5% CO2 until monolayer confluence was reached [20, 27]. Confluent cells were washed with Hank’s balanced salt solution (Corning Cellgro, Manassas, VA, USA) and starved in serum-free medium for 16–18 h before further treatment. For bacterial challenge, the cells were infected with E. coli PCN033, RS218, or HB101 strains each at a multiplicity of infection of 10 for 2 h. In some assays, the cells were pretreated with specific inhibitors prior to bacterial challenge.
Reagents, antibodies, and inhibitors
The p38 inhibitor SB202190, extracellular signal-regulated kinases 1 and 2 (ERK1/2) inhibitor U0126, c-Jun N-terminal kinase (JNK) inhibitor SP600125, NF-κB inhibitor BAY11-7082, and (S, R)-3-(4-hydroxyphenyl)-4, 5-dihydro-5-isoxazole acetic acid methyl ester (ISO-1), an inhibitor of macrophage migration inhibitory factor (MIF), were purchased from MedChem Express (Monmouth, NJ, USA). Recombinant MIF protein was purchased from Novoprotein (Summit, NJ, USA). The nucleic acid dye, 4′-6-diamidino-2-phenylindole (DAPI), was obtained from Solarbio (Beijing, China). Anti-ZO-1, anti-MIF, anti-TATA box-binding protein-like protein 1 (TBPL1), anti-legumain (LGMN), anti-ERK1/2, and anti-phospho-ERK1/2 antibodies (all rabbit) were purchased from ABclonal (Wuhan, Hubei, China). Anti-occludin, anti-dystrophin (DMD), anti-HISTIHIC, anti-JNK, and anti-p38 mitogen-activated protein kinase (MAPK) antibodies (all rabbit) were purchased from Proteintech (Chicago, IL, USA). Anti-phospho-JNK (rabbit) antibody was from R&D Systems (Minneapolis, MO, USA). Anti-phospho-p38, anti-p65, anti-phospho-p65, and anti-IκBα antibodies (all rabbit) were purchased from Cell Signaling Technology (Danvers, MA, USA). Cy3-labeled goat anti-rabbit antibody was purchased from Beyotime Institute of Biotechnology (Shanghai, China). Anti-GAPDH (mouse) antibody was purchased from Beijing Biodragon Immunotechnologies Co., Ltd. (Beijing, China).
Protein isolation, digestion, and labeling with iTRAQ reagents
Bacterial-infected and non-infected cells in 10 cm dishes were collected 2-h post-infection and gently washed with pre-chilled PBS buffer. The cells were lysed in 1 mL lysis buffer, and the soluble protein fraction was harvested by 5 min of ultrasonication treatment (pulse on 2 s, pulse off 3 s, power 180 W) followed by centrifugation at 20000×g for 30 min at 4 °C, and the protein concentration was determined via the Bradford protein assay method with BSA as the standard substance. The proteins were reduced with 10 mM iodoacetamide at room temperature for 45 min in the dark and then precipitated in acetone at − 20 °C for 3 h. After centrifugation at 20000×g for 20 min, the protein pellet was resuspended and ultrasonicated in pre-chilled 50% (w/v) tetraethyl-ammonium bromide (TEAB) buffer supplemented with 0.1% SDS. The proteins were obtained after centrifugation at 20000×g and their concentrations were measured by Bradford assays.
Subsequently, protein (100 μg) in TEAB buffer was incubated with 3.3 μL of trypsin (1 μg/μL) (Promega, Madison, WI, USA) at 37 °C for 24 h in a sealed tube. The tryptic peptides were lyophilized and dissolved in 50% TEAB buffer, and iTRAQ labeling was performed according to the manufacturer’s instructions (AB Sciex, Foster City, CA, USA). Briefly, one unit of iTRAQ reagent was thawed and reconstituted in 24 μL isopropanol and the peptides were incubated at room temperature for 2 h. The peptides from the control, HB101, PCN033, and RS218 groups were designated 114, 115, 116, and 117, respectively. The labeled samples were then mixed and dried with a rotary vacuum concentrator. The labeling efficiency was examined by mass spectrometry (MS).
Strong cation exchange chromatography (SCX) fractionation and liquid chromatography (LC)–MS/MS analysis
The labeled samples were pooled and purified using an SCX column (Phenomenex, USA), and separated by LC using an LC-20AB HPLC pump system (Shimadzu, Japan). The peptides were then mixed with nine times their volume in buffer A (25% ACN, 10 mM KH2PO4, pH = 3) and loaded onto a 4.6 × 250 mm Ultremex SCX column containing 5-μm particles (Phenomenex). The peptides were eluted at a flow rate of 1 ml/min in a buffer B (25% ACN, 2 M KCL, 10 mM KH2PO4, pH = 3) gradient as follows: 0–5% buffer B for 30 min, 5–30% buffer B for 20 min, 30–50% buffer B for 5 min, 50% buffer B for 5 min, 50–100% buffer B for 5 min, and 100% buffer B for 1 min before equilibrating with buffer A for 10 min prior to the next injection. Next, the eluted peptides were desalted with a Strata X C18 column (100 mm × 75 mm, 5-um particles, 300A aperture) (Phenomenex, Torrance, CA, USA) and vacuum dried. The fractions were then dissolved in aqueous solution containing 0.1% formic acid (FA) and 2% ACN and centrifuged at 12000g for 10 min at 4 °C. Five micrograms supernatant was loaded on an LC-20AD nano HPLC (Shimadzu, Kyoto, Japan) by the autosampler onto a 2 cm C18 trap column (inner diameter 200 μm, Waters), and the peptides were eluted onto a resolving 10 cm analytical C18 column (inner diameter 75 μm, Waters). The mobile phases used were composed of solvent A (0.1% FA and 5% ACN) and solvent B (0.1% FA and 95% ACN). The gradient was run at 400 nL/min for 48 min at 5–80% solvent B, followed by running a linear gradient to 80% for 7 min, maintained at 80% B for 3 min, and finally returned to 5% in 7 min.
The peptides were subjected to nano-electrospray ionization followed by tandem mass spectrometry (MS/MS) in a Q EXACTIVE (Thermo Fisher Scientific, San Jose, CA, USA) coupled to the HPLC. Intact peptides were detected in the Orbitrap at a resolution of 70,000 and a mass range of 350–2000 m/z. Peptides were selected for MS/MS using high-energy collision dissociation (HCD), and ion fragments were detected in the Orbitrap at a resolution of 17,500. The electrospray voltage applied was 1.8 kV. MS/MS analysis was required for the 15 most abundant precursor ions, which were above a threshold ion count of 20,000 in the MS survey scan, including a following dynamic exclusion duration of 15 s.
iTRAQ data analysis
The raw data files acquired from the mass spectrometers were converted into MGF files using 5600 MS Converter. Protein identification and quantification were performed using the Mascot Server (http://www.matrixscience.com/search_form_select.html) against the Uniprot_2015_human database (Matrix Science, London, UK; version 2.3.0) and Proteome Discoverer 1.3 (Thermo Fisher Scientific Inc.). To reduce the probability of false peptide identification, only peptides with significance scores at the 95% confidence interval as determined by a Mascot probability analysis were included. The quantitative protein ratios were weighted and normalized by the median ratio in Mascot. Statistical significance analyses were evaluated using two-way ANOVA. The proteins were considered to be differentially expressed if the ratio of mean fold change > 1.2 (or < 0.83) with an Exp pr > 0.05 and a Group pr < 0.05 (Exp pr, three-experiment p value; Group pr, group p value; fold change = experiment + group + error).
The Gene Ontology (GO) annotation of the identified proteins was performed via the online GO program (http://geneontology.org/). The biological functions, networks, and signaling pathways of the differentially expressed proteins (DEPs) were analyzed with Ingenuity Pathways Analysis (IPA) software (version 7.5, http://www.ingenuity.com) (Additional files 8, 9 and 10).
RNA extraction and quantitative real-time PCR
Total RNA from the uninfected or infected cells was extracted with RNAiso Plus reagent according to the manufacturer’s instructions (TakaRa, Japan). Any genomic DNA contamination was eliminated by DNase I treatment, and the RNA was reverse-transcribed into cDNA using the PrimeScript™ RT reagent kit with gDNA Eraser, following the manufacturer’s instructions (Takara, Japan). Quantitative real-time PCR was performed in triplicate using the Power SYBR Green PCR Master Mix (Applied BioSystems, Foster City, CA, USA). The PCR primers for these experiments are listed in Table 1. The expression levels of the target genes were normalized to GAPDH by the 2−ΔΔCT method.
Table 1.
Primers used for real-time PCR in this study
| Primers | Nucleotide sequence(5′-3′) | Gene symbol(s) |
|---|---|---|
| P1 | ACGAATCTCCGACCACT | IL-1β |
| P2 | CCATGGCCACAACAACTGAC | |
| P3 | CTCAGCCTCTTCTCCTTC | TNF-α |
| P4 | GGGTTTGCTACAACATGG | |
| P5 | CCACTCACCTCTTCAGAA | IL-6 |
| P6 | GGCAAGTCTCCTCATTGA | |
| P7 | GACATACTCCAAACCTTTCC | IL-8 |
| P8 | ATTCTCAGCCCTCTTCAAA | |
| P9 | TGCCTCCTGCACCACCAACT | GAPDH |
| P10 | CGCCTGCTTCACCACCTTC |
Western blotting
Uninfected and infected hBMECs were collected and lysed in RIPA buffer supplemented with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA) and then sonicated and centrifuged at 10,000×g for 10 min at 4 °C. The soluble protein concentration in the supernatants was measured using the BCA protein assay kit (Beyotime, China). Aliquots from each sample were separated by 12% SDS-PAGE, and then transferred to polyvinylidene difluoride membranes (Bio-Rad, CA, USA). The blots were blocked with 5% BSA in Tris-buffered saline with Tween 20 at room temperature for 1 h and then incubated overnight at 4 °C with primary antibodies against GAPDH, DMD, MIF, HIST1H1C, TBPL1 or LGMN. The blots were subsequently washed and incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG at 37 °C for 1 h, and visualized with ECL reagents (Bio-Rad, USA). The blots were densitometrically quantified and analyzed with Image Lab software (Bio-Rad).
Immunofluorescence microscopy
Uninfected and infected hBMECs were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. After 2 h of blocking in PBS buffer with 5% BSA, the cells were incubated with the primary antibody (1:100) overnight at 4 °C, washed thrice with PBS, and then incubated with fluorescently labeled anti-mouse or anti-rabbit IgG (1500) for 1 h. Nuclei were stained with DAPI (0.5 μg/mL) for 30 min. Finally, the cells were mounted and then visualized with fluorescence microscopy.
Electric cell substrate impedance sensing (ECIS)
To explore the influence of recombinant MIF on the permeability of the BBB, hBMECs were seeded at 7 × 104 cells on collagen-coated, gold-plated electrodes in 96-well chamber slides (96W1E+) linked to ECIS Zθ equipment (Applied BioPhysics, Troy, NY, USA) and continuously cultured until confluence, and the trans-endothelial electric resistance (TEER) was monitored to reflect the formation of the barrier [28]. After stable maximal TEER was reached, the recombinant human MIF protein was added into the cells at multiple dosages (10, 100, and 200 ng/mL), and the possible TEER alteration of the monolayer cells was automatically recorded by the ECIS system.
Statistical analysis
Data were expressed as the mean ± standard deviation (mean ± SD) from three replicates. Statistical significance of the differences between each group was analyzed by a one-way analysis of variance (ANOVA) or two-way ANOVA embedded in GraphPad Prism, version 6.0 (GraphPad Software Inc., La Jolla, CA, USA). P < 0.05 (*) was considered statistically significant, and p < 0.01 (**), as well as p < 0.001 (***) were all considered extremely significant.
Results
Differential protein profiling of hBMECs in response to E. coli infection
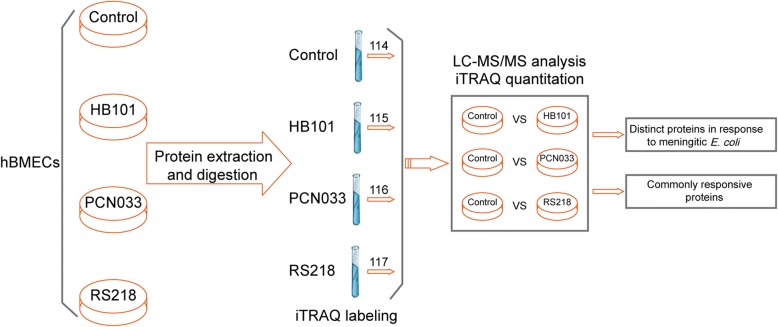
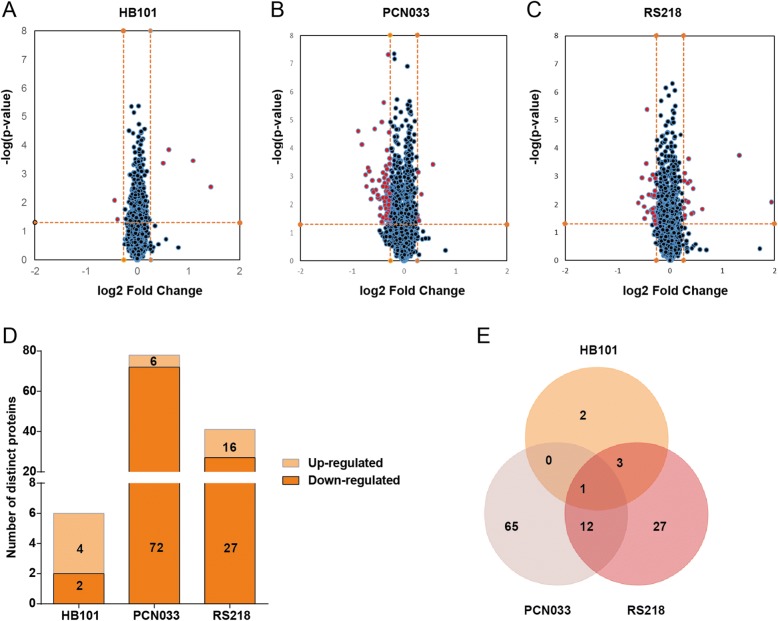
The protein extracts prepared from the hBMECs with or without meningitic E. coli challenge were subjected to the iTRAQ proteomics analysis. The whole work flow was shown in Fig. 1. Approximately 3000 different proteins were identified and quantified by iTRAQ-coupled LC-MS/MS analysis of the hBMECs infected with E. coli HB101, PCN033, or RS218 strains (Additional file 1: Table S1, Additional file 2: Table S2, Additional file 3: Table S3). As shown in Fig. 2a–d, four proteins were identified as being significantly upregulated and two were significantly downregulated upon HB101 infection, six were significantly upregulated, and 72 were significantly downregulated upon PCN033 infection, while 16 significantly upregulated and 27 significantly downregulated proteins were identified in cells challenged with RS218. The details of these differentially expressed proteins (DEPs) are listed in Tables 2, 3, and 4. The meningitic E. coli PCN033 group displayed 65 unique proteins, while the RS218 group displayed 27 unique proteins. They both shared 13 DEPs with 12 of them being distinct proteins in the hBMECs in response to meningitic strains PCN033 and RS218 (Fig. 2e, Table 5). Only one protein, EXOSC4, was shared by the three groups, and it showed a 0.74-, 0.759-, and 0.8-fold decrease in HB101, PCN033 and RS218 groups, respectively (Fig. 2e, Table 5). In contrast, infection with the non-meningitic HB101 strain induced only two unique, differentially altered proteins. Four proteins were shared between HB101 and RS218 groups, and the three of them altered in response to HB101 and RS218 were specific host proteins in both of these human isolates (Fig. 2e).
Fig. 1.
The general work flow for the proteomics analysis in this study
Fig. 2.
An overview of the DEPs in hBMECs in response to infection with meningitic E. coli strains PCN033 and RS218, and non-meningitic E. coli HB101. a–c The volcano plots show the cellular protein profiles in hBMECs after challenge with the three strains. d The number of significantly up- or downregulated proteins in the three infection groups. e Venn diagram showing the overlapping or distinct cellular proteins among the three groups
Table 2.
Significantly changed proteins in HB101-infected hBMECs
| Accession | Description | MW [kDa] | Fold change | P valuea |
|---|---|---|---|---|
| P02656 | Apolipoprotein C-III | 10.8 | 1.544 | 0.000144*** |
| Q07020 | 60S ribosomal protein L18 | 21.6 | 1.422 | 0.000429*** |
| Q96HP4 | Oxidoreductase NAD-binding domain-containing protein 1 | 34.8 | 2.715 | 0.002873** |
| Q9NPD3 | Exosome complex component RRP41 | 26.4 | 0.737 | 0.008415** |
| Q9Y2Q5 | Ragulator complex protein LAMTOR2 | 13.5 | 0.769 | 0.038574* |
| O14556 | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | 44.5 | 2.134 | 0.000347*** |
aP < 0.05 (*) was considered significant, and P < 0.01 (**), as well as < 0.001 (***) were all considered extremely significant
Table 3.
Significantly changed proteins in PCN033-infected hBMECs
| Accession | Description | MW [kDa] | Fold change | P valuea |
|---|---|---|---|---|
| A6ZKI3 | Protein FAM127A | 13.2 | 0.756 | 0.028947* |
| O00625 | Pirin | 32.1 | 1.228 | 0.037445* |
| O43633 | Charged multivesicular body protein 2a | 25.1 | 0.758 | 0.002376** |
| O43752 | Syntaxin-6 | 29.2 | 0.79 | 0.025619* |
| O60524 | Nuclear export mediator factor NEMF | 122.9 | 0.83 | 0.004194** |
| O75190 | DnaJ homolog subfamily B member 6 | 36.1 | 0.799 | 0.020038* |
| O75251 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial | 23.5 | 0.711 | 0.004822** |
| O75817 | Ribonuclease P protein subunit p20 | 15.6 | 0.815 | 0.017814* |
| O95229 | ZW10 interactor | 31.3 | 0.668 | 0.003727** |
| P04004 | Vitronectin | 54.3 | 0.808 | 0.003548** |
| P07305 | Histone H1.0 | 20.9 | 0.776 | 0.008176** |
| P11532 | Dystrophin | 426.5 | 0.691 | 0.003088** |
| P14174 | Macrophage migration inhibitory factor | 12.5 | 1.486 | 0.000377*** |
| P16401 | Histone H1.5 | 22.6 | 0.631 | 0.000673*** |
| P16402 | Histone H1.3 | 22.3 | 0.6 | 0.000896*** |
| P16403 | Histone H1.2 | 21.4 | 0.572 | 7.57E−05*** |
| P35251 | Replication factor C subunit 1 | 128.2 | 0.786 | 0.013936* |
| P35527 | Keratin, type I cytoskeletal 9 | 62 | 0.72 | 0.001431** |
| P39060 | Collagen alpha-1(XVIII) chain | 178.1 | 0.792 | 0.031694* |
| P46013 | Antigen KI-67 | 358.5 | 0.793 | 0.005934** |
| P48651 | Phosphatidylserine synthase 1 | 55.5 | 0.83 | 0.000234*** |
| P49585 | Choline-phosphate cytidylyltransferase A | 41.7 | 0.608 | 0.002306** |
| P50914 | 60S ribosomal protein L14 | 23.4 | 0.71 | 0.005946** |
| P52756 | RNA-binding protein 5 | 92.1 | 0.758 | 0.005815** |
| P56377 | AP-1 complex subunit sigma-2 | 18.6 | 0.765 | 2.4E−06*** |
| P61966 | AP-1 complex subunit sigma-1A | 18.7 | 0.814 | 4.81E−08*** |
| P62277 | 40S ribosomal protein S13 | 17.2 | 0.792 | 0.002441** |
| P62380 | TATA box-binding protein-like protein 1 | 20.9 | 0.621 | 0.000503*** |
| Q13625 | Apoptosis-stimulating of p53 protein 2 | 125.5 | 0.724 | 0.006481** |
| Q14241 | Transcription elongation factor B polypeptide 3 | 89.9 | 0.647 | 0.006556** |
| Q14686 | Nuclear receptor coactivator 6 | 219 | 0.792 | 0.005234** |
| Q15388 | Mitochondrial import receptor subunit TOM20 homolog | 16.3 | 0.823 | 0.007395** |
| Q15629 | Translocating chain-associated membrane protein 1 | 43 | 0.809 | 0.023084* |
| Q17RN3 | Protein FAM98C | 37.3 | 0.821 | 0.010317* |
| Q4V339 | COBW domain-containing protein 6 | 43.9 | 0.747 | 1.2E−05*** |
| Q567U6 | Coiled-coil domain-containing protein 93 | 73.2 | 0.814 | 0.001351** |
| Q5SSJ5 | Heterochromatin protein 1-binding protein 3 | 61.2 | 0.828 | 0.000364*** |
| Q6N069 | N-alpha-acetyltransferase 16, NatA auxiliary subunit | 101.4 | 0.775 | 0.001734** |
| Q709C8 | Vacuolar protein sorting-associated protein 13C | 422.1 | 0.799 | 0.000576*** |
| Q7Z422 | SUZ domain-containing protein 1 | 17 | 0.808 | 0.002542** |
| Q8IXJ9 | Putative Polycomb group protein ASXL1 | 165.3 | 0.807 | 0.007037** |
| Q8N2K0 | Monoacylglycerol lipase ABHD12 | 45.1 | 0.786 | 0.0034** |
| Q8N884 | Cyclic GMP-AMP synthase | 58.8 | 0.82 | 0.013058* |
| Q8NC44 | Protein FAM134A | 57.8 | 0.78 | 0.010814* |
| Q8NC60 | Nitric oxide-associated protein 1 | 78.4 | 0.81 | 0.013637* |
| Q8NEY1 | Neuron navigator 1 | 202.3 | 0.797 | 0.020924* |
| Q8TEM1 | Nuclear pore membrane glycoprotein 210 | 205 | 0.833 | 0.032212* |
| Q8WUP2 | Filamin-binding LIM protein 1 | 40.6 | 0.809 | 0.002633** |
| Q8WVV9 | Heterogeneous nuclear ribonucleoprotein L-like | 60 | 0.804 | 0.013638* |
| Q8WXA3 | RUN and FYVE domain-containing protein 2 | 75 | 0.744 | 0.041934* |
| Q92604 | Acyl-CoA:lysophosphatidylglycerol acyltransferase 1 | 43.1 | 0.789 | 0.033787 * |
| Q96A57 | Transmembrane protein 230 | 13.2 | 0.786 | 0.000449*** |
| Q96LB3 | Intraflagellar transport protein 74 homolog | 69.2 | 0.543 | 2.53E−05*** |
| Q96RU3 | Formin-binding protein 1 | 71.3 | 0.679 | 2.07E−05*** |
| Q96T37 | Putative RNA-binding protein 15 | 107.1 | 0.728 | 0.024086* |
| Q9GZP8 | Immortalization upregulated protein | 10.9 | 1.207 | 0.032624* |
| Q9H074 | Polyadenylate-binding protein-interacting protein 1 | 53.5 | 1.266 | 0.004395** |
| Q9H5N1 | Rab GTPase-binding effector protein 2 | 63.5 | 0.77 | 0.001156** |
| Q9H5X1 | MIP18 family protein FAM96A | 18.3 | 0.8 | 0.000118*** |
| Q9HB40 | Retinoid-inducible serine carboxypeptidase | 50.8 | 1.215 | 0.000733*** |
| Q9HC52 | Chromobox protein homolog 8 | 43.4 | 1.201 | 0.023152* |
| Q9NPD3 | Exosome complex component RRP41 | 26.4 | 0.759 | 0.000746*** |
| Q9NRY4 | Rho GTPase-activating protein 35 | 170.4 | 0.792 | 0.011172* |
| Q9NS87 | Kinesin-like protein KIF15 | 160.1 | 0.785 | 0.010039* |
| Q9NSP4 | Centromere protein M | 19.7 | 0.802 | 0.021316* |
| Q9NTI5 | Sister chromatid cohesion protein PDS5 homolog B | 164.6 | 0.826 | 0.003399** |
| Q9NWU5 | 39S ribosomal protein L22, mitochondrial | 23.6 | 0.812 | 0.016677* |
| Q9NZQ3 | NCK-interacting protein with SH3 domain | 78.9 | 0.661 | 0.00317** |
| Q9P0V3 | SH3 domain-binding protein 4 | 107.4 | 0.797 | 0.001833** |
| Q9UBL6 | Copine-7 | 70.2 | 0.823 | 2.73E−05*** |
| Q9UJW0 | Dynactin subunit 4 | 52.3 | 0.823 | 0.012604* |
| Q9UNP9 | Peptidyl-prolyl cis-trans isomerase E | 33.4 | 0.75 | 0.044207* |
| Q9Y2R0 | Cytochrome c oxidase assembly protein 3 homolog, mitochondrial | 11.7 | 0.792 | 0.003694** |
| Q9Y5Y2 | Cytosolic Fe-S cluster assembly factor NUBP2 | 28.8 | 0.787 | 0.000891*** |
| Q9Y6I9 | Testis-expressed sequence 264 protein | 34.2 | 0.814 | 0.047637* |
| Q9Y3Y2 | Chromatin target of PRMT1 protein | 26.4 | 0.828 | 0.008622** |
| Q9Y4R8 | Telomere length regulation protein TEL2 homolog | 91.7 | 0.735 | 0.013443* |
| P10412 | Histone H1.4 | 21.9 | 0.655 | 0.001429** |
aP < 0.05 (*) was considered significant, and P < 0.01 (**), as well as < 0.001 (***), were all considered extremely significant
Table 4.
Significantly changed proteins in RS218-infected hBMECs
| Accession | Description | MW [kDa] | Fold change | P valuea |
|---|---|---|---|---|
| O00592 | Podocalyxin | 58.6 | 1.214 | 0.001481** |
| O14556 | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | 44.5 | 2.514 | 0.000183*** |
| O43598 | 2′-Deoxynucleoside 5′-phosphate N-hydrolase 1 | 19.1 | 0.8 | 0.020803* |
| O76024 | Wolframin | 100.2 | 0.732 | 0.000347*** |
| O76095 | Protein JTB | 16.3 | 0.815 | 0.026287* |
| O95989 | Diphosphoinositol polyphosphate phosphohydrolase 1 | 19.5 | 0.821 | 0.003332** |
| P05067 | Amyloid beta A4 protein | 86.9 | 0.813 | 0.004913** |
| P10412 | Histone H1.4 | 21.9 | 1.271 | 0.001736** |
| P11532 | Dystrophin | 426.5 | 0.799 | 0.014535* |
| P14174 | Macrophage migration inhibitory factor | 12.5 | 1.276 | 0.008267** |
| P16401 | Histone H1.5 | 22.6 | 1.221 | 0.025445* |
| P16402 | Histone H1.3 | 22.3 | 1.306 | 0.001514** |
| P16403 | Histone H1.2 | 21.4 | 1.332 | 0.021727* |
| P30154 | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform | 66.2 | 0.809 | 0.005474** |
| P35527 | Keratin, type I cytoskeletal 9 | 62 | 0.822 | 0.038701* |
| P42167 | Lamina-associated polypeptide 2, isoforms beta/gamma | 50.6 | 0.826 | 0.0494* |
| P46781 | 40S ribosomal protein S9 | 22.6 | 1.207 | 0.013518* |
| P50402 | Emerin | 29 | 0.8 | 0.000916*** |
| P52756 | RNA-binding protein 5 | 92.1 | 0.74 | 4.23E−06*** |
| P55789 | FAD-linked sulfhydryl oxidase ALR | 23.4 | 1.537 | 0.014932* |
| P61313 | 60S ribosomal protein L15 | 24.1 | 1.286 | 0.000236*** |
| P62380 | TATA box-binding protein-like protein 1 | 20.9 | 0.66 | 0.008696** |
| Q07020 | 60S ribosomal protein L18 | 21.6 | 1.367 | 0.002799** |
| Q4V339 | COBW domain-containing protein 6 | 43.9 | 0.756 | 0.00457** |
| Q8N4H5 | Mitochondrial import receptor subunit TOM5 homolog | 6 | 1.223 | 0.000783*** |
| Q8ND56 | Protein LSM14 homolog A | 50.5 | 0.793 | 0.017061* |
| Q96BZ8 | Leukocyte receptor cluster member 1 | 30.5 | 0.693 | 0.005542** |
| Q96HP4 | Oxidoreductase NAD-binding domain-containing protein 1 | 34.8 | 3.845 | 0.008111** |
| Q96KR1 | Zinc finger RNA-binding protein | 116.9 | 0.811 | 0.01368* |
| Q96LB3 | Intraflagellar transport protein 74 homolog | 69.2 | 0.71 | 0.012006* |
| Q96P47 | Arf-GAP with GTPase, ANK repeat and PH domain-containing protein 3 | 95 | 0.783 | 0.004713** |
| Q99538 | Legumain | 49.4 | 0.692 | 0.001173** |
| Q9BTA9 | WW domain-containing adapter protein with coiled-coil | 70.7 | 0.743 | 0.01886* |
| Q9BZF9 | Uveal autoantigen with coiled-coil domains and ankyrin repeats | 162.4 | 0.824 | 0.031539* |
| Q9H7B2 | Ribosome production factor 2 homolog | 35.6 | 1.367 | 0.020862* |
| Q9HCD5 | Nuclear receptor coactivator 5 | 65.5 | 0.771 | 0.001324** |
| Q9NPD3 | Exosome complex component RRP41 | 26.4 | 0.784 | 0.015931* |
| Q9NZR1 | Tropomodulin-2 | 39.6 | 1.216 | 0.030742* |
| Q9UI10 | Translation initiation factor eIF-2B subunit delta | 57.5 | 0.828 | 0.001214** |
| Q9UIC8 | Leucine carboxyl methyltransferase 1 | 38.4 | 0.811 | 0.0312* |
| Q9UK41 | Vacuolar protein sorting-associated protein 28 homolog | 25.4 | 0.715 | 0.032545* |
| Q9Y4R8 | Telomere length regulation protein TEL2 homolog | 91.7 | 0.807 | 0.019858* |
| Q9Y5V3 | Melanoma-associated antigen D1 | 86.1 | 1.238 | 0.007536** |
aP < 0.05 (*) was considered significant, and P < 0.01 (**), as well as < 0.001 (***), were all considered extremely significant
Table 5.
The distinct differential proteins in hBMECs in response to meningitic E. coli strains PCN033 and RS218
| ID | Name | Protein | Fold Change | ||
|---|---|---|---|---|---|
| RS218 | PCN033 | HB101 | |||
| Q9NPD3 | EXOSC4 | Exosome complex component RRP41 | 0.8 | 0.759 | 0.74 |
| Q96LB3 | IFT74 | Intraflagellar transport protein 74 homolog | 0.7 | 0.543 | / |
| P11532 | DMD | Dystrophin | 0.8 | 0.691 | / |
| P52756 | RBM5 | RNA-binding protein 5 | 0.7 | 0.758 | / |
| Q4V339 | CBWD6 | COBW domain-containing protein 6 | 0.8 | 0.747 | / |
| Q9Y4R8 | TELO2 | Telomere length regulation protein TEL2 homolog | 0.8 | 0.735 | / |
| P35527 | KRT9 | Keratin, type I cytoskeletal 9 | 0.8 | 0.72 | / |
| P62380 | TBOL1 | TATA box-binding protein-like protein 1 | 0.7 | 0.621 | / |
| P16403 | HIST1H1C | Histone H1.2 | 1.3 | 0.572 | / |
| P16402 | HIST1H1D | Histone H1.3 | 1.3 | 0.6 | / |
| P10412 | HIST1H1E | Histone H1.4 | 1.3 | 0.655 | / |
| P16401 | HIST1H1B | Histone H1.5 | 1.2 | 0.631 | / |
| P14174 | MIF | Macrophage migration inhibitory factor | 1.3 | 1.486 | / |
Western blot verification of the DEPs
We next used western blotting to further test the DEPs identified by iTRAQ. We selected several proteins from the iTRAQ results from both PCN033 and RS218 groups. The test proteins were HIST1H1C, TBPL1, and MIF for the PCN033 group (Fig. 3a), and DMD, LGMN, and HIST1H1C for the RS218 group (Fig. 3c). The western blot and densitometry analyses produced the similar expression alteration to those of the iTRAQ results following either PCN033 or RS218 infection (Fig. 3b, d).
Fig. 3.
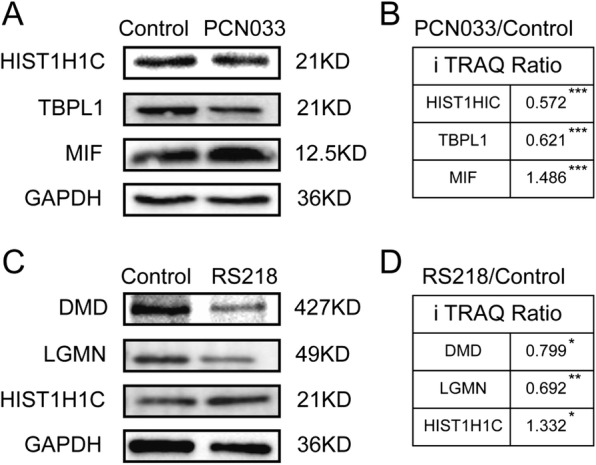
DEPs validation. a Immunoblotting analysis of the DEPs (HIST1H1C, TBPL1, and MIF) in the hBMECs with or without PCN033 infection. b iTRAQ ratios of the DEPs in hBMECs with PCN033 infection. c Immunoblotting analysis of the DEPs (DMD, LGMN, and HIST1H1C) in hBMECs with or without RS218 infection. d iTRAQ ratios of the DEPs in hBMECs with RS218 infection. *(P < 0.05) was considered statistically significant; ** (p < 0.01) and *** (p < 0.001) were extremely significant
Bioinformatic analysis of the DEPs in hBMECs
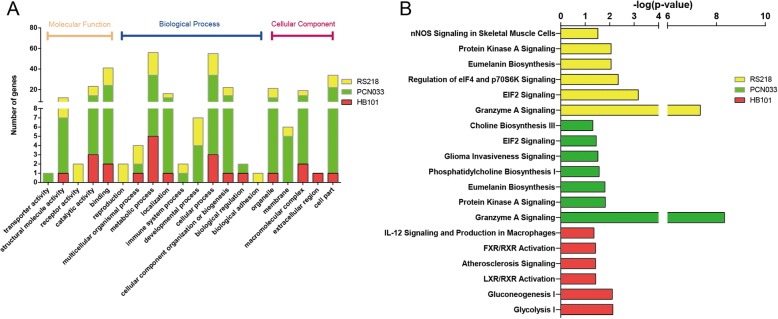
We next investigated and characterized the DEPs by searching the GO and UniProt databases. The DEPs were assigned to the categories of different “biological processes,” “cellular components,” and “molecular functions.” Within the biological processes class, the DEPs from the three groups (RS218, PCN033, and HB101) were mainly divided into metabolic processes, localization, cellular process, and cellular component organization or biogenesis. The immune system process and developmental process classes were found in both RS218 and PCN033 infection groups, but not in the HB101 group. Within the cellular component class, the DEPs were mainly divided into organelle, macromolecular complex, and cell parts, and the membrane-associated ones were only identified in the meningitic strains RS218 and PCN033, not in HB101. As for molecular function, the DEPs were mainly associated with structural molecule activity, catalytic activity, and binding (Fig. 4a, Additional file 4: Table S4).
Fig. 4.
GO annotation and pathway enrichment comparison of DEPs upon meningitic or non-meningitic E. coli infection. a GO annotation characterization of the molecular functions, biological processes, and cellular components based on the DEPs. b Pathway enrichment of cellular DEPs in response to infection with HB101, PCN033, and RS218 strains
We next performed canonical pathway prediction through IPA on the DEPs. The top ranked canonical pathways in each group are shown in Fig. 4b. We found that protein kinase A signaling, eumelanin biosynthesis, EIF2 signaling, and granzyme A signaling were simultaneously enriched in both RS218 and PCN033 infection groups, but not in the HB101 group (Fig. 4b). Noticeably, granzyme A signaling was much more significantly enriched in the DEPs from both meningitic groups, suggesting a potential role for granzyme A in meningitic E. coli invasion of the BBB. Additionally, phosphatidylcholine biosynthesis I, choline biosynthesis III, and glioma invasiveness signaling were only enriched in the PCN033 group, while neuronal NOS signaling and regulation of eIF4 and p70S6K signaling were only identified in the RS218 group, which exhibited distinct signaling pathways that might have strain specificity (Fig. 4b).
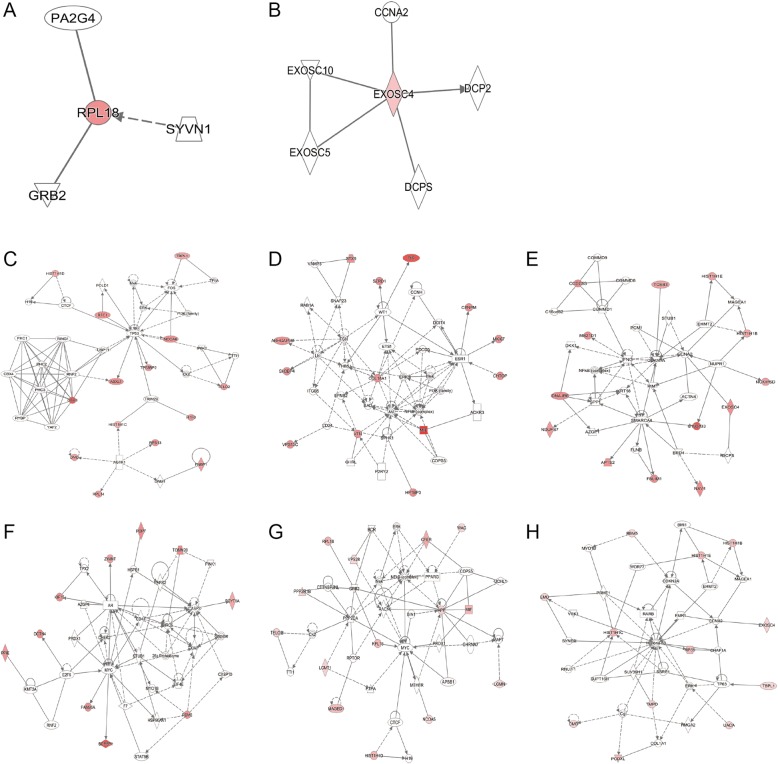
The IPA tool was used to further analyze the potential networks based on the DEPs from the E. coli infections. Two networks were drawn for these differential cellular proteins in response to HB101 infection (Fig. 5a, b, Additional file 5: Table S5). In addition, four networks were generated based on the DEPs from the PCN033 infection (Fig. 5c–f, Additional file 6: Table S6), while two networks were generated from the DEPs upon RS218 infection (Fig. 5g, h, Additional file 7: Table S7). It should be noted that the NF-κB complex, as well as ERK, were included in the networks of both PCN033 and RS218 groups, while they were not observed in the cells in response to the non-meningitic HB101 strain, suggesting that these two essential signaling molecules exert regulatory effects during meningitic E. coli penetration of the BBB.
Fig. 5.
Network analysis of significantly altered proteins in hBMECs upon E. coli infection. For the HB101 infection, two networks were constructed: a cellular assembly and organization, gastrointestinal disease, hepatic system disease; b RNA damage and repair, connective tissue disorders, developmental disorder. For the PCN033 infection, four networks were constructed: c lymphoid tissue structure and development, organ morphology, organismal development; d cellular movement, cancer, organismal injury and abnormalities; e inflammatory disease, inflammatory response, organismal injury and abnormalities; f cell death and survival, cellular development, cellular growth and proliferation. For the RS218 infection two networks were constructed: g neurological disease, organismal injury and abnormalities, cell cycle; h gene expression, cellular assembly and organization, DNA replication, recombination, and repair. The red nodes indicate significantly altered protein expression, and the white ones are those known to be involved in the networks, but not identified in this study. Arrows indicate the interrelationship between two molecules. Solid lines indicate direct interactions and dashed lines indicate indirect interactions
MIF contributes to meningitic E. coli-induced cytokine production and tight junction disruption
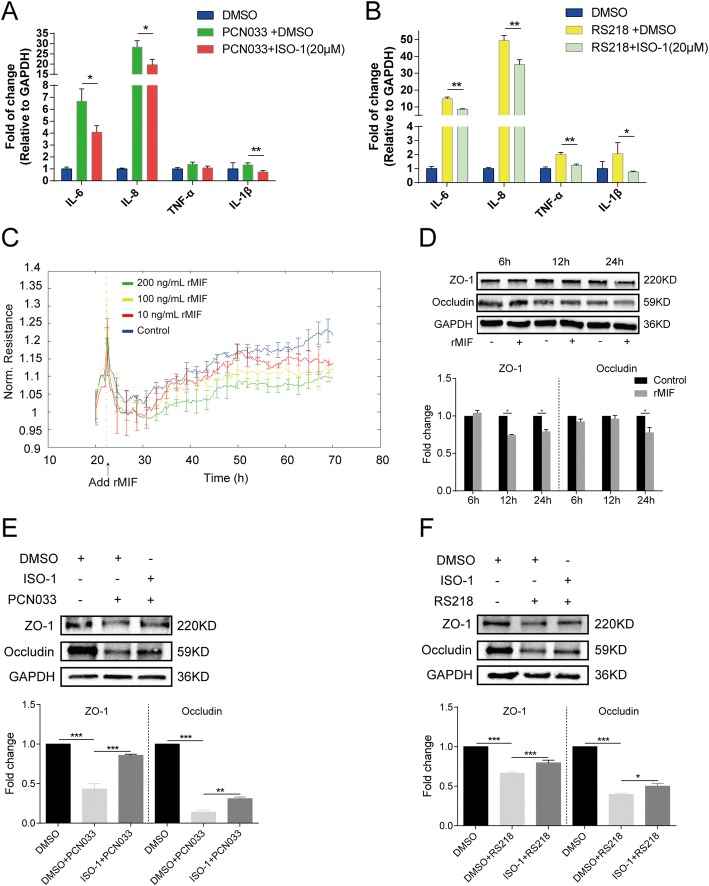
Based on the aforementioned network analysis, we noticed the presence of MIF in the meningitic PCN033 and RS218 strain groups, suggesting potential roles for it in meningitic E. coli invasion of the BBB. Here, by pretreating the hBMECs with 20 μM ISO-1 (a MIF inhibitor), we found that the multiple cytokines [e.g. interleukin (IL)-6, IL-8, tumor necrosis factor (TNF)-α, IL-1β] significantly induced by meningitic E. coli PCN033 or RS218 infection had decreased levels (Fig. 6a, b). Moreover, the ECIS system was applied to evaluate the potential effects of recombinant MIF protein on the barrier function of hBMECs. The results showed that recombinant MIF obviously decreased the resistance formed by the cells in a dose-dependent manner (Fig. 6c). We also observed that treatment with recombinant MIF (200 ng/ml) for 12 and 24 h led to decreased expression of tight junction proteins like ZO-1 and occludin (Fig. 6d); moreover, use of the MIF inhibitor ISO-1 could partially recover the PCN033 or RS218 infection-caused downregulation of tight junction proteins like ZO-1 and Occludin (Fig. 6e, f). Together, these observations support the conclusion that MIF contributes to the induction of proinflammatory cytokines and the decrease in tight junction proteins during meningitic E. coli invasion of the BBB.
Fig. 6.
MIF facilitated the bacteria-induced inflammatory response and tight junction damage in hBMECs. a, b Real-time PCR determination of the expression of cytokines in response to the treatments. The MIF inhibitor ISO-1 (20 μM) significantly attenuated the PCN033- or RS218-induced production of proinflammatory cytokines. c ECIS assay showed a dose-dependent decrease of the hBMECs resistance in response to recombinant MIF protein. d Recombinant MIF protein (200 ng/mL) decreased the expression of tight junction proteins ZO-1 and occludin in hBMECs along with time. The densitometry was performed to quantitatively analyze the Western bands. e, f Western blotting and densitometry analysis showed that ISO-1 treatment partially recovered PCN033- or RS218-mediated downregulation of the tight junction proteins ZO-1 and occludin. Data were expressed as the mean ± standard deviation (mean ± SD) from three replicates or analyses (n = 3). P < 0.05 (*) was considered statistically significant; p < 0.01 (**) and p < 0.001 (***) were extremely significant
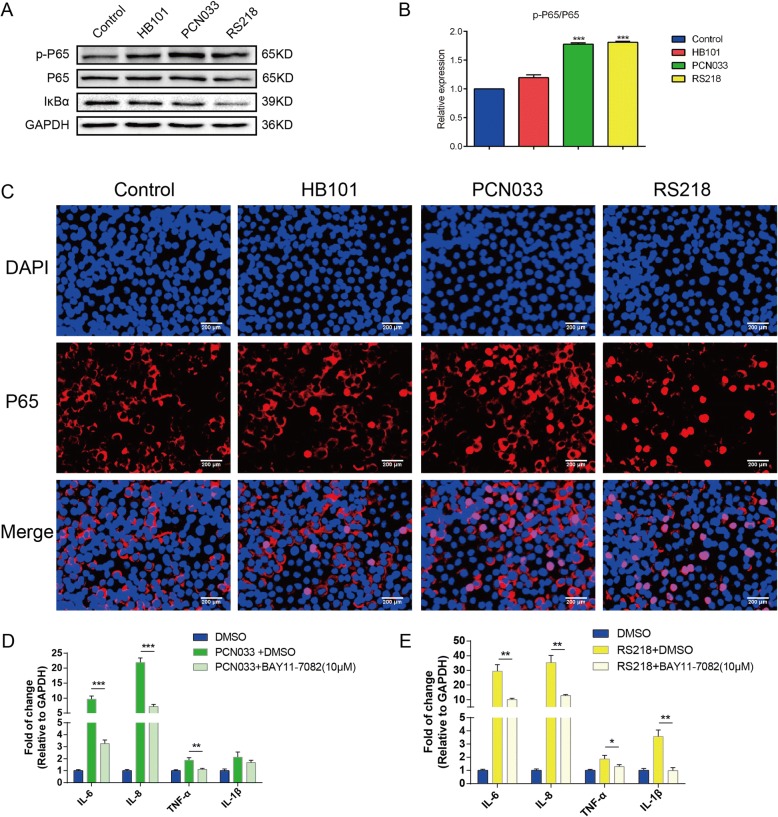
Meningitic E. coli activation of NF-κB signaling mediates the production of cytokines
As mentioned above in the network analysis, involvement of the NF-κB complex was observed in cells following the challenge with meningitic E. coli strains PCN033 and RS218, but not with non-meningitic HB101. Therefore, we investigated NF-κB signaling activation in hBMECs in response to infection. Phosphorylation of the NF-κB p65 subunit increased significantly in response to PCN033 and RS218 infection, and this was much higher than that observed during the response to HB101 infection. Also, degradation of IκBα upon PCN033 or RS218 infection was much greater than that upon HB101 infection (Fig. 7a, b). Using immunofluorescence microscopy, we also observed p65 translocation to the nucleus upon PCN033 and RS218 infection (Fig. 7c), while this nuclear translocation was barely observed in response to HB101 infection (Fig. 7c). These results indicate that the NF-κB signaling pathway is activated during meningitic E. coli interaction with hBMECs. Moreover, by using the NF-κB inhibitor BAY11-7082, we observed that the meningitic E. coli PCN033- or RS218-induced cytokines production (including IL-6, IL-8, TNF-α, and IL-1β) was significantly decreased when compared with DMSO treatment (Fig. 7d, e). Together, these data firmly support our network analysis that the NF-κB signaling pathway is involved in both PCN033 and RS218 infection of hBMECs, and their activation of NF-κB signaling in hBMECs mediates the induction of proinflammatory cytokines.
Fig. 7.
NF-κB signaling is activated in response to meningitic PCN033 or RS218 and mediates the inflammatory response. a, b p65 phosphorylation and IκBα degradation were significantly enhanced upon challenge with PCN033 and RS218, as shown by western blotting and densitometry. c Nuclear translocation of the p65 subunit was apparent in the hBMECs upon infection with PCN033 and RS218, but barely observed in response to infection with HB101. d, e Real-time PCR analysis showed that meningitic E. coli strains PCN033- and RS218-induced proinflammatory cytokines production was significantly decreased via NF-κB signaling inhibition with 10 μM of BAY11-7082. Data were expressed as the mean ± standard deviation (mean ± SD) from three replicates or analyses. P < 0.05 (*) was considered statistically significant; p < 0.01 (**) and p < 0.001 (***) were extremely significant
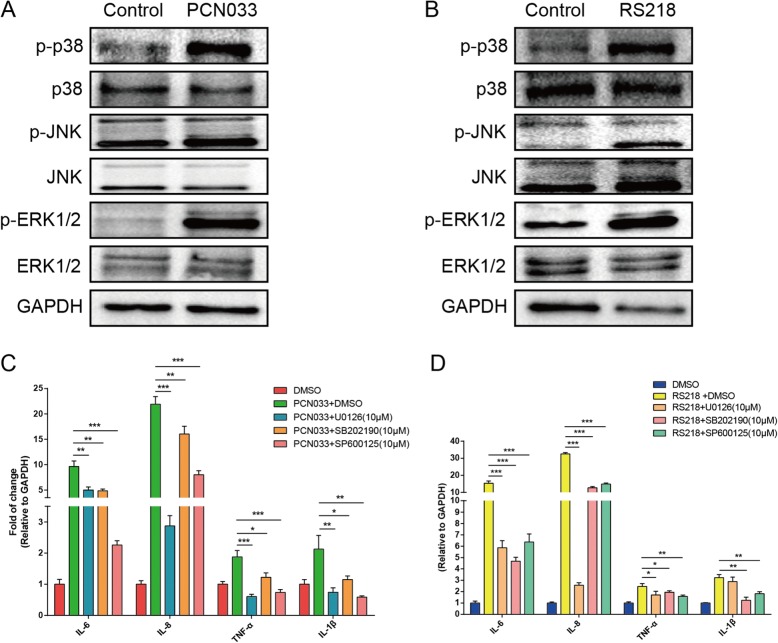
MAPK signaling pathways are involved in proinflammatory cytokine induction by meningitic E. coli strains
Because ERK was assumed to be involved in infections with PCN033 and RS218 based on our network prediction, we next investigated the activation of MAPK pathways in hBMECs in response to meningitic E. coli. The results showed that the phosphorylation of p38, JNK, and ERK1/2 significantly increased in response to meningitic strains PCN033 or RS218 (Fig. 8a, b), indicating the activation of all three MAPK pathways in hBMECs upon meningitic E. coli challenge. After demonstrating the significant induction of several proinflammatory cytokines above, we next investigated whether the MAPK pathways were involved in these cytokines production. Following pretreatment with U0126 (a specific ERK1/2 inhibitor), SB202190 (a selective inhibitor of p38), and SP600125 (a JNK-specific inhibitor), the proinflammatory cytokines (IL-6, IL-8, TNF-α, IL-1β) induced in hBMECs upon PCN033 or RS218 infection were significantly reduced (to different extents), compared with that in each DMSO control group (Fig. 8c). These results indicate that the MAPK signaling pathways, including MAPK-p38, MAPK-ERK1/2, and MAPK-JNK, were all activated and at least participated in meningitic E. coli-induced neuroinflammatory responses.
Fig. 8.
MAPK signaling, which is activated in hBMECs upon meningitic E. coli infection, contributes to the neuroinflammatory response. a, b Phosphorylation of p38, JNK, and ERK1/2 in hBMECs upon challenge with PCN033 and RS218 strains. c, d Blocking the three MAPK signaling pathways through specific inhibitors (U0126, a specific ERK1/2 inhibitor; SB202190, a p38 selective inhibitor; and SP600125, a JNK-specific inhibitor) significantly decreased the infection-induced neuroinflammatory response via real-time PCR analysis. P < 0.05 (*) was considered statistically significant; p < 0.01 (**) and p < 0.001 (***) were extremely significant
Discussion
The iTRAQ-based proteomics, a powerful approach for obtaining comprehensive and quantitative protein expression profiling data, has been used widely to identify and characterize potential cellular targets. In current study, we used iTRAQ to explore the proteomic differences in hBMECs in response to meningitic or non-meningitic E. coli infections. The E. coli strains PCN033 and RS218 were selected for this study because they are representative meningitis-causing strains capable of penetrating the BBB as well as inducing severe neuroinflammation [5, 20], while the E. coli strain HB101 is avirulent and non-meningitic and was therefore used as the negative control.
Based on our data, 13 significantly differentiated proteins in total were found to be shared by PCN033 and RS218 (Fig. 1). They are TELO2, IFT74, CBWD6, EXOSC4, TBOL1, RBM5, KRT9, HIST1H1C, HIST1H1D, HIST1H1B, HIST1H1E, MIF, and DMD (Table 5). Among these, EXOSC4 was the only protein that was also significantly changed in response to non-meningitic E. coli HB101 (Fig. 2, Table 5). EXOSC4, a non-catalytic component of the RNA exosome machinery, has 3′-5′ exoribonuclease activity and participates in a multitude of cellular RNA processing and degradation events [29]. It was reported that EXOSC4 was a potential factor involved in the maintenance of genome stability, by eliminating the RNA processing by-products and non-coding “pervasive” transcripts thereby limiting or excluding their export to the cytoplasm, or by preventing translation of aberrant mRNAs [30–32]. In lung adenocarcinoma, EXOSC4 has been reported to be extremely highly expressed and closely associated with cancer cell proliferation and was, therefore, recognized as a new prognostic marker [30]. Similarly, in patients with liver cancer, the EXOSC4 gene was found to be highly expressed, and its knock-down commonly inhibited cancer cell growth and invasion [33]. Here, we found that EXOSC4 was commonly targeted by the meningitic and the non-meningitic E. coli strains, indicating that this cellular protein is a non-specific infection-related protein. Other than EXOSC4, the remaining 12 proteins were shared by the meningitic strains (PCN033 and RS218) alone, suggesting that these proteins might represent the potential targets hijacked by these meningitic E. coli strains.
Among these 12 meningitic E. coli-specific “cellular responders,” we firstly focused on MIF, which was the only one to exhibit common upregulation in response to both meningitic E. coli PCN033 and RS218 (Table 5). MIF is a proinflammatory cytokine, which has been highlighted as a key player in infection and septic shock [34, 35]. It is reported to be involved in the cytokine storm, which facilitates the uncontrolled release of cytokines into the circulation during pathogen infection or sepsis [36]. As previously evidenced in E. coli-induced meningitis, cytokines and chemokines potentially contribute to BBB damage [5]. The burst of proinflammatory cytokines during infection may lead directly to dysfunction of the endothelial barrier and an increase in vascular permeability in the brain, thus finally leading to severe CNS injury. Moreover, MIF may be secreted by a wide variety of cells upon stimulation, and once MIF binds to its receptors (e.g., CXCR2, CXCR4, and/or CD74 [37, 38]), several downstream signal molecules such as PI3K/Akt or MAPK/ERK become activated, thus mediating the inflammatory response [39, 40]. In the present study, the effects of MIF on meningitic E. coli-induced inflammation were also verified by the observation that the MIF inhibitor ISO-1 significantly decreased meningitic E. coli PCN033- or RS218-induced upregulation of IL-6, IL-8, IL-Iβ, and TNF-α (Fig. 5). Noticeably however, although the ISO-1 inhibitory effects were significant, there was still a significant induction of IL-6 and IL-8 in response to PCN033 and RS218 infection, suggesting that other “switches” for proinflammatory cytokine and chemokine generation commonly exist in response to infection. Except for its role in inflammation, we also observed the involvement of MIF in BBB damage, as evidenced by the fact that recombinant MIF was able to deconstruct the endothelial barrier by inducing a significant decrease in the junction-associated protein ZO-1 and occludin (Fig. 6). Furthermore, when MIF inhibitor ISO-1 was used, the PCN033- and/or RS218-induced downregulation of ZO-1 and occludin was largely restored (Fig. 6). Considering the potential roles of MIF in mediating the neuroinflammatory response as well as in inducing BBB disruption, it is possible that MIF may represent a novel and potential target for clinical prevention and therapy for E. coli meningitis.
Our IPA-based canonical pathways prediction suggested that protein kinase A signaling, eumelanin biosynthesis, EIF2 signaling, and granzyme A signaling were simultaneously enriched in hBMECs upon infection with RS218 and PCN033, but not with HB101. Among these processes, granzyme A signaling was much more significantly enriched. In the RS218 group, HIST1H1B, HIST1H1C, HIST1H1E, and HIST1H1D are included in granzyme A signaling, while in the PCN033 group, HIST1H1B, HIST1H1C, HIST1H1E, HIST1H1D, and H1F0 are involved (Additional file 6: Table S6). Granzyme A was identified as a cytotoxic T lymphocyte protease with multiple roles in infectious diseases. For example, several studies have shown that granzyme A is highly expressed in patients with tuberculosis and may represent a promising diagnostic marker distinct from IFN-γ to discriminate between patients with tuberculosis and other pulmonary diseases [41–43]. Granzyme A is also considered to participate in the host defense response in multiple ways, such as by generating superoxide and inactivating the oxidative defense enzymes that kill intracellular parasites [44], by unfavorably impairing host defenses during Streptococcus pneumoniae pneumonia [45], by performing as a proinflammatory protease that cleaves IL-1β intracellularly into bioactive IL-1β [46, 47], or by causing detachment of alveolar epithelial A549 cells accompanied by promotion of IL-8 release [48]. Here, in the present study, granzyme A signaling was significantly enriched by cellular differentiated proteins in response to both meningitic E. coli strains, but not in non-meningitic E. coli HB101. This result probably indicates that granzyme A could be a potential indicator of E. coli meningitis, but further supportive evidences are needed.
Based on the IPA functional network analysis, we also noticed that the NF-κB complex and MAPK/ERK signaling were involved in both PCN033 and RS218 infection of hBMECs, but barely in the HB101 group. The NF-κB complex comprises a family of closely related transcription factors with important roles in regulating the gene expression involved in inflammation and the immune response [49]. The NF-κB activation process is induced by the phosphorylation of serine residues in IkB proteins, which are subjected to ubiquitination and proteasome degradation and, subsequently, phosphorylation and nuclear translocation of the p65 subunit. Early studies have shown that NF-κB is activated in bacteria-induced CNS infections [50], and NF-κB inhibitors have been found to reduce neuroinflammation [51] as well as protect rat brains from inflammatory injury following transient focal cerebral ischemia [52] and pneumococcal meningitis [53]. In E. coli, it has been evidenced that OmpA+E. coli can induce ICAM-1 expression in hBMECs by activating NF-κB signaling [54] and that the IbeA+E. coli K1 strain can also induce activation and nuclear translocation of NF-κB in hBMECs [55]. In the current study, by western blotting, we also showed that the NF-κB pathway was activated more in hBMECs infected by meningitic strains PCN033 and RS218 compared with that by HB101 infection, where the phosphorylation of p65 and degradation of IκBα were compared, as well as with the immunofluorescence experiments that showed the nuclear translocation of p65. Not unexpectedly, treating hBMECs with the NF-κB inhibitor BAY11-7082 significantly attenuated those cytokines induction during meningitic E. coli infection, suggesting that NF-κB signaling works potently in mediating the neuroinflammatory response.
Likewise, we found that the effects of MAPK signaling were similarly associated with both PCN033 and RS218 infection of hBMECs. MAPK signaling cascades actually involve three major pathways: JNK (which acts as mediator of extracellular stress responses), ERK1/2 (which mediates proliferative stimuli), and p38 (which is also involved in mediating extracellular stress responses, particularly by regulating cytokine expression) [56]. Our IPA network analysis indicated the involvement of ERK during infection with meningitic E. coli PCN033 and RS218, which is consistent with our previous finding that MAPK/ERK signaling is involved in infection and mediates the induction of VEGFA and Snail-1 by the meningitic strain PCN033 [5]; however, via western blotting we showed the activation of all these three signaling molecules in response to PCN033 and RS218 infection. Also, by using specific inhibitors against ERK1/2, p38, and JNK, we observed that inhibition of all three MAPK pathways significantly decreased the infection-induced upregulation of proinflammatory cytokines IL-6, IL-8, IL-Iβ, and TNF-α. Therefore, collectively these data largely support the viewpoint that all three major MAPK signaling pathways play potent roles in meningitic E. coli infection and induce neuroinflammatory responses.
Conclusions
In our study, using the iTRAQ proteomics approach, we compared and analyzed the DEPs in hBMECs infected with meningitic or non-meningitic E. coli strains. Twelve DEPs were identified as the commonly responding proteins in hBMECs upon infection with meningitic E. coli strains PCN033 and RS218, except for only one cellular protein shared by both meningitic and non-meningitic strains. Our data revealed MIF to be an important contributor to meningitic E. coli-induced cytokine production and tight junction disruption, while also showing that the NF-κB and MAPK signaling pathways are involved in the infection process. Comparing and profiling these differential cellular proteins in hBMECs in response to meningitic E. coli strains should open up further research on host responses against meningitic strains and help with the development of more targets for better prevention and therapeutic control of E. coli meningitis.
Additional files
Table S1. Protein profile of HB101-infected hBMECs. (XLSX 287 kb)
Table S2. Protein profile of PCN033-infected hBMECs. (XLSX 290 kb)
Table S3. Protein profile of RS218-infected hBMECs. (XLSX 289 kb)
Table S4. GO term annotation of DEPs. (DOCX 15 kb)
Table S5. Ingenuity Canonical Pathways of HB101-infected group. (XLSX 71 kb)
Table S6. Ingenuity Canonical Pathways of PCN033-infected group. (XLSX 70 kb)
Table S7. Ingenuity Canonical Pathways of RS218-infected group. (XLSX 71 kb)
Table S8. The potential networks in HB101-infected group. (XLSX 70 kb)
Table S9. The potential networks in PCN033-infected group. (XLSX 72 kb)
Table S10. The potential networks in RS218-infected group. (XLSX 71 kb)
Acknowledgements
We would like to thank Prof. Kwang Sik Kim in Johns Hopkins University School of Medicine to kindly provide the hBMECs cells.
Funding
This work was supported by grants from the National Key R&D Program of China (2016YFD0500406), the National Natural Science Foundation of China (NSFC) (Nos. 31772736, 31502062), the Outstanding youth project of Natural Science Foundation in Hubei Province (2018CFA070), and the Fundamental Research Funds for the Central Universities (Program No. 2662018PY032).
Availability of data and materials
There is no data, software, databases, and application/tool available apart from the reported in the present study. All data is provided in manuscript.
Abbreviations
- BBB
Blood-brain barrier
- BMECs
Brain microvascular endothelial cells
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- DEPs
Differentially expressed proteins
- DMD
Dystrophin
- E. coli
Escherichia coli
- ECIS
Electric cell-substrate impedance sensing
- EGFR
Epidermal growth factor receptor
- ERK1/2
Extracellular signal-regulated kinases 1 and 2
- ExPEC
Extraintestinal pathogenic Escherichia coli
- GO
Gene Ontology
- ICAM-1
Intercellular adhesion molecule-1
- IL-1β
Interleukin 1 beta
- IL-8
Interleukin-8
- IPA
Ingenuity Pathways Analysis
- ISO-1
-
(S, R)-3-(4-Hydroxyphenyl)-4, 5-dihydro-5-isoxazole acetic acid methyl ester DAPI
4′-6-Diamidino-2-phenylindole
- iTRAQ
Isobaric tags for relative and absolute quantification
- JNK
c-Jun N-terminal kinase
- KEGG
Kyoto encyclopedia of genes and genomes
- LC-MS/MS
Liquid chromatography tandem mass spectrometry
- LGMN
Legumain
- MAPK
Mitogen-activated protein kinase
- MIF
Macrophage migration inhibitory factor
- NF-κB
Nuclear factor-κB
- NOS
Nitric oxide synthase
- PI3K
Phosphatidylinositol 3-kinase
- S1P
Sphingosine-1-phosphate
- SCX
Strong cation exchange chromatography
- TBPL1
TATA box-binding protein-like protein 1
- TEAB
Tetraethyl-ammonium bromide
- TEER
Trans-endothelial electric resistance
- TNF-α
Tumor necrosis factor-alpha
- VEGFA
Vascular endothelial growth factor A
- ZO-1
Zonula occludens-1, IL-6, interleukin-6
Authors’ contributions
WTL performed all experiments and analyzed the data. YJL drafted the manuscript. RCY, JYF, LL, HW, and QC participated in project planning, and all western blot experiments. XRW conceived of the project, coordinated and supervised the experiments, and revised the manuscript. XRW, CT, and HCC provide technical and administrative support. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wen-Tong Liu, Email: guixiangguo@hotmail.com.
Yu-Jin Lv, Email: yuanhanglyj@126.com.
Rui-Cheng Yang, Email: m15926465282@163.com.
Ji-Yang Fu, Email: fujiyang921021@hotmail.com.
Lu Liu, Email: cofeandsalt@163.com.
Huan Wang, Email: 534837079@qq.com.
Qi Cao, Email: 992514158@qq.com.
Chen Tan, Email: tanchen@mail.hzau.edu.cn.
Huan-Chun Chen, Email: chenhch@mail.hzau.edu.cn.
Xiang-Ru Wang, Email: wangxr228@mail.hzau.edu.cn.
References
- 1.Kim KS. Pathogenesis of bacterial meningitis: from bacteraemia to neuronal injury. Nat Rev Neurosci. 2003;4:376–385. doi: 10.1038/nrn1103. [DOI] [PubMed] [Google Scholar]
- 2.Kim KS. Mechanisms of microbial traversal of the blood-brain barrier. Nat Rev Microbiol. 2008;6:625–634. doi: 10.1038/nrmicro1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim KS. Acute bacterial meningitis in infants and children. Lancet Infect Dis. 2010;10:32–42. doi: 10.1016/S1473-3099(09)70306-8. [DOI] [PubMed] [Google Scholar]
- 4.Candelario-Jalil E, Yang Y, Rosenberg GA. Diverse roles of matrix metalloproteinases and tissue inhibitors of metalloproteinases in neuroinflammation and cerebral ischemia. Neuroscience. 2009;158:983–994. doi: 10.1016/j.neuroscience.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang RC, Liu WT, Miao L, Yang XP, Fu JY, Dou BB, Cai AL, Zong X, Tan C, Chen HC, Wang XR. Induction of VEGFA and Snail-1 by meningitic Escherichia coli mediates disruption of the blood-brain barrier. Oncotarget. 2016;7:63839–63855. doi: 10.18632/oncotarget.11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stins MF, Badger J, Sik Kim K. Bacterial invasion and transcytosis in transfected human brain microvascular endothelial cells. Microb Pathog. 2001;30:19–28. doi: 10.1006/mpat.2000.0406. [DOI] [PubMed] [Google Scholar]
- 7.Stins MF, Gilles F, Kim KS. Selective expression of adhesion molecules on human brain microvascular endothelial cells. J Neuroimmunol. 1997;76:81–90. doi: 10.1016/S0165-5728(97)00036-2. [DOI] [PubMed] [Google Scholar]
- 8.Das A, Asatryan L, Reddy MA, Wass CA, Stins MF, Joshi S, Bonventre JV, Kim KS. Differential role of cytosolic phospholipase A2 in the invasion of brain microvascular endothelial cells by Escherichia coli and Listeria monocytogenes. J Infect Dis. 2001;184:732–737. doi: 10.1086/322986. [DOI] [PubMed] [Google Scholar]
- 9.Kim KS, Itabashi H, Gemski P, Sadoff J, Warren RL, Cross AS. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J Clin Invest. 1992;90:897–905. doi: 10.1172/JCI115965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burkhart A, Thomsen LB, Thomsen MS, Lichota J, Fazakas C, Krizbai I, Moos T. Transfection of brain capillary endothelial cells in primary culture with defined blood-brain barrier properties. Fluids Barriers CNS. 2015;12:19. doi: 10.1186/s12987-015-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eigenmann DE, Xue G, Kim KS, Moses AV, Hamburger M, Oufir M. Comparative study of four immortalized human brain capillary endothelial cell lines, hCMEC/D3, hBMEC, TY10, and BB19, and optimization of culture conditions, for an in vitro blood-brain barrier model for drug permeability studies. Fluids Barriers CNS. 2013;10:33. doi: 10.1186/2045-8118-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbott NJ. Blood-brain barrier structure and function and the challenges for CNS drug delivery. J Inherit Metab Dis. 2013;36:437–449. doi: 10.1007/s10545-013-9608-0. [DOI] [PubMed] [Google Scholar]
- 13.Tajes M, Ramos-Fernandez E, Weng-Jiang X, Bosch-Morato M, Guivernau B, Eraso-Pichot A, Salvador B, Fernandez-Busquets X, Roquer J, Munoz FJ. The blood-brain barrier: structure, function and therapeutic approaches to cross it. Mol Membr Biol. 2014;31:152–167. doi: 10.3109/09687688.2014.937468. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman JA, Badger JL, Zhang Y, Huang SH, Kim KS. Escherichia coli K1 aslA contributes to invasion of brain microvascular endothelial cells in vitro and in vivo. Infect Immun. 2000;68:5062–5067. doi: 10.1128/IAI.68.9.5062-5067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Kim KS. Role of OmpA and IbeB in Escherichia coli K1 invasion of brain microvascular endothelial cells in vitro and in vivo. Pediatr Res. 2002;51:559–563. doi: 10.1203/00006450-200205000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Reddy MA, Prasadarao NV, Wass CA, Kim KS. Phosphatidylinositol 3-kinase activation and interaction with focal adhesion kinase in Escherichia coli K1 invasion of human brain microvascular endothelial cells. J Biol Chem. 2000;275:36769–36774. doi: 10.1074/jbc.M007382200. [DOI] [PubMed] [Google Scholar]
- 17.Khan NA, Wang Y, Kim KJ, Chung JW, Wass CA, Kim KS. Cytotoxic necrotizing factor-1 contributes to Escherichia coli K1 invasion of the central nervous system. J Biol Chem. 2002;277:15607–15612. doi: 10.1074/jbc.M112224200. [DOI] [PubMed] [Google Scholar]
- 18.Reddy MA, Wass CA, Kim KS, Schlaepfer DD, Prasadarao NV. Involvement of focal adhesion kinase in Escherichia coli invasion of human brain microvascular endothelial cells. Infect Immun. 2000;68:6423–6430. doi: 10.1128/IAI.68.11.6423-6430.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung JW, Hong SJ, Kim KJ, Goti D, Stins MF, Shin S, Dawson VL, Dawson TM, Kim KS. 37-kDa laminin receptor precursor modulates cytotoxic necrotizing factor 1-mediated RhoA activation and bacterial uptake. J Biol Chem. 2003;278:16857–16862. doi: 10.1074/jbc.M301028200. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Maruvada R, Morris AJ, Liu JO, Wolfgang MJ, Baek DJ, Bittman R, Kim KS. Sphingosine 1-phosphate activation of EGFR as a novel target for meningitic Escherichia coli penetration of the blood-brain barrier. PLoS Pathog. 2016;12:e1005926. doi: 10.1371/journal.ppat.1005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wijetunge DS, Katani R, Kapur V, Kariyawasam S. Complete genome sequence of Escherichia coli strain RS218 (O18:H7:K1), associated with neonatal meningitis. Genome Announc. 2015;3:e00804-15. [DOI] [PMC free article] [PubMed]
- 22.Tan C, Xu Z, Zheng H, Liu W, Tang X, Shou J, Wu B, Wang S, Zhao GP, Chen H. Genome sequence of a porcine extraintestinal pathogenic Escherichia coli strain. J Bacteriol. 2011;193:5038. doi: 10.1128/JB.05551-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Zheng H, Yang M, Xu Z, Wang X, Wei L, Tang B, Liu F, Zhang Y, Ding Y, et al. Genome analysis and in vivo virulence of porcine extraintestinal pathogenic Escherichia coli strain PCN033. BMC Genomics. 2015;16:717. doi: 10.1186/s12864-015-1890-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan C, Tang X, Zhang X, Ding Y, Zhao Z, Wu B, Cai X, Liu Z, He Q, Chen H. Serotypes and virulence genes of extraintestinal pathogenic Escherichia coli isolates from diseased pigs in China. Vet J. 2012;192:483–488. doi: 10.1016/j.tvjl.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 25.Zhu L, Maruvada R, Sapirstein A, Malik KU, Peters-Golden M, Kim KS. Arachidonic acid metabolism regulates Escherichia coli penetration of the blood-brain barrier. Infect Immun. 2010;78:4302–4310. doi: 10.1128/IAI.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan NA, Kim Y, Shin S, Kim KS. FimH-mediated Escherichia coli K1 invasion of human brain microvascular endothelial cells. Cell Microbiol. 2007;9:169–178. doi: 10.1111/j.1462-5822.2006.00779.x. [DOI] [PubMed] [Google Scholar]
- 27.Yang R, Huang F, Fu J, Dou B, Xu B, Miao L, Liu W, Yang X, Tan C, Chen H, Wang X. Differential transcription profiles of long non-coding RNAs in primary human brain microvascular endothelial cells in response to meningitic Escherichia coli. Sci Rep. 2016;6:38903. doi: 10.1038/srep38903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szulcek R, Bogaard HJ, Amerongen GPV. Electric cell-substrate impedance sensing for the quantification of endothelial proliferation, barrier function, and motility. J Vis Exp. 2014;85. 10.3791/51300. [DOI] [PMC free article] [PubMed]
- 29.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ --> 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Byrne K, Paquet N, Box JK, Adams M, Richard D. 17P examination of EXOSC4 as a new prognostic marker and a novel therapeutic avenue in lung adenocarcinoma. J Thorac Oncol. 2016;11:S63. doi: 10.1016/S1556-0864(16)30131-9. [DOI] [Google Scholar]
- 31.Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Dijk EL, Schilders G, Pruijn GJ. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefanska B, Cheishvili D, Suderman M, Arakelian A, Huang J, Hallett M, Han ZG, Al-Mahtab M, Akbar SM, Khan WA, et al. Genome-wide study of hypomethylated and induced genes in patients with liver cancer unravels novel anticancer targets. Clin Cancer Res. 2014;20:3118–3132. doi: 10.1158/1078-0432.CCR-13-0283. [DOI] [PubMed] [Google Scholar]
- 34.Rosado Jde D, Rodriguez-Sosa M. Macrophage migration inhibitory factor (MIF): a key player in protozoan infections. Int J Biol Sci. 2011;7:1239–1256. doi: 10.7150/ijbs.7.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaloye J, De Bruin IJ, Darling KE, Reymond MK, Sweep FC, Roger T, Calandra T, Cavassini M. Increased macrophage migration inhibitory factor (MIF) plasma levels in acute HIV-1 infection. Cytokine. 2012;60:338–340. doi: 10.1016/j.cyto.2012.07.027. [DOI] [PubMed] [Google Scholar]
- 36.Pang T, Cardosa MJ, Guzman MG. Of cascades and perfect storms: the immunopathogenesis of dengue haemorrhagic fever-dengue shock syndrome (DHF/DSS) Immunol Cell Biol. 2007;85:43–45. doi: 10.1038/sj.icb.7100008. [DOI] [PubMed] [Google Scholar]
- 37.Bernhagen J, Krohn R, Lue H, Gregory JL, Zernecke A, Koenen RR, Dewor M, Georgiev I, Schober A, Leng L, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13:587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz V, Lue H, Kraemer S, Korbiel J, Krohn R, Ohl K, Bucala R, Weber C, Bernhagen J. A functional heteromeric MIF receptor formed by CD74 and CXCR4. FEBS Lett. 2009;583:2749–2757. doi: 10.1016/j.febslet.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lue H, Kapurniotu A, Fingerle-Rowson G, Roger T, Leng L, Thiele M, Calandra T, Bucala R, Bernhagen J. Rapid and transient activation of the ERK MAPK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on JAB1/CSN5 and Src kinase activity. Cell Signal. 2006;18:688–703. doi: 10.1016/j.cellsig.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 40.Lue H, Thiele M, Franz J, Dahl E, Speckgens S, Leng L, Fingerle-Rowson G, Bucala R, Luscher B, Bernhagen J. Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene. 2007;26:5046–5059. doi: 10.1038/sj.onc.1210318. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Laorden MI, Blok DC, Kager LM, Hoogendijk AJ, van Mierlo GJ, Lede IO, Rahman W, Afroz R, Ghose A, Visser CE, et al. Increased intra- and extracellular granzyme expression in patients with tuberculosis. Tuberculosis (Edinb) 2015;95:575–580. doi: 10.1016/j.tube.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 42.Guggino G, Orlando V, Cutrera S, La Manna MP, Di Liberto D, Vanini V, Petruccioli E, Dieli F, Goletti D, Caccamo N. Granzyme A as a potential biomarker of Mycobacterium tuberculosis infection and disease. Immunol Lett. 2015;166:87–91. doi: 10.1016/j.imlet.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Laux da Costa L, Delcroix M, Dalla Costa ER, Prestes IV, Milano M, Francis SS, Unis G, Silva DR, Riley LW, Rossetti ML. A real-time PCR signature to discriminate between tuberculosis and other pulmonary diseases. Tuberculosis (Edinb) 2015;95:421–425. doi: 10.1016/j.tube.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dotiwala F, Mulik S, Polidoro RB, Ansara JA, Burleigh BA, Walch M, Gazzinelli RT, Lieberman J. Killer lymphocytes use granulysin, perforin and granzymes to kill intracellular parasites. Nat Med. 2016;22:210–216. doi: 10.1038/nm.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van den Boogaard FE, van Gisbergen KP, Vernooy JH, Medema JP, Roelofs JJ, van Zoelen MA, Endeman H, Biesma DH, Boon L, Van't Veer C, et al. Granzyme A impairs host defense during Streptococcus pneumoniae pneumonia. Am J Physiol Lung Cell Mol Physiol. 2016;311:L507–L516. doi: 10.1152/ajplung.00116.2016. [DOI] [PubMed] [Google Scholar]
- 46.Pardo J, Simon MM, Froelich CJ. Granzyme A is a proinflammatory protease. Blood. 2009;114:3968. doi: 10.1182/blood-2009-07-231027. [DOI] [PubMed] [Google Scholar]
- 47.Hildebrand D, Bode KA, Riess D, Cerny D, Waldhuber A, Rommler F, Strack J, Korten S, Orth JH, Miethke T, et al. Granzyme A produces bioactive IL-1beta through a nonapoptotic inflammasome-independent pathway. Cell Rep. 2014;9:910–917. doi: 10.1016/j.celrep.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Yoshikawa Y, Hirayasu H, Tsuzuki S, Fushiki T. Granzyme A causes detachment of alveolar epithelial A549 cells accompanied by promotion of interleukin-8 release. Biosci Biotechnol Biochem. 2008;72:2481–2484. doi: 10.1271/bbb.80362. [DOI] [PubMed] [Google Scholar]
- 49.Rothwarf DM, Karin M. The NF-kappa B activation pathway: a paradigm in information transfer from membrane to nucleus. Sci STKE. 1999;1999:RE1. doi: 10.1126/stke.1999.5.re1. [DOI] [PubMed] [Google Scholar]
- 50.Ichiyama T, Isumi H, Yoshitomi T, Nishikawa M, Matsubara T, Furukawa S. NF-kappaB activation in cerebrospinal fluid cells from patients with meningitis. Neurol Res. 2002;24:709–712. doi: 10.1179/016164102101200627. [DOI] [PubMed] [Google Scholar]
- 51.Yang XP, Fu JY, Yang RC, Liu WT, Zhang T, Yang B, Miao L, Dou BB, Tan C, Chen HC, Wang XR. EGFR transactivation contributes to neuroinflammation in Streptococcus suis meningitis. J Neuroinflammation. 2016;13:274. doi: 10.1186/s12974-016-0734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan M, Elango C, Ansari MA, Singh I, Singh AK. Caffeic acid phenethyl ester reduces neurovascular inflammation and protects rat brain following transient focal cerebral ischemia. J Neurochem. 2007;102:365–377. doi: 10.1111/j.1471-4159.2007.04526.x. [DOI] [PubMed] [Google Scholar]
- 53.Koedel U, Bayerlein I, Paul R, Sporer B, Pfister HW. Pharmacologic interference with NF-kappaB activation attenuates central nervous system complications in experimental Pneumococcal meningitis. J Infect Dis. 2000;182:1437–1445. doi: 10.1086/315877. [DOI] [PubMed] [Google Scholar]
- 54.Selvaraj SK, Periandythevar P, Prasadarao NV. Outer membrane protein A of Escherichia coli K1 selectively enhances the expression of intercellular adhesion molecule-1 in brain microvascular endothelial cells. Microbes Infect. 2007;9:547–557. doi: 10.1016/j.micinf.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chi F, Bo T, Wu CH, Jong A, Huang SH. Vimentin and PSF act in concert to regulate IbeA+ E. coli K1 induced activation and nuclear translocation of NF-kappaB in human brain endothelial cells. PLoS One. 2012;7:e35862. doi: 10.1371/journal.pone.0035862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramsauer K, Sadzak I, Porras A, Pilz A, Nebreda AR, Decker T, Kovarik P. p38 MAPK enhances STAT1-dependent transcription independently of Ser-727 phosphorylation. Proc Natl Acad Sci U S A. 2002;99:12859–12864. doi: 10.1073/pnas.192264999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Protein profile of HB101-infected hBMECs. (XLSX 287 kb)
Table S2. Protein profile of PCN033-infected hBMECs. (XLSX 290 kb)
Table S3. Protein profile of RS218-infected hBMECs. (XLSX 289 kb)
Table S4. GO term annotation of DEPs. (DOCX 15 kb)
Table S5. Ingenuity Canonical Pathways of HB101-infected group. (XLSX 71 kb)
Table S6. Ingenuity Canonical Pathways of PCN033-infected group. (XLSX 70 kb)
Table S7. Ingenuity Canonical Pathways of RS218-infected group. (XLSX 71 kb)
Table S8. The potential networks in HB101-infected group. (XLSX 70 kb)
Table S9. The potential networks in PCN033-infected group. (XLSX 72 kb)
Table S10. The potential networks in RS218-infected group. (XLSX 71 kb)
Data Availability Statement
There is no data, software, databases, and application/tool available apart from the reported in the present study. All data is provided in manuscript.