Graphical abstract
Keywords: Antifungal agents, Glucan synthesis, Fks1, Fungal cell wall, Candida species
Highlights
-
•
Poacic acid antifungal activity is both strains and species dependent for a range of Candida species.
-
•
The calcineurin pathway regulates poacic acid sensitivity in C. albicans.
-
•
Point mutations in β-1,3-glucan synthase Fks1 differentially affect poacic acid and echinocandin sensitivity.
Abstract
The rise of widespread antifungal resistance fuels the need to explore new classes of inhibitory molecules as potential novel inhibitors. Recently a plant natural product poacic acid (PA) was shown to inhibit β-1,3-glucan synthesis, and to have antifungal activity against a range of plant pathogens and against Saccharomyces cerevisiae. As with the echinocandins, such as caspofungin, PA targets the synthesis of cell wall β-1,3-glucan and has potential utility in the treatment of medically important fungi. However, the antifungal activity of PA against human pathogenic Candida species has not been explored and the precise mode of action of this compound is not understood. Here, we show that PA sensitivity is regulated by the calcineurin pathway and that susceptibility to PA varied significantly between Candida species, but did not correlate with in vitro β-glucan synthase activity, cell wall β-glucan content or the sensitivity of the species to caspofungin. Strains with point mutations (S645Y or S645P) in the hotspot1 region of the β-1,3-glucan synthase subunit Fks1, had decreased sensitivity to caspofungin but increased sensitivity to PA. C. guilliermondii, C. orthopsilosis, and C. parapsilosis were more sensitive to PA than C. albicans, C. dubliniensis, C. tropicalis, and C. glabrata. These observations suggest that there are significant differences in the mode of action of PA and caspofungin and that PA or PA analogues are not likely to have broad spectrum activity in the treatment of Candida infections.
Introduction
The fungal cell wall is a dynamic organelle that is essential for its viability. The fungal cell wall is a promising antifungal target for the therapeutic treatment of human fungal pathogens because the major components - chitin, glucan, and mannan, are absent from the human body. Depending on the fungus and growth conditions, the proportion and structural composition of cell wall components varies considerably (Free, 2013, Erwig and Gow, 2016, Gow and Netea, 2016, Gow et al., 2017). However, to date all fungi examined have β-1,3-glucan in their cell wall and this plays critical roles as a physical barrier, as a scaffold for the attachment of other cell wall components and in maintaining cell shape (Gow et al., 2017). This makes β-1,3-glucan synthesis an ideal broad-spectrum target for antifungal drugs. However, the cell wall is dynamic and can alter its structure depending on the environment and carbon source the fungi encounter, and in response to cell wall stress. Activation of the Ca2+/calcineurin, HOG and PKC pathways all occur in response to cell wall damage and result in the induction of compensatory mechanisms such as the synthesis of chitin (Munro et al., 2007, Munro, 2013, Gow et al., 2017). Failure to maintain cell wall integrity compromises cell viability and ultimately results in cell death (Negishi and Ohya, 2010, Rodríguez-Pena et al., 2010, Walker et al., 2013a, Walker et al., 2013b), therefore, cell wall integrity is constantly monitored via sensors located in the cell wall and membrane and at critical cell cycle check points (Roberts et al., 1983, Suzuki et al., 2004, Côte et al., 2009, Negishi and Ohya, 2010, Gow and Hube, 2012, Negishi et al., 2016).
The three echinocandins caspofungin (CSF), micafungin, and anidulafungin, are the newest class of antifungal agents on the market and have been clinically approved by the US Food and Drug Administration since 2001 (Pappas et al., 2009, Pound et al., 2010, Pfaller, 2012, Perlin, 2015b). They are non-competitive inhibitors of β-1,3-glucan synthase (Douglas et al., 1997, Odds, 2010, Nett and Andes, 2016) and therefore are not substrate analogues that bind to the enzyme active site. Indeed, the physical binding site of echinocandins to the Fks1 target protein has not been precisely defined. A number of clinical cases of echinocandin resistance have been reported (Imtiaz et al., 2012, Arendrup and Perlin, 2014, Perlin, 2015b, Sanglard, 2016) which is most commonly due to amino acid substitutions in Fks1, one of three major “hotspots” regions in the predicted external face of the β-1,3-glucan synthase transmembrane protein (Kurtz et al., 1996, Park et al., 2005, Garcia-Effron et al., 2008, Garcia-Effron et al., 2009a, Johnson et al., 2011, Johnson and Edlind, 2012, Perlin, 2015b, Kolaczkowska and Kolaczkowski, 2016, Prasad et al., 2016). For example, in C. albicans the serine645 to proline or tyrosine amino acid substitution at of Fks1 is frequently found in echinocandin-resistant strains (Perlin, 2015a, Perlin, 2015b). These mutations are often, but not always, accompanied by elevated cell wall chitin content (Ben-Ami et al., 2011, Ben-Ami and Kontoyiannis, 2012, Lee et al., 2012, Perlin, 2015a, Perlin, 2015b Perlin et al. 2015; Walker et al., 2013a, Walker et al., 2013b). However, resistance to CSF has also been described which is not associated with amino acid substitutions in Fks1. In addition, C. albicans strains that have an elevated chitin content in the cell wall are significantly less susceptible to CSF in vivo and in vitro (Walker et al., 2008, Lee et al., 2012, Walker et al., 2015). Paradoxical growth (also called the “Eagle effect”) of C. albicans at which growth still occurs at supra-MIC concentrations of the echinocandins has also been shown to be correlated with up-regulation of chitin synthesis (Stevens et al., 2006, Stevens, 2009). Furthermore, a number of CSF resistant isolates are found in other “non-albicans” species (Garcia-Effron et al., 2010, Perlin, 2011, Perlin, 2015b), including the emerging pathogen Candida auris, where as many as one third of strains are echinocandin resistant or cross-resistant (Lockhart et al., 2017a, Lockhart et al., 2017b). These health challenges highlight the need for new antifungal agents to augment the antifungal armamentarium.
Poacic acid (diferulate, 8-5-DC), PA, is a natural plant metabolite found in the lignocellulosic hydrolysates of grasses and has been characterised as a promising antifungal agent that targets β-1,3-glucan synthesis (Piotrowski et al., 2015). This compound inhibited growth of Saccharomyces cerevisiae, Alternaria solani and the oomycete Sclerotinia sclerotiorum in vitro. Lesion development on soybean leaves by S. sclerotiorum was also inhibited by PA ex vivo. PA apparently acts by directly binding to β-1,3-glucan polysaccharide, and therefore differs from the mode of action of echinocandins which directly target the β-1,3-glucan synthase enzyme. In this study we contrast the modes of action of two β-glucan synthesis inhibitors, poacic acid (PA) and caspofungin and show that they have distinct mode of actions and different host range activities. We determined the activity of PA against a range of clinically important fungal pathogens and demonstrate that PA is differentially active against a range of Candida species and examine the relationship between species-specificity, β-1,3-glucan synthesis and its sensitivity to PA. Our findings demonstrate that PA and echinocandins inhibit β-1,3-glucan synthesis in different ways and that PA-based pharmacophores may not be suitable as pan-Candida inhibitors.
Results
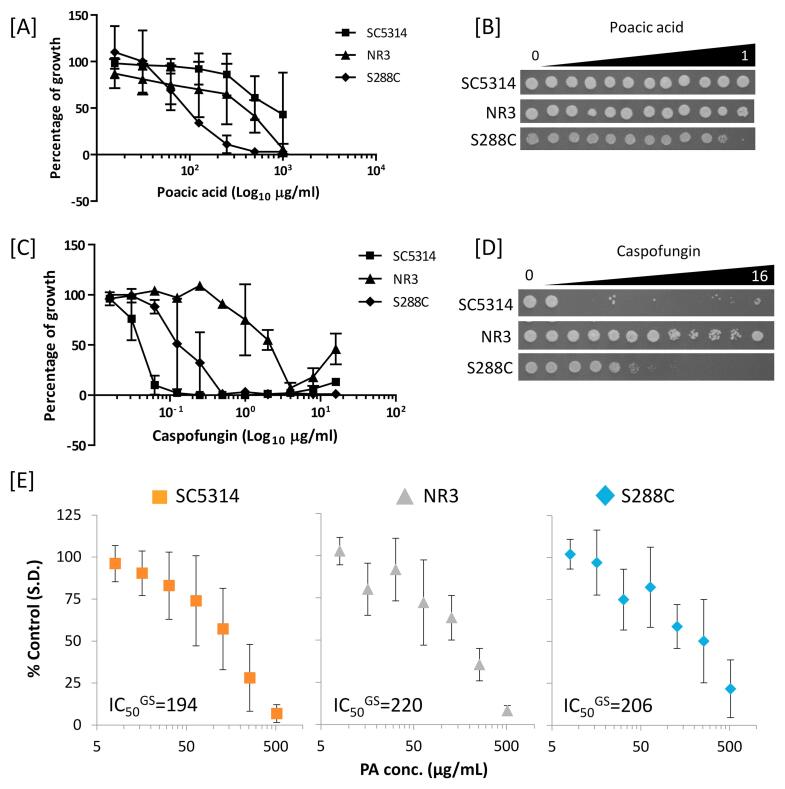
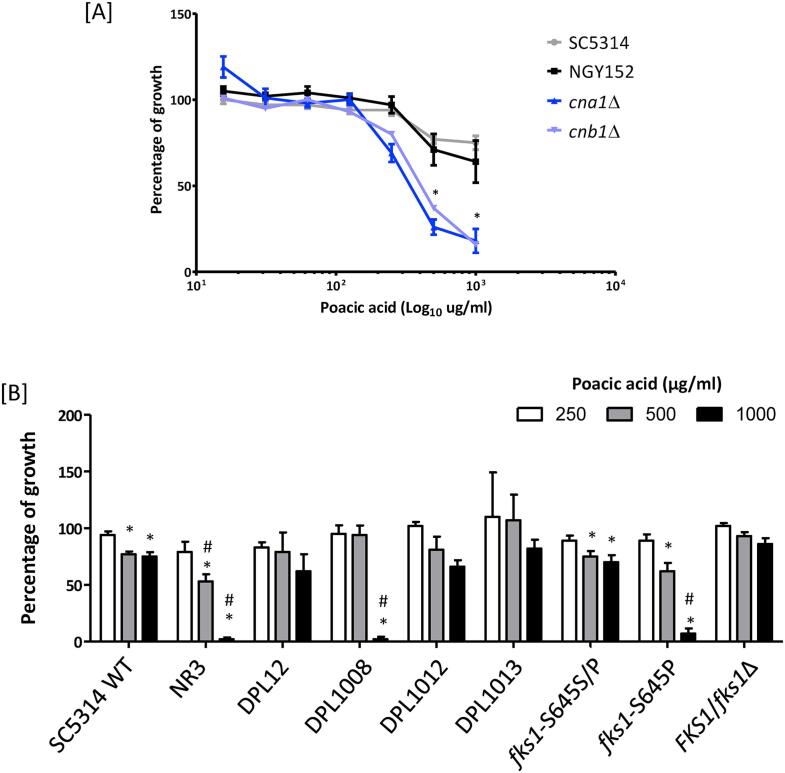
Differential sensitivity of C. albicans and S. cerevisiae to PA. First we investigated the antifungal activity of PA against the C. albicans reference strain SC5314, and S. cerevisiae S288C (shown previously to be sensitive to PA) as well as the echinocandin resistant C. albicans NR3 strain. As shown in Fig. 1A, S. cerevisiae was 5- to 10-fold more sensitive (inhibitory concentration, IC50 = 110 µg/ml) to PA than both C. albicans strains, NR3 (IC50 = 515 µg/ml) and SC5314 (IC50 > 1000 µg/ml). The measured sensitivity against S. cerevisiae corresponds with that cited previously (Piotrowski et al., 2015). Therefore C. albicans SC5314 was significantly less sensitive to PA despite being echinocandin sensitive (Fig. 1C). Unexpectedly, the CSF resistant C. albicans NR3 strain was significantly more sensitive to PA, than SC5314. When the cells from the MIC assays were re-plated on YPD agar plates in the absence of drug the PA-treated C. albicans SC5314 and NR3 strains grew normally, demonstrating that PA had a fungistatic, rather than a fungicidal effect on growth (Fig. 1B and D).
Fig. 1.
PA sensitivity and glucan synthase activity of C. albicans. Sensitivity of C. albicans SC5314 (wild type), NR3 (containing a point mutation in FKS1) and S. cerevisiae S288C to PA [A] and caspofungin [C]. Samples of 2.5 μl from the wells of the MIC plates were spotted on YPD plates and incubated at 30 °C for 24 h [B & D]. Error bars are SEM (n = 3). [E] Membranes of SC5314, NR3, and S288C were prepared from logarithmic-phase cells and glucan synthase (GS) activities were measured after solubilization of purified membrane with 0.4% Brij-35. Final concentrations of PA were 8, 16, 32, 64, 128, 256, and 512 μg/mL. All measurements were performed in triplicate. Plots of S288C, SC5314, and NR3 separately show mean values ± SD (n = 3). The half maximal inhibitory concentration (IC50GS) of S288C, SC5314 and NR3 were 206, 194 and 220 µg/ml, respectively.
Kinetic analyses revealed a correlation between the inhibition of β-1,3-glucan synthase enzyme activity and echinocandin MIC (Garcia-Effron et al., 2009b). In addition, S. cerevisiae β-1,3-glucan synthase activity was reduced when treated with PA and 14C-glucose incorporation in glucan was significantly decreased (Piotrowski et al., 2015). Therefore, we compared the effect of PA on β-glucan synthase activity using microsomal cell membrane preparations as a source of Fks1 enzyme. The inhibitory concentration (IC50GS) was determined for β-1,3-glucan synthases isolated from the total membrane fractions in SC5314, NR3, and S288C. The glucan synthase activity of all three stains was found to be inhibited to a similar extent by PA (Fig. 1E). The PA IC50GS for β-1,3-glucan synthase of SC5314 was 194 µg/ml, which was comparable to NR3 (IC50GS = 220 μg/ml) and S288C (IC50GS = 206 μg/ml) suggesting that this hotspot mutation that confers echinocandin resistance is irrelevant for PA sensitivity. Therefore, in in vitro assays, PA inhibited C. albicans and S. cerevisiae β-1,3-glucan synthases to the same degree suggesting that the differential sensitivity of the C. albicans and S. cerevisiae is not due to differences in the direct action of PA on Fks1 activity.
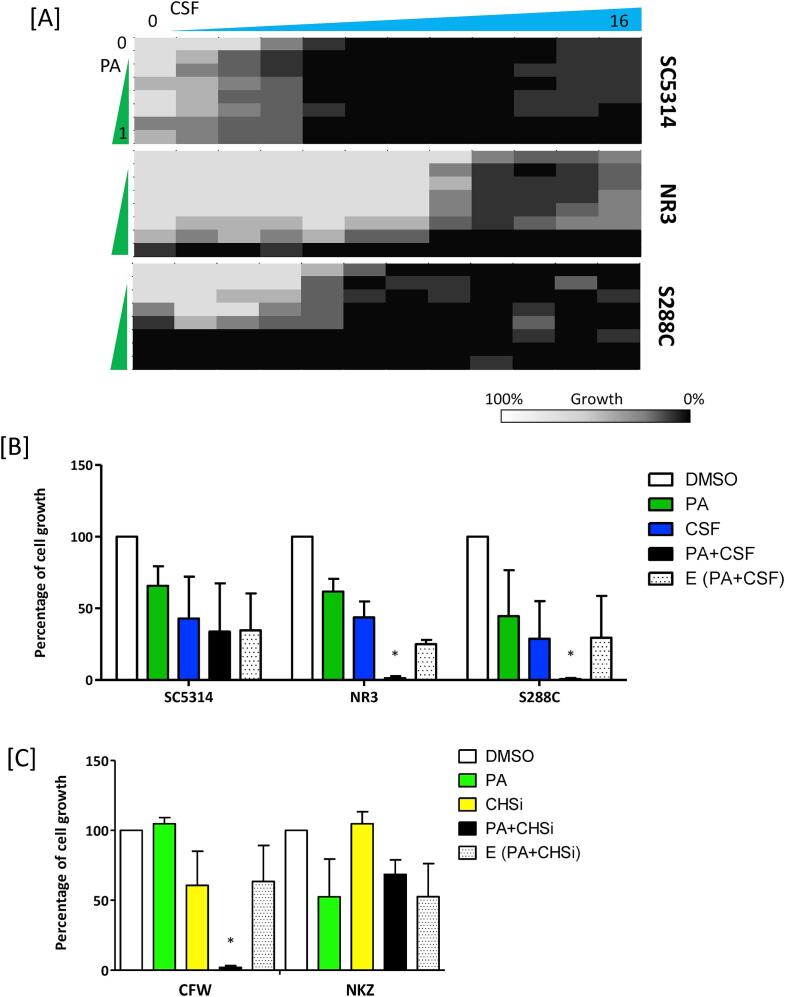
Combinatorial synergy between PA and cell wall synthesis inhibitors. PA could potentially be useful as part of a combinatorial therapy against Candida infections. Piotrowski et al. demonstrated that PA and CSF acted synergistically against S. cerevisiae (Piotrowski et al., 2015). Therefore, we carried out sensitivity assays using combinations of PA and CSF against C. albicans. The results described indicate combination of PA and CSF only had a noticeable inhibitory effect the growth of both C. albicans and S. cerevisiae, compared to treatment with each inhibitor alone (Fig. 2A). Noticeably, when C. albicans SC5314 was treated solely with CSF, it displayed paradoxical growth at >8 μg/ml (Fig. 2A). This paradoxical growth was abolished when SC5314 was treated with both PA and CSF. Combinatorial effects calculated using the sub-MIC/IC50 values of PA and CSF, indicated mild synergist effects of additions of PA and CSF for the NR3 CSF resistant strain and S. cerevisiae S288C (Fig. 2B). However, combinatorial treatments of PA and two other echinocandins, micafungin and anidulafungin, did not demonstrate synergistic inhibition against the C. albicans wild type strain (data not shown).
Fig. 2.
Combinatorial treatment of caspofungin and PA against C. albicans. [A] Sensitivity assays combining PA and caspofungin (CSF) were carried out according to CLSI methodology. Optical densities were measured after 24 h. The heat map is based on the average of three individual values expressed as percentage growth. [B] Growth inhibition due to PA in combination with CSF at sub-MIC concentration calculated as described in the Materials and Methods and presented as percentage growth compared to untreated controls. [C] Inhibition of growth in the presence of PA with or without supplementation with chitin synthesis inhibitors (CHSi) – CFW or nikkomycin Z (NKZ). E = expected percentage growth when both compounds are applied (See Materials and Methods). *p < 0.05 (n = 3) compared to the DMSO control.
Because chitin synthesis protects cells from β-glucan damage we also tested potential synergies between PA and chitin synthesis inhibitors nikkomycin Z (NKZ) (a competitive inhibitor of chitin synthase) and CFW (which can have a cidal effect against fungi by binding to nascent chitin and disrupting chitin chain maturation) (Roncero and Duran, 1985, Gaughran et al., 1994). Similar to previous findings with S. cerevisiae (Piotrowski et al., 2015), we observed that PA and NKZ showed no synergistic effect against C. albicans (Fig. 2C). However, C. albicans SC5314 treated with both PA and CFW exhibited a synergistic effect on growth (Fig. 2C). These results indicate that the combination of PA and CFW may enhance the cell wall polysaccharide instability, leading to enhanced killing.
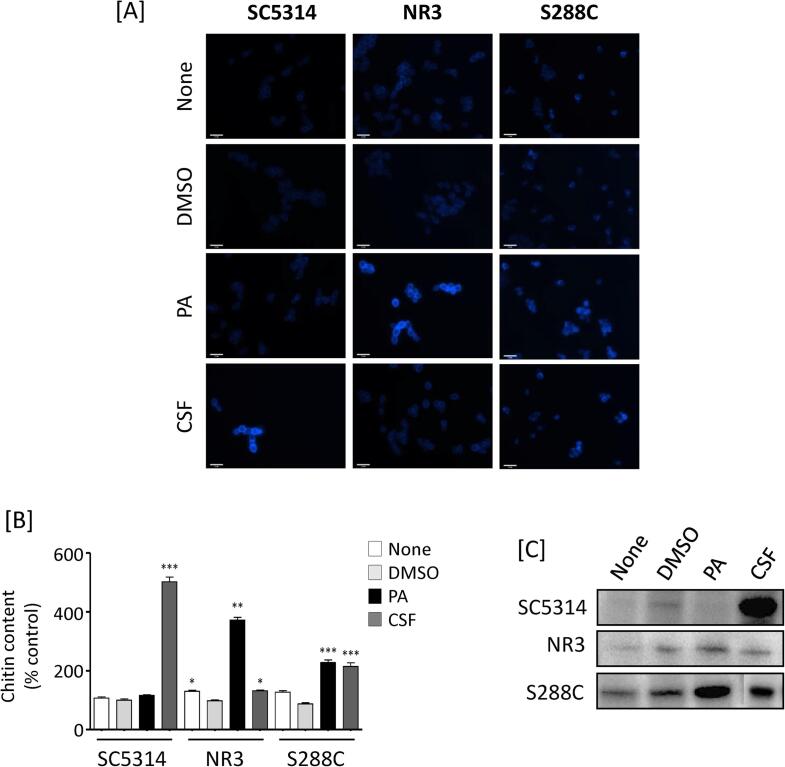
PA has no effect on chitin content of the C. albicans wild type cell wall. In fungi, β-glucan damage often leads to the induction of chitin synthesis (Munro et al., 2007, Walker et al., 2008, Lee et al., 2012). Therefore, we assessed the effect of PA on cell wall chitin measured by staining cells with CFW. DMSO treated cells of all three stains tested had no significant effect on chitin stimulation compared to the untreated controls. For wild type C. albicans, CSF treatments stimulated chitin synthesis, but PA did not (Fig. 3A and B). However, for the CSF resistant C. albicans NR3 mutant both PA and CSF stimulated chitin deposition or contents. Although microscopic observations suggested that the pattern of chitin upregulation between NR3 and S. cerevisiae were not identical, both PA and CSF stimulated chitin synthesis in S. cerevisiae (Fig. 3A and B). These data suggest that both PA and CSF compromised cell wall integrity.
Fig. 3.
Chitin content of C. albicans treated with PA. [A] Cell wall chitin contents of C. albicans SC5314, NR3 and S. cerevisiae S288C treated with either PA or caspofungin (CSF). C. albicans and S. cerevisiae cells were treated with 0.5 mg/ml and 0.1 mg/ml PA, respectively and with 32 ng/ml CSF at 30 °C for 6 h. After treatment, cells were fixed and stained with 25 µg/ml CFW. Scale bars are 10 μm. [B] The fluorescent intensity was measured and expressed as percentage of control values (SC5314, no treatment, none). * p < 0.05. ** p < 0.001, *** p < 0.0001. Error bars = SEM (n > 50). [C] Western blot analysis of phospho-Mkc1p of C. albicans SC5314 and NR3, and phospho-Mpk1p in S. cerevisiae S288C treated with DMSO, PA, and CSF. Cells were grown at 30 °C for 4 h and treated with drugs for 10 min. Proteins were extracted and same amount of protein were loaded per lane. (None = no treatment; DMSO, 0.5% DMSO; PA, 0.5 mg/ml PA; CSF, 32 ng/ml CSF).
Next we investigated whether the PKC/Mkc1 pathway that results in enhanced chitin formation was activated by PA (Fig. 3C). As expected, CSF treatment resulted in activation of Mkc1p/Mpk1p in both C. albicans and S. cerevisiae. Mkc1 phosphorylation was less in NR3 induced compared to SC5314 or S288C when cells were treated with CSF. PA activated Mpk1 phosphorylation in S288C, reflecting its ability to stimulate chitin synthesis in this species (Fig. 3A and B). There was no significant stimulation of Mkc1 phosphorylation in C. albicans SC5314 treated with PA (Fig. 3C). Therefore chitin stimulation by PA may be mediated by alternative cell wall integrity pathways.
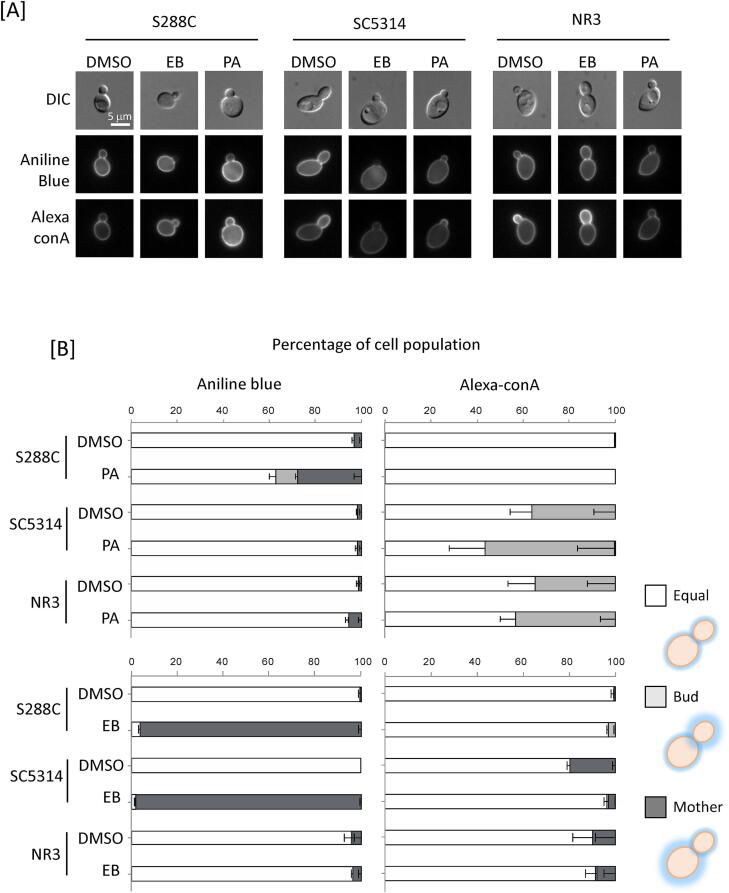
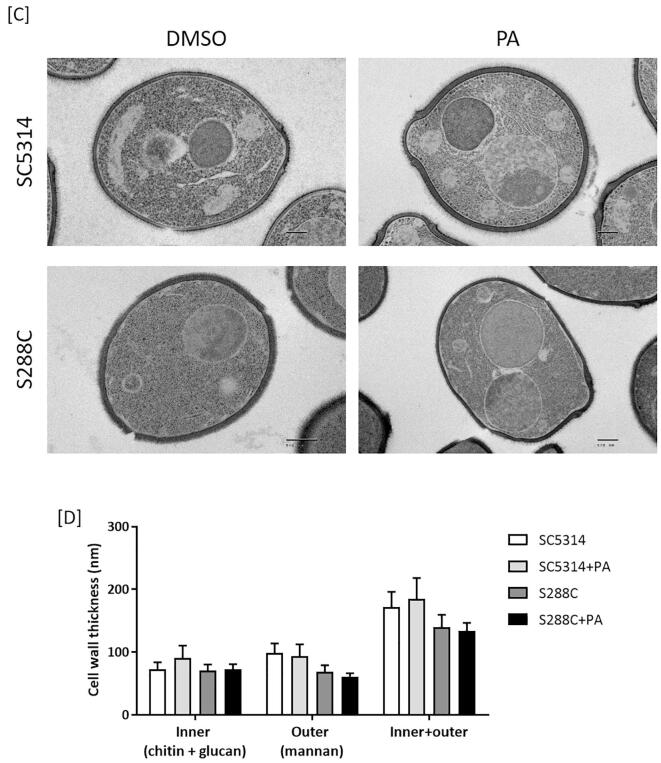
PA does not affect glucan and mannan content of C. albicans. The impact of PA on glucan and mannan content was determined by fluorescence microscopy using Aniline Blue and Alexa-ConA, to stain β-1,3-glucan and mannan respectively. In controls, treatment of S. cerevisiae with PA and echinocandin B significantly decreased β-1,3-glucan staining in buds (Fig. 4A and B). Since de novo synthesis of β-1,3-glucan occurs at sites of budding, the reduced staining in buds implies impairment of β-1,3-glucan synthesis after drug treatment (Piotrowski et al., 2015). In contrast, PA did not alter mannan staining (Fig. 4A and B), confirming the fact that PA acts primarily on the formation of β-1,3-glucan in S. cerevisiae (Piotrowski et al., 2015). PA also decreased β-1,3-glucan staining in C. albicans buds (Fig. 4A), but this was not statistically significant at p > 0.05 (Fig. 4B). The echinocandin-resistant NR3 strain was unaffected by echinocandin B, but the synthesis of β-glucan of buds was slightly affected by PA (Fig. 4B). Neither PA nor echinocandin B significantly affected mannan synthesis in either S. cerevisiae or C. albicans. Reinforcing this, HFP-TEM images did not reveal any gross changes in the outer (mannan) and inner (glucan + chitin) layers of S. cerevisiae and C. albicans cell walls after PA treatment (Fig. 4C-D).
Fig. 4.
Effect on PA on mannan and glucan in S. cerevisiae and C. albicans. [A] Cells were treated with PA (PA) or echinocandin B (EB), and stained with either Aniline Blue to visualise glucan or Alexa-ConA to stain mannan. S288C and SC5314 cells were treated with 500 µg/mL PA. NR3 cells were treated with 125 µg/mL PA at 30 °C for 6 h. The three strains were treated with 4 µg/mL echinocandin B at 30 °C for 2 h. Scale bars represent 5 μm. [B] Semi-quantification of fluorescence intensity of Aniline Blue and Alexa-ConA with or without PA or echinocandin B. “Equal” represents cells where both mother and daughter cells stained uniformly. “Bud” represents cells with strong staining on daughter cells. “Mother” represents heavy staining of mother cells. Data are percentages of three independent groups. (n > 3). [C and D] C. albicans wild type (SC5314) and S. cerevisiae (S288C) cells were treated with PA 0.5 μg/ml or 0.1 μg/ml, respectively for 6 h. Scale bars = 500 nm. The thickness of the inner cell wall (chitin and glucan layer) and outer cell wall (mannoprotein fibrils only) was quantified as described in the Materials and Methods. Mean ± SD (n > 8).
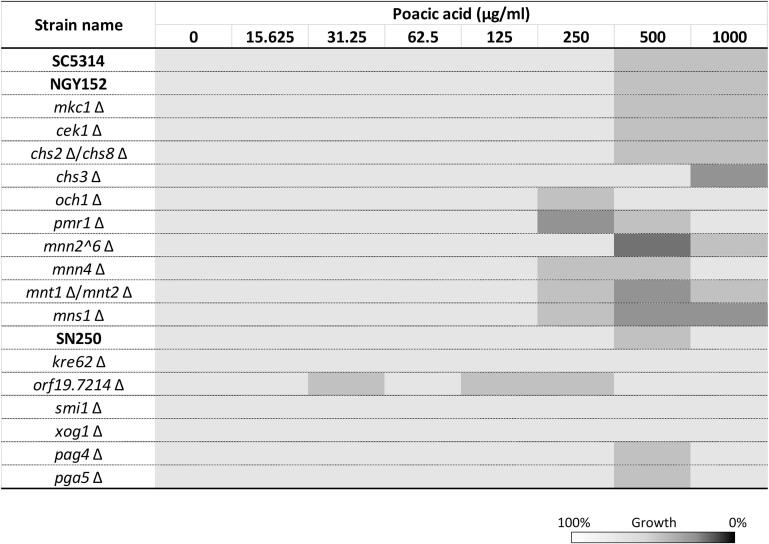
The calcineurin pathway influences sensitivity to PA. Previous studies have demonstrated that the Ca2+/calcineurin pathway is important for the cell remodelling and chitin upregulation of C. albicans in response to Ca2+, CFW, or/and caspofungin (Munro et al., 2007, Walker et al., 2008, Lenardon et al., 2009). To further investigate whether PA effected cell wall remodelling and integrity, a number of C. albicans mutants, that were defective in cell wall synthesis or cell wall integrity pathways were tested for their sensitivity to PA (the strains used are listed in Suppl. Table 1). Four groups of selected mutants were examined: 1) strains lacking genes involved in cell wall integrity pathway genes (MKC1, CEK1, CNA1, and CNB1) (Navarro-Garcia et al., 1995, Csank et al., 1998, Cruz et al., 2002, Sanglard et al., 2003), 2) mutants in cell wall chitin synthase genes (CHS3, CHS2, and CHS8) (Bulawa et al., 1995, Mio et al., 1996, Munro et al., 2003), 3) mannosylation mutants (OCH1, PMR1, MNN2 family, MNN4, MNT1, MNT2, and MNS1) (Hobson et al., 2004, Bates et al., 2005, Munro et al., 2005, Bates et al., 2006, Mora-Montes et al., 2007, Hall et al., 2013), and 4) mutants in genes encoding glucan synthase, glucosidase, or glucosyltransferase (PHR1, PHR2, CWH41, SMI1, KRE62, C1_14060W, XOG1, PGA4 and PGA5) (Fonzi, 1999, Noble et al., 2010). PA sensitivity tests were performed according to the CLSI methods (see Materials and Methods). The phr1Δ mutant was grown in NGY broth at pH 5 and inoculated in RPMI-1640 at pH 5 for MIC tests. Cells lacking either CWH41 or PHR1 showed growth defects in unstimulated conditions, and were not used for PA susceptibility testing. Only cna1Δ and cnb1Δ exhibited significantly increased sensitivity to PA (Fig. 5A). The IC50 values of cna1Δ and cnb1Δ were 332 μg/ml and 430 μg/ml, respectively representing a 3-fold and 2.4-fold increase in susceptibility to PA. None of the other mutants tested had significantly altered sensitivities to PA (Suppl. Fig. 1). Therefore, the calcineurin pathway is involved in PA tolerance in C. albicans.
Fig. 5.
PA susceptibility of C. albicans cell wall associated mutants. [A] PA sensitivity of C. albicans mutants lacking genes encoding the calcineurin subunits (cna1Δ, and cnb1Δ) was measured relative to the control strains SC5314 and NGY152. * p < 0.05 compared to the controls. Error bars = SEM (n > 3). [B] MIC sensitivity of PA against to strains containing a point mutation in Fks1 or the heterozygous mutant FKS1/fks1Δ was shown as percentage of growth the DMSO control. White bars = 250 µg/ml; grey bars = 500 µg/ml; black bars = 1000 µg/ml PA. Error bars = SEM (n > 3),* p < 0.05 in comparison to 100% growth of each strains, #p < 0.05 in comparison to SC5314. 2way ANOVA test.
Point mutations in Fks1 affecting PA sensitivity. The sensitivity to CSF can be decreased by amino acid substitutions in the Fks1 hotspot regions (Ben-Ami et al., 2011, Ben-Ami and Kontoyiannis, 2012). We tested several clinical isolates and laboratory derived strains containing point mutations in Fks1 Hotspot 1 for their sensitivity to PA (Table 1). The laboratory derived strains, fks1-S645S/P (a heterozygous mutant) and fks1-S645P (a homozygous mutation at 645 serine to proline), were generated using CRISPR-CAS9 to introduce this point mutation (see Materials and Methods). A heterozygous FKS1/fks1Δ null mutant was compared displaying marginally reduced sensitivity to PA and increased sensitivity to CSF (Fig. 5B; Table 1). A variety of point mutations in Fks1 result in elevated susceptibility to PA. Particularly strains including NR3 (S645Y), DPL1008 (S645P), and fks1-S645P all had significantly increased susceptibility to PA (Fig. 5B), with IC50s of 515 μg/ml, 767 μg/ml, and 590 μg/ml, respectively (Fig. 5B; Table 1). Amino acid substitutions in Fks1 at 641 (DPL12), 648 (DPL1012), or 689 (DPL1013) did not result in significantly altered PA sensitivity relative to the wild type control (Fig. 5B). Therefore, a variety of point mutations in Fks1 result in elevated susceptibility to PA.
Table 1.
C. albicans containing an amino acid substitution in Fks1 Hotspot1.
| Strain | PA IC50 | CSF IC50 | Fks1 Hotspot1 |
||
|---|---|---|---|---|---|
| μg/ml | μg/ml | Position | Sequence | ||
| Reference | SC5314 | >1000 | 0.032 | WT | FLTLSLRDP |
| Clinical strains | DPL12 | >1000 | 0.545 | F641S | SLTLSLRDP |
| DPL1008 | 767 | 3.500 | S645P | FLTLPLRDP | |
| DPL1012 | >1000 | 0.340 | D648Y | FLTLSLRYP | |
| DPL1013 | >1000 | 0.600 | P649H | FLTLSLRDH | |
| Lab strains | NR3 | 515 | 2.300 | S645Y | FLTLYLRDP |
| fks1 S634 | >1000 | 1.300 | S645S/P | FLTL(S/P)LRDP | |
| fks1 S645P | 590 | 3.700 | S645P | FLTLPLRDP | |
| FKS1/fks1Δ | >1000 | 0.016 | WT/Δ | FLTLSLRDP | |
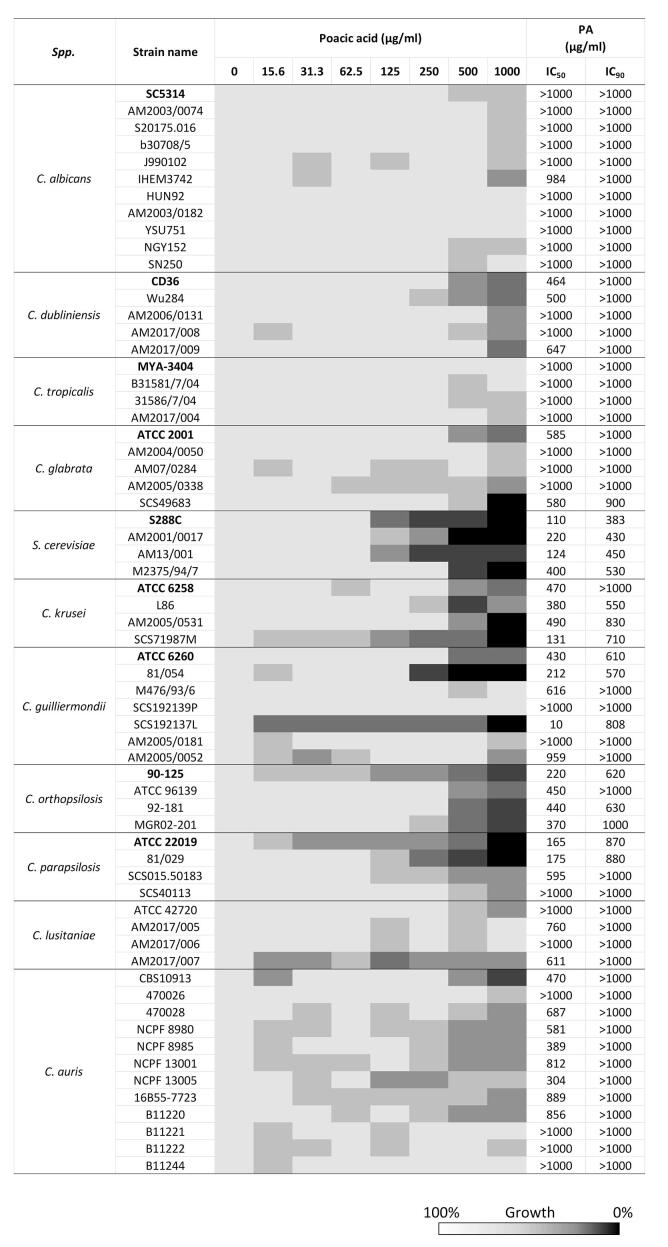
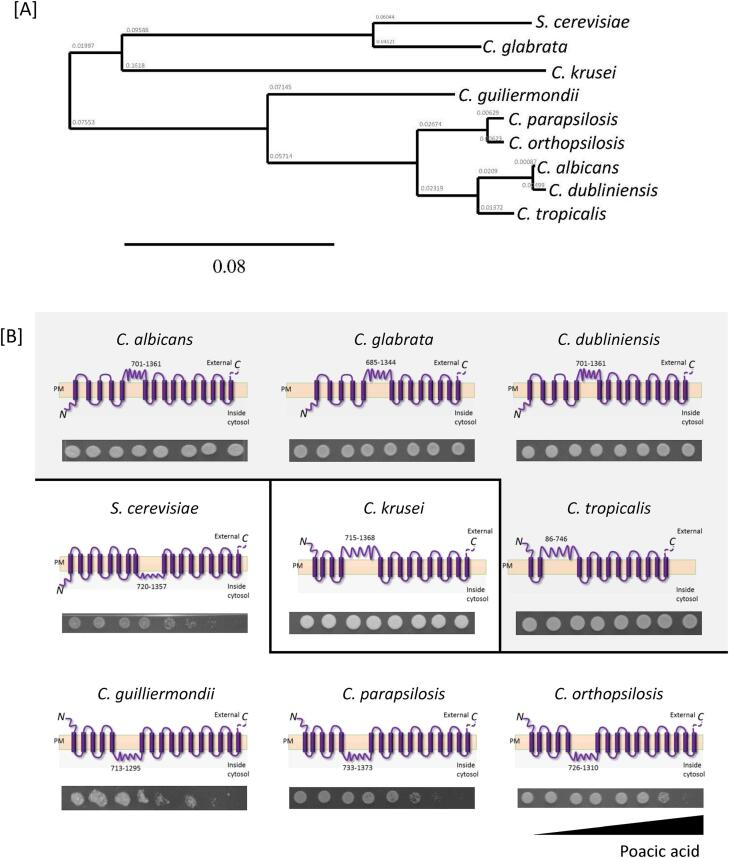
Sensitivity of PA against various Candida species. We next investigated PA susceptibility against 10 different Candida species (63 strains) and four strains of S. cerevisiae (S288C, AM2001/0017, M2375/94/7, and AM13/001) (Fig. 6; Suppl. Table 1). The S. cerevisiae strains were most sensitive to PA. C. krusei, C. parapsilosis, and C. orthopsilosis were more sensitive to PA compared to C. albicans (Fig. 6). Compared to those Candida species, C. guilliermondii showed the most variable PA sensitivity. For example, the C. guilliermondii strains ATCC 6260, 81/054, and SCS192139L were relatively susceptible to PA (IC50 = 10 to 430 μg/ml), whereas M476/93/6, SCS192139P, AM2005/0181, and AM2005/0052 were somewhat less sensitive to PA (IC50 = 616 to > 1000 μg/ml). This species is also variable in its sensitivity to CSF (Walker et al., 2013a, Walker et al., 2013b). Interestingly, C. glabrata was relatively less sensitive to PA compared to S. cerevisiae despite of phylogenetical similarity between two species. Furthermore, C. albicans, C. dubliniensis, and C. tropicalis were relatively less sensitive to PA compared to S. cerevisiae. Of the more uncommon Candida species, C. lusitaniae and C. auris were generally less sensitive to PA. Therefore, PA sensitivity was both species- and strain-dependent.
Fig. 6.
PA susceptibility of Candida species. Ten Candida species plus S. cerevisiae were tested for their relative PA susceptibility. Strains in bold are the reference strain for each species. The inhibitory concentrations (IC) at 50% and 10% growth were determined by analysing a locally weighted scatterplot smoothing (LOWESS) method.
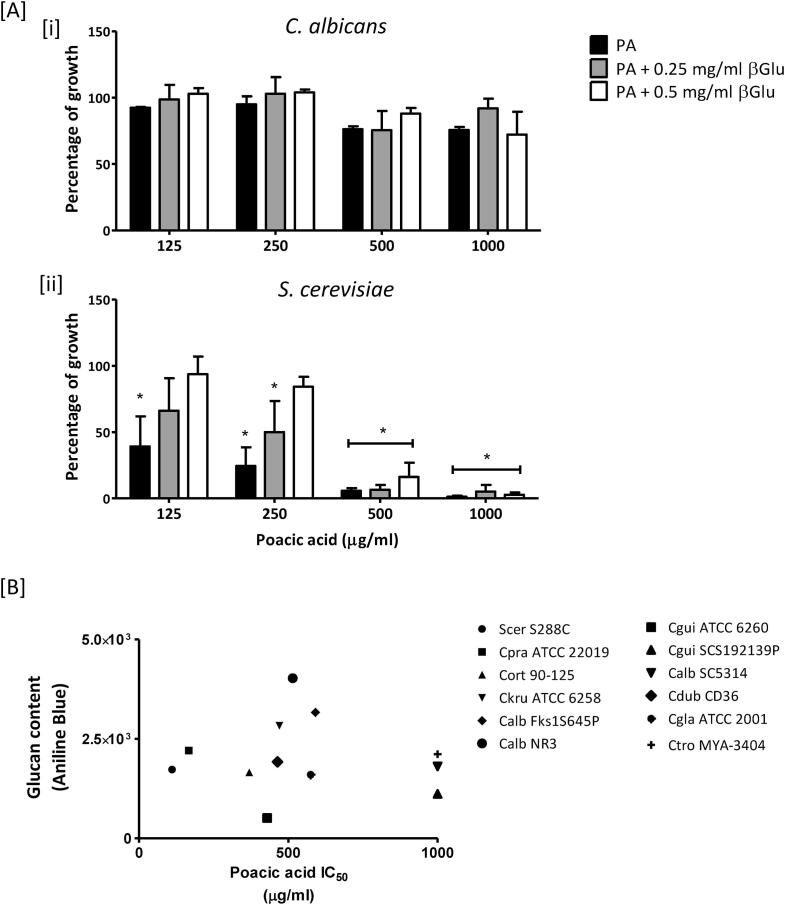
β-glucan content does not correlate with the sensitivity of PA to different Candida species. PA has been reported to bind directly to β-1,3-glucan (Piotrowski et al., 2015). We therefore investigated whether exogenous β-1,3-glucan could influence PA sensitivity (Fig. 7A). As a control, C. albicans and S. cerevisiae were grown in the medium only with exogenous β-1,3-glucan at 0.5 mg/ml. Under these growth conditions, the presence of exogenous β-1,3-glucan (0.5 mg/ml) did not impact cell growth of either C. albicans and S. cerevisiae (data not shown). In C. albicans the PA sensitivity was hardly affected by exogenous β-1,3-glucan at 0.25 mg/ml and 0.5 mg/ml (Fig. 7Ai). On the other hand, the growth of S. cerevisiae was ameliorated by addition of exogenous β-1,3-glucan in a dose-dependent manner (Fig. 7Aii). Therefore, the sensitivity of S. cerevisiae to PA was influenced by exogenous β-1,3-glucan in a PA-concentration-dependent manner (Fig. 7Aii).
Fig. 7.
Cell wall glucan contents in different Candida species and S. cerevisiae. [A and B] MIC of PA was calculated with exogenous β-1,3-glucan (βGlu) at 0.25 mg/ml or 0.5 mg/ml. C. albicans wild type [i] and S. cerevisiae [ii] were treated with PA only or PA with βGlu. Results are expressed as percentage of growth relative to the control (DMSO). Error bars = SEM (n > 3). * p < 0.05. 2way ANOVA test. [B] Correlation between glucan content and PA IC50 values of different Candida species. Cells were grown exponentially for 6 h in YPD and washed with PBS. Cells were then fixed with 4% formaldehyde. Aniline Blue (25 µg/ml) was used to stain cell wall β-glucan. Fluorescence levels of individual cells (1 × 104) were measured by FACS. The IC values were calculated using LOWESS analysis based on the sensitivity assay.
Next we hypothesized that varied basal levels of glucan in different Candida species may influence PA sensitivity. To test this, the β-1,3-glucan content in Candida cells was quantified using Aniline Blue staining by FACS (Fig. 7B). C. albicans NR3 had the highest glucan content and C. guilliermondii ATCC 6260 had the lowest among the strains tested, and the corresponding IC50 values of these strains were 515 μg/ml and 430 μg/ml. Additionally, the glucan contents of C. albicans wild type (SC5314) and S. cerevisiae S288C were comparable, but the IC50 values were 111 μg/ml and > 1000 μg/ml, respectively. Therefore, the PA sensitivity did not correlate simply with the basal β-1,3-glucan content of the cell wall of different yeast species.
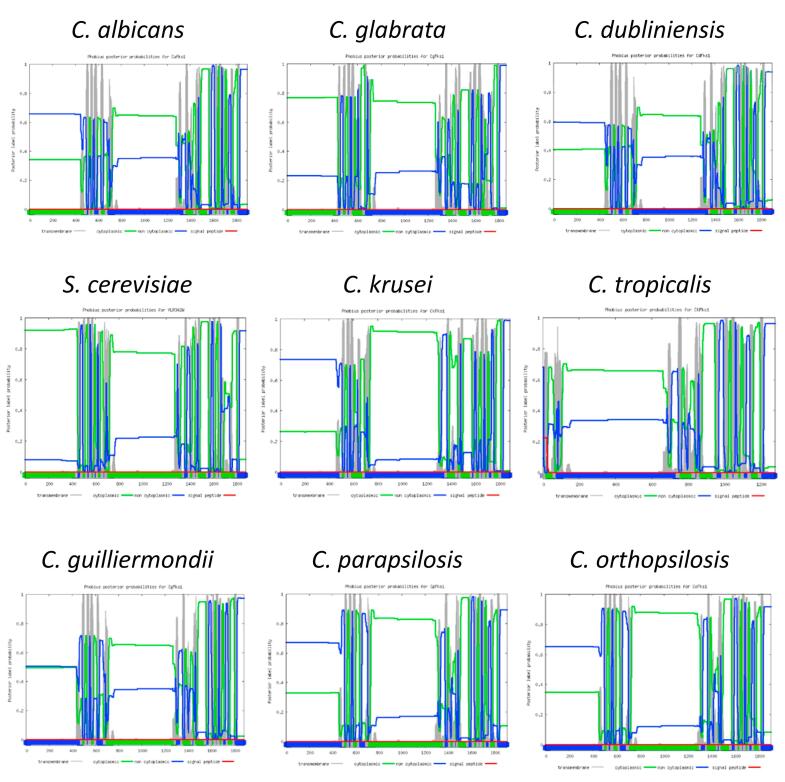
Correlation between computational prediction of β-1,3-glucan topology and PA sensitivity. The predicted transmembrane domains of C. albicans Fks1 were highly conserved in C. dubliniensis and C. tropicalis Fks1 but different in S. cerevisiae Fks1 and C. glabrata Fks1 (Suppl. Figs. 2 and 3). We noted a significant correlation between PA sensitivity and Fks1 active domain topology of five species (C. albicans, C. dubliniensis, C. tropicalis, C. glabrata, C. krusei, and S. cerevisiae (p < 0.05 according to Fisher’s exact test; Suppl. Table 3). One Fks1 domain was predicted to be located on the cytosolic face of the membrane for the most PA sensitive species - C. guilliermondii, C. parapsilosis, C. orthopsilosis, and S. cerevisiae) (Suppl. Fig. 3). However, the topology and function of this domain has yet to be examined experimentally.
Discussion
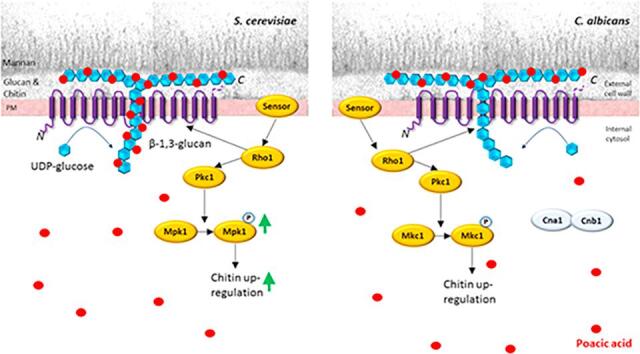
The natural plant metabolite PA has been shown to have antifungal and anti-oomycete activity for a range of plant pathogens. Here we show that PA has antifungal activity against some, but not all, of major species of the human fungal pathogens from the genus Candida. Despite of the demonstrated potency of PA against S. cerevisiae (Piotrowski et al., 2015), our results demonstrated that PA did not have pan-Candida activity, and is therefore unlikely to be useful as a pan-broad spectrum antifungal agent. Although the mode of action of PA is not understood in detail our results support the hypothesis that β-1,3-glucan binding is important for PA action. This was underlined by observations that exogenous β-1,3-glucan attenuated PA inhibition – Most likely by removing PA from solution. β-glucan synthesis involves synthesis of β-1,3-glucan oligomers (nascent short polymers) by the Rho1-dependent β-1,3-glucan synthases Fks1, Fks2 and Gsc1 using UDP-glucose as the substrate (Douglas et al., 2001). The nascent polysaccharide matures into a β-glucan triple helix matures and subsequently undergoes structural modifications and crosslinking via a number of transglycosidase and glucan modifying enzymes such as Phr1, Phr2, the Chr family, Pga4, Pga5, Xog1, Eng1, Bgl2, and Kre62 (González et al., 1997, Plaine et al., 2008, Arroyo et al., 2016, Aimanianda et al., 2017). Our results showed that the β-1,3-glucan synthase activity in in vitro membrane preparations was inhibited to a similar extent by PA in both C. albicans and S. cerevisiae (Fig. 1), suggesting that effect of PA may not influence Fks1 catalytic activity directly and that differences in sensitivity to PA may be dependent on access of this plant metabolite to β-1,3-glucan in the cytosol. Furthermore, mutants lacking XOG1, SMI1, KRE62, PGA4 or PAG5, which are all involved in β-1,3-glucan synthesis, were not altered in PA sensitivity (Suppl. Fig. 1). NMR analysis has demonstrated that the β-glucan of C. albicans hyphal cell walls is a cyclic glucan different from the linear glucans found in yeast cells (Kruppa et al., 2011, Lowman et al., 2014). Although it is unclear where the exact binding site of PA, it is possible that structure of β-1,3-glucan is important for the ability of nascent β-1,3-glucan to mature and that PA binding to this polysaccharide compromises its structural integrity in an analogous way to Calcofluor White binds to, and destabilises, β-1,4-linked N-acetylglucosamine of chitin. However, because PA binds directly to β-glucan rather than the Fks1 protein, it is possible that the Fks domain we have highlighted that correlates with PA sensitivity is involved in β-glucan binding – The product of Fks1 activity. In this model, for PA to efficiently inhibit β-glucan synthesis, binding to this polysaccharide may occur immediately after the nascent polysaccharide is formed. This binding may in turn compromise β-glucan fibril maturation and ultimately its physical strength.
We showed that point mutations in Candida Fks1 impacted differentially on PA and CSF susceptibility and that the transmembrane topology that was determined in silico the position of the predicted catalytic domain of Fks1 correlated with PA sensitivity. In S. cerevisiae, the Fks1 protein is predicted to have 18 transmembrane helixes (TMHs) (Johnson and Edlind, 2012). The central domain (residues ∼715–1300) is highly conserved and required for enzyme function, leading to the hypothesis that it may be associated with UDP-glucose binding and catalysis (Johnson and Edlind, 2012). Computational predictions of topographical models of this domain suggested that its localisation in the plasma membrane may be species-dependent and that it may have either an intracellular or extracellular localisation. Our data, and that published previously, suggest that PA binds β-1,3 glucan directly and that this event cause the polysaccharide instability leading to cell wall damage. High PA sensitivity correlated with a theoretical predicted intracellular location of this central domain – A hypothesis that remains to be tested experimentally. We hypothesize that if PA binding is intracellular it may immediately bind nascent β-glucan leading to more profound perturbation of the structural properties of this structural polysaccharide. Therefore, PA activity may be based on its ability to directly interact with β-glucan rather than its direct effect on β-1,3-glucan synthesis. However, the marked difference in the sensitivity of medically important species to PA suggests that it may have limited application as a lead compound informing new therapeutic strategies in medical mycology.
The structural integrity of the cell wall is monitored by membrane sensors that feed into the PKC/Mkc1 pathway and Hog1 and Ca2+/calcineurin pathways (Munro et al., 2007). In C. albicans, PA did not activate the PKC/Mkc1 cell wall salvage pathway that would be normally associated with upregulation of the chitin content and cell wall remodelling in the cell wall (Fig. 3, Fig. 4). Deletion of MKC1 in C. albicans did not impact on the PA sensitivity compared to the wild type control (Suppl. Fig. 1), although other genes involved in the PKC/Mkc1 pathway were not tested. In a previous study (Piotrowski et al., 2015), S. cerevisiae mutants including rom2Δ, pkc1Δ, and bck1Δ involved in the PKC pathway showed significantly increased sensitivity to PA. Deletion of NBP2 (encoding the Hog1-negative regulator) and PTS1 (encoding a protein phosphatase) resulted in increased resistance to PA. Furthermore, S. cerevisiae Sur1 and Csg2, which are involved in mannosylinositol phosphorylceramide synthesis, activate the PKC pathway and are resistant to PA when the cognate genes are deleted. It is however possible that PA treatment could stimulate cell wall remodelling by via PKC/Mkc1 independent mechanisms (Santiago et al., 2010).
The Ca2+/calcineurin pathway is also important for cell wall remodelling and stress responses to caspofungin (Bader et al., 2006, Munro et al., 2007, Walker et al., 2008, Singh et al., 2009, Kaneko et al., 2010, Lafayette et al., 2010). We showed here that deletion of CNA1 or CNB1 increased the CSF sensitivity in C. albicans. The chitin up-regulation response to caspofungin in C. albicans is also calcineurin-dependent (Walker et al., 2008). Previously a screen of S. cerevisiae mutants did not reveal a role for the Ca2+/calcineurin pathway in determining PA sensitivity (Piotrowski et al., 2015). In contrast we showed in C. albicans that cna1Δ and cnb1Δ were hypersensitive to PA (Fig. 5A).
The accumulation of point mutations in one of three hot spot regions of Fks1 has been shown to confer echinocandin resistance (Garcia-Effron et al., 2009a, Garcia-Effron et al., 2010, Perlin, 2011, Arendrup and Perlin, 2014, Perlin, 2015b, Kolaczkowska and Kolaczkowski, 2016). In addition, inherent caspofungin or micafungin sensitivity is known to be variable among Candida species independent of the presence of Fks1 point mutations (Walker et al., 2013a, Walker et al., 2013b, Arendrup and Perlin, 2014, Perlin et al., 2015, Wanjare et al., 2016). We demonstrate that, PA sensitivity also differed between Candida strains. Cell wall chitin can compensate for β-glucan damage (Walker et al., 2008, Ben-Ami et al., 2011), but again there was no observed correlation between chitin content and PA sensitivity. In addition, our data demonstrated that β-1,3-glucan contents of Candida species varied significantly (Fig. 7), however, PA sensitivity did not correlate with gross glucan content of different Candida species.
In summary, our data suggest that PA and echinocandins inhibit β-1,3-glucan synthesis by different mechanisms and that the extent of PA inhibition is fungal species-specific. It is likely that PA does not directly interfere with the catalytic process of forming β-1,3-glucan at the cytoplasmic face of the membrane, but rather interferes with β-1,3-glucan maturation leading to a weaker cell wall. The variability in the ability of PA to inhibit a range of Candida pathogens limits its potential for development as a novel antifungal agent.
Materials and Methods
Strains, media, and chemicals
The fungal strains used in this study are listed in Suppl. Table 1. All strains of Candida species and S. cerevisiae were grown on an YPD agar plate (1% yeast extract, 2% peptone, 2% glucose, and 2% agar). For overnight cultures, single colonies from the agar plates were inoculated in either YPD or NGY broth (0.1% yeast extract, 0.1% neopeptone, and 0.4% glucose), and incubated at 30 °C with shaking at 200 rpm. Uridine (25 μg/ml) was supplemented in the medium for uridine auxotrophic mutants. CSF was obtained from the Aberdeen Royal Infirmary. Echinocandin B was generous gift from Osamu Kondo (Chugai Pharmaceutical Co., Tokyo, Japan).
Antifungal susceptibility test
The antifungal susceptibility of PA against various Candida species and S. cerevisiae strains was determined according to CLSI M27-A3 guidelines in using modified RPMI-1640 (2 mM L-glutamine, 0.2 M MOPS, 2% glucose (w/v), pH7.2) as described previously (Lee et al., 2012). Briefly, fungal cells were grown in NGY at 30 °C with shaking at 200 rpm overnight to reach a stationary phase. Overnight cultures were washed with distilled water and then inoculated in RPMI-1640 (with/without drugs) to reach approximately up to 5 × 104 cells/ml, and incubated at 37 °C for 24 h in a static incubator. PA concentrations ranged from 0 to 1000 μg/ml, and CSF concentrations from 0 to 16 µg/ml. The glucan extracted from Euglena gracilis (Sigma, 89862-1G-F) was dissolved/gelatinised in DMSO, and used in some experiments in which β-glucan was provided exogenously. After 24 h incubation in microtitre plates, the optical density of wells was measured using a VERSAmax plate reader (Molecular Devices, USA) at 405 nm. For the post-sensitivity assays of cell viability, 2.5 μl samples were taken from the sensitivity assay microtitre plates and were spotted on a fresh YPD and incubated at 30 °C overnight. The inhibitory concentration of MIC plates at 50% cell survival (IC50) or a 10% cell survival (IC90) was calculated from LOWESS curves using GraphPad Prims 5 (v5.04).
Combinatorial sensitivity assays
Combinatorial inhibition assays were performed to investigate synergistic effect of PA with echinocandins. Growth conditions were identical to that used in antifungal susceptibility tests described above. A 12 × 8- or 8 × 8-dose-defined matrix was used in 96-well plates containing PA (0.98 to 1000 μg/ml) in combination with either CSF (0.032–16 μg/ml), NKZ (0.625–40 μM), or CFW (0.2–200 μg/ml). After incubating for 24 h at 37 °C, the final OD at 405 nm was determined. Results are presented as a heatmaps or a bar charts of the percentage of growth inhibition observed. Synergistic effects were calculated as described (Piotrowski et al., 2015) using the formula E = A × B/C (where E is the expected percentage of inhibited growth; A is the percentage growth of when compound A (PA) is applied alone; B is the percentage growth when compound B (CSF) is applied alone; C is the percentage growth DMSO control. Sub-MIC or IC50 concentrations of PA or caspofungin were used for analysis of synergistic effects. Combinations of 1000, 500 and 62.5 µg/ml PA and 0.062, 2, and 0.25 µg/ml CSF for SC5314, NR3 and S288C respectively were used. A combination of 500 µg/ml PA and 25 µg/ml CFW or 40 µM NKZ were used for SC5314.
Semi-quantification of cell wall chitin
Cells treated with either PA or CSF were fixed with 10% neutral buffered formalin and stained with 25 µg/ml CFW to visualise cell wall chitin. Samples were observed by differential interference contrast (DIC) microscopy as described in the previous study (Walker et al., 2008, Lee et al., 2012) using a Zeiss Axioplan 2 microscope to observe samples. The Openlab software (Openlab v 5.02) was used to take images of samples and the CFW fluorescent intensity of individual cells was measured using ImageJ (v1.47).
Mannoprotein and glucan staining
β-1,3-glucan was stained with Aniline Blue (016-21302; Wako Chemicals). Mannoprotein was stained with Alexa594-ConA (C11253; Life Technologies). β-1,3-glucan and mannoprotein were stained as described previously (Piotrowski et al., 2015, Okada et al., 2014, Okada and Ohya, 2016) with slight modification. Briefly, over-night cultured yeast cells were cultured in YPD with PA (500 μg/ml for S288C and SC5314 or 125 μg/ml for NR3) or with echinocandin B (4 μg/ml) at 30 °C. Then, cells were collected at 2 h (for echinocandin B) or 6 h (for PA) after treatment and stained with Alexa594-ConA and Aniline Blue without fixation. Cells mounted on a glass slide were exposed to UV for 30 s to bleach out PA fluorescence before acquiring images.
For fluorescence-activated cell sorting, applied in flow cytometry (FACS) analysis, cells were first grown in YPD at 30 °C for 6 h. Cells were washed twice with PBS, and fixed in 3.7% formaldehyde. Fixed cells were diluted in PBS to reach 1 × 106 cells/ml, 0.5 mM EDTA was added, and samples were sonicated in a water bath for up to 10 min to disrupt cell clumps. Aniline Blue (25 μg/ml) was added immediately before FACS analysis. Unstained controls were used to establish the basal level of fluorescence. A total of 10,000 cells per sample were analysed by FACS according to BD Fontesa manufacturer’s guidelines (BD Biosciences). Data obtained by the BD FACSDiva software were transferred and analysed by FlowJo (v10.02) to semi-quantify the fluorescence intensity representing β-glucan contents.
In vitro β-1,3-glucan synthase activity
To prepare membrane microsomal fractions, cells were grown at 30 °C to a density of 2–4 × 107 cells/ml. The following procedures were carried out at 4 °C. Cells were harvested, washed with 1 mM EDTA, and disrupted by vortexing 4 times for 2 min each with 5 ml of glass beads in 10 ml of breaking solution containing 0.5 M NaCl, 1 mM EDTA with 1 mM phenylmethylsulfonyl fluoride. After centrifugation at 1,200 × g for 5 min, the supernatant was collected and transferred to 33 PC tubes (Hitachi, Japan). Pellets were resuspended in breaking solution, and centrifuged at 1,200 × g for 1 min. The supernatant was also transferred to new 33 PC tubes. Membrane fractions in 33 PC tubes were collected by centrifugation at 100,000×g for 30 min in an RP70T-203 rotor (Hitachi) with Himac CP 65β (Hitachi). The resultant microsomal membrane pellet was suspended in membrane buffer containing 50 mM Tris-HCl, pH 7.5, 10 mM EDTA, 1 mM β-mercaptoethanol, and 33% glycerol, homogenized, and stored at − 80 °C to obtain membrane mixture.
β-1,3-glucan synthase activity was measured as described previously (Piotrowski et al., 2015) with slight modifications. First, a reaction mixture was prepared containing 25 mM Tris·HCl pH 7.5, 25 mM potassium fluoride, 0.5 mM EDTA, 0.4% Brij-35, and 0.2 mM UDP-Glc (with 57 Bq UDP-[Glucose-14C]; NEC403; PerkinElmer). Brij-35 was used for membrane solubilisation was included in the reaction solutions for assays of C. albicans β-1,3-glucan synthase activity (Frost et al., 1994). Membrane fractions containing 50 μg total protein was included in the membrane mixture containing 2.0 μM GTP-γ-S and different concentrations of PA. The membrane fractions (20 μl) were added to 80 μl of the reaction mixture, incubated at 30 °C for 30 min, and stopped by the addition of ethanol. Trapping β-1,3-glucan polymer from the sample was collected by membrane filtration (cellulose ester membranes; 0.2 μm in pore size; ADVANTEC), and washed one time with 1 ml distilled water. Membrane filters were dried at room temperature for 2 h, and measured the radioactivity by a scintillation counter (LSC-6100; Aloka) with 2.5 ml scintillation mixture (Econofluor-2; PerkinElmer). The half maximal inhibitory concentration (IC50GS) was calculated after fitting with Gamlss beta in the R Gamlss package.
High Pressure, freeze substitution transmission electron microscopy (HPF-TEM)
HPF-TEM was performed as described previously (Hall et al., 2013) with some modification. C. albicans SC5314 and S. cerevisiae S288C were grown in YPD at 30 °C overnight. The following day, overnight cultures were diluted in a fresh YPD medium to an OD600 of 0.2 and incubated at 30 °C for 1 h. Then cells were treated for further 6 h with DMSO (0.5% for SC5314 or 0.1% for S288C) or PA (500 μg/ml for SC5314 or 100 μg/ml for S288C). After incubation, cells were collected and washed with mQ-water. Cells were freeze-substituted by using a Leica EM AFS2 automatic freeze substitution system (Leica Micro-systems). Further steps were carried out according to the previous study (Hall et al., 2013). To quantify the length of the inner (chitin + glucan) and outer (mannan fibrils) layers, images were transported into ImageJ (v1.47), and 5–12 cells were randomly selected, and 5–45 measurements made from the different angles of each cell. The results present as a mean (nm) with SEM.
Mkc1 phosphorylation analysis by western analysis
Western analysis was performed using methods described previously with modifications (Munro et al., 2007). Overnight cultures of C. albicans wild type, C. albicans NR3 (Fks1-S645Y), and S. cerevisiae were diluted in fresh YPD and then incubated for 4 h at 30 °C with shaking until they reached mid-log phase. Cells were then treated for 10 min with appropriate treatments, with equivalent non-treated cells used as a control. After treatments, cells were collected, and their total protein complement was extracted.
Proteins (15 µg) were separated by SDS–PAGE (polyacrylamide gel) electrophoresis; NuPAGE®Novex Bis-Tris 4–12% Precast gels (Invitrogen). Blotting was performed according to the manufacturer’s instructions. A mix of the SeeBlue® Plus2 Pre-stained (Invitrogen) and MagicMarkTM standards (Invitrogen) were loaded as markers. After blotting, separated proteins were transferred onto PVDF membranes which were rinsed with PBS and blocked in PBS-T 10% BSA (0.1% Tween-20, 10% BSA in PBS) for 30 min at RT. Membranes were then incubated overnight at 4 °C with Phospho-p44/42 MAP Kinase (Thr202/Tyr204) Antibody (Cell Signalling Technology) in PBS-T 5% BSA (0.1% Tween-20, 5% BSA in PBS). For secondary antibody staining, the membranes were incubated at RT for 60 min in PBS-T 5% BSA containing an anti-rabbit IgG, HRP-linked Antibody (Cell Signalling Technology). Signals were enhanced by SuperSignal® West Femto Maximun Sensitivity Substrate (Thermo Scientific) according to the manufacturer’s instructions. Chemiluminescence signal detection was performed using the Fusion image acquisition system v15.15 (PeQLab Biotechnologie GmbH, Germany).
Creating a mutant containing Fks1 amino acid substitution
Amino acid substitutions in Fks1 (orf19.2929/C1_02420C) were introduced in C. albicans, using the CRISPR-Cas9 system according to a previous study with modifications (Vyas et al., 2015). A single guide RNA was generated using primers of Fks1sgRNA-Fwd and Fks1sgRNA-Rev (Suppl. Table 2). The single guide RNA targeting the FKS1 locus was ligated into the plasmid, pV1200. Positive clones were selected on LB plate containing 100 μg/ml ampicillin and 50 μg/ml nourseothricin. Constructs were confirmed by sequencing. Before, transformation, the plasmid pV1200 + FKS1sgRNA (2–3 µg) for the Cas9 system was digested with KpnI and SacI. The repair template was created using oligos of FKS1rt_Fwd and S645P_Rev. Repair templates (10 μg) were purified and transformed along with linearized pV1200_FKS1sgRNA into C. albicans SC5314. The repair templates contained the desired point mutation (serine to proline at 645), and a silencing mutation at 635 (TTG, Leucine to TTA, Leucine) to prevent the Cas9 system repeatedly cutting the NGG site. A positive colony was selected by plating on an YPD agar 200 μg/ml nourseothricin, and then grown on YPD agar containing 0.5 μg/ml caspofungin. All positive transformations were confirmed by the DNA sequencing analysis as previously described (Lee et al., 2012). To create the dysfunctional fks1 mutant, the repair template was produced by using oligos of FKS1rt_Fwd and fks1del_Rev. The repair template was purified and transformed along with linearized pV1200_FKS1sgRNA into C. albicans SC5314. Positive transformants were selected on an YPD with 200 μg/ml nourseothricin. All mutants were screened by Sanger sequencing with primers of Fks1_HS1_F and _R (Genewiz UK; https://www.genewiz.com/).
Computational analysis of Fks1 or β-1,3-glucan synthase homologue proteins
The amino acid sequences of Fks1 or homologues in all reference strains was obtained from the Candida genome database (http://www.candidagenome.org), the Saccharomyces genome database (http://www.yeastgenome.org) or from the NCBI (http://www.ncbi.nlm.nih.gov) (Table 2). The multiple protein sequence alignment of homologue Fks1s in Candida species was performed by using the web-based analysis tool, MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle) using S. cerevisiae Fks1 as the comparator sequence. The phylogenetic tree of Fks1 was created after MUSCLE analysis, and then re-generated as an unrooted phylogenetic tree, using TreeDyn 198.3 (http://www.phylogeny.fr/). The transmembrane topology prediction of homologue Fks1 was performed via the web-tool, Phobius (http://phobius.sbc.su.se/index.html).
Table 2.
Analysis of Fks1 β-1,3-glucan conserved central domain in Candida species.
| Species | Strain name | Protein sequence info. | Protein length | Chromosome info. | Source | Amino acid position (putative β-1,3-glucan synthase components) | Domain facing |
|---|---|---|---|---|---|---|---|
| C. albicans | SC5314 | C1_02420cp_a | 1897 | Chr1 | CGD | 701-1361 | NON CYTOPLASMIC |
| C. glabrata | CBS 138 | Cagl0k01034gp | 1863 | ChrG | CGD | 685-1344 | NON CYTOPLASMIC |
| C. dubliniensis | CD36 | Cd36_02270p | 1897 | Chr1 | CGD | 701-1361 | NON CYTOPLASMIC |
| C. guilliermondii | ATCC 6260 | EDK37070 | 1882 | Supercontig CH408155 | Ensemble Fungi | 713-1295 | CYTOPLASMIC |
| C. krusei | ATCC 6258 | AAY40291.2 | 1885 | NA | NCBI | 715-1368 | NON CYTOPLASMIC |
| C. orthopsilosis | 90-125 | CCG22470 | 1902 | Chr2 | EnsembleFungi | 726-1310 | CYTOPLASMIC |
| C. parapsilosis | ATCC 22,019 | Cpar2_106400p | 1909 | Chr1 | CGD | 733-1373 | CYTOPLASMIC |
| C. tropicalis | MYA-3404 | EER31878 | 1280 | Supercontig3.6 | EnsembleFungi | 86-746 | NON CYTOPLASMIC |
| S. cerevisiae | S288C | YLR342W | 1876 | Chr XII | SGD | 720-1357 | CYTOPLASMIC |
CGD = Candida Geneome Database (http://www.candidagenome.org/).
EnsembleFungi = http://fungi.ensembl.org/index.html.
NCBI = National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/guide/).
SGD = Saccharomyces Genome Database (http://www.yeastgenome.org/).
Transmembrane domain analysis by using the web-based software, Phobius (http://phobius.sbc.su.se/).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We sincerely thank Jeff Piotrowski and John Ralph for providing poacic acid, and David Perlin for providing C. glabrata fks1Δ and fks2Δ mutant strains and clinical isolates (DPL series) for this study. We thank Carol Munro, Sam Miller and Louise Walker for helpful discussions; and Raif Yuecel, Attila Bebes, and Linda Duncan in the Iain Fraser Cytometry Centre (IFCC) for FACS, and Kevin MacKenzie, Debbie Wilkinson, Gillian Milne, and Lucy Wright for microscopy at the University of Aberdeen core facilities. This work was supported by the Wellcome Trust (101873, 086827, 075470, & 200208) and MRC Centre for Medical Mycology (N006364/1), and Grants-in-Aid for Scientific Research from the Ministry of Education Culture, Sports, Science and Technology, Japan (24370002 and 15H04402 to Y.O.).
Author contributions
KL, JA, IC, XC, HO, AD performed experiments. NG, KL and YO designed experiments and wrote the manuscript. All authors analysed data and reviewed the manuscript. NG and YO provided supporting funding.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.tcsw.2018.09.001.
Appendix A. Supplementary data
Supplementary Fig. 1.
Supplementary Fig. 2.
Supplementary Fig. 3.
References
- Aimanianda V., Simenel C., Garnaud C., Clavaud C., Tada R., Barbin L., Mouyna I., Heddergott C., Popolo L., Ohya Y., Delepierre M., Latge J.P. The dual activity responsible for the elongation and branching of beta-(1,3)-glucan in the fungal cell wall. MBio. 2017;8 doi: 10.1128/mBio.00619-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendrup M.C., Perlin D.S. Echinocandin resistance: an emerging clinical problem? Curr. Opin. Infect. Dis. 2014;27:484–492. doi: 10.1097/QCO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo J., Farkas V., Sanz A.B., Cabib E. Strengthening the fungal cell wall through chitin-glucan cross-links: effects on morphogenesis and cell integrity. Cell. Microbiol. 2016;18:1239–1250. doi: 10.1111/cmi.12615. [DOI] [PubMed] [Google Scholar]
- Bader T., Schröppel K., Bentink S., Agabian N., Köhler G., Morschhäuser J. Role of calcineurin in stress resistance, morphogenesis, and virulence of a Candida albicans wild-type strain. Infect. Immun. 2006;74:4366–4369. doi: 10.1128/IAI.00142-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S., Hughes H.B., Munro C.A., Thomas W.P., MacCallum D.M., Bertram G., Atrih A., Ferguson M.A., Brown A.J., Odds F.C., Gow N.A. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 2006;281:90–98. doi: 10.1074/jbc.M510360200. [DOI] [PubMed] [Google Scholar]
- Bates S., MacCallum D.M., Bertram G., Munro C.A., Hughes H.B., Buurman E.T., Brown A.J., Odds F.C., Gow N.A. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 2005;280:23408–23415. doi: 10.1074/jbc.M502162200. [DOI] [PubMed] [Google Scholar]
- Ben-Ami R., Garicia-Effron G., Lewis R.E., Gamarra S., Leventakos K., Perlin D.S., Kontoyiannis P. Fitness and virulence costs of Candida albicans FKS1 hot spot mutations associated with echinocandin resistance. J. Infec. Dis. 2011;204:626–635. doi: 10.1093/infdis/jir351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami, R. and Kontoyiannis, D.P. 2012. Resistance to echinocandins comes at a cost: The impact of FKS1 hotspot mutations on Candida albicans fitness and virulence. Vol. 3: pp. 95–98. [DOI] [PMC free article] [PubMed]
- Bulawa C.E., Miller D.W., Henry L.K., Becker J.M. Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10570–10574. doi: 10.1073/pnas.92.23.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côte P., Hogues H., Whiteway M. Transcriptional analysis of the Candida albicans cell cycle. Mol. Biol. Cell. 2009;20:3363–3373. doi: 10.1091/mbc.E09-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz M.C., Goldstein A.L., Blankenship J.R., Del Poeta M., Davis D., Cardenas M.E., Perfect J.R., McCusker J.H., Heitman J. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 2002;21:546–559. doi: 10.1093/emboj/21.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank C., Schroppel K., Leberer E., Harcus D., Mohamed O., Meloche S., Thomas D.Y., Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas C.M., D'Ippolito J.A., Shei G.J., Meinz M., Onishi J., Marrinan J.A., Li W., Abruzzo G.K., Flattery A., Bartizal K., Mitchell A., Kurtz M.B. Identification of the FKS1 gene of Candida albicans as the essential target of 1,3-beta-D-glucan synthase inhibitors. Antimicrob. Agents Chemother. 1997;41:2471–2479. doi: 10.1128/aac.41.11.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas C.M. Fungal beta (1,3)-D-glucan synthesis. Med. Mycol. 2001;39(1):55–66. doi: 10.1080/mmy.39.1.55.66. [DOI] [PubMed] [Google Scholar]
- Erwig L.P., Gow N.A. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016;14:163–176. doi: 10.1038/nrmicro.2015.21. [DOI] [PubMed] [Google Scholar]
- Fonzi W.A. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of β-1,3- and β-1,6-glucans. J. Bacteriol. 1999;181:7070–7079. doi: 10.1128/jb.181.22.7070-7079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free S.J. Fungal cell wall organization and biosynthesis. Adv. Genet. 2013;81:33–82. doi: 10.1016/B978-0-12-407677-8.00002-6. [DOI] [PubMed] [Google Scholar]
- Frost D.J., Brandt K., Capobianco J., Goldman R. Characterization of (1,3)-beta-glucan synthase in Candida albicans: microsomal assay from the yeast or mycelial morphological forms and a permeabilized whole-cell assay. Microbiology. 1994;140:2239–2246. doi: 10.1099/13500872-140-9-2239. [DOI] [PubMed] [Google Scholar]
- Garcia-Effron G., Chua D.J., Tomada J.R., DiPersio J., Perlin D.S., Ghannoum M., Bonilla H. Novel FKS mutations associated with echinocandin resistance in Candida species. Antimicrob. Agents Chemother. 2010;54:2225–2227. doi: 10.1128/AAC.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Effron G., Katiyar S.K., Park S., Edlind T.D., Perlin D.S. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 2008;52:2305–2312. doi: 10.1128/AAC.00262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Effron G., Lee S., Park S., Cleary J.D., Perlin D.S. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-β-D-glucan synthase: Implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 2009;53:3690–3699. doi: 10.1128/AAC.00443-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Effron G., Park S., Perlin D.S. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for Candida albicans: implications for interpretive breakpoints. Antimicrob. Agents Chemother. 2009;53:112–122. doi: 10.1128/AAC.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughran J.P., Lai M.H., Kirsch D.R., Silverman S.J. Nikkomycin Z is a specific inhibitor of Saccharomyces cerevisiae chitin synthase isozyme Chs3 in vitro and in vivo. J. Bacteriol. 1994;176:5857–5860. doi: 10.1128/jb.176.18.5857-5860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González M.D.M., Díez-Orejas R., Molero G., Álvarez A.M., Pla J., Nombela C., Sánchez-Pérez M. Phenotypic characterization of a Candida albicans strain deficient in its major exoglucanase. Microbiology. 1997;143:3023–3032. doi: 10.1099/00221287-143-9-3023. [DOI] [PubMed] [Google Scholar]
- Gow N.A., Hube B. Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 2012;15:406–412. doi: 10.1016/j.mib.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Gow N.A., Netea M.G. Medical mycology and fungal immunology: new research perspectives addressing a major world health challenge. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2016;371 doi: 10.1098/rstb.2015.046220150462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow N.A.R., Latge J.P., Munro C.A. The fungal cell wall: structure, biosynthesis, and function. Microbiol. Spectr. 2017;5 doi: 10.1128/microbiolspec.FUNK-0035-2016. [DOI] [PubMed] [Google Scholar]
- Hall R.A., Bates S., Lenardon M.D., Maccallum D.M., Wagener J., Lowman D.W., Kruppa M.D., Williams D.L., Odds F.C., Brown A.J., Gow N.A. The Mnn2 mannosyltransferase family modulates mannoprotein fibril length, immune recognition and virulence of Candida albicans. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson R.P., Munro C.A., Bates S., MacCallum D.M., Cutler J.E., Heinsbroek S.E., Brown G.D., Odds F.C., Gow N.A. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 2004;279:39628–39635. doi: 10.1074/jbc.M405003200. [DOI] [PubMed] [Google Scholar]
- Imtiaz, T., Lee, K.K., Munro, C.A., MacCallum, D.M., Shankland, G.S., Johnson, E.M., Macgregor, M.S., and Bal, A.M. 2012. Echinocandin resistance due to simultaneous FKS mutation and increased cell wall chitin in a Candida albicans bloodstream isolate following brief exposure to caspofungin. 61: pp. 1330–1334. [DOI] [PubMed]
- Johnson M.E., Edlind T.D. Topological and mutational analysis of Saccharomyces cerevisiae Fks1. Eukaryot. Cell. 2012;11:952–960. doi: 10.1128/EC.00082-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.E., Katiyar S.K., Edlind T.D. New Fks hot spot for acquired echinocandin resistance in Saccharomyces cerevisiae and its contribution to intrinsic resistance of Scedosporium species. Antimicrob. Agents Chemother. 2011;55:3774–3781. doi: 10.1128/AAC.01811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y., Ohno H., Kohno S., Miyazaki Y. Micafungin alters the expression of genes related to cell wall integrity in Candida albicans biofilms. Jpn. J. Infect. Dis. 2010;63:355–357. [PubMed] [Google Scholar]
- Kolaczkowska A., Kolaczkowski M. Drug resistance mechanisms and their regulation in non-albicans Candida species. J. Antimicrob. Chemother. 2016;71:1438–1450. doi: 10.1093/jac/dkv445. [DOI] [PubMed] [Google Scholar]
- Kruppa M., Greene R.R., Noss I., Lowman D.W., Williams D.L. C. albicans increases cell wall mannoprotein, but not mannan, in response to blood, serum and cultivation at physiological temperature. Glycobiology. 2011;21:1173–1180. doi: 10.1093/glycob/cwr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz M.B., Abruzzo G., Flattery A., Bartizal K., Marrinan J.A., Li W., Milligan J., Nollstadt K., Douglas C.M. Characterization of echinocandin-resistant mutants of Candida albicans: genetic, biochemical, and virulence studies. Infect. Immun. 1996;64:3244–3251. doi: 10.1128/iai.64.8.3244-3251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafayette S.L., Collins C., Zaas A.K., Schell W.A., Betancourt-Quiroz M., Leslie Gunatilaka A.A., Perfect J.R., Cowen L.E. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of mkc1, calcineurin, and hsp90. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.K., MacCallum D.M., Jacobsen M.D., Walker L.A., Odds F.C., Gow N.A.R., Munro C.A. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob. Agents Chemother. 2012;56:208–217. doi: 10.1128/AAC.00683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardon M.D., Lesiak I., Munro C.A., Gow N.A. Dissection of the Candida albicans class I chitin synthase promoters. Mol. Genet. Genomics. 2009;281:459–471. doi: 10.1007/s00438-009-0423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart S.R., Berkow E.L., Chow N., Welsh R.M. Candida auris for the Clinical Microbiology Laboratory: Not Your Grandfather's Candida Species. Clin. Microbiol. Newsletter. 2017;39:99–103. doi: 10.1016/j.clinmicnews.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart S.R., Etienne K.A., Vallabhaneni S., Farooqi J., Chowdhary A., Govender N.P., Litvintseva A.P. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017;64(2):134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman D.W., Greene R.R., Bearden D.W., Kruppa M.D., Pottier M., Monteiro M.A., Soldatov D.V., Ensley H.E., Cheng S., Netea M.G., Williams D.L. Novel structural features in Candida albicans hyphal glucan provide a basis for differential innate immune recognition of hyphae versus yeast. J. Biol. Chem. 2014;289:3432–3443. doi: 10.1074/jbc.M113.529131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio T., Yabe T., Sudoh M., Satoh Y., Nakajima T., Arisawa M., Yamada-Okabe H. Role of three chitin synthase genes in the growth of Candida albicans. J. Bacteriol. 1996;178:2416–2419. doi: 10.1128/jb.178.8.2416-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Montes H.M., Bates S., Netea M.G., Diaz-Jimenez D.F., Lopez-Romero E., Zinker S., Ponce-Noyola P., Kullberg B.J., Brown A.J., Odds F.C., Flores-Carreon A., Gow N.A. Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot. Cell. 2007;6:2184–2193. doi: 10.1128/EC.00350-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro C.A. Chitin and glucan, the yin and yang of the fungal cell wall, implications for antifungal drug discovery and therapy. Adv. Appl. Microbiol. 2013;83:145–172. doi: 10.1016/B978-0-12-407678-5.00004-0. [DOI] [PubMed] [Google Scholar]
- Munro C.A., Bates S., Buurman E.T., Hughes H.B., Maccallum D.M., Bertram G., Atrih A., Ferguson M.A., Bain J.M., Brand A., Hamilton S., Westwater C., Thomson L.M., Brown A.J., Odds F.C., Gow N.A. Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 2005;280:1051–1060. doi: 10.1074/jbc.M411413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro C.A., Selvaggini S., de Bruijn I., Walker L., Lenardon M.D., Gerssen B., Milne S., Brown A.J., Gow N.A. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 2007;63:1399–1413. doi: 10.1111/j.1365-2958.2007.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro C.A., Whitton R.K., Hughes H.B., Rella M., Selvaggini S., Gow N.A. CHS8-a fourth chitin synthase gene of Candida albicans contributes to in vitro chitin synthase activity, but is dispensable for growth. Fungal Genet. Biol. 2003;40:146–158. doi: 10.1016/s1087-1845(03)00083-5. [DOI] [PubMed] [Google Scholar]
- Navarro-Garcia F., Sanchez M., Pla J., Nombela C. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol. Cell. Biol. 1995;15:2197–2206. doi: 10.1128/mcb.15.4.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi T., Ohya Y. The cell wall integrity checkpoint: Coordination between cell wall synthesis and the cell cycle. Yeast. 2010;27:513–519. doi: 10.1002/yea.1795. [DOI] [PubMed] [Google Scholar]
- Negishi T., Veis J., Hollenstein D., Sekiya M., Ammerer G., Ohya Y. The Late S-Phase transcription Factor Hcm1 Is regulated through phosphorylation by the cell wall integrity checkpoint. Mol. Cell. Biol. 2016;36:941–953. doi: 10.1128/MCB.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett J.E., Andes D.R. Antifungal Agents: Spectrum of activity, pharmacology, and clinical indications. Infect. Dis. Clin. North Am. 2016;30:51–83. doi: 10.1016/j.idc.2015.10.012. [DOI] [PubMed] [Google Scholar]
- Noble S.M., French S., Kohn L.A., Chen V., Johnson A.D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds, F.C. 2010. Editorial: Resistance to antifungal agents. 47: 190. [DOI] [PubMed]
- Okada H., Ohnuki S., Roncero C., Konopka J.B., Ohya Y. Distinct roles of cell wall biogenesis in yeast morphogenesis as revealed by multivariate analysis of high-dimensional morphometric data. Mol. Biol. Cell. 2014;25:222–233. doi: 10.1091/mbc.E13-07-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H., Ohya Y. Fluorescent labelling of yeast cell wall components. Cold Spring Harbor. Protoc. 2016:699–702. doi: 10.1101/pdb.prot085241. [DOI] [PubMed] [Google Scholar]
- P.G. Pappas C.A. Kauffman D. Andes D.K. Benjamin Jr. T.F. Calandra J.E. Edwards Jr. S.G. Filler J.F. Fisher Kullberg, B.-., Ostrosky-Zeichner, L., Reboli, A.C., Rex, J.H., Walsh, T.J., and Sobel, J.D. 2009, Clinical practice guidelines for the management of candidiasis: 2009 Update by the Infectious Diseases Society of America. 48, pp. 503–535. [DOI] [PMC free article] [PubMed]
- Park S., Kelly R., Kahn J.N., Robles J., Hsu M.J., Register E., Li W., Vyas V., Fan H., Abruzzo G., Flattery A., Gill C., Chrebet G., Parent S.A., Kurtz M., Teppler H., Douglas C.M., Perlin D.S. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 2005;49:3264–3273. doi: 10.1128/AAC.49.8.3264-3273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin, D.S. 2011. Echinocandin-resistant Candida: Molecular methods and phenotypes. 5, pp. 113–119.
- Perlin D.S. Echinocandin resistance in Candida. Clin. Infect. Dis. 2015;61(Suppl 6):S612–7. doi: 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin D.S. Mechanisms of echinocandin antifungal drug resistance. Ann. N. Y. Acad. Sci. 2015;1354:1–11. doi: 10.1111/nyas.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin D.S., Shor E., Zhao Y. Update on antifungal drug resistance. Curr. Clin. Microbiol. Rep. 2015;2:84–95. doi: 10.1007/s40588-015-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M.A. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 2012;125:S3–S13. doi: 10.1016/j.amjmed.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Piotrowski J.S., Okada H., Lu F., Li S.C., Hinchman L., Ranjan A., Smith D.L., Higbee A.J., Ulbrich A., Coon J.J., Deshpande R., Bukhman Y.V., McIlwain S., Ong I.M., Myers C.L., Boone C., Landick R., Ralph J., Kabbage M., Ohya Y. Plant-derived antifungal agent poacic acid targets beta-1,3-glucan. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E1490–7. doi: 10.1073/pnas.1410400112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaine A., Walker L., Da Costa G., Mora-Montes H.M., McKinnon A., Gow N.A., Gaillardin C., Munro C.A., Richard M.L. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 2008;45:1404–1414. doi: 10.1016/j.fgb.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pound M.W., Townsend M.L., Drew R.H. Echinocandin pharmacodynamics: review and clinical implications. J. Antimicrob. Chemother. 2010;65:1108–1118. doi: 10.1093/jac/dkq081. [DOI] [PubMed] [Google Scholar]
- Prasad R., Shah A.H., Rawal M.K. Antifungals: mechanism of action and drug resistance. Adv. Exp. Med. Biol. 2016;892:327–349. doi: 10.1007/978-3-319-25304-6_14. [DOI] [PubMed] [Google Scholar]
- Roberts R.L., Bowers B., Slater M.L., Cabib E. Chitin synthesis and localization in cell division cycle mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 1983;3:922–930. doi: 10.1128/mcb.3.5.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Pena J.M., García R., Nombela C., Arroyo J. The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: a yeast dialogue between MAPK routes. Yeast. 2010;27:495–502. doi: 10.1002/yea.1792. [DOI] [PubMed] [Google Scholar]
- Roncero C., Duran A. Effect of Calcofluor White and Congo red on fungal cell wall morphogenesis: In vivo activation of chitin polymerization. J. Bacteriol. 1985;163:1180–1185. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D. emerging threats in antifungal-resistant fungal pathogens. Front. Med. (Lausanne) 2016;3:11. doi: 10.3389/fmed.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D., Ischer F., Marchetti O., Entenza J., Bille J. Calcineurin A of Candida albicans: Involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 2003;48:959–976. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- Santiago R., Malvar R.A. Role of dehydrodiferulates in maize resistance to pests and diseases. Int. J. Mol. Sci. 2010;11:691–703. doi: 10.3390/ijms11020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.D., Robbins N., Zaas A.K., Schell W.A., Perfect J.R., Cowen L.E. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D.A. Frequency of paradoxical effect with caspofungin in Candida albicans. Eur. J. Clin. Microbiol. Infect. Dis. 2009;28:717. doi: 10.1007/s10096-008-0688-y. [DOI] [PubMed] [Google Scholar]
- Stevens D.A., Ichinomiya M., Koshi Y., Horiuchi H. Escape of Candida from caspofungin inhibition at concentrations above the MIC (paradoxical effect) accomplished by increased cell wall chitin; evidence for beta-1,6-glucan synthesis inhibition by caspofungin. Antimicrob. Agents Chemother. 2006;50:3160–3161. doi: 10.1128/AAC.00563-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Igarashi R., Sekiya M., Utsugi T., Morishita S., Yukawa M., Ohya Y. Dynactin is involved in a checkpoint to monitor cell wall synthesis in Saccharomyces cerevisiae. Nat. Cell Biol. 2004;6:861–871. doi: 10.1038/ncb1162. [DOI] [PubMed] [Google Scholar]
- Vyas V.K., Barrasa M.I., Fink G.R. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci. Adv. 2015;1 doi: 10.1126/sciadv.1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.A., Gow N.A., Munro C.A. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob. Agents Chemother. 2013;57:146–154. doi: 10.1128/AAC.01486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.A., Lee K.K., Munro C.A., Gow N.A. Caspofungin treatment of Aspergillus fumigatus results in ChsG-dependent upregulation of chitin synthesis and the formation of chitin-rich micro-colonies. Antimicrob. Agents Chemother. 2015;59(10):5923–5941. doi: 10.1128/AAC.00862-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.A., Lenardon M.D., Preechasuth K., Munro C.A., Gow N.A. Cell wall stress induces alternative fungal cytokinesis and septation strategies. J. Cell. Sci. 2013;126(Pt 12):2668–2677. doi: 10.1242/jcs.118885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.A., Munro C.A., de Bruijn I., Lenardon M.D., McKinnon A., Gow N.A. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanjare S., Gupta R., Mehta P. Caspofungin MIC distribution amongst commonly isolated Candida species in a tertiary care centre – An Indian experience. J. Clin. Diagn. Res. 2016;10:DC11-DC13. doi: 10.7860/JCDR/2016/23731.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.