Abstract
Oaks (Quercus) comprise more than 400 species worldwide and centres of diversity for most sections lie in the Americas and East/Southeast Asia. The only exception is the Eurasian sect. Cerris that comprises about 15 species, most of which are confined to western Eurasia. This section has not been comprehensively studied using molecular tools. Here, we assess species diversity and provide a first comprehensive taxonomic and phylogeographic scheme of western Eurasian members of sect. Cerris using plastid (trnH-psbA) and nuclear (5S-IGS) DNA variation with a dense intra-specific and geographic sampling. Chloroplast haplotypes primarily reflected phylogeographic patterns originating from interspecific cytoplasmic gene flow within sect. Cerris and its sister section Ilex. We identified two widespread and ancestral haplotypes, and locally restricted derived variants. Signatures shared with Mediterranean species of sect. Ilex, but not with the East Asian Cerris oaks, suggest that the western Eurasian lineage came into contact with Ilex only after the first (early Oligocene) members of sect. Cerris in Northeast Asia had begun to radiate and move westwards. Nuclear 5S-IGS diversification patterns were more useful for establishing a molecular-taxonomic framework and to reveal hybridization and reticulation. Four main evolutionary lineages were identified. The first lineage is comprised of Q. libani, Q. trojana and Q. afares and appears to be closest to the root of sect. Cerris. These taxa are morphologically most similar to the East Asian species of Cerris, and to both Oligocene and Miocene fossils of East Asia and Miocene fossils of western Eurasia. The second lineage is mainly composed of the widespread Q. cerris and the narrow endemic species Q. castaneifolia, Q. look, and Q. euboica. The third lineage comprises three Near East species (Q. brantii, Q. ithaburensis and Q. macrolepis), well adapted to continental climates with cold winters. The forth lineage appears to be the most derived and comprises Q. suber and Q. crenata. Q. cerris and Q. trojana displayed high levels of variation; Q. macrolepis and Q. euboica, previously treated as subspecies of Q. ithaburensis and Q. trojana, likely deserve independent species status. A trend towards inter-specific crosses was detected in several taxa; however, we found no clear evidence of a hybrid origin of Q. afares and Q. crenata, as currently assumed.
Keywords: Section cerris, Quercus, Evolution, Plastid DNA, Phylogeography, Western Eurasia, Nuclear Ribosomal 5S-IGS
Introduction
Studies on the genetic diversity of forest species across their distributional ranges are relevant for genetic resource inventories and devising conservation strategies (Pautasso, 2009). Comparative phylogeographic studies may further reveal complex spatial variation patterns within groups of closely related species (sibling lineages, species aggregates), shaped by partly antagonistic evolutionary and ecological processes. The detailed genetic information can be used to address taxonomic questions, assist biodiversity surveys, and implement species conservation and future landscape management strategies (Barak et al., 2016).
Oaks (Quercus L.) are an ideal model for comparative phylogeographic studies. They are common, often (co-)dominant vegetation elements and include several widely distributed and ecologically diverse species (Camus, 1936–1954; Schwarz, 1936–1939). Oaks also have a strong potential for ecological adaptation, accompanied by substantial leaf morphological variability and a high potential for introgression and reticulate evolution (e.g., Burger, 1975; Van Valen, 1976; Petit et al., 2004; McVay, Hipp & Manos, 2017). Therefore, regional estimates about the number of oak species have been strongly deviating (see e.g., http://www.ipni.org). At the same time, taxonomic ambiguity and reticulate evolution during formation of species make sampling and phylogenetic tree inference difficult.
Quercus has recently been formalized as two subgenera with eight sections (Denk et al., 2017). The predominantly Nearctic subgenus Quercus includes the sections Lobatae (Americas), Protobalanus (western North America), Ponticae (two disjunct species in southwestern Georgia/ northeastern Turkey and northern California/southwestern Oregon), Virentes (southeastern US into Mesoamerica), and Quercus (with most species in North America and about 30 species in Eurasia). The Palearctic-Indomalayan subgenus Cerris includes sections Cyclobalanopsis (East Asia), Ilex and Cerris (across Eurasia). For some of these groups, detailed infragroup phylogenies and assessment of main biogeographic patterns have recently been published (e.g., Hipp et al., 2014: North American sect. Quercus; Cavender-Bares et al., 2015: sect. Virentes; Vitelli et al., 2017: western Eurasian members of sect. Ilex; Deng et al., 2018: sect. Cyclobalanopsis). The understudied sect. Cerris (Cerris oaks) currently includes 13 species and a few unresolved taxa (Table 1) occurring from the Atlantic coasts of the Iberian Peninsula and Morocco to Japan. They thrive under a variety of climates (Köppen-Geiger climate types): cold steppe (BSk) and warm temperate or snow climates with different precipitation regimes, including more arid summer-dry and (more) mesic per-humid and winter-dry regimes (Cs, Cf, Cw, Ds, Df, Dw; Kottek et al., 2006; Peel, Finlayson & McMahon, 2007; Rubel et al., 2016). These oaks are deciduous or semi-evergreen (leaf lifespan up to 12–18 months) trees up to 30 m tall, characterized by pollen with scattered verrucate ornamentation, imbricate, recurved and elongated cupule scales, tomentose endocarp, and pointed, elongated styles; their leaves are generally toothed or lobed and usually with a mucronate apex (Denk et al., 2017). Based on the fossil record and molecular differentiation patterns, it has been suggested that sect. Cerris evolved from sect. Ilex, possibly in Europe during the Miocene (Denk & Grimm, 2010; Simeone et al., 2016). However, this scenario needs to be revised as unambiguous fossils of sect. Cerris are now known from early Oligocene deposits of northeastern Russia (Russian Far East), and sect. Ilex appears to have been present in middle Eocene strata of southern China (Denk et al., 2017).
Table 1. Species and taxa included in Quercus Section Cerris.
Nomenclature followed Govaerts & Frodin (1998); species investigated in the present study are bolded. Taxonomic remarks and species distributions according to *Govaerts & Frodin (1998) and **Menitsky (2005).
| Taxon | Taxonomic remarks | Distribution |
|---|---|---|
| Q. afaresPomel | Endemic, Algerian and Tunisian Tell Atlas | |
| Q. brantiiLindl. | S and SE Anatolia to Iran, Lebanon | |
| Q. castaneifoliaC.A. Mey | Endemic; SE Caspian Sea, Azerbaijan to Iran | |
| Q. cerrisL. | E and C Mediterranean, Balkans | |
| Q. crenataLam. | *Poorly known | Endemic, Italian peninsula |
| Q. libaniOliv. | SE Anatolia to Iran | |
| Q. lookKotschy | *Synonym of Q. ithaburensis ssp. ithaburensis; **hybrid Q. ithaburensis×Q. libani | Endemic, Lebanon to Anti-Lebanon mountain range |
| Q. ithaburensisDecne. | **Including subsp. macrolepis (Kotschy), distributed in the European part of the range, and subsp. ithaburensis (Decaisne), in the Middle East | E Mediterranean, SE Italy to Palestine |
| Q. trojanaWebb | *Including subsp. trojana and subsp. euboica (Papaioann.) K.I.Chr., endemic of Euboea (Greece) | Anatolia, Aegean to SE Italy |
| Q. suberL. | C and W Mediterranean | |
| Q. acutissima Carruth. | E and SE Asia | |
| Q. chenii Nakai | E Asia | |
| Q. variabilis Blume | E and SE Asia, Japan |
At present, sect. Cerris is most diverse in western Eurasia, with eight commonly accepted species (Govaerts & Frodin, 1998) and some additional forms (Q. crenata, Q. look, Q. trojana subsp. euboica, Q. ithaburensis subsp. macrolepis) of disputed or unresolved status. The ranges of Cerris oaks vary substantially in size and in the degree of contact with other species of the same section and the sister section Ilex. The Qinghai-Tibet Plateau and the Himalayan front-hills, home of several species of sect. Ilex, separate the East Asian and the western Eurasian taxa. The central and eastern Mediterranean region comprise most of the group’s diversity (Anatolia and Levant to S. Italy and N.E. Algeria, eight species), which decreases westward (Iberian Peninsula and Morocco, one species) and eastward (Iran/ Iraq, three species; Browicz & Zieliński, 1982). Throughout the Mediterranean, the distribution ranges of Cerris oaks overlap with section Ilex, particularly Q. ilex. Two species have broad distributions (Q. suber, the ‘Cork Oak’, partly due to cultivation, and Q. cerris) and three are geographically extremely limited (Q. afares, Q. castaneifolia, Q. look). A hybrid origin has been postulated for Q. crenata (infra-sectional hybrid) and Q. afares (inter-sectional hybrid; Mir et al., 2006; Conte, Cotti & Cristofolini, 2007). Other occasional infra-sectional hybrids have also been described as morphologically intermediates (see Menitsky, 2005). Quercus suber shows interfertility with the partly sympatric Q. ilex of sect. Ilex (Burgarella et al., 2009).
Detailed phylogeographic inferences are so far available only for two East Asian species of sect. Cerris thriving in temperate and subtropical broad-leaved forests in eastern Asia, Q. acutissima and Q. variabilis, based on plastid DNA sequence analyses (Chen et al., 2012; Zhang et al., 2015). In both cases, high genetic diversity but weak phylogeographic structure was found, explained with recent (Pleistocene) speciation and post-glacial re-expansion of lineages. In western Eurasia, the two most widespread species (Q. suber and Q. cerris) were studied using plastid microsatellite variation (Magri et al., 2007; Bagnoli et al., 2016). Geographically structured gene pools were detected and their formation attributed to the Oligocene (Q. suber) and Pleistocene (Q. cerris), respectively. Local investigations focussing on conservation were conducted on Q. trojana in Italy and Q. libani in Iran using nuclear microsatellites (Khadivi-Khub et al., 2015; Carabeo et al., 2017). Finally, species of the entire section were included in DNA barcoding projects and studies on molecular macroevolution (e.g., Denk & Grimm, 2010; Simeone et al., 2013; Simeone et al., 2016), but relied on a limited number of individuals.
At present, firm species delineation and phylogenetic inferences using sequence data are difficult in Quercus. All plastid data assembled so far showed strong disagreement with taxonomy and systematics (Grímsson et al., 2016; Denk et al., 2017) and nuclear regions with sufficient levels of variation, especially when closely related species are involved, are not yet available (Muir, Fleming & Schlotterer, 2001; Oh & Manos, 2008; Hubert et al., 2014). Unrepresentative inter- and (especially) intra-specific samplings constitute additional obstacles (Manos, Zhou & Cannon, 2001; Bellarosa et al., 2005; Chen et al., 2017), that partially reduce the potential for thorough within-lineage species delineation, even with high-resolution phylogenomic approaches (Hipp et al., 2014; Hipp et al., 2018; Cavender-Bares et al., 2015). Nevertheless, the efficacy of the nuclear ribosomal 5S rDNA intergenic spacer (in contrast to the internal spacers of the 35S rDNA, ITS1 and ITS2) to resolve species differentiation in western Eurasian members of Sect. Cerris was demonstrated by Denk & Grimm (2010). Being potentially affected by incomplete lineage sorting, intra-array recombination and intragenomic competition, this marker requires a special analysis framework (cloning and host-associate analysis; cf. Göker & Grimm, 2008) but enables tracking of reticulate evolutionary signatures.
On the other hand, being largely controlled by provenance and decoupled from speciation, plastid data are important to trace the radiation of lineages in space and time (Pham et al., 2017). In this view, the evolutionary trajectories of sect. Cerris and Ilex share some traits that need to be fully addressed. Two recent studies on the Mediterranean members of sect. Ilex (Simeone et al., 2016; Vitelli et al., 2017) found three main plastid haplotype groups, with distinct geographic distribution and phylogenetic features: (1) ‘Euro-Med’, comprising the most distinct haplotypes dominating in the western Mediterranean, a plastid lineage that diverged before the radiation of plastid pools in Subgenus Cerris, (2) ‘WAHEA’ (i.e., West Asian–Himalayan–East Asian), distributed from Anatolia/Levant to East Asia, and sister to (3) ‘Cerris-Ilex’, centred on the Aegean Sea and shared with co-occurring members of sect. Cerris. Haplotype variation of the trnH-psbA intergenic spacer was determinant for the phylogeographic inferences; this marker also showed the highest variation rate among several plastid regions in 35 Chinese oak species (Yang et al., 2017), and in the comprehensively studied Q. acutissima and Q. variabilis (Zhang et al., 2015; Chen et al., 2012).
Clearly, full comprehension of the drivers of speciation of sect. Cerris requires information from both genomes based on extensive geographic and taxonomic sampling. In this work, we investigated 5S-IGS and trnH-psbA molecular diversity in western Eurasian sect. Cerris using a comprehensive intra- and inter-specific sampling. Our objectives were: (i) to assess species coherence and delimitation, (ii) to infer inter-species relationships, (iii) to gain insight into the origin and diversification of the group. We further hypothesize that signals from both markers may combine to reveal potential zones of secondary genetic contact between already established species and to gain insight into the putative hybrid status of Q. crenata and Q. afares. All data will likely contribute to identify biogeographic hotspots for further in-depth studies on species diversity, isolation, introgression and natural hybridisation in oaks.
Materials and Methods
Plant material and DNA sequencing
We combined previously studied (Denk & Grimm, 2010) and new material to develop a sampling design of 221 individuals completely covering the taxonomic breadth and distribution range of Quercus sect. Cerris in western Eurasia (File S1); some individuals with intermediary morphology were labelled as presumed hybrids. 158 individuals were studied for the first time, and new plastid data were generated for the previously and newly studied individuals. DNAs were extracted from silica gel-dried leaf samples with the DNeasy Plant minikit, following the manufacturer’s instructions. The trnH-psbA intergenic spacer was amplified and sequenced following Simeone et al. (2016). The nuclear ribosomal 5S intergenic spacer (5S-IGS) was amplified with the primer pair 5S14a and 5S15 (Volkov et al., 2001; Denk & Grimm, 2010). Individual PCR fragments were ligated into a pGEM-T easy vector (Promega). The ligation mixtures, purified with the Illustra GFX PCR DNA Purification kit (GE Healthcare), were used to transform E. coli strain XL1-Blue electroporation-competent cells (recA1, endA1, gyrA96, thi-1, hsdR17, supE44, relA1, lac, [F’ proAB, lacIqZ ΔM15, Tn10 (tetr)]). The positive clones, selected on LB/Ampicillin plates, were identified by colony PCR using the amplification primers. Five to ten recombinant clones per individual were sequenced with the vector-specific universal primers (SP6/T7) at LGC Genomics (Augsburg, Germany). The GenBank trnH-psbA sequences of several East Asian members of sect. Ilex (Q. baloot, Q. floribunda, Q. phylliraeoides, Q. semecarpifolia, Q. baroni, Q. dolicholepis, Q. spinosa), East Asian species of sect. Cerris (Q. acutissima, Q. variabilis) and further sequence accessions of the investigated group were included in the analyses. In addition, all 5S IGS sequences of Quercus sect. Cerris available on GenBank were included in the final dataset, and the sequences of Q. baloot and Q. floribunda were used as outgroups, based on Denk & Grimm (2010); all GenBank accession numbers are reported in File S1.
Data analyses
Eye-checked electropherograms were aligned in MEGA7 (Kumar, Stecher & Tamura, 2016). Highly dissimilar clone sequences showing no BLAST match with the targeted regions (Altschul et al., 1990) were filtered. Final multiple alignments were obtained with ClustalW 1.81 (Thompson, Higgins & Gibson, 1994) and checked by eye. The diversity of the investigated regions was evaluated with MEGA7 and DnaSP5.1 (Librado & Rozas, 2009). Median-joining (MJ) haplotype networks for the trnH-psbA region were inferred with Network 4.6.1.1 (http://www.fluxus-engineering.com/), treating gaps as fifth state. The MJ algorithm was invoked with default parameters (equal weight of transversion/transition), in order to handle large datasets and multistate characters.
After removal of identical clones, the total 5S-IGS sequences were used to build a Maximum likelihood (ML) tree with RAxML v8.2 (Stamatakis, 2014) using the in-built GTR + Γ model with the ‘extended majority-rule consensus’ criterion as bootstopping option (Pattengale et al., 2009). To infer inter-individual relationships, we applied the approach described by Göker & Grimm (2008) that allows transformation of data matrices of ‘associates’ (here: cloned sequences) into ‘hosts’ (here: individuals). The program g2cef (available at http://www.goeker.org/mg/distance/) was used to transform the primary character matrix (‘associates’, total cloned sequences) into a character consensus matrix of the individuals (‘hosts’) using an association file defining the list of clone sequences belonging to the same individual. The uncorrected pairwise distances of the primary character matrix (‘associates’, total cloned sequences) was calculated and used as input to the program pbc (Göker & Grimm, 2008). This program allows transforming the primary inter-clone pairwise distance matrix into inter-individual distances matrices using different flavours, of which the ‘Phylogenetic Bray-Curtis’ (PBC) transformation performed best in the original study that compared data sets with similar properties than our data set. Here, we applied three of the distance transformations tested by Göker & Grimm (2008), in addition to PBC distances (option -b) also the minimum (MIN; -i) and average (AVG; -a) inter-individual clonal distances. AVG, MIN and PBC distance matrices were generated setting different minimum number of associates per host (-m option); m = 4 (the number of cloned sequences obtained in most individuals) was then used to infer a phylogenetic network using the Neighbour-Net (NN) algorithm (Bryant & Moulton, 2004) implemented in SplitsTree4 (Huson & Bryant, 2006).
Results
In total, 221 individuals effectively covering the taxonomic range of western Eurasian sect. Cerris were analysed (Table 1, File S1). Sequence quality was high for both marker regions and unambiguous electropherograms were obtained for about 90% of the investigated samples. The primary data matrixes comprised 207 plastid (trnH-psbA) and 856 nuclear (5S-IGS) sequence accessions. The nuclear data (192 individuals) included 651 newly sequenced clones and 205 accessions from Denk & Grimm (2010). Ten Q. baloot/Q. floribunda sequences, used here as outgroups (cf. Denk & Grimm, 2010), extended the final dataset to 866 sequenced clones. Individual sequences recovered from positive 5S-IGS clones varied from one (four samples) to 10, with most samples represented by four sequences (70 samples), followed by five and three sequences (44 and 35 samples, respectively). Multiple alignments of both marker regions were straigthforward. A 34-bp inversion occurring in the trnH-psbA region of 14 samples was replaced with its reverse-complementary sequence. Since it did not show further mutations it was deleted and a binary character was inserted to keep record of it.
Plastid trnH-psbA diversity and biogeography
After removing the 34-bp inversion, the trnH-psbA marker showed pairwise uncorrected p-distances ranging between zero and 0.008 (Table 2). The highest intra-specific distance (0.006) was found in Q. suber; four species showed similar values (0.002; Q. cerris, Q. ithaburensis, Q. trojana, Q. libani), while the marker variation in the remaining taxa converged to zero.
Table 2. Diversity values of the trnH-psbA IGS in the investigated dataset.
| Dataset | N | L | p | H | h | Hid | S | PICs |
|---|---|---|---|---|---|---|---|---|
| West Eurasian species | 207 | 503 | 0.000–0.008 (±0.004) | 12 | 0.515 | H1-H12 | 6 (27) | 6 |
| Q. afares | 7 | 491 | 0.000 | 1 | 0.000 | H1 | 0 | 0 |
| Q. brantii | 7 | 487 | 0.000 | 2 | 0.476 | H5, H6 | 0 (1) | 0 |
| Q. cerris | 52 | 493 | 0.000–0.002 | 8 | 0.538 | H1-H3, H5-H7, H9, H10 | 1 (9) | 1 |
| Q. castaneifolia | 2 | 491 | 0.000 | 1 | 0.000 | H1 | 0 | 0 |
| Q. crenata | 6 | 491 | 0.000 | 1 | 0.000 | H1 | 0 | 0 |
| Q. trojanaa | 45 | 493 | 0.000–0.002 | 3 | 0.369 | H1, H2, H4 | 1 (3) | 1 |
| Q. ithaburensisb | 33 | 493 | 0.000–0.002 | 5 | 0.655 | H1, H2, H4, H6, H9 | 1 (8) | 1 |
| Q. look | 3 | 488 | 0.000 | 2 | 0.667 | H9, H10 | 0 (1) | 0 |
| Q. suber | 47 | 501 | 0.000–0.006 | 3 | 0.303 | H1, H11, H12 | 3 (18) | 3 |
| Q. libani | 5 | 493 | 0.000–0.002 | 3 | 0.700 | H1, H3, H8 | 1 (8) | 0 |
| Q. acutissimac | 401 | 564 | 0.000–0.004 | 10 | n.d. | /d | 4 (79) | 0 |
| Q. variabilisc | 528 | 594 | 0.000–0.004 | 11 | n.d. | /d | 2 (99) | 0 |
Notes.
- N
- number of sequences
- L
- Aligned length (bp) with the inversion deleted
- p
- uncorrected p-distance range (STD)
- H
- Number of identified haplotypes (gaps included)
- h
- Haplotype diversity
- Hid
- haplotype code
- S
- Number of polymorphic sites (gaps included)
- PICs
- Number of Parsimony Informative Characters
Including subsp. euboica.
Including subsp. macrolepis.
GenBank haplotype accessions: KT152191, KT152192, KT152193, KT152194, KT152195, KT152196, KT152197, KT152198, KT152199, KT152200, JF753573, JF753574, JF753575, JF753576, JF753577, JF753578, JF753579, JF753580, JF753581, JF753582, JF753583, KM210647, HE585136.
No haplotype shared with the West Eurasian dataset, one haplotype shared between the two East Asian species.
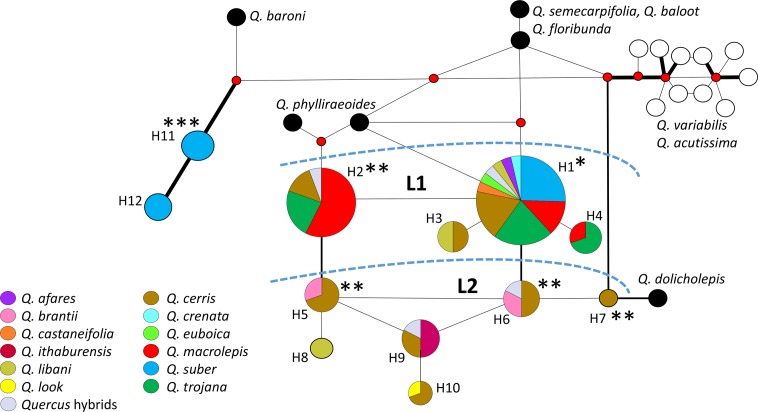
The total matrix was 503-bp characters long, including several indels (1–8 bp) and six polymorphic sites resulting in twelve haplotypes (labelled H1–H12) with a medium overall diversity (h = 0.515). Haplotype H1 hit 100% sequence identity with three non-representative individuals assigned to East Asian species of sect. Ilex in Genbank (haplotype list, occurrence and gene bank matches shown as Files S1 and S2). It was the most common haplotype, occurring in 68.6% of individuals and all taxa except Q. brantii, Q. look, and Q. ithaburensis subsp. ithaburensis (henceforth Q. ithaburensis). Haplotypes H2, H5–H7 and H11 showed 100% sequence identity with Mediterranean members of sect. Ilex (Simeone et al., 2016; Vitelli et al., 2017). H2 is the second most frequent haplotype, found in 10.6% of Q. cerris, Q. trojana, Q. ithaburensis subsp. macrolepis (henceforth Q. macrolepis) samples from Turkey, the Balkans and Italy; H5–H7 were found in Q. brantii, Q. cerris and Q. macrolepis from Turkey, Iran, and Israel. They were all shared with Q. coccifera and Q. ilex of the Aegean ‘Cerris-Ilex’ lineage. Haplotype H11 found in Iberian samples of Q. suber was shared with Q. ilex of the ‘Euro-Med’ lineage. Rare haplotypes restricted to a single species were H7 (one accession of Q. cerris), H8 (three accessions of Q. libani; new ‘Cerris-Ilex’ subtypes) and H11/H12 (eight accessions of Q. suber; ‘Euro-Med’ types); all other haplotypes were shared by more than one species of sect. Cerris. Quercus cerris, the most widespread and ecologically diverse species of sect. Cerris, showed the highest number of haplotypes (eight), followed by Q. brantii (five) and Q. macrolepis (four). In the latter species, haplotype H6, exclusively found in Q. brantii, was also found in a suspected hybrid Q. macrolepis × Q. brantii (sample ml27). All samples of Q. ithaburensis exhibited a single haplotype (H9). The geographically (more) restricted taxa Q. afares, Q. castaneifolia, Q. crenata and Q. trojana subsp. euboica (henceforth: Q. euboica) showed only the most frequent and widespread haplotype (H1).
In comparison (Table 2), the two East Asian members of sect. Cerris (Q. acutissima and Q. variabilis) displayed a higher variation at the trnH-psbA locus, although mostly due to indels. A higher number of haplotypes was found in these species; none of them was shared with any species of sect. Ilex available in gene banks (identity range: 93–99% with Q. baroni, Q. dolicholepis and/or Q. spinosa), and only one haplotype was shared between the two species.
No shared parsimony informative characters (PICs) were found in the East Asian samples. In contrast, three PICs were exclusively shared by haplotypes H11 and H12 (Q. suber from Iberian Peninsula, North Morocco). One further PIC separated H9, including all individuals of Q. ithaburensis, two Q. look, two Israelian and one Italian Q. cerris individuals. A single PIC also defined H4, including three co-occurring Q. trojana and Q. macrolepis accessions from the same locality in western Turkey, and another PIC was limited to two Q. cerris and Q. libani accessions from southern Turkey, corresponding to H3. Table 3 shows that the highest mean intragroup divergence in the West Eurasian dataset was found in Q. suber and Q. libani. The haplotypes of Q. suber and Q. look displayed the highest mean divergence from all other species. Quercus variabilis appeared more similar to its western Eurasian counterparts, while Q. acutissima was highly distinct.
Table 3. Heatmap with the mean estimates of evolutionary divergence of the trnH-psbA IGS over sequence pairs within and between the investigated taxa.
| Dataset | Intra- | Interspecies divergence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q. afares | 0 | 0 | 0.0001 | 0 | 0 | 0.0001 | 0.0001 | 0.0004 | 0.0009 | 0.0006 | 0.0004 | 0.0002 | |
| Q. brantii | 0 | 0 | 0.0001 | 0 | 0 | 0.0001 | 0.0002 | 0.0004 | 0.0015 | 0.0006 | 0.0004 | 0.0002 | |
| Q. cerris | 0.0003 | 0.0002 | 0.0002 | 0.0001 | 0.0001 | 0.0001 | 0.0002 | 0.0004 | 0.0008 | 0.0006 | 0.0004 | 0.0002 | |
| Q, castaneifolia | 0 | 0 | 0 | 0.0002 | 0 | 0.0001 | 0.0001 | 0.0004 | 0.0009 | 0.0006 | 0.0004 | 0.0002 | |
| Q. crenata | 0 | 0 | 0 | 0.0002 | 0 | 0.0001 | 0.0001 | 0.0004 | 0.0009 | 0.0006 | 0.0004 | 0.0002 | |
| Q. trojanab | 0.0001 | 0.0001 | 0.0001 | 0.0003 | 0.0001 | 0.0001 | 0.0001 | 0.0004 | 0.001 | 0.0006 | 0.0004 | 0.0002 | |
| Q. ithaburensisc | 0.0003 | 0.0001 | 0.0003 | 0.0003 | 0.0001 | 0.0001 | 0.0002 | 0.0005 | 0.001 | 0.0006 | 0.0004 | 0.0002 | |
| Q. libani | 0.0008 | 0.0004 | 0.0004 | 0.0006 | 0.0004 | 0.0004 | 0.0005 | 0.0007 | 0.0013 | 0.0007 | 0.0006 | 0.0005 | |
| Q. look | 0 | 0.0014 | 0.0015 | 0.0014 | 0.0014 | 0.0014 | 0.0019 | 0.0018 | 0.0021 | 0.0011 | 0.001 | 0.0009 | |
| Q. suber | 0.0018 | 0.0011 | 0.0011 | 0.0013 | 0.0011 | 0.0011 | 0.0012 | 0.0011 | 0.0015 | 0.0025 | 0.0007 | 0.0006 | |
| Q. acutissimaa | 0.0016 | 0.0008 | 0.0008 | 0.001 | 0.0008 | 0.0008 | 0.0009 | 0.0009 | 0.0013 | 0.0022 | 0.0019 | 0.0005 | |
| Q. variabilisa | 0.0008 | 0.0003 | 0.0003 | 0.0005 | 0.0003 | 0.0003 | 0.0004 | 0.0004 | 0.0007 | 0.0017 | 0.0014 | 0.0012 | |
Notes.
GenBank haplotype accessions as in Table 2
Including subsp. euboica
Including subsp. macrolepis
Standard error estimate are shown above the diagonal.
The haplotype network (Fig. 1) shows the general coherence of the ‘Cerris-Ilex’ lineage, which collects haplotypes typical of the western Eurasian members of sect. Cerris, clearly distinct from the haplotypes found in East Asian members of sect. Cerris and haplotypes H11–H12 (>5 mutations separating each lineage); this latter represents a unique, early diverged plastid lineage, most frequent in the western Mediterranean populations of sect. Ilex (Simeone et al., 2016; Vitelli et al., 2017). Based on the relative number of mutations (1–5) separating each haplotype, the ‘Cerris-Ilex’ lineage can be further subdivided into two groups: (L1) a group of potentially primitive (non-derived) haplotypes (H1–H4) including the most common haplotypes (H1, H2) and still close to haplotypes found in the north-easternmost species of sect. Ilex (Q. phylliraeoides), the plastid sister lineage of ‘Cerris-Ilex’ (Simeone et al., 2016); (L2) a group of derived haplotypes (H5–H10). Haplotypes not shared with sect. Ilex (H3–H4, H8–H10, H12) are derivates of the ‘Cerris-Ilex’ and ‘Euro-Med’ main types (H1–H2, H5-H7, H11), shared by both sections in the Aegean and the western Mediterranean regions. As shown in Figs. 2A–2B, plastid diversity is largely decoupled from species identity and related to geography; the least derived haplotypes within the ‘Cerris-Ilex’ lineage (H1 and H2) occur across the whole distribution range of the investigated group, except the Levant and the western Mediterranean (H2). All other haplotypes are more circumscribed, concentrated in Anatolia (H3–H5, H7–H8), the Levant (H6, H9–H10; the latter two showing single occurrences in Italy), and Iberian Peninsula + Morocco (H11-H12).
Figure 1. Median joining network of the trnH-psbA sequences in western Eurasian section Cerris.
Taxa are indicated with colours (see also File S1); black, Asian species of section Ilex; white, eastern Eurasian species of section Cerris. Line thickness according to 1, <5 and >5 mutations; *, shared with Asian Ilex oaks; **, shared with Cerris-Ilex lineage of section Ilex; ***, shared with West-Med lineage of section Ilex; L1, L2, haplotype lineages identified. All accession numbers are reported in Files S1 and S2.
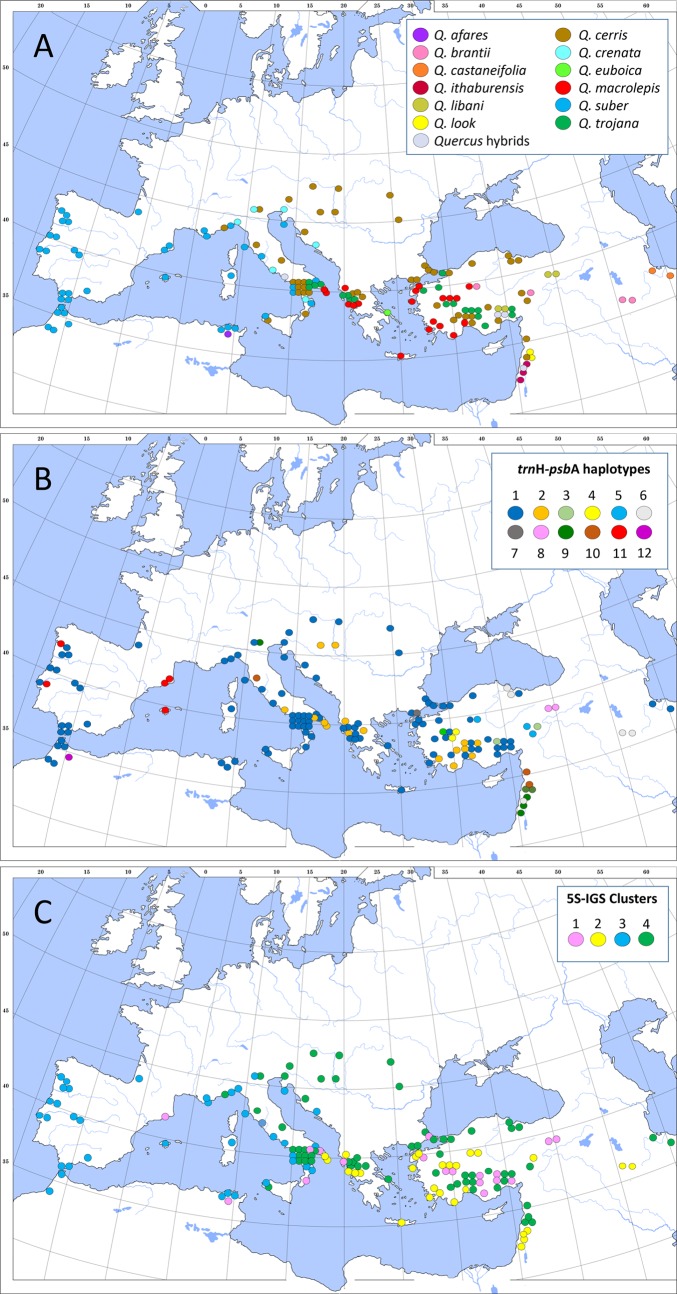
Figure 2. Geographic representation of the investigated dataset and its molecular signatures.
(A) sample distribution. (B) trnH-psbA haplotypes. (C) 5S-IGS clusters; see also File S1.
Nuclear 5S rDNA diversity and species phylogeny
In contrast to trnH-psbA, 5S-IGS sequence variation appeared generally correlated with the taxonomy of the studied individuals, and allowed inferences on potential reticulation and inter-species relationships within the western Eurasian members of sect. Cerris. The 5S-IGS clones varied greatly in sequence features and length (the multiple alignment of the cloned sequences can be viewed in the Online Supplementary Archive at the journal’s homepage). For instance, all Q. brantii clones displayed an intra-specific (ATTT)1−7 simple sequence repeat (SSR) variation. In all the other species, this motif was either absent (replaced by a 5–12 bp long poly-T) or consisting of one to two repeats, with the exception of two clones of the suspected hybrid Q. macrolepis × Q. brantii (individual ml27) that showed four to five repetitions. Two clones of Q. brantii (sample br02; C. Turkey) shared a four-bp insertion with several clones of sympatric Q. macrolepis individuals (ml20–ml22). Three Q. libani individuals (li02, 03, 04; S. and E. Turkey) displayed a long indel (ca. 100 bp) in (nearly) all clones (‘short libani variant’ cf. Denk & Grimm, 2010). The extended sample revealed that the ‘short libani variant’ is not exclusive to Q. libani but is rarely found also in Q. cerris (clones ce2104 and ce4704; Italy, W. Turkey) and Q. trojana (three clones of individual tj33, S. Turkey); the latter, however, is another suspected hybrid (Q. trojana x Q. libani).
Two other deletions were detected in the same region of the ‘short libani variant’. One (22 bp) was shared by single clones of two Q. cerris individuals (ce18, ce22; S.W. and W. Turkey), four clones of a Q. look individual (lk2; Israel) and two clones of Q. trojana (individual tj40; S. Turkey). The second (∼100 bp), largely overlapping with the deletion of the ‘short libani variant’, but beginning a few basepairs downstream, was shared by one clone of Q. cerris (ce34; N. Turkey) and one clone of Q. macrolepis (ml26; S. Greece). An 8-bp deletion occurred exclusively in Q. suber and Q. crenata, with the exception of single clones of samples su07, su09 (N.E. and S. Spain), su37 (Croatia), su53 (S. Italy), cr02 (C. Italy), two clones of sample cr04 (Slovenia) and cr06 (N.E. Italy), and three clones of sample cr05 (Croatia). The same deletion also occurred in two clones of sample tj08, a Q. suber × Q. trojana cultivation hybrid. Further deletions (1–60 bp) were scattered along the alignment and found only in single individuals (e.g., it04, Israel; ml10, N.W. Greece). Finally, an 18-bp highly variable region was exclusively found in some clones of four co-occurring Q. trojana samples (tj03–05, tj16; S.C. Turkey).
The main diversity values of the investigated dataset are reported in Table 4. Identical 5S-IGS sequences typically occur in the same individual and species, and, to a lesser extent, in sympatric, different species (e.g., Q. brantii, Q. cerris, Q. trojana, Q. look; see also File S3). On the contrary, Q. afares, Q. castaneifolia, Q. libani, Q. ithaburensis, Q. macrolepis and Q. euboica showed high species coherence. Quercus suber and Q. macrolepis showed the highest number of intra-individual and intra-specifically shared clones, whereas Q. cerris, Q. trojana and Q. ithaburensis displayed the highest levels of unique variants. No variants were shared between Q. trojana and Q. euboica; Q. suber and Q. crenata (but not Q. cerris) shared 69 identical sequences and are the genetically most similar taxon pair. The pairwise uncorrected p-distance range of the total dataset was much higher than for the plastid marker (0–0.209), with highest values scored by Q. cerris and Q. trojana. The mean intra-specific molecular diversity estimated within sequence pairs (Table 5) was lowest in the two narrow endemics Q. afares and Q. castaneifolia and highest in Q. brantii and Q. ithaburensis. Across the entire dataset, Q. brantii, Q. macrolepis and Q. ithaburensis were the most diverging taxa; the least divergent being Q. afares and Q. castaneifolia. The mean divergence value between Q. macrolepis and Q. ithaburensis (0,0376), treated as subspecies of Q. ithaburensis in current regional floras, was similar to values detected between these taxa and the other species (e.g., Q. afares, Q. brantii, Q. libani). Likewise, the divergence recorded between the putative conspecific Q. trojana and Q. euboica (0.0266) was comparable to the estimates calculated between these and other taxa (e.g., Q. afares, Q. cerris, Q. look, Q. libani). The putative hybrid taxon Q. crenata displayed the lowest divergence (0.0197) with Q. suber, one of the assumed parental species, and a slightly higher estimate (but similar to the values scored with other taxa, e.g., Q. afares, Q. castaneifolia, Q. look) with Q. cerris (0.0266), the other putative parental species.
Table 4. Diversity values of the 5S IGS clones in the investigated dataset.
| Dataset | N | Cs | L | O (u/i/a/s) | D | p | C |
|---|---|---|---|---|---|---|---|
| West Eurasian Cerris oaks | 194 | 856 | 427 | 457/186/121/79 | 0.000–0.209 ± 0.021 | 1–4 | |
| Q. afares | 5 | 17 | 379 | 10/2/5/0 | 0.000–0.019 ± 0.006 | 1 | |
| Q. brantii | 7 | 26 | 403 | 9/11/4/2 | Q. cerris (3)/Q. look, Q. suber (1) | 0.000–0.088 ± 0.014 | 4 |
| Q. castaneifolia | 2 | 2 | 375 | 2/0/0/0 | 0.005 ± 0.003 | 2 | |
| Q. cerris | 48 | 207 | 392 | 157/21/24/5 | Q. brantii (1), Q. trojana (1), Q. suber (1) | 0.000–0.202 ± 0.02 | 1c, 2 |
| Q. crenata | 6 | 29 | 387 | 19/4/2/4 | Q. suber (65) | 0.000–0.054 ± 0.012 | 2d, 3e |
| Q. libani | 5 | 20 | 382 | 13/3/4/0 | 0.000–0.040 ± 0.009 | 1, 2f | |
| Q. look | 3 | 14 | 383 | 10/3/0/1 | Q. brantii/Q. cerris (3) | 0.000–0.032 ± 0.009 | 2 |
| Q. macrolepisa | 28 | 158 | 402 | 44/71/43/0 | 0.000–0.065 ± 0.013 | 4 | |
| Q. ithaburensis | 5 | 21 | 388 | 15/6/0/0 | 0.000–0.079 ± 0.014 | 4g | |
| Q. suber | 38 | 153 | 385 | 30/48/8/67 | Q. brantii (1), Q. cerris (1), Q. crenata (4) | 0.000–0.168 ± 0.019 | 1h, 3 |
| Q. trojanab | 43 | 192 | 391 | 130/30/31/1 | Q. cerris (1) | 0.000–0.198 ± 0.020 | 1, 2, 3i |
| Q. euboica | 4 | 17 | 382 | 17/0/0/0 | 0.000–0.059 ± 0.012 | 2 |
Notes.
- N
- number of individuals
- Cs
- number of clone sequences
- L
- Aligned length (bp)
- O
- occurrence of the IGS variants (u, unique, i, intra-individually identical; a, intra-specifically shared; s, inter-specifically shared)
- D
- distribution of the interspecifically shared variants (no. of variants)
- p
- uncorrected p-distance range (STD)
- C
- clusters identified with the neighbour-net analyses
Including one putative hybrid with Q. brantii.
Including putative hybrids with Q. suber and Q. libani.
Sample ce50 (S Italy).
Sample cr04 (Slovenia).
Including odd-placed sample cr05 (Croatia).
Sample li01 (S Turkey).
Including odd-placed sample it03 (Israel).
Sample su09 (S Spain).
Sample tj08 (Botanical Garden of Naples).
Table 5. Heatmap with the mean estimates of evolutionary divergence of the nuclear 5S IGS over sequence pairs within and between the investigated taxa.
Standard error estimates are shown above the diagonal.
| Dataset | Intra- | Intergroup divergence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q. afares | 0.0057 | 0.0065 | 0.0059 | 0.0058 | 0.0048 | 0.006 | 0.0064 | 0.0058 | 0.0062 | 0.0085 | 0.0052 | 0.0049 | |
| Q. brantii | 0.0357 | 0.0353 | 0.0068 | 0.0066 | 0.006 | 0.0067 | 0.0044 | 0.0063 | 0.0068 | 0.0054 | 0.006 | 0.0061 | |
| Q. cerris | 0.0167 | 0.0244 | 0.0427 | 0.0042 | 0.0043 | 0.0021 | 0.0062 | 0.0046 | 0.0021 | 0.0088 | 0.0056 | 0.0032 | |
| Q. castaneifolia | 0.0053 | 0.0169 | 0.0343 | 0.0166 | 0.0048 | 0.004 | 0.0062 | 0.0045 | 0.0041 | 0.0085 | 0.0055 | 0.0035 | |
| Q. crenata | 0.023 | 0.0241 | 0.0422 | 0.0266 | 0.0216 | 0.0044 | 0.0056 | 0.005 | 0.0046 | 0.0078 | 0.0028 | 0.004 | |
| Q. euboica | 0.0194 | 0.026 | 0.0436 | 0.0193 | 0.0171 | 0.0285 | 0.0061 | 0.0046 | 0.0026 | 0.0088 | 0.0055 | 0.0031 | |
| Q. ithaburensis | 0.0367 | 0.0364 | 0.0386 | 0.0423 | 0.0344 | 0.0422 | 0.0431 | 0.0057 | 0.0062 | 0.0056 | 0.0058 | 0.0056 | |
| Q. libani | 0.0166 | 0.0232 | 0.0382 | 0.0244 | 0.0179 | 0.028 | 0.0255 | 0.037 | 0.0046 | 0.0078 | 0.0054 | 0.0041 | |
| Q. look | 0.0134 | 0.023 | 0.0406 | 0.0159 | 0.0139 | 0.0255 | 0.0175 | 0.0401 | 0.0214 | 0.0089 | 0.0057 | 0.0032 | |
| Q. macrolepis | 0.0194 | 0.0399 | 0.036 | 0.0498 | 0.0399 | 0.0473 | 0.0507 | 0.0376 | 0.0434 | 0.0481 | 0.0079 | 0.0079 | |
| Q. suber | 0.0135 | 0.0211 | 0.0382 | 0.0279 | 0.0201 | 0.0197 | 0.0288 | 0.0385 | 0.0262 | 0.026 | 0.0423 | 0.0045 | |
| Q. trojana | 0.027 | 0.0257 | 0.0433 | 0.0259 | 0.0195 | 0.0304 | 0.0266 | 0.043 | 0.0259 | 0.0235 | 0.0492 | 0.0287 | |
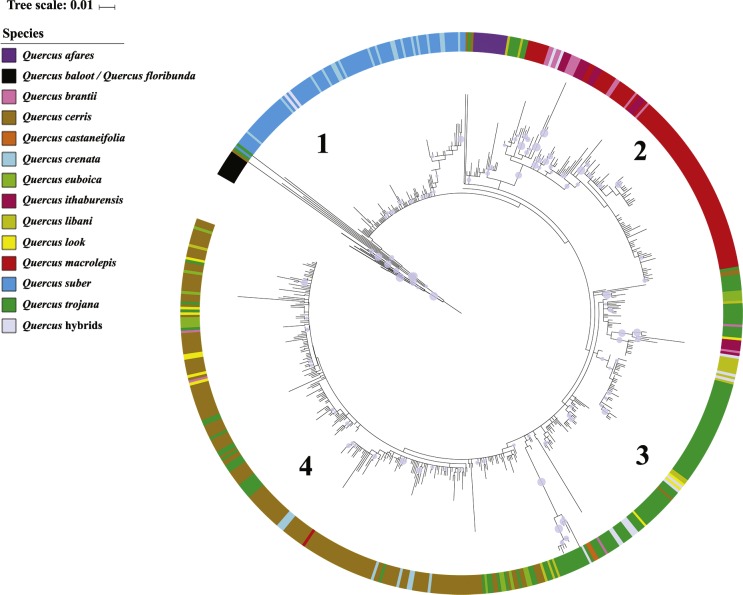
The clone-based ML tree rooted on Q. baloot and Q. floribunda (West-Asian members of sect. Ilex) showed four main topological features (grades/clades) generally coherent with taxonomy (Fig. 3, see also File S4). These grades/clades collected to a large degree clones of (1) Q. crenata and Q. suber (resolved as proximal, weakly differentiated grade), (2) Q. brantii, Q. ithaburensis and Q. macrolepis (the most highly supported clade: BSML = 84), (3) Q. trojana (a large heterogeneous grade), and (4) Q. cerris (the distal, terminal, clade with diminishing support). Quercus libani clones (short and normal-length variants) were present in all clades/grades except grade 1. A moderately supported clade (BS = 63) including all Q. afares clones was placed as sister to the main clade including clades/grades 2–4; Q. castaneifolia clones were placed within grade 3. Clones of Q. ithaburensis also occurred in grade 3, Q. brantii and Q. crenata in clade 4, Q. look and Q. euboica in grade 3 and 4. A few clones of Q. cerris and Q. trojana occurred scattered across the tree (often in proximal positions).
Figure 3. 5S-IGS Clone-based RaxML tree.
The tree was tentatively rooted on Q. baloot and Q. floribunda, two western Asian oaks of Sect. Ilex (cf. Denk & Grimm, 2010; Simeone et al., 2016). Colours as in Figs. 1, 2A and File S1. Numbers 1–4 indicate the four major clades identified. Branch bootstrap support (1–100) is scaled as circles of increasing size (see also File S4 for details on clone labels and bootstrap values).
Of the three clones sequenced from individual ml27, a suspected Q. macrolepis × brantii hybrid, one was identical to another Q. macrolepis clone (individual ml08) and the other two clustered together with Q. brantii. Likewise, three and two of the five clones sequenced in sample tj08, a Q. trojana × Q. suber hybrid, clustered within the respective parental subtrees; the same applies to the five clones of the sample tj33, a supposed Q. libani × Q. trojana hybrid. Conversely, all the three clones sequenced in sample tj02, another tree determined as possible Q. libani × Q. trojana hybrid, clustered with Q. trojana.
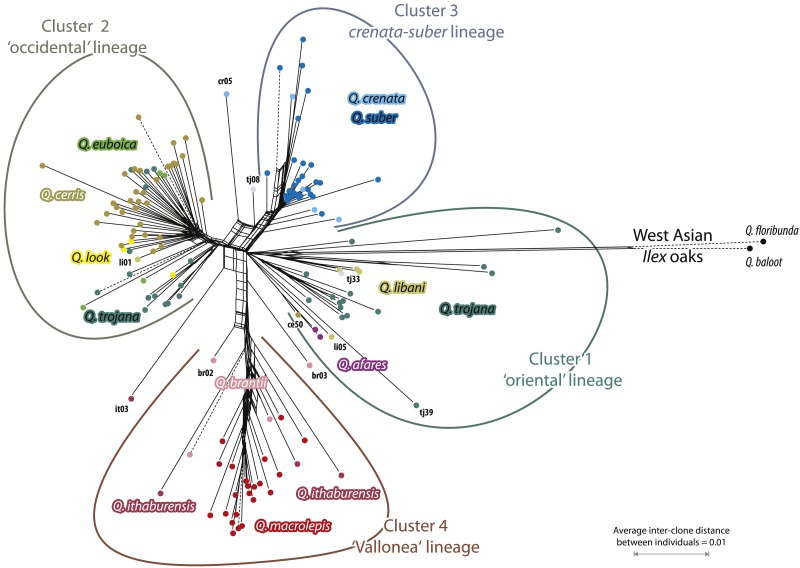
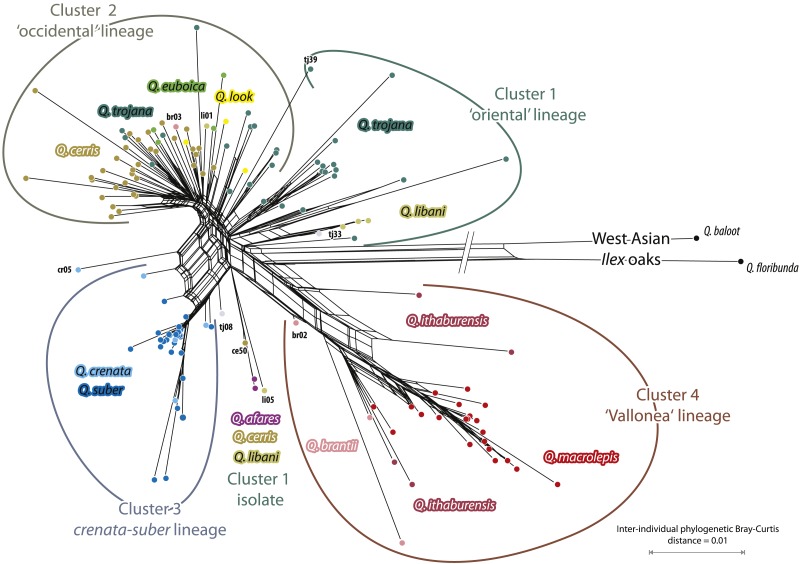
The networks based on transformed 5S-IGS data (Fig. 4 based on AVG-transformed uncorrected distances; Fig. 5 based on PBC-transformed distance matrix; only individuals represented by more than four clones included) largely confirmed the earlier found intra- and inter-species relationships (Denk & Grimm, 2010; because of the amount of shared identical clones, the MIN-transformed networks are largely collapsed, but included in the Online Supporting Archive). Four clusters can be observed: Cluster 1, the ‘oriental’ lineage of sect. Cerris, is the least coherent cluster and equivalent to a grade in a corresponding outgroup-rooted (Q. baloot, Q. floribunda) tree. This lineage included, in the AVG network, Q. afares, four out of five Q. libani individuals and about half of the Q. trojana individuals (Fig. 4). Its counterpart, Cluster 2, the ‘occidental’ lineage, accomodated all Q. look, Q. euboica, the remaining Q. trojana and Q. libani samples, and all but one Q. cerris individual (ce50; Figs. 4 and 5). The PBC network (Fig. 5) reveals a more gradual shift between these two clusters, with Q. afares splitting off with two genetically similar Q. cerris and Q. libani individuals (ce50, li05). The reason for this is that the PBC transformation has a higher chance to capture evolutionary signals (Göker & Grimm, 2008). Cluster 3 included Q. suber and Q. crenata. Here, the only difference is the boxyness inflicted by individuals cr05 and tj08. Cluster 4 included the ‘Vallonea’ (or Aegilops) oaks, Q. brantii, Q. ithaburensis and Q. macrolepis, with two Q. brantii individuals (br02 and br03, with diverging 5S-IGS features and variants; File S4) in proximal (br02 in Figs. 4 and 5; br03 in Fig. 4) or off-cluster (br03 in Fig. 5) position. Thus, the basic structure of the AVG and PBC networks and the ML tree are equivalent, but they differ in placing the outgroup taxa, and the networks refine inter-species relationships.
Figure 4. Network based on transformed 5S-IGS data showing inter-individual average (AVG) clonal distance relationships.
Only individuals represented by more than four clones are included (reconstructions for other cut-offs, m = 2, 3, or 5, are included in the File S5; see also our Online Supplementary Archive).
Figure 5. Network based on transformed 5S-IGS data showing inter-individual PBC clonal distance relationships.
Only individuals represented by more than four clones are included (reconstructions for other cut-offs, m = 2, 3, or 5, are included in our Online Supplementary Archive).
The AVG network (Fig. 4) better captured putative hybrid and intrograded individuals. Strong ambiguous signals came from the hybrid Q. trojana × Q. suber (tj08), one Q. crenata individual (cr05, terminals in the box-like structure connecting the ‘occidental’, cluster 2, and crenata-suber lineage, cluster 3), and one Q. ithaburensis individual (it03, terminal in the box-like structure between clusters 2 and 4). The placement of one Q. libani individual (li01, inserted in cluster 2), with normal-long variants in the clone sample, and one Q. cerris (ce50, in cluster 1 close to the Q. afares subgroup; cf. Fig. 5) does not follow the general trend. Long terminal edges indicative of unique individual clone samples (combinations) are found in each cluster. Besides the outgroup Q. baloot and Q. floribunda, these samples include individuals of Q. cerris (ce29, 44), Q. trojana (tj03, 16, 24, 39, 45), Q. suber (su07, 29, 49), Q. brantii (br06), Q. ithaburensis (it04, 05), and Q. macrolepis (ml10). Some of these samples had unique deletions or highly divergent regions in their clones (e.g., it04, ml10, tj03, tj16; see above). The networks produced with individuals represented by ≥2, ≥3 (and ≥5) clones did not change this structure (File S5); they allowed inclusion of all individuals into the four clusters matching the general scheme and pinpointed a few other (possible) exceptions. Quercus castaneifolia (represented by two clones) and one sample of Q. crenata (cr04; three clones) formed part of the ‘occidental’ lineage, cluster 2; one sample of Q. suber (su09; three clones) was included in cluster 1, the ‘oriental’ lineage of Sect. Cerris (see Table 4); sample br01 (three clones) was placed at the root of cluster 4, similarly to samples br02 and br03. The geographical distribution of the four clusters is shown in Fig. 2C.
In contrast, the PBC network (Fig. 5) provided a better basis for inferring the evolution and differentiation (speciation) processes. The ‘oriental’ (cluster 1) and ‘occidental’ lineages are clearly connected and form a continuum, with the easternbound Q. trojana and Q. libani representing a diverged, differentiated pool from which the other species and western Q. trojana derived. The western Mediterranean crenata-suber lineage is clearly different and only linked to the main pool by occasional introgression or hybridisation with nearby members of the ‘occidental’ lineage (in nature: Q. cerris). The same holds even more for the ‘Vallonea’ oaks (Cluster 4), which appear to have split before the remainder of western Eurasian Cerris (but long-branch/-edge attraction with the extreme long-edged outgroup needs to be considered). A clear signal in the PBC network (Fig. 4) is the uniqueness of Q. afares, a disjunct outpost of the putative ‘oriental’ lineage, genetically closely related to geographically very disjunct (C./S. Anatolian) individuals of Q. cerris and Q. libani (cf. Fig. 3 showing a Q. afares subclade, and File S4, same tree with clones labelled).
Discussion
The western Eurasian members of sect. Cerris exhibit a trnH-psbA diversity well comparable with the Mediterranean oaks of sect. Ilex (Vitelli et al., 2017) and Fagaceae in general (Simeone et al., 2016). As discussed in Grímsson et al. (2016), the plastid genealogy in this genus is largely decoupled from species identity. Nevertheless, the strong geographic signal of plastid data provides useful information to decipher population-area relationships and taxon histories (e.g., isolation, reticulation, introgression; cf. Pham et al., 2017). Conversely, the intergenic spacers of the 5S rDNA were confirmed as the most variable nuclear gene region for a large range of plants (Volkov et al., 2001; Forest et al., 2005; Lehtonen & Myllys, 2008; Denk & Grimm, 2010; Grimm & Denk, 2010). They were highly variable across the entire dataset and displayed inter-individual patterns that allowed circumscription of most of the investigated species; the intra-individual variation in the 5S-IGS further helped to recognize hybridization and infer other reticulation events such as introgression. Insights gained from the combination of data sources were concordant with the known ecology and biogeography of the studied taxa. So far, all other gene regions sequenced in oaks showed (much) less divergence at the intra- and interspecies level (ITS: Denk & Grimm, 2010; LEAFY intron: Oh & Manos, 2008; various single-copy nuclear genes: Hubert et al., 2014; various plastid intergenic spacers and genes: Simeone et al., 2013). Increased resolution may be achieved by using phylogenomic data (e.g., McVay, Hipp & Manos, 2017) but it would be more laborious and cost-intensive for large intra- and interspecific data sets.
Molecular recognition of species and species diversity in Quercus section Cerris
Widespread species such as Q. cerris and (to a lesser extent) Q. libani (Table 1) showed the highest plastid diversity in terms of number of haplotypes and parameters of molecular differentiation (Tables 2 and 3); Q. suber diversity was inflated by the occurrence of few divergent haplotypes linked to—and possibly captured from—the ‘Euro-Med’ lineage of sect. Ilex.
The strikingly high haplotype richness of Q. cerris, especially in the eastern part of this species’ range (cf. Bagnoli et al., 2016), mirrors the high morphological plasticity of this oak (many different varieties and subspecies are traditionally reported; for example, IOPI lists 30 formae and 17 varieties) and its ecological adaptability. Likewise, Q. cerris also displayed a high nuclear (5S-IGS) diversity (Table 4, Fig. 3). This oak has the largest range and broadest climatic envelope (from perhumid Cfa, Cfb via summer-dry warm temperate climates to BSk) and it is the only species of sect. Cerris naturalized on the British Islands (Cfb) and cultivated all over continental Europe (mostly Cfb, sheltered Dfb). Indeed, establishment of a large range across the geologically and ecologically dynamic West Eurasian region might have provided many opportunities for diversification, isolation, drift, conservation of variants and eventual reticulation with sibling species.
Quercus suber, instead, displayed the lowest number of unique 5S-IGS variants (like Q. macrolepis) with a low diversity (Tables 4 and 5), which might indicate ongoing genetic erosion (possibly due to the species domestication for cork, tannins, wood and fruits exploitation). In view of the high species-coherence detected in the other conspecific samples, the few Iberian samples not following the general trend may indicate introgression of Q. suber into Q. ilex (individuals with ‘Euro-Med’ plastid haplotype H11) or reflect ancient reticulation and retention of ancestral signatures (individuals with the widespread, putatively ancestral H1 haplotype).
In Q. libani, the high haplotype diversity coincides with a moderate diversity at the 5S-IGS locus, characterized by interesting variation among the cloned sequences. Two individuals (li01, li05) show potential introgression (e.g., with sympatric Q. trojana samples tj05, tj35; Files S1 and S4). Alternatively, they might represent an ancestral line of diversification within the section (the short 5S-IGS variant so far only known for Q. libani, see Denk & Grimm (2010), also occurs in clones of two non-sympatric Q. cerris individuals). Together with Q. brantii, Q. libani is the easternmost oak among the western Eurasian Cerris, and it is extremely variable at the morphological and ecological level (occurring in climates ranging from Csa to Dsb). For instance, Djavanchir-Khoie (1967) described up to 12 intra-specific taxa within Q. libani, and introgression phenomena with other co-occurring oaks have been postulated (Menitsky, 2005; Khadivi-Khub et al., 2015). Likewise, in the nearby region of Iranian Kurdistan, this oak showed three distinct gene pools based on nuclear microsatellites (Khadivi-Khub et al., 2015).
Based on the phylogenetic reconstructions, the 5S-IGS sequence diversity detected in Q. trojana outmatched the high levels displayed by Q. cerris, rendering it the genetically least-coherent species of the section. Samples with highly diverse clones were detected in both species (see ‘Results’), but individuals of Q. trojana can be found in two different clusters in the 5S-IGS network. Reflecting its more limited distribution and climatic niche (Csa, Csb), the haplotype diversity is substantially lower in Q. trojana s.l. (Q. trojana + Q. euboica) than in Q. cerris (Tables 2 and 3). This finding indicates that the two species retain exceptionally high intra- and inter-individual variability, possibly conserving ancestral variants lost in more homogenized and/or geographically restricted species of sect. Cerris, and corresponds, at the plastid level, to the relative extension of their geographic (and ecological) ranges. Accordingly, the two endemic taxa Q. afares and Q. castaneifolia appeared the least diverse (Tables 4 and 5), although more data are needed for Q. castaneifolia, which was here represented by only two samples. The same holds true for the two other taxa with narrow ranges in our dataset, Q. euboica and Q. look. Both are characterized by low levels of genetic diversity (Tables 4 and 5); however, despite the low number of individuals investigated, our results allow first taxonomic inferences in both cases.
Quercus euboica appears genetically isolated from Q. trojana, based on the number of unique 5S variants (Table 4) and the relative inter-taxa divergence (Table 5). This oak grows isolated from Q. trojana on the Greek island of Euboea and differs morphologically by its coriaceous leaf texture and the conspicuous white tomentum of the abaxial leaf surface that is made up of stellate trichomes (T Denk, pers. obs., 2008 and 2017). In addition, Q. euboica is characterized by special edaphic conditions, growing on serpentine rocks. All these data indicate that the Euboean oak should be better considered as an independent species requiring special protection. Another (hairy) variant of Q. trojana has been locally described at the south-eastern margin of the species’ range, in South-central Turkey (Q. trojana subsp. yaltirikii; Zielinski, Petrova & Tomaszewski, 2006). Some samples collected in the nearby area (tj03, 04, 05, 16) showed 5S-IGS clones with a unique, highly divergent motif, and grouped (mostly) in a specific sublcade (Fig. 3, File S4). However, more (morpho-ecological) data are needed to implement the description of Q. trojana in this part of its range.
The Q. look samples showed a distinct plastid-nuclear signature combination (Tables 2–4; File S4) linking it with Q. cerris (Figs. 3–5). In addition, this species showed the lowest mean estimate of evolutionary divergence of the nuclear 5S IGS together with Q. castaneifolia (Table 5). Although the taxonomic rank of this rare, enigmatic taxon cannot be yet established with certainty (a hybrid origin or a local diversification of an ancestral form of Q. cerris seem equally probable), the two previous assessments of this oak as synonym of Q. ithaburensis or Q. ithaburensis × Q. libani hybrid (Table 1) can be rejected. Additional investigations are required to evaluate if the morphology of Q. look justifies its exclusion from the genetically and morphologically variable Q. cerris.
Finally, a distinct group with medium-high levels of plastid and nuclear diversity includes Q. brantii, Q. ithaburensis, and Q. macrolepis (Tables 2, 4 and 5; ‘Aegilops oaks’) with a range centred in the Csa climates of the central-eastern Mediterranean region, providing a low-land analogue to the situation in Q. trojana-euboica-libani. The Aegilops oaks are a highly specialized group, morphologically and ecologically well distinct from the other oaks in the Cerris section (Menitsky, 2005). The detected genetic diversity at the plastid and, especially, the nuclear markers (Table 5) clearly indicates the genetic isolation from the rest of the Cerris oaks and progressive inter-specific differentiation. Geographic, morphological and ecological differences are also evident in Q. ithaburensis and Q. macrolepis (Dufour-Dror & Ertas, 2002; Dufour-Dror & Ertas, 2004). On these grounds and considering the high inter-taxon 5S-IGS divergence supported by the different haplotypes (Tables 2–4), we suggest these two forms be treated as separate species (cf. Denk et al., 2017, appendix: http://dx.doi.org/10.1101/168146). Interestingly, Q. brantii appeared as the most diverse of the three taxa (Tables 4 and 5), and displayed some 5S-IGS variants shared with Q. cerris and Q. suber (File S3), which might indicate the occurrence of ancestral traits.
Hybrid detection within Quercus Section Cerris
Besides the deletion in Q. libani, some other sequence features, typical of other species (e.g., the SSR motif in Q. brantii, the deletion in Q. suber), confirmed the hybrid identity of a few samples included in our dataset (sample ml27, tj08 and tj33, supposed hybrids Q. macrolepis × brantii, Q. trojana ×Q. suber and Q. trojana × Q. libani, respectively). The haplotypes of these samples (ml27: H6, exclusive of Q. brantii; tj08: H2, never found in Q. suber), also allowed identification of the maternal species. This finding confirms that these oaks can occasionally hybridize in sympatry, and evidence for such hybridization is found in the nuclear genome (see also Fitzek et al., 2018).
Further instances of hybridization and/or introgression events could be inferred from common sequence features, inter-specifically shared variants (see ‘Results’ section, Table 4, Fig. 3, File S3), and the placement of individuals in the AVG Neighbour-Net (Fig. 4), mostly involving Anatolian samples of Q. cerris, Q. brantii, Q. libani, Q. macrolepis and Q. trojana. However, in many cases the involved individuals grew hundreds to a few thousands of kilometres from each other. Based on their (relative) spatial proximity, introgressions could be suggested for samples Q. brantii (br02; C. Anatolia) and Q. macrolepis (e.g., ml20–22; W. and S. Anatolia) sharing sequence features, and South Anatolian Q. libani (li01) and Q. trojana (e.g., tj05, tj35) sharing sequence features and trnH-psbA haplotypes. Outside Anatolia, evidence of reticulation (shared variants) can be traced between Israelian Q. cerris and Q. look (sample ce38, lk03) and between Balkan Q. cerris and Q. suber (sample ce43, Serbia; su37, Croatia). Introgression and past hybridization events between all these species or their precursors is a possible explanation. At the same time, retention of ancestral traits cannot be discarded, as Q. cerris (5S-IGS, trnH-psbA) and Q. trojana (5S-IGS) cover most variability found in sect. Cerris and are highly variable, especially in Anatolia.
In this context, the unresolved taxonomic status of Q. crenata can be discussed in view of the present results (Figs. 3–5). The species is more closely related to Q. suber than to Q. cerris and part of the distinct Q. crenata-suber lineage. The relative distribution of the clones (in the Q. cerris- and in the Q. suber-dominated clades) and the large extent of variants shared with Q. suber can be interpreted as evidence of co-existing both (1) Q. cerris × Q. suber F1 hybrids and (2) introgressive forms into either Q. cerris (North East Italy/Balkans) or Q. suber (Italian peninsula), in partial agreement with Conte, Cotti & Cristofolini (2007). However, all forms would look phenotypically quite similar (intermediacy of habitus, leaf and bark shape between Q. suber and Q. cerris is traditionally used as a diagnostic character of Q. crenata), which seems inconsistent with their presumable different genome composition. Clearly, the hybrid/introgressed phenotypes can be affected by several phenomena such as segregation, epistasis, heterosis, and maternal origin (Rieseberg & Ellstrand, 1993). At the same time, we note that the diagnostic traits used for Q. crenata occur in other species of sect. Cerris (corky bark: Q. afares, Q. variabilis, various forms of Q. cerris; Menitsky, 2005; semi-evergreen habitus: Q. trojana and Q. libani; Yaltırık, 1984; crenate leaves: part of the morphological variation of Q. cerris and Q. suber). Also, Q. cerris shares plastid and nuclear signatures with other species of sect. Cerris, including geographically isolated, morphologically distinct taxa such as Q. afares, Q. castaneifolia, Q. euboica (traditionally included in Q. trojana), and Q. look (traditionally included in Q. ithaburensis), which are part of the ‘occidental’ lineage (see ‘Results’). Besides occasional hybridizations (sample cr05; Fig. 4), the alternative explanation is that Q. crenata represents a less-derived species possessing a limited gene pool within an autonomous evolutionary lineage including Q. suber (Fig. 5). Being closer to the common root, it retained imprints of common origin, possibly ancient reticulation, with the (proto-)Q. cerris (‘occidental’) lineage, representing a geographic-evolutionary gradient (‘oriental’ lineage → ‘occidental’ lineage → crenata-suber lineage). Quercus crenata may then just represent the remainder of the ancestral form from which Q. suber evolved rather than being the product of secondary contact between Q. cerris and Q. suber.
We also found no evidence to support the hybrid origin of Q. afares (Q. suber x Q. canariensis; the latter is a member of sect. Quercus) as suggested by Mir et al. (2006) based on cpDNA-RFLP and allozymes (cf. Welter et al., 2012; Mhamdi et al., 2013). 5S-IGS variants and plastid signatures of western Eurasian white oaks (‘roburoid oaks’ in Denk & Grimm, 2010; see also Simeone et al. 2016, Fig. 1) are very distinct from Cerris types and should be detectable unless the F1 hybrids, with Q. suber as a maternal parent, only backcrossed with the local Q. suber but not Q. canariensis. However, genetic exchange with local Q. suber can be excluded, since no Q. suber-typical 5S-IGS variants, or obvious phylogenetic linkage with the Q. crenata-suber lineage, were found in Q. afares. Ongoing next-generation target sequencing of the 5S-IGS region (producing several 10,000 5S-IGS sequences per sample/individual) showed, so far, no evidence for a clone-sampling artefact in the studied individuals of Q. afares. The possibility of incomprehensive clone-sampling can thus be discarded. Analoguous to Q. crenata, the ancestry level of Q. afares may explain earlier findings interpreted towards a hybrid origin: being much closer to the common ancestor of sect. Cerris than Q. suber, this species may have retained (some) genetic imprints today found in members outside its section. This would explain also the association of Q. afares with two other, geographically very distant, individuals of the ‘oriental’ lineage (ce50, li05, the only Q. libani without the ‘short’ libani 5S-IGS variants but showing variants similar to Anatolian Q. trojana).
Aside from putative hybrid species and swarms (see also the ambiguous placement of sample it03 the 5S-IGS network), our data demonstrates a general permeability of species boundaries in members of Sect. Cerris, allowing occasional crosses. Indeed, our results demonstrate that Q. trojana and Q. libani, Q. brantii and Q. macrolepis, Q. cerris and Q. suber, and Q. trojana and Q. suber are interfertile and hybridize in the wild. Further investigations (e.g., with metagenomics approaches, fine-scale geographic samplings) are needed to distinguish between ancient hybridization with subsequent incomplete lineage sorting and retention of ancestral traits, to clarify the status of several other samples that may represent both phenomena based on shared 5S-IGS variants, the occurrence of unique sequence features, length of terminal branches and odd-placing in the phylogenetic reconstructions. Adequately addressing these issues would be of great relevance to identify relict populations and/or past contact/hybrid zones, to assess the hybridization ability of species growing in sympatry, and further define the evolutionary history of the Cerris oaks.
Taxonomic framework of Quercus section Cerris in western Eurasia
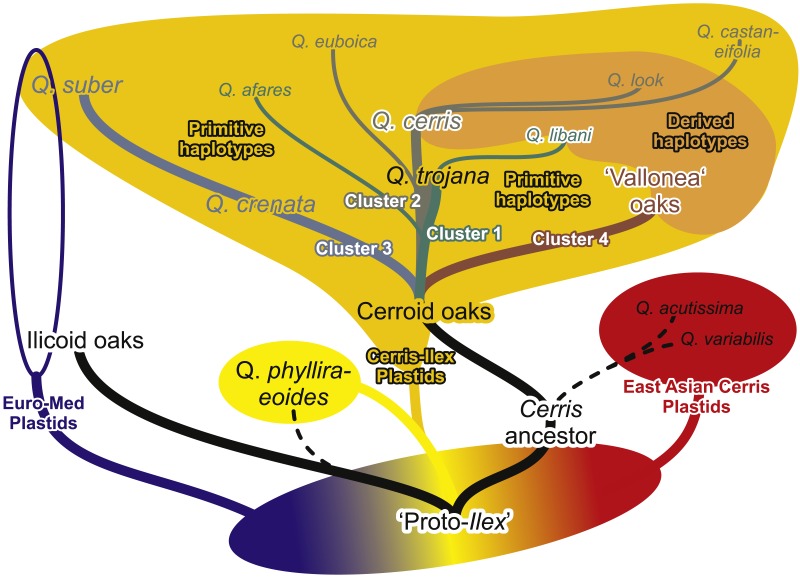
From the clusters identified by the 5S-IGS network based on the average inter-individual clone (Fig. 4) and PBC-transformed distances (Fig. 5), four major groups can be identified representing distinct evolutionary lineages and used as a framework. Figure 6 shows a scheme, a cactus-type branching silhouette (Podani, 2017; Morrison, 2018), based on the 5S-IGS and trnH-psbA differentiation patterns, and with respect to the plastid tree provided in Simeone et al. (2016).
Figure 6. Mixed branching silhouette-tree doodle depicting the molecular differentiation and evolution in Quercus Section Cerris.
The evolutionary or genealogical lineages are indicated by branches (accordingly labelled and coloured), the fields represent shared or unique gene pools. The deep incongruence between plastid genealogies and nuclear-morphological phylogenetic lineages can only be explained by ancient reticulation and incomplete lineage sorting during the formation and isolation of the modern-day lineages following the break-up of the ancestral gene pool (tentatively labelled as ‘proto-Ilex’).
The first, most western lineage includes Q. suber and Q. crenata. Sample cr05 likely represents a F1 hybrid with Q. cerris. The same holds for tj08, a Q. trojana x Q. suber hybrid. A second lineage, tentatively termed the ‘occidentalis’ lineage includes the widespread Q. cerris and the geographically restricted Q. castaneifolia, Q. euboica and Q. look. The third lineage collects the Eastern Mediterranean ‘Vallonea’ oaks Q. brantii, Q. ithaburensis, and Q. macrolepis, with three potential outliers: br02, br03 and it03 (possible recent or ancient hybrids). The last, least coherent group, the ‘oriental’ lineage, includes Q. afares, Q. libani and Q. trojana. Aside from supposed hybrids and introgressed individuals (see section above), these groups almost perfectly match previous taxonomic observations (sect. Heterobalanus subsect. Suber + sect. Cerris subsect. Cerris and Aegilops; Menitsky, 2005; sect. Suber + sect. Eucerris, Aegilops and Erythrobalanus; Schwarz, 1936–1939), with the only exceptions of Q. afares, accomodated by both authors within the Q. cerris-Q. castaneifolia group, and Q. crenata and Q. look that were not included in previous monographs.
Quercus trojana is the only species scattered over two clusters (cluster 1 and 2, Figs. 4 and 5; see also Denk & Grimm, 2010), thus bridging between the ‘oriental’ and ‘occidental’ lineage. No geographic, haplotypic or subspecific relationships could explain this subdivision; it might therefore indicate the occurrence of two different, geographically overlapping but genetically isolated lineages within the species, possibly differentiated in the past (retained ancient polymorphism), especially considering the proximal positions of the sequences of samples tj24, 39 and 45 in the ML tree. Both Q. trojana (s.str.) sublineages occur in Italy and might correspond to the two main nuclear gene pools identified by Carabeo et al. (2017). Indeed, more ecological and molecular data are required to interpret this finding biologically.
Overall, the complex genetic differentiation patterns can only be explained by longer ongoing free genetic exchange or more recent common origin of the ‘oriental’ and ‘occidental’ lineages than in case of the other two lineages within western Eurasian Cerris: the corkish oaks (Q. crenata + Q. suber) and the ‘Vallonea’ (or Aegilops) oaks (Q. brantii, Q. ithaburensis, and Q. macrolepis). Quercus euboica possibly originated by geographical isolation from a proto-trojana still close to the proto-cerris (Fig. 6). Interestingly, all the Q. euboica samples occurred in the Q. cerris- dominated ‘crown’ clade 4 (Fig. 3), hence, are part of the ‘occidentalis’ lineage. Likewise, the microspecies Q. afares, Q. castaneifolia, and Q. look as well as the eastern replacement of Q. trojana, Q. libani, were isolated from the master cerris-trojana genepool(s). It is impossible to provide absolute dates for the final isolation events. Comparison with intra- and inter-species 5S-IGS divergence in the other two main lineages of western Eurasian oaks (‘ilicoid’ oaks of sect. Ilex, 5S-IGS is species-diagnostic; ‘roburoid’ oaks of sections Quercus and Ponticae, 5S-IGS is largely undiagnostic; Denk & Grimm, 2010) indicates that the original split possibly predates diversification in the ‘roburoid’ oaks. The final establishment of species in western Eurasian members of Sect. Cerris is however as young as in the ‘roburoid’ oaks, may be ongoing (Q. trojana), and younger than the main split within the ‘ilicoid’ oaks, i.e., of Q. ilex from Q. aucheri (+Q. coccifera).
Phylogeography of Quercus section Cerris in western Eurasia
The primitive (L1) and derived (L2) haplotype groups of ‘Cerris-Ilex’ lineage, and two haplotypes representing the ‘Euro-Med’ lineage characteristic for western populations of sect. Ilex (Simeone et al., 2016; Vitelli et al., 2017), describe the main evolutionary trajectories of the western Eurasian lineage of sect. Cerris and their contact with already established lineages of sect. Ilex. According to coalescent theory, the most frequent and widespread haplotypes, i.e., H1 and H2, are likely ancestral (Posada & Crandall, 2001; Fig. 1). The close relationship between haplotypes H1 and H2–found all across western Eurasia (Fig. 2B) and in all taxa except the south-eastern species group, Q. brantii, Q. look, and Q. ithaburensis—and Q. phylliraeoides (Fig. 5; cf. Simeone et al., 2016, Fig. 1), the only species of section Ilex extending into Japan—points towards a north-eastern Asian origin of sect. Cerris and a westward migration of a large population into the Mediterranean region. The revised (see Introduction) fossil record of Cerris in (North-)East Asia (starting from early Oligocene) predates earliest records in western Eurasia (Oligocene/Miocene boundary) by ca. 10 Ma, thus rejecting the hypothesis that sect. Cerris evolved from the western stock of sect. Ilex populations with ‘WAHEA’ haplotypes (Denk & Grimm, 2010; Simeone et al., 2016). The effective population size of the early west-migrating Cerris must have been (very) large in contrast to their East Asian siblings. The East Asian species of sect. Cerris are more heterogenous (Chen et al., 2012; Zhang et al., 2015), and differ much more profoundly from Q. phylliraeoides (the Japanese Ilex oak), but also from the ‘WAHEA’ haplotypes of sect. Ilex and Q. baroni (Fig. 1), considered early diverged plastid lineages of subgenus Cerris (‘Old World’ or mid-latitude clade; the earliest diverged plastid lineage being the western Mediterraenan ‘Euro-Med’ type found in Q. ilex; Fig. 5; Simeone et al., 2016). In this context, assessment of the 5S-IGS diversity in (East) Asian members of sections Cerris and Ilex would be needed, to check whether the Asian counterparts are equally coherent or more diverse than their western Eurasian relatives.
Once established in the Mediterranean region (H1, H2; Fig. 2B), local bottlenecks may have contributed to increased genetic drift in the plastome in the eastern part of the range. A likely trigger are the complex orogenies shaping modern-day Turkey and the Levant, areas with an increased haplotype diversity including the most derived ‘Cerris-Ilex’ haplotypes (Figs. 1–2). This, and the general west-east differentiation pattern (see also Figs. 4 and 5), parallels the situation in sect. Ilex, Q. coccifera in particular (Vitelli et al., 2017). A notable difference to sect. Ilex is the lack of plastid structuring (and diversity) in the central and western Mediterranean region, indicating a rather recent, singular colonization by the master population, clearly not affected by Oligocene micro-plate tectonics as suggested for Q. suber by Magri et al. (2007).
The derived L2 ‘Cerris-Ilex’ haplogroup (H5–H10) starts in Anatolia and extends further east (Iran) and south (Levant). In addition to isolation during range establishment, specialization to drier climates (e.g., summer-dry Mediterranean climates: Csa, Csb, Dsb) can be considered as trigger for increased genetic drift, possibly linked to speciation. The Aegilops oaks, Q. brantii, Q. ithaburensis, and Q. macrolepis, a well-circumscribed group based on 5S-IGS differentiation (Figs. 4 and 5) and morphology, are unique by showing only derived ‘Cerris-Ilex’ haplotypes.
A remarkable exception are two Italian Q. cerris individuals showing the derived haplotypes H9/H10, which occur in locations more than 2,000 km apart from other individuals of this Levantine haplotype sublineage. H9/H10 derive from types found in Anatolia and eastwards (Figs. 1 and 2). Long-distance seed dispersal is highly unlikely. The main animal vector for propagation of oaks are the jaybirds, which are sedentary birds, with a short evasion range (<50 km; Pesendorfer et al., 2016). Man-mediated dispersal (in historic times) could be a likely explanation, although we note that haplotypes shared by disjunct central Mediterranean and the Anatolian regions were also found in Q. ilex, and possibly reflect the remnants of a pre-Quaternary continuous range.
In this context, the genetic diversity detected in the Italian Q. trojana populations (both at the nuclear and at the plastid level) and the very limited, amphi-Adriatic distribution of haplotype H2 in Q. macrolepis (Italy, Albania; Fig. 2, File S1) likely confirm that these oaks are native in Italy. Similar close intra-specific phylogeographic relationships have been detected in other plant species on both sides of the Adriatic Sea (Musacchio et al., 2006; Hilpold et al., 2014), including oaks (Lumaret et al., 2002; Fineschi et al., 2002; López de Heredia et al., 2007; Bagnoli et al., 2016). In this case also, the Apulian populations of Q. trojana and Q. macrolepis can be interpreted as the remnants of a once continuous ancestral range (Simeone et al., 2016), or witness a colonization wave that was likely favoured by land connections between the Balkans and southeastern Italy during the Messinian salinity crisis and (or) the Pleistocene glaciations (Nieto Feliner, 2014).
Conclusion
The present study is the first to include all putative species of Quercus sect. Cerris in western Eurasia. Our investigation is based on a dense intra-specific and geographic sampling and makes use of DNA sequence variation of the two most divergent nuclear and plastid regions known for oaks. Although based on just two markers, the obtained results confirm and emend species relationships and the genetic coherence of taxa. An updated subsectional classification of the western Eurasian Cerris oak species is proposed, with the identification of four major lineages, corresponding to subsectional groups that would need to be formalized. Some intraspecific taxa are recognized as distinct species (i.e., Q. macrolepis and Q. euboica) and the systematic relationships of Q. look are clarified. Although we observed the occurrence of occasional F1 hybrids, possible intrograded individuals and several potential outlier individuals across the studied range, we could not confirm the hybrid origin of Q. afares and Q. crenata. The fossil record corroborates major inferences about the origin and diversification of the section.
Characterizing nuclear and plastid differentiation across all species, including numerous individuals and the entire range, can only be the first step. Figure 6 summarizes our results, but also highlights phenomena deserving further investigation. Primarily, 5S-IGS data need to be compiled for (East) Asian members of sections Cerris and Ilex. A future focus should be on all Hindukush to western Himalayan species and the Japanese Q. phylliraeoides, the north-easternmost member of sect. Ilex, which has plastid signatures very similar to the western Eurasian members of sect. Cerris but not to the geographically closer East Asian species of sect. Cerris. The entire fossil record of sections Cerris and Ilex should then be recruited to infer age estimates, following the recent example of the genus Fagus (Renner et al., 2016). Another open question is where to root the nuclear tree (the polytomy in Fig. 6): our incomprehensive outgroup places the root within the crenata-suber portion of the 5S-IGS ML tree, which would mean that the ‘corkish’ oaks represent the first diverging lineage. This rooting hypothesis does not fit well with the structure of the PBC network and is in conflict with plastid and fossil evidence favouring a north-eastern origin of the section. A stepwise East to West invasion of sect. Cerris into the Mediterranean region is also supported by higher species and plastid diversity in the East Mediterranean. It is possible that the westernmost ancestral populations of the cerroid oaks, carrying the common haplotype, went through a relatively recent bottleneck resulting in unique and distinct 5S-IGS variants. These distinct 5S-IGS variants would then be attracted to any possible (distant) outgroup when inferring a tree (ingroup-outgroup branching artefact; cf. position of outgroups in Fig. 4). Finally, Q. cerris should be investigated in detail across its entire range using a combination of morphometric and high-resolution genetic analysis to elucidate its relationships with sympatric species of sect. Cerris and the isolated endemisms. This will allow us to test whether Q. cerris is a primal genetic and ecological resource of the section in western Eurasia and carrier of ancestral signals.
Supplemental Information
We also report the accession numbers of the sequences retrieved from GenBank and used to complement the investigated dataset.
incuding results of the BLAST identity matches on GenBank.
This list reports all the identical 5S-IGS cloned sequences in the investigated dataset. Colors according to species list.
The tree was rooted on Q. baloot and Q. floribunda. Taxa colors as in Fig. 1, 2a and File S1. Branch bootstrap support (1-100) is reported.
Individuals represented by more than 2, 3 and 5 clones are included (cut-offs specimens are indicated).
This Online Supporting Archive includes data and analysis files of Simeone et al. ”Comparative systematics and phylogeography of Quercus Section Cerris in western Eurasia: inferences from plastid and nuclear DNA variation” in several subfolders.
Funding Statement
This project was funded by a Swedish Research Council (VR) grant to Thomas Denk. Sequencing was made possible using funds from a VR ‘ForsAss’ grant (no. 2008–3726) to Guido W. Grimm. Guido W. Grimm has been funded by the Austrian Science Fund FWF ‘Lise-Meitner mobility’ grant (project no. M1751-B16). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Marco Cosimo Simeone conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Simone Cardoni performed the experiments, prepared figures and/or tables, approved the final draft.
Roberta Piredda analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Francesca Imperatori performed the experiments, contributed reagents/materials/analysis tools, approved the final draft.
Michael Avishai and Thomas Denk conceived and designed the experiments, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Guido W. Grimm conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
DNA Deposition
Data Availability
The following information was supplied regarding data availability:
Primary data and analyses are available as Supplemental Files.
References
- Altschul et al. (1990).Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bagnoli et al. (2016).Bagnoli F, Tsuda Y, Fineschi S, Bruschi P, Magri D, Zehlev P, Paule L, Simeone MC, González-Martínez SC, Vendramin GG. Combining molecular and fossil data to infer demographic history of Quercus cerris: insights on European eastern glacial refugia. Journal of Biogeography. 2016;43:679–690. doi: 10.1111/jbi.12673. [DOI] [Google Scholar]
- Barak et al. (2016).Barak RS, Hipp AL, Cavender-Bares J, Pearse WD, Hotchkiss SC, Lynch EA, Callaway JC, Calcote R, Larkin DJ. Taking the long view: integrating recorded, archeological, paleoecological, and evolutionary data into ecological restoration. International Journal of Plant Sciences. 2016;177:90–102. doi: 10.1086/683394. [DOI] [Google Scholar]
- Bellarosa et al. (2005).Bellarosa R, Simeone MC, Papini A, Schirone B. Utility of ITS sequence data for phylogenetic reconstruction of Italian Quercus spp. Molecular Phylogenetics and Evolution. 2005;34:355–370. doi: 10.1016/j.ympev.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Browicz & Zieliński (1982).Browicz K, Zieliński J. Chorology of trees and shrubs in South-West Asia and adjacent regions. Vol. 1. Polish Scientific Publishers; Warsaw: 1982. [Google Scholar]
- Bryant & Moulton (2004).Bryant D, Moulton V. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Molecular Biology and Evolution. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Burgarella et al. (2009).Burgarella C, Lorenzo Z, Jabbour-Zahab R, Lumaret R, Guichoux E, Petit R, Soto A, Gil L. Detection of hybrids in nature: application to oaks (Quercus suber and Q. ilex) Heredity. 2009;102:442–452. doi: 10.1038/hdy.2009.8. [DOI] [PubMed] [Google Scholar]
- Burger (1975).Burger WC. The species concept in Quercus. Taxon. 1975;24:45–50. doi: 10.2307/1218998. [DOI] [Google Scholar]
- Camus (1936–1954).Camus A. Les chênes. Monographie du genre Quercus et monographie du genre Lithocarpus. Lechevalier; Paris: 1936–1954. (Encyclopédie Economique de Sylviculture, vol. VI, VII, VIII). [Google Scholar]
- Carabeo et al. (2017).Carabeo M, Simeone MC, Cherubini M, Mattia C, Chiocchini F, Bertini L, Caruso C, La Mantia T, Villani F, Mattioni C. Estimating the genetic diversity and structure of Quercus trojana Webb populations in Italy by SSRs: implications for management and conservation. Canadian Journal of Forest Research. 2017;47:331–339. doi: 10.1139/cjfr-2016-0311. [DOI] [Google Scholar]
- Cavender-Bares et al. (2015).Cavender-Bares J, Gonzalez-Rodriguez A, Eaton DAR, Hipp AAL, Beulke A, Manos PS. Phylogeny and biogeography of the American live oaks (Quercus subsection Virentes): a genomic and population genetics approach. Molecular Ecology. 2015;24:3668–3687. doi: 10.1111/mec.13269. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2012).Chen DM, Zhang XX, Kang HZ, Sun X, Yin S, Du HM, Yamanaka N, Gapare W, Wu HX, Liu C. Phylogeography of Quercus variabilis based on chloroplast DNA sequence in East Asia: multiple glacial refugia and Mainland-migrated island populations. PLOS ONE. 2012;7:e47268. doi: 10.1371/journal.pone.0047268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2017).Chen J, Zeng Y-F, Liao W-J, Yan P-C, Zhang J-G. A novel set of single-copy nuclear gene markers in white oak and implications for species delimitation. Tree Genetics and Genomes. 2017;13:50. doi: 10.1007/s11295-017-1130-3. [DOI] [Google Scholar]
- Conte, Cotti & Cristofolini (2007).Conte L, Cotti C, Cristofolini G. Molecular evidence for hybrid origin of Quercus crenata Lam. (Fagaceae) from Q. cerris L. and Q. suber L. Plant Biosystems. 2007;141:181–193. doi: 10.1080/11263500701401463. [DOI] [Google Scholar]
- Deng et al. (2018).Deng M, Jiang X-L, Hipp AL, Manos PS, Hahn M. Phylogeny and biogeography of East Asian evergreen oaks (Quercus section Cyclobalanopsis; Fagaceae): insights into the Cenozoic history of evergreen broad-leaved forests in subtropical Asia. Molecular Phylogenetics and Evolution. 2018;119:170–181. doi: 10.1016/j.ympev.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Denk & Grimm (2010).Denk T, Grimm GW. The oaks of western Eurasia: traditional classifications and evidence from two nuclear markers. Taxon. 2010;59:351–366. [Google Scholar]
- Denk et al. (2017).Denk T, Grimm GW, Manos PS, Deng M, Hipp AL. An updated infrageneric classification of the oaks: review of previous taxonomic schemes and synthesis of evolutionary patterns. In: Gil-Pelegrín E, Peguero-Pina J, Sancho-Knapik D, editors. Oaks physiological ecology. Exploring the functional diversity of genus Quercus L. vol. 7. Springer; Cham: 2017. (Tree physiology). [Google Scholar]
- Djavanchir-Khoie (1967).Djavanchir-Khoie K. Ph.D thesis. 1967. Les chenes de l’Iran; p. 221. [Google Scholar]
- Dufour-Dror & Ertas (2002).Dufour-Dror JM, Ertas A. Cupule and acorn basic morphological differences between Quercus ithaburensis Decne. subsp. ithaburensis and Quercus ithaburensis subsp. macrolepis (Kotschy) Hedge & Yalt. Acta Botanica Malacitana. 2002;27:237–242. [Google Scholar]
- Dufour-Dror & Ertas (2004).Dufour-Dror JM, Ertas A. Bioclimatic perspectives in the distribution of Quercus ithaburensis Decne. subspecies in Turkey and in the Levant. Journal of Biogeography. 2004;31:461–474. doi: 10.1046/j.0305-0270.2003.01036.x. [DOI] [Google Scholar]
- Fineschi et al. (2002).Fineschi S, Taurchini D, Grossoni P, Petit RJ, Vendramin GG. Chloroplast DNA variation of white oaks in Italy. Forest Ecology and Management. 2002;156:103–114. doi: 10.1016/S0378-1127(01)00637-5. [DOI] [Google Scholar]
- Fitzek et al. (2018).Fitzek E, Delcamp A, Guichoux E, Hahn M, Lobdell M, Hipp AL. A nuclear DNA barcode for eastern North American oaks and application to a study of hybridization in an Arboretum setting. Ecology and Evolution. 2018;8(11):5837–5851. doi: 10.1002/ece3.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forest et al. (2005).Forest F, Savolainen V, Chase MW, Lupia R, Bruneau A, Crane PR. Teasing apart molecular- versus fossil-based error estimates when dating phylogenetic trees: a case study in the birch family (Betulaceae) Systematic Botany. 2005;30:118–133. doi: 10.1600/0363644053661850. [DOI] [Google Scholar]
- Göker & Grimm (2008).Göker M, Grimm GW. General functions to transform associate data to host data, and their use in phylogenetic inference from sequences with intra-individual variability. BMC Evolutionary Biology. 2008;8:86. doi: 10.1186/1471-2148-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts & Frodin (1998).Govaerts R, Frodin DG. World checklist and bibliography of Fagales (Betulaceae, Corylaceae, Fagaceae and Ticodendraceae) Royal Botanic Gardens; Kew: 1998. [Google Scholar]
- Grimm & Denk (2010).Grimm GW, Denk T. The reticulate origin of modern plane trees (Platanus, Platanaceae)—a nuclear marker puzzle. Taxon. 2010;59:134–147. [Google Scholar]
- Grímsson et al. (2016).Grímsson F, Grimm GW, Zetter R, Denk T. Cretaceous and Paleogene Fagaceae from North America and Greenland: evidence for a Late Cretaceous split between Fagus and the remaining Fagaceae. Acta Palaeobotanica. 2016;56:247–305. doi: 10.1515/acpa-2016-0016. [DOI] [Google Scholar]
- Hilpold et al. (2014).Hilpold A, Vilatersana R, Susanna A, Meseguer AS, Boršić I, Constantinidis T, Filigheddu R, Romaschenko K, Suárez-Santiago VN, Tugay O, Uysal T, Pfeil BE, Garcia-Jacas N. Phylogeny of the Centaurea group (Centaurea, Compositae)—geography is a better predictor than morphology. Mol. Phylogenet. Evol. 2014;77:195–215. doi: 10.1016/j.ympev.2014.04.022. [DOI] [PubMed] [Google Scholar]
- Hipp et al. (2014).Hipp AL, Eaton DAR, Cavender-Bares J, Fitzek E, Nipper R, Manos PS. A framework phylogeny of the American oak clade based on sequenced RAD data. PLOS ONE. 2014;9:e93975. doi: 10.1371/journal.pone.0093975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp et al. (2018).Hipp AL, Manos PS, Gonzalez-Rodriguez A, Hahn M, Kaproth M, McVay JD, Valencia AS, Cavender-Bares J. Sympatric parallel diversification of major oak clades in the Americas and the origins of Mexican oak diversity. New Phytologist. 2018;217:439–452. doi: 10.1111/nph.14773. [DOI] [PubMed] [Google Scholar]
- Hubert et al. (2014).Hubert F, Grimm GW, Jousselin E, Berry V, Franc A, Kremer A. Multiple nuclear genes stabilize the phylogenetic backbone of the genus Quercus. Systematics and Biodiversity. 2014;12:405–423. doi: 10.1080/14772000.2014.941037. [DOI] [Google Scholar]
- Huson & Bryant (2006).Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Khadivi-Khub et al. (2015).Khadivi-Khub A, Shabanian N, Alikhani L, Rahmani M-S. Genotypic analysis and population structure of Lebanon oak (Quercus libani G. Olivier) with molecular markers. Tree Genetics & Genomes. 2015;11:102. doi: 10.1007/s11295-015-0935-1. [DOI] [Google Scholar]
- Kottek et al. (2006).Kottek M, Grieser J, Beck C, Rudolf B, Rubel F. World map of the Köppen-Geiger climate classification updated. Meteorologische Zeitschrift. 2006;15:259–263. doi: 10.1127/0941-2948/2006/0130. [DOI] [Google Scholar]
- Kumar, Stecher & Tamura (2016).Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen & Myllys (2008).Lehtonen S, Myllys L. Cladistic analysis of Echinodorus (Alismataceae): simultaneous analysis of molecular and morphological data. Cladistics. 2008;24:218–239. doi: 10.1111/j.1096-0031.2007.00177.x. [DOI] [Google Scholar]
- Librado & Rozas (2009).Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- López de Heredia et al. (2007).López de Heredia U, Jiménez P, Collada C, Simeone MC, Bellarosa R, Schirone B, Cervera MT, Gil L. Multi-marker phylogeny of three evergreen oaks reveals vicariant patterns in the Western Mediterranean. Taxon. 2007;56:1209–1209. doi: 10.2307/25065912. [DOI] [Google Scholar]
- Lumaret et al. (2002).Lumaret R, Mir C, Michaud H, Raynal V. Phylogeographic variation of chloroplast DNA in holm oak (Q. ilex L.) Molecular Ecology. 2002;11:2327–2336. doi: 10.1046/j.1365-294x.2002.01611.x. [DOI] [PubMed] [Google Scholar]
- Magri et al. (2007).Magri D, Fineschi S, Bellarosa R, Buonamici A, Sebastiani F, Schirone B, Simeone MC, Vendramin GG. The distribution of Quercus suber choloroplast haplotypes matches the palaeogeographic history of the western Mediterranean. Molecular Ecology. 2007;16:5259–5266. doi: 10.1111/j.1365-294X.2007.03587.x. [DOI] [PubMed] [Google Scholar]
- Manos, Zhou & Cannon (2001).Manos PS, Zhou ZK, Cannon CH. Systematics of Fagaceae: phylogenetic tests of reproductive trait evolution. International Journal of Plant Science. 2001;162:1361–1379. doi: 10.1086/322949. [DOI] [Google Scholar]
- McVay, Hipp & Manos (2017).McVay JD, Hipp AL, Manos PS. A genetic legacy of introgression confounds phylogeny and biogeography in oaks. Proceedings of the Royal Society B. 2017;284:20170300. doi: 10.1098/rspb.2017.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menitsky (2005).Menitsky YL. Oaks of Asia. Science Publishers; Enfield: 2005. [Google Scholar]
- Mhamdi et al. (2013).Mhamdi S, Brendel O, Montpied P, Ghouil-Amimi H, Hasnaoui I, Dreyer E. Leaf morphology displays no detectable spatial organisation in the relict Quercus afares Pomel compared to the co-occurring parental species Q. canariensis Willd. and Q. suber L. Annals of Forest Science. 2013;70:675–684. doi: 10.1007/s13595-013-0312-x. [DOI] [Google Scholar]
- Mir et al. (2006).Mir C, Toumi L, Jarne P, Sarda V, Di Giusto F, Lumaret R. Endemic North African Quercus afares Pomel originates from hybridisation between two genetically very distant oak species (Q. suber L. and Q. canariensis Willd.): evidence from nuclear and cytoplasmic markers. Heredity. 2006;96:175–184. doi: 10.1038/sj.hdy.6800782. [DOI] [PubMed] [Google Scholar]
- Morrison (2018).Morrison D. Tree metaphors and mathematical trees. Genealogical World of Phylogenetic Networks. 2018. http://phylonetworks.blogspot.com/2018/02/tree-metaphors-and-mathematical-trees.html http://phylonetworks.blogspot.com/2018/02/tree-metaphors-and-mathematical-trees.html
- Muir, Fleming & Schlotterer (2001).Muir G, Fleming CC, Schlotterer C. Tree divergent rDNA clusters predate the species divergence in Quercus petraea (Matt.) Liebl. and Quercus robur L. Molecular Biology and Evolution. 2001;18:112–119. doi: 10.1093/oxfordjournals.molbev.a003785. [DOI] [PubMed] [Google Scholar]
- Musacchio et al. (2006).Musacchio A, Pellegrino G, Cafasso D, Widmer A, Cozzolino S. A unique A. palustris lineage across the Otranto strait: botanical evidence for a past land-bridge? Plant Systematics and Evolution. 2006;262:103–111. doi: 10.1007/s00606-006-0469-y. [DOI] [Google Scholar]
- Nieto Feliner (2014).Nieto Feliner G. Patterns and processes in plant phylogeography in the Mediterranean Basin. A review. Perspectives in Plant Ecology, Evolution and Systematics. 2014;16:265–278. doi: 10.1016/j.ppees.2014.07.002. [DOI] [Google Scholar]
- Oh & Manos (2008).Oh S-H, Manos PS. Molecular phylogenetics and cupule evolution in Fagaceae as inferred from nuclear CRABS CLAW sequences. Taxon. 2008;57:434–451. doi: 10.2307/25066014. [DOI] [Google Scholar]
- Pattengale et al. (2009).Pattengale ND, Masoud A, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? In: Batzoglou S, editor. RECOMB 2009. Springer-Verlag; Berlin: 2009. pp. 184–200. [Google Scholar]
- Pautasso (2009).Pautasso M. Geographical genetics and conservation of forest tree. Perspectives in Plant Ecology, Evolution and Systematics. 2009;11:157–189. doi: 10.1016/j.ppees.2009.01.003. [DOI] [Google Scholar]
- Peel, Finlayson & McMahon (2007).Peel MC, Finlayson BL, McMahon TA. Updated world map of the Köppen-Geiger climate classification. Hydrology and Earth System Sciences. 2007;11:1633–1644. doi: 10.5194/hess-11-1633-2007. [DOI] [Google Scholar]
- Pesendorfer et al. (2016).Pesendorfer MB, Scott Sillett T, Koenig WD, Morrison SA. Scatter-hoarding corvids as seed dispersers for oaks and pines: a review of a widely distributed mutualism and its utility to habitat restoration. The Condor. 2016;118:215–237. doi: 10.1650/CONDOR-15-125.1. [DOI] [Google Scholar]
- Petit et al. (2004).Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A. Hybridization as a mechanism of invasion in oaks. New Phytologist. 2004;161:151–164. doi: 10.1046/j.1469-8137.2003.00944.x. [DOI] [Google Scholar]
- Pham et al. (2017).Pham KK, Hipp AL, Manos PS, Cronn RC. A time and a place for everything: phylogenetic history and geography as joint predictors of oak plastome phylogeny. Genome. 2017;60:720–732. doi: 10.1139/gen-2016-0191. [DOI] [PubMed] [Google Scholar]
- Podani (2017).Podani J. Different from trees, more than metaphors: branching silhouettes—corals, cacti, and the oaks. Systematic Biology. 2017;66:737–753. doi: 10.1093/sysbio/syx039. [DOI] [PubMed] [Google Scholar]
- Posada & Crandall (2001).Posada D, Crandall KA. Intra-specific gene genealogies: trees grafting into network. Trends in Ecology and Evolution. 2001;16:37–45. doi: 10.1016/S0169-5347(00)02026-7. [DOI] [PubMed] [Google Scholar]
- Renner et al. (2016).Renner SS, Grimm GW, Kapli P, Denk T. Species relationships and divergence times in beeches: new insights from the inclusion of 53 young and old fossils in a birth-death clock model. Philosophical Transactions of the Royal Society B. 2016;371:20150135. doi: 10.1098/rstb.2015.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg & Ellstrand (1993).Rieseberg LH, Ellstrand NC. What can molecular and morphological markers tell us about plant hybridization? Critical Reviews in Plant Sciences. 1993;12:213–241. [Google Scholar]
- Rubel et al. (2016).Rubel F, Brugger K, Haslinger K, Auer I. The climate of the European Alps: shift of very high resolution Köppen-Geiger climate zones 1800–2100. Meteorologische Zeitschrift. 2016;26:115–125. doi: 10.1127/metz/2016/0816. [DOI] [Google Scholar]
- Schwarz (1936–1939).Schwarz O. Monographie der Eichen Europas und des Mittelmeergebietes. Sonderbeiheft D; Berlin-Dahlem: 1936–1939. (Feddes Repertorium regni vegetabilis.). [Google Scholar]
- Simeone et al. (2016).Simeone MC, Grimm GW, Papini A, Vessella F, Cardoni S, Tordoni E, Piredda R, Franc A, Denk T. Plastome data reveal multiple geographic origins of Quercus Group Ilex. PeerJ. 2016;4:e1897. doi: 10.7717/peerj.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone et al. (2013).Simeone MC, Piredda R, Papini A, Vessella F, Schirone B. Application of plastid and nuclear markers to DNA barcoding of Euro–Mediterranean oaks (Quercus, Fagaceae): problems, prospects and phylogenetic implications. Botanical Journal of the Linnean Society. 2013;172:478–499. doi: 10.1111/boj.12059. [DOI] [Google Scholar]
- Stamatakis (2014).Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, Higgins & Gibson (1994).Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen (1976).Van Valen L. Ecological species, multispecies, and oaks. Taxon. 1976;25:233–239. doi: 10.2307/1219444. [DOI] [Google Scholar]
- Vitelli et al. (2017).Vitelli M, Vessella F, Cardoni S, Pollegioni P, Denk T, Grimm GW, Simeone MC. Phylogeographic structuring of plastome diversity in Mediterranean oaks (Quercus Group Ilex, Fagaceae) Tree Genetics and Genomes. 2017;13:3. doi: 10.1007/s11295-016-1086-8. [DOI] [Google Scholar]
- Volkov et al. (2001).Volkov RA, Zanke C, Panchuk I, Hemleben V. Molecular evolution of 5S rDNA of Solanum species (sect. Petota): application for molecular phylogeny and breeding. Theoretical and Applied Genetics. 2001;103:1273–1282. doi: 10.1007/s001220100670. [DOI] [Google Scholar]
- Welter et al. (2012).Welter S, Bracho-Nuñez A, Mir C, Zimmer I, Kesselmeier J, Lumaret R, Schnitzler JP, Staudt M. The diversification of terpene emissions in Mediterranean oaks: lessons from a study of Quercus suber, Quercus canariensis and its hybrid Quercus afares. Tree Physiol. 2012;32:1082–1091. doi: 10.1093/treephys/tps069. [DOI] [PubMed] [Google Scholar]
- Yaltırık (1984).Yaltırık F. Türkiye Meşeleri Teşhis Kılavuzu (Diagnostic Manual of Turkish oaks) Yenilik Press; İstanbul: 1984. [Google Scholar]
- Yang et al. (2017).Yang J, Vázquez L, Chen X, Li H, Zhang H, Liu Z, Zhao G. Development of chloroplast and nuclear DNA markers for Chinese Oaks (Quercus Subgenus Quercus) and assessment of their utility as DNA barcodes. Frontiers in Plant Science. 2017;8:816. doi: 10.3389/fpls.2017.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2015).Zhang XW, Li Y, Liu CY, Xia T, Zhang Q, Fang YM. Phylogeography of the temperate tree species Quercus acutissima in China: inferences from chloroplast DNA variations. Biochemical Systematics and Ecology. 2015;63:190–197. doi: 10.1016/j.bse.2015.10.010. [DOI] [Google Scholar]
- Zielinski, Petrova & Tomaszewski (2006).Zielinski J, Petrova A, Tomaszewski D. Quercus trojana subsp. yaltirikii (Fagaceae), a new subspecies from southern Turkey. Willdenowia. 2006;36:845–849. doi: 10.3372/wi.36.36214. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
We also report the accession numbers of the sequences retrieved from GenBank and used to complement the investigated dataset.
incuding results of the BLAST identity matches on GenBank.
This list reports all the identical 5S-IGS cloned sequences in the investigated dataset. Colors according to species list.
The tree was rooted on Q. baloot and Q. floribunda. Taxa colors as in Fig. 1, 2a and File S1. Branch bootstrap support (1-100) is reported.
Individuals represented by more than 2, 3 and 5 clones are included (cut-offs specimens are indicated).
This Online Supporting Archive includes data and analysis files of Simeone et al. ”Comparative systematics and phylogeography of Quercus Section Cerris in western Eurasia: inferences from plastid and nuclear DNA variation” in several subfolders.
Data Availability Statement
The following information was supplied regarding data availability:
Primary data and analyses are available as Supplemental Files.