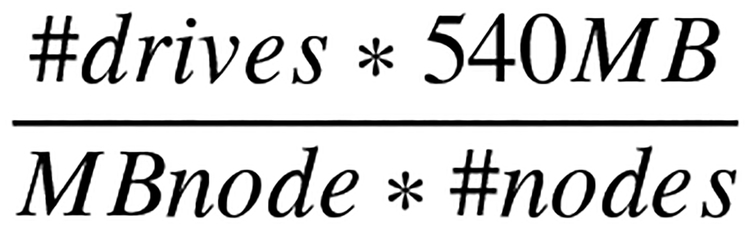Abstract
We explore the feasibility of a database storage engine housing up to 307 billion genetic Single Nucleotide Polymorphisms (SNP) for online access. We evaluate database storage engines and implement a solution utilizing factors such as dataset size, information gain, cost and hardware constraints. Our solution provides a full feature functional model for scalable storage and query-ability for researchers exploring the SNP’s in the human genome. We address the scalability problem by building physical infrastructure and comparing final costs to a major cloud provider.
Keywords: NoSQL, Billion Records, SNP, Big Data, Data Reduction, Distributed Computing, PWM, Edge Computing, Economical Computing, Genomics, Cassandra, MySQL, Elasticsearch
Introduction
Genome-wide association studies (GWAS) have been important to identify significant Single Nucleotide Polymorphisms (SNPs). In particular, regulatory SNPs (rSNPs) are the ones that affect gene regulation by changing transcription factor (TF) binding affinities to genomic sequences. These changes are crucial for understanding disease mechanisms. Affinity Testing SNP (atSNP) [1] is a powerful and efficient R package which implements an importance sampling algorithm coupled with a first-order Markov model for the background nucleotide sequences to test the significance of affinity scores and SNP-driven changes in these scores. A standard in silico approach for identifying rSNPs is done by evaluating how the NP-driven nucleotide changes impacts binding affinity of TFs to the region surrounding the SNP. More specifically, the DNA sequences around each SNP are scored against a library of TF motifs with both the reference and the SNP alleles using Position Weight Matrices (PWMs) of the motifs. The process is very time-consuming and usually needs to handle a large amount of input data and generates huge output files. atSNP is considered the state-of-the-art tool for handling such large-scale task. The output files generated by atSNP can be inspected and analyzed for SNP-motif interactions. However, given the size of the files, it can be unfeasible to browse and visualize the motif information, given current tools. We are tasked with the design, test and implementation of an infrastructure to support the hosting of up to 307 billion SNP-motif records for online web searchability on a researchers budget.
We evaluate the feasibility of three major open source databases for: data loading, storage and search; then implement a solution based on our feasibility testing. Furthermore, we showcase that our implementation: database choice, system hardware, and our final Total Cost of Ownership (TCO); provides an extremely cost effective option even against a major cloud provider.
This paper is organized as follows. In Section I we discuss the problem in more detail and in Section II we discuss solutions in the literature. In Section III we present our data requirements, hardware, software and evaluation method used in our analysis. Section IV presents results. Finally, we close in Section V with concluding remarks.
I. Background
Large scale real time searching is difficult for any significantly large dataset. Genetics and genomics datasets searching domains are significantly larger as small variations within a DNA sequence can produce numerous phenotypic variations [2]. As such, DNA analysis is still a domain ruled by statistically probabilities based on a specific subregion within the DNA strand. The subregions known as Single Nucleotide Polymorphisms [3] (SNPs) are produced using a DNA sequencing technique commonly known as the shotgun approach to DNA sequencing [4]. While the merits of the approach are known and solidly accepted, the probability of any given SNP occurring within a Genome-Wide Association Study (GWAS) is lesser known. The atSNP R package [1] attempts to address the probabilistic significance of affinity scores in SNPs.
atSNP search provides statistical evaluation of impact of SNPs on transcription factor-DNA interactions based on PWMs. The current version is built on SNPs from the dbSNP [5] Build 144 for the human genome assembly GRCh38/hg38 [6]. atSNP search uses in silico calculations based on the statistical method developed by Zuo et al. [1]. atSNP first-time users can start scanning vast amounts of SNP-PWM combinations for potential gain and loss of function with two key features:
p-values quantifying the impact of SNP on the PWM matches (p-value SNP impact)
composite logo plots.
atSNP identifies and quantifies the best DNA sequence matches to the transcription factor (TF) PWMs with both the reference and the SNP alleles in a small window around the SNP location (up to +/− 30 base pairs and considering sub-sequences spanning the SNP position). It evaluates statistical significance of the match scores with each allele and calculates statistical significance of the score difference between the best matches with the reference and SNP alleles.
The discovery of best matches with the reference and SNP alleles are conducted separately. As a result, genomic sub-sequences providing the best match with each allele may be different. The change in goodness of match between the reference and SNP alleles indicates in silico impact of SNPs on transcription factor binding ability to DNA. atSNP search also provides a composite logo plot for easy visualization of the quality of the reference and SNP allele matches to the PWM and the influence of SNPs on the change in match.
Addressing GWAS SNP affinity scores requires addressing magnitudes of data to determine probabilistic significance. After the generation and analysis of the SNP affinity scores, sharing the affinity scores with the community reduces the need to “reinvent the wheel” for any group determining best SNP affinity scores complete with Position Weight Matrix [7] (PWM) motif data [1]. Run once and share with many allows for efficient computational resource allocations for future projects.
The initial generation of the affinity score PWMs was done in silico with the atSNP [1] R [8] package. To complete the full data of JASPAR [9] and ENCODE [10] a distributed divide and conquer method was applied. This research was performed using the compute resources and assistance of the UW-Madison Center For High Throughput Computing (CHTC) in the Department of Computer Sciences. The CHTC is supported by UW-Madison, the Advanced Computing Initiative, the Wisconsin Alumni Research Foundation, the Wisconsin Institutes for Discovery, and the National Science Foundation, and is an active member of the Open Science Grid, which is supported by the National Science Foundation and the U.S. Department of Energy’s Office of Science. Using HTCondor [11] and the CHTC 132,946,852 SNPs combined with 2,270 PWMs generated 3.07 × 1011 SNP-PWM combinations records, some SNP’s where multi-allelic hence the disparity, and required 115,500 CPU hours of computational power. All the records weights are based on the p-values in Table I
TABLE I.
atSNP p-values
| atSNP p-values: | |
|---|---|
| p-value SNP Impact | A significant p-value, e.g. 0.0001, statistically supports the potential gain or loss of function of the genomic region with the SNP in terms of transcription factor binding. |
| p-value Reference | p-value for score with the reference allele (Log Likelihood Reference). A significant p-value indicates that the match to the PWM with the reference allele is statistically supported. |
| p-value SNP | p-value for scores with the SNP allele (Log Likelihood SNP). A significant p-value indicates that the match to the PWM with the SNP allele is statistically supported. |
| p-value Difference | p-value for the difference in scores with the reference and the SNP alleles (Log Likelihood Ratio). This is essentially the result from a likelihood ratio test. While we report this for completeness, the final atSNP results are based on p-value SNP impact which quantifies the significance of the Log Rank Ratio. |
| p-value Condition Ref | Conditional p-value for scores on the reference allele based on Log Enhance Odds. |
| p-value Condition SNP | Conditional p-value for scores on the SNP allele based on Log Reduce Odds. |
The complete details for the data generated are described in the atSNP method [1].
Performance comparisons between RDBMS and NoSQL database [12] show inconclusive results [13]. For our specific use case, searching billions of records, scalability to store billions of records was our primary concern. In Puangsaijai2017 [14], we note that MariaDB [15], a MySQL forked project which is feature complete with MySQL [16], shows ETL insert times to be comparable between NoSQL database Redis [17] to be a >20x faster with pipelines over MariaDB [15]. Interesting [14] also shows select operations to be comparable across many simple and complex queries with the exception of 2 simple equals queries. More interesting are the tested range queries (greater then and less then) [14] shows the NoSQL database to out perform MariaDB by 2x.
II. Related Work
Numerous examples of SNP databases [18] exist across the web with examples ranging from the dbSNP [5] to dbGAP [19]. While these data are provided through online web accessible database, they are a magnitude smaller when compared with the atSNP-generated data. For example, the Genome Variation Map database [20] recently published their current implementation which houses a total of about 4.9 billion variations using a MySQL database [16]. dbSNP [5] also utilizes a Relational Database Management System (RDBMS) [21], an entity relational database [22], which consists of well over 100 relational tables.
Example databases housing hundreds of billion records are found utilizing NoSQL infrastructures. Netflix, for example, has a NoSQL Cassandra [23] deployment consisting of 2,500 nodes hosting 420 TeraBytes (TB) of data. The Netflix database handles over a trillion requests per day [24]. LinkedIn uses Voldemort [25], a key/value store for their critical infrastructure. Google, often credited with starting the big data movement, created their own NoSQL engine BigTable [26]. Therefore, many examples of successful NoSQL database implementations exist and they have proven the design for scale [27]. It has been demonstrated [28] that NoSQL databases have been successfully deployed with hundreds of millions or billions of records and “can result in a dramatic performance difference against RDBMS while dealing with hundreds of millions or billions of records” [28].
Each of the NoSQL databases used by Netflix, Google, and LinkedIn; utilize a different data sharding algorithm. While the algorithmic implementation for each NoSQL sharded database varies, each share a divide and conquer approach through their distributed data storage engine [29].
III. Design
The publicly searchable repository for SNP affinity scores with PWM motifs (over 132,946,852 SNPs) combined with 2,270 PWM generated 3.07 1011records. Each record consumes 1,416 bytes (B)×of data. The specific data, generated by atSNP, used in this work are related with the human genome. The TF libraries used by atSNP came from the JASPAR core motif library [9] and from the ENCODE project [18]. After our analysis the JASPAR [9] dataset comprised SNP 2.7 × 1010 records totaling 35.78 TeraBytes (TB), while the ENCODE [18] SNP’s dataset comprised 2.8 1011 records totaling 360.37 TB. Our total dataset, including multi-allelic SNP’s which added 6.02 × 108 records, comprised 3.07 × 1011 records or 396.14 TB.
Providing 396.14 TB of SNP affinity scores with motif data online with real time query-able parameters requires advanced database techniques. Additionally, data loading and searching for 396.14 TB at 3.07 × 1011 records can cause additional issues as loading time and throughput start to become a bottleneck. As an example, using a gigabit ethernet network interface (gigE) to transfer 1 TB of data takes around two and a half hours. Projecting to the 396.14 TB we can expect around 44 days to perform the transfer.
Our dataset will search based on p-values. Our p-values are represented as floating point and search-ability over ranges of p-values require spatial range queries [30]. Range queries are data searches of number fields within a numeric range, less then or great then are using for range queries.
In addition to the affinity score records, the corresponding motif representation for a given SNP-PWM in graphical representation is a multi-petabyte data storage problem. The graphical representation for a given SNP motif saved as a Portable Network Graphic (PNG) consumes approximately 250 KiloBytes (KB) per image. The full SNP database representation stored in PNG format as a graphical motif requires 3.7 PetaBytes (PB) of data storage.
Single computational systems supporting 396.14 TB of online searching with 3.7 PB of additional storage for images simply do not exist.
The problem domain decomposition starts with the space defined by the SNP-PWM affinity scores. Our full compressed JASPAR [9] dataset at 1.8 TB range seems realistic for ETL (Extraction, Transformation and Loading) into a traditional relational database (e.g. MySQL, Oracle or Postgres). However, our problem expands considerably when considering the 19.8x compression ratio for the datasets. The 1.8 TB data expands to 35.78 TB which stretches the boundaries of current RDBMS technologies. The compressed ENCODE [18] dataset at 18.2 TB motif library expands to 360.37 TB after decompression. Single system RDBMS housing 307 billion records is likely not feasible but we will test and compare feasibility to NoSQL databases. We hypothesize the final implementation to house the SNPPWM data will consist of a database engine utilizing data sharding or the partitioning/breaking apart data for distributed queries against the large datasets.
We utilized custom ETL scripts developed using python for data loading [31]. The code was custom developed for reading the SNP-PWM files and translating them to the specific database ETL format. For our feasible testing we used one system to run our ETL pipeline code. After we refined our process, we utilized HTCondor [11] to distribute our ETL pipeline processes.
A. Cost Evaluations
Hardware and human resources costs were given extra consideration. We evaluated the cost per hardware node and compared a new machine vs a used machines. We consider the cost per TB of a cluster amortizing the cost over 5 years and comparing our costs of buying new vs used equipment vs Cloud hosted. We calculate Total Cost of Ownership (TOC) considering: power/cooling costs and system administration time as factors for a yearly cost per node and further into a cost per GB model. Our cluster requirements are: to support up to 396.14 TB of storage with 3.07 × 1011 records, budget $60k, cost optimize for best hosting environment either cloud of self housed. Additionally, we evaluate costs for self hosting including Total Cost of Ownership (TCO). To calculate TCO we amortizing our hardware costs over 5 years, add power consumed over 5 years, add system administrators time cost over 5 years. We calculate our self hosted data storage engine’s cost on a per Gigabyte (GB) basis.
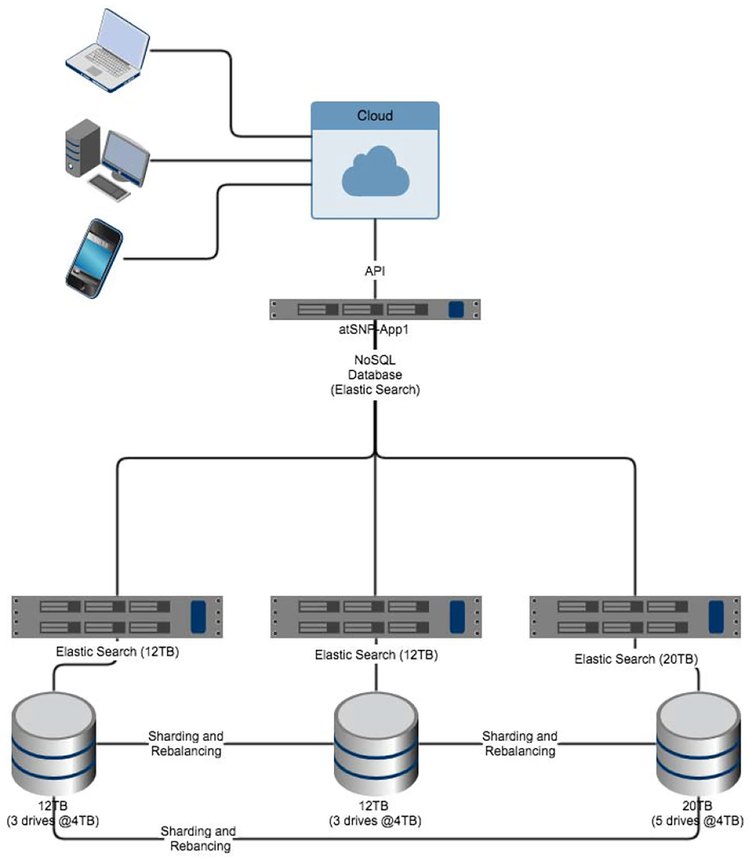
We evaluated the performance of MySQL, Cassandra (a NOSQL database) and Elasticsearch as database engines. In order to run these database engines, we used two computational infrastructures: (1) a customized in-house cluster and a (2) cloud provider. We tested different configurations of clusters varying memory sizes and disk sizes as well as number of disks. We ran the experiments using a trial and refine approach while following agile methodologies. We tested each database for feasibility, scalability and features which could support our data search needs. Each database tested was tested with default configurations using a minimal install of Scientific Linux 6, our test cluster was comprised of 3 nodes described in Table II. Feasible testing evaluation items included: ETL speeds, search retrieval times(within 5 seconds), required on going system administrator time to maintain, scalability and failure recovery. For testing, we initially evaluate databases populated with 1 million records for our feasibility testing.
TABLE II.
Our first generation Elasticsearch cluster composition
| Machine | RAM (GB) | #Processors | Total HDD (TB) |
|---|---|---|---|
| atsnp-db1 | 24 | 4 | 12 |
| atsnp-db2 | 24 | 4 | 12 |
| atsnp-db3 | 32 | 8 | 18 |
| TOTAL | 80 | 16 | 42 |
IV. Results and Evaluation
A. Cassandra
Our first test considered Cassandra [23], a Big Table NoSQL database engine. Our data ETL times to database import were 14,664.2 records per second, which is similar to the University of Toronto NoSQL benchmarks [32]. Our data requires multiple range queries which are not easily indexed within Cassandra. The range queries in the implementation of Cassandra’s instance of NoSQL required every node to query all data housed on the node for any row meeting the range query parameter without indexes [33]. The lack of range queries and the performance penalty for evaluating a data subset proved to be a fatal flaw in our first database evaluation method for genomic SNP data at scale using a NoSQL implementation. Our requirements for multiple range query through p-value scores of the atSNP dataset using realtime search through a user defined parameter space meant that Cassandra was not a feasible long term datastore for our needs.
B. MySQL
Using our data with MySQL, after the first million records our database was responsive (results on the order of milliseconds). As our database started to grow, efficiency started to drop. Once our database was consuming all available ram for indexes any additional record required the use of system swap space which caused response times and ETL times to be unworkable. Based on our ETL estimates approximately 1000 records a second, which is similar to the 1023 records/sec demonstrated in [34]. Therefore, if we continued ETL with database transactions enabled by default to ETL our data is represented in Equation 1:
| (1) |
10 years to ETL our data makes the use of a MySQL database engine unfeasible.
Consideration for using the storage engines InnoDB and NDB with clusters were not even considered, given the limitations of MySQL [35]. Factors against distributing the load across multiple instances of MySQL using partition tables (one per chromosome) in a MySQL cluster were overridden by a concern for the long term support-ability of the database service and system administration time overhead required. Specifically, concerns raised included: status checking, time to maintain and migrate data when needed, updating the application logic after a partition table migration and system monitoring.
C. Elasticsearch
For the third test case, we used a distributed NoSQL since we are validating the hypothesis of using shared NoSQL database for genomic SNP data as a feasible option for large scale storage and query-ability. This test uses a shared NoSQL database engine, but optimized for range queries. Specifically, we consider the Elasticsearch database engine for its focus and utilization of range queries for system log searches. To evaluate Elastic-search lookups of address range queries, we loaded 1,012,032 samples (≈ 1 million records to prove range query feasibility. Our initial tests passed with query times less than 5 ms.
ETL database issues did arise in the ETL phase for our Elasticsearch database. Our ETL times averaged 11,944.5 records per second or 1,040,000,000 records per day which is significantly faster than the MySQL approach but not sufficient for our timeframe to deployment. Given the ETL rates, the ETL process would require to completion 295 days for atSNP entire 3.07 × 1011 records. Additionally, our query search speed degraded significantly while concurrently inserting ETL data; therefore, additional I/O capacity was recommended. Our first configuration utilized 3 recycled from prior production servers augmented with additional storage: 7200 rpm 4 TB hard drives. 2 systems operated with 3 HDD and 1 system operated with 5 drives. Having accomplished a main feasibility objective, we were satisfied with the Elasticsearch option.
A breakdown of our evaluation is described in Table III. Each database is listed in the first column. The second column is our ETL based on records a second (ETL rec/sec). The third column id defined by a boolean yes/no to the database return results of range queries within 5sec for our initial test case. The forth column is the approximate required system administrators time to maintain the final system within our budget. The scalability column defines if the database has been proven to scale to the multi-billion record size. The last column “failure auto recovery” defines if the database engine has built in failure recovery without system administrator input.
TABLE III.
Our evaluation Matrix
| Database | ETL rec/sec | >5sec search | system admin time | scalability | Failure auto recovery |
|---|---|---|---|---|---|
| Cassandra | 14,664 | no | ok | yes | yes |
| MySQL | 1023 | yes | no | no | no |
| Elasticsearch | 11,944 | yes | ok | yes | yes |
D. Refinements
The atSNP output data was computed with high precision p-values for a given SNP evaluation. Addressing each record and reducing the precision of the p-value calculation (64bit to 8bit), we were able to achieve a significant reduction in the record size from 1,416 to 623 bytes of data. The reduction in data record size allowed us to reduce the size of the total data storage impact to 191.26 TB. Our data size reduction; however, did not translate into an overall database size reduction. Elasticsearch, like many sharded NoSQL database engines, utilized internal data partitioning to spread the dataset over the entire cluster. The database internal partition schemas provide cluster redundancy through the cluster replication schema in the case of individual system outages. Using the default Elasticsearch replica factor of 3x our total records size of 191.26 TB ballooned by 3 thus, our database sizing requirements where back to 573.78 TB. Thus, any space saved in individual record size reduction was consumed by the cluster replication redundancy schema.
Further consideration was done for the three key p-values in atsnp search results are p-value SNP impact, p-value reference, and p-value SNP. In a typical application, users would want to identify SNP-TF combinations for which p-value SNP impact is significant (e.g., p-value SNP impact <= 0.05) along with significance of either the p-value reference or the p-value SNP. To enable this in the most liberal way, we included all SNP-TF combinations with a p-value smaller than 0.05 for at least one of the three p-values.
Our final p-value cutoff for at least one of the three p-values at 0.05 enables querying of these statistical evaluations which exhibited a significant score change or a significant match with either the reference or the SNP allele. Resulting in 3.73 108 SNP-PWM records consuming just 10.55 TB. A 10.55 TB dataset size utilize a reduced rplication factor of 2x instead of 3x requires just 21.12 TB. By requiring just 21.12TB we remain well within our 60TB of database disk space limit.
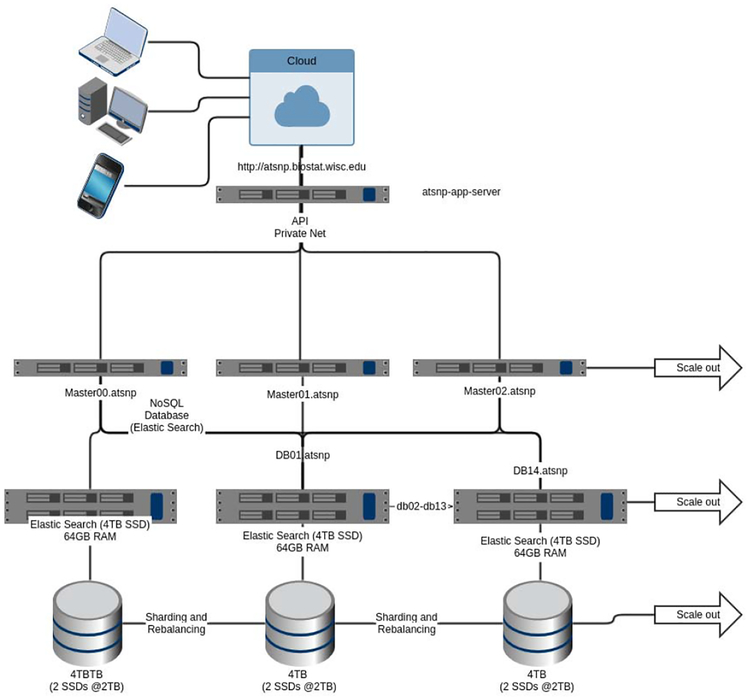
See cost breakdown in the Hardware Cost table IV. To address our ETL times for our dataset, we utilized HTCondor for additional throughput with the ETL scripts. Distributing the ETL tasks in HTCondor proved to be effective; however, our cluster experienced overload conditions. Elasticsearch was a feasible database engine for our atSNP data; however, our ETL times were still beyond functional for our needs. Our initial experiments showed that our first generation cluster running nodes in Table II is not sufficient to query and search the large database at hand. For ETL on our first generation test cluster, we realized a node could maintain 6 ETL scripts per node without experiencing a ‘“Bulk Queue Full’” error exception. Addition considerations were needed for search times. A complete scan all data per node at a rate of 540MB/sec [36] our full node data scan equation in Figure 2. To refine the number of nodes our cluster required, we started with the equation in Figure 2 for a raw Disk I/O. Based on 15 nodes and the cost per gigabyte equation, and using Table VI, we select 2× 2TB SSD’s for the data storage on each node which would give us 60TB of usable search space across 15 nodes. Data storage per node also required knowing the maximum read bandwidth per SSD is 540MB/sec with 90,000 Input/Outputs Per Second (IOPS)/second [36]. We dismissed Hard disk drives as an option due to the required speed to completely scan the data [37].
TABLE IV.
Hardware Cost
| Type | SSD(TB) | Node Cost($) | cost/yr($) | cost/mon($) |
|---|---|---|---|---|
| new | 1 | 5441 | 1088.2 | 90.683 |
| new | 2 | 5731 | 1146.2 | 95.52 |
| new | 4 | 6251 | 1250.2 | 104.183 |
| used | 1 | 1010 | 202 | 16.833 |
| used | 2 | 1300 | 260 | 21.667 |
| used | 4 | 1820 | 364 | 30.333 |
Fig. 2.
Datasearch for cluster based on SSD speed
TABLE VI.
Hosted vs Cloud
| # nodes | GB | Cost/GB ($) | monthly ($) | yearly ($) |
|---|---|---|---|---|
| 15 | 15360 | 0.216 | 3297.30 | 39567.60 |
| 15 | 30720 | 0.109 | 3369.75 | 40437.00 |
| 15 | 61440 | 0.057 | 3499.80 | 41997.60 |
| 15 | 15360 | 0.143 | 2189.55 | 26274.60 |
| 15 | 30720 | 0.078 | 2262.15 | 27145.80 |
| 15 | 61440 | 0.039 | 2392.05 | 28704.60 |
| Hosted | Hosted | $0.135 | varies | varies |
For each node, we followed Elasticsearch heap size recommended to keep below 32GB [38]. Additionally, following Elasticsearch “The standard recommendation is to give 50% of the available memory to Elasticsearch heap, while leaving the other 50% free” [38] this also providing the Operating system with free memory for data page tables. Thus, we concluded our optimized RAM per node at 64 GB of RAM: 30GB for Heap, 30GB system paging, 4GB Kernel/OS.
Our cost per new node, described in Table IV column 1 ‘new’, of $5,151.23 was based on a Dell quote from dell.com for an R630 with 64 GB RAM, 2 × 8 core Intel Xeon E5–2623 v4 2.6GHz, 10 MB Cache, 8.00 GT/s QPI, Turbo, HT, 4C/8T. Our cost per used node, described in column 1 ‘used’ Table IV, of $720 was based on an R620 with 64 GB of RAM, 2×8 core Intel Xeon E5–2650 2.0 GHz, 20 MB Cache, 8.00 GT/s QPI, Turbo, HT, 4C/8T. No extended warranty.
SSDs for Table IV column two were based on crucial MX300 family [36] of SSD, to simplify we are comparing only 1 and 2 TB SSD and consuming a maximum of 2 disk trays per server, the cost for a 1 TB SSD [39] $289.99 and 2 TB SSD [40] $549.99.
Power consumption was addressed by comparing the power cost/cooling costs over 95 days time period a similarly configured system consumed 192.1 KWh or 2.022 KWh/day/node. To include cooling, we doubled this based on cost to cool to 4.044 KWh/day/node at a current Kilo Watt Hour (KWh) price at $0.14KWh [41]. Our final cost per node for power and cooling is $206.64 per year (365 days) or $17.22 per month as reported in column ‘pwr/cool ($)’ for Table V. Systems administration time cost was based from IBM TOC [42] for application server support and administration at $1,343/year for 5 years or $111.92 per month in Table V column adm($).
TABLE V.
Running systems Cost per month
| node($) | pwr/cool ($) | adm ($) | monthly ($) | yearly ($) |
|---|---|---|---|---|
| 90.683 | 17.22 | 111.916 | 219.82 | 2637.84 |
| 95.52 | 17.22 | 111.916 | 224.65 | 2695.84 |
| 104.183 | 17.22 | 111.916 | 233.32 | 2799.84 |
| 16.833 | 17.22 | 111.916 | 145.97 | 1751.64 |
| 21.667 | 17.22 | 111.916 | 150.81 | 1809.64 |
| 30.333 | 17.22 | 111.916 | 159.47 | 1913.64 |
Reducing storage through intelligent runtime
Further scope reductions involved a move away from graphical representation of motif SNP-PWM. As previously stated to store the graphic files for each affinity scored SNP would have required 3.7 PB of storage. Having taken into consideration cost, we reduced our scope to only using 11% of the total records. Therefore, we were able to reduce the graphical storage to 11% of the 3.7PB or 370TB. 370TB of storage for graphics is cost prohibitive as data storage prices, while always decreasing [43], is still expensive. Data storage cost for raw 10TB hard disk storage costs $367.49 [44], keep in mind that a 10TB disk is really just 9.09TB since hard drive manufactures us 1000 instead of 1024 as the calculated bases. Additionally, the cost of data storage is well known to be more than just the raw disk cost per TB [45]. Even without factoring the true cost of storage [45] and using a simplicity price per TB, our 370TB of raw storage with no redundancy would require 41 10TB disk drives. With a real cost per raw TB at $40.41/TB our images storage costs of would still be $15067.09.
Since cost was always an issue and the storage or PNG files for the PWM motifs are computer generated anyway, we decided against storage of the PWM motifs. Instead we chose to build software to generate the a Scalable Vector Graphic (SVG). The software module utilizes javascript and the d3.js [46] library, which uses the PWM data, for generating the Motif at runtime.
To stay consistent and allow us to render the proper PWM motif for a given SNP-PWM record, we chose to store the PWM motif data with each corresponding record. Adjusting our approach from a centralized storage with PNG motif PWM files to distributed edge computing render model, proved fruitacious. The dynamic motif generation by edge computing through client web browser’s javascript engine allowed us to eliminate motif image storage. Thus, we where able to obtain significant data storage savings. For our application after the p-value cutoff was applied, we further were able to reduce our required data storage. Since the prior image data of 370TB was no longer needed and we were able to further reduced costs. Our resulting combined real reduction in data usage from the initial 3.7 PB of storage to just 21.12TB was a significant at a 179x reduction in total required storage.
V. Conclusion
Our use case study had demonstrated the feasibility of using NoSQL database engines for large scale real-time searchable genomic SNP databases. The use of Elasticsearch and the successful implementation is proof that our systems was feasible. Admittedly, our evaluation of database engines is not a conclusive finding. Our goal was to prove database engine feasibility and not be conclusive or exhaustive study case. Factors which impeded a more through database engine study included: cost, personal, speed to implement, and domain knowl edge. Additional factors which played a roll in our final implementation included: support-ability, system administrator time to implement, datacenter rackspace and networking.
Our time-frame for implement was limited and numerous unforeseeable issues arose. At first we expected to build a cluster based on 14 nodes as described in Cassandra for Sysadmins slide 6 [47]. However, we quickly realized our quick turn-a-round to a fully functional web resource was limited by at first range queries, then scalability (MySQL), until we agreed to utilize Elaticsearch through our feasibility testing. Our successful deployment exhibits 11% of the human genome SNP’s with PWM. Barring financial resources constraints, we believe our presented solution would scale and fully support the complete anSNP dataset of 307 billion records.
Our cost structure, based on purchasing and deploying equipment, was validated as a highly competitive cost effective option. For our “do it ourselves” Elasticsearch infrastructure, our cost saving proved significant. When comparing to Amazon’s Elasticsearch [48] implementation, our 3.4x cost savings proved significant; us: $0.039 vs amazon: $0.135 [48]; or a saving of $0.096 per GB.
Fig. 1.
Our test cluster with final selection
Fig. 3.
Final atSNPElasticsearch cluster
Acknowledgments
This work was supported by:
NIH Big Data to Knowledge (BD2K) Initiative under Award Number U54 AI117924
Center for Predictive Computational Phenotyping (CPCP)
University of Wisconsin - Madison References
References
- [1].Zuo C, Shin S, and KeleÅ S§, “atsnp: transcription factor binding affinity testing for regulatory snp detection,” Bioinformatics, vol. 31, no. 20, pp. 3353–3355, 2015. [Online]. Available: 10.1093/bioinformatics/btv328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kreitman M, “Nucleotide polymorphism at the alcohol dehydrogenase locus of drosophila melanogaster,” Nature, vol. 304, pp. 412 EP–, August 1983. [Online]. Available: 10.1038/304412a0 [DOI] [PubMed] [Google Scholar]
- [3].Gray IC, Campbell DA, and Spurr NK, “Single nucleotide polymorphisms as tools in human genetics,” Human Molecular Genetics, vol. 9, no. 16, pp. 2403–2408, 2000. [Online]. Available: + 10.1093/hmg/9.16.2403 [DOI] [PubMed] [Google Scholar]
- [4].Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, and Lander ES, “An snp map of the human genome generated by reduced representation shotgun sequencing,” Nature, vol. 407, pp. 513 EP–, September 2000. [Online]. Available: 10.1038/35035083 [DOI] [PubMed] [Google Scholar]
- [5].“dbsnp short genetic variations,” https://www.ncbi.nlm.nih.gov/SNP/, accessed: 2018-01-03
- [6].“Genome reference consortium human build 38,” https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.26/, accessed: 2016-05-05.
- [7].Stormo GD, Schneider TD, Gold L, and Ehrenfeucht A, “Use of the ‘perceptron’ algorithm to distinguish translational initiation sites in e. coli,” Nucleic Acids Research, vol. 10, no. 9, pp. 2997–3011, 1982. [Online]. Available: + 10.1093/nar/10.9.2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].R Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, 2013, ISBN 3-900051-07-0. [Online]. Available: http://www.R-project.org/ [Google Scholar]
- [9].Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen C.-y., Chou A, Ienasescu H, Lim J, Shyr C, Tan G, Zhou M, Lenhard B, Sandelin A, and Wasserman WW, “Jaspar 2014: an extensively expanded and updated open-access database of transcription factor binding profiles,” Nucleic Acids Research, vol. 42, no. D1, pp. D142–D147, 2014. [Online]. Available: + 10.1093/nar/gkt997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Consortium TEP, “An integrated encyclopedia of dna elements in the human genome,” Nature, vol. 489, no. 7414, pp. 57–74, September 2012, 22955616[pmid]. [Online]. Available: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3439153/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Basney J and Livny M, Deploying a High Throughput Computing Cluster, Buyya R, Ed. Prentice Hall PTR, 1999. [Google Scholar]
- [12].Klein J, Gorton I, Ernst N, Donohoe P, Pham K, and Matser C, “Performance evaluation of nosql databases: A case study,” in Proceedings of the 1st Workshop on Performance Analysis of Big Data Systems, ser. PABS ‘15. New York, NY, USA: ACM, 2015, pp. 5–10. [Online]. Available: http://doi.acm.org/10.1145/2694730.2694731 [Google Scholar]
- [13].Li Y and Manoharan S, “A performance comparison of sql and nosql databases,” in 2013 IEEE Pacific Rim Conference on Communications, Computers and Signal Processing (PACRIM), Aug 2013, pp. 15–19. [Google Scholar]
- [14].Puangsaijai W and Puntheeranurak S, “A comparative study of relational database and key-value database for big data applications,” in 2017 International Electrical Engineering Congress (iEECON), 2017, pp. 1–4. [Google Scholar]
- [15].“Mariadb server,” https://mariadb.com/, accessed: 2018-01-28.
- [16].“Mysql server,” https://www.mysql.com, accessed: 2018-01-28.
- [17].“Redis,” https://redis.io/, accessed: 2018-01-15.
- [18].Mooney SD, Krishnan VG, and Evani US, “Bioinformatic tools for identifying disease gene and snp candidates,” Methods Mol Biol, vol. 628, pp. 307–319, 2010, 20238089[pmid]. [Online]. Available: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3957484/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, and Sherry ST, “The ncbi dbgap database of genotypes and phenotypes,” Nat Genet, vol. 39, no. 10, pp. 1181–1186, October 2007, 17898773[pmid]. [Online]. Available: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2031016/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Song S, Tian D, Li C, Tang B, Dong L, Xiao J, Bao Y, Zhao W, He H, and Zhang Z, “Genome variation map: a data repository of genome variations in big data center,” Nucleic Acids Research, vol. 46, no. D1, pp. D944–D949, 2018. [Online]. Available: + 10.1093/nar/gkx986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Selinger PG, Astrahan MM, Chamberlin DD, Lorie RA, and Price TG, “Access path selection in a relational database management system,” in Proceedings of the 1979 ACM SIGMOD International Conference on Management of Data, ser. SIGMOD ‘79. New York, NY, USA: ACM, 1979, pp. 23–34. [Online]. Available: http://doi.acm.org/10.1145/582095.582099 [Google Scholar]
- [22].Chen PP-S, “The entity-relationship model—toward a unified view of data,” ACM Transactions on Database Systems, vol. 1, no. 1, pp. 9–36, March 1976. [Google Scholar]
- [23].Foundation AS, “Cassandra.” [Online]. Available: http://cassandra.apache.org/
- [24].“7 reasons why netflix uses cassandra databases,” https://www.jcount.com/7-reasons-netflix-uses-cassandra-databases/, accessed: 2017-09-20.
- [25].Voldemort P, “Voldemort.” [Online]. Available: http://www.project-voldemort.com/voldemort/
- [26].Chang F, Dean J, Ghemawat S, Hsieh WC, Wallach DA, Burrows M, Chandra T, Fikes A, and Gruber RE, “Bigtable: A distributed storage system for structured data,” in 7th USENIX Symposium on Operating Systems Design and Implementation (OSDI 06). Seattle, WA: USENIX Association, 2006. [Online]. Available: https://www.usenix.org/conference/osdi-06/bigtable-distributed-storage-system-structured-data [Google Scholar]
- [27].“Relational databases are not designed for scale,” http://www.marklogic.com/blog/relational-databases-scale/, accessed: 2018-01-15.
- [28].Vaish G, Getting Started with Nosql. Packt Publishing, 2013. [Online]. Available: https://books.google.com/books?id=oPiT-V2eYTsC [Google Scholar]
- [29].Cattell R, “Scalable sql and nosql data stores,” SIGMOD Rec, vol. 39, no. 4, pp. 12–27, May 2011. [Online]. Available: http://doi.acm.org/10.1145/1978915.1978919 [Google Scholar]
- [30].Orenstein JA, “Spatial query processing in an object-oriented database system,” in ACM Sigmod Record, vol. 15, no. 2 ACM, 1986, pp. 326–336. [Google Scholar]
- [31].Hudson R, “ss utility scripts.” [Online]. Available: https://github.com/RebeccaHudson/ss_utility_scripts
- [32].“Apache cassandra nosql performance benchmarks,” https://academy.datastax.com/planet-cassandra/nosql-performance-benchmarks, accessed: 2017-01-20.
- [33].Pirzadeh P, Tatemura J, Po O, and Hacıgümüş H, “Performance evaluation of range queries in key value stores,” Journal of Grid Computing, vol. 10, no. 1, pp.109–132, March 2012. [Online]. Available: 10.1007/s10723-012-9214-7 [DOI] [Google Scholar]
- [34].“Improve mysql insert performance,” https://kvz.io/blog/2009/03/31/improve-mysql-insert-performance/, accessed: 2017-06-15.
- [35].“Differences between the ndb and innodb storage engines,” https://dev.mysql.com/doc/refman/5.7/en/mysql-cluster-ndb-innodb-engines.html, accessed: 2017-06-15.
- [36].“Crucial mx300 solid state drive,” http://www.crucial.com/usa/en/storage-ssd-mx300, accessed: 2017-06-18.
- [37].“Comparing iops for ssds and hdds,” http://www.tvtechnology.com/expertise/0003/comparing-iops-for-ssds-and-hdds/276487, accessed: 2017-03-29.
- [38].“Elasticsearch, heap sizing and swapping,” https://www.elastic.co/guide/en/elasticsearch/guide/current/heap-sizing.html, accessed: 2017-09-28.
- [39].“Crucial mx300 2.5” 1tb sata iii 3d nand internal solid state drive (ssd) ct1050mx300ssd1,” https://www.newegg.com/Product/Product.aspx?item=N82E16820156152, accessed: 2017-06-10.
- [40].“Crucial mx300 2tb sata 2.5” 7mm (with 9.5mm adapter) internal ssd,” http://www.crucial.com/usa/en/ct2050mx300ssd1, accessed: 2017-06-10.
- [41].“Commercial and industrial (c&i) electric rates,” https://www.mge.com/customer-service/business/elec-rates-comm/, accessed: 2017-06-30.
- [42].“Tco for application servers: Comparing linux with windows and solaris,” http://www-03.ibm.com/linux/whitepapers/robertFrancesGroupLinuxTCOAnalysis05.pdf, accessed: 2017-06-30.
- [43].“Hard drive cost per gigabyte,” https://www.backblaze.com/blog/hard-drive-cost-per-gigabyte/, accessed: 2017-09-15.
- [44].“Seagate enterprise capacity 3.5” hdd 10tb (helium) 7200 rpm sas 12gb/s 256mb cache standard model 512e internal hard drive st10000nm0096,” https://www.newegg.com/Product/Product.aspx?Item=234-000S-00083&cm_re=10tb_hdd-_-234-000S-00083-_-Product, accessed: 2018-01-29.
- [45].“The true “cost” of enterprise storage -understanding storage management,” https://www.zadarastorage.com/blog/industry-insights/cost-of-enterprise-storage-understanding-storage-management/, accessed: 2017-12-29.
- [46].Bostock Mike, “D3: Data-driven documents - d3.js.” [Online]. Available: https://d3js.org
- [47].“Cassandra 101 for sysem administrators,” https://www.slideshare.net/nmilford/cassandra-for-sysadmins, accessed: 2017-02-02.
- [48].“Amazon elasticsearch service pricing,” https://aws.amazon.com/elasticsearch-service/pricing/, accessed = 2018-02-01.