Abstract
Background/Objectives
Obesity is a pandemic disorder that is characterized by accumulation of adipose tissue and chronic-low grade inflammation that is driven primarily by adipose tissue macrophages (ATMs). While ATM polarization from pro-(M1)to anti-(M2) inflammatory phenotype influences insulin sensitivity and energy expenditure, the mechanisms of such a switch are unclear. In the current study we identified epigenetic pathways including microRNAs (miR) in ATMs that regulate obesity-induced inflammation.
Subjects/Methods
Male C57BL/6J mice were fed normal chow diet (NCD) or high-fat diet (HFD) for 16 weeks to develop lean and diet-induced obese mice respectively. Transcriptome microarrays, microRNA microarrays, and meDIP-Seq were performed on ATMs isolated from visceral fat. Pathway analysis and bone marrow derived macrophage (BMDM) transfections further allowed computational and functional analysis of miRNA-mediated ATM polarization.
Results
ATMs from HFD-fed mice were skewed towards M1 inflammatory phenotype. Concurrently, the expression of miRs 30a-5p, 30c-5p, and 30e-5p was downregulated in ATMs from HFD mice when compared to mice fed NCD. The miR-30 family was shown to target Delta-like-4, a Notch1 ligand, whose expression was increased in HFD ATMs. Inhibition of miR-30 in conditioned BMDM triggered Notch1 signaling, pro-inflammatory cytokine production, and M1 macrophage polarization. In addition, DNA hypermethylation was observed in mir30-associated CpG islands suggesting HFD downregulates miR-30 through epigenetic modifications.
Conclusions
HFD-induced obesity downregulates miR-30 by DNA methylation thereby inducing Notch1 signaling in ATMs and their polarization to M1 macrophages. These findings identify miR-30 as a regulator of pro-inflammatory ATM polarization and suggest miR-30 manipulation could be a therapeutic target for obesity-induced inflammation.
Introduction
Overnutrition causes obesity and adipose tissue macrophage (ATM)-dependent inflammation that enhances risk for type II diabetes, cardiovascular disease, and some cancers. Macrophages are innate immune cells that display remarkable plasticity between pro- and anti-inflammatory phenotypes. They are routinely classified as M1 and M2 respectively, although various subtypes exist 1. At steady state, resident M2-like ATMs contribute to insulin sensitivity and adipose tissue homeostasis through production of anti-inflammatory cytokines such as IL-10 2. During obesity, M1 ATMs dominate the adipose tissue in both phenotype and abundance, promoting insulin resistance and chronic low-grade inflammation 3. Due to vast disparities seen in ATM functionality between lean and obese individuals, ATMs have been suggested to play a substantial role in determining development of obesity-related pathologies. Therapeutic strategies that decrease ATM-dependent inflammation have been heavily investigated due to the tight correlation of macrophage-dependent inflammation and insulin resistance.
Notch signaling plays key roles in metabolic and inflammatory processes 4. It is highly conserved juxtacrine signaling utilized by numerous cell types including macrophages and adipocytes. Binding of Notch receptors (Notch1-4) by Delta-like and Jagged ligands (DLL1, -3, -4 and JAG1, -2) initiates proteolytic release of the Notch intracellular domain (NICD) allowing it to translocate to the nucleus and activate Jκ-Recombination Signal-Binding Protein (RBP-J)-dependent transcription 5. In macrophages, Notch1 signaling promotes pro-inflammatory polarization through IRF8 and NF-κB transcriptional pathways, while in adipocytes, Notch1 signaling inhibits white adipose tissue browning and energy expenditure, and promotes insulin resistance 6–8. Additionally, blockade of the canonical Notch1 ligand DLL4 improves atherosclerosis and metabolic disease indicating DLL4-Notch1 signaling is directly involved in the crosstalk of inflammatory and metabolic pathways 9.
Recent studies in our laboratory have identified epigenetic modifications which regulate immune responses in a variety of diseases including post-traumatic stress disorder, multiple sclerosis, colitis, and liver failure 10–13. MicroRNAs (miRNA, miR) are short (~22 nucleotide long) non-coding RNAs that post-transcriptionally inhibit protein translation by binding the 3′ untranslated region (3′UTR) of target mRNAs 14. Because approximately 60% of protein-coding genes are known conserved targets of miRNAs, they have emerged as important regulators of biological functions such as immune system development and inflammatory responses 15,16. DNA methylation occurs when methyl groups are added to cytosines by DNA methyltransferases (DNMT). These methylated cytosines primarily reside in CpG islands near transcription start sites and repress gene transcription by blocking binding sites for transcription factors through chromatin condensation. Importantly, DNA methylation and miRNAs have been associated with development of aging-associated pathologies including obesity, atherosclerosis, and cancer 17–19.
Numerous miRNAs have already been classified to regulate differentiation and function of both adipocytes and macrophages. However, characterization of miRNA expression specifically in ATMs from lean and obese individuals has not been previously reported. Such investigation warrants merit because ATMs comprise a unique cell type due to their immunometabolic functions and dominance in the adipose tissue of obese individuals. Also, ATMs are thought to be the main initiators of obesity-induced inflammation and insulin resistance 20. In this report, we have identified dysregulated microRNAs in ATMs during obesity, which demonstrates that obese ATMs have distinct miRNA profiles that are likely to contribute to their inflammatory phenotype. Specifically, we discovered downregulation of the miR-30 family in ATMs to modulate macrophage phenotype. Our findings demonstrate that miR-30 regulates pro-inflammatory polarization of ATMs, and is likely due to regulation of DLL4-mediated Notch1 signaling. Our data also suggest therapeutic manipulation of this miRNA family or the epigenetic mechanisms that regulate its expression, may constitute therapeutic modalities for obesity-induced inflammation, insulin resistance, and related cardiometabolic disorders.
Materials and Methods
Mice
Six to 8-week-old male and female C57Bl/6J mice and 22-week-old male C57Bl/6J mice fed either 60% kcal HFD (D12492, Research Diets), NCD (8904, Envigo Teklad), or purified 10% low-fat diet (LFD, D12450J, Research Diets) where indicated, for 16 weeks were obtained from The Jackson Laboratory and housed in a specific-pathogen-free facility. Studies were not blinded and mice were not randomized into experimental groups. At the conclusion of each study, mice were euthanized by overdose isoflurane inhalation. All procedures were performed in accordance with protocols approved by the University of South Carolina Institutional Animal Care and Use Committee.
Analytical Procedures
Body composition of lean mass and fat mass were measured by dual-energy x-ray absorptiometry (DEXA) (LUNAR PIXImus). Mice were placed under isoflurane anesthesia and scanned in the prone position with the head region being excluded. Body weight was monitored using an electronic gram scale with precision ± 0.1g. For glucose tolerance tests, mice underwent a 5hr morning fast before fasting glucose measurement then gavaged with 2 g/kg lean mass glucose (Sigma G7528). Blood glucose was measured 15, 30, 60 and 120m post-glucose bolus by applying approximately 5uL tail-tip blood to a glucose test strip in a glucose meter (Contour Next, Bayer).
Adipose Tissue Dissociation and ATM Isolation
To dissociate cells of the stromal vascular fraction (SVF), epididymal fat pads from 22-week-old mice were dissected, rinsed in phosphate buffered saline (PBS) and homogenized in 5mL digestion media consisting of Hank’s Balanced Salt Solution (HBSS) containing 2% bovine serum albumin (BSA) and 1mg/mL collagenase (Sigma C6885) using a gentleMACs dissociator (Miltenyi Biotec). An additional 5mL digestion media was added to the homogenates and incubated 37°C, 75 RPM, 30–40m until fully dissociated. Next, 5mL complete Dulbecco’s Modified Eagle’s Medium and Ham’s F-12 Nutrient Mixture (DMEM/F12) containing 10% FBS and 1% penicillin/streptomycin was added to the samples before filtering through a 100um nylon mesh. SVF cells were pelleted (1200 RPM, 4°C, 10m), RBC-lysed, filtered through a 70um nylon mesh and washed in complete DMEM/F12 then used immediately for desired application. To purify ATMs, SFV cells were washed twice in FACS buffer consisting of PBS, 2% heat-inactivated fetal bovine serum (FBS), and 1mM EDTA then incubated in FcR-Blocker (StemCell Tech, 18720) followed by PE-conjugated anti-F4/80 (BioLegend, Clone BM8). F4/80+ cells were immunomagnetically selected by EasySep PE positive selection kit according to manufacturer protocol including 4 total wash steps (StemCell Tech, 18557). Flow cytometry was used to verify selection purity, which was greater than 85%.
RNA Purification, cDNA Synthesis, and quantitative RT-PCR
ATMs were lysed in Qiazol and total RNA was purified using Qiagen miRNeasy Micro kit. RNA concentration and purity was measured using a NanoDrop 2000 spectrophotometer. 400ng total RNA was reverse transcribed to cDNA using Qiagen miScript II RT kit with HiFlex buffer. To validate miRNA expression by qRT-PCR, miScript SYBR Green PCR kit and miScript miRNA Primer Assays were used (Qiagen).
ATM MicroRNA and Transcriptome Microarrays
MicroRNA and transcriptome microarrays were performed using 3 biological replicates of total RNA isolated from pools of ATMs (NCD: pools of 20 mice, HFD: pools of 10 mice). For each miRNA microarray, 500 ng total RNA was polyadenylated then labeled using the Affymetrix FlashTag Biotin HSR RNA Labeling Kit. Labeled samples were hybridized to Affymetrix miRNA 4.0 chips overnight (16 h, 48°C, 60 RPM) then washed, stained, and scanned on an Affymetrix GCS 3000 system following manufacturer protocols. For transcriptome microarrays, 100ng total RNA was used as starting material. RNA was prepared for hybridization by using the Affymetrix GeneChip WT PLUS Reagent Kit according to manufacturer protocol. Labeled samples were hybridized to MTA 1.0 chips overnight (16h, 45°C, 60 RPM) then washed, stained, and scanned on an Affymetrix GCS 3000 system. Affymetrix Expression Console Version 1.4.1.46 was used for quality control, data summarization, and normalization. Affymetrix Transcriptome Analysis Console Version 3.1.0.5 was used to perform differential expression analyses. Transcripts or miRNAs were considered differentially expressed between the two groups if linear fold change was greater than ±2 and the ANOVA p-value was less than 0.05. Heatmap figures of differentially expressed microRNAs and mRNAs were made using Genesis Version 1.7.7.21
Immunofluorescence
Epididymal fat was dissected, minced (3mm x 3mm2), washed in PBS, and then fixed in 4% paraformaldehyde for 3h. Fixed tissues were permeabilized with 1% Triton X-100 for 10 min then blocked with 1% BSA and FcR-Blocker for 1h at RT. Samples were incubated with primary antibody (BioLegend: anti-F4/80-AlexaFluor488 clone: BM8, and anti-Notch1 clone: HMN1-12, or anti-DLL4 clone: HMD4-1) overnight at 4°C, then incubated with anti- Hamster IgG-AlexaFluor633 secondary antibody for 1h at RT (Invitrogen SA1-26817, Molecular Probes labeling kit A20170). Tissues were counterstained with 40uM Hoechst 33342 and 5uM BODIPY 558/568 C12 (Molecular Probes H21492 & D3835) for 1h at RT then washed and mounted on slides using a Vaseline boundary and Fluoromount-G (eBioscience, 00-4958-02).
Confocal Microscopy and Image Analysis
Confocal images of whole-mount adipose tissue were acquired on a Zeiss LSM 510 Meta Confocal Scanning Laser Microscope equipped with UV, Argon, green HeNe and red HeNe lasers. Five random images per sample were taken using a 40X water immersion objective. Original .lsm files were imported into Fiji (Fiji Is Just ImageJ, NIH) then split into channels. Thresholds were applied to the Cy5 channel using Fiji’s max Entropy algorithm to identify regions of interest (ROIs) that express either Notch1 or DLL4. Area and intensity (mean gray value) were measured for each ROI. The product of positive signal area and intensity were used to determine total expression per image. Each biological replicate is the mean expression of 5 images. We then divided the expression values for each biological replicate by the mean of the NCD biological replicates. Therefore, data are presented as fold expression in arbitrary units (AU) with mean NCD set as control.
Flow Cytometry
Freshly isolated SVF cells or cultured BMDM were washed in FACS buffer then incubated on ice with FcR-Blocker for 10m followed by appropriate fluorochrome-conjugated antibodies or isotype controls (BioLegend, anti-CD11b-AlexaFluor488 clone: M1/70, anti-F4/80-PE clone: BM8, anti-DLL4-APC clone: HMD4-1, anti-CD45-PECy7 clone: 30-F11, anti-CD11c-APC clone: N418) for 50m. Stained cells were washed 3X in FACS buffer then analyzed on a Beckman Coulter FC500 or BD FACSCelesta flow cytometer. Plots were analyzed with Beckman Coulter CXP Software or FlowJo v10.
In Vitro Locked Nucleic Acid (LNA) Transfection Assays
Bone marrow derived macrophages (BMDM) were differentiated from bone marrow cells (BMC) by flushing the tibia and femur of 6–8 week old female C57Bl/6J mice with PBS. BMCs were filtered through a 70um nylon mesh, RBC-lysed, and washed, then cultured in complete DMEM/F12 supplemented with 10% FBS, 1% penicillin/streptomycin, 2mM L-glutamine, and 1U/mL M-CSF (BioLegend, 576406) for 7–10 days. 3T3-L1 adipocyte-conditioned media (CM-3T3-L1A) was generated by differentiating 3T3-L1 preadipocytes into adipocytes according to the Zen-Bio 3T3-L1 Adipocyte Care Manual (ZBM0009.03). Preadipocyte medium, differentiation medium, and adipocyte maintenance medium were also used (Zen-Bio, PM-1-L1, DM-2-L1, AM-1-L1). Conditioned medium was collected between days 7 and 14 post-differentiation, 0.22um filtered, aliquoted, and stored at −80°C until use. For transfection assays, mature BMDM were plated in poly-D-lysine-coated 6-well plates at a density of 5x105 cells in 2ml CM-3T3-L1A containing 10% FBS without antibiotics. BMDM were incubated 24h (37°C, 5% CO2, 95% humidity) before transfection. Transfection complexes were prepared by diluting Lipofectamine3000 and LNA in Opti-MEM to final concentrations of 2% (v/v) and 0.32μM respectively. Mixtures were incubated 15–20m at RT to allow complexes to form. Meanwhile, conditioned BMDM were washed 3X in pre-warmed Dulbecco’s PBS (DPBS), then media replaced with 2mL Opti-MEM. 500uL transfection complexes were added drop-wise to each well. Cells were incubated (37°C, 5% CO2, 95% humidity) 5–6hr to allow LNA uptake, washed 3X in pre-warmed DPBS, then media replaced with DMEM/F12 containing 10%FBS, 1% penicillin/streptomycin, and 2mM L-glutamine, and cultured for an additional 18h. For inhibition studies, DAPT (5uM), anti-DLL4 antibody (1ug/mL), or appropriate vehicle/isotype antibody controls were added to the culture media. MirCURY LNA oligonucleotides were obtained from Exiqon (LNA Sequences— Ctr LNA: TAACACGTCTATACGCCCA, Anti-30a: TTCCAGTCGAGGATGTTTAC, Anti-30c: CTGAGAGTGTAGGATGTT, Anti-30e: TCCAGTCAAGGATGTTTAC).
Protein Extraction and Western Blotting
Cultured BMDM were washed twice in ice-cold PBS, and then directly scrapped into 100uL blue loading buffer (Cell Signaling Tech 7722) and kept on ice. Protein lysates were sonicated 10s then heated at 95°C for 5m before loading 20uL on Mini-Protean TGX Protein Gels (BioRad 4569034). Precision Plus Protein Dual Color Standards (BioRad 1610374) were loaded for a molecular weight ladder. Samples were run 40V for 30 min followed by 80V for 1.5h. Proteins were transferred to nitrocellulose membranes by using iBlot 2 NC stacks and the ThermoFisher iBlot 2 western transfer system running the P0 protocol (20V 1m, 23V 4m, then 25V 2m). Membranes were blocked in 5% dry milk or 5% BSA for 1h then washed 3X in Tris-buffered saline containing 0.1% Tween-20 (TBS-T). Membranes were incubated in primary antibody overnight at 4°C with gentle shaking then washed 3 x 5m in TBS-T. Membranes were incubated in secondary antibody for 1h at RT, then washed 3 x 5m in TBS-T before addition of ECL substrate and exposure to x-ray film. Films were scanned and densitometry measurements were made using ImageJ gel analysis features (NIH).
Enzyme-linked Immunosorbent Assays (ELISA)
Culture supernatants were aspirated and centrifuged 5000 RPM, 5m, 4°C to rid of debris then aliquoted and stored −80°C before assaying. DLL4 ELISA kits were purchased from Abcam (ab213860). Mouse TNFα and CCL2 ELISA kits were purchased from BioLegend (TNFα 530901 & CCL2 432701). Assays were performed according to manufacturer protocols and plates were read at 450nm. Concentrations were calculated using standard curves.
Methylated DNA Immunoprecipitation Sequencing (MeDIP-seq)
MeDIP-seq libraries were generated from ATM DNA as previously described and sequenced with single-end reads of 75bp on an Illumina NextSeq500 22. Data analysis was performed as previously described 22. Mapped reads were analyzed using MEDIPS software 23. Peaks were visualized in the Integrated Genome Browser 24. The UCSC genome browser was used to locate CpG islands within 10kb of miR-30 gene coding regions 25.
Methylation-specific PCR
Genomic DNA from NCD and HFD ATMs were isolated using Qiagen AllPrep DNA/RNA/miRNA Universial Kit (80224). Bisulfite conversion of DNA was performed using Qiagen EpiTect Fast DNA Bisulfite Kit (59824). PCR using methylated and unmethylated primers specific for the Nfyc CpG island (Methylated: Fwd—TTCGTTAATGGGAGAAAGTTC Rev—CTACCGCCGCCATATTATA Unmethylated: Fwd—TTTTTTGTTAATGGGAGAAAGTTT, Rev—ACTCTACCACCACCATATTATA). Primers were designed using ThermoFisher Methyl Primer Express Software v1.0. BioRad iQ SYBR Green Supermix was used and qRT-PCR was run using the following reaction conditions: initial denaturation- 95°C 5m followed by 31 cycles of - 95°C 15s, 49.8°C 30s, and 70°C 35s. PCR products were run on a 1.5% agarose gel and bands were quantified using FIJI gel analysis features. Methylation ratio was determined by dividing methylated by unmethylated quantities (M/U).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism Version 7.000 for Mac, GraphPad Software, La Jolla California USA, www.graphpad.com. Values are expressed as mean ± standard error. Two-tailed Student’s t tests were performed for paired analyses. One-way ANOVA with a Bonferroni post hoc correction were used for multiple group analyses. The null hypothesis was rejected if p<0.05. All experiments were repeated at least twice, unless otherwise indicated in each figure legend. Detailed sample sizes are provided in each figure legend. Sample sizes were chosen by power analysis based on pilot studies.
Results
Obesity Promotes ATM miRNA Dysregulation and Pro-Inflammatory Phenotype
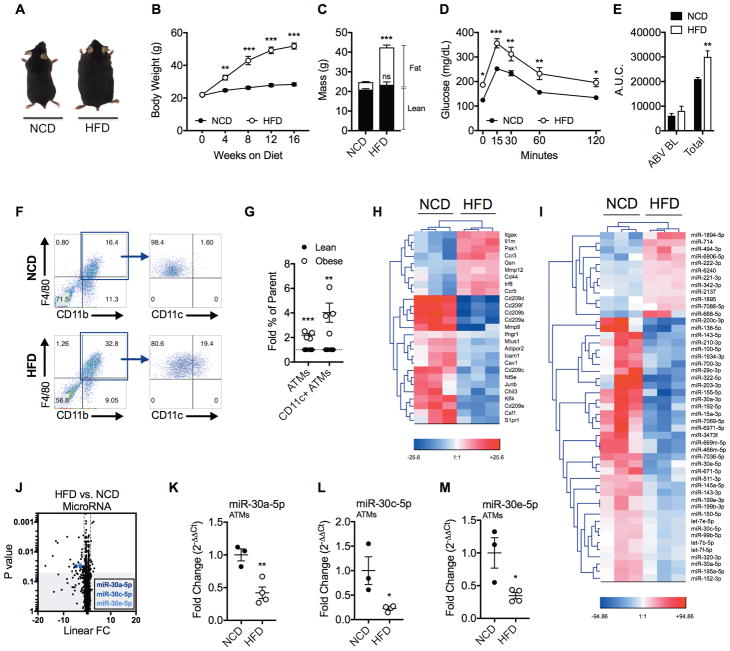
To study ATMs associated with obesity (will be referred to as obese ATMs), we used a high-fat diet (HFD)-induced obesity model whereby 6-week-old male C57BL/6J mice were fed HFD or normal chow diet (NCD) for 16 weeks to generate diet-induced obese and lean control mice. HFD-fed mice more than doubled their body weight during 16 weeks of feeding whereas NCD-fed mice increased their weight by ~3% (Figures 1A–1C), and HFD-induced weight gain occurred due to selective increases in fat mass (Figure 1C). As expected, HFD also caused glucose intolerance measured by glucose tolerance test (GTT) (Figures 1D and 1E).
Fig. 1. HFD-Induced Obesity Stimulates ATM Inflammation and miRNA Dysregulation.
To study HFD-induced obesity, male C57BL/6J mice were fed NCD or HFD for 16 weeks until 22-weeks-old. (A) Representative photo of mice after 16 weeks of NCD or HFD feeding. (B) Weekly measurements of body weight growth. (C) DEXA body composition after 16 weeks of diet. (D) Oral glucose tolerance test (GTT) after 16 weeks of diet. (E) Area under the curve (A.U.C.) for GTT. Represented are A.U.C. above baseline (ABV BL) or total A.U.C. (F) Flow cytometry dot plots of F4/80+/CD11b+/CD11c+ ATMs in the epididymal fat stromal vascular fraction (SVF) of NCD- or HFD-fed mice. (G) Fold percentage increase quantification of F4/80+/CD11b+ cells in the SVF (denoted as “ATMs”) and CD11c+ ATMs. Lean mice were fed either NCD or 10% low-fat diet (LFD). Obese mice were fed 60% HFD. (H–J) Pooled F4/80+ ATMs from epididymal fat were used for transcriptome and miRNA microarrays. (H) Transcriptome microarray heatmap of differentially expressed mRNAs in ATMs related to macrophage polarization. (I) MicroRNA heatmap of differentially expressed miRNAs in ATMs. (J) MicroRNA array volcano plot depicting linear fold change (FC) vs. ANOVA p-value significance. (K–M) qRT-PCR expression validation of miRs 30a-5p, 30c-5p, and 30e-5p. For (A–F), the values are shown as mean ± SEM and are from a single experiment representative of at least 3 independent experiments with 5 mice per experimental group. For (G) data shown as mean ± SEM of 4 independent experiments with 5 mice per experimental group. For (H–M), the data shown are mean ± SEM and are from 3–4 independent experiments with 20 pooled NCD mice and 10 pooled HFD mice per experiment. Statistical differences were determined by using Student’s t-test. *p<0.05, **p<0.01, ***p<0.001. See also Figure S1 and Table S1.
While phenotyping ATMs, we observed percentages of ATMs (F4/80+/CD11b+) and CD11c+ ATMs in epididymal fat of obese mice were more than 2 and 4 fold that of lean mice respectively (Figure 1F and 1G). To identify gene expression alterations in HFD and NCD ATMs we performed transcriptome microarrays using F4/80+ cells from epididymal fat of HFD and NCD mice. Principal component analysis (PCA) displayed HFD and NCD ATMs have distinct transcript expression profiles (Supplemental Figure 1A). HFD ATMs exhibited increased M1- and decreased M2-associated gene expression (Figure 1H). Notably, Irf8, which encodes a transcription factor activated by Notch-RBPJ signaling, as well as Itgax, which encodes the M1 surface marker CD11c, were upregulated in obese ATMs 3,8. Alternatively, Klf4, which encodes Krüppel-like factor 4 that cooperates with STAT6 to promote M2 polarization, and Adipor2, which encodes a receptor for the anti-inflammatory adipokine adiponectin, were downregulated in obese ATMs 26,27. Together these observations suggested that HFD ATM phenotype was skewed towards M1.
To identify differentially expressed miRNAs in ATMs during obesity, we performed miRNA microarrays using F4/80+ cells isolated from epididymal fat of HFD and NCD mice. PCA showed HFD and NCD ATMs have distinct miRNA expression profiles (Supplemental Figure 1B). In total, there were 37 down- and 12 up-regulated miRNAs in HFD versus NCD ATMs (Figures 1I and 1J). Additionally, transcriptome microarrays showed there were 946 down- and 920 up-regulated transcripts in HFD versus NCD ATMs. Of these, 216 and 273 coding genes were up- and down-regulated respectively (Supplemental Figure 1C). We performed core analysis on these dysregulated miRNAs and transcripts using Ingenuity Pathway Analysis and observed significant overlap with canonical pathways including hepatic fibrosis and atherosclerosis signaling, disorders such as cancer and hepatic disease, cellular functions including movement and survival, and toxic effects including cardiotoxicity, hepatotoxicity, and nephrotoxicity (Supplemental Figure 1D–1H).
Upon closer examination of dysregulated miRNAs during obesity, we observed downregulation of miR-322-5p (−17.1 linear FC) and miR-155-5p (−14.19 linear FC), which have been previously characterized for their involvement in macrophage functions 28,29. Interestingly, we also noted downregulation of miRs -30a-5p, -30c-5p, and -30e-5p in HFD ATMs (−12.27 combined linear FC) when compared to NCD ATMs, thereby indicating that the miR-30 family may play a role in macrophage polarization (Figures 1I–1M).
The miR-30 target DLL4 is associated with ATM inflammation
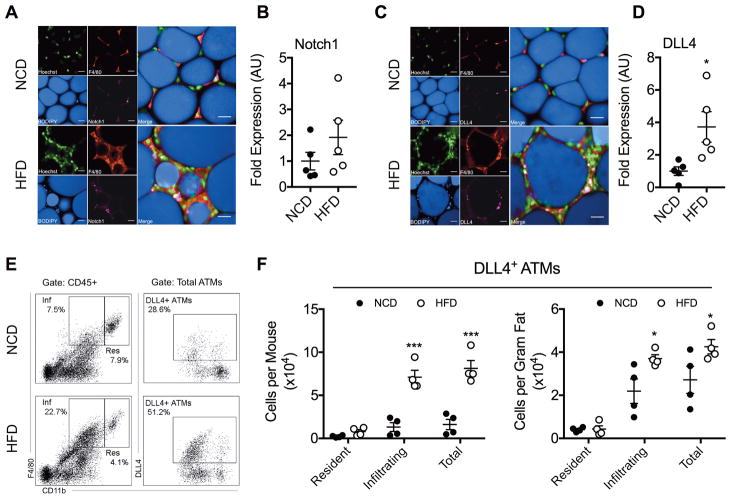
When we looked at potential target molecules for miR-30, we found through in silico analyses that it may target the 3’UTR of Dll4 (Supplemental Figures 2A–2C) Previous studies had confirmed miR-30-Dll4 targeting using luciferase reporter assay 30,31. Interestingly, DLL4 has been shown to be involved in Notch signaling 9,32,33. To that end, we evaluated expression of Notch1 and the miR-30 target DLL4 in adipose tissue. Notch1 and DLL4 were visualized in whole-mounted epididymal fat by confocal microscopy (Figures 2A and 2C). Adipose tissue expression of DLL4 but not Notch1 was elevated in HFD-fed mice (Figures 2B and 2D). Flow cytometry analysis of SVFs was then used to confirm that DLL4 expression was elevated on ATMs (CD45+/CD11b+/F4/80+/DLL4+) (Figures 2E and 2F). Specifically, elevated DLL4 expression was most pronounced in the CD45+/CD11bint/F4/80+ subset of infiltrating ATMs (Figure 2F).
Fig. 2. DLL4 Expression is Elevated in Obese ATMs.
HFD-induced obesity was studied as described in Fig 1 legend. At 22-weeks-old, visceral adipose tissue and adipose SVF were analyzed for DLL4 expression. (A&C) Immunofluorescent staining of whole-mount epididymal fat. Scale bar = 20 μm. Presented are representative confocal micrographs of Notch1 (A) or DLL4 (C). (B&D) Image quantification of Notch1 (B) or DLL4 (D) in adipose tissue. The values are shown as mean ± SEM and are from a single experiment representative of 2 independent experiments with 5 mice per experimental group. (E) Flow cytometry dot plots of DLL4+ ATMs in epididymal fat. CD11bint are denoted as infiltrating (“Inf”) and CD11bhi are denoted as resident (“Res”). (F) Quantification of DLL4+ ATM cell counts represented per mouse and per gram fat. Values are presented as mean ± SEM and are from a single experiment representative of 2 independent experiments with 4 biological replicates (pools of 1–6 mice) per experimental group. Statistical significance was determined by Student’s t-test. *p<0.05, ***p<0.001. See also Figure S2.
MiR-30 Inhibition Promotes DLL4-Notch Signaling-Induced Inflammation in Macrophages
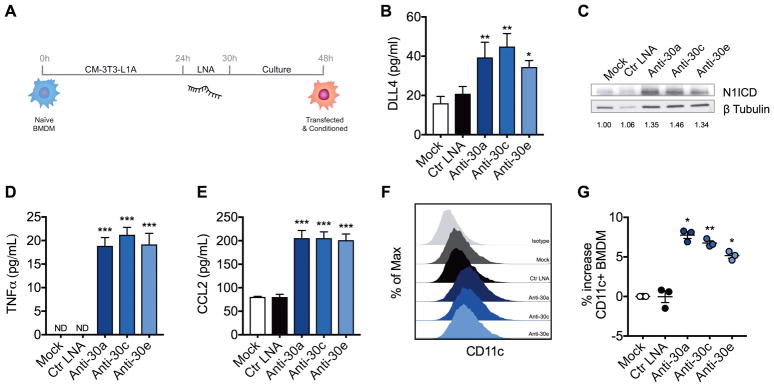
To further demonstrate involvement of miR-30 in regulation of DLL4-mediated Notch signaling and pro-inflammatory response in macrophages, we developed an in vitro assay to mimic the downregulated miR-30 expression observed in vivo in obese ATMs. To that end, naïve bone marrow derived macrophages (BMDM) were cultured in conditioned medium from differentiated 3T3-L1 adipocytes (CM-3T3-L1A) to confer an ATM-like phenotype, and then transfected with miRNA inhibitor locked nucleic acids (LNA) which targeted miRs-30a-5p (Anti-30a), -30c-5p (Anti-30c), and -30e-5p (Anti-30e) (Figure 3A). BMDM transfected with Anti-30a, Anti-30c, and Anti-30e LNAs had decreased expression of miRs -30a-5p, -30c-5p, and -30e-5p relative to Mock and control LNA (Ctr LNA)-transfected controls, although the inhibitors displayed some cross-reactivity (Supplemental Figure 3). Anti-30a, Anti-30c, and Anti-30e transfection increased DLL4 and activated Notch1 intracellular domain (N1ICD) expression compared to Mock and Ctr LNA (Figure 3B–C). Pro-inflammatory cytokines TNFα and CCL2 were also elevated in culture supernatants of inhibitor-transfected cells (Figures 3D and 3E). Moreover, miR-30 inhibitors promoted increased surface expression of CD11c (Figures 3F and 3G). Treatment of transfected cells with the Notch/γ-secretase inhibitor DAPT reduced induction of CD11c in miR-30 inhibitor-transfected cells (Supplemental Figure 4B). Specific blockade of DLL4 signaling using anti-DLL4 antibody also reduced induction of pro-inflammatory cytokines TNFα and CCL2 in miR-30 inhibitor-transfected cells (Supplemental Figure 4C & 4D). Conversely, lentiviral overexpression of miR-30a-5p in the RAW264.7 macrophage cell line reduced M1 polarization evidenced by decreased expression of CD11c and decreased TNFα and CCL2 production (Supplementary Figure 5). Together these data demonstrated that miR-30 plays an anti-inflammatory role in macrophages by regulating DLL4-Notch1 signaling, M1 polarization and pro-inflammatory cytokine production in macrophages.
Fig. 3. miR-30 Inhibition Induces DLL4-Notch1 Signaling and M1 polarization.
BMDM were differentiated from the bone marrow of normal mice then used to study in vitro consequences of miR-30 inhibition on Notch signaling and macrophage polarization. (A) Schematic of in vitro experimental timeline. Differentiated BMDM were incubated in CM-3T3-L1A prior to transfection with microRNA inhibitor LNA and subsequent culture. Cells and supernatants were harvested at 48h. (B) DLL4 detected in culture supernatants by ELISA. (C) Western blot of cleaved/activated Notch1 (N1ICD). Fold induction relative to Mock is represented below each lane. (D–E) Pro-inflammatory cytokines TNFα (D), and CCL2 (E) detected in culture supernatants by ELISAs. (F) Flow cytometry histograms of CD11c expression in transfected BMDM. (G) Percentage increase in CD11c+ macrophages. For figures B–F, values are presented as mean ± SEM and are from a single experiment representative of 2–3 independent experiments. For figure E, data presented are mean ± SEM of 3 independent experiments. Statistical significance was determined by one-way ANOVA with Bonferroni post hoc correction. *p<0.05, **p<0.01, ***p<0.001 vs. Mock. See also Figures S3–S5.
Evidence for DNA Methylation-Dependent Regulation of miR-30
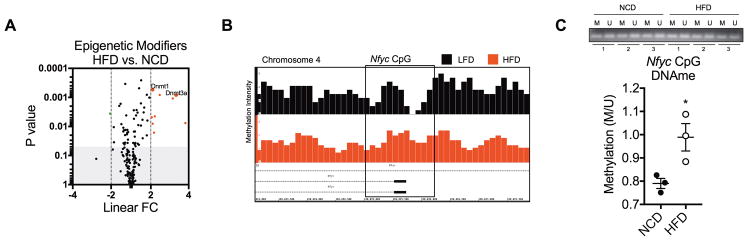
Epigenetic modifications can occur due to various environmental factors such as stress, aging, and diet. Because this study involved diet-induced obesity, we investigated epigenetic mechanisms that may control miR-30 expression in ATMs. Data from transcriptome microarrays enriched for epigenetic modification enzymes and factors revealed that gene expression of Dnmt1 and Dnmt3a were upregulated in ATMs of HFD vs. NCD-fed mice (Figure 4A). Therefore, we investigated DNA methylation intensity of miR-30 gene regions. Mir30c-1 and mir30e are located within the same intron of the Nfyc gene, which contains a CpG island in its promoter region. Mir30a and mir30c-2 are intergenic miRNA genes that do not have any nearby CpG islands, therefore we were not able to identify DNA methylation as a potential epigenetic mechanism regulating their expression. We performed methylated-DNA immunoprecipitation sequencing (MeDIP-seq) to screen genome-wide DNA methylation in ATMs and found DNA hypermethylation in the Nfyc-promoter CpG island in HFD- versus LFD-ATMs indicating expression of miRs -30c and -30e may be regulated by DNA methylation. This was validated in NCD- and HFD- ATMs by methylation-specific PCR (Figure 4C). Together, these data indicated DNA methylation-dependent downregulation of miR-30 may promote pro-inflammatory polarization of adipose tissue macrophages during obesity.
Fig. 4. DNA Methylation-Dependent Regulation of miR-30.
As shown in Figure 1, F4/80+ ATMs were isolated from epididymal fat of 22-week-old HFD-induced obese and lean mice. Gene expression and DNA methylation were evaluated in ATMs. (A) Volcano plot displaying linear FC of genes encoding epigenetic modification enzymes and factors. Fold change and p value observations were extracted from transcriptome microarrays (See Figure 1 and Figure S1). (B) IGB visualization of meDIP-seq peak intensity of DNA methylation in the Nfyc promoter CpG island. (C) Methylation-specific PCR quantification of DNA methylation (DNAme) in the Nfyc promoter CpG island. For (A and C), values presented are representative of 3 independent experiments with 20 pooled NCD mice and 10 pooled HFD mice per experiment. For (B), data are representative of one experiment of 60 pooled LFD and 30 pooled HFD mice. Statistical differences were determined by using Student’s t-test. *p<0.05.
Discussion
There is a clear association between pro-inflammatory ATM polarization and insulin resistance during HFD-induced obesity. However, miRNA-mediated epigenetic mechanisms regulating ATM phenotype have not been well characterized. Our results unveil a role for the miR-30 family in modulating polarization of macrophages in visceral adipose tissue through regulation of the Notch1 ligand DLL4. Furthermore, we identified that DNA methylation may play a role in attenuating miR-30 expression. Jointly, these findings suggest important roles for miRNAs and DNA methylation in regulation of ATM polarization and obesity-induced insulin resistance.
DLL4 is a canonical Notch1 ligand that is linked to macrophage inflammation and metabolic disorders. DLL4 promotes M1 macrophage polarization 32,33. Blockade of DLL4 by administration of anti-DLL4 antibody in mice attenuates atherosclerosis and metabolic disease by decreasing macrophage infiltration and inflammatory response 9. Interestingly, Notch signaling is also linked to insulin resistance 6,7. Conditional knockout of Notch1 or its downstream transcription factor RBP-J in adipocytes ameliorates obesity and insulin resistance by promoting white adipose tissue browning and energy expenditure in mice 6. These findings suggest that downregulation of the DLL4-Notch1 axis may hold significant therapeutic potential for various inflammatory and metabolic disorders. Furthermore, identification of regulatory mechanisms controlling this pathway could reveal therapeutic targets for cardiometabolic disorders. In the current study, we identified epigenetic regulatory mechanisms of this pathway in ATMs.
Growing bodies of evidence suggest miRNA dysregulation and aberrant DNA methylation play pathogenic roles in clinical disorders such as type II diabetes, atherosclerosis, and cancer. Inflammation is many times the underlying cause for these chronic diseases, and macrophages are key mediators of chronic-inflammation. Additionally, obesity is well known to greatly increase risk for these diseases. Because ATMs are a primary source of pro-inflammatory cytokines that initiate chronic sub-clinical inflammation during obesity, ATMs contribute to progression towards comorbid diseases. In the current study, we observed increased infiltration and pro-inflammatory polarization of macrophages in visceral adipose tissue. When we performed core analysis of dysregulated miRNAs and transcripts in ATMs during diet-induced obesity, we found significant overlap with cardiometabolic disease processes and cancer, which suggests that ATM-inflammation is tightly linked to the development of comorbid diseases.
Recent reports have linked specific miRNAs to obesity as well as macrophage polarization. In the context of obesity, expression profiles of human adipose tissue have identified miRNAs such as miR-132 and miR-17-5p to be involved in regulation of adipocyte growth and insulin resistance 34. In macrophage polarization, miR-146a and miR-155-5p are classified to be negative regulators of inflammatory responses to the bacterial endotoxin lipopolysaccharide 35. MicroRNA expression profiles of whole adipose tissue and in vitro polarized macrophages have been previously reported, however dysregulated miRNAs between lean and obese ATMs have not been characterized 19,34,36. ATMs form heterogenous populations and their inflammatory status greatly influences adipose insulin sensitivity. In the current study, the miRNAs that exhibited the most significant fold change in ATMs were miR-322-5p and miR-155-5p. miR-322-5p is well characterized to inhibit pro-inflammatory cytokine production in macrophages, while miR-155-5p is also known to be involved in a variety of macrophage processes 28,29. Most strikingly, we observed downregulation of several miR-30 family miRNAs in ATMs during obesity, yet this miR family has not been previously investigated for its role regulating macrophage phenotype. The miR-30 family is highly conserved and known to target the Notch1 ligand DLL4, which is a ligand that contributes to metabolic disease and macrophage inflammation 9,30–33,37. We observed upregulation of DLL4 on obese ATMs. Moreover, inhibiting miR-30 in vitro triggered DLL4-Notch1 signaling and pro-inflammatory response in macrophages, and blocking DLL4-Notch1 signaling could lessen this effect. Overexpression of miR-30a-5p also decreased M1 polarization in the RAW264.7 macrophage cell line. Thus, we have observed that miR-30 is involved in attenuating M1 macrophage activation through regulation of the DLL4-Notch signaling pathway. These data suggest that miR-30 induction holds therapeutic potential for regulating macrophage-driven inflammatory and metabolic disorders. Furthermore, regulatory mechanisms of the miR-30 family warrant further investigation due to emerging advances in epigenetic therapies and targeted personalized medicine.
To investigate the mechanisms through which obesity downregulates miR-30 in ATMs, we focused on epigenetic regulation. We observed elevated expression of DNMTs and DNA hypermethylation in miR-30-associated CpG islands in obese vs. lean ATMs. Thus, HFD may trigger DNA hypermethylation of miR-30 genes leading to downregulation of miR-30 to allow increased expression of DLL4 and Notch signaling-induced inflammation and insulin resistance during obesity.
Previous studies have suggested that miR-30 induction has metabolic benefits. MicroRNA-30c attenuates hyperlipidemia, hypercholesterolemia, and atherosclerosis in mice, while miR-30b/c promotes thermogenesis and white adipose tissue browning 38–40. These findings suggest that in vivo manipulation of the miR-30 family holds promise for therapeutic intervention of obesity-related disorders. Additionally, macrophage polarization plays key roles in regulation of both brown adipose tissue metabolism and inflammation during atherosclerosis, yet macrophage phenotype was not assessed in the aforementioned studies 41–43. Our data suggest that macrophage-specific miR-30 regulates polarization towards inflammatory phenotype and progression of chronic macrophage-driven inflammatory disorders. Thus, future studies targeting macrophage-specific miR-30 in vivo could shed new light on mechanistic and therapeutic potential of this miR family in attenuating obesity and other metabolic disorders.
Supplementary Material
Acknowledgments
Grant Support: NIH grants R01ES019313, R01MH094755, R01AI123947, R01AI129788, P01AT003961, P20GM103641, and R01AT006888.
The authors would like to thank Dr. Robert Price, Ph.D., and Director of the University of South Carolina Instrumentation Resource Facility for his kind assistance with confocal microscopy acquisition.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
The data discussed in this publication have been deposited in the NCBI’s Gene Expression Omnibus and are accessible through the GEO Series accession numbers GSE114735, GSE114736, and GSE11479244.
Author contributions: Conceptualization, K.M., P.S.N., and M.N.; Methodology, K.M., E.A.M., P.S.N., and M.N.; Validation, K.M.; Formal Analysis, K.M., X.Y.; Investigation, K.M., X.Y., M.B.; Resources, P.S.N. and M.N.; Writing- Original Draft, K.M.; Writing- Review & Editing, K.M., M.B., P.S.N., and M.N.; Visualization, K.M.; Supervision, P.S.N. and M.N.; Funding Acquisition, P.S.N. and M.N.
References and Notes
- 1.Hamilton TA, Zhao C, Pavicic PG, Datta S. Myeloid colony-stimulating factors as regulators of macrophage polarization. Front Immunol. 2014;5:554. doi: 10.3389/fimmu.2014.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boutens L, Stienstra R. Adipose tissue macrophages: going off track during obesity. Diabetologia. 2016;59:879–894. doi: 10.1007/s00125-016-3904-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi P, Kuang S. Notch signaling as a novel regulator of metabolism. Trends Endocrinol Metab. 2015;26:248–255. doi: 10.1016/j.tem.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borggrefe T, Liefke R. Fine-tuning of the intracellular canonical Notch signaling pathway. Cell Cycle. 2012;11:264–276. doi: 10.4161/cc.11.2.18995. [DOI] [PubMed] [Google Scholar]
- 6.Bi P, Shan T, Liu W, Yue F, Yang X, Liang X-R, et al. Inhibition of Notch signaling promotes browning of white adipose tissue and ameliorates obesity. Nat Med. 2014;20:911–918. doi: 10.1038/nm.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pajvani UB, Shawber CJ, Samuel VT, Birkenfeld AL, Shulman GI, Kitajewski J, et al. Inhibition of Notch signaling ameliorates insulin resistance in a FoxO1-dependent manner. Nat Med. 2011;17:961–967. doi: 10.1038/nm.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol. 2012;13:642–650. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuda D, Aikawa E, Swirski FK, Novobrantseva TI, Kotelianski V, Gorgun CZ, et al. Notch ligand delta-like 4 blockade attenuates atherosclerosis and metabolic disorders. Proc Natl Acad Sci USA. 2012;109:E1868–1877. doi: 10.1073/pnas.1116889109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bam M, Yang X, Zumbrun EE, Zhong Y, Zhou J, Ginsberg JP, et al. Dysregulated immune system networks in war veterans with PTSD is an outcome of altered miRNA expression and DNA methylation. Sci Rep. 2016;6:31209. doi: 10.1038/srep31209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busbee PB, Nagarkatti M, Nagarkatti PS. Natural Indoles, Indole-3-Carbinol (I3C) and 3,3’-Diindolylmethane (DIM), Attenuate Staphylococcal Enterotoxin B-Mediated Liver Injury by Downregulating miR-31 Expression and Promoting Caspase-2-Mediated Apoptosis. PLOS ONE. 2015;10:e0118506. doi: 10.1371/journal.pone.0118506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan H, Singh UP, Rao R, Mrelashvili D, Sen S, Hao H, et al. Inverse correlation of expression of microRNA-140-5p with progression of multiple sclerosis and differentiation of encephalitogenic T helper type 1 cells. Immunology. 2016;147:488–498. doi: 10.1111/imm.12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh UP, Murphy AE, Enos RT, Shamran HA, Singh NP, Guan H, et al. miR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology. 2014;143:478–489. doi: 10.1111/imm.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Xiao C, Rajewsky K. MicroRNA Control in the Immune System: Basic Principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Dong C, Yoon W, Goldschmidt-Clermont PJ. DNA methylation and atherosclerosis. J Nutr. 2002;132:2406S–2409S. doi: 10.1093/jn/132.8.2406S. [DOI] [PubMed] [Google Scholar]
- 18.Zhang FF, Cardarelli R, Carroll J, Zhang S, Fulda KG, Gonzalez K, et al. Physical activity and global genomic DNA methylation in a cancer-free population. Epigenetics. 2011;6:293–299. doi: 10.4161/epi.6.3.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge Q, Brichard S, Yi X, Li Q. microRNAs as a New Mechanism Regulating Adipose Tissue Inflammation in Obesity and as a Novel Therapeutic Strategy in the Metabolic Syndrome. J Immunol Res. 2014;2014 doi: 10.1155/2014/987285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 22.Bam M, Yang X, Zhou J, Ginsberg JP, Leyden Q, Nagarkatti PS, et al. Evidence for Epigenetic Regulation of Pro-Inflammatory Cytokines, Interleukin-12 and Interferon Gamma, in Peripheral Blood Mononuclear Cells from PTSD Patients. J Neuroimmune Pharmacol. 2015;11:168–181. doi: 10.1007/s11481-015-9643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics. 2014;30:284–286. doi: 10.1093/bioinformatics/btt650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freese NH, Norris DC, Loraine AE. Integrated genome browser: visual analytics platform for genomics. Bioinformatics. 2016;32:2089–2095. doi: 10.1093/bioinformatics/btw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, et al. Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest. 2011;121:2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Stijn CMW, Kim J, Lusis AJ, Barish GD, Tangirala RK. Macrophage polarization phenotype regulates adiponectin receptor expression and adiponectin anti-inflammatory response. FASEB J. 2015;29:636–649. doi: 10.1096/fj.14-253831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang K, Song F, Lu X, Chen W, Huang C, Li L, et al. MicroRNA-322 inhibits inflammatory cytokine expression and promotes cell proliferation in LPS-stimulated murine macrophages by targeting NF-κB1 (p50) Bioscience Reports. 2017;37:BSR20160239. doi: 10.1042/BSR20160239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bridge G, Monteiro R, Henderson S, Emuss V, Lagos D, Georgopoulou D, et al. The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood. 2012;120:5063–5072. doi: 10.1182/blood-2012-04-423004. [DOI] [PubMed] [Google Scholar]
- 31.Shan T-D, Ouyang H, Yu T, Li J-Y, Huang C-Z, Yang H-S, et al. miRNA-30e regulates abnormal differentiation of small intestinal epithelial cells in diabetic mice by downregulating Dll4 expression. Cell Prolif. 2016;49:102–114. doi: 10.1111/cpr.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fung E, Tang S-MT, Canner JP, Morishige K, Arboleda-Velasquez JF, Cardoso AA, et al. Delta-Like 4 Induces Notch Signaling in Macrophages. Circulation. 2007;115:2948–2956. doi: 10.1161/CIRCULATIONAHA.106.675462. [DOI] [PubMed] [Google Scholar]
- 33.Nakano T, Fukuda D, Koga J, Aikawa M. Delta-Like Ligand 4-Notch Signaling in Macrophage Activation. Arteriosclerosis, Thrombosis, and Vascular Biology. 2016;36:2038–2047. doi: 10.1161/ATVBAHA.116.306926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klöting N, Berthold S, Kovacs P, Schön MR, Fasshauer M, Ruschke K, et al. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS ONE. 2009;4:e4699. doi: 10.1371/journal.pone.0004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taganov KD, Boldin MP, Chang K-J, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. PNAS. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Zhang M, Zhong M, Suo Q, Lv K. Expression profiles of miRNAs in polarized macrophages. Int J Mol Med. 2013;31:797–802. doi: 10.3892/ijmm.2013.1260. [DOI] [PubMed] [Google Scholar]
- 37.Fukuda D, Aikawa M. Expanding role of delta-like 4 mediated notch signaling in cardiovascular and metabolic diseases. Circ J. 2013;77:2462–2468. doi: 10.1253/circj.cj-13-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu F, Wang M, Xiao T, Yin B, He L, Meng W, et al. miR-30 Promotes Thermogenesis and the Development of Beige Fat by Targeting RIP140. Diabetes. 2015;64:2056–2068. doi: 10.2337/db14-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irani S, Pan X, Peck BCE, Iqbal J, Sethupathy P, Hussain MM. MicroRNA-30c Mimic Mitigates Hypercholesterolemia and Atherosclerosis in Mice. J Biol Chem. 2016;291:18397–18409. doi: 10.1074/jbc.M116.728451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13:709–721. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu Y, Nguyen KD, Odegaard JI, Cui X, Tian X, Locksley RM, et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell. 2014;157:1292–1308. doi: 10.1016/j.cell.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao RR, Long JZ, White JP, Svensson KJ, Lou J, Lokurkar I, et al. Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. Cell. 2014;157:1279–1291. doi: 10.1016/j.cell.2014.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002 Jan 1;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.