Abstract
Clostridium perfringens causes many different histotoxic and enterotoxic diseases in humans and animals as a result of its ability to produce potent protein toxins, many of which are extracellular. The current scheme for the classification of isolates was finalized in the 1960s and is based on their ability to produce a combination of four typing toxins - α-toxin, β-toxin, ε-toxin and ι-toxin – to divide C. perfringens strains into toxinotypes A to E. However, this scheme is now outdated since it does not take into account the discovery of other toxins that have been shown to be required for specific C. perfringens-mediated diseases. We present a long overdue revision of this toxinotyping scheme. The principles for the expansion of the typing system are described, as is a mechanism by which new toxinotypes can be proposed and subsequently approved. Based on these criteria two new toxinotypes have been established. C. perfringens type F consists of isolates that produce C. perfringens enterotoxin (CPE), but not β-toxin, ε-toxin or ι-toxin. Type F strains will include strains responsible for C. perfringens-mediated human food poisoning and antibiotic associated diarrhea. C. perfringens type G comprises isolates that produce NetB toxin and thereby cause necrotic enteritis in chickens. There are at least two candidates for future C. perfringens toxinotypes, but further experimental work is required before these toxinotypes can formally be proposed and accepted.
1. Introduction and historical perspective
Clostridium perfringens is a Gram-positive spore-forming anaerobe that is the causative agent of many histotoxic and enterotoxic diseases in humans and animals [1]. The key feature of these diseases is that they are mediated by the production of potent protein toxins, most of which are extracellular. These toxins generally are involved in specific disease syndromes. For example, α-toxin is essential for human clostridial myonecrosis or gas gangrene [2], C. perfringens enterotoxin (CPE) is required for human food poisoning [3], β-toxin is essential for specific enteric C. perfringens infections in several species [4, 5], ε-toxin is the key toxin in many enterotoxemic C. perfringens infections in sheep and goats [6] and NetB toxin is essential for necrotic enteritis in chickens [7].
C. perfringens was first isolated at the end of the nineteenth century from a case of acute articular rheumatism [8] and from a cadaver of a person who died of an aortic aneurism [9]. Welch and Nuttal named the organism as Bacillus aerogenes capsulatus nov. spec. [9] and Fränkel subsequently called it Bacillus phlegmonis emphysematosae [10]. It was subsequently called Bacillus perfringens, from the Latin name perfringere, to break, since the culture in deep agar medium disrupts the agar by abundant production of gas [11], and Bacillus welchii, in honour of William H. Welch [12]. The genus Clostridium, from the Greek name kloster latinized into Clostridium, meaning spindle-shaped [13, 14], was formally proposed in 1920 and both Clostridium perfringens and Clostridium welchii were listed in the Society for American Bacteriologists report on bacterial classification [15].
Pribram [16] distinguished the genus Clostridium (motile and non-capsulated bacteria) and the genus Welchia (non-motile and capsulated bacteria). This classification was used by Prévot and French bacteriologists from 1933 [17], with two species recognized in the genus Welchia, W. perfringens and W. agni on the basis of different toxicity [18, 19]. The older species name, perfringens, was used in 1931 by the Permanent Standards Commission of the Health Organization of the League of Nations (as reported in [20]) and the current name of C. perfringens adopted thereafter [21]. However, for many years the organism was called Clostridium welchii in English-speaking countries, C. perfringens in French-speaking countries and even Fränkel’s bacillus in Germany. C. perfringens was one of the approved codified bacterial names published in 1980 [22, 23]. In this article the organism will be referred to as C. perfringens irrespective of which name was used in the original cited paper.
Initially, C. perfringens was classified into subgroups based on the ability of individual strains to produce acid and gas from the fermentation of inulin and glycerine and to produce spores in media containing these carbohydrates [24–26]. Subsequently, Bull and Pritchard demonstrated that C. perfringens produced lethal exotoxins [27, 28] and Wilsdon developed a typing scheme based upon toxin production ([29] as cited in [30]). He used antisera produced against culture supernatants in toxin-antitoxin neutralisation tests to classify strains into four types based on their ability to produce three toxins, which were called W toxin (now known as α-toxin), X toxin (ε-toxin) and Z toxin (β-toxin). He described type A strains as classical gas gangrene strains that only produced W toxin, type B strains as isolates that produced W, X and Z toxins and caused lamb dysentery, type C strains as ovine Bacillus plaudis strains that produced W and Z toxins and type D strains as W and X toxin-producing Clostridium ovitotoxicus isolates from sheep. Other workers confirmed these observations in 1933 and first used the terms α-toxin, β-toxin and ε-toxin [30]. Wilsdon’s system forms the foundation for the current C. perfringens toxinotyping scheme and toxinotypes A to D are virtually the same as his initial definition, apart from the obvious differences in nomenclature. In 1941, MacFarland and Knight [31] discovered that α-toxin is a phospholipase C that is produced by all C. perfringens strains; it was the first bacterial toxin shown to act as an enzyme.
For epidemiological investigations of C. perfringens strains, notably those involved in food poisoning, a serological typing was developed [32]. This procedure eventually required more than 91 antisera and most of the strains were not typable [33]. Therefore, toxinotyping, and more recently genetic characterization, were preferred to serotyping.
Wilsdon’s toxinotyping scheme has been modified three times since it was introduced; we are now making a fourth modification. In 1943 a strain was isolated from a calf and shown to produce α-toxin and an additional toxin, ι-toxin, which was not neutralized by antiserum against α-, β- or ε-toxins [34]. Such strains were designated as belonging to a new toxin type, C. perfringens type E. Several years later strains that produced α-toxin and β-toxin were isolated from cases of human necrotic enteritis in Germany [35] and based on their spore properties were designated as type F [35, 36]. This type A to F scheme was reported in MacLennan’s classic review on histotoxic clostridial infections of man, which was published in 1962 [37]. Finally, in 1964 it was realised that type F was simply a variant of type C and type F was dropped from the scheme [23, 38]. The toxinotyping scheme has not been altered subsequently and has been reproduced in various formats in almost every major review of the field published since that time [39–44]. In virtually all of these reviews it was pointed out that most C. perfringens isolates, including type A strains, produce numerous other toxins and extracellular hydrolytic enzymes, the latest count is 20 such toxins and putative hydrolytic virulence factors [1, 45].
Finally, it has been known for many years that the genes encoding three of the four typing toxins (β-toxin, ε-toxin and ι-toxin) are encoded on large plasmids [43, 46]. The C. perfringens toxinotyping scheme therefore is fundamentally plasmid-based. More recently these plasmids have been shown to carry the C. perfringens Tcp conjugation locus and hence are highly likely to be conjugative [47–50]. Indeed, conjugative transfer of epsilon toxin plasmids from type D strains to a type A strain has been demonstrated [51], technically leading to the laboratory conversion of a type A strain to type D. Genes encoding other currently non-typing toxins such as CPE [52–54], β2-toxin [55], δ-toxin [56], BEC [57], NetB [58–61], NetE, NetF and NetG [62, 63] also have been shown to be plasmid determined and in several instances conjugative transfer has been demonstrated [51, 54, 60, 62]. Recent studies have provided evidence for in vivo conjugative transfer of the NetB plasmid, within the gastrointestinal tract of chickens [64].
2. The current toxinotyping scheme is outdated
The C. perfringens toxinotyping scheme in its various forms has been valuable for the diagnosis of C. perfringens infections in both humans and animals, but it is outdated and currently does not always serve its original diagnostic and epidemiological purpose. For example, CPE-producing strains of C. perfringens have been recognized for over fifty years and represent one of the world’s major causes of human food poisoning [65]. More recently, NetB toxin was identified and shown to be essential for necrotic enteritis in chickens [7]. These strains both produce very different toxins and cause very different diseases yet they are currently both classified as C. perfringens type A along with gas gangrene-causing strains of C. perfringens. Therefore, we conclude that the current toxinotyping scheme needs to be updated to improve its epidemiological and diagnostic value.
3. Principles for the expansion of the toxinotyping scheme
The expansion of the typing system has been the subject of discussion between many of the authors for several years and was presented and approved in principle at the 10th International Conference on the Molecular Biology and Pathogenesis of the Clostridia held in Ann Arbor, U.S.A. in August 2017. The major principle that has been agreed upon is that what is required is an expansion that builds upon the existing toxin-based typing system rather than a completely new scheme. It is considered that an expanded scheme will be more readily accepted and more widely used by both diagnostic and research focussed laboratories.
How are new toxinotypes to be determined?
The major principle here is that a new toxinotype needs to be unique; it must involve a new typing toxin that is not part of the existing toxinotyping scheme. It is not considered relevant to the typing scheme whether that toxin is encoded on a plasmid or on the chromosome. The established toxinotypes B to E have priority; that is, strains belonging to new toxinotypes cannot produce β-toxin, ε-toxin or ι-toxin. Most importantly, new toxinotypes must be disease based. The new toxinotype strains must have been clearly demonstrated to be associated with a specific disease syndrome, in humans or animals, by either fulfilling molecular Koch’s postulates for the toxin associated with the disease, or by extensive epidemiological analysis if the former is not technically feasible.
How are new toxinotypes to be approved?
A three-stage process is proposed for delineation of a new toxinotype. First, there should be extensive and open discussion between relevant researchers in the C. perfringens field. Second, the new toxinotype should be formally presented for comment and ratification at the biennial clostridial pathogenesis meeting. Third, the formal establishment of the new toxinotype should be published in a relevant peer reviewed journal in an article dedicated to that task. Appropriate leading researchers in the field should be authors of that article so that the new toxinotype is readily accepted by the clostridial community.
4. Two new toxinotypes
To initiate this process we hereby propose the establishment of two new toxinotypes, C. perfringens type F and C. perfringens type G. These new toxinotypes are proposed in accordance with the guidelines established in the previous section. A summary of the latest toxinotyping scheme is presented in Table 1.
Table 1.
The 2018 C. perfringens toxin-based typing schemea
| Toxinotype | α-toxin (plc or cpa) |
β-toxin (cpb) |
ε-toxin (etx) |
ι-toxin (iap and ibp) |
CPE (cpe) |
NetB (netB) |
|---|---|---|---|---|---|---|
| A | + | − | − | − | − | − |
| B | + | + | + | − | − | − |
| C | + | + | − | − | +/− | − |
| D | + | − | + | − | +/− | − |
| E | + | − | − | + | +/− | − |
| F | + | − | − | − | + | − |
| G | + | − | − | − | − | + |
the names of toxin structural genes are shown in parentheses.
C. perfringens type F
Strains belonging to C. perfringens type F are defined as isolates that carry the α-toxin gene and the cpe gene and produce CPE upon sporulation, but do not carry the structural genes for β-toxin, ε-toxin or ι-toxin. These strains have been shown to be responsible for human food-poisoning and non-foodborne C. perfringens-mediated diarrhea, including some instances of antibiotic-associated diarrhea [66, 67]. The epidemiological evidence for the association of these strains with C. perfringens-meditated food poisoning and some cases of non-foodborne diarrhea is very clear and well documented [66, 68]. In addition, molecular Koch’s postulates have been demonstrated for CPE [3]. It was shown that concentrated culture supernatants from two sporulating wild-type strains of what is now designated as C. perfringens type F caused fluid accumulation and mucosal damage in a rabbit intestinal loop model of disease, unlike isogenic cpe mutants isolated by allelic exchange in these different C. perfringens strains. Complementation of the mutants with the wild-type cpe gene restored the ability to cause fluid accumulation and mucosal damage, which provided clear evidence that CPE was essential for disease in a relevant animal model that mimics the human disease. Currently, these strains are referred to as CPE-positive strains of C. perfringens type A. Their designation as C. perfringens type F will provide a sound basis for the clinical and epidemiological analysis of these distinct strains. We suggest that for the next few years researchers mention in their publications that the C. perfringens type F strains were formerly called CPE-positive strains of C. perfringens type A. A consequence of this nomenclature change is that C. perfringens type A food poisoning will be renamed as C. perfringens type F food poisoning, again cross-referencing to the earlier nomenclature would be valuable. As discussed earlier, it is noted that there is a group of type C strains that was previously designated as type F. However, since the original type F nomenclature has not been used for over 50 years we do not consider that there will be any diagnostic confusion.
C. perfringens type G
Strains belonging to C. perfringens type G are defined as isolates that produce α-toxin and NetB toxin, but do not produce β-toxin, ε-toxin or ι-toxin. Note that C. perfringens type A strains are now defined as strains that produce α-toxin, but do not produce β-toxin, ε-toxin, ι-toxin, CPE or NetB.
C. perfringens type G strains have been shown to be responsible for necrotic enteritis in chickens [7]. Once more the genetic evidence for the essential role of NetB in this disease is clear and it is supported by very strong epidemiological evidence [69–71]. Molecular Koch’s postulates again have been fulfilled [7]. NetB-producing strains of C. perfringens cause lesions in a chicken model of subclinical necrotic enteritis, the economically most important form of the disease. Specific mutation of the netB gene eliminates the ability to cause these lesions, which is restored by complementation in trans with the wild-type netB gene. Currently, these strains are referred to as avian necrotic enteritis strains of C. perfringens type A. Their designation as C. perfringens type G isolates again will provide a sound basis for the diagnosis and epidemiological analysis of these distinct isolates.
5. Multiplex PCR toxinotyping test
We propose that the designation of unknown C. perfringens isolates as belonging to toxinotypes A to G should be based on the molecular analysis of their DNA; that is, it should involve the detection of the structural genes encoding the specific typing toxins. Given the rapidly changing nature of molecular methods of genomic analysis we will not specify the method that should be used. However, to assist researchers new to the field we have presented the results of a simple multiplex PCR test carried out on sample isolates from toxinotypes A to G (Fig. 1). This test was done using PCR primers (Table 2) specific for the α-toxin (plc or cpa), β-toxin (cpb), ε-toxin (etx), ι-toxin (iap), CPE (cpe) and NetB (netB) genes and designed so that the PCR products would have different sizes. The value of this approach is shown in Fig. 1, where the distinct toxinotypes, including the new types F and G can readily be distinguished. Note that the C. perfringens type D and E strains that were analysed here also carried the cpe gene.
Fig. 1. Multiplex PCR analysis of representative C. perfringens type A to G strains.
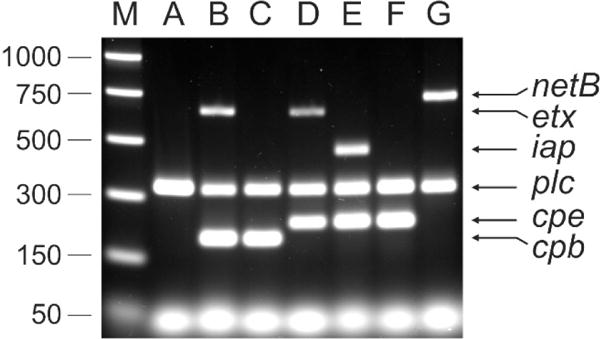
The strains were grown in TGY broth [79] to a turbidity at 600nm between 1.0 and 1.5. Genomic DNA (equivalent to 5mL of culture) was prepared as described previously [80]. DNA preparations were diluted 1 in 50 in sterile distilled water and used as templates in the multiplex toxin PCR based on a previous method [81]. The oligonucleotide primers and their concentrations are listed in Table 2. PCR reactions were prepared using 0.1 units/μL Taq DNA polymerase (Roche) in 1 x supplied buffer (Roche), 2 mM MgSO4 and 0.4 mM dNTPs. The template constituted 0.1 volumes of the final reaction. PCR was performed with an initial denaturation at 95°C for five minutes, followed by 35 cycles of 95°C for 1 minute, 55°C for 1 minute and 72°C for 1 minute. Amplified products were resolved by electrophoresis through a 1.5% (w/v) TAE agarose gel. The multiplex PCR profiles of the following C. perfringens strains are shown: JIR325 (type A)[82], JGS1984 (type B)[83], CN3717 (type C)[84], JGS4138 (type D)[85], ATCC27324 (type E), SM101 (type F)[86], EHE-NE18 (type G)[87]. Size standards were PCR Markers (Promega).
Table 2.
Oligonucleotide primers for multiplex toxin PCR
| Gene | Primers | Sequence (5′-3′) | size | Conc.* | Reference |
|---|---|---|---|---|---|
| netB | JRP6656 JRP6655 |
CTTCTAGTGATACCGCTTCAC CGTTATATTCACTTGTTGACGAAAG |
738 bp | 0.6 0.6 |
V. Adams & J. Rood |
| etx | JRP4234 JRP4235 |
CCACTTACTTGTCCTACTAAC GCGGTGATATCCATCTATTC |
656 bp | 0.44 0.44 |
[78] |
| iap | JRP5507 JRP5508 |
GGAAAAGAAAATTATAGTGATTGG CCTGCATAACCTGGAATGGC |
461 bp | 0.5 0.5 |
V. Adams & J. Rood |
| plc | JRP4232 JRP4233 |
GCTAATGTTACTGCCGTTGACC CCTCTGATACATCGTGTAAG |
324 bp | 0.4 0.4 |
Based on [78] |
| cpe | JRP5179 JRP5180 |
GGAGATGGTTGGATATTAGG GGACCAGCAGTTGTAGATA |
233 bp | 0.3 0.3 |
[78] |
| cpb | JRP5181 JRP5182 |
GCGAATATGCTGAATCATCTA GCAGGAACATTAGTATATCTTC |
196 bp | 0.3 0.3 |
[78] |
Concentration of oligonucleotides in final reaction (μM)
6. Lead candidates for future toxinotypes
C. perfringens produces at least 20 extracellular toxins and hydrolytic enzymes [1, 45], of which only six are typing toxins in the expanded scheme presented here. Although none of the other toxins meet the criteria described earlier for inclusion as typing toxins, two toxins are obvious candidates for inclusion in a future expansion of the scheme, once critical experiments have been carried out. These toxins are NetF [62] and BEC [57, 72].
NetF is an extracellular β-pore-forming toxin that belongs to the same toxin superfamily as NetB, β-toxin and C. perfringens δ-toxin [62]. The netF gene is encoded on a conjugative plasmid that also carries another putative toxin gene, netE. NetEF-positive strains also carry a plasmid that encodes CPE and β2-toxin and a proportion also carry a NetG plasmid. These plasmids all carry the Tcp conjugation locus. The netF gene is preferentially found in C. perfringens strains from cases of acute canine hemorrhagic enteritis and necrotizing enteritis in neonatal foals; these isolates appear to be clonal in origin [62, 73].
Several epidemiological studies have been carried out [62, 74–76] and have shown that the netEF plasmid is associated with isolates from these syndromes. A netF insertional inactivation mutant has been constructed and shown to be no longer toxic for an equine ovarian (EO) cell line [62]. EO toxicity was restored by complementation in trans with the wild-type netF gene. These data, together with conjugation and transformation experiments on these plasmids, clearly showed that NetF was responsible for the cytotoxicity for EO cells. However, molecular Koch’s postulates have not been proven yet for either the canine or equine disease syndromes. The exact role of NetE, NetF and CPE in these diseases remains to be determined [73]. What is required for NetF-positive strains of C. perfringens to be considered as a separate toxinotype is the analysis of isogenic wild-type, netF mutant and complemented strains in an animal model whose pathology resembles that of the canine or equine diseases. If such studies showed that NetF toxin was required for disease causation in this model then the establishment of a new NetF-positive toxinotype clearly would be justified.
Two independent studies have identified a novel binary toxin that appears to be associated with cases of acute foodborne gastroenteritis in Japan [57, 72]. Unfortunately, this toxin has been given two separate names, BEC [57] and CPILE [72]. This situation should not be allowed to continue as inevitably it has caused confusion in the literature. BEC signifies binary enterotoxin of C. perfringens and the two genes encoding the components of the binary toxin are referred to as becA and becB [57]. The term CPILE signifies C. perfringens iota-like enterotoxin. The genes are referred to as cpile-a and cpile-b [72], which does not conform to usual genetic conventions. We have concluded that the BEC terminology provides a simpler and more acceptable nomenclature for the clostridial community. We recommend that all researchers refer to the toxin as BEC, to the individual components as BECa and BECb and to the genes as becA and becB. However, for the next few years it would be helpful if researchers cross-referenced to the CPILE terminology in the abstract and introduction of their papers.
BEC toxin was identified in strains of C. perfringens isolated from several large outbreaks of acute foodborne gastroenteritis in Japan. Since these strains do not produce CPE and do not carry the cpe gene a novel toxin was suspected and subsequently identified as the binary toxin BEC (or CPILE) [57, 72]. A survey of Japanese C. perfringens isolates using a becAB-specific PCR indicates that BEC-positive strains are rare [77]. The becA and becB genes are plasmid determined and encode separate binary toxin components that are distantly related to the components of the clostridial binary toxin family, which includes C. perfringens ι-toxin (43-44% amino acid sequence identity to the individual components). Supernatants from sporulating cultures of these strains cause fluid accumulation in rabbit ileal loop and suckling mouse assays [57], as does purified recombinant toxin [72], and BEC also causes rounding of Vero and L929 cells [72]. Purified BECa has been shown to have ADP-ribosyltransferase activity on purified actin [57, 72], as expected for the enzymatic component of a clostridial binary toxin. Insertional inactivation of the becB gene abrogated the ability of the resultant strain to cause fluid accumulation in the suckling mouse assay, but unfortunately this mutant was not complemented. Therefore, it cannot be said for certain that the loss of fluid accumulation was the direct result of mutation of the becB gene. For this reason, we consider that it would be premature to designate BEC-positive strains as a separate toxinotype at this time.
7. Conclusions
In summary, we have proposed an updated toxinotyping scheme that incorporates two new toxinotypes. C. perfringens type F strains consist of isolates that produce CPE, but do not produce β-toxin, ε-toxin or ι-toxin. These strains are responsible for C. perfringens-mediated human food poisoning and antibiotic associated diarrhea. C. perfringens type G strains comprise isolates that produce NetB toxin and thereby cause necrotic enteritis in chickens. In addition, we describe a mechanism by which new toxinotypes can be formally proposed and subsequently approved.
Highlights.
An expanded C. perfringens toxinotyping scheme is presented.
Two new toxinotypes are proposed.
C. perfringens type F strains produce CPE, but not β, ε or ι toxins.
C. perfringens type G strains produce NetB.
A mechanism for the introduction of new toxinotypes is presented.
Acknowledgments
Research in JIR’s laboratory was supported by Australian Research Council Discovery Grant DP160102680. Research in Bactéries anaérobies et Toxines (MP) was supported by Institut Pasteur funding. Research in SBM’s laboratory was supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R21AI1093. Research in BMcC’s laboratory was supported by grants AI019844-35 and AI125796 from the National Institute of Allergy and Infectious Diseases. The authors thank John Prescott (University of Guelph) for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Uzal FA, Freedman JC, Shrestha A, Theoret JR, Garcia J, Awad MM, et al. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol. 2014;9:361–77. doi: 10.2217/fmb.13.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awad MM, Bryant AE, Stevens DL, Rood JI. Virulence studies on chromosomal α-toxin and θ-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of α-toxin in Clostridium perfringens-mediated gas gangrene. Mol Microbiol. 1995;15:191–202. doi: 10.1111/j.1365-2958.1995.tb02234.x. [DOI] [PubMed] [Google Scholar]
- 3.Sarker MR, Carman RJ, McClane BA. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol Microbiol. 1999;33:946–58. doi: 10.1046/j.1365-2958.1999.01534.x. [DOI] [PubMed] [Google Scholar]
- 4.Garcia JP, Beingesser J, Fisher DJ, Sayeed S, McClane BA, Posthaus H, et al. The effect of Clostridium perfringens type C strain CN3685 and its isogenic beta toxin null mutant in goats. Vet Microbiol. 2012;157:412–9. doi: 10.1016/j.vetmic.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sayeed S, Uzal FA, Fisher DJ, Saputo J, Vidal JE, Chen Y, et al. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol Microbiol. 2008;67:15–30. doi: 10.1111/j.1365-2958.2007.06007.x. [DOI] [PubMed] [Google Scholar]
- 6.Garcia JP, Adams V, Beingesser J, Hughes ML, Poon R, Lyras D, et al. Epsilon toxin is essential for the virulence of Clostridium perfringens type D infection in sheep, goats, and mice. Infect Immun. 2013;81:2405–14. doi: 10.1128/IAI.00238-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, et al. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Path. 2008;4:e26. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Achalme MP. Examen bactériologie d’un cas de rhumatisme articulaire aigu mort de rhumatisme cérébral. Compt Rendu Soc Biol Paris. 1891;43:651–6. [Google Scholar]
- 9.Welch WH, Nuttal GHF. A gas-producing bacillus (Bagillus aerogenes capsulatus, nov. spec.) capable of rapid development in the blood-vessels after death. Bull Johns Hopkins Hosp. 1892;3:81–91. [Google Scholar]
- 10.Fränkel E. Über Gasphlegmone. Hamburg and Leipzig: Leopold Voss; 1893. [Google Scholar]
- 11.Veillon A, Zuber A. Recherches sur quelques microbes strictement anaérobies et léur vole en pathologie. Arch Med Exp Anat Pathol. 1898;10:517–45. [Google Scholar]
- 12.Migula W. System der bakterien: Gustav Fischer, Jena. 1900 [Google Scholar]
- 13.Trécul MA. Production de plantules amylifères dans les cellules végétales pendant la putréfaction. Compt Rendu Soc Biol Paris. 1865;62:432–6. [Google Scholar]
- 14.Dürre P. From Pandora’s Box to cornucopia: Clostridia – a historical perspective. In: Bahl H, Dürre P, editors. Clostridia: biotechnology and medical applications. Weinheim, Germany: Wiley-VCH Verlag GmbH; 20011. pp. 1–17. [Google Scholar]
- 15.Winslow C-EA, Broadhurst J, Buchanan RE, Krumwiede C, Jr, Rogers LA, Smith GH. The families and genera of the bacteria: final report of the committee of the Society of American Bacteriologists on characterization and classification of bacterial types. J Bacteriol. 1920;5:191–229. doi: 10.1128/jb.5.3.191-229.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pribram E. A contribution to the classification of microorganisms. J Bacteriol. 1929;18:361–94. doi: 10.1128/jb.18.6.361-394.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prévot AR. Études de systématique bactérienne. I. Lois generales. - II. Cocci anaérobies. Ann Sci Nat Bot. 1933;15:23–261. [Google Scholar]
- 18.Dalling T, Ross H. Clostridium welchii: notes on the relationship between the types of cultures and the production of toxin. J Comp Pathol Ther. 1938;51:235–49. [Google Scholar]
- 19.PréVot AR, Turpin A, Kaiser P. Les Bactéries anaérobies. Paris: Dunod; 1967. [Google Scholar]
- 20.Breed RS, Murray EGD, Smith NR. Bergy’s manual of determinitive bacteriology. Seventh. Baltimore: Williams & Wilkins Co; 1957. [Google Scholar]
- 21.Hauduroy P, Ehringer G, Urbain A, Guillot G, Magrou J. Dictionnaire des bactéries pathogènes pour l’homme, les animaux et les plantes. Paris: Masson et Cie; 1937. [Google Scholar]
- 22.Skerman VBD, Mcgowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. [Google Scholar]
- 23.Rainey FA, Hollen BJ, Small A. Genus 1. Clostridium prazmowski 1880, 23AL. In: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, et al., editors. Bergy’s Manual of Systematic Bacteriology. Second. Dordrecht: Springer; 2009. [Google Scholar]
- 24.Simonds JP. Classification of the Bacillus welchii group of bacteria. J Infect Dis. 1915;16:31–4. [Google Scholar]
- 25.Esty JR. The biology of Clostridium welchii. J Bacteriol. 1920;5:375–429. doi: 10.1128/jb.5.4.375-429.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergy DH. Bergy’s manual of determinitive bacteriology. Second. London: Bailliere, Tindall & Cox; 1926. [Google Scholar]
- 27.Bull CG, Pritchett IW. Identity of the toxins of different strains of Bacillus welchii and factors influencing their production in vitro. J Exp Med. 1917;26:867–83. doi: 10.1084/jem.26.6.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bull CG, Pritchett IW. Toxin and antitoxin of and protective inoculation against Bacillus welchii. J Exp Med. 1917;26:119–38. doi: 10.1084/jem.26.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilsdon AS. Second Report. University of Cambridge Institute of Animal Pathology. 1931:53–85. [Google Scholar]
- 30.Glenny AT, Barr M, Llewwllyn-Jones M, Dalling T, Ross HE. Multiple toxins produced by some organisms of the Cl. wlechii group. J Pathol Bacteriol. 1933;37:53–74. [Google Scholar]
- 31.MacFarlane MG, Knight BCJG. The biochemistry of bacterial toxins I. The lecithinase activity of Cl. welchii toxins. Biochem J. 1941;35:884–902. doi: 10.1042/bj0350884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hobbs BC, Smith ME, Oakley CL, Warrack GH, Cruickshank JC. Clostridium welchii food poisoning. J Hyg (Lond) 1953;51:75–101. doi: 10.1017/s0022172400015515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labbe R. Clostridium perfringens. In: Doyle MP, editor. Foodborne bacterial pathogens. New York: Marcel Dekker; 1989. pp. 191–234. [Google Scholar]
- 34.Bosworth TJ. On a new type of toxin produced by Clostridium welchii. J Comp Pathol Ther. 1943;53:245–55. [Google Scholar]
- 35.Zeissler J, Rassfeld-Sternberg L. Enteritis necroticans due to Clostridium welchii type F. Br Med J. 1949;1:267–9. doi: 10.1136/bmj.1.4597.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oakley CL. The toxins of Cl. welchii type F. Br Med J. 1949;1:269–70. [Google Scholar]
- 37.MacLennan JD. The histotoxic clostridial infections of man. Bacteriol Rev. 1962;26:177–276. [PMC free article] [PubMed] [Google Scholar]
- 38.Sterne M, Warrack GH. The types of Clostridium perfringens. J Pathol Bacteriol. 1964;88:279–83. [PubMed] [Google Scholar]
- 39.Smith LDS. Virulence factors of Clostridium perfringens. Reviews of Infectious Diseases. 1979;1:251–60. doi: 10.1093/clinids/1.2.254. [DOI] [PubMed] [Google Scholar]
- 40.McDonel JL. Clostridium perfringens toxins (type A, B, C, D, E) Pharmacol Ther. 1980;10:617–35. doi: 10.1016/0163-7258(80)90031-5. [DOI] [PubMed] [Google Scholar]
- 41.Hatheway CL. Toxigenic clostridia. Clin Microbiol Rev. 1990;3:66–98. doi: 10.1128/cmr.3.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rood JI, Cole ST. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol Rev. 1991;55:621–48. doi: 10.1128/mr.55.4.621-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petit L, Gibert M, Popoff MR. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 1999;7:104–10. doi: 10.1016/s0966-842x(98)01430-9. [DOI] [PubMed] [Google Scholar]
- 44.Uzal FA, Shrestha A, Freedman JC, Theoret JR, Garcia J, Awad MM, et al. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol. 2014 doi: 10.2217/fmb.13.168. (in press, accepted 18-12-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Revitt-Mills SA, Rood JI, Adams V. Clostridium perfringens extracellular toxins and enzymes: 20 and counting. Microbiology Australia. 2015;36:114–7. [Google Scholar]
- 46.Canard B, Saint-Joanis B, Cole ST. Genomic diversity and organization of virulence genes in the pathogenic anaerobe Clostridium perfringens. Mol Microbiol. 1992;6:1421–9. doi: 10.1111/j.1365-2958.1992.tb00862.x. [DOI] [PubMed] [Google Scholar]
- 47.Bannam TL, Teng WL, Bulach D, Lyras D, Rood JI. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J Bacteriol. 2006;188:4942–51. doi: 10.1128/JB.00298-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Adams V, Bannam TL, Miyamoto K, Garcia JP, Uzal FA, et al. Toxin plasmids of Clostridium perfringens. Microbiol Mol Biol Rev. 2013;77:208–33. doi: 10.1128/MMBR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams V, Li J, Wisniewski JA, Uzal FA, Moore RJ, Mcclane BA, et al. Virulence plasmids of spore-forming bacteria. Microbiology Spectrum. 2014;2 doi: 10.1128/microbiolspec.PLAS-0024-2014. PLAS-0024-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freedman JC, Theoret JR, Wisniewski JA, Uzal FA, Rood JI, McClane BA. Clostridium perfringens type A-E toxin plasmids. Res Microbiol. 2015;166:264–79. doi: 10.1016/j.resmic.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes ML, Poon R, Adams V, Sayeed S, Saputo S, Uzal FA, et al. Epsilon toxin plasmids of Clostridium perfringens type D are conjugative. J Bacteriol. 2007;189:7531–8. doi: 10.1128/JB.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cornillot E, Saint-Joanis B, Daube G, Katayama SI, Granum PE, Canard B, et al. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol Microbiol. 1995;15:639–47. doi: 10.1111/j.1365-2958.1995.tb02373.x. [DOI] [PubMed] [Google Scholar]
- 53.Collie RE, McClane BA. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with nonfoodborne human gastrointestinal disease. J Clin Microbiol. 1998;36:30–6. doi: 10.1128/jcm.36.1.30-36.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brynestad S, Sarker MR, McClane BA, Granum PE, Rood JI. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect Immun. 2001;69:3483–7. doi: 10.1128/IAI.69.5.3483-3487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibert M, Jolivet-Renaud C, Popoff MR. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene. 1997;203:65–73. doi: 10.1016/s0378-1119(97)00493-9. [DOI] [PubMed] [Google Scholar]
- 56.Manich M, Knapp O, Gibert M, Maier E, Jolivet-Reynaud C, Geny B, et al. Clostridium perfringens delta toxin is sequence related to beta toxin, NetB, and Staphylococcus pore-forming toxins, but shows functional differences. PLoS One. 2008;3:e3764. doi: 10.1371/journal.pone.0003764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yonogi S, Matsuda S, Kawai T, Yoda T, Harada T, Kumeda Y, et al. BEC, a novel enterotoxin of Clostridium perfringens found in human clinical isolates from acute gastroenteritis outbreaks. Infect Immun. 2014;82:2390–9. doi: 10.1128/IAI.01759-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lepp D, Gong J, Songer JG, Boerlin P, Parreira VR, Prescott JF. Identification of accessory genome regions in poultry Clostridium perfringens isolates carrying the NetB plasmid. J Bacteriol. 2013 doi: 10.1128/JB.01032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lepp D, Roxas B, Parreira VR, Marri PR, Rosey EL, Gong J, et al. Identification of novel pathogenicity loci in Clostridium perfringens strains that cause avian necrotic enteritis. PLoS One. 2010;5:e10795. doi: 10.1371/journal.pone.0010795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bannam TL, Yan XX, Harrison PF, Seemann T, Keyburn AL, Stubenrauch C, et al. Necrotic enteritis-derived Clostridium perfringens strain with three closely related independently conjugative toxin and antibiotic resistance plasmids. mBio. 2011;2:e00190–11. doi: 10.1128/mBio.00190-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parreira VR, Costa M, Eikmeyer F, Blom J, Prescott JF. Sequence of two plasmids from Clostridium perfringens chicken necrotic enteritis isolates and comparison with C. perfringens conjugative plasmids. PLoS One. 2012;7:e49753. doi: 10.1371/journal.pone.0049753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mehdizadeh Gohari I, Parreira VR, Nowell VJ, Nicholson VM, Oliphant K, Prescott JF. A novel pore-forming toxin in type A Clostridium perfringens is associated with both fatal canine hemorrhagic gastroenteritis and fatal foal necrotizing enterocolitis. PLoS One. 2015;10:e0122684. doi: 10.1371/journal.pone.0122684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mehdizadeh Gohari I, Kropinski AM, Weese SJ, Parreira VR, Whitehead AE, Boerlin P, et al. Plasmid characterization and chromosome analysis of two netF+ Clostridium perfringens isolates associated with foal and canine necrotizing enteritis. PLoS One. 2016;11:e0148344. doi: 10.1371/journal.pone.0148344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lacey JA, Keyburn AL, Ford ME, Portela RW, Johanesen PA, Lyras D, et al. Conjugation-mediated horizontal gene transfer of Clostridium perfringens plasmids in the chicken gastrointestinal tract results in the formation of new virulent strains. Appl Environ Microbiol. 2017 doi: 10.1128/AEM.01814-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freedman JC, Shrestha A, McClane BA. Clostridium perfringens enterotoxin: action, gnetics, and translational applications. Toxins. 2016;8:73. doi: 10.3390/toxins8030073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McClane BA, Robertson SL, Li J. Clostridium perfringens. In: Doyle MP, Buchanan RA, editors. Food microbiology: fundamentals and frontiers. Washington, DC: ASM Press; 2013. pp. 465–90. [Google Scholar]
- 67.McClane BA, Uzal FA, Fernandez Miyakawa ME, Lyerly D, Wilkins TD. The enterotoxic clostridia. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E, editors. The prokaryotes. Springer US; 2006. pp. 698–752. [Google Scholar]
- 68.Lindstrom M, Heikinheimo A, Lahti P, Korkeala H. Novel insights into the epidemiology of Clostridium perfringens type A food poisoning. Food Microbiol. 2011;28:192–8. doi: 10.1016/j.fm.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 69.Rood JI, Keyburn AL, Moore RJ. NetB and necrotic enteritis: the hole movable story. Avian Pathol. 2016;43:295–301. doi: 10.1080/03079457.2016.1158781. [DOI] [PubMed] [Google Scholar]
- 70.Keyburn AL, Yan XX, Bannam TL, Van Immerseel F, Rood JI, Moore RJ. Association between avian necrotic enteritis and Clostridium perfringens strains expressing NetB toxin. Vet Res. 2010;41:21. doi: 10.1051/vetres/2009069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prescott JF, Parreira VR, Mehdizadeh Gohari I, Lepp D, Gong J. The pathogenesis of necrotic enteritis in chickens: what we know and what we need to know: a review. Avian Pathol. 2016;45:288–94. doi: 10.1080/03079457.2016.1139688. [DOI] [PubMed] [Google Scholar]
- 72.Irikura D, Monma C, Suzuki Y, Nakama A, Kai A, Fukui-Miyazaki A, et al. Identification and characterization of a new enterotoxin produced by Clostridium perfringens isolated from food poisoning outbreaks. PLoS One. 2015;10:e0138183. doi: 10.1371/journal.pone.0138183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mehdizadeh Gohari I, Kropinski AM, Weese SJ, Whitehead AE, Parreira VR, Boerlin P, et al. NetF-producing Clostridium perfringens: clonality and plasmid pathogenicity loci analysis. Infect Genet Evol. 2017;49:32–8. doi: 10.1016/j.meegid.2016.12.028. [DOI] [PubMed] [Google Scholar]
- 74.Finley A, Gohari IM, Parreira VR, Abrahams M, Staempfli HR, Prescott JF. Prevalence of netF-positive Clostridium perfringens in foals in southwestern Ontario. Can J Vet Res. 2016;80:242–4. [PMC free article] [PubMed] [Google Scholar]
- 75.Mehdizadeh Gohari I, Parreira VR, Timoney J, Fallon L, Slovis N, Prescott JF. NetF-positive Clostridium perfringens in neonatal foal necrotising enteritis in Kentucky. Vet Rec. 2016;178:216. doi: 10.1136/vr.103606. [DOI] [PubMed] [Google Scholar]
- 76.Diniz AN, Coura FM, Rupnik M, Adams V, Stent TL, Rood JI, et al. The incidence of Clostridioides difficile and Clostridium perfringens netF-positive strains in diarrheic dogs. Anaerobe. 2017;49:58–62. doi: 10.1016/j.anaerobe.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yonogi S, Kanki M, Ohnishi T, Shiono M, Iida T, Kumeda Y. Development and application of a multiplex PCR assay for detection of the Clostridium perfringens enterotoxin-encoding genes cpe and becAB. J Microbiol Methods. 2016;127:172–5. doi: 10.1016/j.mimet.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 78.Meer RR, Songer JG. Multiplex polymerase chain reaction assay for genotyping Clostridium perfringens. Am J Vet Res. 1997;58:702–5. [PubMed] [Google Scholar]
- 79.Sayeed S, Fernandez-Miyakawa ME, Fisher DJ, Adams V, Poon R, Rood JI, et al. Epsilon-toxin is required for most Clostridium perfringens type D vegetative culture supernatants to cause lethality in the mouse intravenous injection model. Infect Immun. 2005;73:7413–21. doi: 10.1128/IAI.73.11.7413-7421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Connor JR, Lyras D, Farrow KA, Adams V, Powell DR, Hinds J, et al. Construction and analysis of chromosomal Clostridium difficile mutants. Mol Microbiol. 2006;61:1335–51. doi: 10.1111/j.1365-2958.2006.05315.x. [DOI] [PubMed] [Google Scholar]
- 81.Garmory HS, Chanter N, French NP, Bueschel D, Songer JG, Titball RW. Occurrence of Clostridium perfringens beta2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiol Infect. 2000;124:61–7. doi: 10.1017/s0950268899003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lyristis M, Bryant AE, Sloan J, Awad MM, Nisbet IT, Stevens DL, et al. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol Microbiol. 1994;12:761–77. doi: 10.1111/j.1365-2958.1994.tb01063.x. [DOI] [PubMed] [Google Scholar]
- 83.Sawires YS, Songer JG. Multiple-locus variable-number tandem repeat analysis for strain typing of Clostridium perfringens. Anaerobe. 2005;11:262–72. doi: 10.1016/j.anaerobe.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 84.Gurjar A, Li J, McClane BA. Characterization of toxin plasmids in Clostridium perfringens type C isolates. Infect Immun. 2010;78:4860–9. doi: 10.1128/IAI.00715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sayeed S, Li J, McClane BA. Virulence plasmid diversity In Clostridium perfringens type D isolates. Infect Immun. 2007;75:2391–8. doi: 10.1128/IAI.02014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao Y, Melville SB. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J Bacteriol. 1998;180:136–42. doi: 10.1128/jb.180.1.136-142.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sheedy SA, Ingham AB, Rood JI, Moore RJ. Highly conserved alpha-toxin sequences of avian isolates of Clostridium perfringens. J Clin Microbiol. 2004;42:1345–7. doi: 10.1128/JCM.42.3.1345-1347.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


