Abstract
Cells are constantly exposed to endogenous and exogenous chemical and physical agents that damage their genome by forming DNA lesions. These lesions interfere with the normal functions of DNA such as transcription and replication, and need to be either repaired or tolerated. DNA lesions are accurately removed via various repair pathways. In contrast, tolerance mechanisms do not remove lesions but only allow replication to proceed despite the presence of unrepaired lesions. Cells possess two major tolerance strategies, namely translesion synthesis (TLS), which is an error-prone strategy and an accurate strategy based on homologous recombination (homology-dependent gap repair [HDGR]). Thus, the mutation frequency reflects the relative extent to which the two tolerance pathways operate in vivo. In the present paper, we review the present understanding of the mechanisms of TLS and HDGR and propose a novel and comprehensive view of the way both strategies interact and are regulated in vivo.
Keywords: DNA damage tolerance, Translesion synthesis, Recombinational repair
INTRODUCTION
The present paper reviews the various mechanisms by which Escherichia coli cells respond to acute genotoxic stresses such as ultraviolet (UV)-irradiation and summarizes both early work dating as far back as 50 years and on-going research. The isolation of E. coli strains that are highly sensitive to UV irradiation, uvr strains, has led to the discovery of the nucleotide excision repair (NER) pathway (1). A wild-type strain has been shown to endure up to 3000 UV lesions/genome, whereas an isogenic NER deficient (uvrA) strain only tolerates ≈ 50–100 lesions/genome (irradiation at 1–2 J/m2), illustrating the vigorous repair capacity of NER. However, the residual tolerance of 50–100 lesions/genome in an NER deficient strain was found to depend on the recA gene, the master gene in homologous recombination. RecA was proposed to provide a repair function via homologous recombination during replication as previously reviewed (2). As discussed below, the precise mechanisms involved in post-replicative recombinational repair are presently still under active investigation. Early studies involving the incorporation of radio-labelled dNTPs into the genome of UV-irradiated NER-deficient cells led to the surprising discovery that after an initial 15–20 min delay, E. coli cells synthesized approximately the same amount of DNA as un-irradiated control cells did. However, the size of the nascent DNA fragments are shorter and approximately correspond to the inter-lesion distance (2,3). The average delay inflicted on fork progression has been estimated to be in the range of 10–20 s/lesion in two independent studies (3,4). Following further incubation for 45–60 min, the short DNA fragments are converted into larger molecules of the size observed in the un-irradiated control (3). These data are compatible with a model in which the replication fork initially skips over lesions via downstream re-priming leaving gaps opposite lesions. While re-priming is a natural property in lagging strand replication it was shown in vitro that the primase can also prime the leading strand (5). We suggest that these gaps are subsequently repaired by socalled post-replication repair pathways, namely translesion synthesis (TLS) (6) or homology-dependent gap repair (HDGR) using the sister chromatid as the template (2,7,8). Furthermore, UV-irradiated Saccharomyces cerevisiae cells showed uncoupling of leading and lagging strand replication. Electron microscopy (EM) images revealed that small single-stranded DNA (ssDNA) gaps accumulate along replicated duplexes, likely resulting from re-priming events downstream of the lesions on both leading and lagging strands (9). TLS and HDGR counteract gap accumulation without affecting fork progression. Using a genetic tool, it was confirmed that the process by which UV-induced gaps are repaired is separable in time and space from genome replication (10). Compared to the fork skipping reaction, the repair of a single gap is a slow process estimated to take ~30–40 min per lesion (6). Thus, it is critical for these gap-repair events to be processed in parallel after the fork has passed rather than at the fork, which would be extremely time-consuming. Below we will discuss recent findings of the interplay between TLS and HDGR.
INTERPLAY BETWEEN NER AND REPLICATION IN A WILD-TYPE STRAIN FOLLOWING UV IRRADIATION
Compared to a uvrA strain, a wild-type strain can typically tolerate a 30–50-fold higher UV dose (i.e., 60 J/m2 instead of 1–2 J/m2) (1,2). In a wild-type strain, a UV dose of 30–60 J/m2 dramatically reduces the rate of DNA synthesis for at least 30 min before resuming the rate measured in a non-irradiated control strain (4,11). These data are compatible with the following scenario: in the initial 30 min following UV irradiation in a wild-type strain, the bi-directional replication forks skip over lesions at a similar rate as that estimated in an uvrA strain (10–20 s/lesion). This rate corresponds to 90–180 lesions, given that at 60 J/m2 the genome contains ≈ 3000 lesions, the replicated DNA still corresponds to only 3–6% of the genome. During that time NER actively removes lesions throughout the rest of the genome. Indeed, within the initial 30 min post-irradiation, NER removes 90–95% of CPD dimers (4). Thus after 30 min, the residual lesion density encountered by the fork in the wild-type strain becomes similar to the lesion density of the NER-defective strain irradiated at 1–2 J/m2. Consequently, replication restart apparently occurs almost synchronously (4). This model accounts for the higher mutation frequency due to error-prone bypass in the specific regions of the chromosome undergoing DNA replication at the time of irradiation as previously described (12).
POST REPLICATIVE GAP REPAIR: INTERPLAY BETWEEN TLS AND HDGR
As discussed above, while post-replicative gaps were first identified in UV-irradiated E. coli approximately 50 years ago (3,13,14), their repair mechanism is still undergoing extensive investigation (10,15–17). Post-replicative gap repair pathways are not genuine repair pathways, as they do not remove the lesion but only tolerate it to allow replication to progress. Lesion tolerance pathways involve either TLS or homologous recombination using the sister chromatid as a template (18). The process of TLS occurs widely from prokaryotes to eukaryotes (19). This pathway is particularly relevant to human health since a cancer-prone human disease, xeroderma pigmentosum variant (XPV), is caused by mutations in a specialized DNA polymerase gene (20,21). In contrast to HDGR that accurately bypasses the lesions, TLS is intrinsically mutation prone owing to the low replication fidelity of the specialized polymerases. To avoid unnecessary mutagenic risk, TLS polymerases are tightly controlled transcriptionally, post-translationally, or both (19).
INTEGRATED MECHANISM OF TLS IN VIVO
E. coli possesses five DNA polymerases (Pol I to V), and three (II, IV, and V) are involved in TLS (22). One of the specialized DNA polymerases (Pol V) encoded by umuDC (23), is the representative TLS polymerase because it is highly capable of bypassing various DNA damages (24). Indeed, numerous genetic studies dating back approximately 40 years, described the involvement of the umuDC gene products in induced mutagenesis (25,26). Genetic studies have shown that in order to function as a TLS polymerase in vivo, Pol V has to interact with two accessory factors, RecA and β-clamp (27–30). In contrast to the widely accepted genetic model of Pol V activation, two distinct biochemical models referred to as the “cis-activation” and “trans-activation” models have been proposed to describe the biochemical context under which Pol V functions (31–34). While both models similarly indicate that Pol V activation requires its interaction with the RecA filament formed on a ssDNA, the models differ in the provenance of the RecA filament. However, considering that the ssDNA occurs downstream of the lesion when a fork encounters a replication block, the so-called cis-activation model appears to be physiologically more relevant. In addition, it should be stressed that the requirement of the β-clamp for functional activation of Pol V is essential in the cis-but not the trans-activation models [for details see “cis-activation” vs “trans-activation” in (35,36)].
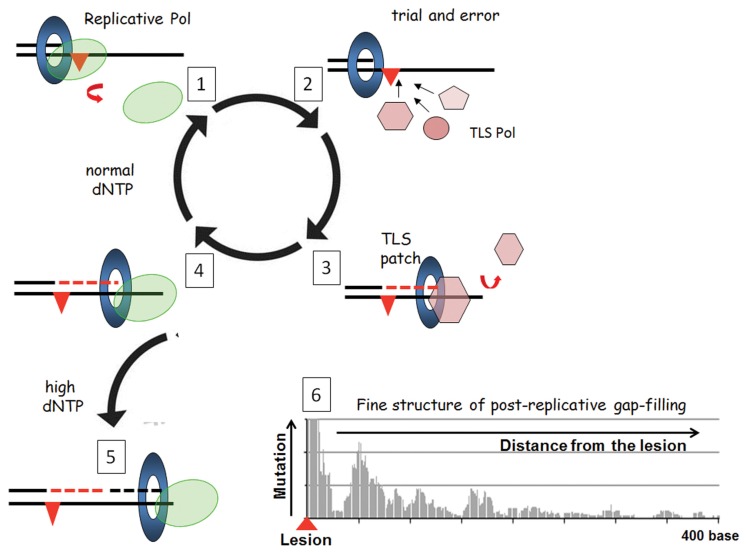
Reconstitution of Pol V-mediated TLS has provided an insight into the biochemical process of lesion bypass, most importantly with respect to length of the patch created by Pol V past the lesion. Pol V is error-prone in contrast to the high-fidelity replicative polymerase and, therefore, the length of the DNA patch created by Pol V (TLS patch, Fig. 1A) is a major determinant of the lesion bypass accuracy because longer TLS patches increase the number of errors. The TLS patch size needs to be above a threshold length to prevent its degradation by the proofreading function of Pol III following its re-loading (37). For a TT pyrimidinepyrimidone (6–4) photoproduct [TT (6–4)], successful bypass is achieved when the TLS patch size is ≥ 6 nucleotides (nt), counting from the nt opposite the 3-T of TT (6–4). The average TLS patch size induced by Pol V at a TT (6–4) in vitro was estimated to be 15 nt, a length compatible with productive bypass (32). The balance between exonucleolytic degradation and polymerization by the replicative DNA polymerase is modulated by the dNTP pool size (38). Genotoxic stress increases the dNTP pool size and, thus, favors elongation over proofreading. In a recent study, we measured the Pol V-mediated TLS patch size at a single TT (6–4) lesion in vivo using a single-molecule deep-sequencing approach to visualize the mutagenic signature of Pol V on undamaged DNA (39). Pol V synthesis was tracked by an increase in mutation events and, unexpectedly, revealed that after completing the initial TLS patch, Pol V was able to re-access the undamaged DNA template downstream from the lesion to synthesize other patch(es) of low fidelity DNA synthesis [referred to as error-prone post-TLS patch(es), Fig. 1, panel 6]. The sequencing data also revealed a patch with increased error frequency upstream of the TT (6–4) lesion that we tentatively attributed to error-prone synthesis following exonuclease-mediated resection of the primer blocked at the lesion site. Taken together, these data underscore the concept of untargeted mutagenesis linked to TLS as a likely contributor to adaptation/evolution within the frame of the SOS response and establish a novel paradigm for post-replicative gap filing in vivo.
Fig. 1.
Integrated view of translesion synthesis (TLS) pathways. Step 1: the replicative DNA polymerase dissociates from the primer template after encountering noncoding template base. Step 2: vacant primer template becomes the substrate for binding by specialized DNA polymerases; there appears to be no active selection process for the binding of a specific polymerase; i.e., binding is stochastic and follows classical mass-action laws. Step 3: a successful specialized polymerase is one that can synthesize a patch long enough to resist proofreading (TLS patch in dotted red) in a single binding event. Interaction of TLS polymerase with the β-clamp left behind on the template following dissociation of replicative DNA polymerase is essential to confer limited processivity to TLS polymerase that are otherwise highly distributive. For all three SOS polymerases (Pol II, Pol IV, and Pol V), mutations that inactivate the β-clamp binding motif abrogate their TLS activity in vivo (30). Step 4: following dissociation of the TLS polymerase, the “TLS patch” is extended after reloading of the replicative polymerase, leading to complete TLS (Step 5). If the TLS patch is too short, the proofreading activity of the replicative DNA polymerase may abort the TLS pathway back to step 1. The balance between exonucleolytic degradation and polymerization by the replicative DNA polymerase is modulated by the dNTP pool size (37). Increased dNTP pools arising because of genotoxic stress favor elongation over proofreading. Recently, deep sequencing of numerous Pol V-mediated TLS events have revealed the fine structure of the post replicative gap-filling process in vivo (39). Pol V synthesis was tracked by an increase in mutation events and, unexpectedly, revealed that after completing the canonical TLS reaction, Pol V reaccessed the undamaged DNA template downstream from the lesion to synthesize other patch(es) of low fidelity DNA synthesis [referred to as error-prone post-TLS patch(es)] over a distance of up to 400 nt downstream of the lesion.
INTERPLAY BETWEEN HDGR AND TLS: HOW HDGR IS MEASURED
The relative proportion to which the two major DNA damage tolerance pathways, TLS and HDGR (also referred to as damage avoidance or copy choice) are used in living cell has received little experimental attention until recently. While plasmid-based probes carrying single lesions have been instrumental to study TLS pathways, they are inappropriate for studying HDGR due to massive replication fork uncoupling (6,40,41). Indeed, fork uncoupling and concomitant progression of the replicative helicase fully unwinds the parental strands and, thus, prevents recombination events with the sister chromatid. To simultaneously monitor TLS and HDGR events in vivo, we developed a method to introduce single lesions into the E. coli chromosome (41). Our experimental system is based on the phage lambda site-specific recombination reaction between a construct that carries the lesion of choice and the E. coli chromosome (41). The lesion-containing construct is fabricated in vitro, and carries a small sequence heterology opposite the lesion that serves as a genetic strand marker (42).
Using this assay, we investigated the mechanism underlying tolerance to three strong replication-blocking lesions, namely G-AAF, TT_CPD, and TT (6–4) (36), are tolerated in the E. coli chromosome (43). All experiments were conducted in an NER deficient genetic background (uvrA) and under mismatch repair (MMR, mutS) conditions to avoid repair of the single lesion and the sequence heterology. Under non-SOS induced conditions, replication-blocking lesions are barely bypassed by TLS (i.e., the TLS ranges from 0.5 to 2%), and most lesion tolerance (> 95%) occurs by recombination. When the SOS response is induced by prior UV irradiation or genetically (lexA[Def] or recA730), there is a robust 10–20-fold increase in TLS across the strong replication-blocking lesions. It should be stressed that even under SOS-induced conditions, the level of TLS (~30% at most) remains low compared to the level of HDGR. Thus, under physiological conditions, TLS across strong replication-blocking lesions appears to be a minor survival pathway compared to recombination-mediated tolerance (Fig. 2). In fact, in the absence of TLS (pol-BumuDC or lexA[Ind-] strains), survival is fully mediated by HDGR without any significant loss in colony formation efficiency. The HDGR pathway is fully proficient in a lexA(Ind-) strain, suggesting that this pathway does not involve induction of any SOS gene. Only recA null mutants were profoundly deficient in the extent of HDGR, indicating that constitutive expression of RecA protein is sufficient to support HR-mediated gap repair. It is also remarkably noteworthy that the level of TLS can be boosted to nearly 100% by raising the expression level of the TLS polymerases (43). In conclusion, under physiological conditions, TLS polymerase concentration acts as a rate-limiting factor for TLS.
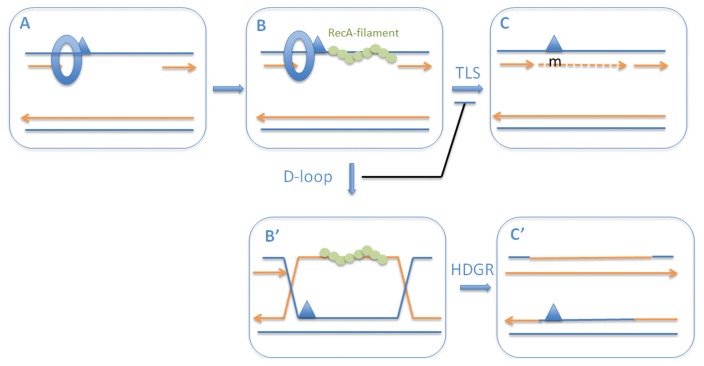
Fig. 2.
Chronological implementation of DNA damage tolerance pathways. When a replication fork encounters a replication-blocking lesion in one of the template strands, the fork skips over the lesion via downstream re-priming leaving a single-stranded gap. These single-stranded DNA gaps are converted into ssDNA.RecA filaments (A ≥ B); loading of RecA to single-stranded DNA gaps is aided by recombination mediator proteins such as RecFOR that help displace SSB from ssDNA. After formation, the ssDNA.RecA filament plays several important roles: i) SOS induction, ii) activation of Pol V, and iii) initiation of homologous recombination. Initially TLS (B ≥ C) is favored by the SOS-mediated increase in the concentration of the specialized DNA polymerase expression as well as by activation of Pol V by the RecA filament. However, formation of a D-loop when the ssDNA.RecA filament invades the homologous sister chromatid (i.e., an early HDGR intermediate) (B ≥ B′), the TLS reaction is shut off by mere substrate sequestration. Completion of the homologous recombination reaction (B′ ≥ C′) leads to an HDGR event. This model defines a chronological switch from TLS to HDGR. The switch from TLS -> HDGR can be modulated genetically as follows. i) Under non-SOS-induced conditions, i.e., at low concentration of TLS Pol’s, TLS is low (1% range); TLS is increased ~10-fold under SOS-induced conditions showing that the concentration of the specialized polymerases is rate-limiting for TLS. ii) Delayed D-loop formation by specific recA alleles extends time window of TLS (i.e., half-life of intermediate B is increased), leading to higher level of TLS at the expense of HDGR (46).
CHRONOLOGICAL IMPLEMENTATION OF TLS AND HDGR
When a replication fork encounters a replication-blocking lesion in one of the template strands, the fork skips over the lesion via downstream re-priming, leaving a single-stranded gap. These single-stranded DNA gaps are converted into ssDNA.RecA filaments with the help of recombination mediator proteins such as RecFOR (and RecBCD to a lesser extent) that aids the displacement of the single-stranded DNA binding protein (SSB) from the ssDNA. Once formed, the ssDNA.RecA filament plays several important roles in DNA damage tolerance pathways (44): i) induction of the SOS response via cleavage of the LexA repressor, ii) activation of Pol V, and iii) initiation of homologous recombination reaction via D-loop formation with the sister chromatid. Both the induction of the SOS response that increases the concentration of the TLS polymerases and the biochemical activation of Pol V trigger TLS. However, as soon as the ssDNA.RecA filament invades the homologous sister chromatid and forms a D-loop, i.e., the early HDGR intermediate, the TLS reaction is shut down by mere substrate sequestration. This process leads to a chronological implementation of the tolerance pathways, with TLS being executed first during a limited time-window defined by the kinetics of D-loop formation. We have experimentally investigated this model using recA point mutants that are partially impaired in the kinetics of D-loop formation but fully competent for ssDNA.RecA filament formation (45). In these strains, the TLS levels are strongly increased (10- to 20-fold) (46). Thus, a partial defect in RecA’s capacity to invade the homologous sister chromatid increases the lifetime of the ssDNA.RecA filament, i.e., the “SOS signal”. The increased lifetime of the ssDNA.RecA filament (B intermediate in Fig. 2) favors TLS by increasing both the TLS polymerase concentration and the lifetime of the TLS substrate before it becomes sequestered by homologous recombination (B′ intermediate in Fig 2). In conclusion, the switch from error-prone TLS to error-free HDGR is controlled by the kinetics of the formation of the early homologous recombination intermediate, i.e., the D-loop between the ssDNA.RecA filament and the sister chromatid (B->B′ transition in Fig. 2).
REFERENCES
- 1.Howard-Flanders P, Theriot L. Mutants of Escherichia coli K-12 defective in DNA repair and in genetic recombination. Genetics. 1966;53:1137–1150. doi: 10.1093/genetics/53.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Courcelle J, Hanawalt PC. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet. 2003;37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- 3.Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 4.Rudolph CJ, Upton AL, Lloyd RG. Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli. Genes Dev. 2007;21:668–681. doi: 10.1101/gad.417607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeeles JTP, Marians KJ. Dynamics of leading-strand lesion skipping by the replisome. Mol Cell. 2013;52:855–865. doi: 10.1016/j.molcel.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pages V, Fuchs RP. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science. 2003;300:1300–1303. doi: 10.1126/science.1083964. [DOI] [PubMed] [Google Scholar]
- 7.Rupp WD, Wilde CE, Reno DL, Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol. 1971;61:25–44. doi: 10.1016/0022-2836(71)90204-X. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: rediscovering old models. DNA Repair (Amst) 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courcelle CT, Chow K-H, Casey A, Courcelle J. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc Natl Acad Sci USA. 2006;103:9154–9159. doi: 10.1073/pnas.0600785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerdá-Olmedo E, Hanawalt PC, Guerola N. Mutagenesis of the replication point by nitrosoguanidine: map and pattern of replication of the Escherichia coli chromosome. J Mol Biol. 1968;33:705–719. doi: 10.1016/0022-2836(68)90315-X. [DOI] [PubMed] [Google Scholar]
- 13.Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J Mol Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-X. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann AR, Kirk-Bell S, Arlett CF, Paterson MC, Lohman PH, de Weerd-Kastelein EA, Bootsma D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc Natl Acad Sci USA. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branzei D, Szakal B. DNA damage tolerance by recombination: Molecular pathways and DNA structures. DNA Repair (Amst) 2016;44:68–75. doi: 10.1016/j.dnarep.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izhar L, Ziv O, Cohen IS, Geacintov NE, Livneh Z. Genomic assay reveals tolerance of DNA damage by both translesion DNA synthesis and homology-dependent repair in mammalian cells. Proc Natl Acad Sci USA. 2013;110:E1462–E1469. doi: 10.1073/pnas.1216894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coulon S, Ramasubramanyan S, Alies C, Philippin G, Lehmann A, Fuchs RP. Rad8Rad5/Mms2-Ubc13 ubiquitin ligase complex controls translesion synthesis in fission yeast. EMBO J. 2010;29:2048–2058. doi: 10.1038/emboj.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs RP. Tolerance of lesions in E. coli: Chronological competition between translesion synthesis and damage avoidance. DNA Repair (Amst) 2016;44:51–58. doi: 10.1016/j.dnarep.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 20.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 21.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 22.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF. UmuD’(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchs RP, Fujii S, Wagner J. Properties and functions of Escherichia coli: Pol IV and Pol V. Adv Protein Chem. 2004;69:229–264. doi: 10.1016/S0065-3233(04)69008-5. [DOI] [PubMed] [Google Scholar]
- 25.Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- 26.Steinborn G. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol Gen Genet. 1978;165:87–93. doi: 10.1007/BF00270380. [DOI] [PubMed] [Google Scholar]
- 27.Blanco M, Herrera G, Collado P, Rebollo JE, Botella LM. Influence of RecA protein on induced mutagenesis. Biochimie. 1982;64:633–636. doi: 10.1016/S0300-9084(82)80102-8. [DOI] [PubMed] [Google Scholar]
- 28.Dutreix M, Moreau PL, Bailone A, Galibert F, Battista JR, Walker GC, Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sweasy JB, Witkin EM, Sinha N, Roegner-Maniscalco V. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol. 1990;172:3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bécherel OJ, Fuchs RPP, Wagner J. Pivotal role of the beta-clamp in translesion DNA synthesis and mutagenesis in E coli cells. DNA Repair (Amst) 2002;1:703–708. doi: 10.1016/S1568-7864(02)00106-4. [DOI] [PubMed] [Google Scholar]
- 31.Fujii S, Gasser V, Fuchs RP. The biochemical requirements of DNA polymerase V-mediated translesion synthesis revisited. J Mol Biol. 2004;341:405–417. doi: 10.1016/j.jmb.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 32.Fujii S, Fuchs RP. Biochemical basis for the essential genetic requirements of RecA and the beta-clamp in Pol V activation. Proc Natl Acad Sci USA. 2009;106:14825–14830. doi: 10.1073/pnas.0905855106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlacher K, Cox MM, Woodgate R, Goodman MF. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature. 2006;442:883–887. doi: 10.1038/nature05042. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF. The active form of DNA polymerase V is UmuD’(2)C-RecA-ATP. Nature. 2009;460:359–363. doi: 10.1038/nature08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs RP, Fujii S. Translesion synthesis in Escherichia coli: lessons from the NarI mutation hot spot. DNA Repair (Amst) 2007;6:1032–1041. doi: 10.1016/j.dnarep.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 36.Fuchs RP, Fujii S. Translesion DNA synthesis and mutagenesis in prokaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012682. doi: 10.1101/cshperspect.a012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujii S, Fuchs RP. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004;23:4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 2011;108:19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isogawa A, Ong JL, Potapov V, Fuchs RP, Fujii S. Pol V-mediated translesion synthesis elicits localized untargeted mutagenesis during post-replicative gap repair. Cell Rep. 2018;24:1290–1300. doi: 10.1016/j.celrep.2018.06.120. [DOI] [PubMed] [Google Scholar]
- 40.Higuchi K, Katayama T, Iwai S, Hidaka M, Horiuchi T, Maki H. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells. 2003;8:437–449. doi: 10.1046/j.1365-2443.2003.00646.x. [DOI] [PubMed] [Google Scholar]
- 41.Pagès V, Mazon G, Naiman K, Philippin G, Fuchs RP. Monitoring bypass of single replication-blocking lesions by damage avoidance in the Escherichia coli chromosome. Nucleic Acids Res. 2012;40:9036–9043. doi: 10.1093/nar/gks675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veaute X, Fuchs RP. Greater susceptibility to mutations in lagging strand of DNA replication in Escherichia coli than in leading strand. Science. 1993;261:598–600. doi: 10.1126/science.8342022. [DOI] [PubMed] [Google Scholar]
- 43.Naiman K, Philippin G, Fuchs RP, Pagès V. Chronology in lesion tolerance gives priority to genetic variability. Proc Natl Acad Sci USA. 2014;111:5526–5531. doi: 10.1073/pnas.1321008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Courcelle J, Donaldson JR, Chow K-H, Courcelle CT. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 2003;299:1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]
- 45.Adikesavan AK, Katsonis P, Marciano DC, Lua R, Herman C, Lichtarge O. Separation of recombination and SOS response in Escherichia coli RecA suggests LexA interaction sites. PLoS Genet. 2011;7:e1002244. doi: 10.1371/journal.pgen.1002244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naiman K, Pagès V, Fuchs RP. A defect in homologous recombination leads to increased translesion synthesis in E. coli. Nucleic Acids Res. 2016;44:7691–7699. doi: 10.1093/nar/gkw488. [DOI] [PMC free article] [PubMed] [Google Scholar]