Abstract
Infection of plants and insects with RNA and DNA viruses triggers Dicer-dependent production of virus-derived small interfering RNAs (vsiRNAs), which subsequently guide specific virus clearance by RNA interference (RNAi). Consistent with a major antiviral function of RNAi, productive virus infection in these eukaryotic hosts depends on the expression of virus-encoded suppressors of RNAi (VSRs). The eukaryotic RNAi pathway is highly conserved, particularly between insects and mammals. This review will discuss key recent findings that indicate a natural antiviral function of the RNAi pathway in mammalian cells. We will summarize the properties of the characterized mammalian vsiRNAs and VSRs and highlight important questions remaining to be addressed on the function and mechanism of mammalian antiviral RNAi.
Introduction
RNA interference (RNAi) refers to homology-dependent gene silencing mechanisms initiated by Dicer-mediated production of small interfering RNAs (siRNAs) and microRNAs (miRNAs) in eukaryotic organisms [1–3]. Three main lines of evidence support a natural antiviral function of the RNAi pathway in fungi, plants and invertebrates [4–8]. First, infection of fungi, plants and insects with RNA and/or DNA viruses triggers the accumulation of abundant virus-derived siRNAs (vsiRNAs) produced specifically by the Dicer family of class 3 RNase III enzymes. Second, fungal, plant and invertebrate viruses frequently replicate to higher levels and become more virulent in host mutants defective in RNAi, including those defective in either the biogenesis or the function of vsiRNAs. Third, virus-encoded suppressors of RNAi (VSRs) are widespread in diverse families of plant and insect viruses. Notably, VSR-deficient mutant viruses are defective in the infection of wildtype hosts, but accumulate to high levels and induce diseases in RNAi-defective host mutants. In this article, we review the recent evidence for an antiviral function of the RNAi pathway in mammals.
Mammalian host cells produce abundant viral siRNAs after infection with distinct positive- and negative-strand RNA viruses
Two studies published in 2013 [9,10] have provided strong evidence for the production of vsiRNAs during authentic infection of mammalian cells by two positive-strand RNA viruses, Nodamura virus (NoV) and encephalomyocarditis virus (EMCV). The vsiRNAs targeting EMCV were detected in mouse embryonic stem cells (mESCs) whereas mESCs, baby hamster kidney 21 cells (BHK-21), and newborn mice all produced vsiRNAs in response to the infection with NoV mutants not expressing a functional VSR, the viral B2 protein. Deep sequencing of small RNAs (sRNA-seq) from the infected mammalian cells revealed that the most abundant population of the total small RNAs mapped to NoV and EMCV is in the 21- to 23-nt size range of Dicer products with a major 22-nt peak for both strands. Importantly, these 22-nt RNAs of both NoV and EMCV are enriched for typical siRNA duplexes with a 20-nt perfectly base-paired duplex region and 2-nt 3′ overhangs. These findings indicate that the infection of mammalian cells by the two positive-strand RNA viruses triggers Dicer processing of viral dsRNA replicative intermediates into vsiRNAs. The Dicer-dependency of EMCV-derived vsiRNAs was further verified using Dicer knockout mESCs [10].
More recently, Dicer-dependent production of vsiRNAs has also been reported in human 293 cells in response to infection with either Influenza A virus (IAV) or human enterovirus 71 (HEV71) not expressing a functional VSR [11,12]. The accumulation of vsiRNAs was also detected in human alveolar basal epithelial cells (A549) and monkey Vero cells after infection with the VSR-deficient mutant IAV and in human rhabdomyosarcoma (RD), primary murine lung fibroblasts (MLFs) and newborn mice following infection with the VSR-deficient mutant HEV71. Table 1 summaries the key properties of the mammalian vsiRNAs in 21 to 23-nt size range re-calculated from the published datasets [9–12] using the small RNAs sequenced from fruit fly infected with a VSR-deficient virus for comparison [13]. Table 1 considered the influenza vsiRNAs only from human A549 cells since they are a better cell culture model than human 293T cells for immune studies on IAVs. EMCV vsiRNAs represent 0.19% of the total mapped reads from the infected mESCs [10] whereas 0.59% and 0.24% of the total mapped reads are the vsiRNAs respectively from human A549 cells and newborn mice after infection with a VSR-deficient virus (Table 1). Interestingly, most of the vsiRNAs targeting each of the four mammalian RNA viruses were processed from the dsRNA precursors corresponding to the terminal regions of the viral genomic or subgenomic RNAs [9–12]. As a comparison, we further analyzed the insect vsiRNAs required for virus clearance in wildtype fruit flies infected with the VSR-deficient mutant Flock house virus (FHV) [13] because levels of vsiRNAs frequently are much higher in insects when RNAi is suppressed by VSRs or in RNAi-defective host mutants to allow robust viral RNA replication and thus to generate abundant dsRNA precursors for dicing [6,8,13]. We found that the total sequenced reads mapped to FHV and fly genomes contained 0.43% vsiRNAs (Table 1). Thus, the relative abundance of the vsiRNAs is similar between insect and mammalian hosts, neither of which encode the cellular RNA-dependent RNA polymerase found in fungi, plants, and nematodes. Notably, the vsiRNAs triggered by each of the four mammalian viruses were readily detectable by Northern blot hybridization [9–12], indicating that the mammalian vsiRNAs are highly abundant according to the criteria used in the annotation of cellular miRNAs [14]. Detection of abundant vsiRNAs in mature mouse, monkey and human cells suggests that the observed decrease in abundance of EMCV vsiRNAs upon the differentiation of mESCs may be specific to stem cell differentiation induced in cell culture [10].
Table 1.
Properties of mammalian viral siRNAs (vsiRNAs) compared to fruit fly vsiRNAs
| Virus (strain/mutant) | Virus family | Viral genome | Host | Experimental system | vsiRNAs* | Ref | |||
|---|---|---|---|---|---|---|---|---|---|
| %of total mapped reads | %of total mapped miRNAs | %of total viral small RNA reads | strand ratio (+/−) | ||||||
| Nodamur a virus (NoVΔB2 ) | Nodaviridae | Segmented positive-strand RNA |
Hamster | Cultured BHK21 cells | 0.06% | 0.13% | 40% | 2.2 | 9 |
| Mice | Embryonic stem cells | 0.02% | 0.04% | 44% | 1.8 | 10 | |||
| Newborn mice | 0.24% | 0.3% | 85.4% | 0.96 | 9 | ||||
| Encephalomyocarditis virus (2887A/91) | Picornaviridae | Positive-strand RNA | Mice | Embryonic stem cells | 0.19% | 0.3% | 33% | 3.1 | 10 |
| Human enterovirus 71 (HEV71D23A) | Picornaviridae | Positive-strand RNA | Human | Cultured 293 cells | 0.04% | 2.4% | 32% | 1.5 | 12 |
| Influenza A Virus (PR8/del NS1) | Orthomyxoviridae | Segmented negative-strand RNA | Human | Cultured A549 cells | 0.59% | 2.4% | 54% | 1.04 | 11 |
| Flock house virus (FHVΔB2) | Nodaviridae | Segmented positive-strand RNA | Fruit fly | Adult flies | 0.43% | 1.9% | 97.4% | 0.74 | 13 |
The sRNA-seq libraries in refs 9, 11, 12 and 13 were re-calculated using the following set of definitions. Total vsiRNAs include only virus reads of 21 to 23 nt in length for mammalian viruses or 20 to 22 nt in length for the fly virus. Total viral small RNA reads include those in the size range of 18 to 28 nt except the small RNAs of EMC V and NoVAB2 from mESCs, which are 19 to 44 nt in length. Total mapped reads in a given sRNA-seq library include all validated reads mapped to both the host and viral genomes.
EMCV and HEV71 are from two different genera in the Picornaviridae whereas NoV and IAV belong to the Nodaviridae and the Orthomyxoviridae (contain a negative-strand RNA genome) respectively. Together, these published studies show that four distinct positive- and negative-strand RNA viruses from the three families trigger the production of abundant 22-nt vsiRNAs in cultured mammalian cells and/or newborn mice, suggesting Dicer-dependent production of vsiRNAs as a conserved mammalian response to RNA virus infection (Fig. 1).
Fig. 1.
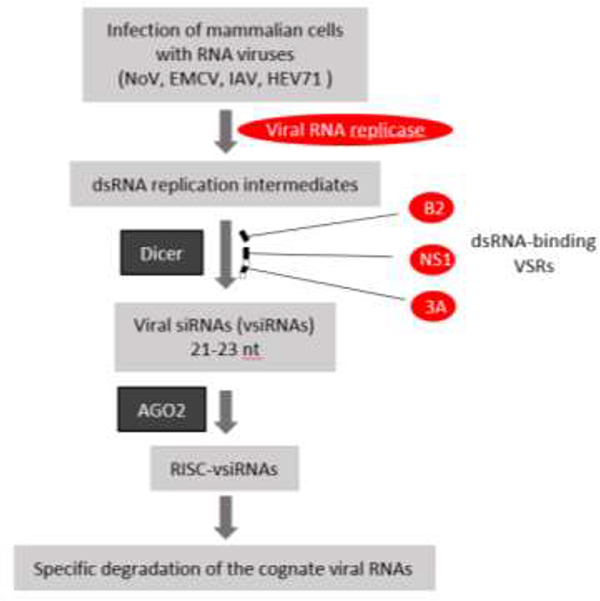
Antiviral RNAi in mammals. During RNA virus infection of mammalian host cells, dsRNA replicative intermediates are processed into viral siRNAs (vsiRNAs) by Dicer, which is the step targeted for the suppression by several dsRNA-binding VSRs. In the absence of VSR expression, specific degradation of the cognate viral RNAs occurs and requires the vsiRNAs and AGO2, both of which are assembled into antiviral RISC in the infected cells.
Three mammalian viruses encode potent dsRNA-binding viral suppressors of RNAi
Among those shown to induce antiviral RNAi in the infected mammalian cells, three viruses each encode a VSR, including the B2 protein of NoV, non-structural protein 1 (NS1) of IAV and the 3A protein of HEV71 [9,11,12] (Table 2). Encoded by viruses from different families, these three mammalian VSRs exhibit no detectable similarity in their primary amino acid sequences. Interestingly, all of the 3 VSRs are dsRNA-binding proteins with NS1 and B2 known to bind long dsRNA by two positively charged antiparallel a helices in homodimer [15–20]. The multifunctional NS1 is best known as a strong antagonist of type I interferons (IFN-I) [21,22]. However, 3A exhibits only a minor IFN-I suppression activity [12] and B2 also is unlikely to be a strong IFN antagonist since B2 expression was associated with higher expression levels of IFN-stimulated genes (ISGs) in both newborn mice [9] and human cancer cells [50].
Table 2.
Properties of the mammalian viral suppressors of RNAi (V SR) verified during authentic infection of mammalian cells
| Virus | VSR | Protein size | Protein structure | dsRNA binding | VSR mechanism | IFN-I antagonist | Virulence factor in vivo | Replication defect of VSR mutants | Rescue of VSR mutants in RNAi-defective cells |
|---|---|---|---|---|---|---|---|---|---|
| Nodamura Virus | B2 | 137 amino acids, 15 kDa | Dimeric, helical bundle structure | Yes | Suppress Dicer processing | nd (not determined) | Yes | Yes | Yes |
| Influenza A Virus | NS1 | 230 amino acids, 26 kDa | Dimeric, helical bundle structure | Yes | Suppress Dicer processing | Strong activity | Yes | Yes | Yes |
| Human Enterovirus 71 | 3A | 86 amino acids, 9.6 kDa | nd | Yes | Suppress Dicer processing | Weak activity | Yes | Yes | Yes |
Both B2 and 3A VSRs inhibit in vitro Dicer processing of long dsRNA into siRNAs and their dsRNA-binding activity is essential for the suppression of Dicer processing [12,19,20,23] (Table 2). Early studies have shown that B2 of NoV inhibits both antiviral RNAi in insect cells [24] and short hairpin RNA-induced RNAi in human 293 cells [20]. Similarly, NS1 encoded by influenza A, B and C viruses was first identified as a VSR because all of them potently suppressed antiviral RNAi induced by viral RNA replication in Drosophila cells [24]. NS1 of IAV also can suppress transgene-induced RNA silencing in plants [25,26] and long dsRNA-induced RNAi in human 293T cells [27]. Similar RNAi suppressor activity has been reported for several other mammalian viral proteins [28,29], including the flaviviral capsid protein that potently suppresses RNAi in the viral mosquito vector [30].
Until recently [9–12], however, it was unknown whether these VSRs indeed can suppress antiviral RNAi induced by authentic virus infection of mammalian cells. Notably, these recent studies have revealed that a dominant population of vsiRNAs was not detectable in mammalian cells either by sRNA-seq or Northern blotting after infection with wildtype NoV, IAV and HEV71, which was in contrast to the production of abundant vsiRNAs triggered by infection with VSR-deficient mutant viruses [9,11,12]. Instead, virus-derived small RNAs (vsRNA) cloned from cells infected with NoV, IAV or HEV71 exhibit random size distribution and strong strand bias [9,11,12] and thus are similar to the vsRNA profiles obtained by simple bulk sRNA-seq from mammalian cells infected with a range of wildtype viruses [31–37]. These findings indicate that expression of B2, NS1 and 3A VSRs suppresses the biogenesis of the vsiRNAs to target the cognate viruses during the infection of mammalian cells. Presumably, VSRs B2, NS1 and 3A all bind to the viral dsRNA replicative intermediates and inhibit Dicer processing of the vsiRNA precursors as has been demonstrated in vitro [12,19,20,23]. Incomplete viral suppression of vsiRNA biogenesis may explain the presence of low abundant vsiRNA duplexes with characteristic 3’ overhangs in the infected mammalian cells [38]. As reported previously in Drosophila cells [39], therefore, use of VSR-deficient mutant viruses may be necessary to characterize the induction and suppression of antiviral RNAi in mammalian cells.
The RNAi pathway confers antiviral activity in mammalian host cells
Mammals possess four Argonaute proteins (AGO 1–4) to mediate miRNA-guided mRNA decay and translational repression in RNA-induced silencing complex (RISC) [3]. Specific cleavage (also known as slicing) of the target RNA occurs when it extensively base-pairs with the miRNA or siRNA loaded in AGO2, the only mammalian AGO that retains the slicer activity [1–3]. Co-immunoprecipitation experiments have demonstrated the presence of mammalian vsiRNAs in mouse and human AGOs, including AGO2 [9–12], indicating a physiological role for mammalian vsiRNAs. The influenza vsiRNAs found in human AGOs exhibit strong preference for those with a 5’-terminal uracil (1U) [11] and thus are similar to cellular miRNAs [3].
NoV is transmissible to mice by mosquitoes and lethal to newborn mice and hamsters as well as insect hosts [40]. Flaccid paralysis of limbs preceding death and neuronal necrosis and degeneration of paravertebral and limb skeletal muscles in infected newborn mice are similar to the symptoms of mice infected with Coxsackie viruses [41]. In contrast to wildtype NoV infection, production of vsiRNAs to target a VSR-deficient mutant of NoV (NoVAB2) in BHK-21 cells and mESCs as well as newborn mice is correlated with a markedly reduced virus accumulation [9,10]. Rapid virus clearance was also observed in newborn mice infected with NoVmB2 [9], which carries a single Arg to Gin mutation at position 59 of B2 known to abolish B2’s activity to bind dsRNA and to suppress Dicer processing [19,23]. The defect of NoVAB2 in infection was efficiently rescued in BHK-21 cells by ectopic expression of homologous and heterologous VSRs and in mESCs by genetic knockout of AGO2 [9,10]. These findings demonstrate the induction of an AGO2-dependent antiviral RNAi response in mammalian cells and indicate that viral suppression of mammalian antiviral RNAi facilitates infection both in vitro and in vivo.
AGO2-knockout mouse embryonic fibroblasts (MEFs) exhibit a marked loss of endogenous miRNAs whereas virtually all miRNAs are present at nearly identical levels in MEFs carrying a genetic mutation, Ago2D597A, which abolishes AGO2’s sheer activity [42–44]. The mutant IAV deleted of its NS1 gene (IAV/delNSl) replicates to significantly higher levels and induces increased cytopathy in primary Ago2D597A MEFs compared to wildtype MEFs [11], indicating restriction of the influenza viral infection by the slicing activity of AGO2 in MEFs. By comparison, abolishing the slicing activity of AGO2 is significantly more effective in enhancing the accumulation of IAV/delNSl than wildtype IAV, which illustrates an increased susceptibility of the VSR-deficient IAV to RNA slicing by antiviral RNAi [11].
Similar to VSR-deficient mutants of NoV and IAV, HEV71 mutants also become defective in the infection of human RD and 293 cells as well as newborn mice after specific mutation (e.g., D23A and R34A) is introduced to abolish 3A’s activity to bind dsRNA and to suppress Dicer processing [12]. Notably, HEV71D23A infection of human 293 cells triggers homology-dependent viral RNA degradation with the specificity determined by Dicer-dependent vsiRNAs [12]. It is unknown whether the RISC loaded with vsiRNAs directs the viral RNA degradation by mRNA decay or slicing. Nevertheless, the findings by Qiu and colleagues show that the vsiRNAs produced during authentic virus infection are active in antiviral RNAi, which is in contrast to cellular miRNAs that become inactive upon induction of the IFN antiviral response by virus infection in human 293 cells [45]. Importantly, HEV71D23A replicates to significantly higher levels after RNAi suppression both in human 293 cells by either Dicer knockout or ectopic expression of homologous and heterologous VSRs and in murine lung fibroblasts (MLFs) by Dicer knockdown [12]. These studies together provide compelling evidence for an antiviral function of the mammalian RNAi pathway and a critical role of viral RNAi suppression in virus infection of cultured cells and newborn mice [9–12].
Antiviral RNAi can act independently of IFN-I antiviral response
Virus infection in mammals triggers potent IFN-regulated antiviral immunity upon the sensing of viral dsRNA, which not only provides the first line of defense against viral pathogens, but also activates the adaptive immunity [46], Levels of IFN-I and induction of ISGs are similar in wildtype MEFs and RNAi-defective Ago2D597A MEFs after virus induction [11]. Importantly, the slicing activity of mouse AGO2 still restricts virus infection of MEFs in the absence of IFN-I signaling, suggesting an IFN-independent antiviral function of RNAi [11]. Qiu and colleagues also investigated the role of IFN-I signaling in the induction and suppression of antiviral RNAi by HEV71 [12]. They found that Dicer-dependent vsiRNAs remain active in antiviral RNAi in human 293 cells treated with Ruxolitinib, an inhibitor of JAK kinases to block IFN-I response. Notably, infection of mutant MLFs and newborn mice lacking a functional IFN-α/β receptor also requires active suppression of RNAi by 3A and Dicer knockdown in the mutant MLFs further enhances the accumulation of HEV71D23A. These findings together indicate that antiviral RNAi can act independently of the defense signaling mediated by IFN-I. Consistently, inactivation of the IFN-I antiviral response facilitates the biogenesis of the siRNAs from the engineered long dsRNA, but not of the vsiRNAs from viral dsRNA replicative intermediates, suggesting differential recognition of the artificial and dsRNA molecules by IFN-I and RNAi responses [27,47–49].
Conclusions
The induction of mammalian antiviral RNAi by distinct positive- and negative-strand RNA viruses in a diverse range of cell types has been reported in recent studies. Three of these viruses each also encode a dsRNA-binding VSR essential for infection. These studies strongly suggest that mammalian antiviral RNAi is not specific to a few viruses or particular cell types (Fig. 1). Indeed, it has been recently shown that antiviral RNAi also provides an IFN-independent protection in human cancer cells so that RNAi suppression by VSR-B2 of NoV further enhances cancer-specific killing of an oncolytic vesicular stomatitis virus variant [50]. However, a recent study showed that the production of abundant vsiRNAs in human 293T cells was not associated with an inhibition of IAV replication [51], which was observed earlier [11]. Thus, suppression of IAV replication by antiviral RNAi is detectable in primary MEFs but not in 293T cells in contrast to the broad activity of antiviral RNAi against NoV and HEV71 in several infection models. Moreover, although primary Ago2D597A MEFs were more susceptible than wildtype MEFs to the infection with all of the three RNA viruses examined, similar enhanced susceptibility was not observed in the immortalized AGO2-knockout MEFs deficient in IFN-I signaling [48]. These observations further highlight an urgent need to develop infection models for the functional characterization of antiviral RNAi under conditions that do not compromise the function of cellular miRNAs.
The existence of a new mammalian antiviral immunity mechanism provides opportunities that may lead to a better understanding of mammalian immunology. For example, mammalian antiviral RNAi provides a genetic pathway for virus clearance without depending on the death of the infected cell; it also describes a defense mechanism that is activated immediately upon infection and programmed with specificity in the form of RNA. Many important questions remain to be addressed on the function and mechanism of mammalian antiviral RNAi. Most importantly, it is unknown whether antiviral RNAi is active and necessary in adult mammals, which activate much more potent IFN-dependent antiviral responses than cultured cells or newborn mice [52]. Does AGO2 confer antiviral defense by mediating RNA slicing or mRNA decay and translational repression? Is antiviral RNAi as widespread in mammals as in plants and insects?
Highlights:
Mammalian cells produce abundant viral siRNAs in response to distinct RNA viruses
Three mammalian viruses encode potent dsRNA-binding viral suppressors of RNAi
The RNAi pathway confers antiviral activity in mammalian host cells
Antiviral RNAi can act independently of IFN-I antiviral response
Acknowledgments
The authors acknowledged the support of NIH grants R01AI52447 and R56AI110579 (to S.W.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
•Refs 5, 6, 7 and 8: These articles provide timely reviews on the studies of antiviral RNAi in plants, insects and nematodes.
••Refs 9 and 10: These two papers together provided the first evidence for an antiviral function of the mammalian RNAi pathway based on the infection of cultured hamster cells and mouse embryonic stem cells as well as newborn mice.
••Refs 11: This study provided the first evidence for the production of viral siRNAs by wildtype human Dicer during virus infection.
••Refs 12: This study provided the first evidence for a critical role of viral siRNAs in homology-dependent degradation of viral RNAs during mammalian virus infection.
••Refs 30: This study provided the first evidence for a critical role of mammalian viral suppressor of RNAi in the mosquito vector of flaviviruses such as yellow fever virus.
•Refs 27 and 47: These two papers show that wildtype and N-terminal-deletion mutant mammalian Dicer proteins can process artificial long dsRNA into functional siRNAs in mammalian cells deficient in IFN responses.
- 1.Baulcombe D: RNA silencing in plants. Nature 2004, 431:356–363. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ: RNA interference. Nature 2002, 418:244–251. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP: Metazoan MicroRNAs. Cell 2018, 173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding SW: RNA-based antiviral immunity. Nat Rev Immunol 2010, 10:632–644. [DOI] [PubMed] [Google Scholar]
- 5.Csorba T, Burgyan J: Antiviral Silencing and Suppression of Gene Silencing in Plants In Current Research Topics In Plant Virology. Edited by Wang A, Zhou X: Springer; 2016:1–34. [Google Scholar]
- 6.Samuel GH, Adelman ZN, Myles KM: Antiviral Immunity and Virus-Mediated Antagonism in Disease Vector Mosquitoes. Trends Microbiol 2018, 26:447–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gammon DB: Caenorhabditis elegans as an Emerging Model for Virus-Host Interactions. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bronkhorst AW, van Rij RP: The long and short of antiviral defense: small RNA-based immunity in insects. Curr Opin Virol 2014, 7:19–28. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Lu J, Han Y, Fan X, Ding SW: RNA interference functions as an antiviral immunity mechanism in mammals. Science 2013, 342:231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O: Antiviral RNA interference in mammalian cells. Science 2013, 342:235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Basavappa M, Lu J, Dong S, Cronkite DA, Prior JT, Reinecker HC, Hertzog P, Han Y, Li WX, et al. : Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat Microbiol 2016, 2:16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu Y, Xu Y, Zhang Y, Zhou H, Deng YQ, Li XF, Miao M, Zhang Q, Zhong B, Hu Y, et al. : Human virus-derived small RNAs can confer antiviral immunity in mammals Immunity 2017, 46:992–1004. [DOI] [PubMed] [Google Scholar]
- 13.Han YH, Luo YJ, Wu Q, Jovel J, Wang XH, Aliyari R, Han C, Li WX, Ding SW: RNA-based immunity terminates viral infection in adult Drosophila in the absence of viral suppression of RNA interference: characterization of viral small interfering RNA populations in wild-type and mutant flies. J Virol 2011, 85:13153–13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. : A uniform system for microRNA annotation. RNA 2003, 9:277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chien CY, Tejero R, Huang Y, Zimmerman DE, Rios CB, Krug RM, Montelione GT: A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat Struct Biol 1997, 4:891–895. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Lynch PA, Chien CY, Montelione GT, Krug RM, Berman HM: Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol 1997, 4:896–899. [DOI] [PubMed] [Google Scholar]
- 17.Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR: Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol 2005,12:952–957. [DOI] [PubMed] [Google Scholar]
- 18.Korber S, Shaik Syed Ali P, Chen JC: Structure of the RNA-binding domain of Nodamura virus protein B2, a suppressor of RNA interference. Biochemistry 2009, 48:2307–2309. [DOI] [PubMed] [Google Scholar]
- 19.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW: Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 2005, 436:1040–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan CS, Ganem D: A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J Virol 2005, 79:7371–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T: Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 1998, 252:324–330. [DOI] [PubMed] [Google Scholar]
- 22.Marc D: Influenza virus non-structural protein NS1: interferon antagonism and beyond. J Gen Virol 2014, 95:2594–2611. [DOI] [PubMed] [Google Scholar]
- 23.Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW: Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 2008, 4:387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, et al. : Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A 2004, 101:1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delgadillo MO, Saenz P, Salvador B, Garcia JA, Simon-Mateo C: Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J Gen Virol 2004, 85:993–999. [DOI] [PubMed] [Google Scholar]
- 26.Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M: The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol 2004, 85:983–991. [DOI] [PubMed] [Google Scholar]
- 27.Kennedy EM, Whisnant AW, Kornepati AV, Marshall JB, Bogerd HP, Cullen B,R,: Production of functional small interfering RNAs by an amino-terminal deletion mutant of human Dicer. Proc Natl Acad Sci U S A 2015, 112:E6945–6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F, Ding SW: Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol 2006, 60:503–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bivalkar-Mehla S, Vakharia J, Mehla R, Abreha M, Kanwar JR, Tikoo A, Chauhan A: Viral RNA silencing suppressors (RSS): novel strategy of viruses to ablate the host RNA interference (RNAi) defense system. Virus Res 2011, 155:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samuel GH, Wiley MR, Badawi A, Adelman ZN, Myles KM: Yellow fever virus capsid protein is a potent suppressor of RNA silencing that binds double-stranded RNA. Proc Natl Acad Sci U S A 2016, 113:13863–13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umbach JL, Yen HL, Poon LL, Cullen BR: Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. MBio 2010, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, Garcia-Sastre A, tenOever BR: Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci U S A 2010, 107:11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girardi E, Chane-Woon-Ming B, Messmer M, Kaukinen P, Pfeffer S: Identification of RNase L-dependent, 3’-end-modified, viral small RNAs in Sindbis virus-infected mammalian cells. MBio 2013, 4:e00698–00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogerd HP, Skalsky RL, Kennedy EM, Furuse Y, Whisnant AW, Flores O, Schultz KL, Putnam N, Barrows NJ, Sherry B, et al. : Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J Virol 2014, 88:8065–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Backes S, Langlois RA, Schmid S, Varble A, Shim JV, Sachs D, tenOever BR: The Mammalian response to virus infection is independent of small RNA silencing. Cell Rep 2014, 8:114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanguy M, Miska EA: Antiviral RNA interference in animals: piecing together the evidence. Nat Struct Mol Biol 2013, 20:1239–1241. [DOI] [PubMed] [Google Scholar]
- 37.Sagan SM, Sarnow P: Molecular biology. RNAi, Antiviral after all. Science 2013, 342:207–208. [DOI] [PubMed] [Google Scholar]
- 38.Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, et al. : Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog 2010, 6:e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li HW, Li WX, Ding SW: Induction and suppression of RNA silencing by an animal virus. Science 2002, 296:1319–1321. [DOI] [PubMed] [Google Scholar]
- 40.Ball LA, Amann JM, Garrett BK: Replication of nodamura virus after transfection of viral RNA into mammalian cells in culture. J Virol 1992, 66:2326–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy FA, Scherer WF, Harrison AK, Dunne HW, Gary GW Jr., : Characterization of Nodamura virus, an arthropod transmissible picornavirus. Virology 1970, 40:1008–1021. [DOI] [PubMed] [Google Scholar]
- 42.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ: A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A: A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 2007, 21:1999–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jee D, Yang JS, Park SM, Farmer DT, Wen J, Chou T, Chow A, McManus MT, Kharas MG, Lai EC: Dual Strategies for Argonaute2-Mediated Biogenesis of Erythroid miRNAs Underlie Conserved Requirements for Slicing in Mammals. Mol Cell 2018, 69:265–278 e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, Hsiang TY, Krug RM, Sullivan CS: Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 2013, 14:435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goubau D, Deddouche S, Reis e Sousa C: Cytosolic sensing of viruses. Immunity 2013, 38:855–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maillard PV, Van der Veen AG, Deddouche-Grass S, Rogers NC, Merits A, Reis ESC: Inactivation of the type I interferon pathway reveals long double-stranded RNA-mediated RNA interference in mammalian cells. Embo J 2016, 35:2505–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuster S, Tholen LE, Overheul GJ, van Kuppeveld FJM, van Rij RP: Deletion of Cytoplasmic Double-Stranded RNA Sensors Does Not Uncover Viral Small Interfering RNA Production in Human Cells. mSphere 2017, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Veen AG, Maillard PV, Schmidt JM, Lee SA, Deddouche-Grass S, Borg A, Kjaer S, Snijders AP, Reis ESC: The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. Embo J 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bastin D, Aitken AS, Pelin A, Pikor LA, Crupi MJ, Huh MS, Bourgeois-Daigneault MC, Bell JC, Ilkow CS: Enhanced susceptibility of cancer cells to oncolytic rhabdo-virotherapy by expression of Nodamura virus protein B2 as a suppressor of RNA interference. Journal for ImmunoTherapy of Cancer 2018, 6:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai K, Courtney DG, Kennedy EM, Cullen BR: Influenza A virus-derived siRNAs increase in the absence of NS1 yet fail to inhibit virus replication. RNA 2018. June 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kollmann TR, Levy O, Montgomery RR, Goriely S: Innate immune function by Toll-like receptors: distinct responses in newborns and the elderly. Immunity 2012, 37:771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]


