Abstract
The embryonic generation of motor neurons is a complex process involving progenitor patterning, fate specification, differentiation, and maturation. Throughout this progression, the differential expression of transcription factors has served as our road map for the eventual cell fate of nascent motor neurons. Recent findings from in-vivo and in-vitro models of motor neuron development have expanded our understanding of how transcription factors govern motor neuron identity and their individual regulatory mechanisms. With the advent of next generation sequencing approaches, researchers now have unprecedented access to the gene regulatory dynamics involved in motor neuron development and are uncovering new connections linking neurodevelopment and neurodegenerative disease.
Introduction
Motor neurons (MNs) are a crucial neuronal subtype responsible for innervating musculature in the periphery and controlling both autonomic and volitional movement. During embryogenesis, combinatorial expression of transcription factors (TFs) guides MN differentiation and diversification [1]. In this review, we survey recent research elucidating the evolutionary origin and broad conservation of these TF programs as well as the DNA-binding mechanics of individual TFs and TF-complexes. We also highlight novel applications of next-generation sequencing technology that have provided valuable genomic and transcriptomic signatures to in-vivo and in-vitro derived MNs.
Motor Neuron Generation
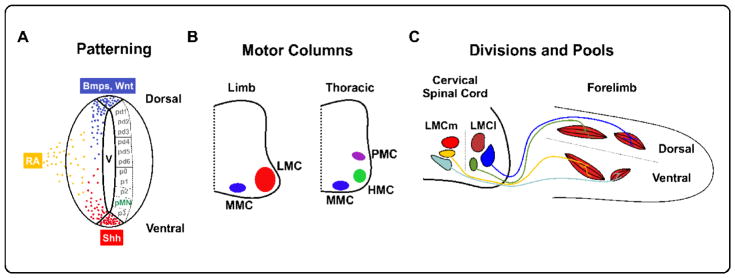
MN generation begins in the embryonic neuroepithelium, wherein opposing gradients of diffusible morphogens (Shh, BMPs, Wnt, RA) pattern proliferating progenitors into discrete domains along the dorsal-ventral body axis. In the ventral spinal cord, MNs are generated from the Olig2+ pMN domain (Figure 1A). Nascent MNs migrate away from the midline and assume positions in distinct motor columns along the rostral-caudal axis that are in register with their target tissues. Motor neurons located within the Medial Motor column (MMC) are found throughout the spinal cord and project to axial muscles. In contrast, Lateral Motor column (LMC) neurons are present at limb levels and innervate target muscles in the fore- and hindlimb, whereas at thoracic regions, Hypaxial Motor column neurons (HMC) and Preganglionic Motor column neurons (PGC) project to body wall muscles and the sympathetic chain ganglia respectively (Figure 1B). Columnar identity is largely defined by Hox proteins, a class of TFs whose clustered 5′ to 3′ chromosomal order maps to their topological expression in the rostral caudal axis [2]. Within a motor column, MNs are further segregated into divisions which delineate broad axonal trajectories. For example, the LMC is divided into medial (LMCm) and lateral (LMCl) divisions that target ventral and dorsal muscles in the limb. Located within each division are motor pools that project to discrete muscles within each area (Figure 1C). Divisional identities are defined in part by the expression of FoxP1 and LIM-homeodomain TFs, whereas motor pools can be distinguished by expression of ETS as well as Hox TFs [3]. Importantly, once generated, MNs themselves are also important players in sculpting the final complement of MNs. MNs within the LMCm are the source of local retinoid signaling via Raldh2 expression that stimulate LMCl generation [4]. Further, GDE2, a retinoid induced GPI-anchor cleaving enzyme expressed in LMC neurons, non-cell autonomously promotes the generation of specific late-born LMC motor pools [5].
Figure 1. Motor Neuron Organization in the CNS.
(A) Embryonic progenitors in the ventricular zone are patterned into discrete dorsal-ventral domains by opposing morphogen gradients. Motor neurons are generated from the ventral pMN domain. V = ventricle, Shh = Sonic Hedgehog, RA = Retinoic Acid. (B) Post-mitotic MNs are organized into motor columns that project to muscles in the limbs (LMC), trunk (MMC), intercostal muscles (HMC), or sympathetic ganglia (PMC). (C) Medial and lateral divisions of the LMC project to ventral and dorsal limb muscles, respectively. Within these divisions, motor pools innervate specific muscle groups.
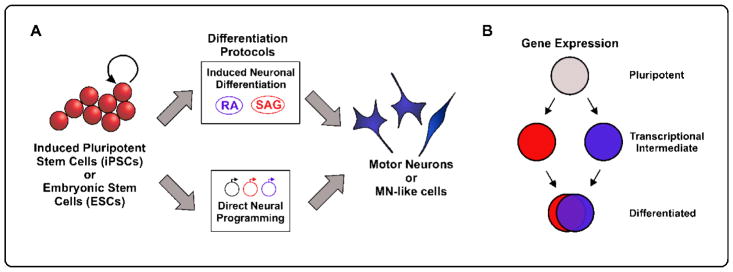
The early stages of MN development can be effectively modeled in-vitro using either undifferentiated Embryonic Stem Cells (ESCs) or induced Pluripotent Stem Cells (iPSCs). iPSCs are differentiated cell types that have been reverted to an unspecified progenitor state. iPSCs afford researchers a large array of starting cell types and enable research on human cells without the ethical restraints of collecting fetal tissue [6–8]. These progenitor cells can be subsequently differentiated by applying exogenous factors that promote neuronal differentiation, including RA, Shh agonists, and Notch antagonists (Induced Differentiation) or by forcing expression of MN TFs such as Lhx1, Isl1, and Ngn2 (Direct Neuronal Programming) (Figure 2A). These reductionist in-vitro platforms allow for the analysis of MNs in an isolated, controlled condition; and they allow access for transcriptomic analysis at the single cell level.
Figure 2. In-vitro Motor Neuron Generation.
(A) Pluripotent ESCs or iPSCs can be differentiated into MNs in-vitro through multiple protocols. Induced neuronal differentiation relies on the delivery of exogenous signaling molecules such as retinoic acid (RA) and Shh pathway agonists (SAG), mirroring embryonic development. MNs can also be differentiated by the forced expression of terminal MN TFs for direct neural programming. (B) RNA sequencing experiments have recently revealed that differentiation via direct neural programming (red circles) versus induced neuronal differentiation (blue circles) can take distinct transcription paths; however, their terminal gene expression patterns are largely convergent.
Motor Neuron Differentiation: Insights from in-vitro platforms
The basic helix-loop-helix (bHLH) TF Olig2 is crucial for specifying MN progenitors in the pMN domain but whether Olig2 promotes neurogenesis or maintains progenitor character is indeterminate. Olig2 induces Ngn2, another bHLH TF required for neuronal differentiation but Olig2 has also been shown to repress terminal MN homeodomain TFs [9,10]. To gain insight into this question, Sagner and colleagues utilized single-cell transcriptomics to map the gene regulatory networks used by Olig2 during ESC-derived MN generation. Distinct transcriptomic profiles can effectively separate early progenitors, MN progenitors, early MNs, and late MNs. Interestingly, the transition into an early post-mitotic MN is accompanied by an increase in Olig2 expression. Chromatin immunoprecipitation sequencing (ChIPseq) revealed that Olig2 acts as a transcriptional repressor for Hes1/Hes5, which are canonical Notch target genes that maintain progenitor character by inhibiting proneuronal TFs [11]. These observations lead to a biphasic model for Olig2 function. Initially, progenitors have low levels of Olig2 expression that permits partial Hes expression. Upon differentiation, Olig2 expression is significantly heightened, repressing Hes1/Hes5 and disinhibiting the expression of proneuronal TFs [12]. Underscoring these changes, unbiased statistical analysis of single cell RNAseq data during the differentiation of murine ESCs into MNs also reveals distinct transcriptional states as cells transition through four phases: pluripotency, neural precursors, MN specified progenitors, and MNs [13]. MN differentiation in-vivo is an asynchronous process, and bulk profiling yields a mélange of cell types in different stages. The single cell resolution of these studies provides a much clearer window into the transcriptional states occupied by differentiating MNs.
Once the correct TFs for terminal MN identity are induced, what are the mechanisms that ensure their expression? Newly born MNs express the LIM-homeodomain proteins Isl1 and Lhx3. They function within a TF complex with nuclear LIM interactor (NLI) to specify MN identity, and these interactions are dependent on key residues within the Lhx3 LIM domain [14,15]. Recent investigations have revealed that the Isl1-Lhx3 complex stabilizes its expression in an autoregulatory manner via binding to enhancers adjacent to the Isl1 and Lhx3 loci. Further, the Isl1-Lhx3 complex upregulates the expression of LIM only Protein 4 (LMO4). LMO4 works in parallel to block the assembly of an Lhx3-only TF complex, which would misdirect the cell towards an interneuron fate [16]. Importantly, sustained Isl1-Lhx3 expression is not a universal feature of all MNs. For example, HMC and LMC MNs lose expression of Lhx3 as they mature, raising the question how expression of terminal MN identity genes regulated by Isl1-Lhx3 is maintained. Rhee et al. performed ChIP-seq from acetylated histone H3 lysine 27 and ATACseq to map genomic regions with an open, accessible chromatin configuration over the course of HMC MN differentiation. They discovered that movement from early to late stages of differentiation is accompanied by highly dynamic changes in enhancer accessibility. Isl1 complexes with Lhx3 to engage the early enhancers, and following Lhx3 downregulation, partners with Onecut1 TFs at late stage enhancers [17]. These relay enhancers thereby ensure stable expression of effector genes over time. The presence of transient enhancers was also observed during the direct neuronal programing of ESCs into spinal MNs; however, contrasting TF-complex dynamics were found. In this paradigm, the same Isl1-Lhx3 TF complex was reported to navigate between early- and late-accessible enhancers without dissociating, and activity at early enhancers repressed progenitor genes rather than activating neuronal genes [18]. This research illustrates how TFs work to control neuronal differentiation amid a transforming chromatin environment in different settings. Importantly, effective TF function is also supported by a range of crucial cofactors working at both the DNA and RNA level (see Box 1).
Box 1. Spatiotemporal control of TF Function is Enabled by Nucleic Acid Regulators.
Multiple DNA/RNA regulators have been recently reported that help TFs fine tune MN identity. At the level of chromatin, the Isl1-Lhx3 TF complexes include single stranded DNA-binding proteins Ssdp1 and Ssdp2. These proteins help recruit histone acetylases, which induce a more open chromatin configuration. In the developing chick spinal cord, genetic knockdown of Sspd1/2 prevents the generation of MNs [35]. At the level of RNA transcription, Topoisomerase IIβ (Top2β) controls the specification of the phrenic MNs by ensuring effective transcription of Pbx and Hox TFs [36]. Top2β null mice exhibit diminished expression of Hoxc6 and Hoxc8 as well as the obligate TALE co-factors Pbx1 and Pbx3. Top2β likely ensures effective transcription of Hox and Pbx transcripts by creating temporary DNA breaks which ameliorate physical stress during RNA synthesis.
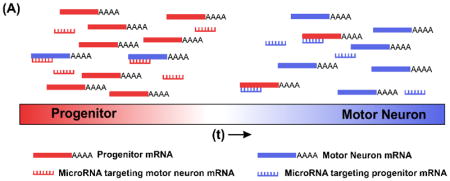
At the level of protein synthesis, microRNAs (miRNA) prevent the translation of specific mRNA transcripts based on sequence homology. Researchers have identified mir-27 as a novel regulator of HoxA5 translation. Using in-silico modeling and genetic approaches, Li et al. reveal how mir-27 dampens transcriptional noise by suppressing HoxA5 translation in MN progenitors. Furthermore, disruption of the miRNA processing machinery causes precocious protein expression of HoxA5 in ventricular zone (VZ) progenitors and newly differentiating intermediate zone (IZ) cells and blurs the transition from rostral HoxA5 to caudal HoxC8 domains [37]. Conversely, mir-375 is upregulated in newly differentiated MNs and dampens translation of progenitor proteins Pax6 and CCND2, a cyclin dependent kinase that promotes proliferation [34]. Thus, miRNAs can prevent the ectopic expression of proneuronal genes during proliferation and progenitor genes during differentiation, sharpening the transition between developmental stages (Panel A). These results illustrate how accessory proteins/molecules facilitate TF function during enhancer binding, RNA transcription, and protein translation.
In-vivo Mechanisms of Motor Neuron Diversification
LMCl versus LMCm axonal targeting is regulated by differential expression of Eph-Ephrin guidance cues whose expression is downstream of LIM-homeodomain TFs [19,20]. Do TFs directly influence the expression of axon guidance factors in other MN populations? New studies have uncovered a similar system regulating axon guidance in the hindbrain. In the absence of Islet1, branchiomotor neuron innervation in the face and jaw is disrupted due to diminished expression of the guidance factor Slit2. Importantly, ChIPseq shows that the Isl1-Lhx3 and Isl1-Lhx4 TF-complexes can directly bind Slit2 enhancers [21]. These results suggest that the same TFs that separate subtype identity can directly promote the expression of axon guidance cues instead of functioning through intermediate transcriptional networks.
Recent findings indicate that Hox proteins play an analogous role in determining MN peripheral connectivity. In the developing forelimb, Catela et al. elegantly describe how combinatorial expression of Hoxc6 and Hoxc8 TFs along with Meis and Pbx co-factors segregate brachial MN projection subtypes via regulation of Ret and Gfrα receptors [22]. Similarly, phrenic motor neurons fail to fully innervate diaphragm muscles in the absence of Hoxa5 expression [23]. Interestingly, strong Hoxa5 expression was found in the diaphragm itself. Does corresponding Hox gene expression in MNs and their respective target tissue aid in topographic axon targeting? In support of this idea, recent experiments in the zebrafish hindbrain reveal a gradient of Hox5 expression in cranial MNs along the anterior-posterior axis. Posterior MNs express high Hox5 and preferentially innervate posterior pharyngeal arches that also express high Hox5 [24]. Analogous mechanisms have also been described in the larval Drosophila melanogaster. In the fly system, somatic MNs expressing the Hox ortholog Deformed (Dfd) are specifically targeted to head muscles that also express Dfd, to create the motor circuits necessary to control feeding [25].
As illustrated by the bi-phasic activity of Olig2 in MN differentiation [12], the degree of TF expression within the same cell type can also influence aspects of MN identity. Mendelsohn and colleagues have recently identified a role for differential HoxC TF expression in separating digit-innervating motor neurons within the LMC. FoxP1+ LMC MNs express Raldh2 and innervate the forelimb; however, the authors find that MNs that project to the digits do not express Raldh2 [26]. They postulate that low (but not zero) levels of Hoxc9, which normally inhibits both FoxP1 and Raldh2, gives digit innervating MNs their unique phenotype that distinguishes them from LMC MNs.
Positional coding by Hox genes is widely conserved among many organisms but the evolutionary origins of limb-innervating LMC neurons have been unclear. Jung and colleagues explore this question in the little skate Leucorja erinacea, which displays an ambulatory gate using its pectoral and pelvic fins. They find that the LMC Hox code is conserved in L. erinacea. Interestingly, these organisms lack a thoracic HoxC gene cluster creating a continuous FoxP1+ LMC, which conforms to the juxtaposition of pectoral and pelvic fins [27]. Their results illustrate the broad conservation of Hox genes and columnar identity, and also indicate that LMC Hox programs evolved far before tetrapods moved to land, in a common ancestor to all vertebrates with paired appendages.
Taken together, these in-vivo data reinforce a broad evolutionary conservation of Hox and LIM-homeodomain TFs in establishing and maintaining diverse MN subtypes. In these efforts, tractable model systems have proven invaluable in decoding MN identity—this is seen in the nematode worm C. elegans, where investigators have elucidated a nuanced combination of transcriptional repressors and positional Hox coding to diversify MNs (see Box 2). It is now apparent that TFs, once considered broad cell fate determinants, directly contribute to the diverse terminal phenotypes of MNs across the hindbrain and spinal cord.
Box 2. MN diversification in Caenorhabditis elegans.
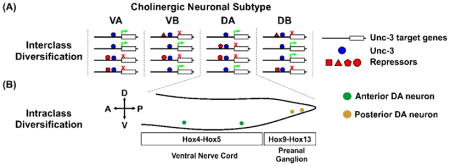
In contrast to vertebrates and insects, the motor system of the nematode C. elegans is highly defined with 75 discrete, identifiable MNs distributed throughout the ventral nerve cord. MNs are first grouped by their neurotransmitter system, either cholinergic or GABA-ergic. Within these groups, MNs are further delineated into 8 classes based on morphology, position, and the combinatorial expression of effector genes. This invariant topology allows researchers to precisely define the mechanisms of MN diversification. The terminal selector gene Unc-3 encodes a COE-type transcription factor necessary for the cholinergic MN lineage. Using forward genetics, Kerk and colleagues demonstrate that cholinergic MN subtypes are diversified through the co-expression of selective repressor TFs that block subsets of UNC-3 enhancers (interclass diversification). They also determine that continued expression of the repressors is required to maintain individual MN identities after development [38] (Panel A). Within a given MN class, additional subtypes exist depending on their somal position along the anterior-posterior body axis. In contrast to the inter-class co-repressor scheme, this intra-class diversification is dependent on region-specific Hox gene expression [39] (Panel B). Importantly, several of the repressor TFs are evolutionarily conserved, raising the possibility that this nuanced combination of subtype repressors and positional Hox coding may operate in vertebrates.
Motor Neurons in Development and Disease
As more and more groups begin to utilize in-vitro differentiation platforms, it becomes critically important to evaluate whether the same types of MNs arise from separate induction protocols. For instance, the disparate results of the Isl1-Lhx3 transient enhancer experiments may arise from the inherent differences between the differentiation of MNs with standard exogenous factors [17] versus direct programming [18]. To address these questions, investigators have generated single cell transcriptomic profiles of ESCs undergoing standard or directed MN induction. Notably, they find that each programming method takes a unique transcriptomic pathway but produces comparable differentiated neurons [28]. These findings shed light on why divergent intermediate transcriptional states can yield similar end-products (Figure 2B). Additional experiments also show promising congruency between in-vitro derived MNs and primary MNs. Similar transcriptomic profiles have been generated comparing MNs derived from genetically unmatched human ESCs versus iPSCs. Here too, investigators find largely overlapping gene expression among the two conditions [29]. These exciting results lend credence to in-vitro derived neurons as representative models of in-vivo MN generation.
MNs are particularly vulnerable in multiple degenerative diseases, and their loss and/or dysfunction produces a gamut of debilitating behavioral consequences [30,31]. Accordingly, a research niche has emerged to understand whether in-vitro derived MNs offer an appropriate model of disease and/or a suitable substrate for stem cell replacement therapy. iPSC induction and in-vitro MN differentiation can occur over the course of weeks while certain neurodegenerative conditions develop over the span of decades. A critical question becomes: do patient iPSC derived MNs resemble their in-vivo counterparts? To address this question, Ho et al. generated transcriptomic profiles of human iPSC differentiated MNs using exogenous factors. They compared these profiles to the transcriptome of in-vivo isolated fetal and adult stage MNs. They discovered that the iPSC MNs more closely resembled the transcriptional state of fetal MNs, suggesting that these neurons may be inappropriate to model late onset neurodegenerative diseases such as Amyotrophic Lateral Sclerosis (ALS) [32]. However, hierarchical clustering identified gene modules that were associated with embryonic development and maturation, and gene expression within these modules was significantly disrupted in ALS derived MNs. Is this perturbation of development related genes merely the result of studying the effect of disease mutations on insufficiently mature MNs; or could the same pathways that ensure proper generation of neurons be subsequently repurposed to safeguard neuronal survival? In support of the latter, we have recently shown how GDE2, which promotes neuronal differentiation embryonically, is essential for MN survival postnatally [33]. In addition, expression of mir-375, which helps secure neuronal identity during differentiation (see Box 1), reduces apoptosis following DNA damage [34]. As we accrue more comprehensive pictures of the degenerating transcriptome and proteome, leveraging these possible connections to developmental programs may help identify new survival pathways that are compromised in disease.
Conclusion
Understanding how TFs and TF-complexes regulate MN identity is integral to decrypt the pathways that govern the precise differentiation, maturation, and diversification of MNs. Combining insights from increasingly reductionist model systems (moving from vertebrates to invertebrates to in-vitro ESC/iPSC derived MNs), we now have a better understanding of how TFs dictate specific MN identities and their regulatory mechanisms. Based on the broad expression of these TF classes, it is likely that principles gleaned from MN development will apply to the development of other neuronal subtypes in the nervous system.
Highlights.
Hox transcription factors are upstream of motor neuron axon guidance cues necessary for proper peripheral innervation.
New findings in C. elegans reveal an elegant system of gene repressors and activators controlling motor neuron diversification.
Single-cell RNA sequencing reveals how different motor neuron induction protocols can utilize distinct gene transcription pathways to converge on a shared motor neuron identity.
Acknowledgments
Research in the laboratory was supported by grants from the Muscular Dystrophy Association and the National Institutes of Health (NINDS, RO1NS046336).
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Alaynick WA, Jessell TM, Pfaff SL. SnapShot: Spinal Cord Development. Cell. 2011;146:178–178e1. doi: 10.1016/j.cell.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dasen JS, Jessell TM. Chapter Six Hox Networks and the Origins of Motor Neuron Diversity. Current topics in developmental biology. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- 3.Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- 4.Sockanathan S, Jessell TM. Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell. 1998;94:503–514. doi: 10.1016/s0092-8674(00)81591-3. [DOI] [PubMed] [Google Scholar]
- 5.Sabharwal P, Lee C, Park S, Rao M, Sockanathan S. GDE2 regulates subtype-specific motor neuron generation through inhibition of Notch signaling. Neuron. 2011;71:1058–70. doi: 10.1016/j.neuron.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdullah RH, Yaseen NY, Salih SM, Al-Juboory AA, Hassan A, Al-Shammari AM. Induction of mice adult bone marrow mesenchymal stem cells into functional motor neuron-like cells. J Chem Neuroanat. 2016;77:129–142. doi: 10.1016/j.jchemneu.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Darvishi M, Tiraihi T, Mesbah-Namin SA, Delshad A, Taheri T. Motor Neuron Transdifferentiation of Neural Stem Cell from Adipose-Derived Stem Cell Characterized by Differential Gene Expression. Cell Mol Neurobiol. 2017;37:275–289. doi: 10.1007/s10571-016-0368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moghaddam SA, Yousefi B, Sanooghi D, Faghihi F, Hayati Roodbari N, Bana N, Joghataei MT, Pooyan P, Arjmand B. Differentiation potential of human CD133 positive hematopoietic stem cells into motor neuron- like cells, in vitro. J Chem Neuroanat. 2017;86:35–40. doi: 10.1016/j.jchemneu.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 10.Lee S-K, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–94. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Dev Growth Differ. 2008;50(Suppl 1):S97–103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- 12••.Sagner A, Gaber Z, Delile J, Kong JH, Rousso DL, Pearson CA, Weicksel SE, Melchionda M, Gharavy NM, Briscoe J, et al. Olig2 and Hes regulatory dynamics during motor neuron differentiation revealed by single cell transcriptomics. PLoS Biol. 2018;16:e2003127. doi: 10.1371/journal.pbio.2003127. The authors examine Olig2 expression over the course of in-vitro MN differentiation. They discovered that a surge of Olig2 expression occurs in nascent MNs that represses the Notch targets Hes1 and Hes 5, thereby promoting differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizvi AH, Camara PG, Kandror EK, Roberts TJ, Schieren I, Maniatis T, Rabadan R. Single-cell topological RNA-seq analysis reveals insights into cellular differentiation and development. Nat Biotechnol. 2017;35:551–560. doi: 10.1038/nbt.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- 15.Seo SY, Lee B, Lee S. Critical Roles of the LIM Domains of Lhx3 in Recruiting Coactivators to the Motor Neuron-Specifying Isl1-Lhx3 Complex. Mol Cell Biol. 2015;35:3579–3589. doi: 10.1128/MCB.00335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erb M, Lee B, Yeon Seo S, Lee JW, Lee S, Lee S-K. The Isl1-Lhx3 Complex Promotes Motor Neuron Specification by Activating Transcriptional Pathways that Enhance Its Own Expression and Formation. Eneuro. 2017;4 doi: 10.1523/ENEURO.0349-16.2017. ENEURO.0349-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Rhee HS, Closser M, Guo Y, Bashkirova EV, Tan GC, Gifford DK, Wichterle H. Expression of Terminal Effector Genes in Mammalian Neurons is Maintained by a Dynamic Relay of Transient Enhancers. Neuron. 2016;92:1252–1265. doi: 10.1016/j.neuron.2016.11.037. Using multiple deep sequencing approaches, the authors find that Isl1 maintains neuronal effector gene expression by utilizing transient enhancers during early and late stages of MN differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velasco S, Ibrahim MM, Kakumanu A, Garipler G, Aydin B, Al-Sayegh MA, Hirsekorn A, Abdul-Rahman F, Satija R, Ohler U, et al. A Multi-step Transcriptional and Chromatin State Cascade Underlies Motor Neuron Programming from Embryonic Stem Cells. Cell Stem Cell. 2017;20:205–217e8. doi: 10.1016/j.stem.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma K, Leonard AE, Lettieri K, Pfaff SL. Genetic and epigenetic mechanisms contribute to motor neuron pathfinding. Nature. 2000;406:515–519. doi: 10.1038/35020078. [DOI] [PubMed] [Google Scholar]
- 20.Luria V, Krawchuk D, Jessell TM, Laufer E, Kania A. Specification of Motor Axon Trajectory by Ephrin-B:EphB Signaling: Symmetrical Control of Axonal Patterning in the Developing Limb. Neuron. 2008;60:1039–1053. doi: 10.1016/j.neuron.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Kim KT, Kim N, Kim HK, Lee H, Gruner HN, Gergics P, Park C, Mastick GS, Park HC, Song MR. ISL1-based LIM complexes control Slit2 transcription in developing cranial motor neurons. Sci Rep. 2016;6:36491. doi: 10.1038/srep36491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catela C, Shin MM, Lee DH, Liu JP, Dasen JS. Hox Proteins Coordinate Motor Neuron Differentiation and Connectivity Programs through Ret/Gfrα Genes. Cell Rep. 2016;14:1901–1915. doi: 10.1016/j.celrep.2016.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landry-Truchon K, Houde N, Boucherat O, Joncas F-H, Dasen JS, Philippidou P, Mansfield JH, Jeannotte L. HOXA5 plays tissue-specific roles in the developing respiratory system. Development. 2017;144:3547–3561. doi: 10.1242/dev.152686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barsh GR, Isabella AJ, Moens CB. Vagus Motor Neuron Topographic Map Determined by Parallel Mechanisms of hox5 Expression and Time of Axon Initiation. Curr Biol. 2017;27:3812–3825e3. doi: 10.1016/j.cub.2017.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich J, Sorge S, Bujupi F, Eichenlaub MP, Schulz NG, Wittbrodt J, Lohmann I. Hox Function Is Required for the Development and Maintenance of the Drosophila Feeding Motor Unit. Cell Rep. 2016;14:850–860. doi: 10.1016/j.celrep.2015.12.077. [DOI] [PubMed] [Google Scholar]
- 26.Mendelsohn AI, Dasen JS, Jessell TM. Divergent Hox Coding and Evasion of Retinoid Signaling Specifies Motor Neurons Innervating Digit Muscles. Neuron. 2017;93:792–805e4. doi: 10.1016/j.neuron.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Jung H, Baek M, D’Elia KP, Boisvert C, Currie PD, Tay B-H, Venkatesh B, Brown SM, Heguy A, Schoppik D, et al. The Ancient Origins of Neural Substrates for Land Walking. Cell. 2018;172:667–682e15. doi: 10.1016/j.cell.2018.01.013. Uncovers a conserved Hox code that specifies LMC identity in an aquatic skate that displays bi-pedal motion of its pectoral and pelvic fins, indicating these programs evolved millions of years before tetrapods moved to land. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Briggs JA, Li VC, Lee S, Woolf CJ, Klein A, Kirschner MW. Mouse embryonic stem cells can differentiate via multiple paths to the same state. Elife. 2017;6:e26945. doi: 10.7554/eLife.26945. Utilizing the superior resolution of single-cell RNAsequencing, the authors describe how two separate in-vitro neuronal differentiation paradigms (factor induced versus direct programming) can take alternate transcriptional paths but arrive at a similar MN mRNA profile. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marei HE, Althani A, Lashen S, Cenciarelli C, Hasan A. Genetically unmatched human iPSC and ESC exhibit equivalent gene expression and neuronal differentiation potential. Sci Rep. 2017;7:17504. doi: 10.1038/s41598-017-17882-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burghes AHM, Beattie CE. Spinal muscular atrophy: Why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig. Nat Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 32.Ho R, Sances S, Gowing G, Amoroso MW, O’Rourke JG, Sahabian A, Wichterle H, Baloh RH, Sareen D, Svendsen CN. ALS disrupts spinal motor neuron maturation and aging pathways within gene co-expression networks. Nat Neurosci. 2016;19:1256–1267. doi: 10.1038/nn.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cave C, Park S, Rodriguez M, Nakamura M, Hoke A, Pletnikov M, Sockanathan S. GDE2 is essential for neuronal survival in the postnatal mammalian spinal cord. Mol Neurodegener. 2017;12:8. doi: 10.1186/s13024-017-0148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhinge A, Namboori SC, Bithell A, Soldati C, Buckley NJ, Stanton LW. MiR-375 is Essential for Human Spinal Motor Neuron Development and May Be Involved in Motor Neuron Degeneration. Stem Cells. 2016;34:124–134. doi: 10.1002/stem.2233. [DOI] [PubMed] [Google Scholar]
- 35.Lee B, Lee S, Agulnick AD, Lee JW, Lee S-K. Single-stranded DNA binding proteins are required for LIM complexes to induce transcriptionally active chromatin and specify spinal neuronal identities. Development. 2016;143:1721–1731. doi: 10.1242/dev.131284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edmond M, Hanley O, Philippidou P. Topoisomerase IIβ selectively regulates motor neuron identity and peripheral connectivity through Hox/Pbx-dependent transcriptional programs. eNeuro. 2017;4 doi: 10.1523/ENEURO.0404-17.2017. ENEURO.0404-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37••.Li CJ, Hong T, Tung YT, Yen YP, Hsu HC, Lu YL, Chang M, Nie Q, Chen JA. MicroRNA filters Hox temporal transcription noise to confer boundary formation in the spinal cord. Nat Commun. 2017;8:14685. doi: 10.1038/ncomms14685. The authors combine in-vitro, in-vivo, and in-silico approaches to identify microRNA dependent circuits that control the spatiotemporal expression of Hox genes during development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38••.Kerk SY, Kratsios P, Hart M, Mourao R, Hobert O. Diversification of C. elegans Motor Neuron Identity via Selective Effector Gene Repression. Neuron. 2017;93:80–98. doi: 10.1016/j.neuron.2016.11.036. Identifies a unique method for motor neuron diversification in C. elegans, not by varying the expression of activating TFs, but by the combinatorial expression of supressor TFs which inhibit specific Unc-3 loci among cholinergic MNs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kratsios P, Kerk SY, Catela C, Liang J, Vidal B, Bayer EA, Feng W, De La Cruz ED, Croci L, Giacomo Consalez G, et al. An intersectional gene regulatory strategy defines subclass diversity of C. Elegans motor neurons. Elife. 2017;6:1–31. doi: 10.7554/eLife.25751. [DOI] [PMC free article] [PubMed] [Google Scholar]