Abstract
We have produced Csf1r-deficient rats by homologous recombination in embryonic stem cells. Consistent with the role of Csf1r in macrophage differentiation, there was a loss of peripheral blood monocytes, microglia in the brain, epidermal Langerhans cells, splenic marginal zone macrophages, bone-associated macrophages and osteoclasts, and peritoneal macrophages. Macrophages of splenic red pulp, liver, lung, and gut were less affected. The pleiotropic impacts of the loss of macrophages on development of multiple organ systems in rats were distinct from those reported in mice. Csf1r−/− rats survived well into adulthood with postnatal growth retardation, distinct skeletal and bone marrow abnormalities, infertility, and loss of visceral adipose tissue. Gene expression analysis in spleen revealed selective loss of transcripts associated with the marginal zone and, in brain regions, the loss of known and candidate novel microglia-associated transcripts. Despite the complete absence of microglia, there was little overt phenotype in brain, aside from reduced myelination and increased expression of dopamine receptor-associated transcripts in striatum. The results highlight the redundant and nonredundant functions of CSF1R signaling and of macrophages in development, organogenesis, and homeostasis.
Introduction
Resident macrophages are abundant in all tissues, where they adapt to distinct niches and environments, expressing specific genes to perform tissue-specific homeostatic functions (reviewed in Ref. 1, 2). Macrophage differentiation from progenitor cells, and many aspects of their mature function, is controlled by two ligands, CSF1 and IL-34, which both signal through the CSF1R. The biology of CSF1R and its ligands is conserved from birds to mammals (3). Csf1r expression in adults is restricted to cells of the myeloid lineages, and transcriptional regulation of the gene has been studied extensively, both in vitro and with the use of transgenic mice, chickens, and sheep (4–6).
In humans, mutations in the tyrosine kinase domain of CSF1R have been associated with a neurodegenerative disease, autosomal dominant, adult-onset leukoencephalopathy with axonal spheroids and pigmented glia (ALSP, previously known as hereditary diffuse leukoencephalopathy with spheroids) (7). Disease-associated mutant CSF1R isoforms can be expressed on the cell surface but lack ligand-dependent kinase activity and probably act as dominant negative repressors of the wild type allele (8).
Targeted mutation of Csf1r in mice depleted tissue macrophages from most organs and had pleiotropic impacts on growth and development (9). The more penetrant phenotype of the receptor mutation, compared with the natural mutation of the Csf1 ligand in the Csf1op/op mouse, predicted the existence of the second ligand, IL-34 (9). Phenotypes of the Csf1 or Csf1r mutant mice include increased bone density (osteopetrosis), abnormalities of the sensory nervous system, global defects in brain development, infertility, failure of pancreatic β cell development, and severe postnatal growth retardation (reviewed in Ref. 10, 11). On the original cross-bred background, around 50% of Csf1r−/− mice survived to weaning. On an inbred background, survival beyond 3 weeks was rare (12).
Rat models have been used extensively in the study of human disease and are preferred to mice in areas of inflammation, development, physiology, neurobiology, pharmacology, and behavior (reviewed in Ref. 13). The use of rats has recently been accelerated by the expanding availability of genetic modification technologies via homologous recombination in embryonic stem cells (ESC) and, more recently, CRISPR–Cas9 and other targeted nucleases. This has led to a comeback of the rat in biomedical research (14). Rats provide many experimental advantages over mice such as larger sample volumes, higher resolution in vivo imaging, improved performance in learning/memory behavioral assays, easier surgical procedures, and physiological similarities with humans (15). They are the model of choice in areas such as diabetes, breast cancer, and chronic inflammatory and cardiovascular diseases as well as age-related illnesses (reviewed in Ref. 16). Current knowledge of CSF1R biology in rats has been based upon limited analysis of the toothless (tl/tl) mutation in the ligand CSF1, focused largely upon the osteopetrotic phenotype in bone (17). In this article, we report on the generation Csf1r−/− rats by homologous recombination in ESC and their detailed phenotypic characterization. The results highlight significant differences in CSF1R between species and suggest the rat may be a better predictive model for human macrophage biology.
Materials and Methods
Production of Csf1r−/− rats
An EGFP-PGK-Neo cassette flanked by 2.2 kb 5′ and 5 kb 3′ homology arms (Supplemental Fig. 1A) was introduced into ESC derived from a male Dark Agouti rat [clone DAK31 (18) provided by Austin Smith] by electroporation. Positive selection was provided by a PGK-Neo cassette, and negative selection was provided by a diphtheria toxin-A chain cassette. The diphtheria toxin-A chain cassette was placed upstream of the 5′ homology arm and so was not incorporated into the genome. Csf1r gene-targeted rat ESC clones were identified by 5′ PCR screening and 3′ Southern blot analysis. Of the 24 clones analyzed, 20 were positive at the 5′ end for the targeted allele by PCR, and 18 were positive at the 3′ end for the targeted allele by Southern blot (Supplemental Fig. 1D, 1E). Fifty percent of clones analyzed had normal karyotypes (data not shown), and clone DAK31-C2 was used to generate the colony (Supplemental Fig. 1B).
All experiments were carried out under the authority of a U.K. Home Office Project License under the regulations of the Animals (Scientific Procedures) Act 1986. Approval was obtained from ethics committees of The Roslin Institute and The University of Edinburgh. Csf1r−/− rats were produced by blastocyst injection as described in (19), except that Sprague–Dawley females were used as both embryo donors and pseudo-pregnant recipients. Genotyping and the 5′ PCR screen of ESC clones was performed by PCR using MyTaq HS DNA polymerase (Bioline) with the primers forward: 5′-GCTGCAGTCCCTTACATAGGTCTAC-3′, reverse 1: 5′-GGAGGTGCAGATGAACTTCAGG-3′, and reverse 2: 5′-AGCTTCCCCTGCCCTGAGAAG-3′. Expected products were 3.5 and 2.9 kb for the wild type and mutant Csf1r, respectively (Supplemental Fig. 1C).
3′ Southern blot analysis of ESC clones
Genomic DNA (7.5 μg) from ESC clones were digested with Xbal and used in Southern blot analysis as per standard procedures. The probe was 503 bp and spanned the third exon of rat Csf1r (Rat Genome Sequencing Consortium 6.0/rn6, chr18:56,437,991–56,438,493).
Gene expression analysis
Livers and spleens were preserved in RNAlater (Invitrogen). Total RNA was isolated in TRIzol (Invitrogen), followed by purification with an RNeasy Mini kit (QIAGEN) according to instructions. cDNA synthesis and quantitative real-time PCR (qRT-PCR) were performed as described in (6). The oligonucleotides used are listed in Table I.
Table I. Oligonucleotides.
| Gene | Forward Oligonucleotide | Reverse Oligonucleotide |
|---|---|---|
| Adgre4 | 5′-TGCCCTTATTGTTGCTGTGTCTGC-3′ | 5′-CACTGGCCCCAAGAAGCTCCA-3′ |
| Cd209b | 5′-TCCAAGATCCCCAGCCTCCAG-3′ | 5′-GCAGAGTCGACACAGGCGGA-3′ |
| Csf1r (genotyping) | 5′-GCTGCAGTCCCTTACATAGGTCTAC-3′ | R1, 5′-GGAGGTGCAGATGAACTTCAGG-3′ |
| R2, 5′-AGCTTCCCCTGCCCTGAGAAG-3′ | ||
| Csf1 | 5′-AGTCTTGCTGGCTGTCGGGG-3′ | 5′-GGTCGCCCCACAGAAGAATCCA-3′ |
| Csf1r | 5′-ACGGCCACCATGAACTTCCA-3′ | 5′-CGCAGGGTGAGCTCAAAGGT-3′ |
| Fmod | 5′-CCCTCCCGTCAACACCAACCT-3′ | 5′-AAGTTCATGACGTCCACCACCG-3′ |
| Gal3st2 | 5′-GCTGGCTGTGCTCCTGTTGG-3′ | 5′-GCGGTTCCTGGGCCTTGTCC-3′ |
| Gapdh | 5′-ATGACTCTACCCACGGCAAG-3′ | 5′-TGGGTTTCCCGTTGATGACC-3′ |
| Ghr | 5′-CGGGTGTTCTTAACCCTGGCACT-3′ | 5′-GCAGAACCGGGGAAGCTTTGC-3′ |
| Total Igf1a | 5′-TGTGTGGACCAAGGGGCTTT-3′ | 5′-GTCTGTGGTGCCCTCCGAAT-3′ |
| Il22ra2 | 5′-GGACACCCCGCTTCACTCCA-3′ | 5′-CCCTCAAAGATGCATTAACTCGGGT-3′ |
| Mpeg1 | 5′-GGTTTGCCGGGTCCCTTGGT-3′ | 5′-ACATTCGTGCAGCCAGGGTG-3′ |
| Nr1h3 (Lxrα) | 5′-GCAGAGACCCTCCCAGAGCCTA-3′ | 5′-ACACTGCATAGCTCGTTCCCCAG-3′ |
Spans local and circulating transcripts.
R, reverse.
Radiographs and micro–computed tomography analysis of bone architecture
Radiographs were produced at the Hospital for Small Animals located at the Royal (Dick) School of Veterinary Studies (Easter Bush, U.K). Micro–computed tomography (μCT) analysis was performed on formalin fixed tibias and skulls using a SkyScan 1172 (Bruker) at a resolution of 17.22 μm with a 0.5-mm aluminum filter and four-frame averaging to improve the signal to noise ratio of the images. The tibias were scanned using a source voltage of 71 kV and a source current of 139 μA. To accommodate the larger dimensions of the skulls, a double width acquisition mode was used with a source voltage of 81 kV and a source current of 122 μA. An exposure time of 1180 ms was used for all scans. The images were reconstructed using NRecon v1.6.9 (Bruker) and analyzed using CTAn v1.13.5 (Bruker) software.
Complete blood count analysis
Whole EDTA blood was analyzed as described in (20).
Serum biochemistry analysis
Serum was analyzed using the ILab 650 automated biochemistry analyzer (Instrumentation Laboratory).
Flow cytometry
Blood was collected into EDTA tubes via cardiac bleeds and prepared using Dako Uti-Lyse erythrocyte lysing solution according to instructions. Blood was stained with AF647-labeled CSF1-Fc (6) and the following Abs: CD32APC (Clone REA256, 1:40) and CD161APC (Clone REA227, 1:40) from Miltenyi Biotec, B220PE (Clone His24, 1:400; eBioscience), CD4FITC (Clone W3/25, 1:10; Bio-Rad Laboratories), CD3AF488 (Clone 1F4, 1:200), and SIRPαPE (Clone OX-41, 1:400) from BioLegend. Myelin-depleted single-cell suspensions of brain were prepared as described in (21), except rats were perfused with physiological saline and heparin and then stained with CD45EF450 (Clone OX1, 1:200; eBioscience) and CD11b/cAF647 (Clone OX-42, 1:500; BioLegend). Peritoneal cavity cells were isolated as described in (22) and stained with SIRPαPE and CD11b/cAF488 as described above. Isotype controls were used for all flow cytometry data. Cells were analyzed by flow cytometry on a FACSCalibur, LSRFortessa X-20 or LSRFortessa (BD Biosciences). Analysis was performed with FlowJo software (FlowJo).
Western blot
Plasma was separated from EDTA whole blood by centrifugation at 2000 G for 15 min at 4°C. A 1:50 dilution was prepared using SDS-PAGE loading buffer. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes for immunoblotting. Membranes were probed with polyclonal primary Abs: rabbit anti-CSF1 (1:500, MBS551037; MyBioSource) and 1:2000 sheep anti-Transferrin (1:2000, ab9033; Abcam) in PBS/3% milk powder/0.1% (v/v) Tween 20 at 4°C overnight. The secondary Abs used were goat anti-rabbit HRP (1:5000, no. 7074; Cell Signaling Technologies) and rabbit anti-sheep HRP (1:10,000, ab97130; Abcam). Blots were visualized using Pierce ECL Western blotting solution (Thermo Fisher Scientific).
Histology and immunohistochemistry
Formalin fixed organs were processed into paraffin using standard procedures. For histological examination, sections were stained with H&E or Luxol fast blue. For immunohistochemical analysis, details of Ag retrieval, Ab concentrations, and detection systems used are listed in Table II. Femurs were decalcified in EDTA, and sections were stained using an acid phosphatase and tartrate-resistant acid phosphatase (TRAP) kit (Sigma-Aldrich) according to instructions, except incubation time was increased to 2 h, and a counterstain was not used. Epidermal sheets were obtained after incubating the dorsal face of both ears in 20 mM EDTA for 2 h (37°C), followed by fixation in 4% PFA for 20 min and washing in PBS. Tissues were blocked for 1 h (room temperature [RT]) in PBS/0.1% BSA/5% goat serum, then stained with anti-rat MHC class II (MHC-II) (clone OX-6, 1:100; Abcam). Secondary Ab was F(ab′)2-Goat anti-Mouse IgG (H+L)AF647 (1:500, A-21053; Invitrogen). Epidermal sheets were washed in HBSS (Thermo Fisher Scientific), and nuclei were stained for 5 min (RT) with Hoechst 33258 (1:1000, 861405; Sigma-Aldrich). Stained epidermal sheets were mounted with ProLong Gold (Life Technologies) and cured for 24 h (RT) before imaging.
Table II. Immunohistochemistry conditions.
| Target (Clone) | Supplier | Ag Retrieval | Dilution | Detection |
|---|---|---|---|---|
| CD68 (ED1) | Bio-Rad | Proteinase K 20 min, RT | 1:500 | ImmPRESS anti-mouse 15 min |
| 45 min | DAB, hematoxylin counterstain | |||
| CD163 (ED2) | Thermo Fisher Scientific | Proteinase K 20 min, RT | 1:100 | ImmPRESS anti-mouse 10 min |
| 60 min | DAB, hematoxylin counterstain | |||
| IBA1 rabbit polyclonal | Wako Pure Chemical Corp. | 0.01 M citrate buffer, pH 6.0 | 1:500 | EnVision anti-rabbit 40 min |
| 110°C 5 min | 30 min | DAB, hematoxylin counterstain | ||
| MRC1 rabbit polyclonal | Abcam | 1 mM EDTA, 0.1% Tween 20, pH 8 | 1:1000 | EnVision anti-rabbit 40 min |
| 16 h 60°C | 60 min | Vector NovaRED, hematoxylin counterstain | ||
| SIGLEC1 (ED3) | Bio-Rad | 0.01 M citrate buffer, pH 6 | 1:200 | ImmPRESS anti-mouse 30 min |
| 110°C 20 min | 2 h | Vector VIP, no counterstain. |
DAB, 3,3-diaminobenzidine.
Microarrays
Total RNA was prepared from snap frozen brain regions (hippocampus, striatum, olfactory bulbs, and pituitary gland) and spleens as described above from nonperfused rats (two female rats per genotype). Library preparation and hybridization to the Affymetrix Rat Gene 2.1 ST array was performed by Edinburgh Genomics, The University of Edinburgh. CEL files were normalized [RMA (23)] and annotated in R/Bioconductor. The data from the microarrays are available at Gene Expression Omnibus, National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/), under accession number GSE100696. To identify spleen-specific genes downregulated by the loss of Csf1r, the normalized transcriptomic data were loaded into Graphia Pro (Kajeka, U.K.). Using a Pearson correlation threshold cutoff of R = 0.98, a graph was obtained comprising 20,515 nodes (individual probe sets). Clustering of the graph using the Markov clustering algorithm (MCL) was used with an MCL inflation value of 1.5 to determine the granularity of clusters.
Evaluation of blood–brain barrier permeability
A 2% solution of Evans blue in PBS (4 μl/g body weight) was injected i.p. in postnatal day 17 rats. The stain was allowed to circulate for 3 h before rats were culled by cervical dislocation. Spleens and brains were removed, then weighed. The organs were dried overnight in a 55°C oven, and Evans blue was extracted by addition of formamide (16 μl/mg spleen and 8 μl/mg brain). After 48 h at 55°C, Evans blue stain was measured by spectrophotometer at 620 nm and quantified according to a standard curve. The results are presented as microgram of Evans blue per milligram of dried tissue.
Statistical analysis
Analysis was performed with GraphPad Prism version 6.04, and statistical significance was assessed using Student two-tailed t test. Resulting values were considered statistically significant at p < 0.05.
Results
Generation and gross phenotype of Csf1r−/− rats
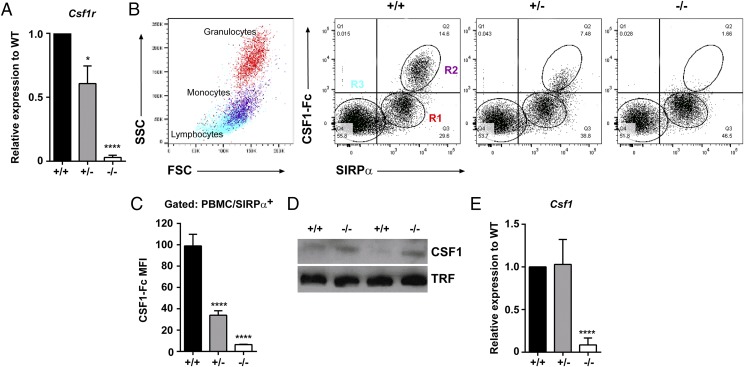
Csf1r−/− rats were produced using ESC in which the first coding exon of rat Csf1r was disrupted with a drug selection cassette by homologous recombination (Supplemental Fig. 1A). qRT-PCR demonstrated partial (∼50%) and complete loss of Csf1r mRNA in spleens from Csf1r+/− and Csf1r−/− rats, respectively (Fig. 1A), indicating that there was no dosage compensation in the heterozygote. The loss of functional CSF1R was demonstrated using a labeled CSF1R ligand [porcine CSF1-Fc (6)]. There was reduced (>60%) binding activity on monocytes (SIRPα+, SSClo cells) from Csf1r+/− heterozygous rats. There was no CSF1 binding activity on any blood cells from Csf1r−/− rats (Fig. 1B, 1C). The loss of CSF1R activity was associated with an increase in the ligand in the circulation, as observed in Csf1r−/− mice (9), confirmed by Western blot analysis (Fig. 1D). By contrast, Csf1r−/− rats displayed a significant reduction in Csf1 mRNA in the spleen (Fig. 1E). This likely reflects the depletion of CSF1-dependent macrophages, which in rats (unlike mice, see data on www.biogps.org or www.immgen.org) express abundant Csf1 mRNA (see below).
FIGURE 1.
Expression of Csf1r and Csf1 in rats. (A) cDNA was prepared from total splenic RNA and Csf1r expression analyzed by qRT-PCR (n = 7+/+, 7+/−, and 3−/−). (B) Whole EDTA blood was used to analyze SIRPα expression and binding of CSF1R to its ligand (CSF1-Fc) by flow cytometry. Regions R1–R3 highlighted in Csf1r+/+ rat blood were back-gated onto a forward versus side scatter (FSC/SSC) dot plot to highlight the three main cell populations in blood. (C) Combined mean fluorescent intensity of CSF1-Fc binding in SIRPα+ monocytes (n = 6+/+, 8+/−, and 5−/−). (D) Protein lysates were prepared from EDTA plasma (female adults) and assessed for CSF1 expression by Western blot. An anti-transferrin (TRF) Ab was used as the loading control. (E) cDNA was prepared from total spleen RNA and Csf1 expression analyzed by qRT-PCR (n = 7+/+, 7+/−, and 3−/−). Graphs show the mean + SEM. Significance compared with wild type is indicated by *p = 0.015 and ****p < 0.0001 using a t test.
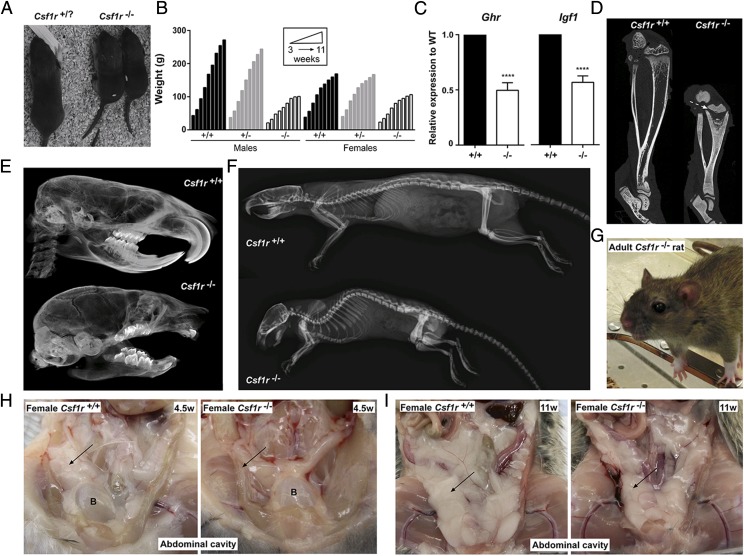
Csf1r−/− rats were indistinguishable from their littermates at birth but were identified from P9–11 by the absence of tooth eruption, smaller body size, and shorter snout (Fig. 2A), similar to the reported phenotype of the Csf1 mutant (tl/tl) rat (24). The frequencies of wild type, heterozygous, and homozygous genotypes at birth were 27, 52, and 21% (n = 266), respectively, not significantly different from the expected 1:2:1 ratio expected under Mendelian inheritance (p = 0.3). All female Csf1r−/− rats survived beyond weaning (n = 22), but 23% of male Csf1r−/− rats were lost postnatally (n = 8) without obvious clinical signs. Csf1r−/− rats weighed less than their littermates at weaning, and their body weight plateaued after 10 wk. As in Csf1r−/− mice, the differential increase in growth/body size normally seen in wild type males was abolished in the Csf1r−/− mutant animals, most likely reflecting a lack of testosterone production (9) (Fig. 2B). Tl/tl rats lack circulating insulin-like growth factor 1 (IGF1; responsible for body growth) in the postnatal period, implying a link between CSF1R and growth hormone receptor (GHR) signaling (25). Mouse macrophages grown in CSF1 express high levels of Igf1 mRNA and are likely to be the major extrahepatic source (25). We have confirmed this finding in rats (see below). In the Csf1r−/− rats, the mutation also impacted upon the major source of Igf1 in the postpubertal growth surge. Both Ghr and total Igf1 mRNA levels were consistently reduced by around 50% in the liver of mutant rats compared with age- and sex-matched litter mate controls (Fig. 2C).
FIGURE 2.
Gross phenotype of Csf1r−/− rats. (A) Two Csf1r−/− rat pups and a littermate control at postnatal day 11. (B) Rats were weighed weekly following weaning at P21. For males, n = 5+/+, 8+/−, and 3−/−. For females, n = 7+/+, 4+/−, and 8−/−. Animals culled prior to 11 wk of age were included in the analysis. Graph shows the mean + SEM. (C) Total RNA was isolated from livers of wild type (+/+) and Csf1r−/− rats (n = 6) for analysis of Ghr and total Igf1 expression via qRT-PCR. There were equal numbers of males and females, four animals were adults, and two were 18 d old in each group. Graph shows the mean + SEM for pairwise comparison with age- and sex-matched littermate controls. Absolute values did not differ markedly between males and females or ages. Significance compared with wild type is indicated by ****p < 0.0001 using a t test. (D) Lower limbs from 7-wk males were fixed in 10% buffered formalin and analyzed for bone density by μCT. Dotted arrow indicates growth plate. (E) Skulls from 7-wk males were scanned by μCT. (F) Rats were analyzed by radiography at 6 mo of age. μCT images are from females, which are representative of both sexes. (G) An adult female Csf1r−/− rat with bulging eyes. Representative photograph of the abdominal cavities of 4.5-wk-old (H) and 11-wk-old (I) females. Arrow points to visceral fat in the wild type (left) and absence or reduction of visceral fat in Csf1r−/− rat (right). Images are also representative of males. B, bladder.
Csf1r−/− rats were osteopetrotic, with reduced bone marrow (BM) cavities and disrupted growth plates in the tibia (Fig. 2D). Although Csf1r−/− rats lack any visibly erupted teeth, μCT analysis of the skull clearly showed molars and rudimentary incisors as well as a domed skull (Fig. 2E). By 6 mo of age, a severe curvature of the spine was apparent (Fig. 2F). Ageing Csf1r−/− rats developed further phenotypes, including bulging eyes (Fig. 2G), that secreted porphyrin. The secretions were likely a consequence of the inability to blink properly, causing corneal dryness and increased porphyrin production. Postmortem analysis revealed that both male and female Csf1r−/− rats had an almost complete absence of visceral white adipose tissue at 4.5 wk (Fig. 2H) yet still developed interscapular brown adipose tissue (Supplemental Fig. 2A). White adipose tissue was detected in older Csf1r−/− rats but was still greatly reduced when compared with littermate controls (Fig. 2I). As the rats aged, they developed increased respiratory rates, most likely a result of cranial bone defects limiting the space available for soft tissues such as the soft palate and tongue, similar to brachycephalic syndrome in certain dog breeds (26). Indeed, the tongues of Csf1r−/− rats were the same size as littermate controls, despite their much smaller body size (Supplemental Fig. 2B). A subset of males exhibited a protruding penis, without any adverse effects on behavior or urination, with osteopetrosis of the penile bone being the most probable cause.
Aside from the obvious difference in viability between mutant rats and mice, many pleiotropic impacts of macrophage deficiency described in Csf1 and Csf1r mutant mice (9, 27, 28) were not observed in the Csf1r−/− rats. For example, they showed no evidence of sensory defects (sight, hearing, and smell were apparently normal), produced insulin in histologically normal pancreatic islets (Supplemental Fig. 2C), and retained Paneth cells in the intestines, which also showed normal villous architecture (Supplemental Fig. 2D).
Analysis of blood from Csf1r−/− rats
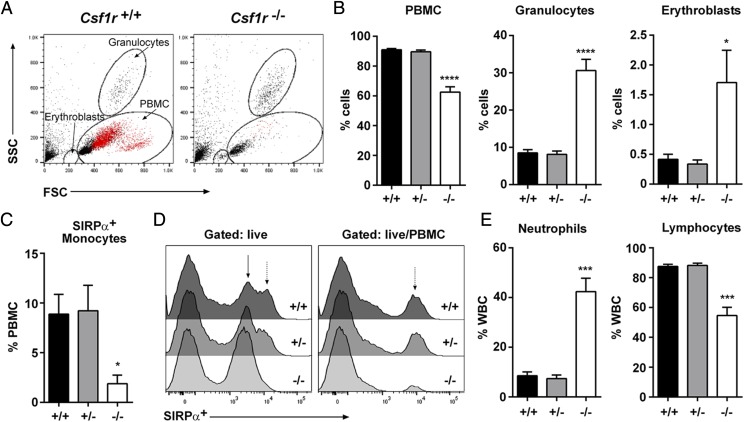
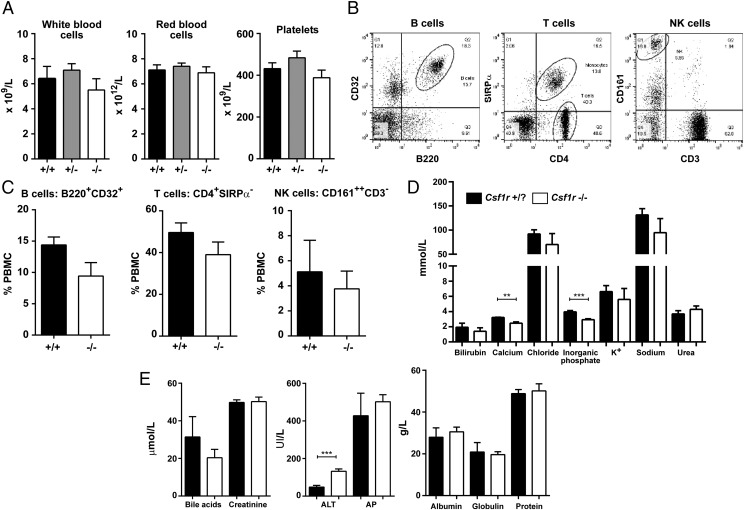
Csf1r−/− rats had a significant decrease in PBMCs, whereas the percentages of granulocytes and erythroblasts were increased (Fig. 3A, 3B). By contrast with the CSF1 ligand–deficient Csf1tl/tl rat (29), there was no evidence of thrombocytopenia, nor any change in total RBC or WBC count (Fig.4A). SIRPα+ monocytes were unaffected in the heterozygote but were almost undetectable in the homozygous mutant (Fig. 3C, 3D). Conversely, neutrophils were increased and lymphocytes were decreased (Fig. 3E) as reported previously in the Csf1r−/− mice (9). There was no change in relative abundance of B, T, or NK cells within the lymphoid compartment (Fig. 4B, 4C). Blood albumin and electrolyte levels were unaffected by the mutation (Fig. 4D, 4E), with the exception of calcium and inorganic phosphate, which were both significantly reduced, most likely associated with deficient bone resorption.
FIGURE 3.
Analysis of whole blood. (A) Whole EDTA blood from adult rats was used to analyze forward versus side scatter (FSC/SSC) profiles by flow cytometry. SIRPα+ monocytes are colored red. (B) The percentages of PBMCs, granulocytes, and erythroblasts were determined by FSC/SSC profiles (n = 5 per genotype). (C) Cells were gated on SIRPα+ PBMC to determine the percentage of monocytes (n = 5 per genotype). (D) SIRPα expression was analyzed in whole EDTA blood gating on total live cells and live PBMC. Solid arrow highlights SIRPαlow granulocytes. Dotted arrow highlights SIRPαhigh monocytes (n = 5 per genotype). (E) Whole EDTA blood was analyzed on an automated counter to determine the percentage of neutrophils and lymphocytes in WBCs (n = 6+/+, 10+/−, and 8−/−). Graphs show the mean + SEM. Significance compared with wild type is indicated by *p = 0.046 (erythroblasts) and 0.015 (monocytes), ***p < 0.0003, and ****p < 0.0001 using a t test.
FIGURE 4.
Further analysis of Csf1r-deficient rat blood. (A) Whole EDTA blood was used to determine the total number of WBCs, RBCs, and platelets from male and female rats aged between 6 and 12 wk (n = 6+/+, 11+/−, and 8−/−). (B) Whole EDTA blood was analyzed by flow cytometry to identify B, T, and NK cells. Dot plots show gating strategy for each cell type using Csf1r +/+ blood. Cells were gated on the PBMC population by forward versus side scatter (FSC/SSC). Quadrants were determined with isotype controls (n = 5+/+ and 6−/−). (C) Graphs shows the mean + SEM. p = 0.09, 0.23, and 0.63 for B, T, and NK cells, respectively. (D and E) Serum from male and female rats aged 2–4 wk was analyzed (n = 5 per genotype). Graphs shows the mean + SEM. **p = 0.0018, ***p = 0.0010 (inorganic phosphate) and 0.0007 (ALT). ALT, alanine aminotransferase; AP, alkaline phosphatase, K+, potassium.
Fertility of Csf1r−/− rats
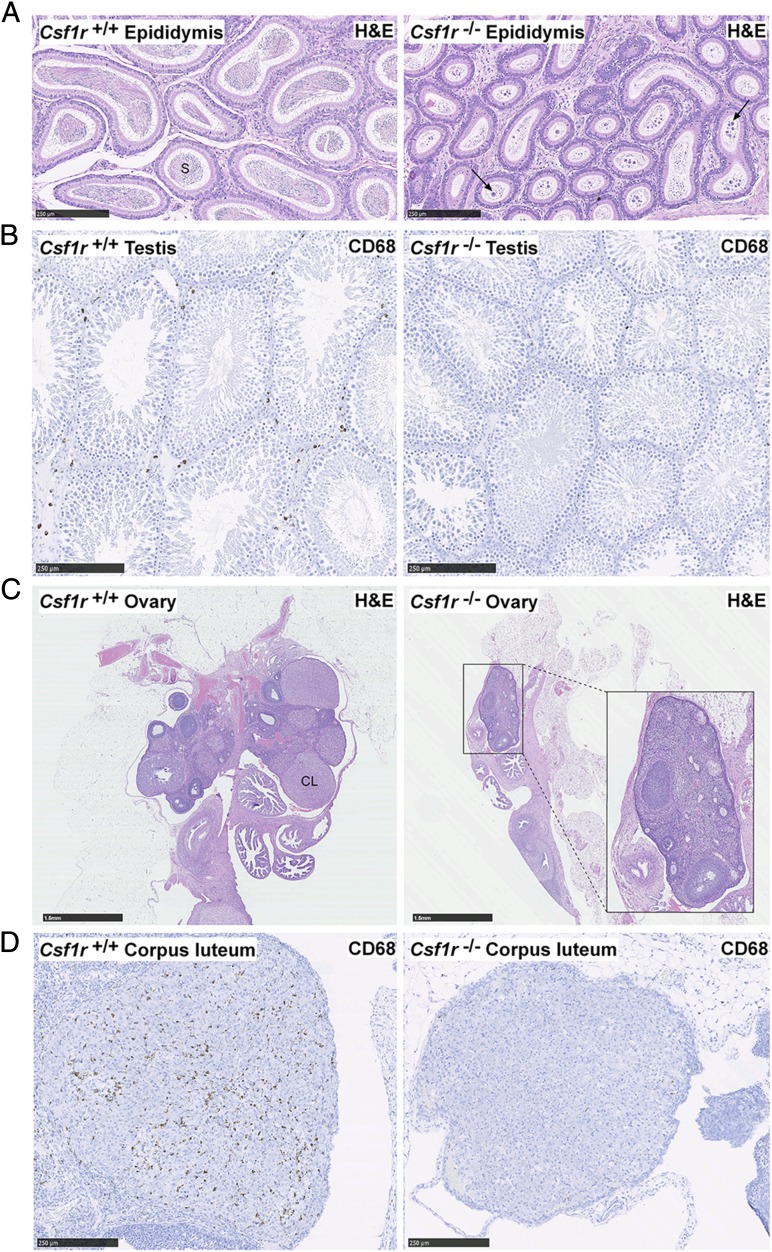
Csf1tl/tl male rats reach sexual maturity and were reported to be fertile (17). To date, male Csf1r−/− rats have produced no pregnancies when placed with wild type females. Histological analysis revealed a severe reduction in the number of mature sperm cells in the lumen of the epididymis and a large number of atypical residual bodies, suggesting impaired maturation of spermatids (30) (Fig. 5A). The mAb ED1 recognizes rat CD68 and is commonly used to identify rat macrophages (31). In the testis of Csf1r−/− males, there was a decrease in CD68+ interstitial macrophages (Fig. 5B), similar to that shown in mice treated with anti-CSF1R (32). The size difference between Csf1r−/− females and adult male wild type rats precluded test mating on welfare grounds, but the females are also likely to be infertile. Their ovaries were greatly reduced in size, and corpora lutea were either absent or greatly reduced (Fig. 5C). The few corpora lutea that were observed lacked CD68+ macrophages (Fig. 5D), which are known to regulate corpus luteum development in mice (33).
FIGURE 5.
Histology of reproductive organs. Formalin-fixed and paraffin-embedded epididymides, testes, and ovaries were stained with H&E or an Ab against CD68. (A) Longitudinal sections of epididymides and (B) testes. Images are representative of six rats aged between 7 and 11 wk. Arrows point to atypical residual bodies. Scale bar, 250 μm. (C and D) Longitudinal sections of ovaries stained with H&E and CD68. Images are representative of seven rats aged 11–12 wk. Scale bar, 1.5 mm (C) and 250 μm (D). In wild type and Csf1r+/− rats, an average of 5.8 (range 3–11) corpora lutea were observed per ovary in H&E-stained sections (n = 10) compared with only 0.7 (range 0–3) in Csf1r−/− rats (n = 7). Whole slide images were produced with a NanoZoomer slide scanner and images were exported with NDP.view2 software (Hamamatsu Photonics). CL, corpus luteum, S, sperm.
Analysis of tissue macrophages in Csf1r−/− rats
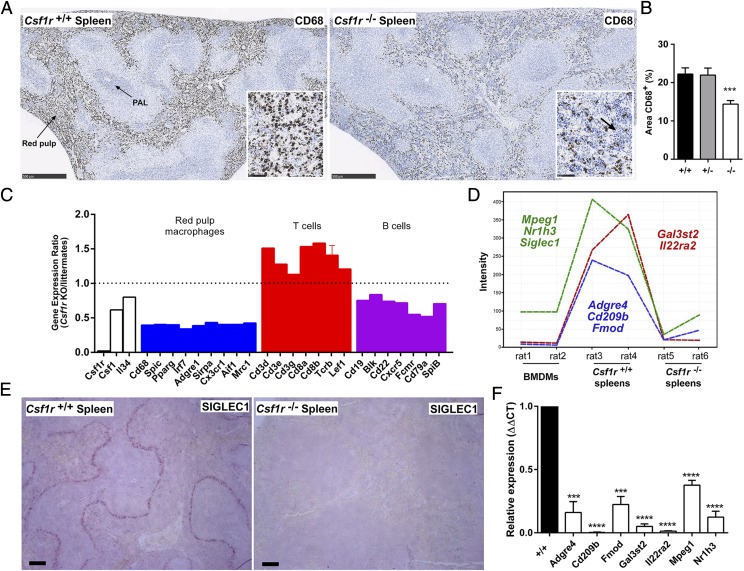
There was a reduction of CD68+ macrophages in both the red pulp and the periarteriolar lymphoid sheath in spleen compared with wild type and heterozygous rats (Fig. 6A, 6B). This decrease was partly due to extensive infiltration of the red pulp by small mononuclear cells. There were no nucleated RBCs and no obvious increase in the number of megakaryocytes (Fig. 6A). Hence, there was no evidence of the extramedullary hematopoiesis that occurs in Csf1r mutant mice in the absence of a BM cavity (9). To evaluate the impact of the mutation on spleen cell populations, we compared gene expression profiles from Csf1r+/+ and Csf1r−/− rats with BM-derived macrophages (BMDM). The full data are provided in Supplemental Table I. The set of 543 transcripts with expression ratios from 0.02 (Csf1r) to 0.5 (calculated as the ratio of expression in Csf1r−/− to expression in littermates) (Supplemental Table I) includes CD68 (consistent with Fig. 6A), transcription factors (SpiC, Pparg), and the majority of markers of splenic red pulp macrophages including Adgre1 (F4/80) (Fig. 6C). The reduction in the relative abundance of these transcripts (and by inference, the red pulp macrophages) may be partly attributable to the overall expansion of the lymphoid populations. The increased relative expression of T cell–associated genes (Fig. 6C) indicates that this expansion is biased toward T lymphocytes, whereas B cell markers were all reduced (Fig. 6C). A similar bias toward T cells was reported when Csf1 was disrupted in mice (34). The spleen contains multiple different mononuclear phagocyte populations occupying specific niches, including the red pulp macrophages, marginal zone macrophages and metallophils, tingible body macrophages, and classical dendritic cells (35). The network analysis tool Graphia Pro (formerly BioLayout Express3D) was used to identify spleen-specific, macrophage-related genes that were downregulated with the loss of Csf1r. The Csf1r-dependent genes identified included markers of the separate marginal zone (Siglec1) and marginal zone metallophilic (CD209b) macrophages (Fig. 6D). Complete loss of the former population was confirmed via immunohistochemistry with an anti-SIGLEC1 Ab (Fig. 6E). The selective loss of CD209b, as well as a number of other candidate marginal zone-associated transcripts discussed below, was confirmed by qRT-PCR (Fig. 6F). The selective loss of the marginal zone macrophages may explain the disrupted splenic architecture and the migration of lymphocytes into the red pulp.
FIGURE 6.
Analysis of spleens in Csf1r−/− rats. (A) Formalin-fixed and paraffin-embedded spleens were stained with an Ab against CD68. Scale bar, 500 μm. Inset shows an area of red pulp. Scale bar, 50 μm. Arrow points to infiltration of lymphocytes in Csf1r−/− inset. Whole slide images were produced with a NanoZoomer slide scanner and jpeg files exported with NDP.view2 software. PAL, periarteriolar lymphoid sheath. (B) The area of CD68+ cells in the red pulp was determined from 10 images per spleen (at original magnification ×80) using ImageJ (Fiji) (n = 10+/+, 8+/−, and 13−/−). Graph shows mean + SEM. Significance compared with wild type is indicated by ***p = 0.0002 using a t test. (C) Microarray data from spleens were used to determine expression ratios for genes associated with red pulp macrophages and T and B cells. Graph shows mean + SEM for genes with multiple probes. (D) Gene expression plot generated in Graphia Pro highlighting splenic macrophage-specific genes which were downregulated in Csf1r−/− rats. (E) Formalin-fixed and paraffin-embedded adult spleens were stained with an Ab against SIGLEC1. Scale bar, 100 μm. Images are representative of 2+/+ and 3−/− rats and two repeat experiments. (F) Total RNA was isolated from spleens of adult wild type (+/+) and Csf1r−/− rats (n = 3) for analysis of candidate marginal zone macrophage-associated gene expression identified in (D) via quantitative PCR. Graph shows mean + SEM. Significance compared with wild type is indicated by ***p = 0.003 and ****p < 0.0001 using a t test.
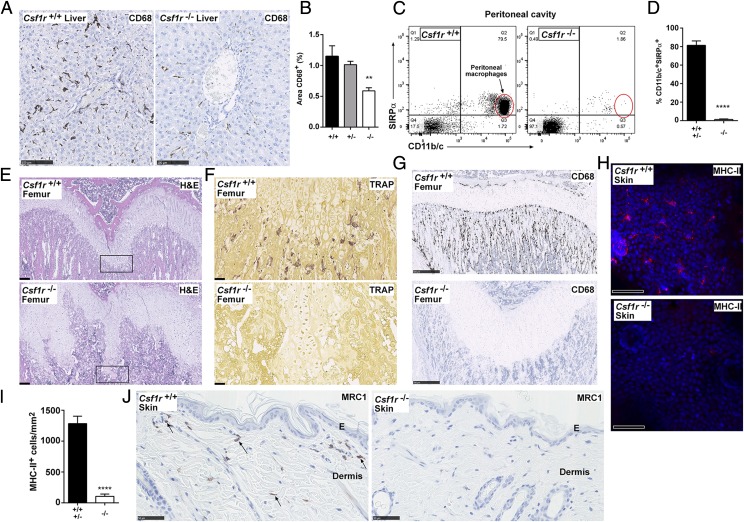
In the liver, CD68+ Kupffer cells were unaffected in the heterozygotes but partially depleted in Csf1r−/− rats. The residual CD68+ cells appeared smaller and less ramified, but there was otherwise no gross change in liver size (relative to body weight) or architecture (Fig. 7A, 7B). Liver enzymes, alanine aminotransferase, and alkaline phosphatase were marginally increased in the circulation, but bile acids and bilirubin were unchanged and there was no histological evidence of injury (Fig. 4D, 4E). As in the Csf1r−/− mice (9), peritoneal macrophages were absent in the Csf1r−/− rats (Fig. 7C).
FIGURE 7.
Analysis of tissue macrophages in Csf1r-deficient rats. (A) Formalin-fixed and paraffin-embedded livers were stained with an Ab against CD68. Whole slide images were produced with a NanoZoomer slide scanner and exported as jpeg files with NDP.view2 software. Scale bar, 100 μm. (B) The area of CD68+ cells was determined from 10 images per liver (at original magnification ×20) using ImageJ (n = 7+/+, 4+/−, and 9−/−). Graph shows mean + SEM. Significance compared with wild type is indicated by **p = 0.0035 using a t test. (C) Peritoneal cavity cells were analyzed by flow cytometry for expression of SIRPα and CD11b/c. Dead cells were excluded with propidium iodide, and quadrants were set with isotype controls. (D) Graph shows mean + SEM of CD11b/c+SIRPα+ peritoneal macrophages. Significance compared with wild type/heterozygotes is indicated by ****p < 0.0001 using a t test (n = 3). Decalcified femurs from adult rats were either stained with (E) H&E or stained for expression of (F) TRAP and (G) CD68. The boxed growth plate in (E) shows the area of TRAP staining. Images are representative of five rats per genotype. Scale bar, 200 μm (H&E), 500 μm (CD68), or 50 μm (TRAP). Whole slide images were produced with a NanoZoomer slide scanner and images exported with NDP.view2 software. (H) Epidermal sheets were prepared from the ears of adult rats and immunostained for MHC-II (red). Nuclear staining was performed using Hoechst 33258 (blue). Epidermal sheets were imaged as z-stacks (25 μm) using a Zeiss LSM710 confocal microscope. MHC-II and nuclei signals were acquired with 633 and 405 nm lasers, respectively. (I) Maximum intensity projections were produced from the z-stacks and MHC-II+ cells were quantified as per (102). Graph shows mean +SEM. Significance compared with wild type/heterozygotes is indicated by ****p < 0.0001 using a t test. Images are representative of four rats per genotype, two z-stacks analyzed per rat. Scale bar, 50 μm. (J) Formalin-fixed and paraffin-embedded dorsal skin was immunostained for MRC1 expression. Image is representative of 2+/+ and 3−/− adult rats per genotype and two repeat experiments. Arrows point to MRC1+ dermal macrophages. Whole slide images were produced with a NanoZoomer slide scanner and images exported with NDP.view2 software. Scale bar, 50 μm. E, epidermis.
To examine effects on bone architecture, femurs were stained for TRAP (also known as ACP5) and CD68. As noted also in Fig. 2D, despite the cortical thickening and increased trabecular bone, the Csf1r−/− rats retained a substantial BM compartment. This likely explains the lack of extramedullary hematopoiesis in the spleen. Osteoclasts were entirely deficient in Csf1r−/− rats (Fig. 7F), as reported in Csf1tl/tl rats (17), whereas in Csf1r−/− mice TRAP+ cells are retained, albeit at a lower frequency (9). The surface of bone in mice and humans contains a population of specialized osteal macrophages, intercalated among the osteoblasts (36). Abundant osteal macrophages were detected with CD68 staining in rat bone, and they were completely absent in bones from the Csf1r−/− animals (Fig. 7G).
In mice, IL-34 is also expressed in the epidermis and is required for the development of the MHC-II–positive epidermal Langerhans cells (37). In the Csf1r−/− rats. MHC-II+ Langerhans cells, visualized in epidermal sheets, were almost completely depleted (Fig. 7H, 7I); the few remaining cells were only weakly MHC-II+. Dermal macrophages were also absent in Csf1r−/− rats as detected by CD206 (mannose receptor, MRC1) expression (Fig. 7J).
Analysis of Csf1r−/− rat brains
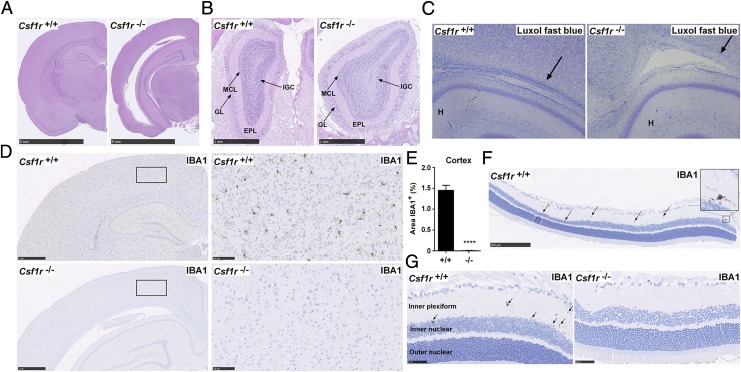
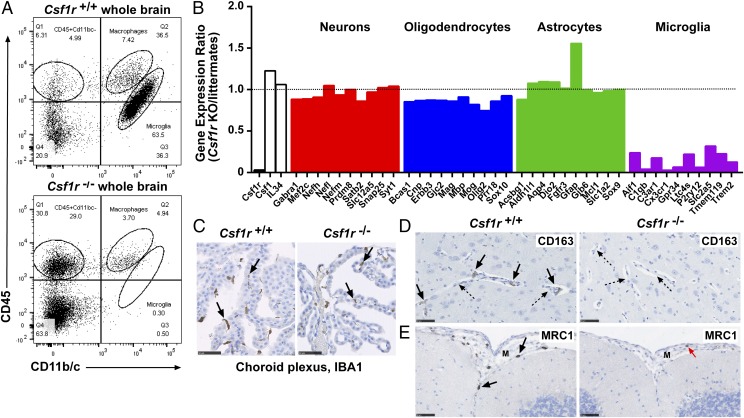
The brains of Csf1r−/− rats at 11 wk of age revealed few abnormalities apart from some enlargement of the lateral ventricles (Fig. 8A). Whereas Csf1r−/− mice had hollow and fragile olfactory bulbs (27), these were intact and apparently normal in the rat (Fig. 8B). Minor loss of myelin was apparent at the level of the corpus callosum (Fig. 8C). Csf1r-deficient mice had a gross deficiency of microglia, detected with the microglial marker, IBA1 (27). IBA1+ cells with ramified morphology were similarly not detectable in any region of the brain (Fig. 8D) or in the plexiform layers of the retina (Fig. 8F, 8G) in adult Csf1r−/− rats. To confirm that this deficit was due to a loss of microglia, rather than loss of the marker, cells extracted from total brain tissue were analyzed (21). CD11b/c+CD45low microglia were entirely absent in Csf1r−/− rats (Fig. 9A), and CD11b/c+CD45hi macrophages (normally a minor component in brain digests) were decreased. The population of CD11b/c−CD45hi cells was increased in Csf1r−/− rats. We attribute this to blood contamination as a result of incomplete perfusion (discussed below).
FIGURE 8.
Analysis of brains from Csf1r−/− rats. All brains were formalin fixed and paraffin embedded for histology and immunohistochemistry. (A) Brains from 11-wk-old rats were stained with H&E. Images are representative of seven rats per genotype. Scale bar, 5 mm. (B) Adult heads were fixed in formalin and stained with H&E following EDTA decalcification. Image shows olfactory bulbs in situ. Scale bar, 1 mm. Image representative of seven rats per genotype. EPL, external plexiform layer; GL, glomerular layer; IGC, internal granular cell layer of the olfactory bulbs; MCL, mitral cell layer. (C) Formalin-fixed brains from 3-wk-old rats were stained with Luxol fast blue. Arrows point to myelin preservation and myelin pallor in wild type and mutant rats, respectively. Image of three rats per genotype. (D) Adult brains (8–14 wk) were stained with an Ab against IBA1. Scale bar, 1 mm (left) or 100 μm (right). (E) For each rat, 10 20× images of the cortex were analyzed for the percentage of IBA1+ staining using ImageJ (n = 7 per genotype). Graphs show mean + SEM. Significance compared with wild type is indicated by ****p < 0.0001 using a t test. (F and G) Formalin-fixed eyes were stained with an Ab against IBA1. Arrows point to IBA1+ microglia in the inner plexiform layer of the retina. Images are representative of two rats per genotype, two repeat experiments. Scale bar, 250 μm (F) and 50 μm (G).
FIGURE 9.
Further analysis of brains from Csf1r−/− rats. (A) Single-cell suspensions of brain were depleted of myelin and analyzed by flow cytometry for CD11b/c and CD45 expression. Dead cells were excluded with propidium iodide, and blood granulocytes were excluded from Csf1r−/− samples via forward versus side scatter (FSC/SSC). Quadrants were determined using isotype controls. Image is representative of six rats per genotype. (B) Microarray data from dissected striatum, hippocampus, olfactory bulbs, and pituitary gland were used to determine expression ratios for genes associated with the major cell populations of the brain. (C) Choroid plexuses from adult brains stained with an Ab against IBA1. Images are representative of seven rats per genotype. Arrows point to IBA1+ choroid plexus macrophages. Sclae bar, 50 μm (D) Adult brains were stained with an Ab against CD163. Images are representative of seven rats per genotype. Solid arrows point to CD163+ perivascular macrophages. Dotted arrows point to blood vessels. Scale bar, 50 μm. (E) Adult brains were stained with an Ab against MRC1. Images are representative of seven rats per genotype. Solid arrows point to MRC1+ meningeal macrophages in the Csf1r+/+ rat brain. Red arrow points to a monocyte in the Csf1r−/− meninges (M). Scale bar, 50 μm. Whole slide images were produced with a NanoZoomer slide scanner and analyzed with NDP.view2 software (Hamamatsu Photonics).
The brain, like the spleen, contains several macrophage populations in addition to microglia (38). To further analyze the impact of Csf1r mutation on brain myeloid populations and on other brain cells, we profiled gene expression in the striatum, hippocampus, olfactory bulbs, and pituitary gland. The full dataset is provided in Supplemental Table I. There was no global change in expression of generic neuron-associated genes in Csf1r−/− rats shared by the four brain regions examined, but there was some evidence of region-specific impacts, discussed below.
The set of 131 transcripts with expression ratios from 0.03 (Ctss, Cx3cr1, and Csf1r) to 0.5 (Supplemental Table I) contains many microglia-enriched transcripts identified in mice (Fig. 9B) (39–41) and rats (42). By contrast to the loss of microglia-specific markers, many macrophage-associated marker genes remained readily detectable in Csf1r−/− brains (e.g., Cd14 and Fcgr1a encoding CD64). We attempted to locate residual macrophages with known markers. As in the Csf1r-deficient mouse (27), there were no detectable CD68+ cells in Csf1r−/− rat brains via immunohistochemistry (data not shown). Csf1r−/− rats retained IBA1+ macrophages in the choroid plexus (Fig. 9C) but lacked detectable CD163+ perivascular macrophages (43) and MRC1+ meningeal macrophages (38) (Fig. 9D, 9E).
The blood–brain barrier was intact in Csf1r−/− rats, as evident from Evans blue exclusion. Interestingly, there was an obvious reduction in Evans blue staining in the periphery of Csf1r−/− rats, suggesting a subtle vascular phenotype. Evans blue staining in the spleen was also reduced (Fig. 9A), despite no differences in levels of the carrier albumin (Fig. 4E), potentially highlighting the known role of macrophages in angiogenesis (reviewed in Ref. 44).
Network analysis of gene expression in the spleen of Csf1r-deficient rats
In previous studies of the effect of CSF1 treatment on macrophage numbers and gene expression profiles in the liver, BMDM were used as an external reference to identify genes that were enriched in macrophages relative to the tissue (45). These genes provided a signature of CSF1-dependent expansion of the macrophage population detectable in total mRNA from the liver (45). To provide a similar reference for the rat, BMDM were generated by cultivation in a medium containing recombinant human CSF1 for a period of 7 d as described for mouse (45), mRNA was isolated, and transcript expression was profiled in parallel with tissue mRNA from adult rats.
To identify the set of genes that were spleen enriched, based upon comparison with BMDM grown in CSF1, the wild type, heterozygous, and homozygous Csf1r mutant expression profiles were combined with BMDM and clustered using Graphia Pro. Surprisingly, one of the two heterozygous mutant animals had much lower expression of Csf1r, albeit higher than in the knockout. (Supplemental Table I).
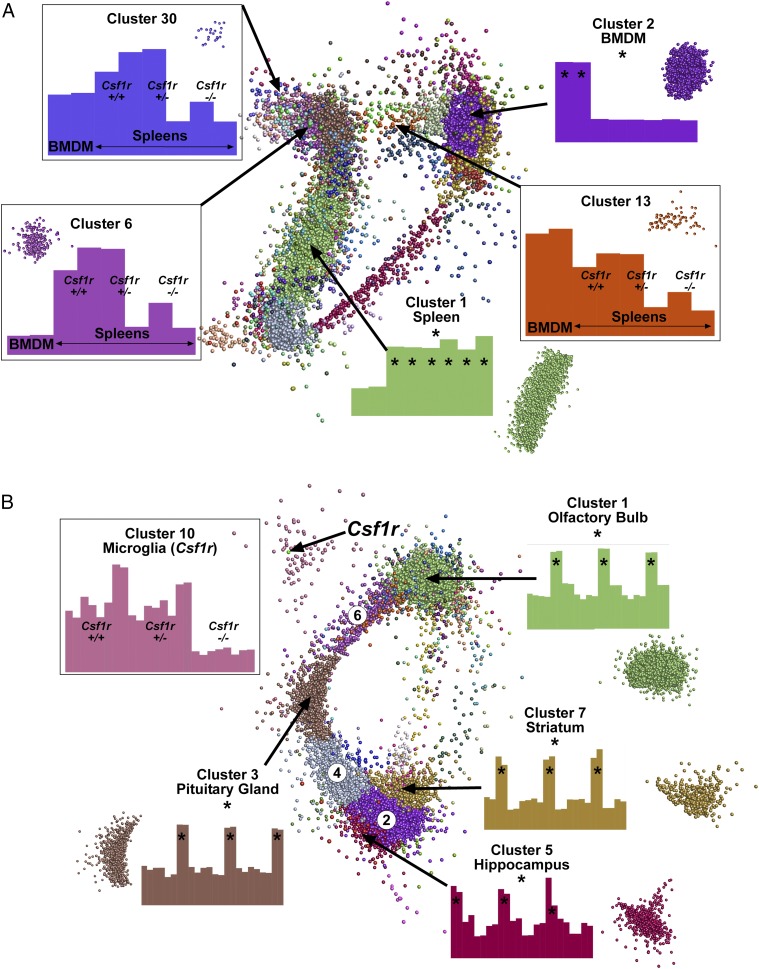
The network graph divided into two major aggregates, essentially comprising clusters of macrophage-enriched and spleen-enriched transcripts. Fig. 10A shows the clustered nodes for the main element in the layout graph.
FIGURE 10.
Network analysis of gene expression in the spleen and brains of Csf1r deficient rats. RMA-normalized microarray data from Supplemental Table I was analyzed with Graphia Pro. Edges have been removed for ease of visualization. Nodes allocated to the same cluster are the same color. Histograms show the averaged expression patterns of all genes in the cluster. Boxed clusters refer to genes affected by loss of Csf1r. Unboxed clusters refer to genes that are tissue specific (*). (A) Key clusters from spleen. All known genes in which no sample reached an intensity of 20 were excluded. Analysis was performed at a Pearson correlation coefficient ≥0.95 (12,305 nodes making 1,746,925 edges). Clustering was performed at an inflation of 2.0 with a minimum cluster size of 10. (B) Key clusters from brain. All known genes in which no sample reached an intensity of 20 were excluded. Analysis was performed at a Pearson correlation coefficient ≥0.85 (11,833 nodes making 3,617,804 edges). Clustering was performed at an inflation of 2.0 with a minimum cluster size of 10. Three clusters (circled numbers) shared gene expression with multiple brain regions: cluster 2 (striatum and hippocampus), cluster 4 (pituitary gland, striatum, and hippocampus), and cluster 6 (olfactory bulb and pituitary gland). Histograms for these clusters are shown in Supplemental Table I.
Five clusters of specific interest are among those listed in Supplemental Table I, together with their average expression profile. The largest cluster, cluster 1 (2171 genes; green nodes), was strongly enriched in the spleen compared with BMDM and not sensitive to Csf1r mutation. The second largest cluster (cluster 2, 1986 genes; purple nodes) was strongly enriched in the BMDM and also was not affected by the loss of Csf1r. This list is a complex mixture of genes associated with phagocyte biology and include Adgre1 (F4/80), Itgam (CD11b), components of the vacuolar ATPase (Atp6v family), Fcgr, and Clec families. The existence of this cluster indicates that there is a substantial residual macrophage population in the spleen of Csf1r−/− rats, and they share with BMDM the majority of the generic macrophage transcriptome. That conclusion is consistent with the residual CD68 staining in Fig. 6A and 6B. Also within cluster 2, there were more general anabolic/metabolism-associated genes involved in mitochondria (e.g., Nduf, Cox, Mrpl families) and RNA/protein synthesis. The enrichment of these genes in cluster 2 reflects the difference between a pure growing macrophage population and the splenic lymphoid population, in which most of the cells are relatively metabolically inactive.
Csf1r-dependent clusters varied in their level of spleen enrichment. The expression profile of cluster 6 (238 genes) was much higher in spleen than in BMDM and downregulated in the knockout and in the low Csf1r-expressing heterozygote. Cluster 30 (24 genes) had a similar expression profile to cluster 6, but with higher relative expression in BMDM. The spleen-specific cluster 6 contained the signature gene for the marginal zone macrophages (Siglec1) which was among the most downregulated with the loss of Csf1r (Supplemental Table I). The IL-22–binding protein, encoded by Il22ra2, has recently been studied in detail in Peyer patch, where it is highly expressed by myeloid cells underlying follicle-associated epithelium, regulates IL-22 signaling, and indirectly affects bacterial uptake (46). The loss of this transcript was confirmed by qRT-PCR (Fig. 6F).
The gene expression profile in the spleen of Csf1r-deficient rats is consistent with almost complete loss of the marginal zone macrophage populations. These cells, and the related Ag capture cells in the subcapsular sinus of lymph nodes, were also depleted in op/op mice (47). The data suggest that relatively few Csf1r-dependent genes are absolutely restricted to the marginal zone.
Genes that are less spleen specific but are nevertheless Csf1r dependent (e.g., those in cluster 13, including Sirpa) may be shared by macrophages of the marginal zone and red pulp, or specifically associated with red pulp macrophages. Overall, the main impact of Csf1r-deletion is to compromise spleen-specific macrophage differentiation.
Network analysis of gene expression in the brain of Csf1r-deficient rats
As with the spleen, the first pass analysis of gene expression data for two brain regions was to determine the expression ratios (ratio of Csf1r−/−/littermates) for each of the transcripts on the arrays in each of the tissues. There was no global change in expression of generic neuron-associated genes in Csf1r−/− rats shared by the brain regions examined, but there was some evidence of region-specific impacts (Supplemental Table I). There were several overlaps between the Csf1r-dependent genes in the brain regions and spleen, including Gpr31, Gpr34, Cd33, Tmem119, Tmem176a, P2ry13, Pld4, Clec4a1, and C1q genes, but most were not shared, consistent with the tissue-specific adaptation of Csf1r-dependent macrophages in the two locations. Fig. 10B shows the network graph for the combined analysis of the four brain regions; gene lists are provided in Supplemental Table I. The largest cluster was cluster 1 (4283 genes), containing olfactory bulb–associated genes. Other region-specific clusters were cluster 3 (pituitary gland; 878 genes), cluster 5 (hippocampus; 513 genes), and cluster 7 (striatum; 384 genes). The second largest cluster (cluster 2; 1253 genes) contained genes that were more highly expressed in both hippocampus and striatum. Two other main clusters also shared highly expressed genes between tissues: cluster 4 (pituitary gland, striatum and hippocampus; 757 genes) and cluster 6 (pituitary gland and olfactory bulb; 471 genes). None of these clusters showed any evidence of genotype association. Cluster 10 (107 genes) contained genes that were downregulated in the Csf1r−/− rats in all brain regions, including Csf1r itself (Fig. 10B). This set contains most known microglia-specific transcripts and is presumably attributed to the loss of microglia.
Discussion
The Csf1r knockout rats described in this article join a small set of rat knockout lines that have been produced via homologous recombination in ESC [(48) and reviewed in Ref. 13]. The comparison between rat and mouse mutant phenotypes supports the view that the rat, in many cases, provides a superior model for studying human genetic disease. For example, p53–deficient rats exhibited a more human-like tumor spectrum than mice (49), and dystrophin mutant rats displayed heart defects which were more similar to the human condition Duchenne muscular dystrophy (50, 51). The phenotype of Csf1r−/− rats is clearly very different from mice, and because of the improved viability, we have assayed phenotypes that could not be accessed in mice. In any insertional mutagenesis, or deletion, there is always the possibility of cis-acting impacts on neighboring genes. The two genes neighboring Csf1r, namely Hmgxb3 and Pdgrb, are both robustly expressed in all three tissues examined by gene expression profiling, and there was no significant change in their expression in response to insertional mutagenesis of Csf1r.
A common trait between Csf1r−/− rats and mice is increased bone density (osteopetrosis) attributed to either a reduction (mice) or complete absence (rats) of bone-resorbing osteoclasts. In humans, genetic susceptibility to disordered bone turnover in Paget disease has been linked to variation at the Csf1 locus (52). In Csf1tl/tl rats, there is a severe loss of osteoclasts in addition to the loss of osteoblasts (53). Unlike Csf1tl/tl rats (54), the Csf1r−/− rats developed a BM cavity, albeit reduced, and showed no evidence of extramedullary hematopoiesis in the spleen. Nevertheless, only monocytes were greatly reduced in the blood. We speculate that the loss of macrophages in liver and spleen increases the half-life of red cells, platelets, and neutrophils, compensating for reduced production by the BM. The receptor mutants also do not display the early onset deafness seen in Csf1tl/tl rats, attributed to auditory ossicle abnormalities (55). The loss of osteoclasts in Csf1r−/− rats is probably balanced in part by deficient calcification by osteoblasts. Osteoblasts on the bone surface interact intimately with a population of osteal macrophages, which are essential for intramembranous and endochondral ossification (36, 56). In the Csf1r−/− rat, there were no CD68+ osteal macrophages detectable on the bone surfaces (Fig. 7F). The difference between ligand and receptor mutations presumably reflects the role of the second ligand, IL-34. Both CSF1 and IL-34 (57) are produced by osteoblasts and contribute to osteoclastogenesis. It appears likely that IL-34 is essential for rat osteal macrophage development. Conversely, there is a lack of sperm development and fertility in male Csf1r−/− rats, whereas Csf1tl/tl male animals are fertile. This suggests that the alternative ligand, IL-34, has a specific function in testis development in the rat. Consistent with that hypothesis, Il34 mRNA is highly expressed in rat testis and downregulated by candidate male contraceptives (58).
Other phenotypes shared between Csf1r−/− rats and mice include postnatal growth retardation, lack of tooth eruption, reduced circulating monocyte numbers, defects in fertility, reduction in liver and peritoneal macrophages, and loss of Langerhans cells. The most obvious difference between Csf1r−/− rats and mice is the greatly increased viability, relative lack of a severe brain phenotype, and absence of effects on the sensory nervous system, insulin/pancreatic islet development, and gastrointestinal tract in the rats, as noted above. These species-specific differences, especially the latter two, probably contribute to the improved postweaning survival of the rat knockout compared with the mouse. The gut phenotype in mice has been attributed to Csf1r expression by Paneth cells, which appeared to be absent in mutant mice, and Csf1 signaling is proposed to indirectly control the differentiation of intestinal epithelial cells (59, 60). Treatment of mice with anti-CSF1R Ab completely depleted intestinal macrophage populations and lysozyme expression in the intestinal crypts and was associated with an increase in goblet cells (32). More recently, we demonstrated that Csf1r mRNA is not expressed in mouse Paneth cells, but macrophages intimately associated with the crypt control the differentiation of these cells and Lgr5+ stem cells. Whereas lysozyme expression was Csf1r dependent, other Paneth cell markers, including defensins, were not. In addition to increased goblet cell formation, anti-CSF1R treatment produced a complete loss of Ag-sampling M cells (61). In the rat Csf1r knockout, there was no effect on the cellularity of the lamina propria, no loss of Paneth cells, no increase in goblet cells, and no discernible change in villus architecture (Supplemental Fig. 2). In mice, the large intestinal macrophage population apparently depends upon continuous replenishment from circulating monocytes (62). In the Csf1r mutant rat, monocytes were greatly reduced, yet there was no apparent impact on the intestine.
In contrast, the almost complete loss of visceral fat in Csf1r−/− rats at an early age is a novel phenotype that has not previously been noted in mice. The Csf1op/op mouse is not deficient in adipose tissue but does have a reduction of macrophages in this tissue (63). Adipose tissue growth is related to growth hormone and IGF1 production (64). Hence, the loss of adipose tissue is most likely linked to the reduced GHR and IGF1 we observed in the liver and the consequent loss of circulating IGF1, as noted previously in Csf1tl/tl rats (45). A previous study also noted the loss of GHR expression in the bone of Csf1tl/tl rats (65).
Injection of Evans blue stain highlighted a deficiency in development of the peripheral vasculature in Csf1r−/− rats. Notably, the apparent leakiness in the spleen (Supplemental Fig. 3B) appears to be associated with the marginal zone and may be an indirect consequence of the loss of the marginal zone macrophages. The tl/tl CSF1 mutant rats display impaired capillary proliferation in the femur, which can be rescued with CSF1 treatment (66). However, the peripheral blood vessels have not been examined in these rats.
The gene expression analysis in Fig. 6C and 6D allowed us to identify spleen-specific genes that were downregulated with the loss of Csf1r. The large majority of genes enriched in BMDM relative to spleen were not affected. Transcripts associated with classical dendritic cell differentiation, such as Itgax, Zbtb46, Ly75, or Clec9a, were also unaffected, suggesting that these cells are not Csf1r dependent. Those genes that were both spleen enriched and Csf1r dependent provide a surrogate indicator of the loss of specific cell populations. The marginal zone macrophages of the mouse have resisted isolation and characterization (41). The commonly studied marker genes Siglec1 (marginal zone macrophages) and CD209b (marginal metallophils) (35) were almost ablated in Csf1r−/− rats, and coregulated genes are implicated, by association, in marginal zone macrophage differentiation and function (Supplemental Table I). Very few genes show the same level of Csf1r dependence. One example is the IL-22–binding protein, encoded by Il22ra2. This transcript is highly expressed by myeloid cells underlying mouse follicle-associated epithelium, regulates IL-22 signaling, and indirectly affects bacterial uptake (46).
The marginal zone macrophage population is also absent from Csf1op/op mice (47). Marginal zone macrophages in mice are distinguished from red pulp macrophages by expression of the widely used macrophage marker, F4/80, encoded by Adgre1 (previously Emr1) (67). Adgre1 is also highly expressed by rat macrophages (Supplemental Table I), and the relative loss of expression in the knockout spleen (∼70%) is consistent with expression in the red pulp macrophages. The related gene, Adgre4 (previously Emr4), was strongly spleen enriched and highly Csf1r dependent (Fig. 6F). ADGRE4 was shown to mediate binding to B lymphocytes (68), supporting the hypothesis that it may be a novel functional marginal zone marker.
The set of candidate marginal zone–enriched transcripts includes the transcription factor Nr1h3 (aka Lxrα) (Fig. 6F), which is essential for both marginal zone macrophage populations in mouse (69). Other transcription factors coregulated with Lxrα (Supplemental Table I, cluster 6) included Nfe2, Nr1d1, Rara, Klf2, and Tcf21.
One report on Nr1d1 in macrophages confirms the enriched expression in mouse splenic macrophages and a circadian oscillation (70). Regulated expression of the retinoic acid receptor, Rara, has also been associated with myeloid differentiation (71), and treatment of mice with retinoids led to an expansion of the marginal zone in spleen (72). Tcf21 is required during development for the formation of a spleen (73). This Csf1r-dependent cluster also contained multiple growth factors of the TGFB (Tgfb2, Tgfb3) and BMP (Bmp2, 3, 4, 6) families, which could contribute to the differentiated phenotype of splenic macrophages (74, 75). Expression of the transcription factors Tfec and Tcf7l2 was downregulated with the loss of Csf1r (Supplemental Table I). Tfec is a member of the MITF transcription factor family and is known to be macrophage enriched and PU.1 dependent (76). Analysis of the Tfec−/− mouse suggested a role in IL-4–inducible expression of genes, including Csf3r (77), which was also downregulated in the Csf1r−/− spleen. Our finding suggests that Tfec could have a specific function in splenic macrophages. Tcf7l2 is associated with differentiation of plasmacytoid dendritic cells (78). Plasmacytoid dendritic cells express Csf1r and were found to be 70% reduced in the spleen of op/op (CSF1-deficient) mice (79). The alteration of the gene expression profile in the spleen of Csf1r-deficient rats is consistent with the immunohistochemistry (IHC) data and indicates almost complete loss of the marginal zone macrophage populations. These cells, and the related Ag capture cells in the subcapsular sinus of lymph nodes, were also depleted in op/op mice (47). Genes that were less spleen specific, but are nevertheless Csf1r dependent (e.g., those in cluster 13 and 30), may be shared by macrophages of the marginal zone and red pulp or specifically associated with red pulp macrophages.
Csf1r−/− rats had no microglia detected by staining for IBA1 in the brain and retina or in flow cytometry profiles of brain digests (Figs. 8D–G, 9A). This observation was strongly supported by the network analysis of gene expression in the four brain regions shown in Fig. 10B. Cluster 10 (Supplemental Table I) provides a list of 107 rat microglia-enriched genes that have been previously published as expressed in microglia from mouse (21, 39, 40, 80, 81) and human (81, 82). Cluster 10 also includes 18 of the top 35 genes downmodulated in the brains of mice treated with a CSF1R kinase inhibitor to deplete microglia (83). As expected, Csf1r was almost undetectable in the homozygotes and was the only transcript that was also reproducibly reduced, by 40–50%, in the heterozygous mutants, in all brain regions. Expression of Cx3cr1, as well as cathepsin S (Ctss), the enzyme required for CX3CL1 cleavage (84), was also almost completely ablated, to the same extent as Csf1r. Microglia-associated transcripts are of particular interest given the emerging consensus that these cells are central players in the pathologic condition of Alzheimer disease. Genes including Csf1r and others within cluster 10, including Trem2, C1q, Tyrobp, Abi3, and Spi1, are associated with disease susceptibility (85–87).
Many known macrophage-associated transcripts were less affected by Csf1r mutation, including Cd14, Csf2ra, Mertk, Fcgr2a (CD32), Stab1, Msr1, Marco, Gpr84, Icam1, Clec7a, and several TLR (Tlr2,4,8,9,11) (average 25% less expression; Supplemental Table I). Among macrophage-expressed transcription factors, Spi1 (PU.1) was reduced by around 75%, but Irf8, Cebpa, Cebpb, Runx1, Tfec, and Stat5A, all of which were readily detectable and are expressed by microglia in the mouse (39, 41), were reduced by <37%. One interpretation is that these transcripts are more highly expressed in the nonmicroglial macrophage population, which is also less Csf1r dependent. The CD45hi brain macrophage population in rats were isolated and characterized by Ford et al. (88). They were larger than microglia, with a lower nucleus/cytoplasm ratio. A subset expressed MHC-II; notably, aside from RT1-DMb, the expression of MHC-II genes (RT1-DMa, RT1-Da, RT1-Db1, RT-1DB2, RT1-DOb) was not affected by the Csf1r mutation. There was no detectable reduction in lysosome/endosome-associated genes (e.g., Gpnmb, Ctsb, Lipa, Tcirg1) that in mice are very strongly enriched in macrophages and microglia compared with total brain (see www.biogps.org). An alternative interpretation is that the loss of microglial endocytic activity is compensated by increased activity in other nonmacrophage cells.
Few impacts on overall brain architecture are shared with the Csf1r knockout mouse (27). There was little overt phenotype and specifically no impact on the olfactory bulb, where the mouse mutation had the greatest effect (Fig. 8B). A recent study described a specific microglial population associated with myelination in the brain in the postnatal period (89). These cells were proposed to be the major source of IGF1 required for myelination. The loss of these cells could explain the minor deficiency of myelination and myelination-associated transcripts (Figs. 8C, 9B) observed in the mutant rats. However, in the gene expression profiles of rat brain regions, there was no deficiency in Igf1 mRNA associated with the Csf1r mutation. Similarly, microglia have been considered an important source of brain-derived neutrotrophic factor (BDNF), although conditional microglial depletion did not reduce the total level of Bdnf mRNA in the mouse cortex or hippocampus (90). Bdnf mRNA was not reduced in the Csf1r mutant rat brain. Presumably, the other cellular sources of these trophic factors can compensate for the absence of microglia. It remains to be seen whether the loss of microglia produces the alterations in synaptic proteins and synaptic plasticity observed in mouse microglia depletion studies (90, 91). Microglia in mice have been associated with the outgrowth of dopaminergic neurons, and their depletion produced an imbalance in dopaminergic innervation of the striatum (92). In the striatum of Csf1r-deficient rats (Supplemental Table I), we observed a 30–40% increase in mRNA encoding dopamine receptors (Drd1, 2, and 3), the dopamine transporter Slc41a3, and other genes correlated with dopamine receptor expression across brain regions, Gnal, Dgkb, Pcp4l1, Kcna, and Tac1 (93). More subtle brain phenotypes may emerge from further investigation. The advantage of the rat model is that these phenotypes can be studied in adult animals in the absence of gross changes in brain architecture. The flow cytometry analysis and gene expression profiling indicate that there is no compensatory monocyte recruitment or inflammatory cytokine production in the Csf1r−/− brain, consistent with the absence of circulating monocytes. Monocyte recruitment is the hallmark of brain pathologic conditions (38, 94) and has been observed in other models of microglial deficiency (40). We suggest that the impact of the loss of the neuroprotective functions of microglia in the Csf1r−/− rat may be mitigated by the absence of monocytes.
The human disease ALSP [formerly known as hereditary diffuse leukoencephalopathy with spheroids (95)] is associated with mutations affecting conserved amino acids in the kinase domain of CSF1R (96), which are likely to have a dominant-negative impact on signaling by forming inactive heterodimers with the wild type receptor (8). Fifty-eight different mutations have been described in ALSP patients (7), and within the human exome database (exac.broadinstitute.org), there are a further 38 variants, each identified only as a singleton, that affect amino acids conserved from humans to chickens. One report claimed that the disease could be associated with haploinsufficiency arising from a frame-shift mutation (7), but their data indicate >50% loss of CSF1R protein in the brains of these patients. Chitu and colleagues (97) have characterized Csf1r+/− inbred mice as a model for ALSP. Like these authors, we found that there is no dosage compensation for loss of one Csf1r allele, which is surprising because CSF1 signaling regulates Csf1r mRNA levels (98). However, despite the lack of dosage compensation, we have found no significant impact of the Csf1r heterozygous mutation on gene expression in the adult brain (Supplemental Table I). We have maintained heterozygous mutant rats for more than 12 months and observed no overt phenotype. Accordingly, the impact of dominant-acting mutations in patients is likely to depend in part upon the level of expression of the other allele and on the activity of the compensatory mechanisms that permit relatively normal brain function when microglial differentiation is compromised.
In overview, analysis of Csf1r−/− rats highlights selective impacts of the mutation on macrophage populations and clear differences in the cell populations affected and the pleiotropic consequences compared with mutant mice. The effect of Csf1 mutation in mice on macrophages and osteoclast populations is gradually corrected with age. The macrophage phenotype can be corrected with exogenous GM-CSF (CSF2), but endogenous CSF2 is not required (99). Conversely, mutation of Flt1 prevents the age-dependent development of osteoclasts (100). As noted above, one major difference between rats and mice is that rat macrophages themselves express high levels of Csf1 mRNA. This explains the loss of Csf1 mRNA in the spleen of Csf1r mutant rats. In the brain of rats, Csf1 and Il34 were both highly expressed and not notably region specific. The Csf1r mutation did not reduce the level of expression, suggesting that microglia are not the major source. We suggest that differences between the rodent species derive from differences in expression or function of alternative growth factors that can compensate for the lack of Csf1r. Although female Csf1r mutant rats appear infertile, hormone-dependent expression of CSF2 and other factors by the uterus (101) might also mitigate the impact of the Csf1r mutation on macrophage numbers and function in females relative to males.
The increased longevity and plethora of phenotypes found in Csf1r−/− rats provide a unique model for studying the role of CSF1R during development and adulthood, the functions of microglia, and other tissue macrophage populations and the molecular basis for adult onset disease associated with human CSF1R mutations. Many of the macrophage phenotypes are shared between mice and rats, so the differences mainly lie in the relative redundancy of macrophages in developmental processes in the two species.
Supplementary Material
Acknowledgments
We thank the assistance provided by animal technicians at the Roslin Institute, particularly Christine Marshall. We thank Prof. V. Hugh Perry for providing feedback prior to submission.
This work was supported by Medical Research Council Grant MR/M019969/1.
The sequences presented in this article have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE100696.
The online version of this article contains supplemental material.
- ALSP
- adult-onset leukoencephalopathy with axonal spheroids and pigmented glia
- BM
- bone marrow
- BMDM
- BM-derived macrophage
- μCT
- micro–computed tomography
- ESC
- embryonic stem cell
- GHR
- growth hormone receptor
- IGF1
- insulin-like growth factor 1
- MCL
- Markov clustering algorithm
- MHC-II
- MHC class II
- qRT-PCR
- quantitative real-time PCR
- RT
- room temperature
- tl/tl
- toothless
- TRAP
- tartrate-resistant acid phosphatase.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Hoeffel G., Ginhoux F. 2015. Ontogeny of tissue-resident macrophages. Front. Immunol. 6: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okabe Y., Medzhitov R. 2016. Tissue biology perspective on macrophages. Nat. Immunol. 17: 9–17. [DOI] [PubMed] [Google Scholar]
- 3.Garceau V., Smith J., Paton I. R., Davey M., Fares M. A., Sester D. P., Burt D. W., Hume D. A. 2010. Pivotal advance: avian colony-stimulating factor 1 (CSF-1), interleukin-34 (IL-34), and CSF-1 receptor genes and gene products. J. Leukoc. Biol. 87: 753–764. [DOI] [PubMed] [Google Scholar]
- 4.Sasmono R. T., Oceandy D., Pollard J. W., Tong W., Pavli P., Wainwright B. J., Ostrowski M. C., Himes S. R., Hume D. A. 2003. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood 101: 1155–1163. [DOI] [PubMed] [Google Scholar]
- 5.Balic A., Garcia-Morales C., Vervelde L., Gilhooley H., Sherman A., Garceau V., Gutowska M. W., Burt D. W., Kaiser P., Hume D. A., Sang H. M. 2014. Visualisation of chicken macrophages using transgenic reporter genes: insights into the development of the avian macrophage lineage. Development 141: 3255–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pridans C., Davis G. M., Sauter K. A., Lisowski Z. M., Corripio-Miyar Y., Raper A., Lefevre L., Young R., McCulloch M. E., Lillico S., et al. 2016. A Csf1r-EGFP transgene provides a novel marker for monocyte subsets in sheep. J. Immunol. 197: 2297–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konno T., Yoshida K., Mizuno T., Kawarai T., Tada M., Nozaki H., Ikeda S. I., Nishizawa M., Onodera O., Wszolek Z. K., Ikeuchi T. 2017. Clinical and genetic characterization of adult-onset leukoencephalopathy with axonal spheroids and pigmented glia associated with CSF1R mutation. Eur. J. Neurol. 24: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pridans C., Sauter K. A., Baer K., Kissel H., Hume D. A. 2013. CSF1R mutations in hereditary diffuse leukoencephalopathy with spheroids are loss of function. Sci. Rep. 3: 3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai X. M., Ryan G. R., Hapel A. J., Dominguez M. G., Russell R. G., Kapp S., Sylvestre V., Stanley E. R. 2002. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99: 111–120. [DOI] [PubMed] [Google Scholar]
- 10.Wynn T. A., Chawla A., Pollard J. W. 2013. Macrophage biology in development, homeostasis and disease. Nature 496: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenkins S. J., Hume D. A. 2014. Homeostasis in the mononuclear phagocyte system. Trends Immunol. 35: 358–367. [DOI] [PubMed] [Google Scholar]
- 12.Li J., Chen K., Zhu L., Pollard J. W. 2006. Conditional deletion of the colony stimulating factor-1 receptor (c-fms proto-oncogene) in mice. Genesis 44: 328–335. [DOI] [PubMed] [Google Scholar]
- 13.Meek S., Mashimo T., Burdon T. 2017. From engineering to editing the rat genome. Mamm. Genome 28: 302–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homberg J. R., Wöhr M., Alenina N. 2017. Comeback of the rat in biomedical research. ACS Chem. Neurosci. 8: 900–903. [DOI] [PubMed] [Google Scholar]
- 15.Iannaccone P. M., Jacob H. J. 2009. Rats! Dis. Model. Mech. 2: 206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang G., Tong C., Kumbhani D. S., Ashton C., Yan H., Ying Q. L. 2011. Beyond knockout rats: new insights into finer genome manipulation in rats. Cell Cycle 10: 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Wesenbeeck L., Odgren P. R., MacKay C. A., D’Angelo M., Safadi F. F., Popoff S. N., Van Hul W., Marks S. C., Jr 2002. The osteopetrotic mutation toothless (tl) is a loss-of-function frameshift mutation in the rat Csf1 gene: evidence of a crucial role for CSF-1 in osteoclastogenesis and endochondral ossification. Proc. Natl. Acad. Sci. USA 99: 14303–14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blair K., Leitch H. G., Mansfield W., Dumeau C. E., Humphreys P., Smith A. G. 2012. Culture parameters for stable expansion, genetic modification and germline transmission of rat pluripotent stem cells. Biol. Open 1: 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong C., Li P., Wu N. L., Yan Y., Ying Q. L. 2010. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature 467: 211–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauter K. A., Waddell L. A., Lisowski Z. M., Young R., Lefevre L., Davis G. M., Clohisey S. M., McCulloch M., Magowan E., Mabbott N. A., et al. 2016. Macrophage colony-stimulating factor (CSF1) controls monocyte production and maturation and the steady-state size of the liver in pigs. Am. J. Physiol. Gastrointest. Liver Physiol. 311: G533–G547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabert K., Michoel T., Karavolos M. H., Clohisey S., Baillie J. K., Stevens M. P., Freeman T. C., Summers K. M., McColl B. W. 2016. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 19: 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasmono R. T., Williams E. 2012. Generation and characterization of MacGreen mice, the Cfs1r-EGFP transgenic mice. Methods Mol. Biol. 844: 157–176. [DOI] [PubMed] [Google Scholar]
- 23.Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 24.Cotton W. R., Gaines J. F. 1974. Unerupted dentition secondary to congenital osteopetrosis in the Osborne-Mendel rat. Proc. Soc. Exp. Biol. Med. 146: 554–561. [DOI] [PubMed] [Google Scholar]
- 25.Gow D. J., Sester D. P., Hume D. A. 2010. CSF-1, IGF-1, and the control of postnatal growth and development. J. Leukoc. Biol. 88: 475–481. [DOI] [PubMed] [Google Scholar]
- 26.Dupré G., Heidenreich D. 2016. Brachycephalic syndrome. Vet. Clin. North Am. Small Anim. Pract. 46: 691–707. [DOI] [PubMed] [Google Scholar]
- 27.Erblich B., Zhu L., Etgen A. M., Dobrenis K., Pollard J. W. 2011. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One 6: e26317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huynh D., Dai X. M., Nandi S., Lightowler S., Trivett M., Chan C. K., Bertoncello I., Ramsay R. G., Stanley E. R. 2009. Colony stimulating factor-1 dependence of paneth cell development in the mouse small intestine. Gastroenterology 137: 136–144, 144.e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiede M. A., Smock S. L., Mason-Savas A., MacKay C. A., Odgren P. R., Marks S. C., Jr 1996. Thrombocytopenia in the toothless (osteopetrotic) rat and its rescue by treatment with colony-stimulating factor-1. Exp. Hematol. 24: 722–727. [PubMed] [Google Scholar]
- 30.Creasy D., Bube A., de Rijk E., Kandori H., Kuwahara M., Masson R., Nolte T., Reams R., Regan K., Rehm S., et al. 2012. Proliferative and nonproliferative lesions of the rat and mouse male reproductive system. Toxicol. Pathol. 40(6 Suppl.): 40S–121S. [DOI] [PubMed] [Google Scholar]
- 31.Damoiseaux J. G., Döpp E. A., Calame W., Chao D., MacPherson G. G., Dijkstra C. D. 1994. Rat macrophage lysosomal membrane antigen recognized by monoclonal antibody ED1. Immunology 83: 140–147. [PMC free article] [PubMed] [Google Scholar]
- 32.Sauter K. A., Pridans C., Sehgal A., Tsai Y. T., Bradford B. M., Raza S., Moffat L., Gow D. J., Beard P. M., Mabbott N. A., et al. 2014. Pleiotropic effects of extended blockade of CSF1R signaling in adult mice. J. Leukoc. Biol. 96: 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Care A. S., Diener K. R., Jasper M. J., Brown H. M., Ingman W. V., Robertson S. A. 2013. Macrophages regulate corpus luteum development during embryo implantation in mice. J. Clin. Invest. 123: 3472–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris S. E., MacDougall M., Horn D., Woodruff K., Zimmer S. N., Rebel V. I., Fajardo R., Feng J. Q., Gluhak-Heinrich J., Harris M. A., Abboud Werner S. 2012. Meox2Cre-mediated disruption of CSF-1 leads to osteopetrosis and osteocyte defects. Bone 50: 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies L. C., Jenkins S. J., Allen J. E., Taylor P. R. 2013. Tissue-resident macrophages. Nat. Immunol. 14: 986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang M. K., Raggatt L. J., Alexander K. A., Kuliwaba J. S., Fazzalari N. L., Schroder K., Maylin E. R., Ripoll V. M., Hume D. A., Pettit A. R. 2008. Osteal tissue macrophages are intercalated throughout human and mouse bone lining tissues and regulate osteoblast function in vitro and in vivo. J. Immunol. 181: 1232–1244. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Szretter K. J., Vermi W., Gilfillan S., Rossini C., Cella M., Barrow A. D., Diamond M. S., Colonna M. 2012. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 13: 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prinz M., Erny D., Hagemeyer N. 2017. Ontogeny and homeostasis of CNS myeloid cells. Nat. Immunol. 18: 385–392. [DOI] [PubMed] [Google Scholar]
- 39.Hickman S. E., Kingery N. D., Ohsumi T. K., Borowsky M. L., Wang L. C., Means T. K., El Khoury J. 2013. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16: 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butovsky O., Jedrychowski M. P., Moore C. S., Cialic R., Lanser A. J., Gabriely G., Koeglsperger T., Dake B., Wu P. M., Doykan C. E., et al. 2014. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. [Published erratum appears in 2014 Nat. Neurosci. 17: 1286.] Nat. Neurosci. 17: 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gautier E. L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K. G., Gordonov S., et al. Immunological Genome Consortium 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13: 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bohlen C. J., Bennett F. C., Tucker A. F., Collins H. Y., Mulinyawe S. B., Barres B. A. 2017. Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron 94: 759–773.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabriek B. O., Van Haastert E. S., Galea I., Polfliet M. M., Döpp E. D., Van Den Heuvel M. M., Van Den Berg T. K., De Groot C. J., Van Der Valk P., Dijkstra C. D. 2005. CD163-positive perivascular macrophages in the human CNS express molecules for antigen recognition and presentation. Glia 51: 297–305. [DOI] [PubMed] [Google Scholar]
- 44.Pollard J. W. 2009. Trophic macrophages in development and disease. Nat. Rev. Immunol. 9: 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gow D. J., Sauter K. A., Pridans C., Moffat L., Sehgal A., Stutchfield B. M., Raza S., Beard P. M., Tsai Y. T., Bainbridge G., et al. 2014. Characterisation of a novel Fc conjugate of macrophage colony-stimulating factor. Mol. Ther. 22: 1580–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jinnohara T., Kanaya T., Hase K., Sakakibara S., Kato T., Tachibana N., Sasaki T., Hashimoto Y., Sato T., Watarai H., et al. 2017. IL-22BP dictates characteristics of Peyer’s patch follicle-associated epithelium for antigen uptake. J. Exp. Med. 214: 1607–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witmer-Pack M. D., Hughes D. A., Schuler G., Lawson L., McWilliam A., Inaba K., Steinman R. M., Gordon S. 1993. Identification of macrophages and dendritic cells in the osteopetrotic (op/op) mouse. J. Cell Sci. 104: 1021–1029. [DOI] [PubMed] [Google Scholar]
- 48.Lan H., Li S., Guo Z., Men H., Wu Y., Li N., Bryda E. C., Capecchi M. R., Wu S. 2016. Efficient generation of selection-gene-free rat knockout models by homologous recombination in ES cells. FEBS Lett. 590: 3416–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan H. X., Wu H. P., Ashton C., Tong C., Ying Q. L. 2012. Rats deficient for p53 are susceptible to spontaneous and carcinogen-induced tumorigenesis. Carcinogenesis 33: 2001–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura K., Fujii W., Tsuboi M., Tanihata J., Teramoto N., Takeuchi S., Naito K., Yamanouchi K., Nishihara M. 2014. Generation of muscular dystrophy model rats with a CRISPR/Cas system. Sci. Rep. 4: 5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larcher T., Lafoux A., Tesson L., Remy S., Thepenier V., François V., Le Guiner C., Goubin H., Dutilleul M., Guigand L., et al. 2014. Characterization of dystrophin deficient rats: a new model for Duchenne muscular dystrophy. PLoS One 9: e110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albagha O. M., Visconti M. R., Alonso N., Langston A. L., Cundy T., Dargie R., Dunlop M. G., Fraser W. D., Hooper M. J., Isaia G., et al. 2010. Genome-wide association study identifies variants at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget’s disease of bone. Nat. Genet. 42: 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shalhoub V., Jackson M. E., Lian J. B., Stein G. S., Marks S. C., Jr 1991. Gene expression during skeletal development in three osteopetrotic rat mutations. Evidence for osteoblast abnormalities. J. Biol. Chem. 266: 9847–9856. [PubMed] [Google Scholar]
- 54.Marks S. C., Jr., Wojtowicz A., Szperl M., Urbanowska E., MacKay C. A., Wiktor-Jedrzejczak W., Stanley E. R., Aukerman S. L. 1992. Administration of colony stimulating factor-1 corrects some macrophage, dental, and skeletal defects in an osteopetrotic mutation (toothless, tl) in the rat. Bone 13: 89–93. [DOI] [PubMed] [Google Scholar]
- 55.Aharinejad S., Grossschmidt K., Franz P., Streicher J., Nourani F., MacKay C. A., Firbas W., Plenk H., Jr., Marks S. C., Jr 1999. Auditory ossicle abnormalities and hearing loss in the toothless (osteopetrotic) mutation in the rat and their improvement after treatment with colony-stimulating factor-1. J. Bone Miner. Res. 14: 415–423. [DOI] [PubMed] [Google Scholar]
- 56.Batoon L., Millard S. M., Wullschleger M. E., Preda C., Wu A. C., Kaur S., Tseng H. W., Hume D. A., Levesque J. P., Raggatt L. J., Pettit A. R. 2017. CD169(+) macrophages are critical for osteoblast maintenance and promote intramembranous and endochondral ossification during bone repair. Biomaterials. DOI: 10.1016/j.biomaterials.2017.10.033. [DOI] [PubMed]
- 57.Chen Z., Buki K., Vääräniemi J., Gu G., Väänänen H. K. 2011. The critical role of IL-34 in osteoclastogenesis. PLoS One 6: e18689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tash J. S., Attardi B., Hild S. A., Chakrasali R., Jakkaraj S. R., Georg G. I. 2008. A novel potent indazole carboxylic acid derivative blocks spermatogenesis and is contraceptive in rats after a single oral dose. Biol. Reprod. 78: 1127–1138. [DOI] [PubMed] [Google Scholar]
- 59.Akcora D., Huynh D., Lightowler S., Germann M., Robine S., de May J. R., Pollard J. W., Stanley E. R., Malaterre J., Ramsay R. G. 2013. The CSF-1 receptor fashions the intestinal stem cell niche. Stem Cell Res. 10: 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huynh D., Akçora D., Malaterre J., Chan C. K., Dai X. M., Bertoncello I., Stanley E. R., Ramsay R. G. 2013. CSF-1 receptor-dependent colon development, homeostasis and inflammatory stress response. PLoS One 8: e56951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sehgal A., Donaldson D. S., Pridans C., Sauter K. A., Hume D. A., Mabbott N. A. 2018. The role of CSF1R-dependent macrophages in control of the intestinal stem-cell niche. Nat. Commun. 9: 1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bain C. C., Bravo-Blas A., Scott C. L., Perdiguero E. G., Geissmann F., Henri S., Malissen B., Osborne L. C., Artis D., Mowat A. M. 2014. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15: 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weisberg S. P., McCann D., Desai M., Rosenbaum M., Leibel R. L., Ferrante A. W., Jr 2003. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 112: 1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lecomte V., Maloney C. A., Wang K. W., Morris M. J. 2017. Effects of paternal obesity on growth and adiposity of male rat offspring. Am. J. Physiol. Endocrinol. Metab. 312: E117–E125. [DOI] [PubMed] [Google Scholar]
- 65.Symons A. L., MacKay C. A., Leong K., Hume D. A., Waters M. J., Marks S. C., Jr 1996. Decreased growth hormone receptor expression in long bones from toothless (osteopetrotic) rats and restoration by treatment with colony-stimulating factor-1. Growth Factors 13: 1–10. [DOI] [PubMed] [Google Scholar]
- 66.Aharinejad S., Marks S. C., Jr., Böck P., Mason-Savas A., MacKay C. A., Larson E. K., Jackson M. E., Luftensteiner M., Wiesbauer E. 1995. CSF-1 treatment promotes angiogenesis in the metaphysis of osteopetrotic (toothless, tl) rats. Bone 16: 315–324. [DOI] [PubMed] [Google Scholar]
- 67.Saunderson S. C., Dunn A. C., Crocker P. R., McLellan A. D. 2014. CD169 mediates the capture of exosomes in spleen and lymph node. Blood 123: 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stacey M., Chang G. W., Sanos S. L., Chittenden L. R., Stubbs L., Gordon S., Lin H. H. 2002. EMR4, a novel epidermal growth factor (EGF)-TM7 molecule up-regulated in activated mouse macrophages, binds to a putative cellular ligand on B lymphoma cell line A20. J. Biol. Chem. 277: 29283–29293. [DOI] [PubMed] [Google Scholar]
- 69.A-Gonzalez N., Guillen J. A., Gallardo G., Diaz M., de la Rosa J. V., Hernandez I. H., Casanova-Acebes M., Lopez F., Tabraue C., Beceiro S., et al. 2013. The nuclear receptor LXRα controls the functional specialization of splenic macrophages. Nat. Immunol. 14: 831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silver A. C., Arjona A., Hughes M. E., Nitabach M. N., Fikrig E. 2012. Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav. Immun. 26: 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu J., Heyworth C. M., Glasow A., Huang Q. H., Petrie K., Lanotte M., Benoit G., Gallagher R., Waxman S., Enver T., Zelent A. 2001. Lineage restriction of the RARalpha gene expression in myeloid differentiation. Blood 98: 2563–2567. [DOI] [PubMed] [Google Scholar]
- 72.Katz D. R., Drzymala M., Turton J. A., Hicks R. M., Hunt R., Palmer L., Malkovský M. 1987. Regulation of accessory cell function by retinoids in murine immune responses. Br. J. Exp. Pathol. 68: 343–350. [PMC free article] [PubMed] [Google Scholar]
- 73.Lu J., Chang P., Richardson J. A., Gan L., Weiler H., Olson E. N. 2000. The basic helix-loop-helix transcription factor capsulin controls spleen organogenesis. Proc. Natl. Acad. Sci. USA 97: 9525–9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prabagar M. G., Do Y., Ryu S., Park J. Y., Choi H. J., Choi W. S., Yun T. J., Moon J., Choi I. S., Ko K., et al. 2013. SIGN-R1, a C-type lectin, enhances apoptotic cell clearance through the complement deposition pathway by interacting with C1q in the spleen. Cell Death Differ. 20: 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ravishankar B., Shinde R., Liu H., Chaudhary K., Bradley J., Lemos H. P., Chandler P., Tanaka M., Munn D. H., Mellor A. L., McGaha T. L. 2014. Marginal zone CD169+ macrophages coordinate apoptotic cell-driven cellular recruitment and tolerance. Proc. Natl. Acad. Sci. USA 111: 4215–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rehli M., Lichanska A., Cassady A. I., Ostrowski M. C., Hume D. A. 1999. TFEC is a macrophage-restricted member of the microphthalmia-TFE subfamily of basic helix-loop-helix leucine zipper transcription factors. J. Immunol. 162: 1559–1565. [PubMed] [Google Scholar]
- 77.Rehli M., Sulzbacher S., Pape S., Ravasi T., Wells C. A., Heinz S., Söllner L., El Chartouni C., Krause S. W., Steingrimsson E., et al. 2005. Transcription factor Tfec contributes to the IL-4-inducible expression of a small group of genes in mouse macrophages including the granulocyte colony-stimulating factor receptor. J. Immunol. 174: 7111–7122. [DOI] [PubMed] [Google Scholar]
- 78.Cisse B., Caton M. L., Lehner M., Maeda T., Scheu S., Locksley R., Holmberg D., Zweier C., den Hollander N. S., Kant S. G., et al. 2008. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.MacDonald K. P., Rowe V., Bofinger H. M., Thomas R., Sasmono T., Hume D. A., Hill G. R. 2005. The colony-stimulating factor 1 receptor is expressed on dendritic cells during differentiation and regulates their expansion. J. Immunol. 175: 1399–1405. [DOI] [PubMed] [Google Scholar]
- 80.Orre M., Kamphuis W., Osborn L. M., Melief J., Kooijman L., Huitinga I., Klooster J., Bossers K., Hol E. M. 2014. Acute isolation and transcriptome characterization of cortical astrocytes and microglia from young and aged mice. Neurobiol. Aging 35: 1–14. [DOI] [PubMed] [Google Scholar]
- 81.Gosselin D., Skola D., Coufal N. G., Holtman I. R., Schlachetzki J. C. M., Sajti E., Jaeger B. N., O’Connor C., Fitzpatrick C., Pasillas M. P., et al. 2017. An environment-dependent transcriptional network specifies human microglia identity. Science. DOI: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Galatro T. F., Holtman I. R., Lerario A. M., Vainchtein I. D., Brouwer N., Sola P. R., Veras M. M., Pereira T. F., Leite R. E. P., Möller T., et al. 2017. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 20: 1162–1171. [DOI] [PubMed] [Google Scholar]
- 83.Elmore M. R., Najafi A. R., Koike M. A., Dagher N. N., Spangenberg E. E., Rice R. A., Kitazawa M., Matusow B., Nguyen H., West B. L., Green K. N. 2014. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82: 380–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seo Y., Kim H. S., Kang I., Choi S. W., Shin T. H., Shin J. H., Lee B. C., Lee J. Y., Kim J. J., Kook M. G., Kang K. S. 2016. Cathepsin S contributes to microglia-mediated olfactory dysfunction through the regulation of Cx3cl1-Cx3cr1 axis in a Niemann-Pick disease type C1 model. Glia 64: 2291–2305. [DOI] [PubMed] [Google Scholar]
- 85.Salter M. W., Stevens B. 2017. Microglia emerge as central players in brain disease. Nat. Med. 23: 1018–1027. [DOI] [PubMed] [Google Scholar]
- 86.Sarlus H., Heneka M. T. 2017. Microglia in Alzheimer’s disease. J. Clin. Invest. 127: 3240–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Efthymiou A. G., Goate A. M. 2017. Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol. Neurodegener. 12: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ford A. L., Goodsall A. L., Hickey W. F., Sedgwick J. D. 1995. Normal adult ramified microglia separated from other central nervous system macrophages by flow cytometric sorting. Phenotypic differences defined and direct ex vivo antigen presentation to myelin basic protein-reactive CD4+ T cells compared. J. Immunol. 154: 4309–4321. [PubMed] [Google Scholar]
- 89.Wlodarczyk A., Holtman I. R., Krueger M., Yogev N., Bruttger J., Khorooshi R., Benmamar-Badel A., de Boer-Bergsma J. J., Martin N. A., Karram K., et al. 2017. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 36: 3292–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parkhurst C. N., Yang G., Ninan I., Savas J. N., Yates J. R., III, Lafaille J. J., Hempstead B. L., Littman D. R., Gan W. B. 2013. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155: 1596–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Werneburg S., Feinberg P. A., Johnson K. M., Schafer D. P. 2017. A microglia-cytokine axis to modulate synaptic connectivity and function. Curr. Opin. Neurobiol. 47: 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Squarzoni P., Oller G., Hoeffel G., Pont-Lezica L., Rostaing P., Low D., Bessis A., Ginhoux F., Garel S. 2014. Microglia modulate wiring of the embryonic forebrain. Cell Rep. 8: 1271–1279. [DOI] [PubMed] [Google Scholar]
- 93.Walker J. R., Su A. I., Self D. W., Hogenesch J. B., Lapp H., Maier R., Hoyer D., Bilbe G. 2004. Applications of a rat multiple tissue gene expression data set. Genome Res. 14: 742–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baufeld C., O’Loughlin E., Calcagno N., Madore C., Butovsky O. 2018. Differential contribution of microglia and monocytes in neurodegenerative diseases. J. Neural Transm. (Vienna). 125: 809–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nicholson A. M., Baker M. C., Finch N. A., Rutherford N. J., Wider C., Graff-Radford N. R., Nelson P. T., Clark H. B., Wszolek Z. K., Dickson D. W., et al. 2013. CSF1R mutations link POLD and HDLS as a single disease entity. Neurology 80: 1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rademakers R., Baker M., Nicholson A. M., Rutherford N. J., Finch N., Soto-Ortolaza A., Lash J., Wider C., Wojtas A., DeJesus-Hernandez M., et al. 2011. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat. Genet. 44: 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chitu V., Gokhan S., Gulinello M., Branch C. A., Patil M., Basu R., Stoddart C., Mehler M. F., Stanley E. R. 2015. Phenotypic characterization of a Csf1r haploinsufficient mouse model of adult-onset leukodystrophy with axonal spheroids and pigmented glia (ALSP). Neurobiol. Dis. 74: 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Himes S. R., Tagoh H., Goonetilleke N., Sasmono T., Oceandy D., Clark R., Bonifer C., Hume D. A. 2001. A highly conserved c-fms gene intronic element controls macrophage-specific and regulated expression. J. Leukoc. Biol. 70: 812–820. [PubMed] [Google Scholar]
- 99.Nilsson S. K., Lieschke G. J., Garcia-Wijnen C. C., Williams B., Tzelepis D., Hodgson G., Grail D., Dunn A. R., Bertoncello I. 1995. Granulocyte-macrophage colony-stimulating factor is not responsible for the correction of hematopoietic deficiencies in the maturing op/op mouse. Blood 86: 66–72. [PubMed] [Google Scholar]
- 100.Niida S., Kondo T., Hiratsuka S., Hayashi S., Amizuka N., Noda T., Ikeda K., Shibuya M. 2005. VEGF receptor 1 signaling is essential for osteoclast development and bone marrow formation in colony-stimulating factor 1-deficient mice. Proc. Natl. Acad. Sci. USA 102: 14016–14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Robertson S. A., Mayrhofer G., Seamark R. F. 1996. Ovarian steroid hormones regulate granulocyte-macrophage colony-stimulating factor synthesis by uterine epithelial cells in the mouse. Biol. Reprod. 54: 183–196. [DOI] [PubMed] [Google Scholar]
- 102.Chorro L., Sarde A., Li M., Woollard K. J., Chambon P., Malissen B., Kissenpfennig A., Barbaroux J. B., Groves R., Geissmann F. 2009. Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. J. Exp. Med. 206: 3089–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.