Abstract
The combined inhibition of histone deacetylases (HDAC) and the proteins of the bromodomain and extraterminal (BET) family have recently shown therapeutic efficacy against melanoma, pancreatic ductal adenocarcinoma, testicular, and lymphoma cancers in murine studies. However, in such studies, the role of the immune system in therapeutically controlling these cancers has not been explored. We sought to investigate the effect of the HDAC inhibitor romidepsin (RMD) and the BET inhibitor IBET151, both singly and in combination, on vaccine-elicited immune responses. C57BL/6 mice were immunized with differing vaccine systems (adenoviral, protein) in prime-boost regimens under treatment with RMD, IBET151, or RMD+IBET151. The combined administration of RMD+IBET151 during vaccination resulted in a significant increase in the frequency and number of Ag-specific CD8+ T cells. RMD+IBET151 treatment significantly increased the frequency of vaccine-elicited IFN-γ+ splenic CD8+ T cells and conferred superior therapeutic and prophylactic protection against B16-OVA melanoma. RNA sequencing analyses revealed strong transcriptional similarity between RMD+IBET151 and untreated Ag-specific CD8+ T cells except in apoptosis and IL-6 signaling–related genes that were differentially expressed. Serum IL-6 was significantly increased in vivo following RMD+IBET151 treatment, with recombinant IL-6 administration replicating the effect of RMD+IBET151 treatment on vaccine-elicited CD8+ T cell responses. IL-6 sufficiency for protection was not assessed. Combined HDAC and BET inhibition resulted in greater vaccine-elicited CD8+ T cell responses and enhanced therapeutic and prophylactic protection against B16-OVA melanoma. Increased IL-6 production and the differential expression of pro- and anti-apoptotic genes following RMD+IBET151 treatment are likely contributors to the enhanced cancer vaccine responses.
Introduction
The combined inhibition of two classes of epigenetic regulators, histone deacetylases (HDACs) and bromodomain and extraterminal proteins (BET), has recently shown therapeutic efficacy against multiple cancer types, including pancreatic ductal adenocarcinoma, testicular, lymphoma, and melanoma (1–4). Proposed mechanisms for the enhanced tumor killing by the combination of HDAC and BET inhibition include the suppression of AKT and YAP signaling (3), the induction of apoptosis over cell cycle arrest in Myc-induced cells (2), the transcriptional induction of p57 (1), and anti-angiogenesis (4). In addition to intrinsic tumor cell apoptosis, it is now established that CD8+ T cells of the mammalian immune system can recognize and kill tumor cells (5). This has resulted in a paradigm shift in cancer therapy, whereby immunotherapies exploiting this T cell ability have resulted in better clinical outcomes than some conventional cancer therapies (6). CD8+ T cell differentiation and functionality is tightly regulated under epigenetic controls (7–9), suggesting potential immunotherapeutic involvement in the HDAC and BET inhibitor cancer therapy.
In this study, we use the ability of vaccines to induce Ag-specific T cell responses to assess changes to T cell differentiation and function following HDAC and BET inhibition. We report that the combination of these two inhibitors significantly increases vaccine-elicited CD8+ T cell number and confers enhanced prophylactic and therapeutic protection against melanoma when used with vaccination. HDAC and BET inhibition increase elicited CD8+ T cells through transcriptionally altering their apoptosis and IL-6/JAK/STAT–related genes following cancer vaccination.
Materials and Methods
Mice, immunizations, and Listeria challenge
Female 6–10-wk-old C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME) were kept in accordance with the Institutional Animal Care and Use Committee guidelines of Beth Israel Deaconess Medical Center. Mice received immunizations of E1/E3-deleted adenovirus (Ad) serotype 26 (Ad26) and Ad serotype 5 (Ad5) expressing either SIVmac239 Gag (Gag), SIVmac239 Env (Env), OVA, or the OVA immunodominant epitope SIINFEKL as described previously (10, 11). Vectors were administered at 109 or 107 viral particles (vp) i.m. in the quadricep in 50 μl volume. OVA (Low Endo, Worthington Biochemical) and CpG (ODN 1826; InvivoGen) or CDN (c-di-AMP VacciGrade; InvivoGen) were administered at 10 and 10 μg, respectively, in 100 μl s.c. at the tail base. Mice were challenged with either 1 × 105 or 5 × 105 CFU of recombinant Listeria monocytogenes expressing the OVA epitope SIINFEKL by i.v. injection. L. monocytogenes bacterial load was calculated by plated splenocytes on brain-heart infusion agar plates as described previously (12).
Epigenetic inhibitors
Romidepsin (RMD; Selleckchem) was suspended in sterile PBS plus 0.5% DMSO and injected i.p. at 1 mg/kg dose (13, 14) with a single dose at time of vaccination. IBET151 (GSK1210151A; Selleckchem) was suspended in sterile PBS plus 30% DMSO and injected i.p. at 30 mg/kg (15) with a single dose at time of vaccination. Combined administration of RMD and IBET151 was performed i.p. at the same dosages above in a final volume of 250 μl at a single dose at time of vaccination. Vehicle control mice received 250 μl of sterile PBS plus 30% DMSO i.p.
mAb and cytokine administration
The monoclonal anti-CD8a (clone 53-6.72; BioXCell) was administered i.p. at 500 μg every 7 d from d−1 prior to tumor injection to maintain CD8+ T cell depletion. Anti–PD–ligand 1 (clone 10F.9G2; BioXCell) or isotype IgG2b (clone LTF-2; BioXCell) were administered by 200 μg i.p. injections as described previously (16, 17) at d−1, d0, d1, and d3 with respect to vaccine boost. Mouse rIL-6 (R&D Systems) suspended in PBS was given at 60 ng i.p. daily from d0 to d3 at boost (18).
Luminex assay
Serum samples were collected 6 h postvaccination and assayed using a Luminex bead-based multiplex ELISA (MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel; Millipore) according to manufacturer’s instructions. Sample data were acquired on a MAGPIX instrument running xPONENT software (Luminex) and analyzed using a five-parameter logistic model.
Tumor cell culturing and inoculation
B16-OVA cells were kindly provided by D. Irvine (Massachusetts Institute of Technology) and cultured in complete DMEM (DMEM; GE Healthcare Life Sciences) supplemented with 10% FBS and 200 μg/ml G418 (InvivoGen). Cells were maintained at 37°C, 5% CO2. An inoculum of 5 × 105 B16-OVA tumor cells was injected s.c. on the flank of mice in 50 μl of sterile PBS. Tumor size was measured as area (longest dimension × perpendicular dimension) and mice euthanized when tumor area exceeded 100 mm2.
Flow cytometry, intracellular cytokine staining, and cell sorting
Lymphocytes were isolated from either the blood or spleen, stained, and analyzed by flow cytometry as previously described (19). Abs to CD8α (53-6.7), CD4 (RM4-5), CD44 (IM7), CD127 (A7R34), KLRG1 (2F1), PD-1 (RMPI-30), IFN-γ (XMG1.2), and Foxp3 (FJK-16S) were purchased from BD Biosciences (Myrtle, U.K.), eBioscience, or BioLegend (San Diego, CA). Cell viability was assessed by LIVE/DEAD Fixable Aqua (Life Technologies). Ag-specific cells were identified by MHC class I tetramer staining using either the AL11 peptide of Gag (AAVKNWMTQTL, H-2Db) (20) or the immunodominant OVA257–264 (SIINFEKL, H-2Kb) epitope from OVA protein (21), with monomers provided by the National Institutes of Health Tetramer Core Facility (Emory University, Atlanta, GA). For intracellular cytokine staining, splenocytes were incubated for 5 h at 37°C with 1 μg/ml SIVmac239Gag peptide pool. During peptide incubation, brefeldin A and monensin (BioLegend) were added. Cells were subsequently washed, stained, and permeabilized with Cytofix/Cytoperm (BD Biosciences). Cells were acquired on an LSR II flow cytometer (BD Biosciences) or sorted on a FACSAria III Special Order (BD Biosciences) with >98% purity. Data were analyzed using FlowJo v.9.8.5 (Tree Star).
RNA sequencing data acquisition and analysis
Splenic CD8+ T cells were enriched by negative selection (CD8+ T Cell Isolation Kit; Stemcell Technologies) prior to Db/SIINFEKL+CD44+CD8+ T cell sorting to >98% purity (data not shown). Sorted cells were stored at −80°C in 1 ml of TRIzol. RNA extraction was performed using the RNeasy 96 QIAcube HT Kit (QIAGEN) per manufacturer’s instructions. RNA concentration and quality were confirmed by Agilent’s 2100 Bioanalyzer (Agilent Technologies). A Low-Input Total RNA library (Clontech SMARTer) was constructed and sequenced on Illumina NS500 Paired-End 75 bp at the Molecular Biology Core Facility at Dana-Farber Cancer Institute. Raw sequences in fastq format were aligned to the mouse reference genome (version mm10) using STAR (22) with default settings, which include soft clipping using the following options: sjdbOverhang: 74; SortedByCoordinate: –quantMode; and TranscriptomeSAM: GeneCounts. Illumina adapters were cut using Trimmomatic (23). Only unique and concordant alignments were used for downstream analysis. Quantification and normalization of the mapped reads at the level of gene model were performed with edgeR (24). A p value cutoff of 0.05 was used to detect significant differential gene expression. Pathway analyses were performed using gene set enrichment analyses (GSEA) (25) with C2 gene sets. The enrichment of chromatin remodeling and IL-6/JAK/STAT3 pathways was assessed using the Fisher exact test and odds ratios calculated using the background population of all annotated mouse genes. All p values were adjusted for multiple testing using a cutoff of 0.05.
Quantitative Ig ELISA
A quantitative Ig ELISA protocol described previously (26) was followed. Briefly, 0.5 μg/ml of either SIVmac239 Env or OVA protein–coated ELISA plates were blocked with PBS/1% BSA/0.05% Tween-20. After washing, diluted serum samples were incubated with the plates for 1 h prior to further washing and the addition of 1:4000 dilution of either anti-mouse IgG-, IgG1-, or IgG2a-HRP (Southern Biotech). The standard wells of anti–mouse κ (1:3200) and Λ (1:3200) L chain (Serotec) coating were blocked and washed prior to adding purified IgG, IgG1, or IgG2a (Southern Biotech) starting at 1000 ng/ml. Samples and standards were developed with TMB, and the reaction stopped after 5 min with TMB Stop Solution (KPL). Absorbance was read on a spectrophotometer at OD450–500 nm with SoftMax Pro GxP v5 software.
Influenza challenge
Six- to eight-week-old female BALB/c mice were obtained from Harlan UK (Littlemore, Oxford, U.K.) and kept in specific pathogen-free conditions in accordance with the United Kingdom’s Home Office guidelines. All work was approved by the Animal Welfare and Ethical Review Board at Imperial College London. Mice were immunized i.m. with 0.005 μg of purified surface Ag from influenza strain H1N1 A/California/7/2009 (GSK Vaccines, Siena, Italy) twice with a minimum of 3-wk interval between immunizations. Mice received RMD+IBET151 i.p. during boosting vaccination. For infections, H1N1 influenza strain A/California/7/2009 was grown in Madin-Darby canine kidney (MDCK) cells in serum-free DMEM supplemented with 1 μg/ml trypsin. Mice were anesthetized using isoflurane and infected intranasally with 3 × 104 PFU influenza, and weight loss was monitored daily.
Immune cells were assessed as described in Materials and Methods but with the following Abs: influenza A H-2Kd HA533-541 pentamer R-PE (ProImmune, Oxford, U.K.), CD3-FITC (BD Biosciences), CD4-PE/Cy7 (BioLegend), and CD8-APC-H7 (BD Biosciences). Analysis was performed on an LSRFortessa flow cytometer (BD Biosciences) after collecting data on at least 50,000 live CD3+ events.
Serum Abs specific to influenza HA1 were measured as described in above, with the exception that plates were coated with 1 μg/ml H1N1 surface protein, and absorbance was read at 450 nm on a FLUOstar spectrophotometer (BMG LABTECH GmbH, Ortenberg, Germany), and absorbance values for the standard titration were fitted with a four- parameter logistic curve, and unknown values were interpolated using BMG Ω software.
Statistical analysis
Statistical analyses were performed using Prism 6.0 (GraphPad Software). Normality of the data was assessed using the Kolmogorov–Smirnov normality test, with nonparametric data analyzed by the Kruskal–Wallis test with Dunn multiple comparison posttest (more than two groups) or the two-tailed Mann–Whitney U test (for two groups). For parametric data, a one-way ANOVA with Bonferroni multiple comparison posttest (more than two groups) or an unpaired t test (for two groups) was used. A p value < 0.05 was considered significant (*p < 0.05, **p < 0.01, ***p < 0.001).
Results
RMD+IBET151 treatment improves therapeutic adenoviral vaccination and reduces B16 melanoma burden
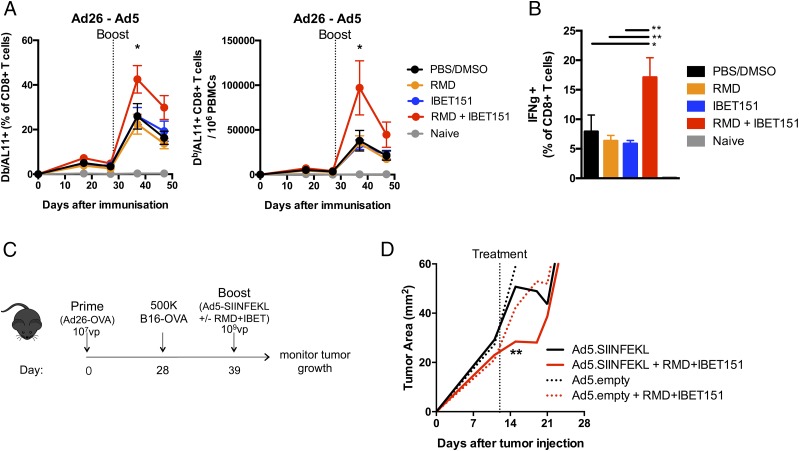
To investigate the role of HDAC (RMD) and BET inhibition (IBET151) on cellular immune responses, the inhibitors were administered singly and in combination during Ad vector immunization. Mice were immunized with vectors expressing the HIV Gag gene in an Ad26 prime Ad5 boost regimen, which has been shown to generate robust, measurable, Ag-specific CD8+ T cell responses (27). Mice were administered RMD, IBET151, RMD+IBET151 mixture or PBS/DMSO vehicle control by the i.p. route just prior to i.m. boost vaccination with Ad5.Gag. The combined administration of RMD+IBET151 resulted in a significant (*p < 0.05) doubling of the frequency and number of Gag- (epitope AL11) specific CD8+ T cells circulating in the blood at day 7 after boost compared with the inhibitors administered individually or the PBS/DMSO control (Fig. 1A). In addition, there was a significant increase in the frequency of CD8+ T cells that produced IFN-γ upon ex vivo restimulation in mice that were treated with the RMD+IBET151 combination compared with RMD or IBET151 alone (**p < 0.01; Fig. 1B). RMD+IBET151 treatment did not significantly change the total number of circulating CD8+ T cells (Supplemental Fig. 1A) or the percentage of regulatory Foxp3+ CD4 T cells (Supplemental Fig. 1B). RMD+IBET151 treatment during priming and boosting vaccination provided the same Ag-specific CD8+ T cell increase as RMD+IBET151 treatment at boosting alone (Supplemental Fig. 1C). Therefore, subsequent vaccinations only received RMD+IBET151 treatment at boost.
FIGURE 1.
RMD+IBET151 increases adenoviral vector–elicited T cell responses and confers superior protection against B16 melanoma. (A and B) C57BL/6 mice were primed with Ad26 (109 vp) and boosted with Ad5 (109 vp) in combination with RMD, IBET151, RMD+IBET151, or PBS/DMSO. (A) Frequency and number of Gag-specific CD8+ T cells in the blood. (B) Frequency of Gag-specific IFN-γ+ CD8+ T cells in the spleen. (C) Experimental outline of B16-OVA challenge. (D) Tumor area measurements following B16-OVA tumor injection into primed mice and during therapeutic vaccination treatments. Each dot represents an individual mouse. n = 4–10 per group per experiment. Data are presented as means ± SEM. *p < 0.05, **p < 0.01, Student t test.
To evaluate the therapeutic efficacy of RMD+IBET151 treatment during vaccination against melanoma tumor progression, mice were primed with Ad26.OVA (107 vp) followed by the s.c. injection of 500,000 B16-OVA cells 28 d later. Eleven days after melanoma injection, mice were therapeutically treated with Ad5.SIINFEKL or Ad5.empty (109 vp) with or without RMD+IBET151 (Fig. 1C). RMD+IBET151 administration resulted in a significant increase in OVA-specific SIINFEKL+CD8+ T cells at day 7 after Ad5.SIINFEKL treatment measured by tetramer staining (*p < 0.05; Supplemental Fig. 1D). The combined treatment of Ad5.SIINFEKL with RMD+IBET151 resulted in a significant regression in B16 melanoma tumor burden compared with Ad5.SIINFEKL treatment alone (**p < 0.01; Fig. 1D) at day 4 after treatment. Although tumor load remained lower following Ad5.SIINFEKL + RMD+IBET151 therapy, this statistical significance was lost by day 7, and all treated mice succumbed to tumor growth and death (Supplemental Fig. 1H). With a greater Ad26.OVA priming dose (109 vp), all mice exhibited protection against B16-OVA melanoma following treatment with Ad5.SIINFEKL (109 vp) with or without RMD+IBET151 (Supplemental Fig. 1E–G).
Increased protein vaccine–elicited cellular immune response following RMD+IBET151 treatment provides CD8+ T cell–mediated melanoma protection
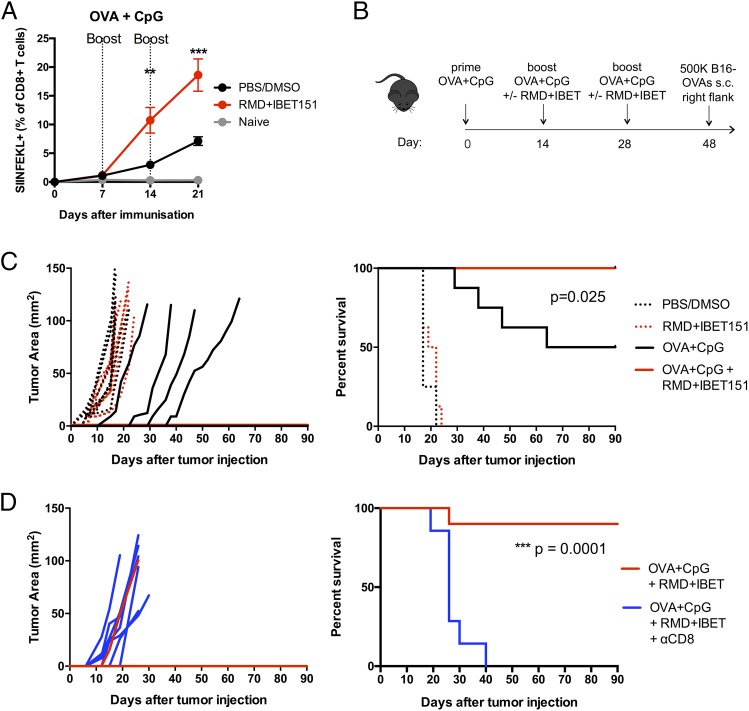
To determine whether the doubling in frequency and number of Ag-specific CD8+ T cells following the combined administration of RMD+IBET151 was applicable to a range of vaccination platforms and regimens, we tested its effect on a protein-adjuvant–based immunization regimen. Following the s.c. immunization of mice with OVA protein adjuvanted with CpG in an accelerated prime-boost-boost regimen, mice that received RMD+IBET151 at boost exhibited a significantly greater (>2-fold, **p < 0.01 after first boost, ***p < 0.001 after second boost) frequency of circulating SIINFEKL+CD8+ T cells in the blood (Fig. 2A). This effect was also observed with OVA protein adjuvanted with the STING agonist CDN (Supplemental Fig. 2A). To evaluate the efficacy of RMD+IBET151 treatment upon vaccination against melanoma tumor establishment, mice were primed s.c. with OVA+CpG and boosted days 14 and 28 with OVA+CpG ± RMD+IBET151 followed by the s.c. injection of 500,000 B16-OVA cells at day 48 (Fig. 2B; SIINFEKL+CD8+ T cell frequency, Supplemental Fig. 2B). The treatment of mice with RMD+IBET151 or PBS/DMSO alone, without vaccination, had no effect on tumor establishment, with all mice bearing tumors by day 8 and all mice sacrificed by day 24 (after tumor injection) because of tumor area exceeding 100 mm2 (Fig. 2C). Only 50% of OVA+CpG–vaccinated mice established melanoma and were subsequently sacrificed, with the tumors appearing between days 15 and 38 after their injection (Fig. 2C). Strikingly, vaccination with OVA+CpG in combination with RMD+IBET151 treatment resulted in complete prevention of melanoma establishment for the length of the study (90 d). Zero out of eight of the OVA+CpG + RMD+IBET151–treated mice presented any measurable tumor load, resulting in a significant survival benefit over OVA+CpG vaccination alone (*p = 0.025; Fig. 2C).
FIGURE 2.
RMD+IBET151 increases protein-elicited T cell responses in an accelerated prime-boost regimen, conferring superior CD8+ T cell–mediated melanoma protection. (A–D) C57BL/6 mice were vaccinated s.c. with OVA protein adjuvanted with CpG in combination with RMD+IBET151 or PBS/DMSO vehicle control. (A) Frequency of OVA-specific CD8+ T cells in the blood. (B) Experimental outline of B16-OVA challenge. Tumor area measurements and survival curves following B16-OVA tumor injection into vaccinated mice (C) and CD8+ T cell–depleted vaccinated mice (D). Each dot represents an individual mouse. n = 6–10 per group per experiment. Data are presented as means ± SEM. **p < 0.01, ***p < 0.001, Student t test.
We examined the involvement of CD8+ T cells in the antitumor effects observed following the OVA+CpG + RMD+IBET151 combination therapy. After the vaccination regimen, and 1 d prior to injection of 500,000 B16-OVA tumor cells, CD8+ T cells were depleted in vivo using a specific mAb against murine CD8a. This CD8+ T cell depletion was maintained weekly. All CD8+ T cell–depleted mice developed tumors, and all were sacrificed by day 40 because of excessive tumor size (***p = 0.0001), revealing a major role for CD8+ T cells in the tumor protection conferred following OVA+CpG + RMD+IBET151 combination therapy (Fig. 2D).
Increased vaccine-elicited CD8+ T cell responses following RMD+IBET151 treatment confer superior L. monocytogenes protection
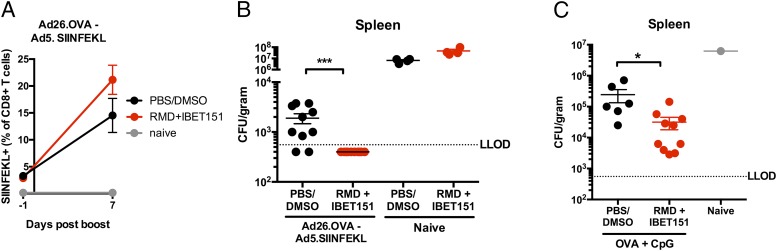
Having observed an effect of RMD+IBET151 treatment on antitumor immunity, we wanted to determine the effect on antipathogen immunity. As such, we assessed the protective role of RMD+IBET151 treatment on vaccination against L. monocytogenes, a bacterial model for CD8+ T cell–meditated protection. Mice were vaccinated with Ad26.OVA–Ad5.SIINFEKL with or without RMD+IBET151 treatment at boost (Fig. 3A) and challenged i.v. with 5 × 105 CFU of recombinant L. monocytogenes–expressing OVA. RMD+IBET151 treatment during boosting vaccination afforded a significant reduction in L. monocytogenes load in the spleen of challenged mice (***p < 0.001; Fig. 3B). In addition to boosting the response to adenoviral vaccination, mice that received RMD+IBET151 during protein (OVA+CpG) vaccination exhibited a significant reduction in L. monocytogenes bacterial load in the spleen at day 2 following infection (*p < 0.05; Fig. 3C).
FIGURE 3.
RMD+IBET151 with vaccination provides enhanced protection against L. monocytogenes. C57BL/6 mice were either (A and B) primed with Ad26 (109 vp) and boosted with Ad5 (109 vp) or (C) vaccinated s.c. with OVA protein adjuvanted with CpG in combination with RMD, IBET151, RMD+IBET151, or PBS/DMSO. (B and C) L. monocytogenes titers in the spleen 2 d after challenge. Each dot represents an individual mouse. n = 4–10 per group per experiment. Data are presented as means ± SEM. *p < 0.05, ***p < 0.001, Student t test.
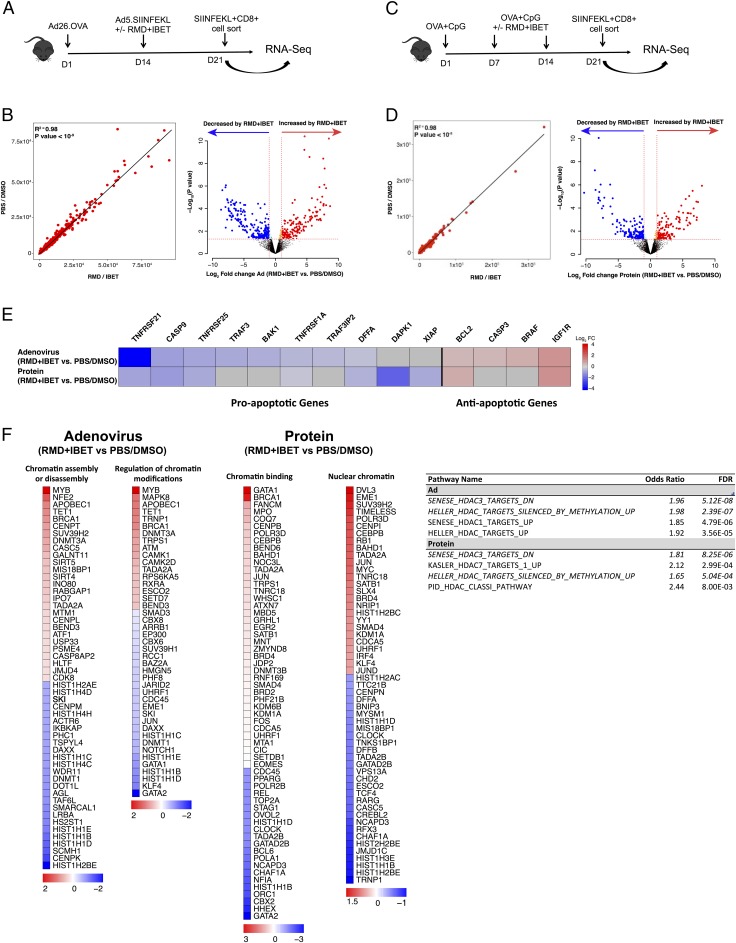
RMD+IBET151 treatment alters limited genes in adenoviral and protein vaccine–elicited CD8+ T cells
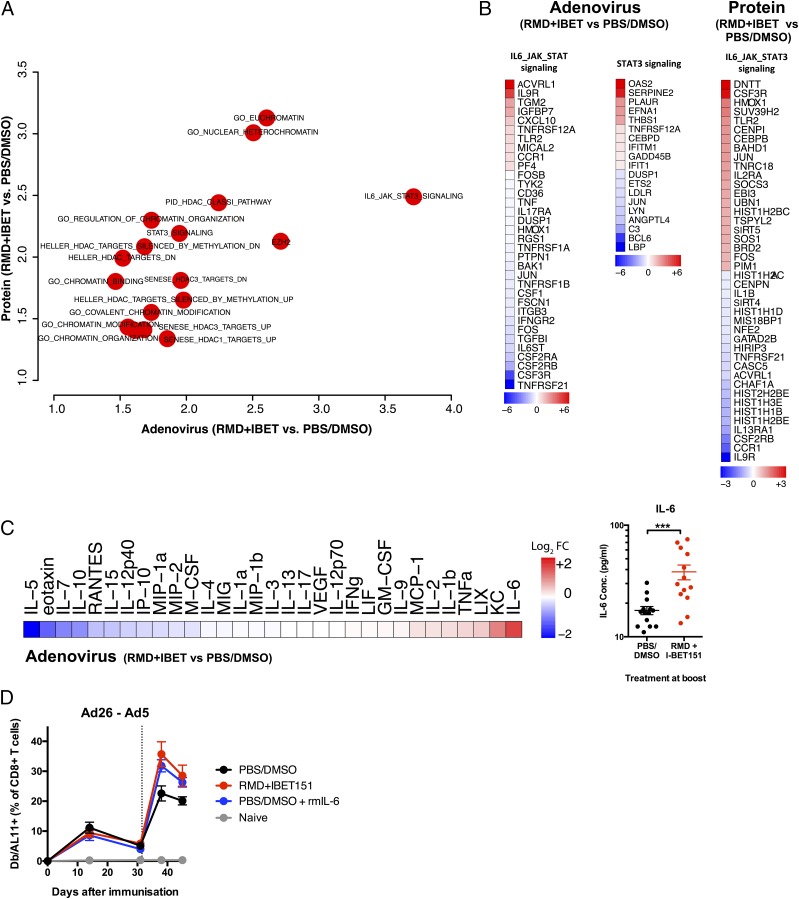
Given the multifaceted involvement of epigenetic control and thus the broad transcriptional impact that inhibition of HDAC and BET could result in, we sought to understand the transcriptional state of the vaccine-elicited CD8+ T cells following RMD+IBET151 treatment. To examine this, mice were vaccinated by either the Ad26-Ad5 prime-boost (Fig. 4A) or the OVA+CpG prime-boost-boost (Fig. 4C) regimen with either RMD+IBET151 or PBS/DMSO treatment during boost. Seven days after final vaccine boost, SIINFEKL+CD8+ T cells from the spleen of vaccinated mice were sorted and their transcriptome profiling performed by RNA sequencing (RNA-Seq). Multidimensional analysis of the full transcriptomic profile of RMD+IBET151–treated and –untreated groups revealed a high level of similarity between the two (Fig. 4B, 4D), indicating RMD+IBET151 treatment is not affecting the global gene expression profile of CD8+ T cell. Although the global pattern was similar, there were differences observed in the expression of pro- and anti-apoptotic genes. The addition of RMD+IBET151 to either protein or adenoviral vaccination regimens led to a downregulation of the proapoptotic genes Tnfrsf21 (Ad: fold change [FC] = −7.422; protein: FC = −0.865), Casp9 (Ad: FC = −0.974; protein: FC = −1.052), and Tnfrsf25 (Ad: FC = −0.851; protein: FC = −0.771) and the upregulation of the anti-apoptotic genes Igf1r (Ad: FC = 1.205; protein: FC = 1.251) and Bcl2 (Ad: FC = 0.519; protein: FC = 0.724) relative to untreated controls (Fig. 4E).
FIGURE 4.
Differential expression of apoptosis- and chromatin-related genes in CD8+ T cells following RMD+IBET151 treatment. (A and C) Experimental outline prior to Ag-specific CD8+ T cell sorting and RNA-Seq. (B and D) Correlation graph and volcano plot of differentially expressed genes between RMD+IBET151–treated and PBS/DMSO–treated SIINFEKL+CD8+ T cells. (E) Differentially expressed genes related to apoptosis. (F) Differentially enriched pathways following RMD+IBET151 treatment on adenoviral and protein vaccination related to chromatin modification and binding. n = 5 per group, C57BL/6 mice.
Likewise, there was differential expression of genes related to chromatin modification and the enrichment of HDAC-related pathways (Fig. 4F), consistent with the fact that RMD and IBET151 are HDAC and BET domain inhibitors, respectively.
RMD+IBET151 treatment increases IL-6 production and enhances CD8+ T cell proliferation
Pathways enrichment analysis revealed the differential expression of genes in the IL-6/JAK/STAT3 pathway following RMD+IBET151 treatment (Fig. 5A), and this was consistent for both adenoviral and protein vaccination (Fig. 5B). In addition, Luminex analysis on serum samples from 6 h after Ad5.Gag vaccination revealed a significant increase in IL-6 concentration in RMD+IBET151–treated mice over PBS/DMSO control–treated mice (***p < 0.001) (Fig. 5C). Subsequent daily high-dose administration of rIL-6 was capable of augmenting CD8+ T cell numbers to that of RMD+IBET151 treatment alone during Ad26-Ad5 vaccination (Fig. 5D). Together, these data indicate a role for IL-6 and its subsequent intracellular signaling cascade in enhancing CD8+ T cell proliferation following treatment with RMD+IBET151.
FIGURE 5.
Increased production of IL-6 and JAK/STAT3 signaling following RMD+IBET151 treatment at vaccination. (A) Odds ratio plot of pathways altered by RMD+IBET151 treatment on adenoviral and protein vaccination regimens. (B) Differentially expressed genes within IL-6/JAK/STAT3 signaling following RMD+IBET151 treatment. (C) Heat map of cytokine/chemokine FC of RMD+IBET151 treatment from PBS/DMSO treatment as measured by Luminex. Graph of individual mouse IL-6 serum concentrations (picograms per milliliter) following either RMD+IBET151 treatment or PBS/DMSO treatment. Each dot represents an individual mouse. n = 14 per group. Data are presented as means ± SEM. ***p < 0.001, Student t test. (D) Frequency of Gag-specific CD8+ T cells in the blood following Ad5.Gag (109 vp) boost in combination with RMD+IBET151, recombinant IL-6, or PBS/DMSO (n = 8–10 per group).
RMD+IBET151 administration augments vaccine-elicited Ab responses and confers superior protection against influenza infection
Having characterized the vaccine-elicited cellular immune responses following RMD+IBET151, we sought to investigate any effect by the treatment upon vaccine-elicited humoral immune responses. Mice were vaccinated i.m. with Ad5.SIV Env (109 vp) and Ad26.SIV Env (109 vp) in heterologous prime-boost regimen with or without RMD+IBET151 treatment at boost. Fourteen days after final boost, mice were bled, and serum IgG specific to the vaccine Ag SIV Env was quantified by quantitative ELISA. For both Ad26-Ad5 and Ad5-Ad26 regimens, the treatment with RMD+IBET151 significantly (*p < 0.05) increased the specific serum IgG concentrations elicited (Supplemental Fig. 3A). The dual combination of RMD+IBET151 elicited greater serum IgG concentrations than either RMD or IBET151 treatment alone (Supplemental Fig. 3A). To assess whether this was applicable to protein and adjuvant vaccination as well, mice were vaccinated s.c. with OVA+CpG in a prime-boost-boost regimen with or without RMD+IBET151 treatments at boost. As with adenoviral vaccination, the treatment with RMD+IBET151 significantly (*p < 0.05) increased the OVA-specific serum IgG concentrations elicited from a mean of 404–816 ng/ml (Supplemental Fig. 3B). Further characterization of the elicited IgG response revealed a significant shift in the IgG isotype following RMD+IBET151 treatment. With both adenoviral (Supplemental Fig. 3C) and protein in adjuvant (Supplemental Fig. 3D) vaccination, RMD+IBET151 treatment preferentially increased the induction of IgG1 while not affecting the induction of IgG2a. Mice vaccinated with HA protein in combination with RMD+IBET151 conferred greater protection against H1N1 influenza (strain A/California/7/2009) infection, as observed by a reduction in weight loss compared with HA vaccination alone (day 5, ***p < 0.001; day 6 and 7, ****p < 0.0001) (Supplemental Fig. 3E). This correlated with an increase in both the serum HA-specific IgG concentration (Supplemental Fig. 3F) and HA-specific CD8+ T cell frequency in mice that received RMD+IBET151 (Supplemental Fig. 3G).
Discussion
In this study, we demonstrate that the combined inhibition of the epigenetic regulators HDAC and BET augment vaccine-elicited cellular and humoral immune responses. This effect translates across different vaccine platforms and confers superior antitumor efficacy against melanoma both therapeutically and prophylactically. We show that CD8+ T cells mediate this antitumor efficacy, and RMD+IBET151 treatment enhances protection against the bacteria L. monocytogenes and against the virus influenza H1N1. RNA-Seq analysis of CD8+ T cells reveals only limited transcriptional differences following RMD+IBET151 treatment. These include apoptotic- and IL-6/JAK/STAT3–related genes, concordant with IL-6 being preferentially expressed following RMD+IBET151 administration. The exogenous administration of IL-6 following vaccination recapitulates the increase in Ag-specific CD8+ T cell number to that of RMD+IBET151 treatment.
The control of cellular immune responses, including the differentiation of naive T cells into memory or effector cells, is choreographed by the epigenetic regulation of transcription factors (28). It has been demonstrated that epigenetic modifications (in particular, histone modifications and DNA methylation) control the transcription of effector molecules (e.g., IFN-γ, IL-2, granzyme b, and perforin) through restricting chromatin access to transcription factors and polymerase (29–31). We observed no adverse effects on the vaccine-elicited CD8+ T cells following the inhibition of HDAC and BET during vaccination, with their cell-killing ability maintained and their transcriptional profiles near-identical. This may be due to the epigenetic inhibition being provided alongside multiple signals from vaccination or due to the specific nature of the epigenetic inhibitor drugs RMD and IBET151. Previous research has revealed that HDAC and BET inhibitors induce similar genes, corroborating our observations of only few differentially expressed genes following RMD+IBET151 treatment (2).
We report that the combination of RMD+IBET151 during vaccination increases the production of IL-6, as measured in the serum by Luminex. Furthermore, we observe a greater gene expression signature for IL-6/JAK/STAT3 signaling in CD8+ T cells following vaccination with RMD+IBET151. However, previous use of IBET151 has shown it to selectively block the production of IL-6 (32). Although not in vivo and not in combination with HDAC inhibition or vaccine stimulation, the previous study revealed that IBET151 interacts with the binding of BRD4 to the IL-6 promoter. Therefore, there is a known molecular interaction between IBET151 and the IL-6 promoter as well as with the BRD-controlled promoters of TNF and IL-1b (33), for which we also saw increased production following RMD+IBET151 treatment. Future work will be needed to tease out the role of IL-6 and other pathways after RMD+IBET151 treatment. This may include depletion or supplementation of IL-6 prior to melanoma, Listeria, or influenza challenges to fully assess the cytokine’s involvement in their protection.
IL-6 has been previously shown to have pleiotropic activity on the differentiation and function of T cells. IL-6 promotes the specific differentiation of naive CD4 T cells, performing a key function in linking the innate and acquired immune response (34) and is indispensable for Th17 differentiation (35). Relevant to the findings of this study, Okada et al. (36) previously revealed that IL-6 induced the differentiation of CD8+ T cells into cytotoxic T cells. It has been further revealed that IL-6 also promotes T follicular helper cell differentiation, which regulates Ig synthesis and IgG4 production in particular (37) and can induce the differentiation of activated B cells into Ab-producing plasma cells.
Combined HDAC and BET inhibitor therapy has also shown superiority over monotherapy for testicular germ cell cancer (4) and melanoma (3). However, these observations were in nude mice lacking an adaptive immune system. This indicates that alternate modes of action of combined HDAC and BET inhibition, independent of immunological mechanisms, are also at play. These may include anti-angiogenesis or the activation of proapoptotic pathways in the tumor cells themselves (3). We see indication of this with therapeutic RMD+IBET151 treatment (without vaccination) slightly reducing melanoma growth compared with vehicle alone. However, in all cases, the combined administration of RMD+IBET151 with vaccination greatly improved melanoma protection over RMD+IBET151 treatment alone.
Following our observations that the HDAC and BET inhibitor need to be administered in combination, future experiments should build upon this. One could use the recently synthesized dual HDAC/BET small molecule inhibitor DUAL946 (38). The dual inhibition of BET and HDAC proteins has been confirmed in vitro and has shown cellular activity in cancer cells, suggesting that our observation may be recapitulated with this single inhibitor in concert with vaccination.
The effect of epigenetic inhibition on vaccination warrants further investigation. We believe this study to be the first assessment, to our knowledge, of epigenetic inhibition on the immunogenicity of viral vectored– and protein-based vaccines, with HDAC inhibition previously investigated with autologous tumor cell–based vaccines. Such HDAC inhibition was shown to enhance MHC class II, CD40, and B7-1/2 expression on B16 cell–based vaccines and correspondingly promoted the immune killing of autologous B16 melanoma cells via CD8+ T cell induction (39–41). Our data indicate that this epigenetic enhancement expands beyond cellular vaccines to both protein- and viral vectored–based vaccines, allowing greater translational use of the strategy.
This is the first time, to our knowledge, that these epigenetic modulators have been shown to increase vaccine efficacy via augmentation of Ag-specific CD8+ T cell responses. These data suggest a greater role for CD8+ T cell responses for melanoma than previously appreciated and open up new avenues to increasing vaccine-elicited immune responses for improving cancer vaccine efficacy.
Supplementary Material
Acknowledgments
We thank the Center for Virology and Vaccine Research Flow Cytometry Core for assistance with cell sorting, the NIH Tetramer Core Facility (Emory University) for provision of MHC class I monomers, and the Dana-Farber Cancer Institute for RNA-Seq.
This work was supported by the Howard Hughes Medical Institute (D.J.I. is an investigator). P.P.-M. was supported by National Institutes of Health (NIH) Grant 1K22 AI118421, American Cancer Society Grant IRG-15-173-21, and Center for AIDS Research Grant P30 AI117943. D.H.B. was supported by NIH Grants AI096040, AI124377, AI126603, AI128751, AI129797, and OD024917 and Ragon Institute grants.
The online version of this article contains supplemental material.
- Ad
- adenovirus
- Ad5
- Ad serotype 5
- Ad26
- Ad serotype 26
- BET
- bromodomain and extraterminal
- FC
- fold change
- HDAC
- histone deacetylase
- RMD
- romidepsin
- RNA-Seq
- RNA sequencing
- vp
- viral particle.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Mazur P. K., Herner A., Mello S. S., Wirth M., Hausmann S., Sánchez-Rivera F. J., Lofgren S. M., Kuschma T., Hahn S. A., Vangala D., et al. 2015. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat. Med. 21: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhadury J., Nilsson L. M., Muralidharan S. V., Green L. C., Li Z., Gesner E. M., Hansen H. C., Keller U. B., McLure K. G., Nilsson J. A. 2014. BET and HDAC inhibitors induce similar genes and biological effects and synergize to kill in Myc-induced murine lymphoma. Proc. Natl. Acad. Sci. USA 111: E2721–E2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinemann A., Cullinane C., De Paoli-Iseppi R., Wilmott J. S., Gunatilake D., Madore J., Strbenac D., Yang J. Y., Gowrishankar K., Tiffen J. C., et al. 2015. Combining BET and HDAC inhibitors synergistically induces apoptosis of melanoma and suppresses AKT and YAP signaling. Oncotarget 6: 21507–21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jostes S., Nettersheim D., Fellermeyer M., Schneider S., Hafezi F., Honecker F., Schumacher V., Geyer M., Kristiansen G., Schorle H. 2017. The bromodomain inhibitor JQ1 triggers growth arrest and apoptosis in testicular germ cell tumours in vitro and in vivo. J. Cell. Mol. Med. 21: 1300–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bethune M. T., Joglekar A. V. 2017. Personalized T cell-mediated cancer immunotherapy: progress and challenges. Curr. Opin. Biotechnol. 48: 142–152. [DOI] [PubMed] [Google Scholar]
- 6.Couzin-Frankel J. 2013. Breakthrough of the year 2013. Cancer immunotherapy. Science 342: 1432–1433. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J. A., Mortazavi A., Williams B. A., Wold B. J., Rothenberg E. V. 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149: 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scharer C. D., Barwick B. G., Youngblood B. A., Ahmed R., Boss J. M. 2013. Global DNA methylation remodeling accompanies CD8 T cell effector function. J. Immunol. 191: 3419–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russ B. E., Olshanksy M., Smallwood H. S., Li J., Denton A. E., Prier J. E., Stock A. T., Croom H. A., Cullen J. G., Nguyen M. L., et al. 2014. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. [Published erratum appears in 2014 Immunity 41: 1064.] Immunity 41: 853–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufman D. R., De Calisto J., Simmons N. L., Cruz A. N., Villablanca E. J., Mora J. R., Barouch D. H. 2011. Vitamin A deficiency impairs vaccine-elicited gastrointestinal immunity. J. Immunol. 187: 1877–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts D. M., Nanda A., Havenga M. J., Abbink P., Lynch D. M., Ewald B. A., Liu J., Thorner A. R., Swanson P. E., Gorgone D. A., et al. 2006. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 441: 239–243. [DOI] [PubMed] [Google Scholar]
- 12.Shen H., Slifka M. K., Matloubian M., Jensen E. R., Ahmed R., Miller J. F. 1995. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc. Natl. Acad. Sci. USA 92: 3987–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishton M. J., Gardiner E. E., Harrison S. J., Prince H. M., Johnstone R. W. 2013. Histone deacetylase inhibitors reduce glycoprotein VI expression and platelet responses to collagen related peptide. Thromb. Res. 131: 514–520. [DOI] [PubMed] [Google Scholar]
- 14.Wilson A. J., Lalani A. S., Wass E., Saskowski J., Khabele D. 2012. Romidepsin (FK228) combined with cisplatin stimulates DNA damage-induced cell death in ovarian cancer. Gynecol. Oncol. 127: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson M. A., Prinjha R. K., Dittmann A., Giotopoulos G., Bantscheff M., Chan W. I., Robson S. C., Chung C. W., Hopf C., Savitski M. M., et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barber D. L., Wherry E. J., Masopust D., Zhu B., Allison J. P., Sharpe A. H., Freeman G. J., Ahmed R. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439: 682–687. [DOI] [PubMed] [Google Scholar]
- 17.Tsukamoto H., Senju S., Matsumura K., Swain S. L., Nishimura Y. 2015. IL-6-mediated environmental conditioning of defective Th1 differentiation dampens antitumour immune responses in old age. Nat. Commun. 6: 6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knudsen J. G., Murholm M., Carey A. L., Biensø R. S., Basse A. L., Allen T. L., Hidalgo J., Kingwell B. A., Febbraio M. A., Hansen J. B., Pilegaard H. 2014. Role of IL-6 in exercise training- and cold-induced UCP1 expression in subcutaneous white adipose tissue. PLoS One 9: e84910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Provine N. M., Larocca R. A., Aid M., Penaloza-MacMaster P., Badamchi-Zadeh A., Borducchi E. N., Yates K. B., Abbink P., Kirilova M., Ng’ang’a D., et al. 2016. Immediate dysfunction of vaccine-elicited CD8+ T cells primed in the absence of CD4+ T cells. J. Immunol. 197: 1809–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barouch D. H., Pau M. G., Custers J. H., Koudstaal W., Kostense S., Havenga M. J., Truitt D. M., Sumida S. M., Kishko M. G., Arthur J. C., et al. 2004. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J. Immunol. 172: 6290–6297. [DOI] [PubMed] [Google Scholar]
- 21.Rötzschke O., Falk K., Stevanović S., Jung G., Walden P., Rammensee H. G. 1991. Exact prediction of a natural T cell epitope. Eur. J. Immunol. 21: 2891–2894. [DOI] [PubMed] [Google Scholar]
- 22.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolger A. M., Lohse M., Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson M. D., McCarthy D. J., Smyth G. K. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badamchi-Zadeh A., McKay P. F., Holland M. J., Paes W., Brzozowski A., Lacey C., Follmann F., Tregoning J. S., Shattock R. J. 2015. Intramuscular immunisation with chlamydial proteins induces Chlamydia trachomatis specific ocular antibodies. PLoS One 10: e0141209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan W. G., Jin H. T., West E. E., Penaloza-MacMaster P., Wieland A., Zilliox M. J., McElrath M. J., Barouch D. H., Ahmed R. 2013. Comparative analysis of simian immunodeficiency virus gag-specific effector and memory CD8+ T cells induced by different adenovirus vectors. J. Virol. 87: 1359–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen M. L., Jones S. A., Prier J. E., Russ B. E. 2015. Transcriptional enhancers in the regulation of T cell differentiation. Front. Immunol. 6: 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Northrop J. K., Thomas R. M., Wells A. D., Shen H. 2006. Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J. Immunol. 177: 1062–1069. [DOI] [PubMed] [Google Scholar]
- 30.Kersh E. N., Fitzpatrick D. R., Murali-Krishna K., Shires J., Speck S. H., Boss J. M., Ahmed R. 2006. Rapid demethylation of the IFN-gamma gene occurs in memory but not naive CD8 T cells. J. Immunol. 176: 4083–4093. [DOI] [PubMed] [Google Scholar]
- 31.Araki Y., Fann M., Wersto R., Weng N. P. 2008. Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B). J. Immunol. 180: 8102–8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett E., Brothers S., Wahlestedt C., Beurel E. 2014. I-BET151 selectively regulates IL-6 production. Biochim. Biophys. Acta 1842: 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicodeme E., Jeffrey K. L., Schaefer U., Beinke S., Dewell S., Chung C. W., Chandwani R., Marazzi I., Wilson P., Coste H., et al. 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468: 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka T., Narazaki M., Kishimoto T. 2014. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 6: a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korn T., Bettelli E., Oukka M., Kuchroo V. K. 2009. IL-17 and Th17 cells. Annu. Rev. Immunol. 27: 485–517. [DOI] [PubMed] [Google Scholar]
- 36.Okada M., Kitahara M., Kishimoto S., Matsuda T., Hirano T., Kishimoto T. 1988. IL-6/BSF-2 functions as a killer helper factor in the in vitro induction of cytotoxic T cells. J. Immunol. 141: 1543–1549. [PubMed] [Google Scholar]
- 37.Ma C. S., Deenick E. K., Batten M., Tangye S. G. 2012. The origins, function, and regulation of T follicular helper cells. J. Exp. Med. 209: 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atkinson S. J., Soden P. E., Angell D. C., Bantscheff M., Chung C., Giblin K. A., Smithers N., Furze R. C., Gordon L., Drewes G., et al. 2014. The structure based design of dual HDAC/BET inhibitors as novel epigenetic probes. MedChemComm 5: 342–351. [Google Scholar]
- 39.Murakami T., Sato A., Chun N. A., Hara M., Naito Y., Kobayashi Y., Kano Y., Ohtsuki M., Furukawa Y., Kobayashi E. 2008. Transcriptional modulation using HDACi depsipeptide promotes immune cell-mediated tumor destruction of murine B16 melanoma. J. Invest. Dermatol. 128: 1506–1516. [DOI] [PubMed] [Google Scholar]
- 40.Khan A. N., Magner W. J., Tomasi T. B. 2004. An epigenetically altered tumor cell vaccine. Cancer Immunol. Immunother. 53: 748–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan A. N., Magner W. J., Tomasi T. B. 2007. An epigenetic vaccine model active in the prevention and treatment of melanoma. J. Transl. Med. 5: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.