Abstract
Purpose:
To evaluate the error in segmented tissue images and to show the usefulness of the brain image in voxel-based morphometry (VBM) using Statistical Parametric Mapping (SPM) 12 software and 3D T1-weighted magnetic resonance images (3D-T1WIs) processed to simulate idiopathic normal pressure hydrocephalus (iNPH).
Materials and Methods:
VBM analysis was performed on sagittal 3D-T1WIs obtained in 22 healthy volunteers using a 1.5T MR scanner. Regions of interest for the lateral ventricles of all subjects were carefully outlined on the original 3D-T1WIs, and two types of simulated 3D-T1WI were also prepared (non-dilated 3D-T1WI as normal control and dilated 3D-T1WI to simulate iNPH). All simulated 3D-T1WIs were segmented into gray matter, white matter, and cerebrospinal fluid images, and normalized to standard space. A brain image was made by adding the gray and white matter images. After smoothing with a 6-mm isotropic Gaussian kernel, group comparisons (dilated vs non-dilated) were made for gray and white matter, cerebrospinal fluid, and brain images using a paired t-test.
Results:
In evaluation of tissue volume, estimation error was larger using gray or white matter images than using the brain image, and estimation errors in gray and white matter volume change were found for the brain surface.
Conclusion:
To our knowledge, this is the first VBM study to show the possibility that VBM of gray and white matter volume on the brain surface may be more affected by individual differences in the level of dilation of the lateral ventricles than by individual differences in gray and white matter volumes. We recommend that VBM evaluation in patients with iNPH should be performed using the brain image rather than the gray and white matter images.
Keywords: brain volumetry, idiopathic normal pressure hydrocephalus, lateral ventricles, missegmentation, voxel-based morphometry
Introduction
Voxel-based morphometry (VBM)1 with 3D T1-weighted magnetic resonance images (3D-T1WI) has been used to evaluate the gray and white matter volume changes associated with various diseases (e.g., Alzheimer’s disease,2 epilepsy,3 diabetes,4 Parkinson’s disease,5 and panic disorder.6 Voxel-based morphometry is also established as a useful tool for evaluating gray and white matter volume; however, VBM is prone to estimation error by missegmentation of the gray and white matter, and cerebrospinal fluid (CSF) images, with the level of estimation error being influenced by the quality of the 3D-T1WIs.7–14 Until recently, estimation error due to missegmentation was worse for CSF images than for gray and white matter images, but accuracy improved following the introduction of Statistical Parametric Mapping (SPM; Wellcome Trust Centre, London, United Kingdom) 12 software in 2015.15,16
Some previous studies have used VBM to estimate gray and white matter, and CSF volumes in idiopathic normal pressure hydrocephalus (iNPH).17–20 For example, in 2008, Ishii et al. reported that morphological change occurs in the front parietal high convexity, with ventricular dilatations, dilated sylvian fissures, and tight sulci in the medial parietal lobes.17 In 2010, Yamashita et al. reported that VBM using a CSF image can detect characteristic morphological changes; i.e., the coexistence of a dilated lateral ventricle or Sylvian fissure with a narrowed high convexity or midline CSF space in iNPH patients.20 Voxel-based morphometry is a useful tool for understanding volume changes in subjects with iNPH, but few reports have evaluated iNPH using VBM. We consider that there are two main reasons for the lack of such studies. First, the estimation accuracy of CSF was probably lower with the older versions of SPM software compared with SPM12.15 Second, the lateral ventricles of patients with iNPH are much more dilated than those of normal subjects. Transformation of the brain becomes a source of estimation error during spatial normalization in the VBM process.12 However, no previous report has identified dilated lateral ventricles as the cause of estimation error. The first aim of the present study is to evaluate the estimation error for tissue (gray matter, white matter, and CSF) images. The previous study reported missegmentation between gray and white matter.7 Based on the study, we think that VBM with brain image21 (combination image of gray and white matter) is robust for influence by source of missegmentation. The second is to show the usefulness of the brain image21 in VBM using SPM12 software and 3D-T1WIs processed to simulate iNPH.
Materials and Methods
Subjects
A total of 22 healthy volunteers participated in this study (17 males, 5 females; mean age, 31.1 ± 7.3 years; age range, 23–47 years). The MR images were inspected by a board-certified radiologist, who found none of the following findings in any subject: brain tumor, infarction, hemorrhage, brain atrophy, or white matter lesions graded higher than grade 2 of Fazekas’s classification.22 The protocol was approved by the Ethical Committee of our institution. After the study had been explained to each subject, written informed consent was obtained from all participants. To protect subject confidentiality, patient information was stripped from all data.
MRI scanning protocol
MRI data were obtained using a 1.5T scanner (Magnetom Vision, Siemens Healthineers, Erlangen, Germany). A circularly polarized head coil was used as the transmit-receive coil. For the VBM analysis, 3D magnetization-prepared rapid gradient echo was used to obtain sagittal 3D-T1WI with a slice thickness of 1.25 mm (TR/TE = 9.7/4 ms; inversion time = 300 ms; flip angle = 12°; FOV = 22 cm; number of excitations = 1; 256 × 200 pixel matrix). The voxel dimensions were 0.8594 × 1.1 × 1.25 mm.
Preparation of 3D-T1WI to simulate iNPH
Regions of interest for the lateral ventricles (V-ROIs) were carefully outlined on the original 3D-T1WIs of all 22 subjects, and set to show optimal contrast between brain and CSF using Mango (Multi-Image Analysis GUI) software developed at the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC; https://www.nitrc.org/projects/mango). We measured the mean signal intensities in the 22 V-ROIs of the original 3D-T1WIs.
We then prepared two sets of simulated 3D-T1WIs.
Non-dilated 3D-T1WI as normal control: the signal intensity of the V-ROI in the non-dilated 3D-T1WI was rewritten as the measured mean signal intensity inside the V-ROI of the corresponding original 3D-T1WI, for each of the 22 subjects.
Dilated 3D-T1WI to simulate iNPH: the V-ROIs of the non-dilated 3D-T1WIs were individually processed to achieve dilation (dilation size, 5), using Mango software. Details of the dilation processing can be found on the Mango software web site (http://ric.uthscsa.edu/mango/usingmango_roi.html).
Image preprocessing for VBM
A total of 44 3D-T1WIs (22 subjects × non-dilated and dilated) were processed using SPM12. In the segmentation process, the affine regularization space template for the International Consortium for Brain Mapping was changed from ‘European brain’ to ‘East Asian brain’ because all the subjects in our study were Japanese, and Diffeomorphic Anatomical Registration Through Exponentiated Lie Algebra (DARTEL) imported data were saved to spatially normalization with DARTEL; default settings were used for all other parameters. The images were segmented according to tissue type into gray matter, white matter, and CSF images, and spatially normalized with DARTEL (DARTEL templates were created from the present data). Finally, the normalized data were intensity-modulated to the Montreal Neurological Institute (MNI) space with ‘Preserve amount’. Brain images were then constructed by adding gray and white matter, and the 176 images (gray, white, CSF, and brain × non-dilated and dilated × 22 subjects) were smoothed with a 6-mm isotropic Gaussian kernel and analyzed with SPM12 software. The segmentation algorithm on single generative model was used in our segmentation process because target in the present study is cross-sectional study as group comparison between control and iNPH group.
Statistical analyses for VBM
Group comparisons between smoothed tissue images in the non-dilated and dilated 3D-T1WIs with VBM were performed by paired t-test with SPM12 software using mask images. We made four types of mask image (for gray matter, white matter, cerebrospinal fluid, and brain image). The mask images were constructed from the spatially normalized images of all 22 subjects, for each tissue type, and included voxels that had a value of >0.01. In the group comparisons, a P value of <0.05, corrected with family-wise error in voxel difference and a cluster size greater than 30 voxels, was considered to be statistically significant. We did not have to input covariates (such as age, gender, and intracranial volume) in the statistical analyses for VBM because we used a paired t-test model.
Results
Preparation of 3D-T1WI to simulate iNPH
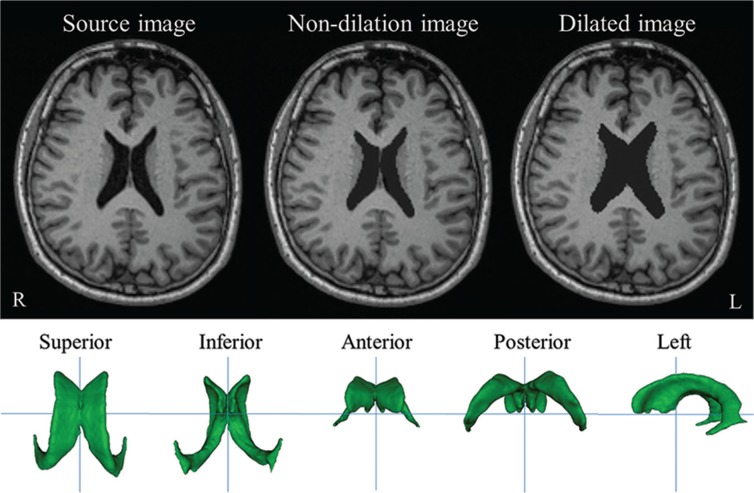
The V-ROI volumes in the non-dilated and dilated 3D-T1WIs are listed in Table 1. Figure 1 shows examples of original, non-dilated, and dilated 3D-T1WIs, and various rotations of the 3D V-ROI.
Table 1.
Region of interest for the lateral ventricles (V-ROI) volumes in non-dilated and dilated 3D-T1-weighted magnetic resonance images (T1WI)
| Subject no. | V-ROI volume (cm3) | |
|---|---|---|
| Non-dilated 3D-T1WI | Dilated 3D-T1WI | |
| 1 | 22.9 | 45.4 |
| 2 | 15.8 | 35.9 |
| 3 | 13.6 | 32.0 |
| 4 | 12.4 | 30.7 |
| 5 | 29.3 | 53.6 |
| 6 | 13.5 | 30.7 |
| 7 | 11.8 | 29.2 |
| 8 | 14.3 | 30.8 |
| 9 | 12.0 | 29.6 |
| 10 | 8.8 | 23.7 |
| 11 | 7.4 | 20.1 |
| 12 | 10.6 | 25.4 |
| 13 | 7.9 | 21.5 |
| 14 | 16.5 | 36.8 |
| 15 | 13.3 | 30.0 |
| 16 | 11.3 | 27.2 |
| 17 | 10.3 | 25.5 |
| 18 | 10.4 | 26.5 |
| 19 | 9.5 | 23.8 |
| 20 | 7.0 | 19.5 |
| 21 | 16.5 | 34.8 |
| 22 | 23.5 | 44.9 |
|
| ||
| Mean | 13.6 | 30.8 |
Fig. 1.
Original, non-dilated, and dilated 3D-T1-weighted magnetic resonance images (T1WI), and 3D views of the region of interest for the lateral ventricles (V-ROI) in original image of subject 1. R and L indicate the right and left sides of the subject, respectively.
Group comparisons with VBM
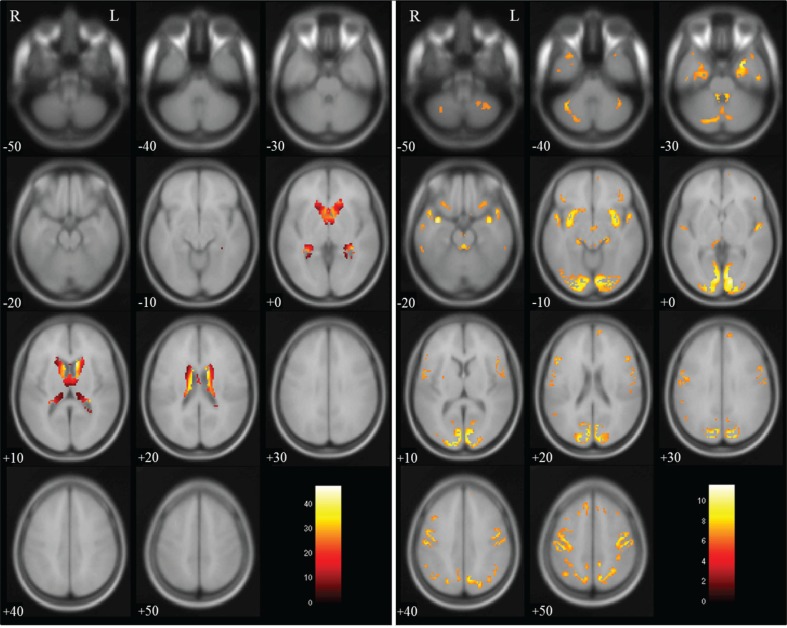
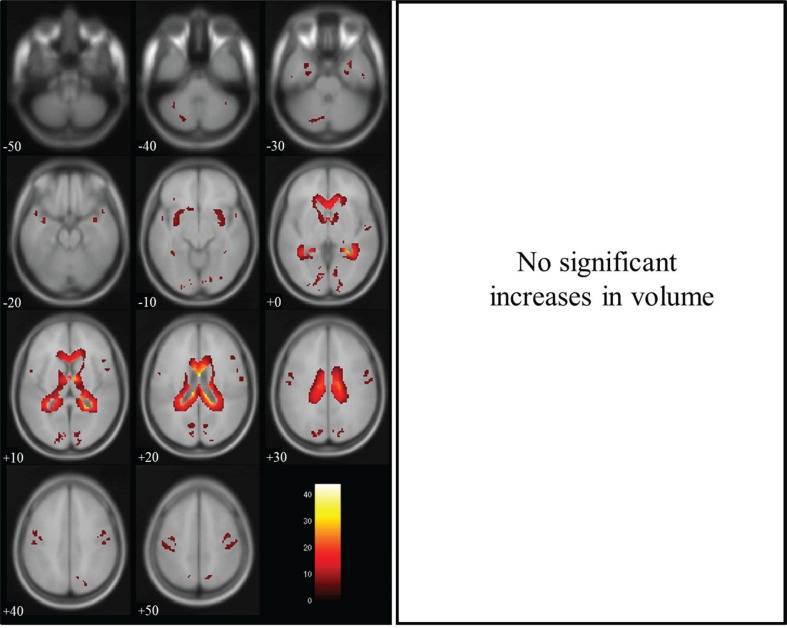
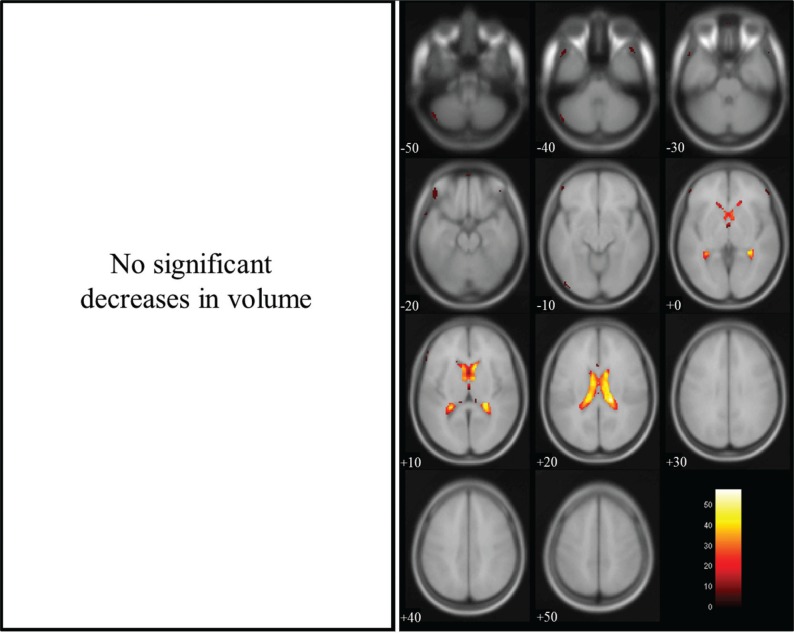
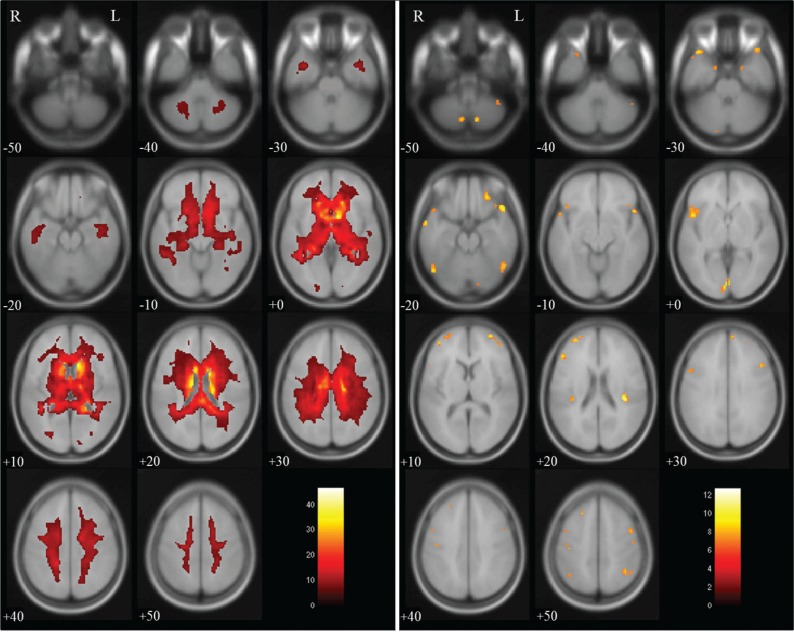
Group comparisons for tissue volume change between non-dilated and dilated 3D-T1WIs by paired t-test revealed a significant decrease in gray matter volume in regions near the lateral ventricles, and a significant increase in volume in brain surface regions, as shown in Fig. 2. There were significant decreases in white matter volume in regions near the lateral ventricles and on the brain surface, as shown in Fig. 3, and significant increases in CSF volume in regions within the lateral ventricles, as shown in Fig. 4. In the brain images, there was a significant decrease in volume in the deep white matter, and a significant increase in volume in brain surface regions, as shown in Fig. 5.
Fig. 2.
Gray matter volume in dilated 3D-T1-weighted magnetic resonance images (T1WI) compared with that in non-dilated 3D-T1WI. Regions of significantly decreased volume (max cluster size is 8260 and max T value is 47.29 as true volume change near the lateral ventricles) are shown in the left panel, and regions of significantly increased volume (max cluster size is 12,946 and max T value is 11.62 as the estimation error for the brain surface) are shown in the right panel. Significant volume differences are superimposed on the T1 template image. The color bar (red to white) represents the T score. R and L indicate the right and left sides of the subject, respectively. (The color version is available online.)
Fig. 3.
White matter volume in dilated 3D-T1-weighted magnetic resonance images (T1WI) compared with that in non-dilated 3D-T1WI. Regions of significantly decreased volume (max cluster size is 21882 and max T value is 43.93 as true volume change near the lateral ventricles; and max cluster size is 2912 and max T value is 11.32 as the estimation error for the brain surface) are superimposed on the T1 template image; no region showed a significant increase in volume. The color bar (red to white) represents the T score. R and L indicate the right and left sides of the subject, respectively. (The color version is available online.)
Fig. 4.
Cerebrospinal fluid (CSF) volume in dilated 3D-T1-weighted magnetic resonance images (T1WI) compared with that in non-dilated 3D-T1WI. Regions of significantly increased volume (max cluster size is 6934 and max T value is 57.70 as true volume change within the lateral ventricles; and max cluster size is 357 and max T value is 15.43 as the estimation error for the brain surface) are superimposed on the T1 template image; no region showed a significant decrease in volume. The color bar (red to white) represents the T score. R and L indicate the right and left sides of the subject, respectively. (The color version is available online.)
Fig. 5.
Brain volume in dilated 3D-T1-weighted magnetic resonance images (T1WI) compared with that in non-dilated 3D-T1WI. Regions of significantly decreased volume (max cluster size is 103039 and max T value is 46.62 as true volume change within the lateral ventricles) are shown in the left panel, and regions of significantly increased volume (max cluster size is 530, max T value is 12.69 as the estimation error for the brain surface) are shown in the right panel. Significant volume differences are superimposed on the T1 template image. The color bar (red to white) represents the T score. R and L indicate the right and left sides of the subjects, respectively. (The color version is available online.)
Discussion
Non-dilated and dilated 3D-T1WIs were analyzed with SPM12 software to evaluate the estimation error for segmented tissue (gray matter, white matter, CSF) images with VBM, using 3D-T1WIs processed to simulate iNPH. The results showed a significant difference in volume between the segmented gray and white matter images from the dilated 3D-T1WIs compared with those from the non-dilated 3D-T1WIs. Because the only difference between the non-dilated and dilated 3D-T1WIs is the change in V-ROI size, decreases in gray and white matter volume in areas near the lateral ventricles are true changes in volume; similarly, increases in CSF volume within the lateral ventricles are true changes in volume. In contrast, changes in tissue volume in locations distant to the lateral ventricles (e.g., areas on the brain surface) are caused by estimation error in the SPM12 software, as no change has been made to tissue volume located distant to the lateral ventricles during processing of the dilated 3D-T1WIs. Based on this theory, we interpreted our results as follows: evaluation of tissue volume using the gray and white matter images had large estimation errors compared with that using the brain image, and estimation errors for gray and white matter volume change were found on the brain surface (Figs. 2, 3 and 5). These results indicate the possibility that gray and white matter volume measured with VBM on the brain surface may be more affected by individual differences in the level of dilation of the lateral ventricles than by individual differences in gray/white matter volumes. Therefore, we propose that in the case of a brain with dilated lateral ventricles, as in iNPH, evaluation should be performed with VBM using the brain image rather than the gray and white matter images.
We found estimation errors in gray and white matter volume change on the brain surface (Figs. 2 and 3), and consider that the main cause of estimation error for gray and white matter volume with VBM is segmentation process. We checked segmented gray matter images in native space, and found increased gray matter volume in the global brain area with dilated 3D-T1WI compared with non-dilated 3D-T1WI. Signal intensity profile of 3D-T1WI was used in the segmentation process with SPM12 software. We think that the signal intensity profile was changed by dilation of lateral ventricles. As a result, the increased gray matter volume was found. Other cause of estimation error is the spatial normalization process. A brain form is deformed to the template image and voxel-value was modulated for spatially transform level in our study. If the lateral ventricle on the 3D-T1WI is large (i.e., if the lateral ventricle volume makes up a high proportion of the total intracranial volume) compared with that of the template, gray matter on the surface of the brain is stretched in the direction of the lateral ventricle. As the ventricle on the dilated 3D-T1WI is larger than that on the non-dilated 3D-T1WI, gray matter on the surface of the brain is more strongly stretched on the dilated 3D-T1WI than on the non-dilated 3D-T1WI. In addition, stretched gray matter on the brain surface of the dilated 3D-T1WI overlaps with white matter located close to the gray matter on the brain surface of the non-dilated 3D-T1WI. Accordingly, in a group comparison with VBM, the overlap area (voxels) showed increased gray matter volume and decreased white matter volume in the dilated 3D-T1WIs relative to the non-dilated 3D-T1WIs. We ran additionally examination for checking the effect of modulation. As a result, effect of missegmentation in brain image decrease compared with our result. However, unmodulated data cannot show tissue volume, and modulated data can show tissue volume with atlas ROI volumetry. We often use atlas ROI volumetry in our research, and the understanding of effect on modulated data is important. If ROI volumetry is not necessary in a study, VBM evaluation in patients with iNPH should be performed using the unmodulated image rather than the modulated images.
There are some differences between the brain morphology of patients with iNPH and that of normal subjects,17–20 and signal change has been previously observed in the periventricular white matter on MR images of patients with iNPH.23–25 Tissue signal change in the 3D-T1WIs is a cause of missegmentation in VBM.7 In the segmentation process with SPM12, the signal intensity in each voxel is important information because the mean signal intensity of the tissue determines the existence of that tissue in the segmented image, and signal intensity arising from elsewhere decreases the probability that the tissue type will be correctly represented in the segmented image. In patients with iNPH, we found that periventricular white matter with reduced signal intensity had been segmented into the gray matter image. Therefore, as well as for the reason associated with the cause of estimation error mentioned in the first paragraph of the Discussion, we propose that patients with iNPH be evaluated with VBM using the brain image rather than the gray and white matter images.
Increased CSF volume in the lateral ventricles of the dilated 3D-T1WI is a true volume change, but that on the brain surface is an estimation error, according to the design of the present study. A very small region of increased CSF volume (estimation error) is seen on the brain surface of the dilated 3D-T1WI (Fig. 4). The cause of this estimation error may be the same as that of the gray and white matter estimation errors. However, it is a very small region (cluster size is 357) compared with that of gray matter (cluster size is 12946) and white matter (cluster size is 2912). There is a case that a small cluster size with significant difference does not mean that the estimation error is smaller. For example, a smaller cluster is found for significant difference if the area with the estimation error is widely distributed than if the area with the estimation error is concentrated at a point. However, we believe that the design of the present study has ensured that the estimation error is similarly concentrated for all images (gray, white, CSF, and brain image) because we used the same deformation field with DARTEL during the spatial normalization process for images of all tissue types. Therefore, we conclude that estimation error is smaller in CSF than in gray and white matter in VBM with SPM12.
A major limitation of the present study is that in the simulation of the dilated 3D-T1WIs, only the size of the lateral ventricles was changed. It is likely that other morphological changes associated with clinical iNPH would also influence VBM results. Also, lateral ventricle dilatation in our study is too conservative to simulate iNPH, subject with iNPH have larger lateral ventricles compared with images in our study. Therefore, the effects of missegmentation as our result increase in VBM with clinical iNPH subject. A second limitation is that we reported the effects using the same scanner, same scanning parameters, and the same VBM method. Changing the MR scanner, the scanning parameters, and the VBM method would change the profile curve (mean signal intensity in gray matter, white matter, and cerebrospinal fluid) of the 3D-T1WIs; accordingly, the areas of significant volume change would also differ slightly from the present results. In addition, if target in a study is estimation of longitudinal volume change, using pairwise longitudinal registration implemented in SPM12 might disappear observation of missegmentaion as our results.
Conclusion
To our knowledge, this is the first VBM study to show an association between dilation of the lateral ventricles and estimation errors in gray and white matter images. Our results indicate the possibility that brain surface gray and white matter volume measured with VBM may be more affected by individual differences in the level of dilation of the lateral ventricles than by individual differences in gray and white matter volumes. Missegmentation in brain surface was decreased in VBM using brain image. We recommend that VBM evaluation in patients with iNPH should be performed using the brain image rather than the gray and white matter images.
Acknowledgments
This work was supported by JSPS Kakenhi Grant 15K19774 and JP16H06280.
Footnotes
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Ashburner J, Friston KJ. Voxel-based morphometry—the methods. Neuroimage 2000; 11:805–821. [DOI] [PubMed] [Google Scholar]
- 2.Baron JC, Chételat G, Desgranges B, et al. In vivo mapping of gray matter loss with voxel-based morphometry in mild Alzheimer’s disease. Neuroimage 2001; 14:298–309. [DOI] [PubMed] [Google Scholar]
- 3.Keller SS, Roberts N. Voxel-based morphometry of temporal lobe epilepsy: an introduction and review of the literature. Epilepsia 2008; 49:741–757. [DOI] [PubMed] [Google Scholar]
- 4.Musen G, Lyoo IK, Sparks CR, et al. Effects of type 1 diabetes on gray matter density as measured by voxel-based morphometry. Diabetes 2006; 55:326–333. [DOI] [PubMed] [Google Scholar]
- 5.Fioravanti V, Benuzzi F, Codeluppi L, et al. MRI correlates of Parkinson’s disease progression: a voxel based morphometry study. Parkinsons Dis 2015; 2015: 378032. doi: 10.1155/2015/378032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayano F, Nakamura M, Asami T, et al. Smaller amygdala is associated with anxiety in patients with panic disorder. Psychiatry Clin Neurosci 2009; 63:266–276. [DOI] [PubMed] [Google Scholar]
- 7.Goto M, Abe O, Miyati T, et al. Association between iron content and gray matter missegmentation with voxel-based morphometry in basal ganglia. J Magn Reson Imaging 2013; 38:958–962. [DOI] [PubMed] [Google Scholar]
- 8.Goto M, Abe O, Miyati T, Yamasue H, Gomi T, Takeda T. Head motion and correction methods in resting-state functional MRI. Magn Reson Med Sci 2016; 15:178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto M, Suzuki M, Mizukami S, et al. Repeatability of brain volume measurements made with the atlas-based method from T1-weighted images acquired using a 0.4 Tesla low field MR scanner. Magn Reson Med Sci 2016; 15:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jack CR, Bernstein MA, Fox NC, et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging 2008; 27:685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doran SJ, Charles-Edwards L, Reinsberg SA, Leach MO. A complete distortion correction for MR images: I. Gradient warp correction. Phys Med Biol 2005; 50:1343–1361. [DOI] [PubMed] [Google Scholar]
- 12.Goto M, Abe O, Kabasawa H, et al. Japanese Alzheimer’s disease neuroimaging initiative Effects of image distortion correction on voxel-based morphometry. Magn Reson Med Sci 2012; 11:27–34. [DOI] [PubMed] [Google Scholar]
- 13.Goto M, Abe O, Miyati T, et al. Japanese Alzheimer’s disease neuroimaging initiative Influence of signal intensity non-uniformity on brain volumetry using an atlas-based method. Korean J Radiol 2012; 13:391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis EB, Fox NC. Correction of differential intensity inhomogeneity in longitudinal MR images. Neuroimage 2004; 23:75–83. [DOI] [PubMed] [Google Scholar]
- 15.Malone IB, Leung KK, Clegg S, et al. Accurate automatic estimation of total intracranial volume: a nuisance variable with less nuisance. Neuroimage 2015; 104:366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goto M, Abe O, Miyati T, et al. Accelerated hippocampal volume reduction in post-menopausal women: an additional study with Atlas-based method. Radiol Phys Technol 2011; 4:185–188. [DOI] [PubMed] [Google Scholar]
- 17.Ishii K, Kawaguchi T, Shimada K, et al. Voxel-based analysis of gray matter and CSF space in idiopathic normal pressure hydrocephalus. Dement Geriatr Cogn Disord 2008; 25:329–335. [DOI] [PubMed] [Google Scholar]
- 18.DeVito EE, Salmond CH, Owler BK, Sahakian BJ, Pickard JD. Caudate structural abnormalities in idiopathic normal pressure hydrocephalus. Acta Neurol Scand 2007; 116:328–332. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita F, Sasaki M, Saito M, et al. Voxel-based morphometry of disproportionate cerebrospinal fluid space distribution for the differential diagnosis of idiopathic normal pressure hydrocephalus. J Neuroimaging 2014; 24:359–365. [DOI] [PubMed] [Google Scholar]
- 20.Yamashita F, Sasaki M, Takahashi S, et al. Detection of changes in cerebrospinal fluid space in idiopathic normal pressure hydrocephalus using voxel-based morphometry. Neuroradiology 2010; 52:381–386. [DOI] [PubMed] [Google Scholar]
- 21.Abe O, Yamasue H, Aoki S, et al. Aging in the CNS: comparison of gray/white matter volume and diffusion tensor data. Neurobiol Aging 2008; 29:102–116. [DOI] [PubMed] [Google Scholar]
- 22.Fazekas F, Chawluk JB, Alavi A, Hurtig HI, Zimmerman RA. MR signal abnormalities at 1.5 T in Alzheimer’s dementia and normal aging. AJR Am J Roentgenol 1987; 149:351–356. [DOI] [PubMed] [Google Scholar]
- 23.Akiguchi I, Ishii M, Watanabe Y, et al. Shunt-responsive parkinsonism and reversible white matter lesions in patients with idiopathic NPH. J Neurol 2008; 255:1392–1399. [DOI] [PubMed] [Google Scholar]
- 24.Alperin N, Oliu CJ, Bagci AM, et al. Low-dose acetazolamide reverses periventricular white matter hyperintensities in iNPH. Neurology 2014; 82:1347–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivkovic M, Reiss-Zimmermann M, Katzen H, et al. MRI assessment of the effects of acetazolamide and external lumbar drainage in idiopathic normal pressure hydrocephalus. Fluids Barriers CNS 2015; 12:9. [DOI] [PMC free article] [PubMed] [Google Scholar]