Abstract
AIM
To investigate the accuracy of fungal dysbiosis in mucosa and stool for predicting the diagnosis of Crohn’s disease (CD).
METHODS
Children were prospectively enrolled in two medical centers: one university hospital and one private gastroenterology clinic in the city of Riyadh, Kingdom of Saudi Arabia. The children with confirmed diagnosis of CD by standard guidelines were considered cases, and the others were considered non-inflammatory bowel disease controls. Mucosal and stool samples were sequenced utilizing Illumina MiSeq chemistry following the manufacturer’s protocols, and abundance and diversity of fungal taxa in mucosa and stool were analyzed. Sparse logistic regression was used to predict the diagnosis of CD. The accuracy of the classifier was tested by computing the receiver operating characteristic curves with 5-fold stratified cross-validation under 100 permutations of the training data partition and the mean area under the curve (AUC) was calculated.
RESULTS
All the children were Saudi nationals. There were 15 children with CD and 20 controls. The mean age was 13.9 (range: 6.7-17.8) years for CD children and 13.9 (3.25-18.6) years for controls, and 10/15 (67%) of the CD and 13/20 (65%) of the control subjects were boys. CD locations at diagnosis were ileal (L1) in 4 and colonic (L3) in 11 children, while CD behavior was non-stricturing and non-penetrating (B1) in 12 and stricturing (B2) in 3 children. The mean AUC for the fungal dysbiosis classifier was significantly higher in stools (AUC = 0.85 ± 0.057) than in mucosa (AUC = 0.71 ± 0.067) (P < 0.001). Most fungal species were significantly more depleted in stools than mucosal samples, except for Saccharomyces cerevisiae and S. bayanus, which were significantly more abundant. Diversity was significantly more reduced in stools than in mucosa.
CONCLUSION
We found high AUC of fungal dysbiosis in fecal samples of children with CD, suggesting high accuracy in predicting diagnosis of CD.
Keywords: Fungiome, Mycobiome, Crohn’s disease, Inflammation, Saudi children
Core tip: We found high accuracy of fungal dysbiosis in predicting diagnosis of Crohn’s disease (CD), a finding similar to bacterial dysbiosis. However, the higher area under the curve for the fungal dysbiosis classifier in stool (0.85 ± 0.057) than in mucosa (0.71 ± 0.067) (P < 0.001), contrasts with bacterial studies, suggesting higher accuracy of stool samples. Although the clinical application of this finding is limited at present by the high cost of fungal analysis, such information is important from a scientific viewpoint, to increase the understanding of the role of fungal flora in CD and to stimulate further studies.
INTRODUCTION
Inflammatory bowel diseases (IBDs), including Crohn’s disease (CD) and ulcerative colitis, are chronic conditions. Their incidence is highest, with an increasing trend, in Western populations[1,2]. However, their incidence is increasing in non-Western populations as well[3,4]. The cause of CD remains unknown despite extensive research, and a multifactorial etiology has been suggested. In genetically-susceptible individuals, environmental triggering factors play a major role and diet and microbiota are the most relevant causative factors for children[5]. Dietary components may act directly or through alteration of gut microbiota to initiate and maintain inflammation in susceptible subjects[6,7]. Significant fungal dysbiosis has been demonstrated in adults and children with CD[8-10]. Recent reports found high accuracy of bacterial dysbiosis in predicting the diagnosis of IBD in general and CD in particular[11-13].
Despite the demonstration of fungal dysbiosis in adults and children with CD, there are no similar reports on the potential role of fungal dysbiosis in the diagnosis of CD. The objective of this report was, therefore, to evaluate the accuracy of fungal dysbiosis in stool and mucosal samples, for the diagnosis of CD in a cohort of non-Western children with new onset disease.
MATERIALS AND METHODS
Ethical considerations
This report is a portion of the main study project titled “Characteristics of inflammatory bowel disease in Saudi children”. The study was reviewed and approved by the Institutional Review Board of the College of Medicine, King Saud University. Riyadh, Kingdom of Saudi Arabia (Approval number: 10/2647/IRB).
Study population, sample collection, storage, and processing
Children were prospectively enrolled in two medical centers: One university hospital and one private gastroenterology clinic in the same city of Riyadh, Kingdom of Saudi Arabia. The children were referred to these clinics for investigation of suspected IBD. The children with confirmed diagnosis of CD by standard guidelines[14] were considered cases and those in whom the diagnosis of CD was excluded were considered non-IBD controls. The most common final diagnoses in non-IBD controls were functional abdominal pain and polyps. Mucosal and fecal samples were collected from 15 children with confirmed CD and 20 controls without inflammation or infection. A total of 78 samples (58 from CD children and 20 from non-IBD controls) were obtained. Stool samples were collected before bowel preparation, and none afterward. Mucosal forceps biopsies were taken from various parts of the colon and ileum. All samples were put into cryovials without preservatives and transported immediately in ice to the laboratory and stored at -80 °C. At the end of the study, all samples were shipped by express mail in dry ice to MRDNA Laboratories (Shallowater, TX, United States) for microbiome analysis.
Fungal DNA extraction and sequencing
Fungal DNA was extracted using the Mobio PowerSoil kit as per the manufacturer’s instructions (Mobio, Carlsbad, CA, United States). Amplicon sequencing service (bTEFAP®) was performed at MRDNA Laboratories and used for the fungal analysis[15]. The internal transcribed spacer primers, ITS1F (CTTGGTCATTTAGAGGAAGTAA) and ITS2R (GCTGCGTTCTTCATCGATGC), were used. A single-step 30-cycle polymerase chain reaction (PCR with HotStart Taq Plus Master Mix kit (Qiagen, Valencia, CA, United States) was employed. Samples were sequenced utilizing Illumina MiSeq chemistry following the manufacturer’s protocols.
The Q25 sequence data derived from the sequencing process was processed using the MRDNA ribosomal and functional gene analysis pipeline (www.mrdnalab.com). Sequences were depleted of barcodes and primers, short sequences of < 150 bp were removed, and sequences with ambiguous base calls were removed. Operational taxonomic units were defined by clustering at 3% divergence (97% similarity), followed by removal of singleton sequences and chimeras. Final operational taxonomic units were taxonomically classified using BLASTn top hit analysis against a curated database derived from RDPII and NCBI (http://rdp.cme.msu.edu and www.ncbi.nlm.nih.gov, respectively) and compiled into each taxonomic level as both “counts” and “percentage” files.
Statistical analysis
All analyses were performed using Python and scikit-learn[16]. Custom functions implementing the permutation test were written to detect the taxa with abundances significantly different between CD and control samples. When more than one sample was available from the same patient for analysis, the log relative abundances from these samples were averaged.
It has been shown that variations in species abundance are better captured by a log-transformed than a linear scale, improving the statistical power[13]. Therefore, we followed this approach. In addition, rare taxa (< 1% abundance or absent from > 50% of the samples) were removed to improve the statistical power. Statistical significance was assessed via a permutation test (Fisher’s exact test), which yielded raw, uncorrected P-values. These were transformed into q-values (corrected P-value) that measure the probability of false discovery following the Benjamini Hochberg procedure[17]. We considered associations statistically significant only when the corrected P-values were less than 0.05.
A linear logistic regression classifier (linearmodel.LogisticRegression) in scikit-learn, Machine Learning in Python[16], was used to predict CD based on the subject’s microbiota. The accuracy of the classifier was tested by computing the receiver operating characteristic (ROC) curve with 5-fold stratified cross-validation under 100 permutations of the training data partition. We partitioned the data into randomly assigned training and test sets 100 times; in each case, the classifier was trained on 4/5 of the data and tested on 1/5 of the data (i.e. 5-fold cross-validation).
Alpha diversity, a measure of genera richness (number of genera), was evaluated using the Shannon index. We used Fisher’s t-test to determine P-values for alpha diversity.
The difference in community composition (Beta diversity) was quantified by the Bray-Curtis distance, which accounts for both patterns of presence-absence of taxa and changes in their relative abundances. Nonparametric multidimensional scaling (NMDS) was applied to visualize the distance between mucosa and stool samples taken from CD and control subjects. NMDS quantifies the dissimilarity in community composition between samples via a combination of presence-absence and absolute abundance of taxa. For the data shown, the separations were analyzed by the ANOSIM or analysis of (dis)similarity. The ANOSIM statistic compares the mean of ranked dissimilarities between groups to the mean of ranked dissimilarities within groups.
The statistical analyses in this article were performed and reviewed by the coauthors Kirill S Korolev, PhD and Rajita Menon, PhD from the Bioinformatics Program, Boston University, Boston, MA, United States.
RESULTS
Demographic data
All children were Saudi nationals. There were 15 CD children and 20 controls; mean age was 13.9 (range: 6.7-17.8) years for CD children and 13.9 (3.25-18.6) years for controls, and 10/15 (67%) of the CD and 13/20 (65%) of the control subjects were boys. At diagnosis, CD locations were ileal (L1) in 4 and ileocolonic (L3) in 11 children, while CD behavior was non-stricturing and non-penetrating (B1) in 12 and stricturing (B2) in 3 children. Controls included all patients with no evidence of IBD or other causes of inflammation. The final diagnoses in control subjects were juvenile polyps, recurrent abdominal pain, recurrent cyclic vomiting or other functional gastrointestinal disorders.
Fungal dysbiosis
In a previous report, description of fungal community structure in mucosa and stool in children with CD relative to controls indicated significantly-abundant CD-associated taxa included Psathyrellaceae (P = 0.01), Cortinariaceae (P = 0.04), Psathyrella (P = 0.003), and Gymnopilus (P = 0.03). Monilinia was significantly depleted (P = 0.03), whereas other depleted taxa, although not statistically significant, included Leotiomycetes (P = 0.06), Helotiales (P = 0.08), and Sclerotiniaceae (P = 0.07)[10].
Prediction analysis
The mean area under the ROC curve (AUC) for the fungal dysbiosis classifier is illustrated in Figure 1, indicating a significantly-higher AUC in stools (0.85 ± 0.057) than in mucosa (0.71 ± 0.067) (P < 0.001).
Figure 1.
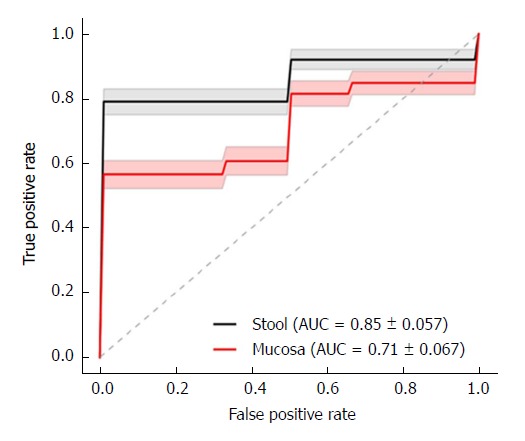
Dysbiosis score classification curve to distinguish between children with Crohn’s disease and controls. The receiver operating characteristic (ROC) curve demonstrated the logistic regression dysbiosis classifier. The mean ROC curve for stool (solid black line) and mucosal (solid red line) dysbiosis scores in fungi cohorts is shown. The standard deviation from 100 permutations is shown in gray and light red shading. The area under the curve for stool dysbiosis is significantly higher than in mucosal dysbiosis, using a Fisher’s t-test computed P < 0.001.
This analysis was further expanded to demonstrate the difference in abundance and diversity between mucosa and stool in controls and children with CD separately. Table 1 shows a comparison of fungal abundance between mucosa and stools in controls, indicating that only two species, Volvariella dunensis (P = 0.03) and Lepraria humida (P = 0.04), were significantly less abundant in stool than in mucosal samples. In contrast, about 50 species were significantly less abundant in stools of children with CD (P < 0.05) and only two species, Saccharomyces cerevisiae (P = 0.02) and S. bayanus (P = 0.001), were significantly more abundant in stools than in mucosal samples (Table 2). Alpha diversity, as measured by the Shannon Index and illustrated in Figure 2, was different in mucosa and stool. The stool community for children with CD was more than 5 times less diverse than that of mucosa (P = 0.0001), whereas in controls the reduction in stool diversity was statistically not significant (P = 0.35). Beta diversity, as measured by the Bray-Curtis distance and visualized by the NMDS in Figure 3, shows a significant difference in fungal community separation between mucosal and stool samples (P = 0.005).
Table 1.
Fungal species abundance in mucosa and stool of controls
| Fungal species | Mucosa abundance, % | Stool abundance, % | Ratio | P value |
| Volvariella dunensis | 0.027 | 0.0013 | 0.047 | 0.036 |
| Lepraria humida | 0.015 | 0.0010 | 0.063 | 0.042 |
Table 2.
Fungal species abundance in mucosa and stool of children with Crohn’s disease
| Fungal species | Mucosa abundance, % | Stool abundance, % | Ratio | P value |
| Volvariella dunensis | 0.06 | 0.0014 | 0.025 | < 0.001 |
| Malassezia restricta | 0.09 | 0.0044 | 0.049 | < 0.001 |
| Ceriporia lacerate | 0.03 | 0.0022 | 0.065 | < 0.001 |
| Cl. Cladosporioides | 0.11 | 0.004 | 0.035 | < 0.001 |
| Trametes hirsute | 0.048 | 0.0043 | 0.089 | < 0.001 |
| Psathyrella artemisiae | 0.44 | 0.028 | 0.065 | < 0.001 |
| Amyloporia sp | 0.027 | 0.0012 | 0.046 | < 0.001 |
| Irpex sp | 0.095 | 0.0019 | 0.02 | < 0.001 |
| Bjerkandera adusta | 0.022 | 0.0011 | 0.049 | < 0.001 |
| Lepista sordida | 0.041 | 0.0015 | 0.037 | < 0.001 |
| Cerrena sp | 0.022 | 0.0011 | 0.05 | < 0.001 |
| Coprinellus radians | 0.034 | 0.0011 | 0.032 | < 0.001 |
| Phlebia acanthocystis | 0.022 | 0.0011 | 0.048 | < 0.001 |
| Leptosphaerulina sp | 0.037 | 0.0013 | 0.036 | < 0.001 |
| Coprinus sp | 0.085 | 0.0019 | 0.022 | < 0.001 |
| Malassezia globose | 0.028 | 0.0026 | 0.095 | < 0.001 |
| Alternaria alternate | 0.054 | 0.0037 | 0.069 | < 0.001 |
| Ramalinopsis mannii | 0.066 | 0.0018 | 0.027 | < 0.001 |
| Saccharomyces bayanus | 2.5 | 53 | 21 | < 0.001 |
| Trichoderma hypocrea | 0.022 | 0.0014 | 0.067 | < 0.001 |
| Aspergillus penicillioides | 0.11 | 0.0066 | 0.062 | < 0.001 |
| Psathyrella candolleana | 0.063 | 0.0059 | 0.093 | < 0.001 |
| Cladosporium sp | 0.072 | 0.0053 | 0.074 | < 0.001 |
| Aspergillus sp | 0.064 | 0.0049 | 0.076 | < 0.001 |
| Galactomyces geotrichum | 0.079 | 0.0063 | 0.08 | < 0.001 |
| Peniophora incarnate | 0.011 | 0.0027 | 0.25 | < 0.001 |
| Eutypella sp | 0.038 | 0.0044 | 0.12 | < 0.001 |
| Ophiocordyceps sinensis | 0.059 | 0.004 | 0.068 | < 0.001 |
| Nakaseomyces candida | 0.12 | 0.0058 | 0.049 | 0.019 |
| Hypocrea ceramic | 0.02 | 0.0037 | 0.18 | 0.019 |
| Saccharomyces cerevisiae | 0.043 | 0.23 | 5.3 | 0.024 |
Figure 2.
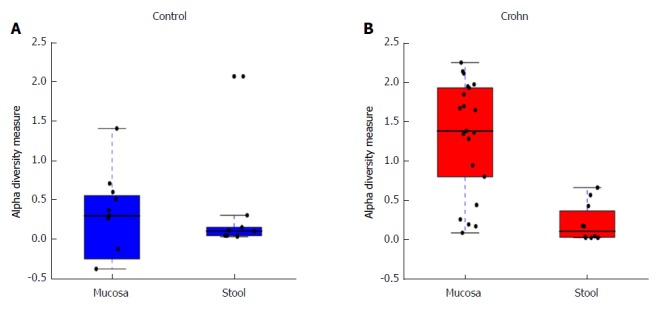
Alpha diversity evaluated by the Shannon Index. Comparison between mucosa and stool in children with Crohn’s disease (CD) and controls. Alpha diversity is significantly lower in stool than in mucosa in CD samples (P = 0.0001), while the difference is not significant in control samples (P = 0.35).
Figure 3.
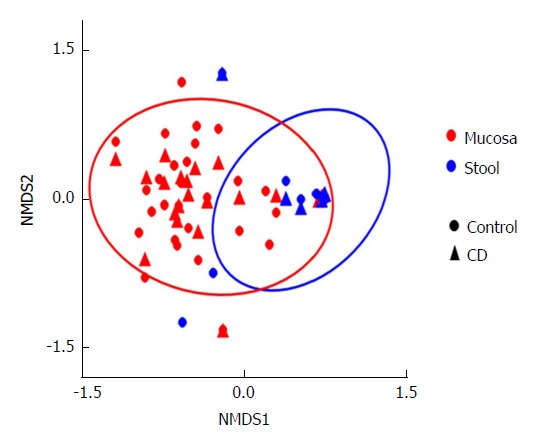
Beta diversity evaluated by Bray-Curtis distance and illustrated by the NMDS plot. This is a measure of beta diversity, and quantifies the similarity in abundance of taxa between samples. The ovals contain 95% of the probabilities for mucosa and stool. This plot shows a clear separation of mucosa and stool samples (P = 0.005). NMDS: Nonparametric multidimensional scaling; CD: Crohn’s disease.
DISCUSSION
Microbial dysbiosis in the form of depletion of beneficial organisms, expansion of harmful organisms, and reduced microbial diversity may occur independently or concurrently and result in significant effects on immune responses[18]. Fungal dysbiosis demonstrated in Saudi children with CD is in line with previous studies[19,20].
Key findings
This is the first report on the accuracy of fungal dysbiosis in mucosa and stools in predicting the diagnosis of CD in children. The main finding in this study is the high AUC for the fungal dysbiosis classifier, suggesting high accuracy in predicting the diagnosis of CD, which is in line with the high AUC for bacterial dysbiosis[11-13]. However, the higher AUC for the fungal dysbiosis classifier is in stool (0.85 ± 0.057) rather than in mucosa (0.71 ± 0.067) (P < 0.001), in contrast with the higher AUC for bacterial dysbiosis in mucosal samples[12-13]. Since this is the first report on fungal dysbiosis accuracy in predicting the diagnosis of CD, further studies are needed to clarify this result.
The finding of significant differences in a large number of species between stool and mucosa in CD children compared to much smaller numbers of species in controls reflects the degree of disturbance of the fungal community in children with CD. In this study, except for S. cerevisiae and S. bayanus, which were significantly more abundant in stool samples (P = 0.02 and P = 0.003, respectively), the significantly lower abundance of most fungal species in stool than in mucosal samples indicates variation in dysbiosis between stools and mucosa and supports the variable accuracy in predicting the diagnosis of CD.
It has been suggested that the presence of some fungi in the gut may reflect environmental exposure, rather than colonization of the mucosa[21]. However, reports indicate that many fungi, such as Candida, Cryptococcus, Malassezia, Saccharomyces and Trichosporon spp, have been isolated from the stool of humans[22,23]. Furthermore, the role of fungi in the pathogenesis of IBD has been suggested based on animal models of colitis. Pattern recognition receptors in the innate immune system cells include dectin-1, dectin-2, DC-SIGN, mannose receptor, and mannose receptor lecithin[24], and mice lacking dectin-1 had increased susceptibility to experimental colitis[25]. In addition, treatment with antifungal drugs may reduce the inflammation[26].
Strengths and weaknesses of this study
The strengths of this study comprise the facts that it is the first to investigate the subject and provides inclusion of newly-diagnosed, treatment-naïve children with CD and controls from a well-defined population. Weaknesses include the relatively small number of cases which may be adequate for a first report. The controls were not completely healthy. Obviously, performing endoscopy and biopsies to exclude IBD in healthy children is unethical. Therefore, children who are free of IBD, infection and inflammation have been considered appropriate non-IBD controls.
In conclusion, the most important finding in this study is the high AUC in fecal samples of children with CD, suggesting high accuracy in predicting the diagnosis. Although, the clinical application of this finding is limited at present by the high cost of fungal analysis, such information is important from a scientific viewpoint, to increase the understanding of the role of fungal flora in CD and to stimulate further research, possibly leading to a “dysbiosis test” as a noninvasive screening tool for CD.
ARTICLE HIGHLIGHTS
Research background
Bacterial dysbiosis has been reported to predict the diagnosis of Crohn’s disease (CD), but no similar reports for fungal dysbiosis exist. The study is of scientific significance to stimulate further research important for further clarification of the role of fungi in CD.
Research motivation
The role of microbiota in CD, bacterial or fungal, is of worldwide research interest. However, a key problem to be solved is whether dysbiosis is the cause or the result of inflammation. Solving this problem may facilitate discovery of new microbiota-based treatment options. Regarding the accuracy of fungal dysbiosis in predicting the diagnosis, the main problem would be the current high cost of fungal analysis. Solving this problem could lead to development of a dysbiosis screening test for CD.
Research objectives
The objective of this study was to evaluate the accuracy of intestinal fungal dysbiosis as a predictor of CD. High accuracy was found. Future research is needed to confirm this finding and to develop low-cost fungal dysbiosis tests.
Research methods
Mucosal and stool samples were collected from children with CD at presentation and controls. Fungal DNA was extracted from theses samples and sequencing was performed. Fungal abundance and diversities were determined. Fungal dysbiosis in children with CD was demonstrated. This is the first study of the accuracy of fungal dysbiosis in predicting the diagnosis of CD in children.
Research results
The main finding was the high accuracy of fungal dysbiosis in predicting diagnosis of CD. This should stimulate further research to confirm our findings and to develop a low-cost dysbiosis test.
Research conclusions
The high accuracy of fungal dysbiosis in predicting the diagnosis of CD is a new finding. The finding could lead to further research in the role of fungal dysbiosis in CD. A new theory suggests the possibility of design of a noninvasive fungal dysbiosis screening test for CD.
Research perspectives
The role of microbiota in CD may include development of a noninvasive screening test. Further research is needed to confirm the findings and to develop low-cost fungal analysis.
ACKNOWLEDGMENTS
The authors extend their appreciations to the Deanship of Scientific Research at King Saud University in Riyadh, Kingdom of Saudi Arabia for funding this work through Research Group No [RGP-1436-007]. This work was also supported by a grant from the Simons Foundation [No.409704] to Kirill Korolev) and by the startup fund from Boston University to Kirill Korolev. Simulations were carried out on Shared Computing Cluster at Boston University. Rajita Menon was partially supported by a Hariri Graduate Fellowship from Boston University. Harland Winter, MD received support from Martin Schlaff and the Diane and Dorothy Brooks Foundation.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Saudi Arabia
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: The manuscript is part of the study of the characteristics of inflammatory bowel disease in Saudi children approved by the IRB of the College of Medicine, King Saud University (No: 10/2647/IRB dated 29/6/2010).
Informed consent statement: Parents and/or children gave consent and/or assent to participate in the study before enrollment.
Conflict-of-interest statement: All authors declare no conflict of interest related to this study.
STROBE statement: The authors have read the STROBE Statement-checklist of items, and the manuscript was prepared and revised according to the STROBE Statement - checklist items
Peer-review started: July 2, 2018
First decision: July 12, 2018
Article in press: October 5, 2018
P- Reviewer: Day AS, Gazouli M S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Huang Y
Contributor Information
Mohammad I El Mouzan, Department of Pediatrics, King Saud University, Riyadh 11461, Saudi Arabia. drmouzan@gmail.com.
Kirill S Korolev, Bioinformatics Program, Boston University, Boston, MA 02215, United States.
Mohammad A Al Mofarreh, Al Mofarreh PolyClinic, Riyadh 11423, Saudi Arabia.
Rajita Menon, Bioinformatics Program, Boston University, Boston, MA 02215, United States.
Harland S Winter, MassGeneral Hospital for Children, Boston, MA 02114, United States.
Ahmad A Al Sarkhy, Department of Pediatrics, King Saud University, Riyadh 11461, Saudi Arabia.
Scot E Dowd, MRDNA, Shallowater, TX 79363, United States.
Ahmad M Al Barrag, Department of Microbiology, King Saud University, Riyadh 11461, Saudi Arabia.
Asaad A Assiri, Department of Pediatrics, Supervisor of Prince Abdullah Bin Khalid Celiac Disease Research Chair, King Saud University, Riyadh 11461, Saudi Arabia.
References
- 1.Kappelman MD, Moore KR, Allen JK, Cook SF. Recent trends in the prevalence of Crohn’s disease and ulcerative colitis in a commercially insured US population. Dig Dis Sci. 2013;58:519–525. doi: 10.1007/s10620-012-2371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burisch J, Pedersen N, Čuković-Čavka S, Brinar M, Kaimakliotis I, Duricova D, Shonová O, Vind I, Avnstrøm S, Thorsgaard N, et al. East-West gradient in the incidence of inflammatory bowel disease in Europe: the ECCO-EpiCom inception cohort. Gut. 2014;63:588–597. doi: 10.1136/gutjnl-2013-304636. [DOI] [PubMed] [Google Scholar]
- 3.Benchimol EI, Fortinsky KJ, Gozdyra P, Van den Heuvel M, Van Limbergen J, Griffiths AM. Epidemiology of pediatric inflammatory bowel disease: a systematic review of international trends. Inflamm Bowel Dis. 2011;17:423–439. doi: 10.1002/ibd.21349. [DOI] [PubMed] [Google Scholar]
- 4.El Mouzan MI, Saadah O, Al-Saleem K, Al Edreesi M, Hasosah M, Alanazi A, Al Mofarreh M, Asery A, Al Qourain A, Nouli K, et al. Incidence of pediatric inflammatory bowel disease in Saudi Arabia: a multicenter national study. Inflamm Bowel Dis. 2014;20:1085–1090. doi: 10.1097/MIB.0000000000000048. [DOI] [PubMed] [Google Scholar]
- 5.Leone V, Chang EB, Devkota S. Diet, microbes, and host genetics: the perfect storm in inflammatory bowel diseases. J Gastroenterol. 2013;48:315–321. doi: 10.1007/s00535-013-0777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman-Kiddell CA, Davies PS, Gillen L, Radford-Smith GL. Role of diet in the development of inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:137–151. doi: 10.1002/ibd.20968. [DOI] [PubMed] [Google Scholar]
- 7.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liguori G, Lamas B, Richard ML, Brandi G, da Costa G, Hoffmann TW, Di Simone MP, Calabrese C, Poggioli G, Langella P, et al. Fungal Dysbiosis in Mucosa-associated Microbiota of Crohn’s Disease Patients. J Crohns Colitis. 2016;10:296–305. doi: 10.1093/ecco-jcc/jjv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Mouzan M, Wang F, Al Mofarreh M, Menon R, Al Barrag A, Korolev KS, Al Sarkhy A, Al Asmi M, Hamed Y, Saeed A, et al. Fungal Microbiota Profile in Newly Diagnosed Treatment-naïve Children with Crohn’s Disease. J Crohns Colitis. 2017;11:586–592. doi: 10.1093/ecco-jcc/jjw197. [DOI] [PubMed] [Google Scholar]
- 11.Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, Giannoukos G, Ciulla D, Tabbaa D, Ingram J, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One. 2012;7:e39242. doi: 10.1371/journal.pone.0039242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Kaplan JL, Gold BD, Bhasin MK, Ward NL, Kellermayer R, Kirschner BS, Heyman MB, Dowd SE, Cox SB, et al. Detecting Microbial Dysbiosis Associated with Pediatric Crohn Disease Despite the High Variability of the Gut Microbiota. Cell Rep. 2016;14:945–955. doi: 10.1016/j.celrep.2015.12.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine A, Koletzko S, Turner D, Escher JC, Cucchiara S, de Ridder L, Kolho KL, Veres G, Russell RK, Paerregaard A, et al. ESPGHAN revised porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents. J Pediatr Gastroenterol Nutr. 2014;58:795–806. doi: 10.1097/MPG.0000000000000239. [DOI] [PubMed] [Google Scholar]
- 15.Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- 18.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukhopadhya I, Hansen R, Meharg C, Thomson JM, Russell RK, Berry SH, El-Omar EM, Hold GL. The fungal microbiota of de-novo paediatric inflammatory bowel disease. Microbes Infect. 2015;17:304–310. doi: 10.1016/j.micinf.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD. Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015;21:1948–1956. doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suhr MJ, Hallen-Adams HE. The human gut mycobiome: pitfalls and potentials--a mycologist’s perspective. Mycologia. 2015;107:1057–1073. doi: 10.3852/15-147. [DOI] [PubMed] [Google Scholar]
- 22.Hallen-Adams HE, Kachman SD, Kim J, Legg RM, Martinez I. Fungi inhabiting the healthy human gastrointestinal tract: A diverse and dynamic community. Fungal Ecol. 2015;15:9–17. [Google Scholar]
- 23.Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 2017;5:153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang ZK, Yang YS, Stefka AT, Sun G, Peng LH. Review article: fungal microbiota and digestive diseases. Aliment Pharmacol Ther. 2014;39:751–766. doi: 10.1111/apt.12665. [DOI] [PubMed] [Google Scholar]
- 25.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zwolinska-Wcislo M, Brzozowski T, Budak A, Kwiecien S, Sliwowski Z, Drozdowicz D, Trojanowska D, Rudnicka-Sosin L, Mach T, Konturek SJ, et al. Effect of Candida colonization on human ulcerative colitis and the healing of inflammatory changes of the colon in the experimental model of colitis ulcerosa. J Physiol Pharmacol. 2009;60:107–118. [PubMed] [Google Scholar]


