Summary
Plants can re‐programme their transcriptome, proteome and metabolome to deal with environmental and biotic stress. It has been shown that the rhizosphere microbiome has influence on the plant metabolome and on herbivore behaviour. In the present study, Trichoderma gamsii was isolated from Arabidopsis thaliana rhizosphere soil. The inoculation of roots of Arabidopsis thaliana with T. gamsii significantly inhibited the feeding behaviour of Trichoplusia ni and affected the metabolome as well as the content of phytohormones in Arabidopsis leaves. T. gamsii‐treated plant leaves had higher levels of amino acids and lower concentrations of sugars. In addition, T. gamsii‐treated plant leaves had more abscisic acid (ABA) and lower levels of salicylic acid (SA) and indole‐3‐acetic acid (IAA) in comparison with the untreated plants. Furthermore, the inoculation with T. gamsii on different signalling mutants showed that the induction of defences were SA‐dependent. These findings indicate that T. gamsii has potential as a new type of biocontrol agent to promote plant repellence to insect attacks.
Introduction
The change of metabolite profile (Walling, 2000; War et al., 2012) is one of the strategies used by plants to defend against biotic and abiotic stresses. Many studies have indicated that plant‐beneficial microbe interactions can alter the plant metabolite profile and improve plant defence to insect attacks (van de Mortel et al., 2012; Estrada et al., 2013). Trichoderma species play an important role in suppression of plant disease by directly antagonizing with root and foliar pathogens as well as by inducing resistance in plants. The Trichoderma‐induced resistance in plants leads to physiological and biochemical changes including enhanced defence enzymatic activity, upregulation of defence‐related genes, and changes in metabolic profiles (Mastouri et al., 2010; Contreras‐Cornejo et al., 2011; Carreras‐Villasenor et al., 2012). A recent study showed that Trichoderma harzianum T22 can enhance the production of volatile organic compounds (VOCs) in tomato leading to an increased attractiveness towards aphid parasitoids (Coppola et al., 2017). A similar study showed that T. atroviride triggers maize defences against the insect herbivore S. frugiperda by the increased emission of volatile terpenes and accumulation of jasmonic acid (JA) (Contreras‐Cornejo et al., 2018).
Phytohormones, including JA, ethylene (ET), salicylic acid(SA), abscisic acid (ABA), auxin, cytokinins (CK), brassinosteroids (BR) and gibberllins (GB), have been reported to play important roles in both plant‐insect and plant‐microbe interactions (Erb et al., 2012; Lazebnik et al., 2014). These phytohormones function as signal transducers in plant‐induced defence pathways. For instance, JA and SA are the two major regulators of local and systemic defence responses (Pieterse et al., 2012; Soler et al., 2013). Other hormones, such as ET and ABA also modify the plant defence responses (van Loon et al., 2006; Ton et al., 2009; Robert‐Seilaniantz et al., 2011). Recently, it was found that application of the combination of JA and SA led to metabolic changes in the leaves and phloem of Plantago lanceolata and altered herbivory behaviour (Schweiger et al., 2014). Similar studies have shown that the phytohormone profiles of Trichoderma‐treated plants were modified leading to defence and growth promotion activity (Contreras‐Cornejo et al., 2011; Mathys et al., 2012; Martinez‐Medina et al., 2013). Co‐inoculation of T. virens and T. atroviride to Arabidopsis roots accumulated the canonical defence hormones SA and JA as well as camalexin, which conferred resistance against Botrytis cinerea (Contreras‐Cornejo et al., 2011).
Trichoplusia ni (Lepidoptera: Noctuidae), also known as the cabbage looper moth, has a broad range of host plants (Sutherland and Greene, 1984) and is a major and devastating pest in vegetable growing areas worldwide. To date, the control of T. ni excessively depends on the use of chemical insecticides which can damage the environment. In addition, the development of resistance to insecticides is increasing (Soares and Porto, 2007; Weddle et al., 2009). Application of biocontrol agents is an alternative to control this insect and other pests. Therefore, the purpose of this work was to characterize the components of a rhizosphere microbiome that was previously reported to have potential impact on the metabolome profile of Arabidopsis and negatively affected T. ni larvae feeding (Badri et al., 2013). In the present study, we isolated a fungus (Trichoderma gamsii) from Arabidopsis thaliana rhizosphere soil that altered the metabolome of Arabidopsis and negatively affected Trichoplusia ni larvae feeding when it was inoculated to the soil in close proximity to roots. We further studied the levels of phytohormones and plant metabolites in Arabidopsis to unravel the mechanisms behind the change of larvae feeding behaviour by T. gamsii.
Results
Fungal isolates altered herbivore feeding behaviour
A previous study showed that the microbiome from the Arabidopsis soil enhanced the plant resistance to T. ni herbivore (Badri et al., 2013). To determine if fungi and/or bacteria from the Arabidopsis soil played a role in changing the metabolome of Arabidopsis leaves, we treated the plants with whole slurry (containing all microbes), fungi‐enrichment‐slurry or filtered slurry (all microbes removed). We found the T. ni larval weights were significantly reduced when fed on plants treated with whole slurry and fungi‐enrichment‐slurry compared to those treated with filtered slurry and control plants (no treatment) (Fig. 1). Moreover, plants treated with whole slurry and fungi‐enrichment‐slurry did not have significant differences on gained larval weights. These findings indicate that fungi in the Arabidopsis soil played a key role in modulating the feeding behaviour of T. ni. Thus, we isolated fungal cultures from the soil slurry.
Figure 1.
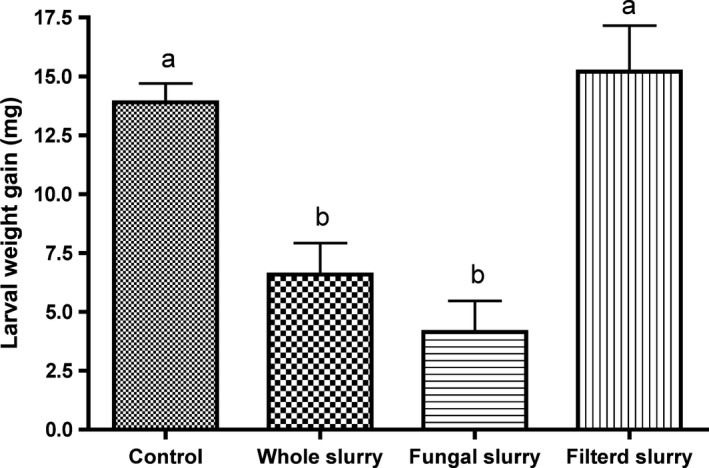
Larval gained weight of Trichoplusia ni fed for 24 h on four‐week‐old Arabidopsis plants amended with soil microbiomes. The control treatment received only Hoagland's solution. Each treatment had 18 pots (repetitions) and each pot contained one plant. The values represent the means ± SEM. Bars with dissimilar letters are significantly different (P < 0.05; Tukey's honest significance test).
In total, 13 fungal isolates were obtained from the Arabidopsis soil and were screened by applying them individually to Arabidopsis roots to test the effects on the herbivore behaviour of T. ni. The relative gained weight of T. ni fed on Arabidopsis plants, inoculated with these fungal isolates individually, revealed that these fungal isolates have different effects on herbivore behaviour of T. ni (Fig. S1). Some isolates (e.g. F4‐2, F7, F14, and F16) showed positive effects on the gained weight of T. ni; while the isolate F18 had negative effects on the gained weight of T. ni. Meanwhile, the rest of the fungal isolates (e.g. F1, F3‐1, F10, F13, F15, F17 and F19) didn't have significant effects on the gained weight of T.ni. The fungal isolate F18 was selected for further experimentation because it consistently showed significant decrease of the larval weight gain of T. ni (Fig. S1).
We repeated the experiment four times to confirm the effects of F18 on the larval weight gain of T. ni. The results from these experiments showed the treatment with F18 had a significantly lower (40% decrease) larval weight gain compared to that of the control treatment (Fig. 2).
Figure 2.
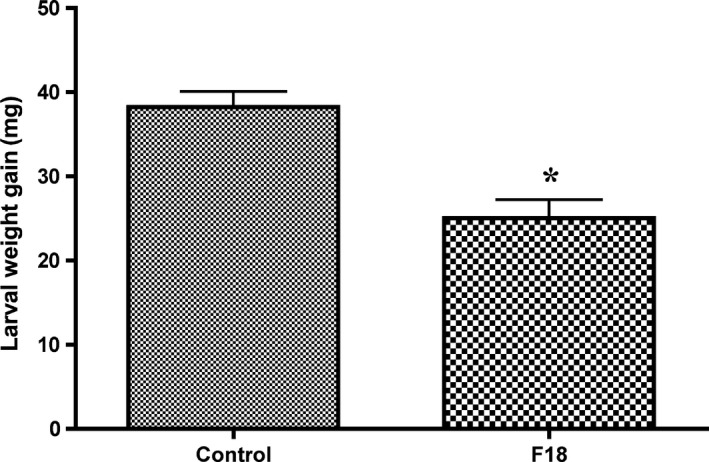
Larval gained weight of Trichoplusia ni fed for 24 h on four‐week‐old Arabidopsis Col‐0 plants amended with fungal isolate F18. The control treatment received only Hoagland's solution. Each treatment had 18 pots and each pot contained four plants. This experiment was repeated four times. All experiments showed the same trend. Data in this figure represent the results of one experiment. The values represent the means ± SEM. The asterisk above the bar indicates significance relative to the control at p < 0.05 level (t‐test).
Identification of the selected fungus by ITS sequencing
The highly conserved region of fungal rRNA gene of F18 was amplified and sequenced. The sequence was used in a BLAST (blastn) search using the NCBI nucleotide databases. The fungal isolate F18 was identified as Tricoderma gamsii with 100% sequence identity to Trichoderma gamsii strain DAOM 231637 (GenBank Accession: EU280129). Thus, we name the selected fungal isolate F18 as Trichoderma gamsii F18.
Influence of T. gamsii F18 on the leaf metabolome
The influence of T. gamsii F18 on the leaf metabolites of Arabidopsis was analysed by GC‐MS. Based on GC‐MS analyses, 524 putative compounds were detected, and 177 compounds were annotated. Principle component analysis (PCA) showed that the metabolome of plants inoculated with T. gamsii F18 formed a distinct cluster in comparison with that of control plants (Fig. 3A). There were no significant differences in the contents of phenolic (t‐test, P = 0.23) and organic acids (t‐test, P = 0.27) between T. gamsii F18‐treated and untreated plants (Fig. 3B). However, the inoculation with T. gamsii F18 showed significantly lower levels of total sugars, sugar alcohols, other compounds including amide and amine, and unknown compounds (t‐test, P < 0.05). On the contrary, the control plants exhibited less amino acids and their derivatives (t‐test, P < 0.05) than plants inoculated with T. gamsii F18. Among the 27 detected amino acids, thirteen amino acids (tyrosine, tryptophan, serine, oxoproline, ornithine, phenylalanine, lysine, glutamine, glutamic acid, cyanoalanine, citrulline, aspartic acid and asparagine) increased their contents by more than 50% after T. gamsii F18 inoculation, while five amino acids (valine, N‐methylalanine, homoserine, beta‐alanine and alanine) were reduced by 25%–73% compared with the control plants (Fig. 3C).
Figure 3.
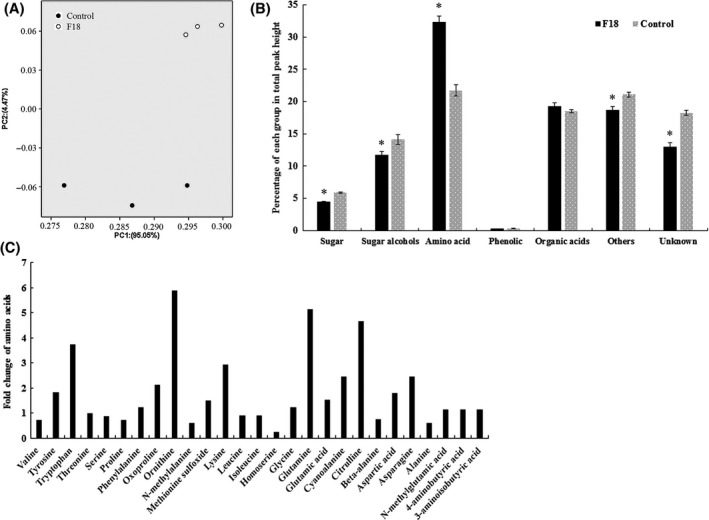
Leaf metabolites of Arabidopsis analysed by GC‐MS.
A. Principal Component Analysis (PCA) of the leaf metabolites of Arabidopsis inoculated with and without T. gamsii F18.
B. Metabolomics features detected by GC‐MS were categorized into six groups to generate a cumulative peak height for each group. The six groups included sugar, sugar alcohols, amino acid, phenolics, other compounds (fatty acid, amide and amine) and unknown (uncategorized compounds). The values represent the means SEM. The asterisks above the bars indicate significance relative to the control at P < 0.05 level (t‐test).
C. Fold change of amino acids in T. gamsii F18‐treated leaves compared with control plants.
Influence of T. gamsii F18 on the content of plant phytohormones
Five phytohormones including indole‐3‐carboxylic acid (ICOOH), indole‐3‐acetic acid (IAA), jasmonic acid (JA), abscisic acid (ABA) and salicylic acid (SA) in Arabidopsis plants were detected and analysed by LC‐MS. As shown in Table 1, the inoculation with T. gamsii F18 resulted in different levels of phytohormones in Arabidopsis plants. For instance, the inoculation with T. gamsii F18 significantly increased the content of ABA (28% higher than that of control), but significantly decreased the content of IAA and SA (19% and 21% less than that of control, respectively). Furthermore, no significant effects on the contents of ICOOH and JA were found by the inoculation of T. gamsii F18.
Table 1.
Phytohormones in Arabidopsis leaf tissue inoculated with T. gamsii F18 and control
| Indole‐3‐carboxylic acid (ng in extract) | Indole‐3‐acetic acid (ng in extract) | Jasmonic acid (ng in extract) | Abscisic acid (ng in extract) | Salicylic acid (ng in extract) | |
|---|---|---|---|---|---|
| Control | 33.78 ± 2.30 | 77.74 ± 5.03 | 26.91 ± 6.33 | 42.71 ± 1.72 | 629.01 ± 1.80 |
| F18 | 36.10 ± 8.55 | 56.18 ± 1.70* | 21.01 ± 2.92 | 50.77 ± 4.59* | 498.16 ± 7.90* |
The values represent the means ± SEM. The asterisks above numbers indicate significance relative to the control at the P < 0.05 level (t‐test).
Insect performance on signalling mutants
We compared the difference of larval‐herbivore performance on ein2‐1 (impaired in ET signalling pathway), NahG and sid2‐1 (impaired in SA signalling pathway) plants after treatment with T. gamsii F18. The inoculation of ein2‐1 with T. gamsii F18 resulted in 35% reduction in larval weight gain of T. ni compared to that of control (ein2‐1 without T. gamsii F18 inoculation). In contrast, no significant difference in larval weight gain was found on NahG and sid2‐1 plants inoculated with T. gamsii F18 compared to un‐inoculated controls (Fig. 4).
Figure 4.
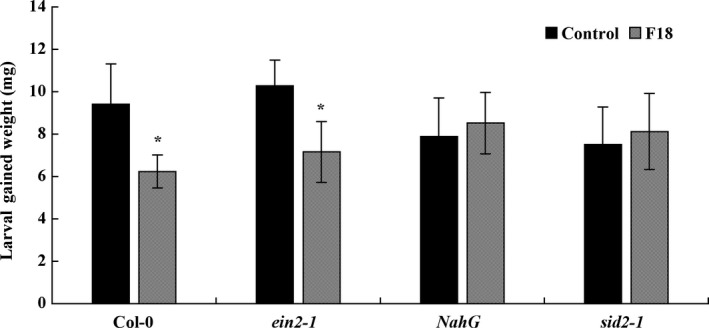
Larval gained weight of Trichoplusia ni fed for 24 h on four‐week‐old Arabidopsis Col‐0, ein2‐1, NahG and sid2‐1 plants amended with T. gamsii F18 a week prior to insect herbivory. The control treatment received only Hoagland's solution. The values represent the means ± SEM. The asterisks above the bars indicate significance relative to the control at P < 0.05 level (t‐test).
Discussion
In the present study, we isolated 13 culturable fungal isolates from Arabidopsis soil and selected the fungus T. gamsii F18 which was found to modify the metabolome of Arabidopsis plants to make them less attractive to insects. Although other fungi present in the soil could have the potential to induce similar defence responses against T. ni, we only studied the culturable fungal isolates in this publication. Previous studies have shown that Trichoderma sp. have beneficial effects on plants. For instance, there are commercial agricultural products containing T. atroviride, T. harzianum and T. hamatum (Ha, 2010) that promote plant growth (Chang et al., 1986; Gravel et al., 2007; Contreras‐Cornejo et al., 2009), reduce the severity of plant diseases (Naseby et al., 2000; Nawrocka and Malolepsza, 2013) and enhance tolerance to abiotic stresses (Bae et al., 2009; Montero‐Barrientos et al., 2010; Dixit et al., 2011; Contreras‐Cornejo et al., 2014). In addition, Trichoderma sp. can control phytopathogenic nematodes such as Rotylenchulus reniformis, Meloidogyne javanica and Meloidogyne cionopagum (Sahebani and Hadavi, 2008; Bokhari, 2009). It also has been documented as a bio‐fungicide on flowers, ornamentals, vegetables, aromatic herbs and strawberries by the European Food Safety Authority (European Food Safety Authority, 2013). Our findings showed that the inoculation of Arabidopsis roots with T. gamsii F18 affected T. ni herbivory feeding behaviour on Arabidopsis leaves.
T. gamsii F18 altered plant metabolome
Plant metabolites are modified by associated microbes such as endophytic fungi, beneficial soil‐borne microbes and plant growth promoting rhizobacteria (Pineda et al., 2010; van de Mortel et al., 2012; Estrada et al., 2013). In the Trichoderma‐plant interaction, it has been shown that the fungus re‐programmes the plant's transcriptome, proteome and metabolome leading to enhancement of the plant's ability to tolerate different types of stresses (Alfano et al., 2007; Shoresh et al., 2010).
In the present study, we observed that the T. gamsii F18‐treated leaves had less sugars than control plants. Sugars are essential for plant growth and development due to their roles as carbon and energy sources, signals and putative defence priming agents (Koch, 2004; Gomez‐Ariza et al., 2007). T. gamsii F18 may mediate the T. ni feeding behaviour by diminishing the attractiveness of the insect for primary metabolites such as sugars (Juniper and Richard, 1986). Another possibility is that T. gamsii F18 may be able to intensify the plant's immune reactions by reducing the sugar pools in favour of defence. Plant defence responses require the primary metabolites to support cellular energy and impose a fitness cost (Heil and Baldwin, 2002; Kangasjarvi et al., 2012).
Increasing evidence shows that amino acid signalling pathways may crosstalk with biotic and abiotic signalling networks in plants. Transcriptomics and metabolomics approaches used in Arabidopsis revealed that amino acids are differentially produced in Arabidopsis in response to pathogen and herbivory attack (Rojas et al., 2014). The biosynthesis of tryptophan, glutamine and glutamic acid are induced in herbivore‐infested plants. The overproduction of these amino acids seems to serve as precursors for inducible defence metabolites (Brotman et al., 2012; Rojas et al., 2014; Zhou et al., 2015). In our study, the leaves of T. gamsii F18‐treated plants had more tyrosine, tryptophan, serine, oxoproline, ornithine, phenylalanine, lysine, glutamine, glutamic acid, cyanoalanine, citrulline, aspartic acid and asparagine compared to the control plants. These amino acids are likely to account for plant defence in multiple manners.
In plant–herbivore interactions, amino acids function both as major growth‐limiting nutrients and as precursors for the generation of plant defence compounds. For example, tryptophan plays a critical role in regulating plant responses to biotic and abiotic stresses (Radwanski and Last, 1995; Zhao and Last, 1996; Sangha et al., 2011). Transcript profiling data generally show herbivory‐induced changes in the expression of genes related to the production of glutamine, glutamic acid, asparagine and aspartic acid (Thompson and Goggin, 2006; Caldana et al., 2011; Appel and Cocroft, 2014; Zhou et al., 2015), which are involved in nitrogen assimilation (Coruzzi and Last, 2000). Amino acids also play a role in signal transduction to induce defence gene expression. Glutamine plays a signalling role in the synthesis of other amino acids and regulates the expression of genes involved in plant immunity (Kan et al., 2015). Other amino acids induced in the leaves of T. gamsii F18‐treated plants, such as ornithine and oxoproline, are important intermediates in primary metabolism, and their roles in plant defence are unknown (Zeier, 2013). Decreased levels of valine, homoserine and alanine were observed in T. gamsii F18‐treated plants. There is limited information on the role of these amino acids on plant–insect interactions. Taken together, the induction of plant resistance to T. ni by T. gamsii F18 is related to the perturbation in particular amino acid homeostasis.
Phenolic compounds play important roles in plant defence responses during pathogen infection and herbivore attack because of their antioxidant properties (Kulbat, 2016). In our study, no significant difference was observed in total phenolics content between T. gamsii F18‐treated and untreated plants. However, it should be noted that SA and shikimic acid were dramatically lower in T. gamsii F18‐treated plants than in the control plants. The shikimic acid pathway provides important intermediates in terms of chorismic acid for the synthesis of aromatic acids (Tzin and Galili, 2010). The reduction of shikimic acid results in low levels of its derivatives. SA levels are indeed decreased in T. gamsii F18‐treated plants because its formation is through the side‐chain degradation of chorismic acid, which is an important intermediate of the shikimic acid pathway. SA is a key player in the regulation of plant‐induced resistance. Inoculation of Trichoderma has been observed to increase SA levels in cucumber plants (Segarra et al., 2007). However, SA levels were decreased by T. gamsii F18 in Arabidopsis; this might be due to a specific strain effect of T. gamsii F18.
Although there was no significant difference in organic acids levels between treatments, some were increased by T. gamsii F18 such as malic acid, citric acid and 2‐hydroxyglutaric acid. These acids are intermediate compounds in tricarboxylic acid cycle and are essential to generate cell energy to fuel metabolic reactions during plant defence responses (Agut et al., 2016; Groen, 2016).
T. gamsii F18 affected phytohormones content in Arabidopsis
In addition to the metabolome alterations, Trichoderma isolates can also induce phytohormonal network changes on the host plant (Contreras‐Cornejo et al., 2011; Mathys et al., 2012; Martinez‐Medina et al., 2013, 2014). Studies have shown that the levels of defence‐ and stress‐related compounds such as JA, SA and ABA were enhanced in Trichoderma‐inoculated plants (Contreras‐Cornejo et al., 2011; Martinez‐Medina et al., 2011). Another study showed an increase in IAA induced by Trichoderma inoculation of Prunus and cherry rootstocks (Sofo et al., 2011). Consistent with previous studies, we also found that the application of T. gamsii F18 to the roots altered the content of IAA, SA and ABA in Arabidopsis leaves (Table 1).
IAA is the most common and important natural auxin in plants and plays an important role in plant development such as cell division, root initiation, emergence and shoot development (Phillips et al., 2011; Simon and Petrasek, 2011). However, unlike other reported Trichoderma strains that induced IAA levels in Arabidopsis and had growth promotion capacity (Contreras‐Cornejo et al., 2009; Martinez‐Medina et al., 2011), T. gamsii F18 did not increase the IAA content in our studies. This result might explain the fact that no plant growth promotion effect was observed with the inoculation of this strain (data not shown) indicating a different mode of action.
ABA is a major regulator of abiotic stress responses caused by drought, salt, cold and wounding (Christmann et al., 2006; Raghavendra et al., 2010), and also plays a prominent role in plant resistance to biotic stresses such as pathogen attack (Ton et al., 2009; Denance et al., 2013) and herbivorous insects (Bodenhausen and Reymond, 2007; Verhage et al., 2011; Pineda et al., 2012; Lazebnik et al., 2014). In our study, ABA content was increased in the Arabidopsis leaves when it was inoculated with T. gamsii F18. This result is consistent with previous studies that Trichoderma species increase ABA levels of host plants (Contreras‐Cornejo et al., 2011, 2015; Martinez‐Medina et al., 2011). Additionally, ABA has been shown to have positive regulatory effect on plants against insect feeding (Thaler and Bostock, 2004; Bodenhausen and Reymond, 2007). For example, Thaler and Bostock (2004) found that ABA‐deficient tomato plants were more susceptible to a chewing insect Spodoptera exigua. Bodenhausen and Reymond (2007) found that Spodoptera littoralis larvae performed better on Arabidopsis ABA‐biosynthetic mutant aba2‐1 and revealed a new role for ABA in defence against insects. In our case, the accumulation of ABA induced by T. gamsii F18 may be related to the negative regulation of the T. ni feeding behaviour.
JA and SA are dominant regulators of defence responses to herbivorous insects in plants (Glazebrook, 2005; Howe and Jander, 2008; Pieterse et al., 2009). In plant–insect interactions, the SA‐dependent pathway is induced by phloem‐feeding insects, whereas the JA‐dependent pathway is induced by chewing insects (Walling, 2000; De Vos et al., 2005; Glazebrook, 2005; Pieterse et al., 2012). Studies have reported that Trichoderma can activate both JA‐ and SA‐related signalling pathways in host plants (Segarra et al., 2007; Contreras‐Cornejo et al., 2011; Martinez‐Medina et al., 2011). The larval‐herbivore performance on SA mutants was not affected by T. gamsii F18 treatment further confirming that the resistance to T. ni attack induced by T. gamsii F18 is SA‐dependent.
Ethylene is a wound‐response regulator that is involved in plant defence against insects and also is involved in pathogen defence. For example, the Arabidopsis ein2‐1 mutant had more damage than wild‐type plants upon S. littoralis attack (Stotz et al., 2000). In the present study, the inoculation of ein2‐1 plants with T. gamsii F18 showed a significant decrease of larva weight gain compared with the untreated ein2‐1 plants. This result suggests that T. gamsii F18 induces other defence pathways to affect T. ni feeding that are not related to ethylene. In this study, due to the reduced growth and plant size of ABA mutant aba2‐1, the seedlings were unable to be fed to T. ni. Thus, at this time, we cannot give an exact explanation on the role ABA related to the resistance induced by T. gamsii F18.
In conclusion, this study reports a newly isolated Trichoderma gamsii strain that could trigger changes on primary metabolites and alter the phytohormones especially SA and ABA in plant leaf tissues leading to enhanced resistance to insect attack. However, further studies are necessary to determine the effect of T. gamsii on multiple plant hosts. It should be noted that some PGPR have growth promotion effects on multiple plant species while others have effects on specific host plants (Herde and Howe, 2014).
Experimental procedures
Isolation and characterization of fungi
The microbiome was isolated from the A. thaliana soil reported in the study of Badri et al. (2013). Three different soil slurries including whole slurry (containing all microbes), fungi‐enrichment‐slurry or filtered slurry (all microbes removed) were applied to A. thaliana plants to determine what bio‐components were responsible for changing the metabolome of Arabidopsis leaves. The whole soil slurry was prepared according to the method described by Badri et al. (2013). The fungi‐enrichment‐slurry was prepared by incubating the whole slurry with an antibiotic cocktail containing four kinds of antibiotics (Rifampicin, 100 μg/ml; Kanamycin, 100 μg/ml; Erythromycin, 10 μg/ml; Spectinomycin, 100 μg/ml) for 48 h to eliminate the bacteria. The fungi‐enrichment‐slurry was applied to LB agar plates for 24 h to make sure there were no bacteria in the slurry. Then, the fungi‐enrichment‐slurry was filtered through Whatman Grade No. 1 filter paper to collect the fungal mycelia. The mycelia were washed with fresh Hoagland's solution twice to get rid of the antibiotic solution and re‐suspended in 500 ml Hoagland's solution. The filtered slurry was prepared by centrifuging the whole slurry at 12 000 rpm for 15 min and then passed through a 0.2 μm filter to remove all the microbes. The filtered soil slurry was plated on LB media to ascertain no microbial colonies were presented.
Surface‐sterilized A. thaliana (Col‐0) seeds were germinated on Murashige and Skoog medium (MS) (Murashige and Skoog, 1962) agar plates containing 1% sucrose for a week in a growth chamber (25°C, continuous light). Seven‐day‐old Arabidopsis seedlings were transferred into pots (6 cm × 3.6 cm × 6 cm) containing approximately 80 g of a mix of sterile sand and vermiculite (1:1 = v:v). Transferred seedlings grew in a growth chamber (25°C, 16 h/8 h light/dark, relative humidity of 80‐85%). Ten ml of the whole soil slurry, fungi‐enrichment‐slurry, the filtered slurry or the Hoagland's solution (as control) was inoculated to plants after 2 weeks of transplanting (three‐week‐old plants), respectively.
One week after inoculation, Trichoplusia ni larvae in the 5th‐instar stage were placed on the leaves of Arabidopsis plants. Each pot contained one plant and one larva. Larvae were weighed individually before feeding (initial weight) and after 1 day feeding (final weight). The gained weight of each larva was calculated by the final weight minus the initial weight. The experiment was conducted with 18 replicates per treatment.
Isolation and screening of fungal isolates for inhibition of herbivore behaviour
The fungi‐enrichment‐slurry was prepared as the method described above. The fungi‐enrichment‐slurry was gradient‐diluted from 10−1 to 10−4 and streaked onto Potato Dextrose Agar (PDA) plates, and the plates were subsequently incubated for 5 days at 28°C. Single colonies of fungal isolates with different morphology were transferred to new PDA plates. Each fungal isolate was cultured on new PDA plates for at least three times to make sure it was a pure colony and stored at 4°C on PDA plates for use. In total, 13 fungal isolates were obtained.
The fungal isolates were cultured on PDA agar plates at 28°C for 5 days, and mycelia blocks were suspended in Hoagland's solution to get spore solution at a concentration of 1 × 106 CFU/ml. Three‐week‐old A. thaliana (Col‐0) seedlings were root‐inoculated with the fungal spores of the 13 isolates, respectively, suspended in 2 ml Hoagland's solution at a concentration 1 × 106 CFU/ml. The same amount of Hoagland's solution was applied to control plants. T. ni larvae in the 5th‐instar stage were transferred on the leaves of Arabidopsis plant after 1 week of fungal inoculation. Each pot contained one plant and one larva. Larvae were weighed individually before feeding (initial weight) and after 1 day feeding (final weight). The gained weight of each larva was calculated by the final weight minus the initial weight. The experiment was conducted with 18 replicates per treatment.
Based on the results of the screening study, fungal isolates which had significant effects on the herbivore behaviour of T. ni larvae were selected for further experimentation. To study the effect of those selected fungi on herbivore behaviour of T. ni, 2 ml spores at a concentration 1 × 106 CFU/ml of the selected fungi were applied to three‐week‐old A. thaliana (Col‐0) seedlings in pots (9.0 cm × 9.0 cm × 9.0 cm) using the experimental conditions described above. After a week of inoculation, larvae in the 5th‐instar stage were placed onto the leaves. There were nine pots for each treatment. Each pot contained four plants that were fed to one larva. The gained weight of each larva was calculated by the final weight minus the initial weight. This experiment was repeated four times.
Identification of selected fungus
Total DNA of selected fungal isolates was extracted using a method described by Liu et al. (2000). The highly conserved fungal rRNA gene was PCR amplified using primers ITS1F (5′‐ TCCGTAGGTGAACCTGCGG‐3′) and ITS4 (5′‐ TCCTCCGCTTATTGATATGC‐3′) (White et al., 1990). The PCR products were sequenced at the Proteomics and Metabolomics Facility at Colorado State University. The ITS sequences were compared against the NCBI nucleotide databases using the standard nucleotide–nucleotide BLAST (blastn) search algorithm.
Leaf metabolome
Twenty mg of freeze‐dried leaf tissue was finely grounded and extracted in 1 ml of 80% methanol. The cleaned extract was aliquoted into two equal portions, and the supernatant was dried down for further analysing. Internal standards C08‐C30 fatty acid methyl esters (FAMEs) were added, and the sample was derivatized by methoxyamine hydrochloride in pyridine and subsequently by N‐methyl‐N‐trimethylsilyltrifluoroacetamide for trimethylsilylation of acidic protons (Broeckling et al., 2005). Samples were analysed according to the methods of Feihn (Fiehn et al., 2008) by GC‐TOF primary metabolite analysis at West Coast Metabolomics Center, UC‐DAVIS. Mass spectrometry parameters were used as follows: a Leco Pegasus IV mass spectrometer was used with unit mass resolution at 17 spectra s−1 from 80–500 Da at −70 eV ionization energy and 1800 V detector voltage with a 230°C transfer line and a 250°C ion source. ChromaTOF versus 2.32 was used for data preprocessing. Apex masses were reported for using in the BinBase algorithm. Result files were exported to a data server with absolute spectra intensities and further processed by a filtering algorithm implemented in the metabolomics BinBase database. Quantification was reported as peak height using the unique ion as default, unless a different quantification ion is manually set in the BinBase administration software BinView. Raw data files were secured at the NIH Metabolomics database, http://www.metabolomicsworkbench.org.
Phytohormone extraction
The phytohormone was extracted and analysed using the methods described by Huang et al. (2017). Briefly, 100 mg of finely ground plant tissue was transferred into a 1.5 ml tube. One mL cold extraction solvent (20:79:1, methanol: isopropanol: acetic acid, v:v:v) was added to the tube. The tubes were vortexed on medium speed for 120 min at 4°C. Then, the tubes were centrifuged at 4°C (13 000 g, 15 min), and the supernatants were transferred into a new 1.5 ml tube. The samples were dried under gentle nitrogen stream and dissolved in 100 μl solvent (50:50, acetonitrile: water with 0.1% formic acid). Tubes were stored at −80°C overnight and centrifuged at 4°C (13 000 g, 15 min) to remove precipitation. The supernatants were transferred into HPLC vial and analysed by LC‐MS/MS.
LC‐MS/MS
Phytohormones were chromatographically separated using a Waters nanoAcquity UPLC system on a Waters Atlantis dC18 column (3 μM, 300 μM × 150 mm) held at 40°C. Samples were held at 4°C in the auto‐sampler. Water (A) and acetonitrile (B), both with 0.1% formic acid, were used as buffers. The flow rate was 11.5 μl/min, and injection volume was 1 μl. Each sample was injected twice and hormone levels averaged. Phytohormones were analysed by selected reaction monitoring (SRM) on a Waters Xevo TQ‐S mass spectrometer in both negative and positive ion modes. The UPLC gradient was as follows: time (t) = 0 min, 10% B; t = 0.5 min, 10% B; t = 5.5 min, 95% B; t = 7.5 min, 95% B; t = 8 min, 10% B. The column was equilibrated for three minutes before each injection.
Insect performance on signalling mutants
Seeds of ein2‐1 (an ethylene‐insensitive mutant) and sid2‐1(SA induction‐deficient mutant) were obtained from P. Reymond (University of Lausanne, Lausanne, Switzerland) (Bodenhausen and Reymond, 2007), and seeds of transgenic NahG lines (salicylic acid‐deficient NahG transgenic lines) (Delaney et al., 1994) were obtained from J. Vivanco's lab. Seven‐day‐old seedlings of all the mutants were transferred to pots (6 cm × 3.6 cm × 6 cm) containing sterile vermiculite and sand (1:1) and were grown in a growth chamber at the same conditions as above studies. Two ml of fungal mycelia solution (1 × 106 CFU/ml) was applied to each plant two times when they were 14‐day and 21‐day old, respectively. The larvae were randomly placed onto the leaves after 7 days of the second inoculation. There were nine pots for each treatment. Each pot contained four plants and one larva. The gained weight of each larva was calculated by the final weight minus the initial weight. This experiment was repeated two times.
Statistical analysis
Tukey's HSD (Honestly Significant Difference) test and t‐test were conducted to compare the difference in larva weight gain of the treatments and controls (SPSS Student Version 16.0 18.). Statistical significant differences of identified leaf metabolites between treatment and control were determined by t‐test. Clustering of the leaf metabolites (based on the peak height) was performed by principal component analysis (PCA) using Vegan package in R (R Core Team, 2013).
Conflict of interest
We have no declaration of conflict of interest.
Supporting information
Fig. S1. Larval weight gain of Trichoplusia ni fed for 24 h on 4‐week‐old Arabidopsis Col‐0 plants amended with fungal isolates. The control treatment received only Hoagland's solution. Larval weight gain of T. ni feeding for 24 h on 4‐w‐old Arabidopsis Col‐0 plants amended with F18. A, first experiment; B, second experiment; C, third experiment. The values represent the means ± SDEM. The asterisk above the bar indicates significance relative to the control at P < 0.05 level (t‐test).
Table S1. List of compounds identified from T. gamsii F18‐ treated and un‐treated Arabidopsis leaves based on GC‐MS analyses.
Acknowledgement
We thank members of Professors Vivanco's and Guo's groups for technical assistance and helpful discussions. We are thankful to Dr. Shusheng Zhu (Yunnan Agricultural University, China), Dr. Daniel K. Manter (USDA‐ARS Soil Management and Sugar Beet Research Unit, Fort Collins, CO, USA) and Prof. Isgouhi Kaloshian (University of California, Riverside, CA, USA) for reviewing the manuscript and providing valuable comments. This work was in part supported by China Scholarship Council (No. 201206850028) and by Colorado State University Agricultural Experiment Station.
Microbial Biotechnology (2018) 11(6), 1195–1206
Funding Information
This work was in part supported by China Scholarship Council (No. 201206850028) and by Colorado State University Agricultural Experiment Station.
References
- Agut, B. , Gamir, J. , Jaques, J.A. , and Flors, V. (2016) Systemic resistance in citrus to Tetranychus urticae induced by conspecifics is transmitted by grafting and mediated by mobile amino acids. J Exp Bot 67: 5711–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, G. , Ivey, M.L. , Cakir, C. , Bos, J.I. , Miller, S.A. , Madden, L.V. , et al (2007) Systemic modulation of gene expression in tomato by Trichoderma hamatum 382. Phytopathology 97: 429–437. [DOI] [PubMed] [Google Scholar]
- Appel, H.M. , and Cocroft, R.B. (2014) Plants respond to leaf vibrations caused by insect herbivore chewing. Oecologia 175: 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri, D.V. , Zolla, G. , Bakker, M.G. , Manter, D.K. , and Vivanco, J.M. (2013) Potential impact of soil microbiomes on the leaf metabolome and on herbivore feeding behavior. New Phytol 198: 264–273. [DOI] [PubMed] [Google Scholar]
- Bae, H. , Sicher, R.C. , Kim, M.S. , Kim, S.H. , Strem, M.D. , Melnick, R.L. , and Bailey, B.A. (2009) The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao . J Exp Bot 60: 3279–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhausen, N. , and Reymond, P. (2007) Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis . Mol Plant Microbe Interact 20: 1406–1420. [DOI] [PubMed] [Google Scholar]
- Bokhari, F.M. (2009) Efficacy of some Trichoderma species in the control of Rotylenchulus reniformis and Meloidogyne javanica . Arch Phytopathol Plant Protect 42: 361–369. [Google Scholar]
- Broeckling, C.D. , Huhman, D.V. , Farag, M.A. , Smith, J.T. , May, G.D. , Mendes, P. , et al (2005) Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J Exp Bot 56: 323–336. [DOI] [PubMed] [Google Scholar]
- Brotman, Y. , Lisec, J. , Meret, M. , Chet, I. , Willmitzer, L. , and Viterbo, A. (2012) Transcript and metabolite analysis of the Trichoderma‐induced systemic resistance response to Pseudomonas syringae in Arabidopsis thaliana . Microbiology 158: 139–146. [DOI] [PubMed] [Google Scholar]
- Caldana, C. , Degenkolbe, T. , Cuadros‐Inostroza, A. , Klie, S. , Sulpice, R. , Leisse, A. , et al (2011) High‐density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J 67: 869–884. [DOI] [PubMed] [Google Scholar]
- Carreras‐Villasenor, N. , Sanchez‐Arreguin, J.A. , and Herrera‐Estrella, A.H. (2012) Trichoderma: sensing the environment for survival and dispersal. Microbiology 158: 3–16. [DOI] [PubMed] [Google Scholar]
- Chang, Y.C. , Chang, Y.C. , Baker, R. , Kleifeld, O. , and Chet, I. (1986) Increased growth of plants in the presence of the biological‐control agent Trichoderma harzianum . Plant Dis 70: 145–148. [Google Scholar]
- Christmann, A. , Moes, D. , Himmelbach, A. , Yang, Y. , Tang, Y. , and Grill, E. (2006) Integration of abscisic acid signalling into plant responses. Plant Biol 8: 314–325. [DOI] [PubMed] [Google Scholar]
- Contreras‐Cornejo, H.A. , Macias‐Rodriguez, L. , Cortes‐Penagos, C. , and Lopez‐Bucio, J. (2009) Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin‐dependent mechanism in Arabidopsis . Plant Physiol 149: 1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras‐Cornejo, H.A. , Macias‐Rodriguez, L. , Beltran‐Pena, E. , Herrera‐Estrella, A. , and Lopez‐Bucio, J. (2011) Trichoderma‐induced plant immunity likely involves both hormonal‐ and camalexin‐dependent mechanisms in Arabidopsis thaliana and confers resistance against necrotrophic fungi Botrytis cinerea . Plant Signal Behav 6: 1554–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras‐Cornejo, H.A. , Macias‐Rodriguez, L. , Alfaro‐Cuevas, R. , and Lopez‐Bucio, J. (2014) Trichoderma spp. improve growth of Arabidopsis seedlings under salt stress through enhanced root development, osmolite production, and Na+ elimination through root exudates. Mol Plant Microbe Interact 27: 503–514. [DOI] [PubMed] [Google Scholar]
- Contreras‐Cornejo, H.A. , Macias‐Rodriguez, L. , Vergara, A.G. , and Lopez‐Bucio, J. (2015) Trichoderma modulates stomatal aperture and leaf transpiration through an abscisic acid‐dependent mechanism in Arabidopsis . J Plant Growth Regul 34: 425–432. [Google Scholar]
- Contreras‐Cornejo, H.A. , Macias‐Rodriguez, L. , Del‐Val, E. , and Larsen, J. (2018) The root endophytic fungus Trichoderma atroviride induces foliar herbivory resistance in maize plants. Appl Soil Ecol 124: 45–53. [Google Scholar]
- Coppola, M. , Cascone, P. , Chiusano, M.L. , Colantuono, C. , Lorito, M. , Pennacchio, F. , et al (2017) Trichoderma harzianum enhances tomato indirect defense against aphids. Insect Sci 24: 1025–1033. [DOI] [PubMed] [Google Scholar]
- Coruzzi, G.M. , and Last, R.L. (2000) Amino acids In Biochemistry and Molecular Biology of Plants. Buchanan R.B., Gruissem W., and Jones R. (eds). Rockville, MD: Am. Soc. Plant Physiol, Wiley Press, pp. 358–410. [Google Scholar]
- De Vos, M. , Van Oosten, V.R. , Van Poecke, R.M.P. , Van Pelt, J.A. , Pozo, M.J. , Mueller, M.J. , et al (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18: 923–937. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P. , Uknes, S. , Vernooij, B. , Friedrich, L. , Weymann, K. , Negrotto, D. , et al (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250. [DOI] [PubMed] [Google Scholar]
- Denance, N. , Sanchez‐Vallet, A. , Goffner, D. , and Molina, A. (2013) Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front Plant Sci 4: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit, P. , Mukherjee, P.K. , Sherkhane, P.D. , Kale, S.P. , and Eapen, S. (2011) Enhanced tolerance and remediation of anthracene by transgenic tobacco plants expressing a fungal glutathione transferase gene. J Hazard Mater 192: 270–276. [DOI] [PubMed] [Google Scholar]
- Erb, M. , Meldau, S. , and Howe, G.A. (2012) Role of phytohormones in insect‐specific plant reactions. Trends Plant Sci 17: 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada, C. , Wcislo, W.T. , and Van Bael, S.A. (2013) Symbiotic fungi alter plant chemistry that discourages leaf‐cutting ants. New Phytol 198: 241–251. [DOI] [PubMed] [Google Scholar]
- European Food Safety Authority (2013) Conclusion on the peer review of the pesticide risk assessment of the active substance Trichoderma gamsii ICC080. EFSA J 11: 3042. [Google Scholar]
- Fiehn, O. , Wohlgemuth, G. , Scholz, M. , Kind, T. , Lee, D.Y. , Lu, Y. , et al (2008) Quality control for plant metabolomics: reporting MSI‐compliant studies. Plant J 53: 691–704. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Gomez‐Ariza, J. , Campo, S. , Rufat, M. , Estopa, M. , Messeguer, J. , San Segundo, B. , and Coca, M. (2007) Sucrose‐mediated priming of plant defense responses and broad‐spectrum disease resistance by overexpression of the maize pathogenesis‐related PRms protein in rice plants. Mol Plant Microbe Interact 20: 832–842. [DOI] [PubMed] [Google Scholar]
- Gravel, V. , Antoun, H. , and Tweddell, R.J. (2007) Growth stimulation and fruit yield improvement of greenhouse tomato plants by inoculation with Pseudomonas putida or Trichoderma atroviride: possible role of indole acetic acid (IAA). Soil Biol Biochem 39: 1968–1977. [Google Scholar]
- Groen, S.C. (2016) Signalling in systemic plant defence‐roots put in hard graft. J Exp Bot 67: 5585–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, T.N. (2010) Using Trichoderma species for biological control of plant pathogen in Vietnam. J ISSAAS 16: 17–21. [Google Scholar]
- Heil, M. , and Baldwin, I. T. (2002) Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci 7: 61–67. [DOI] [PubMed] [Google Scholar]
- Herde, M. , and Howe, G.A. (2014) Host plant‐specific remodeling of midgut physiology in the generalist insect herbivore Trichoplusia ni . Insect Biochem Mol Bio 50: 58–67. [DOI] [PubMed] [Google Scholar]
- Howe, G.A. , and Jander, G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66. [DOI] [PubMed] [Google Scholar]
- Huang, X.F. , Zhou, D.M. , Lapsansky, E.R. , Reardon, K.F. , Guo, J.H. , Andales, M.J. , et al (2017) Mitsuaria sp and Burkholderia sp from Arabidopsis rhizosphere enhance drought tolerance in Arabidopsis thaliana and maize (Zea mays L.). Plant Soil 419: 523–539. [Google Scholar]
- Juniper, B. E. , and Richard, S. (1986) Insects and the Plant Surface. London, UK: Edward Arnold. [Google Scholar]
- Kan, C.C. , Chung, T.Y. , Juo, Y.A. , and Hsieh, M.H. (2015) Glutamine rapidly induces the expression of key transcription factor genes involved in nitrogen and stress responses in rice roots. BMC Genom 16: 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangasjarvi, S. , Neukermans, J. , Li, S. , Aro, E.M. , and Noctor, G. (2012) Photosynthesis, photorespiration, and light signalling in defence responses. J Exp Bot 63: 1619–1636. [DOI] [PubMed] [Google Scholar]
- Koch, K. (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246. [DOI] [PubMed] [Google Scholar]
- Kulbat, K. (2016) The role of phenolic compounds in plant resistance. Biotechnol Food Sci 80: 97–108. [Google Scholar]
- Lazebnik, J. , Frago, E. , Dicke, M. , and van Loon, J.J.A. (2014) Phytohormone mediation of interactions between herbivores and plant pathogens. J Chem Ecol 40: 730–741. [DOI] [PubMed] [Google Scholar]
- Liu, D. , Coloe, S. , Baird, R. , and Pedersen, J. (2000) Rapid mini‐preparation of fungal DNA for PCR. J Clin Microbiol 38: 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon, L.C. , Geraats, B.P.J. , and Linthorst, H.J.M. (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11: 184–191. [DOI] [PubMed] [Google Scholar]
- Martinez‐Medina, A. , Roldan, A. , Albacete, A. , and Pascual, J.A. (2011) The interaction with arbuscular mycorrhizal fungi or Trichoderma harzianum alters the shoot hormonal profile in melon plants. Phytochemistry 72: 223–229. [DOI] [PubMed] [Google Scholar]
- Martinez‐Medina, A. , Fernandez, I. , Sanchez‐Guzman, M.J. , Jung, S.C. , Pascual, J.A. , and Pozo, M.J. (2013) Deciphering the hormonal signaling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front Plant Sci 4: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Medina, A. , Del Mar Alguacil, M. , Pascual, J.A. , and Van Wees, S.C. (2014) Phytohormone profiles induced by trichoderma isolates correspond with their biocontrol and plant growth‐promoting activity on melon plants. J Chem Ecol 40: 804–815. [DOI] [PubMed] [Google Scholar]
- Mastouri, F. , Bjorkman, T. , and Harman, G.E. (2010) Seed treatment with Trichoderma harzianum alleviates biotic, abiotic, and physiological stresses in germinating seeds and seedlings. Phytopathology 100: 1213–1221. [DOI] [PubMed] [Google Scholar]
- Mathys, J. , De Cremer, K. , Timmermans, P. , Van Kerckhove, S. , Lievens, B. , Vanhaecke, M. , et al (2012) Genome‐wide characterization of ISR induced in Arabidopsis thaliana by Trichoderma hamatum T382 against Botrytis cinerea infection. Front Plant Sci 3: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero‐Barrientos, M. , Hermosa, R. , Cardoza, R.E. , Gutierrez, S. , Nicolas, C. , and Monte, E. (2010) Transgenic expression of the Trichoderma harzianum hsp70 gene increases Arabidopsis resistance to heat and other abiotic stresses. J Plant Physiol 167: 659–665. [DOI] [PubMed] [Google Scholar]
- van de Mortel, J.E. , de Vos, R.C. , Dekkers, E. , Pineda, A. , Guillod, L. , Bouwmeester, K. , et al (2012) Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol 160: 2173–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T. , and Skoog, F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497. [Google Scholar]
- Naseby, D.C. , Pascual, J.A. , and Lynch, J.M. (2000) Effect of biocontrol strains of Trichoderma on plant growth, Pythium ultimum populations, soil microbial communities and soil enzyme activities. J Appl Microbiol 88: 161–169. [DOI] [PubMed] [Google Scholar]
- Nawrocka, J. , and Malolepsza, U. (2013) Diversity in plant systemic resistance induced by Trichoderma . Biol Control 67: 149–156. [Google Scholar]
- Phillips, K.A. , Skirpan, A.L. , Liu, X. , Christensen, A. , Slewinski, T.L. , Hudson, C. , et al (2011) vanishing tassel2 encodes a grass‐specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Leon‐Reyes, A. , Van der Ent, S. , and Van Wees, S.C.M. (2009) Networking by small‐molecule hormones in plant immunity. Nat Chem Biol 5: 308–316. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. , and Van Wees, S.C.M. (2012) Hormonal modulation of plant immunity. Annu Rev Cell Develop Biol 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Pineda, A. , Zheng, S.J. , van Loon, J.J.A. , Pieterse, C.M.J. , and Dicke, M. (2010) Helping plants to deal with insects: the role of beneficial soil‐borne microbes. Trends Plant Sci 15: 507–514. [DOI] [PubMed] [Google Scholar]
- Pineda, A. , Zheng, S.J. , van Loon, J.J.A. , and Dicke, M. (2012) Rhizobacteria modify plant‐aphid interactions: a case of induced systemic susceptibility. Plant Biol 14: 83–90. [DOI] [PubMed] [Google Scholar]
- R Core Team (2013) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Radwanski, E.R. , and Last, R.L. (1995) Tryptophan biosynthesis and metabolism – biochemical and molecular‐genetics. Plant Cell 7: 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra, A.S. , Gonugunta, V.K. , Christmann, A. , and Grill, E. (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401. [DOI] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. , and Jones, J.D.G. (2011) Hormone crosstalk in plant disease and defense: more than just JASMONATE‐SALICYLATE antagonism. Annu Rev Phytopathol 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Rojas, C.M. , Senthil‐Kumar, M. , Tzin, V. , and Mysore, K.S. (2014) Regulation of primary plant metabolism during plant‐pathogen interactions and its contribution to plant defense. Front Plant Sci 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahebani, N. , and Hadavi, N. (2008) Biological control of the root‐knot nematode Meloidogyne javanica by Trichoderma harzianum . Soil Biol Biochem 40: 2016–2020. [Google Scholar]
- Sangha, J.S. , Khan, W. , Ji, X. , Zhang, J. , Mills, A.A.S. , Critchley, A.T. , and Prithiviraj, B. (2011) Carrageenans, sulphated polysaccharides of red seaweeds, differentially affect Arabidopsis thaliana resistance to Trichoplusia ni (Cabbage Looper). PLoS ONE 6: e26834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger, R. , Heise, A.M. , Persicke, M. , and Muller, C. (2014) Interactions between the jasmonic and salicylic acid pathway modulate the plant metabolome and affect herbivores of different feeding types. Plant, Cell Environ 37: 1574–1585. [DOI] [PubMed] [Google Scholar]
- Segarra, G. , Casanova, E. , Bellido, D. , Odena, M.A. , Oliveira, E. , and Trillas, I. (2007) Proteome, salicylic acid, and jasmonic acid changes in cucumber plants inoculated with Trichoderma asperellum strain T34. Proteomics 7: 3943–3952. [DOI] [PubMed] [Google Scholar]
- Shoresh, M. , Harman, G.E. , and Mastouri, F. (2010) Induced systemic resistance and plant responses to fungal biocontrol agents. Annu Rev Phytopathol 48: 21–43. [DOI] [PubMed] [Google Scholar]
- Simon, S. , and Petrasek, J. (2011) Why plants need more than one type of auxin. Plant Sci 180: 454–460. [DOI] [PubMed] [Google Scholar]
- Soares, W.L. , and Porto, M.F. (2007) Agricultural activity and environmental externality: an analysis of the use of pesticides in the Brazilian savannah. Cien Saude Colet 12: 131–143. [DOI] [PubMed] [Google Scholar]
- Sofo, A. , Scopa, A. , Manfra, M. , De Nisco, M. , Tenore, G. , Troisi, J. , et al (2011) Trichoderma harzianum strain T‐22 induces changes in phytohormone levels in cherry rootstocks (Prunus cerasus x P.canescens). Plant Growth Regul 65: 421–425. [Google Scholar]
- Soler, R. , Erb, M. , and Kaplan, I. (2013) Long distance root‐shoot signalling in plant‐insect community interactions. Trends Plant Sci 18: 149–156. [DOI] [PubMed] [Google Scholar]
- Stotz, H.U. , Pittendrigh, B.R. , Kroymann, J. , Weniger, K. , Fritsche, J. , Bauke, A. , and Mitchell‐Olds, T. (2000) Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant Physiol 124: 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, D.W.S. , and Greene, G.L. (1984) Suppression and management of Cabbage Looper populations: cultivated and wild host plants In Suppression and Management of Cabbage Looper Populations, Technical Bulletin. Lingren P.D., and Green G.L. (eds). Washington, DC: United States Department of Agriculture, no. 1694, pp. 1–13. [Google Scholar]
- Thaler, J.S. , and Bostock, R.M. (2004) Interactions between abscisic‐acid‐mediated responses and plant resistance to pathogens and insects. Ecology 85: 48–58. [Google Scholar]
- Thompson, G.A. , and Goggin, F.L. (2006) Transcriptomics and functional genomics of plant defence induction by phloem‐feeding insects. J Exp Bot 57: 755–766. [DOI] [PubMed] [Google Scholar]
- Ton, J. , Flors, V. , and Mauch‐Mani, B. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci 14: 310–317. [DOI] [PubMed] [Google Scholar]
- Tzin, V. , and Galili, G. (2010) New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant 3: 956–972. [DOI] [PubMed] [Google Scholar]
- Verhage, A. , Vlaardingerbroek, I. , Raaymakers, C. , Van Dam, N.M. , Dicke, M. , Van Wees, S.C.M. , and Pieterse, C.M.J. (2011) Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front Plant Sci 2: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling, L.L. (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19: 195–216. [DOI] [PubMed] [Google Scholar]
- War, A.R. , Paulraj, M.G. , Ahmad, T. , Buhroo, A.A. , Hussain, B. , Ignacimuthu, S. , and Sharma, H.C. (2012) Mechanisms of plant defense against insect herbivores. Plant Signal Behav 7: 1306–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weddle, P.W. , Welter, S.C. , and Thomson, D. (2009) History of IPM in California pears–50 years of pesticide use and the transition to biologically intensive IPM. Pest Manag Sci 65: 1287–1292. [DOI] [PubMed] [Google Scholar]
- White, T.J. , Bruns, T. , Lee, S. , and Taylor, J. (1990) Amplification and direct seqencing of fungal ribosomal RNA genes for phylogenetics PCR Protocols. San Diego, CA: Academic Press, pp. 315–322. [Google Scholar]
- Zeier, J. (2013) New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant, Cell Environ 36: 2085–2103. [DOI] [PubMed] [Google Scholar]
- Zhao, J.M. , and Last, R.L. (1996) Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis . Plant Cell 8: 2235–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, S. , Lou, Y.R. , Tzin, V. , and Jander, G. (2015) Alteration of plant primary metabolism in response to insect herbivory. Plant Physiol 169: 1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Larval weight gain of Trichoplusia ni fed for 24 h on 4‐week‐old Arabidopsis Col‐0 plants amended with fungal isolates. The control treatment received only Hoagland's solution. Larval weight gain of T. ni feeding for 24 h on 4‐w‐old Arabidopsis Col‐0 plants amended with F18. A, first experiment; B, second experiment; C, third experiment. The values represent the means ± SDEM. The asterisk above the bar indicates significance relative to the control at P < 0.05 level (t‐test).
Table S1. List of compounds identified from T. gamsii F18‐ treated and un‐treated Arabidopsis leaves based on GC‐MS analyses.


