Summary
The expanding aquaculture industry plays an important role in feeding the growing human population and with the expansion, sustainable bacterial disease control, such as probiotics, becomes increasingly important. Tropodithietic acid (TDA)‐producing Phaeobacter spp. can protect live feed, for example rotifers and Artemia as well as larvae of turbot and cod against pathogenic vibrios. Here, we show that the emerging live feed, copepods, is unaffected by colonization of the fish pathogen Vibrio anguillarum, making them potential infection vectors. However, TDA‐producing Phaeobacter inhibens was able to significantly inhibit V. anguillarum in non‐axenic cultures of copepod Acartia tonsa and the copepod feed Rhodomonas salina. Vibrio grew to 106 CFU ml−1 and 107 CFU ml−1 in copepod and R. salina cultures, respectively. However, vibrio counts remained at the inoculum level (104 CFU ml−1) when P. inhibens was also added. We further developed a semi‐strain‐specific qPCR for V. anguillarum to detect and quantify the pathogen in non‐axenic systems. In conclusion, P. inhibens efficiently inhibits the fish larval pathogen V. anguillarum in the emerging live feed, copepods, supporting its use as a probiotic in aquaculture. Furthermore, qPCR provides an effective method for detecting vibrio pathogens in complex non‐axenic live feed systems.
Introduction
Aquaculture provides high‐quality protein for approximately three billion people worldwide and with the estimated increase in the human population combined with overfishing, this number is likely to increase (WWF and ZSL, 2015; FAO, 2016). Larvae of several commercially important species including cod, halibut and turbot are fed using live feed due to the lack of suited artificial feed formulations. Traditionally, marine fish larvae have been fed using rotifers and Artemia, although copepods are more representative of the natural diet (Turner, 2004). Copepods have several advantages over traditional aquaculture live feed including desirable amino acid profiles, a high fatty acid content and swimming patterns that promote feeding in fish larvae (Støttrup et al., 1986; Støttrup and Norsker, 1997; Bell et al., 2003; Turingan et al., 2007). Furthermore, substituting rotifers with copepods (Acartia tonsa) in the initial larval feeding stage has a long‐term positive effect on survival, growth and viability of Atlantic cod and ballan wrasse fish larvae (Øie et al., 2017). So far, the use of copepods as live feed in larval rearing has not been cost‐effective; however, new approaches to copepod breeding are improving the cost balance (Abate et al., 2016), thus making copepods a relevant live feed in commercial aquaculture.
A major bottleneck in aquaculture is disease outbreaks where bacterial infections account for approx. 55% (Kibenge et al., 2012). One of the most prominent bacterial genera causing disease in aquaculture is Vibrio including Vibrio anguillarum, V. harveyi, V. vulnificus and V. splendidus (Thompson et al., 2004; Toranzo et al., 2005). Live feed used in aquaculture is a potential point of entry and vector for pathogenic bacteria (Olafsen, 2001), and several Vibrio species, including the human and fish pathogens V. cholerae, V. vulnificus and V. splendidus, are found in association with zooplankton, for example copepods, which in the marine environment seem to serve as natural Vibrio reservoirs (Sochard et al., 1979; Heidelberg et al., 2002; Colwell et al., 2003; Vezzulli et al., 2015). The role of copepods as hosts for V. cholerae has especially been studied extensively, and V. cholerae has repeatedly been isolated from the surface, gut and exuviae of these small crustaceans (Huq et al., 1983; Tamplin et al., 1990; Gugliandolo et al., 2008). Thus, although vibrios such as Vibrio parahaemolyticus, V. alginolyticus and V. mimicus have been reported to be unable to colonize the copepod species Temora stylifera, Acartia clausi, Centropages typicus and Paracalanus parvus (Dumontet et al., 1996), it is a valid concern that Acartia tonsa may function as infection vectors for other Vibrio species including the fish pathogenic V. anguillarum in aquaculture systems.
Historically, antibiotics have been used to control bacterial infections in aquaculture. However, risk of antibiotic resistance development and the associated health risk have led to a search for sustainable alternatives (Skjermo and Vadstein, 1999; Sommerset et al., 2005; Defoirdt et al., 2011). The deployment of vaccines has limited the use of antibiotics especially in Europe and North America (Defoirdt et al., 2011; Ringø et al., 2014). However, antibiotics are still used in many countries and fish larviculture as their undeveloped immune system does not allow for vaccination (Sommerset et al., 2005; Defoirdt et al., 2011). An alternative to antibiotics in such systems is probiotics, that is live microorganisms which exert beneficial effects on the host health (FAO and WHO, 2001), for example by inhibiting growth of pathogenic bacteria (Gatesoupe, 1999; Skjermo and Vadstein, 1999; Kesarcodi‐Watson et al., 2008). Members of the genera Phaeobacter and Ruegeria which produce the broad‐spectrum antimicrobial compound tropodithietic acid (TDA) have proven probiotic properties (Ruiz‐Ponte et al., 1999; Hjelm et al., 2004; Planas et al., 2006; D'Alvise et al., 2012). Phaeobacter spp. inhibit the growth of pathogenic vibrios and protect live feed such as rotifers and Artemia (D'Alvise et al., 2012; Grotkjær et al., 2016a,b). Furthermore, Phaeobacter spp. can protect larvae of turbot and cod in challenge trials (Planas et al., 2006; D'Alvise et al., 2012, 2013). Phaeobacter spp. have repeatedly been isolated from different aquaculture facilities (Ruiz‐Ponte et al., 1998; Hjelm et al., 2004; Porsby et al., 2008; Gram et al., 2015; Grotkjær et al., 2016b), which indicate that they are part of the natural microbiota in these aquaculture units. Most studies on the probiotic effect of Phaeobacter spp. and Ruegeria spp. have been carried out in axenic live feed or fish larvae systems (Planas et al., 2006; D'Alvise et al., 2012, 2013; Grotkjær et al., 2016b). Although non‐axenic algae and Artemia systems have been tested (Grotkjær et al., 2016a), the probiotic effect of Phaeobacter spp. and Ruegeria spp. in these more ‘natural’ systems requires further studies.
The purpose of this study was to investigate whether the copepod species Acartia tonsa would potentially function as an infection vector for fish pathogenic vibrio, and whether the TDA‐producing P. inhibens could antagonize V. anguillarum in non‐axenic copepod systems as well as in algae used as copepod feed. Furthermore, we aimed at developing a strain‐specific quantitative PCR (qPCR) protocol for detection and quantification of pathogens in complex systems without the dependency on selection markers, such as antibiotic resistance that potentially could affect pathogen behaviour.
Results
Invasion of copepods by GFP‐tagged V. anguillarum
To test whether V. anguillarum could invade the copepod Acartia tonsa and make the crustacean a potential infection vector in larviculture, a GFP‐tagged V. anguillarum strain NB10 was added to non‐axenic A. tonsa feed, that is Rhodomonas salina. After 4 days of incubation, the GFP signal of NB10 was detected on the surface and inside the copepods (Fig. 1). Specifically, the vibrios appeared to be concentrated within the intestinal system of the crustacean. No GFP signal was detected in non‐inoculated copepods. Throughout the experiment, Vibrio abundances in the surrounding medium were stable around the inoculum level of 105 CFU ml−1 (data not shown). Furthermore, A. tonsa seemed unaffected by the addition of vibrios, exhibiting mortalities after 4 days of 1.47 ± 0.72%.
Figure 1.
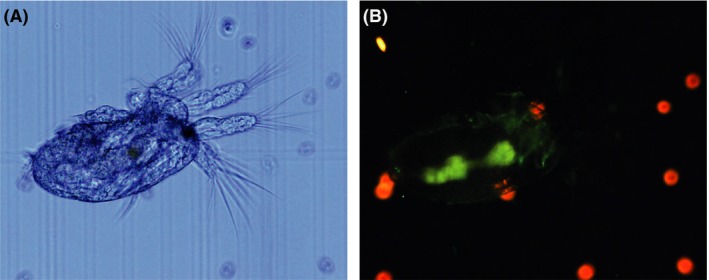
Microscopy of A. tonsa nauplii colonized by GFP‐tagged V. anguillarum NB10. Red fluorescent cells are R. salina. Magnification 100 × A. Phase‐contrast microscopy. B. Fluorescence microscopy (WIB excitation 460–490 nm, emission > 515 nm).
Co‐culture in non‐axenic Rhodomonas salina
As A. tonsa was found to be a potential vector for V. anguillarum, we investigated whether P. inhibens could antagonize vibrios in cultures of copepod and in the copepod live feed R. salina. We conducted a co‐culture experiment in non‐axenic R. salina cultures. Counts of R. salina were stable during the co‐culture experiment, and no significant difference was seen between the differently treated cultures (data not shown). Phaeobacter inhibens established itself and remained at approx. 107 CFU ml−1 in all set‐ups, independently of addition of V. anguillarum strain 90‐11‐286 (Fig. 2). TSA counts showed that V. anguillarum was significantly reduced when grown in the presence of P. inhibens. Without P. inhibens, V. anguillarum grew to approx. 107 CFU ml−1, while in the presence of P. inhibens, V. anguillarum numbers were significantly reduced with approximately three orders of magnitude (P < 0.0001) and never exceeded the inoculum level of 104 CFU ml−1.
Figure 2.
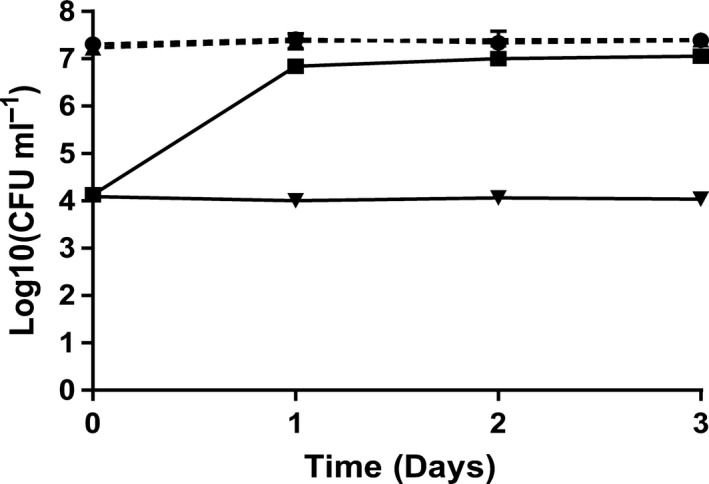
Colony counts of V. anguillarum 90‐11‐286 and P. inhibens DSM 17395 during a co‐culture experiment in non‐axenic R. salina cultures. V. anguillarum without P. inhibens (■, solid line), V. anguillarum with P. inhibens (▼, solid line), P. inhibens without V. anguillarum (●, dashed line) and P. inhibens with V. anguillarum (▲, dashed line). The two P. inhibens lines are situated on top of one another. Error bars show the standard deviation of three biological replicates.
Co‐culture in non‐axenic A. tonsa, and detection and quantification of V. anguillarum with qPCR
Non‐axenic A. tonsa were not affected by the presence of either the probiont or the pathogen resulting in mortalities after 3 days of 5.7 ± 2.52%, 7.3 ± 1.15%, 5.7 ± 2.08% and 6.3 ± 3.51% in cultures with P. inhibens, V. anguillarum, a combination of P. inhibens and V. anguillarum, or no addition (control), respectively. As for the R. salina co‐culture experiment, no differences were observed between P. inhibens abundances in the presence or absence of V. anguillarum (Fig. 3) with counts being stable around the inoculum level of 106–107 CFU ml−1. The abundance of V. anguillarum was significantly reduced by P. inhibens (P = 0.0004). With the addition of P. inhibens, Vibrio counts remained at the inoculum level around 104 CFU ml−1, approx. two orders of magnitude below the abundances observed in cultures without P. inhibens. Numbers of V. anguillarum were estimated both by counts on TSA plates and by qPCR resulting in similar abundances (P > 0.05; Table 1).
Figure 3.
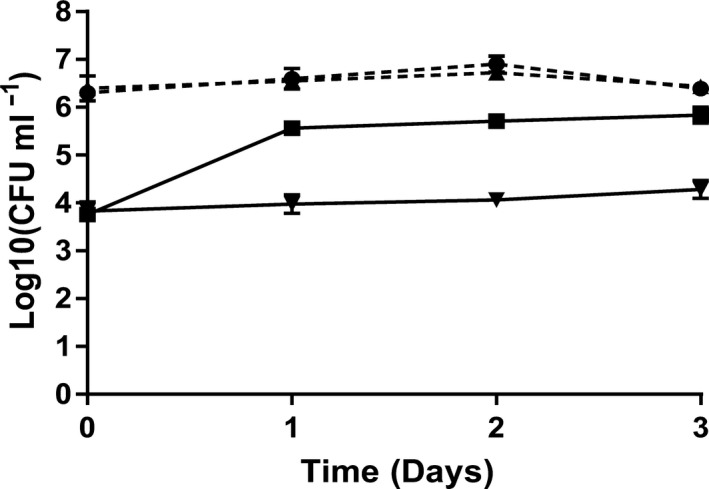
Colony counts of V. anguillarum 90‐11‐286 and P. inhibens DSM 17395 during co‐culture in non‐axenic A. tonsa cultures. V. anguillarum without P. inhibens (■, solid line), V. anguillarum with P. inhibens (▼, solid line), P. inhibens without V. anguillarum (●, dashed line) and P. inhibens with V. anguillarum (▲, dashed line). Error bars show the standard deviation of three biological replicates.
Table 1.
Quantification of V. anguillarum strain 90‐11‐286 using colony counts and qPCR. Values are presented as log10‐transformed counts. Standard deviations are based on three biological replicates. P values were calculated using t‐test (alpha = 0.05) and describe differences between TSA plate counts and C t based counts
| Time (days) | V. anguillarum counts without P. inhibens | V. anguillarum counts with P. inhibens | ||||
|---|---|---|---|---|---|---|
| TSA | C t based | P value | TSA | C t based | P value | |
| 0 | 3.76 ± 0.10 | 3.82 ± 0.14 | 0.6114 | 3.83 ± 0.20 | 3.70 ± 0.16 | 0.4416 |
| 1 | 5.57 ± 0.11 | 5.70 ± 0.08 | 0.1657 | 3.98 ± 0.20 | 3.93 ± 0.04 | 0.7095 |
| 2 | 5.71 ± 0.13 | 5.72 ± 0.12 | 0.9621 | 4.06 ± 0.03 | 3.83 ± 0.18 | 0.1016 |
| 3 | 5.84 ± 0.17 | 5.88 ± 0.17 | 0.7626 | 4.28 ± 0.18 | 3.85 ± 0.21 | 0.0561 |
Verification of primer specificity and standard curve
Primer‐BLAST was used to design and test the specificity of the V. anguillarum 90‐11‐286 primers in silico, and this indicated that the primers were strain‐specific. The primers were subsequently tested in vitro against 11 V. anguillarum strains and one Vibrio harveyi strain (Table 2). The primers amplified the DNA of three strains resulting in cycle threshold (C t) values within the limits of detection (C t ≤ 30). Furthermore, one strain, NB10, was close to the limit of detection with a C t value of 30.29 ± 0.13. Strains 90‐11‐286 and PF7 showed similar C t values of 19.56 ± 0.05 and 19.16 ± 0.11, respectively, and a higher C t value of 23.53 ± 0.08 was seen for PF4. For quantification, a standard curve was made with strain 90‐11‐286 overnight culture that was 10‐fold serial diluted from approx. 107 to 103 (Fig. 4). The standard curve was used to compare C t values to bacterial density (CFU ml−1) for Vibrio samples from the non‐axenic A. tonsa co‐culture.
Table 2.
Vibrio strains used to test the specificity of the V. anguillarum strain 90‐11‐286 primers. Standard deviations are based on technical duplicates
| Strain | Species | Isolation | Serotype | Plasmid pJM1 | Accession no. | Mean C t ‐value | ||
|---|---|---|---|---|---|---|---|---|
| Host | Country | Genome | Plasmid | |||||
| 4299 | V. anguillarum | Unknown | Norway | O2b | − | CP011458/CP011459 | 35.57 ± 1.51 | |
| 90‐11‐286 | Rainbow trout | Denmark | O1 | − | CP011460/CP011461 | 19.56 ± 0.05 | ||
| 90‐11‐287 | Rainbow trout | Denmark | O1 | + | CP011475/CP011476 | CP016254 | 32.31 ± 0.02 | |
| 775 | Coho salmon | Norway | O1 | + | CP002284/CP002285 | AY312585 | 33.41 ± 0.78 | |
| H610 | Atlantic cod | USA | O2a | − | CP011462/CP011463 | 37.06 ± 2.11 | ||
| NB10 | Unknown | Sweden | O1 | + | LK021130/LK021129 | LK021128 | 30.29 ± 0.13 | |
| PF4 | Atlantic salmon | Chile | O3 | − | CP010080/CP010081 | 23.53 ± 0.08 | ||
| PF7 | Atlantic salmon | Chile | O3 | − | CP011464/CP011465 | 19.16 ± 0.11 | ||
| VIB243 | Sockeye salmon | USA | O1/VaNT1 | + | CP010030/CP010031 | CP016261 | 35.18 ± 0.30 | |
| DSM21597 | Atlantic cod | Norway | O2 | − | CP010084/CP010085 | 34.31 ± 0.76 | ||
| S2 2/9 | Rainbow trout | Denmark | O1 | − | CP011472/CP011473 | 36.61 ± 2.17 | ||
| DSM19623 | V. harveyi | Amphipod | USA | – | − | BAOD01000001 | 39.58 | |
Figure 4.
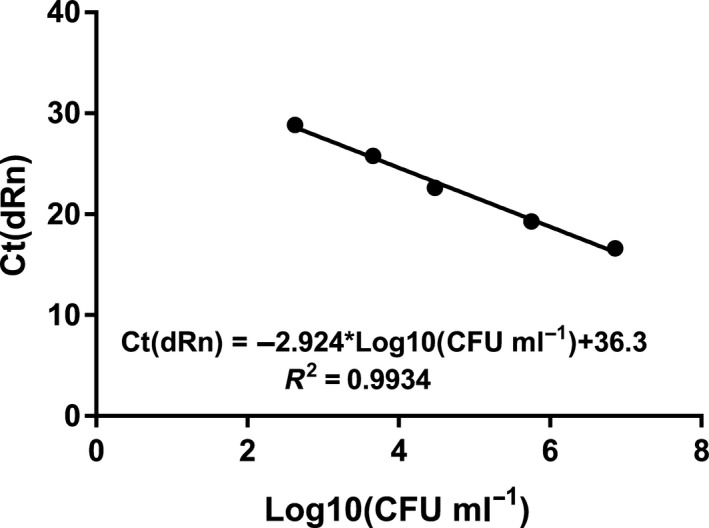
qPCR standard curves for V. anguillarum 90‐11‐286 in non‐axenic copepod cultures. Standard curve was obtained by running, in triplicates, DNA extracted from one sample.
Discussion
Live feed is essential at the larval stage in several commercially important species of finfish, and controlling pathogenic bacteria in the live feed is important as these may act as vectors for the pathogens. Here, we demonstrate that P. inhibens can inhibit the highly virulent V. anguillarum strain 90‐11‐286 in two different non‐axenic live feed systems. Specifically, we demonstrate the probiotic potential in copepods as these are superior as larval feed compared to Artemia and rotifers, and future aquaculture is expected to rely increasingly on a copepod‐based live feed.
Live feed is known as a point of entry and vector for pathogenic bacteria (Olafsen, 2001). Copepods have previously been shown to host some Vibrio spp. (Huq et al., 1983; Tamplin et al., 1990; Heidelberg et al., 2002; Gugliandolo et al., 2008; Vezzulli et al., 2015); however, some Vibrio spp. seem to be unable to colonize some copepod species (Dumontet et al., 1996). Using a GFP‐tagged V. anguillarum NB10, we found that V. anguillarum was able to colonize the outside and the gut of the feeding copepods without inducing notable mortality (1.47 ± 0.72% after 4 days) hence making them potential infection vectors in larviculture.
Vibrio spp. are part of the established microbiota of live feed used in larviculture (Berland et al., 1970; Austin and Allen, 1982; Salvesen et al., 2000; López‐Torres and Lizárraga‐Partida, 2001) and when introduced, these opportunistic pathogens can proliferate rapidly leading to great losses (Reid et al., 2009). In the co‐culture experiment, no differences were seen in mortality of the unfed copepods regardless of the presence of P. inhibens or V. anguillarum (5.7 ± 2.52% to 7.3 ± 1.15% mortality). In contrast, V. anguillarum has been shown to cause high mortality (92%) in non‐axenic Artemia cultures. Here, the addition of P. inhibens reduced mortality of the infected Artemia to 11% (Grotkjær et al., 2016b). The observed differences in mortality potentially give copepods an advantage over Artemia as live feed, as it has been reported that homogenized Artemia or sea bass larval biomass, simulating the presence of dead Artemia or sea bass larvae, increase the virulence of V. anguillarum, resulting in a significant increase in mortality in challenged sea bass larvae (Li et al., 2014). Thus, copepods with a higher survival rate than Artemia could result in fewer and potentially less virulent vibrios. The differences in mortality between Artemia and copepods can be explained by several factors. A notable difference between the studies is the vibrio abundances observed at the end of the co‐culture experiments. Although inoculated at same level (104 CFU ml−1), abundances of V. anguillarum in cultures without P. inhibens were 10‐fold higher in the Artemia experiment (107 CFU ml−1) (Grotkjær et al., 2016a) as compared to the copepod experiment conducted in the present study (106 CFU ml−1). This suggests that the presence of copepods, or the background microbiota of the copepods, could have an inhibiting effect on vibrio growth, especially as vibrio abundances in the algae cultures of both studies, Tetraselmis suecica and R. salina, respectively, were similar to that of the Artemia cultures, that is 107 CFU ml−1. The immune systems of invertebrates are poorly understood (Loker et al., 2004). However, there are differences between copepods and Artemia, and for instance, the heat shock protein hsp70, which is associated with pathogen resistance, is induced in Artemia when infected with Vibrio (Norouzitallab et al., 2016). In contrast, an analog of this stress responder was not induced in the copepod Eurytemora affinis when exposed to Vibrio spp. (Almada and Tarrant, 2016). Different copepod species are colonized by different bacteria, and some Vibrio spp. are unable to colonize some copepod species (Dumontet et al., 1996). Vibrios colonizing copepods have been reported to trigger upregulation of saposin‐like genes and C‐type lectins (Almada and Tarrant, 2016) that have antimicrobial properties (Hoeckendorf and Leippe, 2012; Wang et al., 2014). This suggests that copepods have a potential inhibiting effect on the growth of V. anguillarum, which could explain the lower Vibrio abundance and mortality in copepod cultures relative to Artemia cultures. It is also possible that the V. anguillarum strain 90‐11‐286 is avirulent against A. tonsa. The virulence of strains within the same species can vary immensely as seen for thirty V. anguillarum strains tested in three different hosts (turbot, halibut and cod larvae), causing mortality from 100% to 9.1% with the same infection dose (Rønneseth et al., 2017). Thus, if other V. anguillarum strains had been tested in the Artemia and A. tonsa systems, the outcome might have been different.
Several co‐culture studies of P. inhibens and V. anguillarum in axenic systems have shown significant inhibition of vibrios, resulting in lowered vibrio counts relative to the inoculum level (D'Alvise et al., 2010, 2012; Grotkjær et al., 2016b). In contrast, P. inhibens kept V. anguillarum at the inoculum level in both the non‐axenic co‐culture experiments conducted in the present study and in similar experiments with non‐axenic Tetraselmis and Artemia (Grotkjær et al., 2016a). This suggests that the microbiota of the non‐axenic systems have a protective effect on V. anguillarum; however, this is not necessarily a problem in an aquaculture context as it is not essential to completely eradicate all vibrios to get a protective effect. Rønneseth et al. (2017) found a significant decrease in virulence for several V. anguillarum strains tested in turbot, halibut and cod larvae challenge trials when added at low density (104 CFU ml−1) as compared to high density (106 CFU ml−1). Hence, although V. anguillarum was not reduced to below inoculum levels in the presence of P. inhibens, our findings support the use of Phaeobacter spp. as probiotics in copepod systems. In both co‐culture experiments, P. inhibens was able to establish itself at a constant level, which is consistent with its reported association with micro‐ and macroalgae, zooplankton, squids, copepods and fish larvae (Hjelm et al., 2004; Rao et al., 2005; Venmathi Maran et al., 2007; Porsby et al., 2008; Collins et al., 2012; Gram et al., 2015; Grotkjær et al., 2016b). This supports the proposal that one way of utilizing Phaeobacter spp. in aquaculture is by introducing them directly into the live feed systems (Planas et al., 2006; Prol et al., 2009; Grotkjær et al., 2016b).
Previously, non‐axenic co‐culture experiments have used a chloramphenicol‐resistant V. anguillarum as target organism and estimated vibro abundances on solid substrates supplemented with antibiotics (Grotkjær et al., 2016a). However, using antibiotic as selection marker is not without challenges. The natural microbiome may harbour bacteria resistant to the antibiotic marker, the antibiotic marker may be unstable, and the marker may affect the fitness of the tagged strain, all of which can result in over‐ or underestimation of the actual abundances (Andersson and Levin, 1999; Allen et al., 2010). Several non‐growth dependent methods have been developed for detection and/or quantification of vibrios (Eiler and Bertilsson, 2006; Prol et al., 2009; Saulnier et al., 2009; Kim and Lee, 2014). However, as previously described, vibrios are part of the established microbiota of live feed, thus genus‐ or species‐specific primers will potentially amplify vibrios from the background microbiota. In this study, we attempted to develop strain‐specific primers for quantification of the pathogenic V. anguillarum 90‐11‐286. Three of the 12 tested vibrio strains, including the target strain, were amplified within the limits of detection (C t ≤ 30). The two non‐target strains were the closely related V. anguillarum PF7 and PF4 (Table 2). All three strains are highly virulent and cluster closely when compared by 163 concatenated virulence factors and by core genome phylogeny (Castillo et al., 2017; Rønneseth et al., 2017). However, the semi‐strain‐specific primers developed provide an effective method for detecting few specific vibrios in non‐axenic systems. As they were made for research and not diagnostic purposes, it is possible to test the applicability of the primer set prior to experimental use.
In conclusion, our study shows that the emerging live feed copepod, A. tonsa, potentially can function as an infection vector for pathogenic V. anguillarum strains. However, Vibrio counts in the copepod cultures were lower than what has previously been seen for Artemia and algae cultures, supporting the use of A. tonsa as fish larvae feed. P. inhibens efficiently inhibited V. anguillarum in both non‐axenic copepod and algae cultures supporting its use as a probiotic in live feed systems. Lastly, we have described direct qPCR methodology that provides an effective means of detecting vibrios in complex non‐axenic live feed systems without the use of selection markers, which potentially could affect pathogen behaviour.
Experimental procedures
Bacterial strain and culture conditions
Two fish pathogenic V. anguillarum strains were used in this study. Strain NB10, used for the adhesion/invasion experiment, was isolated from the Gulf of Bothnia and is pathogenic towards turbot and halibut larvae (Rønneseth et al., 2017). The strain has been tagged by insertion of plasmid pNQFlaC4‐gfp27 (cat, gfp) into the chromosome (Croxatto et al., 2007) and was provided by Prof. Debra L. Milton (Department of Molecular Biology, Umeå University). The highly virulent V. anguillarum strain 90‐11‐286 (Rønneseth et al., 2017) was used in the co‐culture experiments and was isolated from diseased rainbow trout (Oncorhynchus mykiss) from a Danish aquaculture (Pedersen and Larsen, 1993; Skov et al., 1995). P. inhibens DSM 17395 was used for the co‐culture experiments (Martens et al., 2006; Vandecandelaere et al., 2008; Buddruhs et al., 2013). NB10 and 90‐11‐286 were grown and counted on TSA (Tryptone Soy Agar, Difco 212185) supplemented with 2.5 mg l−1 chloramphenicol for NB10. The vibrios have very distinct colony morphology on TSA. Phaeobacter inhibens DSM 17395 is unable to grow on TSA due to its low salinity, thus TSA functions as a semiselective medium. Phaeobacter inhibens was grown and counted on MA (Marine Agare, Difco 2216) where it grows as distinct brown colonies due to Fe‐TDA complex (D'Alvise et al., 2016). Bacterial stock cultures were stored at −80°C in 20% (vol/vol) glycerol. Two to three days prior to use, stock cultures were streaked on agar plates and incubated at 25°C. The purity of the bacteria was checked by colony morphology, and single colonies were used for inoculation of each preculture. All bacterial precultures were grown in 20 ml of ½YTSS (2 g Bacto Yeast extract, 1.25 g Bacto Tryptone, 20 g Sigma Sea Salts, 1 L deionized water) (Sobecky et al., 1997) at 25°C and 200 rpm for 24 h.
Preparation of non‐axenic algae and copepods cultures
The non‐axenic R. salina K‐1487 originates from Denmark and was provided by Prof. Thomas Kiørboe (National Institute of Aquatic Resources, Technical University of Denmark) (Nielsen and Kiørboe, 2015). A R. salina K‐1487 stock culture was maintained in f/2 medium (Guillard and Ryther, 1962; Guillard, 1975) without Na2SiO3 but with 5 mM NH4Cl in 3% IO (Instant Ocean salts, Aquarium Systems Inc.) (f/2‐Si+NH4) at 18°C and 24 μmol photons m−2 s−1, photosynthetically active radiation (PAR). The algal density was determined by counting in a Neubauer‐improved counting chamber, and the culture was adjusted to approx. 100 000 cell ml−1 in f/2‐Si+NH4 and 25 ml was distributed into 50‐ml falcon tubes. A. tonsa eggs were provided by Prof. Benni W. Hansen (Department of Science and Environment, Roskilde University) (Drillet et al., 2006; Hansen et al., 2010) and were kept at 5°C until use. Two days before the experiment, eggs were inoculated in 3% IO and incubated at 18°C and 24 μmol photons m−2 s−1, photosynthetically active radiation (PAR). For bacterial mono‐ and co‐culture experiments, 50‐ml falcon tubes were set up with 25 ml 3% IO with 2–3 Acartia tonsa nauplii per millilitre. For the invasion experiment, 50‐ml falcon tubes containing 20 ml R. salina culture were supplemented with approx. 5 Acartia tonsa nauplii per millilitre.
A. tonsa invasion experiment
Precultures of V. anguillarum NB10 were 10‐fold serially diluted in 3% IO, and 250 μl dilutions were used to inoculate non‐axenic cultures of A. tonsa fed R. salina, aiming at an initial concentration of 105 CFU ml−1. Experiments were carried out over 96 h where bacterial concentrations and copepod mortality were determined every 24 h. At the end of the experiment, A. tonsa from cultures with and without NB10 was investigated for GFP signal using phase‐contrast and fluorescence microscopy at 100× magnification using an Olympus BX51 fluorescence microscope (WIB excitation 460–490 nm, emission >515 nm).
Co‐culture experiment in non‐axenic R. salina and unfed A. tonsa cultures
Precultures of V. anguillarum 90‐11‐286 were 10‐fold serially diluted in 3% IO, and 250 μl diluted culture was used to inoculate non‐axenic R. salina and A. tonsa cultures, aiming at an initial concentration of 104 CFU ml−1. Precultures of P. inhibens DSM 17395 were added undiluted to the non‐axenic R. salina and A. tonsa cultures, aiming at an initial concentration of 107 CFU ml−1. 250 μl ½YTSS was added to uninoculated controls and cultures where only 90‐11‐286 had been added. The cultures were incubated at 18°C, lying horizontally on a rotary shaker at 60 rpm in an algae growth cabinet. Experiments were carried out over 72 h where bacterial and algal abundances, and copepod mortality were determined every 24 h. Samples for DNA extraction for qPCR‐based quantification of 90‐11‐286 were taken every 24 h (see below). Bacterial abundances were determined by CFU counts. 90‐11‐286 counts were determined on TSA, and DSM 17395 counts were determined on MA. Plates were incubated at 25°C. TSA plates were counted after 1 and 2 days and MA plates after 2‐3 days. R. salina abundances were counted in a Neubauer‐improved counting chamber. The number of dead A. tonsa was determined using a Sedgewick‐Rafter counting chamber. The number of surviving A. tonsa was determined after the experiment was terminated. All combinations were made in biological triplicates.
Primer design and quantification of V. anguillarum 90‐11‐286 by qPCR
The primers Fw_90‐11‐286 (5′ ‐ CAACTTAGGCGTGCAATGGG ‐ 3′) and Rev_90‐11‐286 (5′‐ ACCGCTTTACTGGTGGTGG ‐ 3′) were designed using Primer‐BLAST from NCBI (Ye et al., 2012). Standard settings were used except for the ‘PCR product size’ which was set to min 75 bp and max 200 bp and the database specifications which was set to Genomes and Bacteria (taxid:2). V. anguillarum 90‐11‐286 chromosome I, complete sequence (NZ_CP011460.1) was used as template. For qPCR detection and quantification of the pathogen, genomic DNA was extracted from 1 ml samples using the NucleoSpin Tissue kit (M740952; Macherey‐Nagel, Düren, Germany) as described by the manufacturer. Extracted DNA was stored at ‐20° C until use. Amplification reaction mixtures contained 12.5 μl 2 × SYBR® Green PCR Master Mix (4309155; Applied Biosystems), 1 μl (10 μM) Fw primer, 1 μl (10 μM) Rev primer, 2 μl template DNA and 8.5 μl H2O (DNA grade). Reactions were run on a Mx3000P (Stratagene) qPCR System, using the program 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 15 s, 56°C for 1 min and 72°C for 1 min followed by a dissociation curve 95°C for 1 min, 55°C for 30 s and 95°C for 30 s. DNA grade water was included as non‐template controls (NTC), and positive controls consisted of genomic DNA from the target strain. For in vivo testing of primer specificity against 11 V. anguillarum strains and one V. harveyi strain (Table 2), precultures were diluted to 107 CFU ml−1 in 3% IO from which TSA plate counts were made and DNA was extracted. For quantification, a standard curve relating C t (cycle threshold) values to bacterial density (CFU ml−1) was made with 90‐11‐286 preculture. The preculture was 10‐fold serial diluted in 3% IO and inoculated into non‐axenic A. tonsa cultures aiming for 107 to 103 CFU ml−1. Vibrio counts of the cultures were made using TSA plates, and DNA was extracted from the samples. The measurements were analysed using GraphPad Prism 7.04 (GraphPad Software, San Diego CA). The limit of detection was estimated based on C t values of the control samples with only A. tonsa and background microbiota.
Statistical analysis
CFU ml−1 values and CFU ml−1 of the corresponding C t values for each biological replicate were log10‐transformed prior to statistical analysis. Statistical analyses of differences were performed using t‐test (alpha = 0.05) in GraphPad Prism 7.04 (GraphPad Software, San Diego CA).
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This study was funded by grants from The Danish Council for Strategic Research|Programme Commission on Health, Food and Welfare (12‐132390; ProAqua) and by grant VKR023285 from the Villum Fonden.
Microbial Biotechnology (2018) 11(6), 1070–1079
Funding Information
This study was funded by grants from The Danish Council for Strategic Research|Programme Commission on Health, Food and Welfare (12‐132390; ProAqua) and by grant VKR023285 from the Villum Fonden.
References
- Abate, T.G. , Nielsen, R. , Nielsen, M. , Jepsen, P.M. , and Hansen, B.W. (2016) A cost‐effectiveness analysis of live feeds in juvenile turbot Scophthalmus maximus (Linnaeus, 1758) farming: copepods versus Artemia . Aquac Nutr 22: 899–910. [Google Scholar]
- Allen, H.K. , Donato, J. , Wang, H.H. , Cloud‐Hansen, K.A. , Davies, J. , and Handelsman, J. (2010) Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8: 251–259. [DOI] [PubMed] [Google Scholar]
- Almada, A.A. , and Tarrant, A.M. (2016) Vibrio elicits targeted transcriptional responses from copepod hosts. FEMS Microbiol Ecol 92: 1–11. [DOI] [PubMed] [Google Scholar]
- Andersson, D.I. , and Levin, B.R. (1999) The biological cost of antibiotic resistance. Curr Opin Microbiol 2: 489–493. [DOI] [PubMed] [Google Scholar]
- Austin, B. , and Allen, D.A. (1982) Microbiology of laboratory‐hatched brine shrimp (Artemia). Aquaculture 26: 369–383. [Google Scholar]
- Bell, J.G. , McEvoy, L.A. , Estevez, A. , Shields, R.J. , and Sargent, J.R. (2003) Optimising lipid nutrition in first‐feeding flatfish larvae. Aquaculture 227: 211–220. [Google Scholar]
- Berland, B.R. , Bonin, D.J. , and Maestrini, S.Y. (1970) Study of bacteria associated with marine algae in culture. Mar Biol 3: 68–76. [Google Scholar]
- Buddruhs, N. , Pradella, S. , Göker, M. , Päuker, O. , Pukall, R. , Spröer, C. , et al (2013) Molecular and phenotypic analyses reveal the non‐identity of the Phaeobacter gallaeciensis type strain deposits CIP 105210T and DSM 17395. Int J Syst Evol Microbiol 63: 4340–4349. [DOI] [PubMed] [Google Scholar]
- Castillo, D. , Alvise, P.D. , Xu, R. , Zhang, F. , and Middelboe, M. (2017) Comparative genome analyses of Vibrio anguillarum strains reveal a link with pathogenicity traits. mSystems 2: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, A.J. , LaBarre, B.A. , Won, B.S.W. , Shah, M.V. , Heng, S. , Choudhury, M.H. , et al (2012) Diversity and partitioning of bacterial populations within the accessory nidamental gland of the squid Euprymna scolopes . Appl Environ Microbiol 78: 4200–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell, R.R. , Huq, A. , Islam, M.S. , Aziz, K.M.A. , Yunus, M. , Khan, N.H. , et al (2003) Reduction of cholera in Bangladeshi villages by simple filtration. Proc Natl Acad Sci USA 100: 1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxatto, A. , Lauritz, J. , Chen, C. , and Milton, D.L. (2007) Vibrio anguillarum colonization of rainbow trout integument requires a DNA locus involved in exopolysaccharide transport and biosynthesis. Environ Microbiol 9: 370–382. [DOI] [PubMed] [Google Scholar]
- D'Alvise, P.W. , Melchiorsen, J. , Porsby, C.H. , Nielsen, K.F. , and Gram, L. (2010) Inactivation of Vibrio anguillarum by attached and planktonic Roseobacter cells. Appl Environ Microbiol 76: 2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alvise, P.W. , Lillebø, S. , Prol‐Garcia, M.J. , Wergeland, H.I. , Nielsen, K.F. , Bergh, Ø. , and Gram, L. (2012) Phaeobacter gallaeciensis reduces Vibrio anguillarum in cultures of microalgae and rotifers, and prevents vibriosis in cod larvae. PLoS ONE 7: e43996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alvise, P.W. , Lillebø, S. , Wergeland, H.I. , Gram, L. , and Bergh, Ø. (2013) Protection of cod larvae from vibriosis by Phaeobacter spp.: a comparison of strains and introduction times. Aquaculture 384–387: 82–86. [Google Scholar]
- D'Alvise, P.W. , Phippen, C.B.W. , Nielsen, K.F. , and Gram, L. (2016) Influence of iron on production of the antibacterial compound tropodithietic acid and its noninhibitory analog in Phaeobacter inhibens . Appl Environ Microbiol 82: 502–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defoirdt, T. , Sorgeloos, P. , and Bossier, P. (2011) Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr Opin Microbiol 14: 251–258. [DOI] [PubMed] [Google Scholar]
- Drillet, G. , Iversen, M.H. , Sørensen, T.F. , Ramløv, H. , Lund, T. , and Hansen, B.W. (2006) Effect of cold storage upon eggs of a calanoid copepod, Acartia tonsa (Dana) and their offspring. Aquaculture 254: 714–729. [Google Scholar]
- Dumontet, S. , Krovacek, K. , Baloda, S.B. , Grottoli, R. , Pasquale, V. , and Vanucci, S. (1996) Ecological relationship between Aeromonas and Vibrio spp. and planktonic copepods in the coastal marine environment in southern Italy. Comp Immunol Microbiol Infect Dis 19: 245–254. [DOI] [PubMed] [Google Scholar]
- Eiler, A. , and Bertilsson, S. (2006) Detection and quantification of Vibrio populations using denaturant gradient gel electrophoresis. J Microbiol Methods 67: 339–348. [DOI] [PubMed] [Google Scholar]
- FAO (2016) The State of World Fisheries and Aquaculture 2016. Rome: FAO. [Google Scholar]
- FAO and WHO (2001) Report of a joint FAO/WHO expert consultation on evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria.
- Gatesoupe, F.J. (1999) The use of probiotics in aquaculture. Aquaculture 180: 147–165. [Google Scholar]
- Gram, L. , Rasmussen, B.B. , Wemheuer, B. , Bernbom, N. , Ng, Y.Y. , Porsby, C.H. , et al (2015) Phaeobacter inhibens from the Roseobacter clade have a natural environmental niche as surface colonizer in Danish harbors. Syst Appl Microbiol 38: 483–493. [DOI] [PubMed] [Google Scholar]
- Grotkjær, T. , Bentzon‐Tilia, M. , D'Alvise, P. , Dierckens, K. , Bossier, P. , and Gram, L. (2016a) Phaeobacter inhibens as probiotic bacteria in non‐axenic Artemia and algae cultures. Aquaculture 462: 64–69. [Google Scholar]
- Grotkjær, T. , Bentzon‐Tilia, M. , D'Alvise, P. , Dourala, N. , Nielsen, K.F. , and Gram, L. (2016b) Isolation of TDA‐producing Phaeobacter strains from sea bass larval rearing units and their probiotic effect against pathogenic Vibrio spp. in Artemia cultures. Syst Appl Microbiol 39: 180–188. [DOI] [PubMed] [Google Scholar]
- Gugliandolo, C. , Irrera, G.P. , Lentini, V. , and Maugeri, T.L. (2008) Pathogenic Vibrio, Aeromonas and Arcobacter spp. associated with copepods in the Straits of Messina (Italy). Mar Pollut Bull 56: 600–606. [DOI] [PubMed] [Google Scholar]
- Guillard, R.R.L. (1975) Culture of phytoplankton for feeding marine invertebrates In Culture of marine invertebrate animals. Smith W.L. and Chanley M.H. (ed.). Boston, MA: Springer, pp. 29–60. [Google Scholar]
- Guillard, R.R.L. , and Ryther, J.H. (1962) Studies of marine planktonic diatoms: Cyclotella nana hustedt, and Detonula confervacea (cleve) gran. Can J Microbiol 8: 229–239. [DOI] [PubMed] [Google Scholar]
- Hansen, B.W. , Drillet, G. , Kozmér, A. , Madsen, K.V. , Pedersen, M.F. , and Sørensen, T.F. (2010) Temperature effects on copepod egg hatching: does acclimatization matter? J Plankton Res 32: 305–315. [Google Scholar]
- Heidelberg, J.F. , Heidelberg, K.B. , and Colwell, R.R. (2002) Bacteria of the γ‐subclass Proteobacteria associated with zooplankton in Chesapeake Bay. Appl Environ Microbiol 68: 5498–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelm, M. , Bergh, O. , Riaza, A. , Nielsen, J. , Melchiorsen, J. , Jensen, S. , et al (2004) Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst Appl Microbiol 27: 360–371. [DOI] [PubMed] [Google Scholar]
- Hoeckendorf, A. , and Leippe, M. (2012) SPP‐3, a saposin‐like protein of Caenorhabditis elegans, displays antimicrobial and pore‐forming activity and is located in the intestine and in one head neuron. Dev Comp Immunol 38: 181–186. [DOI] [PubMed] [Google Scholar]
- Huq, A. , Small, E.B. , West, P.A. , Huq, M.I. , and Colwell, R.R. (1983) Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol 45: 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarcodi‐Watson, A. , Kaspar, H. , Lategan, M.J. , and Gibson, L. (2008) Probiotics in aquaculture: the need, principles and mechanisms of action and screening processes. Aquaculture 274: 1–14. [Google Scholar]
- Kibenge, F.S.B. , Godoy, M.G. , Fast, M. , Workenhe, S. , and Kibenge, M.J.T. (2012) Countermeasures against viral diseases of farmed fish. Antiviral Res 95: 257–281. [DOI] [PubMed] [Google Scholar]
- Kim, J.Y. , and Lee, J.L. (2014) Multipurpose assessment for the quantification of Vibrio spp. and total bacteria in fish and seawater using multiplex real‐time polymerase chain reaction. J Sci Food Agric 94: 2807–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Defoirdt, T. , Yang, Q. , Laureau, S. , Bossier, P. , and Dierckens, K. (2014) Host‐induced increase in larval sea bass mortality in a gnotobiotic challenge test with Vibrio anguillarum . Dis Aquat Organ 108: 211–216. [DOI] [PubMed] [Google Scholar]
- Loker, E.S. , Adema, C.M. , Zhang, S.‐M. , and Kepler, T.B. (2004) Invertebrate immune systems ‐ not homogeneous, not simple, not well understood. Immunol Rev 198: 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Torres, M.A. , and Lizárraga‐Partida, M.L. (2001) Bacteria isolated on TCBS media associated with hatched Artemia cysts of commercial brands. Aquaculture 194: 11–20. [Google Scholar]
- Martens, T. , Heidorn, T. , Pukall, R. , Simon, M. , Tindall, B.J. , and Brinkhoff, T. (2006) Reclassification of Roseobacter gallaeciensis Ruiz‐Ponte et al. 1998 as Phaeobacter gallaeciensis gen. nov., comb. nov., description of Phaeobacter inhibens sp. nov., reclassification of Ruegeria algicola (Lafay et al. 1995) Uc. Int J Syst Evol Microbiol 56: 1293–1304. [DOI] [PubMed] [Google Scholar]
- Nielsen, L.T. , and Kiørboe, T. (2015) Feeding currents facilitate a mixotrophic way of life. ISME J 9: 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norouzitallab, P. , Baruah, K. , Biswas, P. , Vanrompay, D. , and Bossier, P. (2016) Probing the phenomenon of trained immunity in invertebrates during a transgenerational study, using brine shrimp Artemia as a model system. Sci Rep 6: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Øie, G. , Galloway, T. , Sørøy, M. , Holmvaag Hansen, M. , Norheim, I.A. , Halseth, C.K. , et al (2017) Effect of cultivated copepods (Acartia tonsa) in first‐feeding of Atlantic cod (Gadus morhua) and ballan wrasse (Labrus bergylta) larvae. Aquac Nutr 23: 3–17. [Google Scholar]
- Olafsen, J.A. (2001) Interactions between fish larvae and bacteria in marine aquaculture. Aquaculture 200: 223–247. [Google Scholar]
- Pedersen, K. , and Larsen, J. (1993) rRNA gene restriction patterns of Vibrio anguillarum serogroup O1. Dis Aquat Organ 16: 121–126. [Google Scholar]
- Planas, M. , Pérez‐Lorenzo, M. , Hjelm, M. , Gram, L. , Uglenes Fiksdal, I. , Bergh, Ø. , and Pintado, J. (2006) Probiotic effect in vivo of Roseobacter strain 27‐4 against Vibrio (Listonella) anguillarum infections in turbot (Scophthalmus maximus L.) larvae. Aquaculture 255: 323–333. [Google Scholar]
- Porsby, C.H. , Nielsen, K.F. , and Gram, L. (2008) Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish Turbot (Scophthalmus maximus)‐rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl Environ Microbiol 74: 7356–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prol, M.J. , Bruhn, J.B. , Pintado, J. , and Gram, L. (2009) Real‐time PCR detection and quantification of fish probiotic Phaeobacter strain 27‐4 and fish pathogenic Vibrio in microalgae, rotifer, Artemia and first feeding turbot (Psetta maxima) larvae. J Appl Microbiol 106: 1292–1303. [DOI] [PubMed] [Google Scholar]
- Rao, D. , Webb, J.S. , and Kjelleberg, S. (2005) Competitive interactions in mixed‐species biofilms containing the marine bacterium Pseudoalteromonas tunicata . Appl Environ Microbiol 71: 1729–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, H.I. , Treasurer, J.W. , Adam, B. , and Birkbeck, T.H. (2009) Analysis of bacterial populations in the gut of developing cod larvae and identification of Vibrio logei, Vibrio anguillarum and Vibrio splendidus as pathogens of cod larvae. Aquaculture 288: 36–43. [Google Scholar]
- Ringø, E. , Olsen, R.E. , Jensen, I. , Romero, J. , and Lauzon, H.L. (2014) Application of vaccines and dietary supplements in aquaculture: possibilities and challenges. Rev Fish Biol Fish 24: 1005–1032. [Google Scholar]
- Rønneseth, A. , Castillo, D. , D'Alvise, P. , Tønnesen, Ø. , Haugland, G. , Grotkjaer, T. , et al (2017) Comparative assessment of Vibrio virulence in marine fish larvae. J Fish Dis 40: 1373–1385. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Ponte, C. , Cilia, V. , Lambert, C. , and Nicolas, J.L. (1998) Roseobacter gallaeciensis sp. nov., a new marine bacterium isolated from rearings and collectors of the scallop Pecten maximus . Int J Syst Bacteriol 48: 537–542. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Ponte, C. , Samain, J.J.F. , Sánchez, J.L.J. , and Nicolas, J.J.L. (1999) The benefit of a Roseobacter species on the survival of scallop larvae. Mar Biotechnol 1: 52–59. [DOI] [PubMed] [Google Scholar]
- Salvesen, I. , Reitan, K.I. , Skjermo, J. , and Øie, G. (2000) Microbial environments in marine larviculture: impacts of algal growth rates on the bacterial load in six microalgae. Aquac Int 8: 275–287. [Google Scholar]
- Saulnier, D. , De Decker, S. , and Haffner, P. (2009) Real‐time PCR assay for rapid detection and quantification of Vibrio aestuarianus in oyster and seawater: a useful tool for epidemiologic studies. J Microbiol Methods 77: 191–197. [DOI] [PubMed] [Google Scholar]
- Skjermo, J. , and Vadstein, O. (1999) Techniques for microbial control in the intensive rearing of marine larvae. Aquaculture 177: 333–343. [Google Scholar]
- Skov, M.N. , Pedersen, K. , and Larsen, J.L. (1995) Comparison of pulsed‐field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum Serovar O1. Appl Environ Microbiol 61: 1540–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobecky, P.A. , Mincer, T.J. , Chang, M.C. , and Helinski, D.R. (1997) Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl Environ Microbiol 63: 888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sochard, M.R. , Wilson, D.F. , Austin, B. , and Colwell, R.R. (1979) Bacteria associated with the surface and gut of marine copepods. Appl Environ Microbiol 37: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerset, I. , Krossøy, B. , Biering, E. , and Frost, P. (2005) Vaccines for fish in aquaculture. Expert Rev Vaccines 4: 89–101. [DOI] [PubMed] [Google Scholar]
- Støttrup, J.G. , and Norsker, N.H. (1997) Production and use of copepods in marine fish larviculture. Aquaculture 155: 231–247. [Google Scholar]
- Støttrup, J.G. , Richardson, K. , Kirkegaard, E. , and Pihl, N.J. (1986) The cultivation of Acartia tonsa Dana for use as a live food source for marine fish larvae. Aquaculture 52: 87–96. [Google Scholar]
- Tamplin, M.L. , Gauzens, A.L.L. , Huq, A. , Sack, D.A.A. , and Colwell, R.R.R. (1990) Attachment of Vibrio cholerae serogroup 01 to zooplankton of Bangladesh waters. Appl Environ Microbiol 56: 1977–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, F.L. , Iida, T. , and Swings, J. (2004) Biodiversity of Vibrios. Microbiol Mol Biol Rev 68: 403–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toranzo, A.E. , Magariños, B. , and Romalde, J.L. (2005) A review of the main bacterial fish diseases in mariculture systems. Aquaculture 246: 37–61. [Google Scholar]
- Turingan, R.G. , Beck, J.L. , Krebs, J.M. , and Licamele, J.D. (2007) Development of feeding mechanics in marine fish larvae and the swimming behavior of zooplankton prey: implications for rearing marine fishes In Copepods in aquaculture. Lee C.S., O'Bryen P.J. and Marcus N.H. (ed.). Ames, IA: Blackwell Publishing Professional, pp. 119–132. [Google Scholar]
- Turner, J.T. (2004) The importance of small pelagic planktonic copepods and their role in pelagic marine food webs. Zool Stud 43: 255–266. [Google Scholar]
- Vandecandelaere, I. , Segaert, E. , Mollica, A. , Faimali, M. , and Vandamme, P. (2008) Leisingera aquimarina sp. nov., isolated from a marine electroactive biofilm, and emended descriptions of Leisingera methylohalidivorans Schaefer et al. 2002, Phaeobacter daeponensis Yoon et al. 2007 and Phaeobacter inhibens Martens et al. 2006. Int J Syst Evol Microbiol 58: 2788–2793. [DOI] [PubMed] [Google Scholar]
- Venmathi Maran, B.A. , Iwamoto, E. , Okuda, J. , Matsuda, S. , Taniyama, S. , Shida, Y. , et al (2007) Isolation and characterization of bacteria from the copepod Pseudocaligus fugu ectoparasitic on the panther puffer Takifugu pardalis with the emphasis on TTX. Toxicon 50: 779–790. [DOI] [PubMed] [Google Scholar]
- Vezzulli, L. , Pezzati, E. , Stauder, M. , Stagnaro, L. , Venier, P. , and Pruzzo, C. (2015) Aquatic ecology of the oyster pathogens Vibrio splendidus and Vibrio aestuarianus . Environ Microbiol 17: 1065–1080. [DOI] [PubMed] [Google Scholar]
- Wang, X.W. , Xu, J.D. , Zhao, X.F. , Vasta, G.R. , and Wang, J.X. (2014) A shrimp C‐type lectin inhibits proliferation of the hemolymph microbiota by maintaining the expression of antimicrobial peptides. J Biol Chem 289: 11779–11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WWF and ZSL (2015) Living Blue Planet Report – Species, habitats and human well‐being.
- Ye, J. , Coulouris, G. , Zaretskaya, I. , Cutcutache, I. , Rozen, S. , and Madden, T.L. (2012) Primer‐BLAST: a tool to design target‐specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]


