Summary
The biological detoxification of mycotoxins, including deoxynivalenol (DON), represents a very promising approach to address the challenging problem of cereal grain contamination. The recent discovery of Devosia mutans 17‐2‐E‐8 (Devosia spp. 17‐2‐E‐8), a bacterial isolate capable of transforming DON to the non‐toxic stereoisomer 3‐epi‐deoxynivalenol, along with earlier reports of bacterial species capable of oxidizing DON to 3‐keto‐DON, has generated interest in the possible mechanism and enzyme(s) involved. An understanding of these details could pave the way for novel strategies to manage this widely present toxin. It was previously shown that DON epimerization proceeds through a two‐step biocatalysis. Significantly, this report describes the identification of the first enzymatic step in this pathway. The enzyme, a dehydrogenase responsible for the selective oxidation of DON at the C3 position, was shown to readily convert DON to 3‐keto‐DON, a less toxic intermediate in the DON epimerization pathway. Furthermore, this study provides insights into the PQQ dependence of the enzyme. This enzyme may be part of a feasible strategy for DON mitigation within the near future.
Introduction
Deoxynivalenol (DON, also known as vomitoxin) is one of the most commonly detected mycotoxins during the course of infection by Fusarium graminearum, which causes head blight and ear rot diseases. DON is secreted by the fungal pathogen and acts as a potent virulence factor, aiding in disease spread (Proctor et al., 1995; Maier et al., 2006). These diseases result in considerable economic losses due to reduced yields and diminished grain quality from mycotoxin contamination (Lee and Ryu, 2015; Zhang et al., 2016). More importantly, DON has negative consequences on both human and animal health, (Streit et al., 2013; Hassan et al., 2015) causing gastrointestinal issues especially in mono‐gastric animals (Pestka, 2010a). Acute exposure leads to emesis associated with a total loss of appetite (Rotter et al., 1996) while long‐term exposure leads to lack of weight gain, reduced immunity and increased sensitivity towards disease (Pestka, 2010b; Streit et al., 2013; Payros et al., 2016). Furthermore, DON contamination is widespread within the cereal industry with reports showing its presence in 55% of samples tested (Streit et al., 2013).
Grains contaminated with DON must be sold at deeply discounted rates for alternative uses, such as the fuel ethanol industry, where high levels of DON can still contaminate the animal feed produced (Wu et al., 2016). Due to the large economic losses (estimated at billions of dollars within the North American economy during an outbreak in the 1990s) (Bhat et al., 2010), DON mitigation strategies have been intensely explored in the past three decades (Zhu et al., 2016), but no solution has been found. Several attempts to reduce DON levels through chemical means have been deemed impractical due to the associated costs or their detrimental effect on the final grain quality (Karlovsky et al., 2016). The microbial detoxification of DON by various species has been demonstrated using a wide variety of microorganisms and been extensively reviewed (Karlovsky, 2011; Zhu et al., 2016). Earlier reported mechanisms of DON microbial detoxification have focused on de‐epoxidation, transformation of the C3 carbon (or attached groups), or the complete mineralization of DON (Karlovsky, 2011; Zhu et al., 2016). Reported modifications of C3 include oxidation and epimerization of the attached hydroxyl group, which was demonstrated to reduce toxicity (He et al., 2015a,b). Devosia mutans 17‐2‐E‐8 (Devosia spp. 17‐2‐E‐8) has been reported to epimerize DON to 3‐epi‐DON (He et al., 2016), which was recently shown to be a two‐step process: the first step oxidizes DON to 3‐keto‐DON and the second step stereo‐selectively reduces 3‐keto‐DON to 3‐epi‐DON (Hassan et al., 2017). The reduced toxicity of both reported enzymatic products (3‐keto‐DON intermediate and the final 3‐epi‐DON stereoisomer), demonstrated in in vitro and in vivo experimental models, opens up the potential of finding an enzymatic solution to address DON contamination. Enzymes that play a role in DON detoxification through epimerization have been under intense investigation by the mycotoxin community, but their identities still remain elusive (Karlovsky, 2011). Identifying the enzymes responsible will enable the development of multiple technologies that could prove immediately useful. For example, the purified enzymes could be applied directly to contaminated animal feed, incorporated into industrially important microorganisms such as lactic acid bacteria or yeasts, or inserted directly into plants to detoxify DON in situ and enhance the resistance against Fusarium diseases.
We hypothesized that the DON epimerization (Dep) pathway in D. mutans 17‐2‐E‐8 contains at least two putative enzymes, designated as DepA and DepB, which are responsible for the oxidation of DON to 3‐keto‐DON (Fig. 1) and the reduction of 3‐keto‐DON to 3‐epi‐DON respectively. Our research group has extensively characterized the D. mutans 17‐2‐E‐8 isolate and, through a combination of RNA‐sequencing and comparative genomics, found evidence to suggest that the redox cofactor pyrroloquinoline quinone (PQQ) plays a significant role in its physiology (unpublished data). Based on this realization, we also posited that the Dep pathway may be PQQ dependent. The presented work describes the identification of DepA from D. mutans 17‐2‐E‐8, the first reported enzyme capable of oxidizing DON to 3‐keto‐DON. This report should help to significantly increase the pace at which DON mitigation strategies are developed.
Figure 1.

Devosia mutans 17‐2‐E‐8 transforms DON into 3‐keto‐DON.
Results and discussion
Partial purification of DepA from Devosia mutans 17‐2‐E‐8
Epimerization activity was initially assayed in crude lysates from D. mutans 17‐2‐E‐8 so an effort was made to purify the first enzyme in the pathway. A frozen glycerol stock of D. mutans 17‐2‐E‐8 was used to inoculate LB media containing 34 μg ml−1 of kanamycin, shaking at 200 RPM at 28°C. After two weeks, 40 ml of this culture was used to inoculate four litres of LB, and the cultures were grown for six days, centrifuged to pellet the cells and the pellets were stored at −20°C. Frozen pellets were thawed and cells resuspended in 50 mM Tris, pH 8.0 with 150 mM NaCl and were lysed by extensive sonication. The lysate was centrifuged and filtered through a 0.45 μm syringe filter and Halt Protease Inhibitor Cocktail, (Thermo Fisher, Mississauga, Canada) without EDTA, was added. To ensure DepA was still active and soluble, the clarified lysate was tested for activity. One part lysate was combined with four parts buffer (Tris, pH 7.5) with a final concentration of DON at 100 μg ml−1. After an overnight incubation, the reaction was subjected to HPLC. Samples and standards (20 μl) were separated by a Proteo Phenomenex column using a 15‐min isocratic elution of 23% acetonitrile in water. 3‐keto‐DON was detected by monitoring the absorbance at 218 nm and quantified by comparing to a known commercial 3‐keto‐DON standard (TripleBond, Guelph, Canada). Compared to control reactions, in which no 3‐keto‐DON was detected, the treated samples had a peak corresponding to 3‐keto‐DON with a corresponding decrease in the amount of DON.
In an attempt to isolate the responsible enzyme, a series of protein purification steps were undertaken. After ammonium sulfate precipitation (Duong‐Ly and Gabelli, 2014), the vast majority of the DepA activity was found among the proteins that precipitated between 50 and 70% of saturating conditions. This fraction was then dialysed to remove excess ammonium sulfate and heated at 60°C to precipitate less stable proteins.
The protein solution was then subjected to anion exchange chromatography to further purify the enzymes responsible for the DepA activity (Fig. S1A). Fractions (and flow through) were assessed for DepA activity, and those with high activity were pooled and concentrated. Fractions were also subjected to SDS‐PAGE revealing a band of approximately 62 kDa, the intensity of which positively correlated with DepA activity (Fig. S1B). The pooled fractions were then subjected to size exclusion column chromatography (Fig. S2A). The resulting fractions were again assessed for DepA activity and were pooled and concentrated. SDS‐PAGE was performed, and the abundance of the band representing one or more proteins of approximately 62 kDa had a positive correlation with DepA activity (Fig. S2B). The protein corresponding to this band was thought to be a prime candidate for DepA.
DepA activity was stable for at least one month at 4°C and after a heat treatment of 60°C in the partially purified samples. The stability of this enzyme is quite important as some of its potential downstream applications, such as corn wet‐milling, require maintaining this activity at moderately high temperatures (~50°C) (Jackson and Shandera, 1995).
Initial cofactor analysis and protein sequencing
An initial cofactor analysis was performed to help determine the required cofactors for the reaction. Various cofactors such as NADH, NADPH, NAD+, NADP+ and ATP, which are commonly involved in enzymatic reactions, as well as PQQ, which is believed to play an important role in D. mutans 17‐2‐E‐8 metabolism, as mentioned above, were individually added to reactions to test their effect on DepA activity. Only PQQ increased DepA activity; previously published reports have shown that PQQ can only be dialysed out of certain PQQ‐dependant dehydrogenases in the presence of EDTA (Görisch et al., 1989). This strong binding may explain why even after repeated rounds of dialysis and column chromatography there was still some remaining enzymatic activity when exogenous PQQ was not added to the reaction. PQQ appeared to increase the rate at which DON was oxidized suggesting it likely plays some role in the activity of DepA.
To confirm this hypothesis and test the involvement of metal cofactors, EDTA was added to the protein solution. EDTA stopped DepA activity, indicating that the protein was likely metal dependent. After the EDTA was removed by dialysis, various metal ions were added; Ca2+, Mg2+ and Mn2+ (in the presence of PQQ) appeared to restore activity in overnight reactions (Table S1). Further results showed that, after the addition of Ca2+, in the absence of exogenous PQQ, no detectable activity was observed after 2 h and only minimal activity after ~60 h of incubation (Fig. 2). The addition of exogenous PQQ, however, drastically enhanced the amount of 3‐keto‐DON formed after both 2 and ~60 h. These results demonstrate that DepA is likely a PQQ‐ and metal‐dependant enzyme. Based on these results, we hypothesized that the metal stabilizes the binding of PQQ, as it was not until the metal was removed that a substantial amount of PQQ appeared to dissociate from the enzyme.
Figure 2.
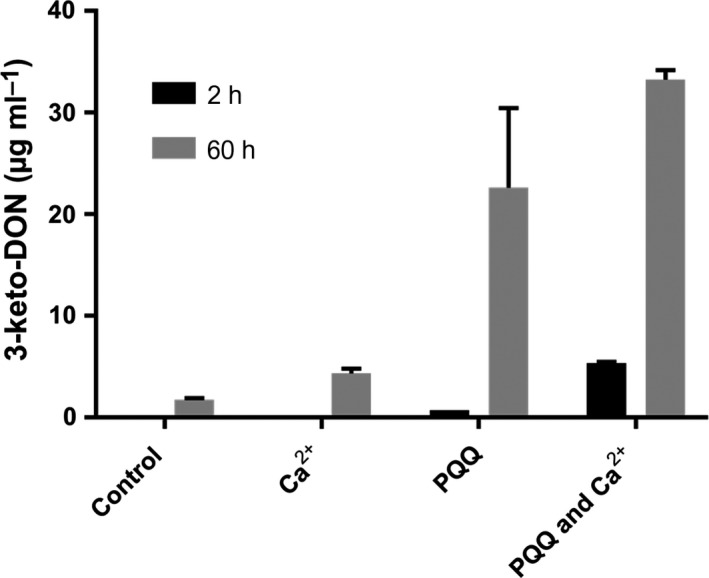
Effects of Ca2+ and PQQ on partially purified DepA from Devosia mutans 17‐2‐E‐8. Reactions contained 50 μg ml−1 DON in Tris, pH 7.5, with 1 mM Ca2+ and 100 μM PQQ. Buffers and water used to dilute substrates have been treated with Chelex 100 (Sigma) to remove any metal ion. This experiment was replicated with independently purified protein with similar results.
Aliquots of fractions 15 and 16 from size exclusion chromatography (Fig. S2) were subsequently sent for protein sequencing to the Advanced Analysis Centre at the University of Guelph (Guelph, ON, Canada), and the results were compared against the reference genome (Hassan et al., 2014) (Table S2). Fractions 15 and 16 contained 70 and 83 proteins, 51 and 57 of which contained at least two unique peptides respectively. An analysis of the protein sequencing results revealed that six of the proteins were annotated as PQQ‐dependent dehydrogenases and predicted to be of similar size to the band observed on the PAGE gel, thus prime targets for having DepA activity. The protein with the greatest number of unique peptides (fig|6666666.163324.CDS.378) had been annotated as an ATP‐binding protein (KFL25551.1) in the D. mutans 17‐2‐E‐8 genome sequence by NCBI and as a hypothetical protein by the RAST annotation server. However, a BLASTp search revealed a similar protein in Devosia spp. DBB001 (CDP53119.1) annotated as a PQQ‐dependent dehydrogenase, and furthermore, a PQQ‐specific beta‐propeller repeat domain was predicted by the NCBI Conserved Domain Database (CDD) in this sequence. This protein was therefore also included as a candidate. The subcellular localization of the genes was predicted using PSORTb 3.0 (Yu et al., 2010), and signal sequences, as predicted by SignalP 4.0 (Petersen et al., 2011), were removed prior to synthesis of the gene. The DNA sequences of the candidate genes were codon‐optimized for E. coli overexpression, synthesized and cloned into pET28a, by Genscript (Piscataway, NJ, USA). Candidate protein sequences are found in Table S3.
DepA activity of candidate genes
Each of the candidate genes in pET28a was transformed into E. coli BL21, expressed and purified by Ni‐NTA chromatography (Fig. S3). The expressed proteins were then each tested for DepA activity by separately incubating crude lysate and/or purified protein with DON in the presence of PQQ and CaCl2. The only protein able to transform DON to 3‐keto‐DON was fig|6666666.163324.CDS.378 (GenBank KFL25551.1), hereafter termed DepA. To confirm that DepA is a PQQ‐dependent enzyme, the activity of Apo‐DepA (E. coli does not produce PQQ so all DepA heterologously expressed would be in the Apo form) was compared to PQQ‐DepA (Fig. 3A). No 3‐keto‐DON was detected in reactions that lacked exogenous PQQ, confirming PQQ is necessary for activity. Also of note, there was no detectable DON after an overnight incubation with DepA, demonstrating that DON can be completely transformed (Fig. 3B). When using deoxynivalenol as a substrate and in the presence of PQQ and phenazine (to regenerate the PQQ) in Tris at pH 7.5, DepA has a k cat (s−1) of 4.18 ± 0.12 and a K m,app of 32 ± 4 μM and a catalytic efficiency (k cat/K m) of 13.0 × 104 M−1 s−1.
Figure 3.
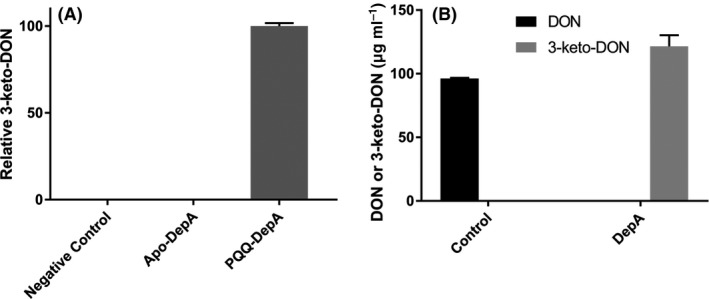
DON transformation ability of heterologously expressed DepA.
A. PQQ is essential for DepA activity. Reactions contained 100 μg ml−1 DON, in Tris pH 7.5 with 15 μg of purified DepA in the presence or absence of 100 μM PQQ.
B. DepA completely transforms DON to 3‐keto‐DON. Each reaction consisted of 100 μg ml−1 DON, 40 μM phenazine and 100 μM PQQ with or without 5 μg of DepA. Reactions were incubated overnight at room temperature and replicated with an independent purification.
The current approach successfully used a combination of protein purification and standard biochemical techniques, whereas many other groups looking for novel enzymes/functions regularly use genomic library screening techniques. In this case, that approach would not have revealed the function of this gene in most common expression systems, such as E. coli and yeast, which do not produce PQQ natively. We have demonstrated that the Apo form of DepA is not active and therefore without previous knowledge of the importance of PQQ, the activity could have been missed. The importance of host‐specific cofactors should not be overlooked when screening for novel enzyme activity from organisms that may produce unique or less common cofactors.
A BLASTp search of DepA against the NCBI nr database also revealed that genes with more than 90% identity exist in Devosia sp. DBB001, Devosia sp. H5989 (AKR58401.1) and interestingly, Pseudogluconobacter saccharoketogenes (BAB62258.1), which has not been reported to have DON detoxification activity. Surprisingly this Type I quinoprotein alcohol dehydrogenase (PQQ‐ADH) from P. saccharoketogenes has previously been crystalized and some biochemical characterization performed (Rozeboom et al., 2015). The structure (PDB: 4CVB) shows an eight‐bladed β‐propeller fold and contains a bound PQQ and Ca2+ ion near an open and accessible active site. The tight binding of PQQ to DepA can be explained by the number of hydrogen bonds seen between PQQ and PQQ‐ADH and hydrophobic stacking between a leucine and tryptophan. Additionally, the O5, N6 and 07A atoms of PQQ are bound to the Ca2+ ion. PQQ‐ADH has broad substrate specificity, including sugars, oligosaccharides and alcohols, but its ability to oxidase DON has not yet been tested. However, due to the close sequence similarity, conserved active site (Fig. S4) and broad substrate specificity, it is expected that this enzyme will possess this activity.
Conclusions
The identification of an enzyme that can oxidize DON to 3‐keto‐DON is a significant step towards identifying a complete pathway of DON detoxification. The capability to transform DON to the less toxic 3‐keto‐DON has important applications for many industries, including livestock production, corn milling and fuel ethanol fermentation. If this enzyme, or a microorganism containing the enzyme, can be incorporated into production lines, it could drastically reduce DON levels in the animal feed leaving the facilities. In seasons with highly contaminated corn or wheat, producers may also be able to accept grain with certain levels of DON without jeopardizing the quality of their final products. This will add value to both farmers supplying the grain and to the processors selling the feed. This enzyme may also have applications at the farm level, where it could be applied directly to the feed. Furthermore, as DON is a virulence factor during wheat infection by Fusarium, incorporating this gene into plants may provide significant resistance against Fusarium infection. Although there may still be challenges to face, such as the requirement for PQQ (or a PQQ‐like cofactor), the discovery of this enzyme's capability to degrade DON demonstrates the potential that soil bacteria offer for detoxifying mycotoxins and provides exciting opportunities for future work.
Conflict of interest
None declared.
Supporting information
Fig. S1. (A) Elution profile of the protein solution separated by anion exchange.
Fig. S2. (A) Elution profile of the protein solution separated by size exclusion.
Fig. S3. Ni‐NTA purification of potential DepA candidates.
Fig. S4. Sequence alignment of DepA and PQQ‐ADH from Pseudogluconobacter saccharoketogenes.
Table S1. Metal Specificity of partially purified DepA.
Table S2. Proteins identified by peptide sequencing in partially purified fractions.
Table S3. Candidate genes selected for protein expression to assess putative DepA activity.
Acknowledgements
The authors would like to thank Dyanne Brewer and Armen Charchoglyan of the University of Guelph Advanced Analysis Centre and Honghui Zhu and Xiu‐Zhen Li of Guelph Research and Development Center for technical assistance. The financial support from Agriculture and Agri‐Food Canada for this research is much appreciated (Project ID J‐001498).
Microbial Biotechnology (2018) 11(6), 1106–1111
Funding Information
The financial support from Agriculture and Agri‐Food Canada for this research is much appreciated (Project ID J‐001498).
References
- Bhat, R. , Rai, R.V. , and Karim, A.A. (2010) Mycotoxins in food and feed: present status and future concerns. Compr Rev Food Sci Food Saf 9: 57–81. [DOI] [PubMed] [Google Scholar]
- Duong‐Ly, K.C. , and Gabelli, S.B. (2014) Salting out of proteins using ammonium sulfate precipitation. Methods Enzymol 541: 85–94. [DOI] [PubMed] [Google Scholar]
- Görisch, H. , Geiger, O. and Adler, M. (1989) Mode of binding of pyrroloquinoline quinone to glucose dehydrogenase from acinetobacter calcoaceticus In PQQ and Quinoproteins: Proceedings of the First International Symposium on PQQ and Quinoproteins, Delft, The Netherlands, 1988. Jongejan, J.A. and Duine, J.A. (eds). Dordrecht: Springer Netherlands, pp. 100–102. [Google Scholar]
- Hassan, Y.I. , Lepp, D. , He, J. and Zhou, T. (2014) Draft genome sequences of Devosia sp. Strain 17‐2‐E‐8 and Devosia riboflavina Strain IFO13584. Genome Announc 2(5), e00994–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan, Y.I. , Watts, C. , Li, X.Z. , and Zhou, T. (2015) A novel Peptide‐binding motifs inference approach to understand deoxynivalenol molecular toxicity. Toxins 7: 1989–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan, Y.I. , He, J.W. , Perilla, N. , Tang, K. , Karlovsky, P. , and Zhou, T. (2017) The enzymatic epimerization of deoxynivalenol by Devosia mutans proceeds through the formation of 3‐keto‐DON intermediate. Sci Rep 7: 6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J.W. , Bondy, G.S. , Zhou, T. , Caldwell, D. , Boland, G.J. , and Scott, P.M. (2015a) Toxicology of 3‐epi‐deoxynivalenol, a deoxynivalenol‐transformation product by Devosia mutans 17‐2‐E‐8. Food Chem Toxicol 84: 250–259. [DOI] [PubMed] [Google Scholar]
- He, J.W. , Yang, R. , Zhou, T. , Boland, G.J. , Scott, P.M. , and Bondy, G.S. (2015b) An epimer of deoxynivalenol: purification and structure identification of 3‐epi‐deoxynivalenol. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 32: 1523–1530. [DOI] [PubMed] [Google Scholar]
- He, J.W. , Hassan, Y.I. , Perilla, N. , Li, X.Z. , Boland, G.J. , and Zhou, T. (2016) Bacterial epimerization as a route for deoxynivalenol detoxification: the influence of growth and environmental conditions. Front Microbiol 7: 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D.S. , and Shandera, D.L. Jr (1995) Corn wet milling: separation chemistry and technology. Adv Food Nutr Res 38: 271–300. [DOI] [PubMed] [Google Scholar]
- Karlovsky, P. (2011) Biological detoxification of the mycotoxin deoxynivalenol and its use in genetically engineered crops and feed additives. Appl Microbiol Biotechnol 91: 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlovsky, P. , Suman, M. , Berthiller, F. , De Meester, J. , Eisenbrand, G. , Perrin, I. , et al (2016) Impact of food processing and detoxification treatments on mycotoxin contamination. Mycotoxin Res 32: 179–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenna, R. , McGettigan, P.A. , McWilliam, H. , et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics (Oxford, England) 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee, H.J. , and Ryu, D. (2015) Advances in mycotoxin research: public health perspectives. J Food Sci 80: T2970–T2983. [DOI] [PubMed] [Google Scholar]
- Maier, F.J. , Miedaner, T. , Hadeler, B. , Felk, A. , Salomon, S. , Lemmens, M. , et al (2006) Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol Plant Pathol 7: 449–461. [DOI] [PubMed] [Google Scholar]
- Payros, D. , Alassane‐Kpembi, I. , Pierron, A. , Loiseau, N. , Pinton, P. , and Oswald, I.P. (2016) Toxicology of deoxynivalenol and its acetylated and modified forms. Arch Toxicol 90: 2931–2957. [DOI] [PubMed] [Google Scholar]
- Pestka, J.J. (2010a) Deoxynivalenol: mechanisms of action, human exposure, and toxicological relevance. Arch Toxicol 84: 663–679. [DOI] [PubMed] [Google Scholar]
- Pestka, J.J. (2010b) Deoxynivalenol‐induced proinflammatory gene expression: mechanisms and pathological sequelae. Toxins 2: 1300–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, T.N. , Brunak, S. , von Heijne, G. , and Nielsen, H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Proctor, R.H. , Hohn, T.M. , and McCormick, S.P. (1995) Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. MPMI 8: 593–601. [DOI] [PubMed] [Google Scholar]
- Robert, X. , and Gouet, P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter, B.A. , Prelusky, D.B. , and Pestka, J.J. (1996) Toxicology of deoxynivalenol (vomitoxin). J Toxicol Environ Health 48: 1–34. [DOI] [PubMed] [Google Scholar]
- Rozeboom, H.J. , Yu, S. , Mikkelsen, R. , Nikolaev, I. , Mulder, H.J. , and Dijkstra, B.W. (2015) Crystal structure of quinone‐dependent alcohol dehydrogenase from Pseudogluconobacter saccharoketogenes. A versatile dehydrogenase oxidizing alcohols and carbohydrates. Protein Sci 24: 2044–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit, E. , Naehrer, K. , Rodrigues, I. , and Schatzmayr, G. (2013) Mycotoxin occurrence in feed and feed raw materials worldwide: long‐term analysis with special focus on Europe and Asia. J Sci Food Agric 93: 2892–2899. [DOI] [PubMed] [Google Scholar]
- Wu, L. , Li, J. , Li, Y. , Li, T. , He, Q. , Tang, Y. , et al (2016) Aflatoxin B(1), zearalenone and deoxynivalenol in feed ingredients and complete feed from different Province in China. J Anim Sci Biotechnol 7: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, N.Y. , Wagner, J.R. , Laird, M.R. , Melli, G. , Rey, S. , Lo, R. , et al (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics (Oxford, England) 26: 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Wong, J.W. , Krynitsky, A.J. , and Trucksess, M.W. (2016) Perspective on advancing FDA regulatory monitoring for mycotoxins in foods using liquid chromatography and mass spectrometry (review). J AOAC Int 99: 890–894. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Hassan, Y.I. , Watts, C. , and Zhou, T. (2016) Innovative technologies for the mitigation of mycotoxins in animal feed and ingredients—A review of recent patents. Anim Feed Sci Technol 216: 19–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (A) Elution profile of the protein solution separated by anion exchange.
Fig. S2. (A) Elution profile of the protein solution separated by size exclusion.
Fig. S3. Ni‐NTA purification of potential DepA candidates.
Fig. S4. Sequence alignment of DepA and PQQ‐ADH from Pseudogluconobacter saccharoketogenes.
Table S1. Metal Specificity of partially purified DepA.
Table S2. Proteins identified by peptide sequencing in partially purified fractions.
Table S3. Candidate genes selected for protein expression to assess putative DepA activity.


