Summary
Microsclerotia (MS) are pseudoparenchymatous aggregations of hyphae of fungi that can be induced in liquid culture for biocontrol applications. Previously, we determined that the high‐osmolarity glycerol (HOG) signalling pathway was involved in regulating MS development in the dimorphic insect pathogen Metarhizium rileyi. To further investigate the mechanisms by which the signalling pathway is regulated, we characterized the transcriptional factor MrMsn2, a homologue of the yeast C2H2 transcriptional factor Msn2, which is predicted to function downstream of the HOG pathway in M. rileyi. Compared with wild‐type and complemented strains, disruption of MrMsn2 increased the yeast‐to‐hypha transition rate, enhanced conidiation capacity and aggravated pigmentation in M. rileyi. The ▵MrMsn2 mutants were sensitive to stress, produced morphologically abnormal clones and had significantly reduced MS formation and decreased virulence levels. Digital expression profiling revealed that genes involved in antioxidation, pigment biosynthesis and ion transport and storage were regulated by MrMsn2 during conidia and MS development. Taken together, our findings confirm that MrMsn2 controlled the yeast‐to‐hypha transition, conidia and MS formation, and virulence.
Introduction
DNA‐binding and multimerization domains are often used to classify transcriptional factors (TFs) into basic leucine zipper, zinc finger motif, helix–turn–helix and helix–loop–helix types (Park et al., 2008; Chai et al., 2012; Jung et al., 2015; Yin et al., 2017). As the core of signalling pathway, fungal TFs are important for transcriptional regulation of gene expression during cellular growth, secondary metabolism, stress responses and pathogenesis (Klug, 2010; Hong et al., 2013; Liu et al., 2013; Marinho et al., 2014; Zhang et al., 2014; Huang et al., 2015; Shelest, 2017; Yin et al., 2017; Song et al., 2018).
Filamentous fungal Msn2/4 homologues are C2H2‐like zinc finger TFs that regulate the general stress response, pathogenicity, secondary metabolism and cellular growth. They are similar to ScMsn2/4 of Saccharomyces cerevisiae (Schmitt and Mcentee, 1996) and have been characterized in Aspergillus parasiticus, A. flavus, Beauveria bassiana, Magnaporthe oryzae, Metarhizium robertsii and Verticillium dahliae (Chang et al., 2011; Liu et al., 2013; Zhang et al., 2014; Tian et al., 2017). Under abiotic and biotic stresses, Msn2/4 is phosphorylated for translocation from the cytoplasm to the nucleus, where it drives the transcription of stress‐induced genes (Hansen et al., 2015; Yi and Huh, 2015; Li et al., 2017). The underlying mechanism of the regulation of Msn2/4 activity by protein kinase A (PKA), the rapamycin signalling pathway, the Snf1 protein kinase pathway and the high‐osmolarity glycerol (HOG) pathway have been identified (Liu et al., 2013; Zhang et al., 2014; Li et al., 2017).
Microsclerotia (MS) are pseudoparenchymatous aggregations of hyphae with a diameter of 50–600 μm and become melanized during their development. As promising fungal propagules, MS can be induced in liquid culture and used for biocontrol applications such as biofungicides, bioherbicides, bionematicides and mycoinsecticides (Shearer, 2007; Jackson et al., 2010; Song et al., 2014, 2016a). To enhance the liquid fermentation efficiency of Metarhizium rileyi MS, we previously investigated the molecular mechanism of MS formation and demonstrated that internal oxidative stress triggers MS differentiation (Song et al., 2013, 2015, 2016b, 2018; Jiang et al., 2014). We found that HOG and cell wall integrity (CWI) pathways cooperate to regulate MS formation (Song et al., 2016b). We also found that M. rileyi MrMsn2 was predicted to function downstream of the HOG pathway and was upregulated during MS formation in comparative transcriptome analysis (Song et al., 2013). Furthermore, a bioinformatics analysis found no Msn4 orthologues in any public genome databases of M. rileyi (Song et al., 2013; Shang et al., 2016). These results imply a possible involvement of MrMsn2 in the regulation of MS development. However, this function has not been studied clearly.
Moreover, M. rileyi is a well‐known dimorphic entomopathogenic fungus with yeast‐like hyphal bodies and a true filamentous growth phase (Boucias et al., 2000; Fronza et al., 2017), which occurs synchronously in vivo and in vitro (Pendland and Boucias, 1997; Boucias et al., 2016). The yeast‐to‐hypha transition is critical for the pathogenesis and life cycle of dimorphic fungi (Wanchoo et al., 2009; Boyce and Adrianopoulos, 2015; Gauthier, 2015; Marcos et al., 2016). Although signalling pathways related to dimorphic transition are well characterized in the model yeast Candida albicans, the mechanisms are not well defined (Noble et al., 2017). Thus, studies on M. rileyi are useful model for understanding the dimorphic transition mechanism.
This study seeks to further elucidate the role of MrMsn2 in dimorphism transition, conidiation, virulence and MS formation by phenotypic analyses of deletion/rescue mutants constructed previously (Shao et al., 2015; Song et al., 2016b). We found that the absence of MrMsn2 resulted in increased yeast‐to‐hypha transition rate, enhanced conidiation capacity, aggravated pigmentation and induced or suppressed expression of target genes involved in the important phenotypes of M. rileyi, as presented below.
Results
Molecular characterization of MrMsn2
The full‐length sequence of MrMsn2 (GenBank Accession No.: MG641237) is 1752 bp, including one intron, with a calculated molecular weight of 56.7 kDa and an isoelectric point of 5.06 (http://expasy.org/tools/protparam.html). Furthermore, MrMsn2 contains a zinc finger double domain (Hu et al., 2014). In this study, a phylogenetic tree analysis demonstrated that the MrMsn2 protein from M. rileyi was closely related to other Metarhizium spp. proteins (Fig. S1). In addition, the amino acid sequence of MrMsn2 showed similarities (79–81% identity) to a cutinase G‐box binding protein of Metarhizium spp. (Hu et al., 2014; Shang et al., 2016) and a zinc finger protein (66% identity) of Tolypocladium ophiglossoides (Quandt et al., 2015).
To characterize the functions of the MrMsn2 gene in M. rileyi, gene replacement mutants and complementary transformants were generated. All recombinant strains were verified by polymerase chain reaction (PCR) and quantitative real‐time PCR (RT–qPCR) screening (Fig. S2). The confirmed ▵MrMsn2 mutants and the complemented (▵MrMsn2 + Msn2) strains were used in further experiments.
MrMsn2 negatively regulates yeast‐to‐hypha transition and conidiation
The M. rileyi CQNr01 (wild‐type, WT) strain was grown on solid Sabouraud maltose agar fortified with yeast extract (SMAY). Compared with the initial results at day 0, expression of MrMsn2 was found to be downregulated in the yeast‐to‐hypha transition at days 2 and 4 and conidiation initiation at day 6 (Fig. 1A). These results indicate that MrMsn2 may be involved in the yeast‐to‐hypha transition and conidiation.
Figure 1.
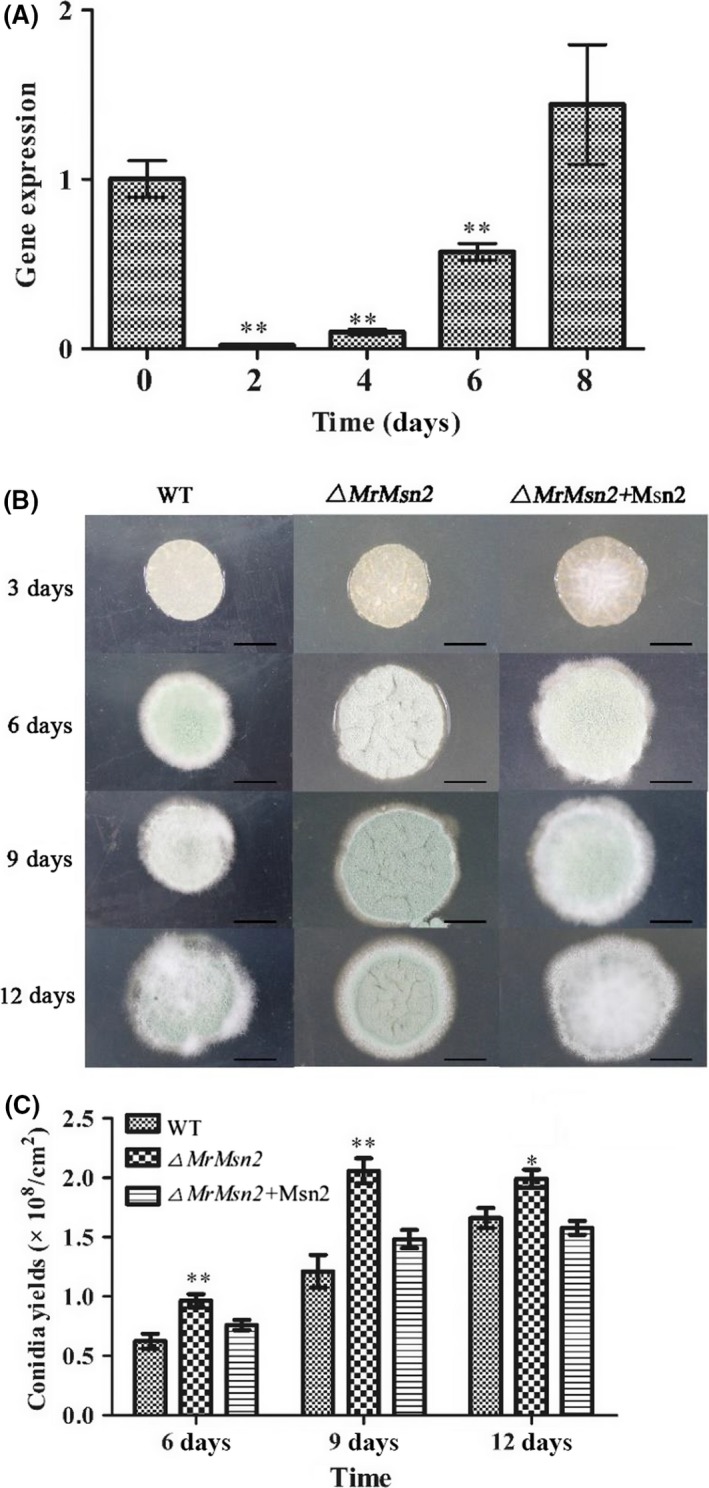
Mycelia growth, conidial morphology and yield of wild‐type, complemented and ▵MrMsn2 mutants strains on SMAY media and RT–qPCR for MrMsn2 expression during conidiation.
A. Transcription of MrMsn2 during conidiation on different cultivation days. 3 μl of conidial suspensions (1 × 107 conidia ml−1) was spotted on SMAY plates and cultured under continuous light at 25°C for 8 days. Stages of conidial development: inoculated conidia at initial culture time (day 0), blastospores (day 2), hyphal period (day 4), conidiation initiation (day 6) and conidia at start of maturation (day 8).
B. Images of colonies on SMAY plates for 3, 6, 9 and 12 days. 3 μl of conidial suspensions (1 × 107 conidia ml−1) was spotted on SMAY plates and cultured under continuous light at 25°C for 12 days. Bar = 5 mm.
C. Conidial yield of tested strains on SMAY medium after 6, 9 and 12 days of incubation. Error bars are standard error. * P < 0.05, ** P < 0.01, significantly different compared to wild type or 0 day incubation.
Further investigations showed that at day 3, the yeast‐to‐hyphae transition was advanced in ▵MrMsn2 mutants compared to the WT and complemented (CP) strains (Fig. 1B). Furthermore, colony surfaces of ▵MrMsn2 mutants were more convoluted compared to the normal smooth colony surfaces of WT and CP strains. After 6 days, the diameter of the mutant colonies was larger compared to that of the WT (Fig. 1B). Additionally, the ▵MrMsn2 mutants had significantly increased conidial yields compared to the WT and CP strains (P < 0.001) (Fig. 1C). After 9 and 12 days, conidial yield had increased by 1.5‐ to 2.2‐fold in ▵MrMsn2 mutants compared to the WT and CP strains. Taken together, these transcription and phenotype growth analysis suggest that MrMsn2 is involved in negative control of conidia production and yeast‐to‐hypha transition.
To further analyse the effect of MrMsn2 on the dimorphic transition, yeast cells of test strains was grown on SMAY medium. The investigations into the switching rates and median transition time required for 50% transition of blastospores to hyphae (TT50) showed a significantly difference among the ▵MrMsn2 mutants (TT50 = 5.4 ± 0.2 days), WT (TT50 = 7.3 ± 0.1 days) and CP (TT50= 7.0 ± 0.2 days) strains (P < 0.001) (Fig. 2). This suggests that deletion of MrMsn2 promoted the yeast‐to‐hypha transition.
Figure 2.
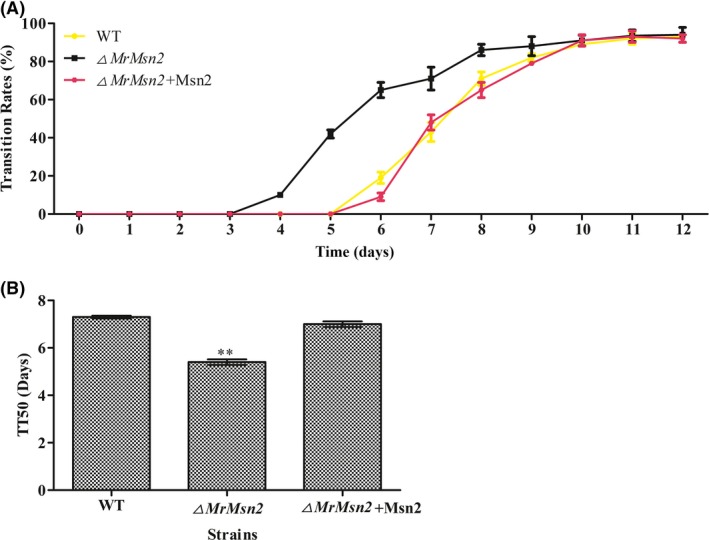
Quantitative analysis dimorphic transition of test strains.
A. Quantitative analysis of dimorphic transition (from yeast cells to hypha) rate of wild‐type, complemented and ▵MrMsn2 mutants with approximately 100 single yeast cells plated on SMAY medium. The growth morphology was observed every day.
B. Median transition time required for 50% transition of blastospores to hyphae (TT 50) of wild‐type, complemented and ▵MrMsn2 mutants was compared. TT 50 was calculated using a probit analysis with the SPSS program. Error bars are standard error. * P < 0.05, ** P < 0.01, significantly different compared with wild type.
Absence of MrMsn2 leads to aggravated pigmentation
After incubation on SMAY medium for 6 days, ▵MrMsn2 mutants were found to have altered aggravated pigmentation (Fig. 3A). After 9 and 12 days, the tergal pigment of ▵MrMsn2 mutants had increased compared to WT and CP strains respectively. Furthermore, the pigment concentration in ▵Mrsn2 mutants was found to be significantly increased by factors of 1.8‐ to 4.2‐fold, compared to that of WT and CP strains respectively (Fig. 3B). These data suggest that MrMsn2 had a negative influence on clone pigment biosynthesis.
Figure 3.
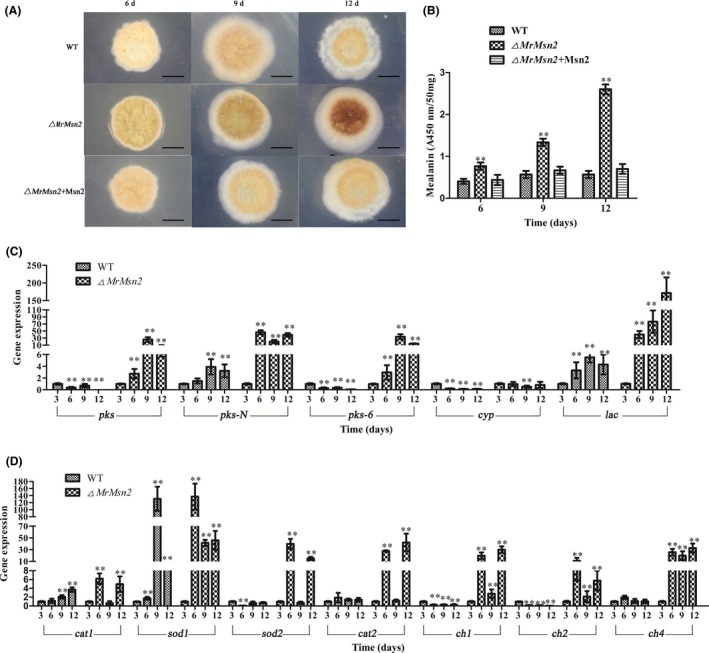
Pigment produced by wild‐type, complemented and ▵MrMsn2 mutant strains and RT–qPCR of pigment biosynthesis, chitin synthase and antioxidant enzyme genes.
A. Bottom of morphology clone of wild‐type, omplemented and ▵MrMsn2 mutants. Bar = 5 mm.
B. Quantitative analysis pigment produced. Pigment was calculated from three independent experiments and measured spectrophotometrically by absorbance at 459 nm. RT–qPCR of pigment biosynthesis‐related genes (C) and (D) antioxidation genes and chitin synthase genes during conidia development of wild‐type or ▵MrMsn2 mutants. At 3, 6, 9 and 12 days, clones of wild‐type or ▵MrMsn2 mutants were collected for RT–qPCR analysis. Mrtub and Mrtef genes were used as reference. Error bars are standard error. * P < 0.05, ** P < 0.01, significantly different compared with the results at day 3.
To investigate the mechanism of dimorphic transition, pigment biosynthesis and conidiation regulated by MrMsn2, genes that were potentially involved were selected from transcriptome libraries (Song et al., 2013, 2018) and examined by transcriptional analysis. The following genes were selected: pigment biosynthesis‐related genes (polyketide synthase, pks; polyketide synthase–non‐ribosomal peptide synthetase, pks‐N; polyketide synthase 6, pks‐6; conidial yellow pigment biosynthesis, cyp; and laccase, lac), several chitin synthase genes (ch1, ch2 and ch4, for class I, II and IV chitin synthases respectively) and antioxidant enzyme genes (cat1 for catalase‐1 and cat2 for catalase‐2, sod1 for superoxide dismutase‐1 and sod2 for superoxide dismutase‐2). It was found that the lac, pks‐N, cat1 and sod1 genes were upregulated, whereas pks, pks‐6, cyp, ch1 and ch2 genes were downregulated in the WT strain during conidiation (Figs. 3C and D). Compared with the WT, all pigment biosynthesis‐related gene and chitin synthase genes were significantly upregulated during conidiation in the ▵Mrsn2 mutants (Fig. 3C). In addition, the antioxidant enzyme genes were upregulated in aggravated pigmentation after 6 days for ▵Mrsn2 mutants (Fig. 3D).
MrMsn2 contributes to tolerance to abiotic stress
To examine the function of MrMsn2 on the abiotic stress response, strains were cultured under various abiotic stress conditions. Convoluted colony surfaces were more apparent, especially under cell wall perturbation and oxidative stress, for ▵MrMsn2 mutants compared to the normal smooth colony surfaces of the WT and CP strains, after 3 day incubation (Fig. S3A). Furthermore, compared to WT strain, smaller colonies were present in the ▵MrMsn2 mutants (Fig. S3A). After 12 days, the conidial yield of ▵MrMsn2 was significantly reduced between 26.6 and 91.9% on SMAY medium, under osmosensitivity, cell wall perturbation or oxidative stress (P < 0.001). Interestingly, the conidial yield of ▵MrMsn2 mutants was found to be significantly reduced under KCl stress, however, it was significantly increased under NaCl stress compared to the WT and CP strains (P < 0.001) (Fig. S3B). These results indicate MrMsn2 contributes to tolerance of abiotic stress.
MrMsn2 is needed for MS development
An expression analysis showed that the relative transcriptional of MrMsn2 peaked with MS initiation (72 h) (Fig. 4A) and MrMsn2 were upregulated in liquid amended medium (AM) or MM (AM without basal salts) medium, cultured with exogenous oxidative stress (Fig. 4B). These results suggest that MrMsn2 may be involved in the regulation of MS formation.
Figure 4.
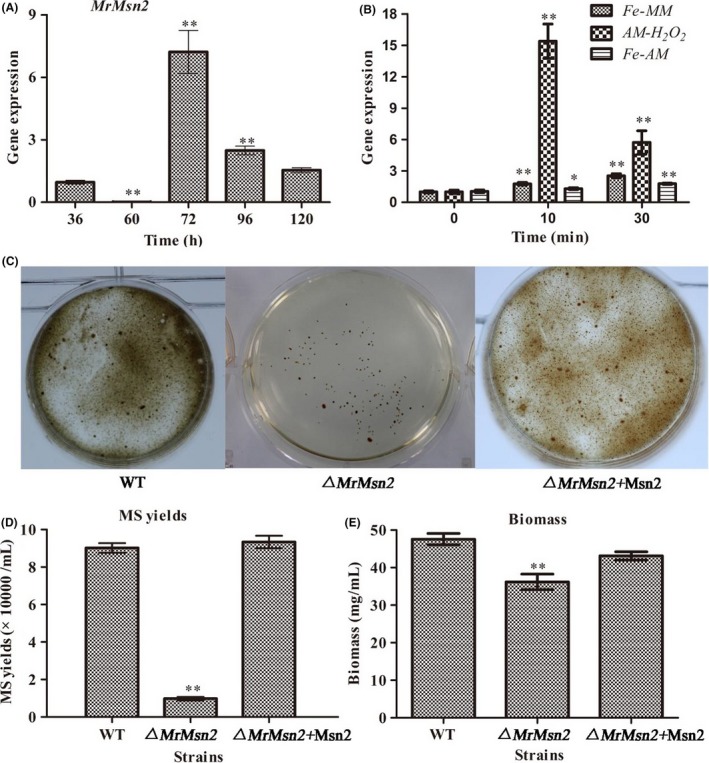
MS development in AM and RT–qPCR of MrMsn2 during MS development and independent treatment for exogenous oxidative stress.
A. Transcription of MrMsn2 during MS development.
B. Relative expression of MrMsn2 following independent treatments for exogenous iron and oxidative stress. Mrtub and Mrtef genes were used as reference.
C. Phenotypic characterization of MS of tested strains. AM cultures were inoculated with conidial suspensions of tested strains and cultured for 6 days. MS yield (D) and biomass (E) of tested strains. Error bars are standard error. * P < 0.05, ** P < 0.01, significantly different compared with wild type.
After incubation in liquid AM for 144 h, MS produced by WT and CP strains matured and were accompanied by secondary mycelia growth, whereas the density of the induced MS in the ▵MrMsn2 mutants was significantly decreased, with the ▵MrMsn2 culture broth exhibiting a low degree of pigmentation (Fig. 4C). Compared to the WT and CP strains, the MS yield of ▵MrMsn2 mutants was reduced by approximately 88.9% (Fig. 4D), and the biomass was decreased by 23.6% in the AM culture (Fig. 4E). These results indicate that MrMsn2 is needed for MS development.
Expression analysis during MS development
To investigate the genes regulated by MrMsn2 during MS formation, several groups of genes were analysed by qRT–PCR. It was found that antioxidation genes such as cat1, cat2, sod1, sod2 and the monooxygenase (mon) gene were upregulated in the ▵MrMsn2 mutants, while the flavoprotein–ubiquinone oxidoreductase (fuo) gene was downregulated (Fig. 5A). Pigment biosynthesis genes pks, pks‐6 and cyp were found to also be downregulated and pks‐N and lac genes were upregulated in the ▵MrMsn2 mutants (Fig. 5A). Additionally, chitin synthase genes ch2 and ch4 were significantly upregulated in the ▵MrMsn2 mutants and ch1 was significantly downregulated (Fig. 5B). Interestingly, Slt2, the core gene of the CWI signalling pathway, and hog1, the core gene of the HOG signalling pathway, were significantly upregulated in the ▵MrMsn2 mutants (Fig. 5B). Finally, transport and storage genes for major salts, sidA (siderophore iron transporter), ct‐1 (calcium‐transporting ATPase 1), ct‐2 (calcium‐transporting p‐type ATPase) and ccca (vacuolar Fe2+/Mn2+ transporter) were all found to be significantly upregulated in the ▵MrMsn2 mutants (Fig. 5B).
Figure 5.
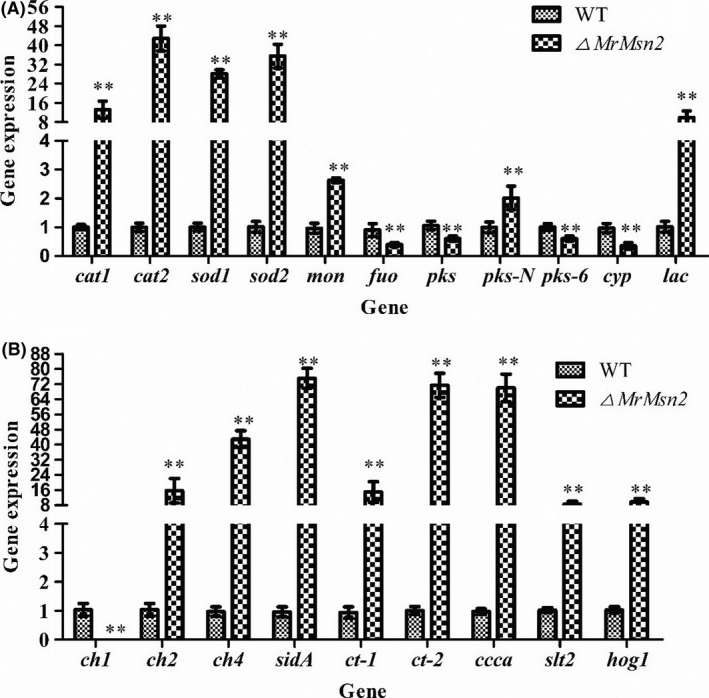
Relative transcripts of MS development‐related genes after 72‐h incubation in AM culture. Transcription of (A) antioxidation genes and pigment biosynthesis‐related genes, (B) chitin synthase genes and major salts transport and store genes, and other genes were carried out during MS development. Wild‐type and ▵MrMsn2 mutants were incubated for 72 h in AM cultures. Relative transcript abundances of genes were measured by RT–qPCR. Mrtub and Mrtef genes were used as reference. Error bars are standard error. * P < 0.05, ** P < 0.01, significantly different compared with wild type.
MrMsn2 is required for the virulence of M. rileyi
Pathogenicity assays were conducted using third‐instar Spodoptera litura larvae. These assays showed that the virulence of the ▵MrMsn2 mutants was significantly lower than of the WT and CP strains (Fig. 6). The mean lethal time (LT50) for the WT strain was 6.2 ± 0.4 days in a topical bioassay and 4.3 ± 0.5 days in the injection bioassays. The LT50 values for the CP strains were 6.3 ± 0.5 days in topical bioassays and 4.9 ± 0.5 days in injection bioassays, whereas the LT50 values for the ▵MrMsn2 mutants were 10.1 ± 0.5 (P < 0.001) in the topical bioassay and 8.6 ± 0.4 (P < 0.001) in the injection bioassays. These results show that MrMsn2 is required for virulence.
Figure 6.
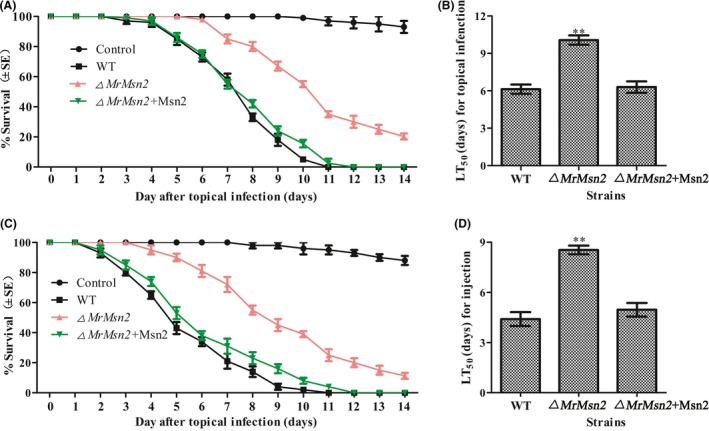
Insect bioassays. Insect survival after (A) topical application or (C) injection of conidia of tested strains. Mean lethal time for (B) topical infections and (D) injection applications. Topical infections were generated by immersing of conidial suspensions of tested strains (5 μl of 1 × 107 conidia ml−1 in cotton seed oil). For injection assays, insects were injected conidial suspensions of tested strains (5 μl of 1 × 106 conidia ml−1 in sterile water with 0.01% Tween 80). Three replicate groups had 30 larvae each. Controls were treated with pure cotton seed oil or sterile water containing 0.01% Tween 80 only. Error bars are standard error of three trials. * P < 0.05, ** P < 0.01, significantly different compared with wild type.
Discussion
In our previous investigation, we found that the HOG signalling pathway regulated the dimorphic transition and MS development (Song et al., 2016b). To better define the mechanisms of regulation, in this study, we identified and characterized the transcription factor MrMsn2, which is predicted to be downstream of the HOG pathway in M. rileyi. The results were unexpected because the gene was important for conidiation, the yeast‐to‐hypha transition and MS formation.
MrMsn2 belongs to a group of proteins containing C2H2‐like Zn finger domains that are important in development, secondary metabolism and stress responses (Chang et al., 2011; Liu et al., 2013; Zhang et al., 2014; Tian et al., 2017). In this study, ▵MrMsn2 mutants negatively controlled the yeast‐to‐hypha transition (Figs 1 and 2). This result was unlike the observation in Yarrowia lipolytica yeast, in which disruption of Mhy1p, an Msn2/4‐like protein, restricted the dimorphic transition, and in C. albicans where it had no significant role in CaMsn2/CaMsn4 mutations (Hurtado and Rachubinski, 1999; Nicholls et al., 2004). These studies show distinct strategies for regulating the yeast‐to‐hypha transition using Msn2 in different fungi.
Moreover, similar to A. parasiticus and A. flavus (Chang et al., 2011), deletion of MrMsn2 inhibited growth but increased production of conidia (Fig. 1). This was unlike in ▵MoMsn2 in M. oryzae, ▵Bbmsn2 in B. bassiana or ▵Mrmsn2 in M. robertsii, which decreased conidia production (Liu et al., 2013; Zhang et al., 2014). Msn2‐like proteins do not appear to be important in transcriptional regulation of the stress response in C. albicans and V. dahliae (Nicholls et al., 2004; Tian et al., 2017), but are important in the stress response in C. glabrata, and S. cerevisiae (Schmitt and Mcentee, 1996; Roetzer et al., 2008). Our research found that ▵MrMsn2 mutants were defective in response to osmotic, cell wall perturbation and oxidative stress (Fig. S3). These studies show distinct strategies for regulating the stress response and conidiation by Msn2 in different fungi.
In eukaryotes microorganism, cellular developmental processes are reported to correlate with increased reactive oxygen species (ROS) levels (Georgiou et al., 2006; Takemoto et al., 2007). Fungi have evolved effective antioxidant mechanisms that include enzyme families that act as ROS scavengers. Previous studies have shown members of antioxidant enzymes families such as SODs and CATs may have complementary effects during the cellular developmental processes (Xie et al., 2012; Youseff et al., 2012; Wang et al., 2013; Zhang and Feng, 2018). Our investigations confirmed that antioxidant enzyme genes cat1, sod1, cat2 and sod2 had different expressions during conidia development in M. rileyi (Fig. 3). In response to ROS stress, Msn2/4 accumulated in the nucleus, where they promoted transcriptional activation of stress‐responsive genes (Hansen et al., 2015; Yi and Huh, 2015). However, deletion of MrMsn2 increased production of ROS (data not shown) and upregulated expression of antioxidant genes to protect against ROS. Upregulation of these antioxidant genes in ▵MrMsn2 mutants suggests that there may be regulated by other signalling network (Zhang and Feng, 2018).
Secondary metabolism, such as pigments production, is triggered and intensified by ROS build‐up, with pigments being important for protecting the fungi against stress conditions (Cho et al., 2012; Hong et al., 2013). In the absence of MrMsn2 aggravated pigmentation and pigment synthesis‐associated genes were significantly upregulated during conidiation in ▵MrMsn2 mutants. Chitin is a main component of cell walls and is related to morphogenesis and adaptation to ecological niches (Roncero, 2002; Liu et al., 2017). Expression analysis showed that genes from the class I and II of chitin synthases were downregulated during morphogenesis and conidiation, and then in the absence of MrMsn2, these were upregulated (Fig. 3). Based on this data, we propose a link between chitin biosynthesis and MrMsn2, however, the molecular mechanism remains unknown.
Current conidia mass production methods are not cost‐effective, limiting M. rileyi commercialization. MS can be used as an alternative fungal propagule for mycoinsecticide (Song et al., 2014) and has been used in large‐scale production in submerged fermentation (Song et al., 2017). As for solid culture, ▵MrMsn2 mutants were defective in hyphal growth in liquid AM (data not shown). Furthermore, vegetative hyphae are the prerequisite for MS formation (Song et al., 2013; Jiang et al., 2014). Consistent with the defective MS formation in the ▵MrHog1 mutants, the ▵MrMsn2 mutants had limited ability to form MS (Fig. 4). This result was unlike the observation in V. dahliae, in which VdMsn2 deletion mutants produce more MS than wild type on solid media (Tian et al., 2017). Previously, it was found that the CWI and HOG signalling pathways cooperate to regulate MS development (Song et al., 2016b). This has been confirmed in this study, with the core genes of the CWI and HOG signalling pathway both being upregulated in the ▵MrMsn2 mutants.
Our previous study demonstrated that intracellular H2O2 levels fluctuated during MS development and peak at MS initiation stage (Song et al., 2018). Antioxidant enzyme genes were significantly upregulated in ▵MrMsn2 mutants (Fig. 5), indicating that intracellular H2O2 levels were not equilibrated in the MS‐initiating formation. This result confirmed our previous results that oxidative stress triggered MS formation (Song et al., 2013, 2015, 2016b, 2018; Jiang et al., 2014). As mentioned, the signalling network that regulates the chitin synthesis‐ and pigment synthesis‐associated genes in solid SMAY and liquid AM culture is highly complicated. Major basal salts such as iron and calcium cations are necessary for MS formation (Song et al., 2014) and SidA is the major pathway of cellular iron uptake for MS formation (Li et al., 2016). We demonstrated that the iron importer and calcium transports, Ct‐1 and Ct‐2, were important in MS formation (Wang and Yin, unpublished data). In addition to similar transcriptional mechanisms by Msn2/4 for regulating ccc1 in yeast (Li et al., 2017), we found multiple transcriptional mechanisms for regulating genes for iron and calcium cation transport and storage by MrMsn2 during MS development (Fig. 5). However, the transcriptional mechanisms for regulating cation transport are not clear and further experiments are needed to elucidate the multiple mechanisms.
Research on the function of Msn‐like transcriptional factors in pathogenicity is widespread for entomopathogenic, human pathogenic and phytopathogenic fungi (Roetzer et al., 2008; Liu et al., 2013; Zhang et al., 2014). In M. rileyi, our data indicated that ▵MrMsn2 mutants were significantly less pathogenic than WT by both topical infection and injection assays (Fig. 6). Similar results were reported for ▵MoMsn2 mutants of M. oryzae, ▵Bbmsn2 mutants of B. bassiana, and ▵Mrmsn2 mutants of M. robertsii in which gene deletions cause decreased virulence (Liu et al., 2013; Zhang et al., 2014). In contrast, in C. glabrata, Msn2 was found to have no effect on virulence (Roetzer et al., 2008). One explanation for this is that ▵MrMsn2 mutants counter oxidative stress from hosts in vivo (Song et al., 2016a,b) and were hypersensitive to stress. Another explanation involves morphogenic defects in the mutants. This investigation revealed that vegetative growth of ▵MrMsn2 mutants was defective in haemocoel (data not shown). These results could be reasons why the mutants had decreased pathogenicity in vivo.
In summary, this study revealed the MrMsn2 had negative effects on the dimorphic transition and conidiation and was required for abiotic stress resistance, virulence, and MS formation. Furthermore, the current transcriptional networks of MrMsn2 during conidia and MS development will enhance our ability to comprehensively understand the molecular mechanism of yeast‐to‐hypha transition and conidia and MS development.
Experimental procedures
Strains, media and culture conditions
The M. rileyi CQNr01 strain was from the Engineering Research Center for Fungal Insecticides, Chongqing, China. WT and engineered strains were cultured on SMAY (Sabouraud maltose agar, fortified with 1% (w/v) yeast extract) under continuous light at 25°C for 12 days for the conidiation assays or in liquid AM (comprising of 40 g l−1 glucose, 2.5 g l−1 peptone, 5 g l−1 yeast extract, 4.0 g l−1 KH2PO4, 0.8 g l−1 CaCl2.2H2O, 0.6 g l−1 MgSO4.7H2O, 0.1 g l−1 FeSO4.7H2O, 37 mg l−1 CoCl2.6H2O, 16 mg l−1 MnSO4.H2O and 14 mg l−1 ZnSO4.7H2O) for the MS incubation assays according to previous methods (Song et al., 2016b). Escherichia coli DH5α (Invitrogen, Shanghai, China) was used for recombinant DNA manipulation and Agrobacterium tumefaciens AGL‐1 (Invitrogen, Shanghai, China) for fungal transformations. Both were cultured as previously described (Shao et al., 2015; Song et al., 2016b).
Gene cloning and bioinformatics analyses
To study the potential function of MrMsn2, the full genomic DNA sequence of MrMsn2 was amplified using primers MrMsn2‐L/MrMsn2‐R (Table S1) based on sequences of a previous transcriptomic library (Song et al., 2013). The protein sequence and potentially homologues from other fungal species were aligned with Blastp (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequences were aligned with DNAMAN software (http://www.lynnon.com). Neighbour‐joining tree was generated using the software MEGA 6.0 (http://www.megasofware.net) (Tamura et al., 2013).
Generation of deletion and complementation mutants
The M. rileyi genome was not annotated when we constructed the targeted gene deletion plasmid. Therefore, fusion primer and nested integrated PCR (Wang et al., 2011) with primers in Table S1 were used to obtain flanking regions (data not shown). Upstream and downstream flanking sequences were amplified using primers Ms‐LF/Ms‐LR and Ms‐RF/Ms‐RR (Table S1), respectively, digested with restriction endonucleases and inserted into the plasmid pPZP‐Hph‐Knock, a hygromycin B‐resistance vector. The resultant plasmid was named pPZP‐Hph‐msn2. For the mutant complementation strains, the open reading frame (ORF) of MrMsn2 with the promoter and terminator regions, was amplified based on the subsequently public annotated of the M. rileyi genome (Shang et al., 2016) using the primers Ms‐HF/Ms‐HR (Table S1). PCR products were digested by restriction endonucleases and ligated into the sulfonylurea resistance vector pPZP‐Sur‐Knock to generate the plasmid, pPZP‐Sur‐msn2. Disruption and complementation vectors were transformed into Agrobacterium and transformants were screened as described previously (Song et al., 2018).
Phenotypic analyses of test strains on SMAY media
To analyse the function of MrMsn2 in yeast‐to‐hyphae transition, vegetative growth, conidial development and abiotic stress tolerance, conidial suspensions of the tested strains were plated on SMAY as previously described (Song et al., 2016b). Colony morphology was investigated, and images were collected using a digital camera (60‐mm Macro lens; Canon Inc., Tokyo, Japan) and microscope. Conidia numbers were counted as previously described (Song et al., 2015).
In vitro, M. rileyi was grown in yeast cell form for 2–4 days on SMAY to transformed into the filamentous form. The switching rates of the tested strains were counted as previously described (Li et al., 2016). Approximately 100 simple yeast cells were pipetted onto SMAY medium and grown at 25°C. Switching rates at indicated time points were recorded and TT50 was estimated.
Purification and measurement of clone pigments
Clone pigments of tested strains were counted as described previously (Li et al., 2016). Clones from 6‐day‐old, 9‐day‐old or 12‐day‐old SMAY cultures were isolated with 2% NaOH. Collected clones were ground with liquid nitrogen, suspended in NaOH solution and boiled at 100°C for 2 h. Subsequently, solutions were acidified to pH 2.0 with HCl. Precipitates after centrifugation at 6000 × g for 15 min, were dissolved in 2% NaOH and measured on a spectrophotometer at the wavelength of 459 nm (Babitskaya et al., 2000).
MS formation assay in liquid AM media
Conidial suspensions of tested strains were inoculated in liquid AM culture for 6 days. Biomass and MS yield were quantified for the AM cultures and determined as previously described (Song et al., 2015). MS morphologies were observed using the digital camera.
Gene expression by qRT–PCR
To assess the effect of exogenous agents on MrMsn2 expression, AM or MM was supplemented with exogenous 1 M iron or 3 mM H2O2 as previously described (Song et al., 2016b, 2018). Mycelia were subsequently harvested for RNA extraction. For time‐specific expression patterns during conidia development, samples of WT inoculated on SMAY were collected at 0, 2, 4, 6 and 8 days for RNA extraction. To explore the impact on other genes related to dimorphic transition and melanin production during conidiation, WT or ▵MrMsn2 mutants were incubated on SMAY media and 3‐, 6‐, 9‐ or 12‐day‐old clones were collected independently for transcriptional analysis. For time‐specific expression patterns during MS development, samples of WT inoculated in AM were collected at 36, 60, 72, 96 and 120 h for RNA extraction. To investigate the regulation of other genes during MS formation, WT and ▵MrMsn2 mutants were incubated in AM cultures. After 72 h, mycelia were collected and total RNA was prepared. Gene expression patterns were confirmed for samples of WT, ▵MrMsn2 or CP mycelia cultured in AM for 72 h.
Total RNA was collected according to previous methods (Song et al., 2015). RT–qPCR was performed using SYBR Green (Invitrogen, Shanghai, China), as per the manufacturer's instructions. β‐tubulin (Mrtub) and translation elongation factor (Mrtef) genes were used as internal standards. Relative expression levels were evaluated using the 2−ΔΔCt method (Vandesompele et al., 2002).
Insect virulence assays
Topical infections tests and injection assays were conducted as previously described (Song et al., 2016b). Three replicate groups had 30 larvae each, and after treatment, the larvae were reared as described previously (Song et al., 2015). Larval mortality was recorded daily, and LT50 values were calculated using probit analysis with the software SPSS 17.0 (SPSS Inc., Chicago, IL, USA).
Data analysis
All assays were repeated three times. Data were analysed by one‐way analysis of variance (ANOVA), followed by Duncan's multiple range tests using SPSS 17.0 software. Graphs were constructed in GraphPad Prism 5 software (GraphPad Software Inc., La Jolla, CA, USA). Error bars represent the standard error.
Conflict of interest
None declared.
Supporting information
Fig. S1 Phylogenetic analysis of MrMsn2 protein.
Fig. S2 Confirmation of gene disruption and complementation.
Fig. S3 Morphology analysis and conidial yield of wild‐type (WT), ▵MrSwi6 mutants and complemented (CP) strains mutants under abiotic stress.
Table S1. Oligonucleotide primers used in this study.
Acknowledgements
This research was supported financially by the National Science Foundation of the People's Republic of China (No. 31701127; No. 31570073), Educational Commission of Sichuan Province of China (18ZA0515) and Joint project on Luzhou City and Southwest Medical University (2017LZXNYD‐T06).
Microbial Biotechnology (2018) 11(6), 1157–1169
Funding Information
This research was supported financially by the National Science Foundation of the People's Republic of China (No. 31701127; No. 31570073), Educational Commission of Sichuan Province of China (18ZA0515) and Joint project on Luzhou City and Southwest Medical University (2017LZXNYD‐T06).
Contributor Information
Zhangyong Song, Email: szy83529@163.com.
Zhongkang Wang, Email: w-zk@163.com.
References
- Babitskaya, V.G. , Shcherba, V.V. , Filimonova, T.V. , and Grigorchuk, E.A. (2000) Melanin pigments from the fungi Paecilomyces variotii and Aspergillus carbonarius . Appl Biochem Microbiol 36: 128–133. [PubMed] [Google Scholar]
- Boucias, D.G. , Tigano, M.S. , Sosa‐Gomez, D.R. , Glare, T.R. , and Inglis, P.W. (2000) Genotypic properties of the entomopathogenic fungus Nomuraea rileyi . Biol Control 19: 124–138. [Google Scholar]
- Boucias, D. , Liu, S. , Meagher, R. , and Baniszewski, J. (2016) Fungal dimorphism in the entomopathogenic fungus Metarhizium rileyi: detection of an in vivo quorum‐sensing system. J Invertebr Pathol 136: 100–108. [DOI] [PubMed] [Google Scholar]
- Boyce, K.J. , and Adrianopoulos, A. (2015) Fungal dimorphism: the switch from hyphae to yeast is a specialized morphogenetic adaption allowing colonization of a host. FEMS Microbiol Rev 39: 797–811. [DOI] [PubMed] [Google Scholar]
- Chai, G.H. , Hu, R.B. , Zhang, D.Y. , Qi, G. , Zuo, R. , Cao, Y.P. , et al (2012) Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genom 13: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, P.K. , Scharfenstein, L.L. , Luo, M. , Mahoney, N. , Molyneux, R.J. , Yu, J. , et al (2011) Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid. Toxins 3: 82–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y. , Srivastava, A. , Ohm, R.A. , Lawrence, C.B. , Wang, K.H. , Grigoriev, I.V. , et al (2012) Transcription factor Amr1 induces melanin biosynthesis and suppresses virulence in Alternaria brassicicola . PLoS Pathog 8: e1002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronza, E. , Specht, A. , Heinzen, H. , and de Barros, N.M. (2017) Metarhizium (Nomuraea) rileyi as biological control agent. Biocontrol Sci Techn 2: 1–22. [Google Scholar]
- Gauthier, G.M. (2015) Dimorphism in fungal pathogens of mammals, plants, and insects. PLoS Pathog 11: e1004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou, C.D. , Patsoukis, Ν. , Papapostolou, Ι. , and Zervoudakis, G. (2006) Sclerotial metamorphosis in filamentous fungi is induced by oxidative stress. Integr Comp Biol 46: 691–712. [DOI] [PubMed] [Google Scholar]
- Hansen, A.S. , Hao, N. , and O'Shea, E.K. (2015) High‐throughput microfluidics to control and measure signaling dynamics in single yeast cells. Nat Protoc 10: 1181–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.Y. , Roze, L.V. , and Linz, J.E. (2013) Oxidative stress‐related transcription factors in the regulation of secondary metabolism. Toxins 5: 683–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X. , Xiao, G.H. , Zheng, P. , Shang, Y.F. , Su, Y. , Zhang, X.Y. , et al (2014) Trajectory and genomic determinants of fungal‐pathogen speciation and host adaptation. Proc Natl Acad Sci USA 111: 16796–16801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W. , Shang, Y.F. , Chen, P.L. , Cen, K. , and Wang, C.S. (2015) Basic leucine zipper (bZIP) domain transcription factor MBZ1 regulates cell wall integrity, spore adherence, and virulence in Metarhizium robertsii . J Biol Chem 290: 8218–8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado, C.A. , and Rachubinski, R.A. (1999) MHY1 encodes a C2H2‐type zinc finger protein that promotes dimorphic transition in the yeast Yarrowia lipolytica . J Bacteriol 181: 3051–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, M.A. , Dunlap, C.A. , and Jaronski, S.T. (2010) Ecological considerations in producing and formulating fungal entomopathogens for use in insect biocontrol. Biocontrol 55: 129–145. [Google Scholar]
- Jiang, S.S. , Yin, Y.P. , Song, Z.Y. , Zhou, G.L. , and Wang, Z.K. (2014) RacA and Cdc42 regulate polarized growth and microsclerotium formation in the dimorphic fungus Nomuraea rileyi . Res Microbiol 165: 233–242. [DOI] [PubMed] [Google Scholar]
- Jung, K.O. , Yang, D.H. , Maeng, S. , Lee, K.T. , So, Y.S. , Hong, J. , et al (2015) Systematic functional profiling of transcription factor networks in Cryptococcus neoformans . Nat Commun 6: 6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug, A. (2010) The discovery of zinc fingers and their development for practical application. Annu Rev Biochem 43: 213–231. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Wang, Z.K. , Liu, X.E. , Song, Z.Y. , Li, R. , Shao, C.W. , et al (2016) Siderophore biosynthesis but not reductive iron assimilation is essential for the dimorphic fungus Nomuraea rileyi conidiation, dimorphism transition, resistance to oxidative stress, pigmented microsclertium formation, and virulence. Front Microbiol 7: 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Kaplan, J. , and Ward, D.M. (2017) The glucose sensor Snf1 and the transcription factors Msn2 and Msn4 regulate transcription of the vacuolar iron importer gene CCC1 and iron resistance in yeast. J Biol Chem 292: 15577–15586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Ying, S.H. , Li, J.G. , Tian, C.G. , and Feng, M.G. (2013) Insight into the transcriptional regulation of Msn2 required for conidiation, multi‐stress responses and virulence of two entomopathogenic fungi. Fungal Genet Biol 54: 42–51. [DOI] [PubMed] [Google Scholar]
- Liu, R. , Xu, C. , Zhang, Q.Q. , Wang, S.Y. , and Fang, W.G. (2017) Evolution of the chitin synthase gene family correlates with fungal morphogenesis and adaption to ecological niches. Sci Rep 7: 44527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos, C.M. , de Oliveira, H.C. , de Melo, W.M. , da J.S., A. , P.A., S. and L., etal. (2016) Anti‐immune strategies of pathogenic fungi. Front Cell Infect Microbiol 6, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinho, H.S. , Real, C. , Cyrne, L. , Soares, H. , and Antunes, F. (2014) Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol 2: 535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls, S. , Straffon, M. , Enjalbert, B. , Nantel, A. , Macaskill, S. , Whiteway, M. , et al (2004) Msn2‐and Msn4‐like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans . Eukaryot Cell 3: 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, S.M. , Gianetti, B.A. , and Witchley, J.N. (2017) Candida albicans cell‐type switching and functional plasticity in the mammalian host. Nat Rev Microbiol 15: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Park, J. , Jang, S. , Kim, S. , Kong, S. , Choi, J. , et al (2008) FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 24: 1024–1025. [DOI] [PubMed] [Google Scholar]
- Pendland, J.C. , and Boucias, D.G. (1997) In vitro growth of the entomopathogenic hyphomycete Nomuraea rileyi . Mycologia 89: 66–71. [Google Scholar]
- Quandt, C.A. , Bushley, K.E. , and Spatafora, J.W. (2015) The genome of the truffle‐parasite Tolypocladium ophioglossoides and the evolution of antifungal peptaibiotics. BMC Genom 16: 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roetzer, A. , Gregori, C. , Jennings, A.M. , Quintin, J. , Ferrandon, D. , Butler, G. , et al (2008) Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors. Mol Microbiol 69: 603–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero, C. (2002) The genetic complexity of chitin synthesis in fungi. Curr Genet 41: 367–378. [DOI] [PubMed] [Google Scholar]
- Schmitt, A.P. , and Mcentee, K. (1996) Msn2p, a zinc finger DNA‐binding protein, is the transcriptional activator of the multistress response in Saccharmyces cerevisiae . Proc Natl Acad Sci USA 93: 5777–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, Y.F. , Xiao, G.H. , Zheng, P. , Cen, K. , Zhan, S. , and Wang, C.S. (2016) Divergent and convergent evolution of fungal pathogenicity. Genome Biol Evol 8: 1374–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, C.W. , Yin, Y.P. , Qi, Z.R. , Li, R. , Song, Z.Y. , and Wang, Z.K. (2015) Agrobacterium tumefaciens mediated transformation of the entomopathogenic fungus Nomuraea rileyi . Fungal Genet Biol 83: 19–25. [DOI] [PubMed] [Google Scholar]
- Shearer, J.F. (2007) Some observations concerning microsclerotia and spore production of Mycoleptodiscus terrestris in culture. Mycologia 99: 88–90. [DOI] [PubMed] [Google Scholar]
- Shelest, E. (2017) Transcription factors in fungi: TFome dynamics, three major families, and dual‐specificity TFs. Front Genet 8: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Z.Y. , Yin, Y.P. , Jiang, S.S. , Liu, J.J. , Chen, H. , and Wang, Z.K. (2013) Comparative transcriptome analysis of microsclerotia development in Nomuraea rileyi . BMC Genom 14: 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Z.Y. , Yin, Y.P. , Jiang, S.S. , Liu, J.J. , and Wang, Z.K. (2014) Optimization of culture medium for microsclerotia production by Nomuraea rileyi and analysis of their viability for use as a mycoinsecticide. Biocontrol 59: 597–605. [Google Scholar]
- Song, Z.Y. , Shen, L. , Yin, Y.P. , Tan, W.Y. , Shao, C.W. , Xu, J.M. , et al (2015) Role of two Nomuraea rileyi transmembrane sensors Sho1p and Sln1p in adaptation to stress due to changing culture conditions during microsclerotia development. World J Microb Biot 31: 477–485. [DOI] [PubMed] [Google Scholar]
- Song, Z.Y. , Shen, L. , Zhong, Q. , Yin, Y.P. , and Wang, Z.K. (2016a) Liquid culture production of microsclerotia of Purpureocillium lilacinum for use as bionematicide. Nematology 18: 719–726. [Google Scholar]
- Song, Z.Y. , Zhong, Q. , Yin, Y.P. , Shen, L. , Li, Y. , and Wang, Z.K. (2016b) The high osmotic response and cell wall integrity pathways cooperate to regulate morphology, microsclerotia development, and virulence in Metarhizium rileyi . Sci Rep 6: 38765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Z.Y. , Lin, Y.L. , Du, F. , Yin, Y.P. , and Wang, Z.K. (2017) Statistical optimisation of process variables and large scale production of Metarhizium rileyi (Ascomycetes: Hypocreales) microsclerotia in submerged fermentation. Mycology 8: 39–47. [Google Scholar]
- Song, Z.Y. , Yin, Y.P. , Lin, Y.L. , Du, F. , Ren, G.W. , and Wang, Z.K. (2018) The bZip transcriptional factor activator protein‐1 regulates Metarhizium rileyi morphology and mediates microsclerotia formation. Appl Microbiol Biot 102: 4577–4588. [DOI] [PubMed] [Google Scholar]
- Takemoto, D. , Tanaka, A. , and Scott, B. (2007) NADPH oxidases in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet Biol 44: 1065–1076. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. , and Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L. , Yu, J. , Wang, Y. , and Tian, C. (2017) The C2H2 transcription factor VdMsn2 controls hyphal growth, microsclerotia formation, and virulence of Verticillium dahlia . Fungal Biol 121: 1001–1010. [DOI] [PubMed] [Google Scholar]
- Vandesompele, J. , De Preter, K. , Pattyn, F. , Poppe, B. , Roy, N.V. , Paepe, A.D. , et al (2002) Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, Research 0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanchoo, A. , Lewis, M.W. , and Keyhani, N.O. (2009) Lection mapping reveals stage‐specific display of surface carbohydrates in in vitro and haemolymph‐derived cell of the entomopathogenic fungus Beauveria bassiana . Microbiology 155: 3131–3133. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Ye, S.F. , Li, J.J. , Zheng, B. , Bao, M.Z. , and Ning, G.G. (2011) Fusion primer and nested integrated PCR (FPNI‐PCR): a new high‐efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol 11: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.L. , Zhang, L.B. , Ying, S.H. , and Feng, M.G. (2013) Catalases play differentiated roles in the adaptation of a fungal entomopathogen to environmental stress. Environ Microbiol 15: 409–418. [DOI] [PubMed] [Google Scholar]
- Xie, X.Q. , Li, F. , Ying, S.H. , and Feng, M.G. (2012) Additive contributions of two manganese‐cored superoxide dismutases (MnSODs) to antioxidation, UV tolerance and virulence of Beauveria bassiana . PLoS ONE 7: e30298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, D.G. , and Huh, W.K. (2015) PKA, PHO and stress response pathways regulate the expression of UDP‐glucose pyrophosphorylase through Msn2/4 in budding yeast. FEBS Lett 589: 2409–2416. [DOI] [PubMed] [Google Scholar]
- Yin, W.X. , Cui, P. , Wei, W. , Lin, Y. , and Luo, C.X. (2017) Genome‐wide identification and analysis of the basic leucine zipper (bZIP) transcription factor gene family in Ustilaginoides virens . Genome 60: 1051–1059. [DOI] [PubMed] [Google Scholar]
- Youseff, B.H. , Holbrook, E.D. , Smolnycki, K.A. , and Rappleye, C.A. (2012) Extracellular superoxide dismutase protects histoplasma yeast cells from host‐derived oxidative stress. PLoS Pathog 8: e1002713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L.B. and Feng, M.G. (2018) Antioxidant enzymes and their contributions to biological control potential of fungal insect pathogens. Appl Microbiol Biot 102, 4995–5004. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Zhao, Q. , Guo, X. , Guo, M. , Qi, Z. , Tang, W. , et al (2014) Pleiotropic function of the putative zinc‐finger protein MoMsn2 in Magnaporthe oryzae . Mol Plant Microbe Interact 27: 446–460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Phylogenetic analysis of MrMsn2 protein.
Fig. S2 Confirmation of gene disruption and complementation.
Fig. S3 Morphology analysis and conidial yield of wild‐type (WT), ▵MrSwi6 mutants and complemented (CP) strains mutants under abiotic stress.
Table S1. Oligonucleotide primers used in this study.


