Abstract
Lipid asymmetries between the outer and inner leaflet of the lipid bilayer exist in nearly all biological membranes. Although living cells spend great effort to adjust and maintain these asymmetries, little is known about the biophysical phenomena within asymmetric membranes and their role in cellular function. One reason for this lack of insight into such a fundamental membrane property is the fact that the majority of model-membrane studies have been performed on symmetric membranes. Our aim is to overcome this problem by employing a targeted, enzymatic reaction to prepare asymmetric liposomes with phosphatidylserine (PS) primarily in the inner leaflet. To achieve this goal, we use a recombinant version of a water soluble PS decarboxylase from Plasmodium knowlesi, which selectively decarboxylates PS in the outer leaflet, converting it to phosphatidylethanolamine. The extent of decarboxylation is quantified using high-performance thin-layer chromatography, and the local concentration of anionic PS in the outer leaflet is monitored in terms of the ζ potential. Starting, for example, with 21 mol % 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine sodium salt, the assay leads to liposomes with 21 mol % in the inner and 6 mol % PS in the outer leaflet. This asymmetry persists virtually unchanged for at least 4 days at 20°C and at least 2 days at 40°C. The use of a highly specific enzyme carries the advantage that a minor component such as PS can be adjusted without affecting or being affected by the other lipid species present in the model membrane. The phenomena governing the residual outside PS content are addressed but warrant further study.
Introduction
Virtually all biological membranes show a distinct lipid asymmetry between the outer and inner leaflet of the lipid bilayer, including organelle membranes except those of the endoplasmic reticulum (1). The eukaryotic plasma membrane has most of its phosphatidylcholine (PC) and sphingomyelin (SM) in the extracellular leaflet, whereas phosphatidylethanolamine (PE) and phosphatidylserine (PS) are preferentially in the inner leaflet. Among these different types of lipids, the asymmetry of PS is most pronounced. The local PS content in the outer leaflet can be as little as 0–2 mol % but also up to 3.2 mol % (referring to all lipids in the outer leaflet) (2, 3). In the inner leaflet, PS represents ∼20 mol % (4, 5, 6). During apoptosis, this lipid asymmetry is lost, and PS is exposed in the outer leaflet, leading to physiological consequences such as the recognition and removal of apoptotic cells by scavenging phagocytes (7). In cancer cells, the amount of PS on the extracellular leaflet is increased because the PS asymmetry is lost, potentially making them a suitable target for host defense peptides specific for PS (8, 9).
Substantial efforts have been made to investigate how cells generate, preserve, and employ this asymmetry (10). Nevertheless, it is not fully understood how lipid asymmetry influences embedded proteins on a structural and functional level. One reason for this lies in the lack of widely accessible and well-defined asymmetric model membranes, which are essential for quantitative analyses. Pautot et al. (11) proposed an inverted emulsion technique producing asymmetric liposomes from different monolayers at oil-water interfaces. This approach has recently been further developed utilizing microfluidics (12). It joins separate monolayers without a need for subsequent lipid exchange, but its yield is limited, and some oil typically remains in the membrane. Cyclodextrins (13, 14, 15) or lipid transport proteins (16) are used as lipid carriers or complexing agents allowing for the exchange of outer-leaflet lipids after liposome formation. Cyclodextrins are specific for sterols compared to diacyl phospholipids, with α-cyclodextrin excluding cholesterol (17) and methyl-β-cyclodextrin preferring to form complexes with cholesterol as compared to with phospholipids (18, 19, 20, 21). However, these carriers show little or no preference for one phospholipid over another. This is an advantage when a minor component is to be added to the outer leaflet, for example, to produce liposomes with outside only, 30 mol % phosphatidylglycerol (PG) to mimic bacterial membranes (22). However, to reduce the amount of a low-abundant lipid even further, for example, to achieve the PS asymmetry of eukaryotic cells, virtually all outside lipid has to be exchanged without compromising liposome stability and multiple exchange cycles would be necessary. The more straightforward strategy in this case is to selectively replace a lipid species such as PS in the outer layer.
This strategy is pursued here using PS decarboxylase (PSD), an enzyme converting outer-leaflet PS into PE. This idea was first presented and tested in 1986 by Denkins and Schroit (23). In a proof-of-concept study, they showed that an Escherichia coli extract containing E. coli PSD was able to selectively convert the outer-leaflet PS in liposomes containing a trace amount of 1% PS, which was fluorescently labeled (nitrobenzoxadiazole-PS/PSD molar ratio of around 2). However, these authors did not report any attempts to produce liposomes that mimic typical cell membranes with their 20 times larger PS concentration in the inner leaflet. It turns out that it is not trivial to scale up the Denkins and Schroit (23) protocol to produce such liposomes with a high and stable asymmetry and controlled composition. A key problem here is to use purified PSD at sufficient activity without compromising asymmetry.
The PSD enzymes are an integral part of the PE synthesis pathway, which is evolutionary conserved in prokaryotes and eukaryotes. Mature PSDs are typically membrane proteins, consisting of an α- and β-subunit which are produced by autoendoproteolytic cleavage of a single proenzyme precursor. The proenzyme is cleaved to the active form to yield distinct α- and β-subunits, which are separate molecular entities but still physically associated with each other. The enzyme’s active site is located in the smaller α-subunit, which harbors a pyruvoyl prosthetic group (24, 25). As membrane proteins, most PSDs depend on detergents to be active in vitro. Dowhan et al. (26), for example, described in 1973 the purification and properties of an E. coli PSD in which the activity was strongly dependent on the nonionic detergent Triton X-100. Similar results were obtained for a Clostridium butyricum PSD (27). Detergents, however, tend to promote the flip-flop of lipids and, hence, prevent an asymmetry from building up and persisting (28).
We used a modified, soluble form of engineered parasite PSD (MBP-His6-Δ34PkPSD), which is expressed in E.coli and purified from cell extracts (29). PkPSD heterologously expressed in yeast was found in both membrane and soluble fractions. The solubility of the enzyme has further been increased by deletion of the hydrophobic N-terminal 34 amino acids and fusion to maltose-binding protein. The soluble PSD construct used here is active without detergent, weakly interacting with the lipid headgroups in a membrane bilayer and thus suitable for the preparation of PS-asymmetric liposomes.
Overall, our strategy was to first prepare symmetric liposomes that have PS in both membrane leaflets and then treat the outer leaflet with PSD to render them asymmetric. The decreasing local PS content in the outer leaflet was monitored in a label-free manner in terms of a less negative ζ potential (14). The decrease of PS and the increase of PE were quantified by high-performance thin-layer chromatography (HPTLC), which yields the overall lipid composition of the bilayer. With the help of the bilayer PS contents (HPTLC) as well as the outside-only PS contents (ζ), we calculated the asymmetry of PS localization. To assess the stability of PSD-generated asymmetry, we also monitored outer-leaflet PS contents as a function of time.
Materials and Methods
Materials
Egg PC (EPC) and egg SM were kindly provided by Lipoid (Ludwigshafen, Germany). 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine sodium salt (POPS) was purchased from Avanti Polar Lipids (Alabaster, AL) and cholesterol was purchased from Sigma-Aldrich (Munich, Germany).
Ni-NTA (nickel-nitrilotriacetic acid) agarose was purchased from Qiagen (Venlo, the Netherlands), and all other chemicals were obtained from Carl Roth (Karlsruhe, Germany) and were of analytical grade.
Preparation of large unilamellar vesicles
For vesicle preparation, the thin-film method was used (30). Briefly, the desired amounts of lipids were dissolved in chloroform and mixed in a round-bottom flask. The solvent was removed using a rotary evaporator, and the resulting lipid film was submitted to further drying under a high vacuum for at least 2 h.
Liposomes used for the PSD assay were prepared in decarboxylation buffer (10 mM KH2PO4, 5 mM K2HPO4 (pH 6.8)) with a total lipid concentration of 10 or 20 mM. The liposomes were extruded 51 times through polycarbonate membranes with pores of 80 nm (Nuclepore Whatman; Little Chalfont, UK). Lipid content was determined by a phosphorous assay (using a buffer blank to correct for the phosphate in the buffer) (31), and the hydrodynamic diameter (100–110 nm) was confirmed by dynamic light scattering (DLS) (Zetasizer ZS, Malvern Instruments; Worcestershire, UK).
DLS
A Zetasizer Nano ZS (Malvern) equipped with a 633-nm He-Ne laser was used to measure particle size distributions of liposomes by DLS using a detection angle of 173 at 25°C. The effects of buffer components on refractive index and viscosity were taken into account.
ζ potential
ζ potential measurements were performed with the Zetasizer Nano ZS (Malvern) using a flow-through, high-concentration ζ potential cell (high concentration cell [HCC]) (Malvern), as described previously (14). Briefly, the kinetics of PS decarboxylation by PSD were recorded at 28°C. A new measurement was started every 3–5 min for 80–120 min. The sample was pushed forward a few millimeters in the tubing of the HCC for each measurement. Doing so, a fresh sample was in the measuring chamber for every measurement. ζ potential was measured in triplicates if possible. If an SD of 3 mV was exceeded, data were considered of low quality. In such a case, the HCC was refilled, and another series of triplicates was obtained. Before and after the PSD assay, a ζ standard of −42 mV was measured to assess the quality of the setup. If the standard did not meet its specification (−42 ± 4.2 mV), the HCC was disassembled and cleaned (double-distilled H2O and/or 1% Hellmanex III), and the assay was repeated.
HPTLC
The lipid composition of liposomes was obtained by HPTLC as described previously (14, 32). Briefly, a mixture of 60 mL chloroform, 30 mL methanol, 6.5 mL formic acid, 4.5 mL acetic acid, and 0.1 mL aqueous 0.1% MgCl2 solution was used as a mobile phase. A sample applicator, a developing chamber, and a TLC scanner for densitometry analysis were used (CAMAG; Berlin, Germany). For analysis, liposomes were diluted with methanol to the desired concentration, and EPC, POPS, and POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine) concentrations were quantified separately.
After development with CuSO4 followed by a derivatization step at 140°C for 10 min, the absorbance of lipid spots on the HPTLC plate was detected by optical densitometry using a mercury lamp emitting at 365 nm. Calibration curves were fitted empirically by second-order polynomial functions.
PSD assay
For decarboxylation of outer bilayer leaflets, POPS to POPE, liposomes were diluted to 1 mM total lipid in decarboxylation buffer, and PSD was added to a final protein concentration of 16 μg/mL. The mixture was incubated applying slight agitation for 80–120 min at 28°C.
Ni-NTA pull-down assay
The PSD was removed using Ni-NTA (Ni-PD) agarose and selective adsorption of the enzyme to the matrix. After PSD treatment, vesicle preparations were incubated twice with Ni-NTA beads. The first adsorption of enzyme to agarose was conducted for 3–4 h at 20°C on a rotating wheel and the second one overnight with fresh beads. 10-μL bead slurry was used for the 100-μL assay mixture. Beads were removed by a brief centrifugation using a mini tabletop centrifuge for ∼1–2 min.
Stability test
For stability studies, the PSD was removed via pull-down assay, and the asymmetric liposomes were stored at 20°C.
To record ζ potential and perform HPTLC, samples were taken after 1, 2, and 4 days.
Results
Preparation of asymmetric liposomes: ζ potential monitoring
We aimed at creating asymmetric liposomes with a PS content of 20% in the inner leaflet and little PS on the outside. To this end, we first prepared symmetric liposomes consisting of EPC and the enzyme substrate (PO)PS in an 80:20 molar ratio. Then, the liposomes were diluted in decarboxylation buffer to 1 mM total lipid concentration, and the PSD was added to a final protein concentration of 16 μg/mL.
The sample was then incubated for 80 min at 28°C with slight agitation.
Because the PSD is soluble and cannot pass the liposomal membrane, only the outer PS was decarboxylated to PE, whereas PS inside the liposomes was not accessible to the enzyme.
The decarboxylation process is accompanied by a loss of negative charge because negatively charged PS is converted to neutral PE. Thus, the decarboxylation can be monitored by ζ-potential measurements (Fig. 1 A).
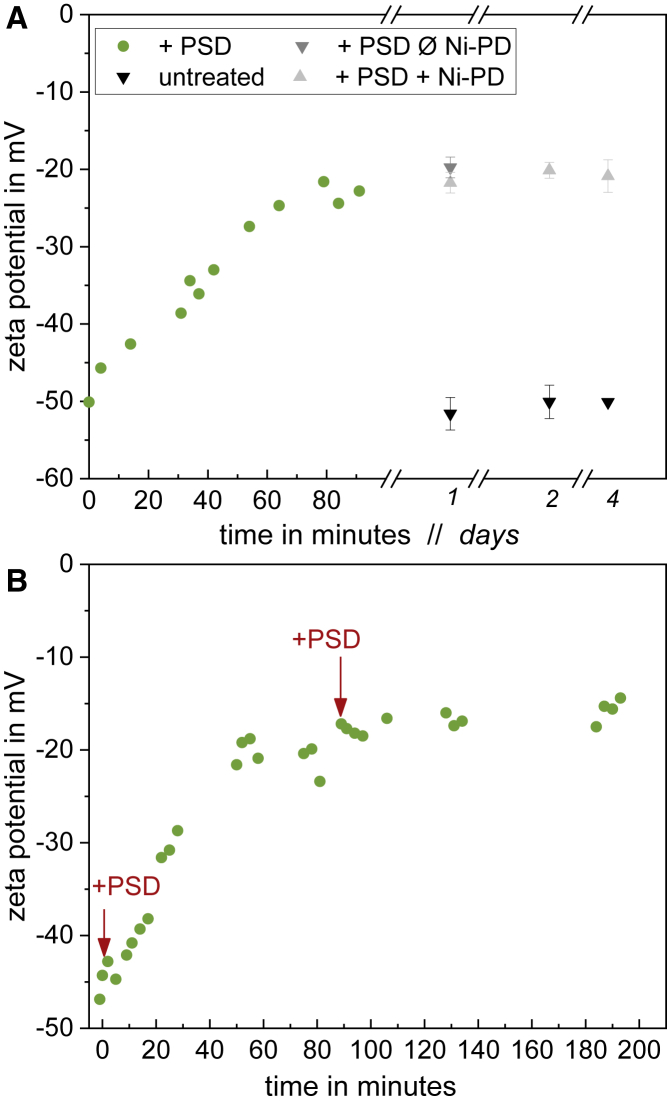
Figure 1.
Kinetics of PS decarboxylation by PSD monitored by ζ potential. PSD was added to a final protein concentration of 16 μg/mL to symmetric liposomes consisting of 80 mol % PC and 20 mol % PS with 1 mM total lipid. The assay mixture was incubated at 28°C with slight agitation. Measurements were performed at 28°C, ζ potential was measured in regular time intervals to observe the decarboxylation of PS (green circles). (A) Light and dark gray triangles compare ζ potential before (Ø Ni-PD) and after PSD removal (+Ni-PD), respectively, incubated at 20°C for 4 days. Black triangles show the ζ potential of untreated symmetric liposomes. Error bars indicate the SD of three measurements of the same sample. (B) Repetition of the assay as shown in (A); after 90 min, a second dose of 16-μg/mL PSD is added. To see this figure in color, go online.
When using symmetric liposomes containing 20 mol % PS, the ζ potential started at −50 mV, and during a decarboxylation of 80 min, the ζ potential dropped to −23 mV (Fig. 1 A). At this point, the curve leveled off, and no further decarboxylation took place, even after 4 days of incubation. Full decarboxylation would lead to liposomes with an outer-leaflet composition of 80 mol % PC and 20 mol % PE. Such vesicles have a ζ potential of around −3 mV.
The leveling off of ζ at about −23 mV does not seem to be due to a loss of enzyme activity with time or substrate conversion. Adding a second, fresh dose of PSD to the assay after conversion had essentially come to a halt did not allow for a significant further reduction of outside PS either (Fig. 1 B).
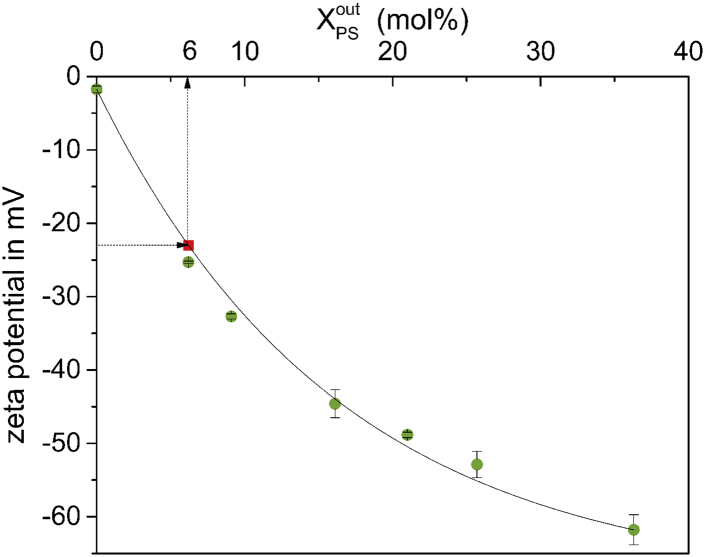
To quantify the remaining PS on the liposomal surface, a calibration curve was established to relate the measured ζ potential to a certain PS percentage in the outer layer of the liposomes (Fig. 2). It was generated by producing individual samples of symmetric liposomes with different PS contents, ranging from 0- to 36 mol % PS. The ζ potentials of these liposomes were determined to range from −2 mV with no PS to −62 mV with 36 mol % PS.
Figure 2.
ζ potential calibration curve. ζ potential of PC-PS liposomes as a function of the mole fraction of PS is shown. Measurements were done with symmetrical membranes at which the local outside PS content, XPSout determining ζ agrees with the overall average, 〈XPS〉 set gravimetrically and validated via HPTLC. The empirical fit is provided as in Eq. 1. The red square relates the post-PSD ζ obtained in Fig. 1 to 6 mol % PS in the outer leaflet. To see this figure in color, go online.
For a quick numerical conversion, the data were fitted empirically by the following:
| (1) |
The red square represents the ζ potential of mixed PC-PS liposomes after PSD treatment for 80 min, corresponding to 6 mol % PS in the outer layer. Error bars indicate the SD of three measurements of the same sample.
Summarizing this section, PSD treatment has reduced the PS content in the outer leaflet from 21 to 6 mol %.
Quantifying asymmetry based on HPTLC and ζ potential
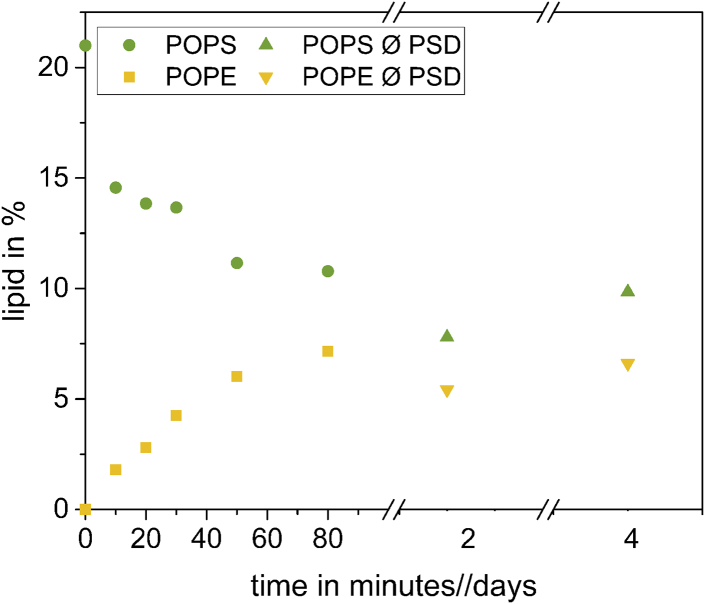
What cannot be assessed from ζ potential is the composition of the inner leaflet of the liposomes. To demonstrate that the content of PS in the inner leaflet had remained unchanged after PSD treatment, we quantified the total PS content by HPTLC (Fig. 3).
Figure 3.
Time-dependent conversion of PS to PE by PSD as seen by HPTLC: symmetric liposomes consisting of 80 mol % EPC and 20 mol % PS at a total lipid concentration of 1 mM were mixed with PSD to a final protein concentration of 16 μg/mL. The assay mixture was incubated at 28°C with slight agitation. HPTLC samples were taken in regular intervals to observe the decarboxylation of PS (green) to PE (yellow). Day 2 and 4 indicate the stability of the lipid ratio after PSD removal (Ø PSD) and incubation at 20°C for 4 days (green and yellow triangles). To see this figure in color, go online.
HPTLC determines the total content of PE, averaged over both the inner and outer leaflets as indicated by angular brackets:
| (2) |
and analogously for PS. Here, nPE, nPS, and nPC represent the total mole number of these lipids in the sample. Decarboxylation converts one PS into one PE and keeps the total mole number of lipids constant so that the changes upon enzyme action must obey the following:
| (3) |
and information on Δ and Δ should be mutually redundant. In our example illustrated in Fig. 3, this criterion was fulfilled within error with the total average decreasing from 21 to 10 mol % (Δ = −(11 ± 5) mol %) and increasing from 0 to 7 mol % (Δ = (7 ± 1) mol %. The larger estimated error of results from a considerably less steep calibration curve of the assay for PS compared to PE. Therefore, total changes in PS were practically quantified by measuring Δ and taking into account Eq. 3.
In the large liposomes studied here, the two membrane leaflets contain essentially the same number of lipid molecules, and the average mole fraction defined in Eq. 2 represents the average of the local mole fractions in the inner and outer leaflets, and (5). Hence, averaging can be written as follows:
| (4) |
and analogously for Δ〈X〉, yielding for the changes upon decarboxylation
| (5) |
and analogously for PE.
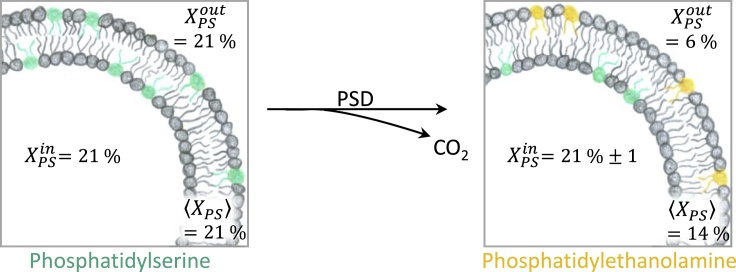
In our example, HPTLC quantification of the overall PE content after decarboxylation resulted in Δ = −Δ = −7 mol %. With this value and Δ = (6–21) mol % = −15 mol % as established above using ζ potential, Eq. 5 yielded for a change in local PS content in the inner leaflet upon outside decarboxylation: Δ = 2 × −7 mol % + 15 mol % = 1 mol %. That means PSD treatment left the inner leaflet essentially unchanged (Δ ≈ 0), as expected (Fig. 4).
Figure 4.
Schematic overview of decarboxylation process by PSD to illustrate terminology: the left panel refers to the starting symmetric liposomes with 21 mol % in the outer and inner leaflet and, hence, as the total average. After decarboxylation, the total ≈ 14 mol %, inner-leaflet PS remained unchanged: ≈ 21 mol % (Δ ≈ 0), whereas outside PS was decarboxylated to ≈ 6 mol % (Δ ≈ −15). To see this figure in color, go online.
We emphasize that we quantify PS content in terms of the local outside, local inside, and average fraction of PS referred to all lipids in the respective compartment. Note that in the literature, different specifications of asymmetric compositions are used and sometimes confused with each other.
The asymmetry of the liposomal membrane was between = 6 mol % and = 21 mol %. We may define an asymmetry parameter, a as follows:
| (6) |
that ranges from −1 for all-inside localization of PS via zero for symmetric distribution to one for outside-only PS. For our example, we found a = −0.5 for PS (strong preference for the inner leaflet), and a ≈ 1 for PE (virtually exclusively in the outer leaflet). The good agreement of the data obtained from different methods could be considered a consistence criterion for the approach.
Note that whenever the (symmetrical) starting content of PS, , was known from the preparation and asymmetric, outside-only decarboxylation could be assumed, a measurement of the ζ potential was sufficient for obtaining and a. For the latter, it was assumed in Eq. 6 that the final agrees with the initial, overall PS content in the sample.
Long-term stability of lipid asymmetry
After establishing the PS asymmetry in the liposomes, the PSD could be removed. This was necessary to determine the long-term stability of lipid asymmetry. The presence of a His tag facilitated PSD removal using Ni-NTA agarose beads. Affinity adsorption and centrifugation provided a relatively gentle method for separating PSD from liposomes.
After removal of the PSD, a flop of PS from the inner to the outer leaflet would reestablish a more negative ζ potential of the liposomes. Black and gray triangles (Fig. 1 A) demonstrated that no such effect could be detected over 4 days, implying that the PS asymmetry remained stable at least for this period of time. This is long enough for most applications including, for example, a functional assay of a reconstituted protein.
Furthermore, the removal of the PSD and further incubation of the liposomes at 20°C did not affect the lipid composition because no change in relative lipid amounts was detectable as shown by the yellow and green triangles in Fig. 3.
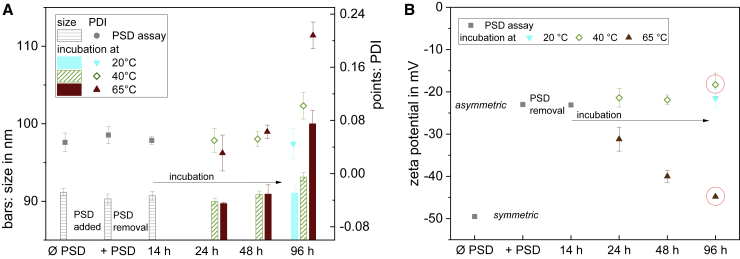
Next, we tested whether a transmembrane flop would become detectable at least at higher temperatures. For up to 48 h of incubation, no trend was found also at 40°C (asymmetry stable; Fig. 5 B). In contrast, at 65°C, the asymmetry decreased with a half-life of the order of 14 h, with ζ suggesting XPSout ≈ 9 mol % after 10 h, and XPSout ≈ 〈XPS〉 ≈ 14 mol % (asymmetry ≈ 0) after 48 h. Under these conditions, the liposomes as such showed stable size and polydispersity index (PDI) in DLS so that the change in ζ can be tentatively assigned to molecular flip-flop. After 96 h, the liposomes stored at 40 and 65°C showed increased polydispersity and size. That means the liposomes fuse or collapse and the ζ potentials should not be interpreted as a measure of flip-flop (indicated by red circles around symbols in Fig. 5 B).
Figure 5.
Z-average size, polydispersity index (PDI), and ζ potential of liposomes at increased storage temperatures. First measurement (Ø PSD) was done with symmetrical liposomes of 80 mol % PC and 20 mol % PS. Then, liposomes were incubated with PSD at 28°C for 120 min and then DLS and ζ measured again (+PSD). Then, the PSD was removed via Ni-NTA agarose overnight at 20°C (measurement 14h). Aliquots of this sample were then stored at 20°C, 40°C, or 65°C and measured at the time points indicated. (A) Size and PDI of the samples were determined by DLS measurements. (B) ζ potential of the samples was determined. 96-h measurements at 40°C and 65°C are encircled to indicate that they refer to poor quality samples with large particle size and PDI. Error bars indicate three measurements of the same sample. To see this figure in color, go online.
Variation of substrate abundance
To assess whether the amount of PS had an influence on the PSD kinetics or the outcome of the assay, we prepared liposomes with different amounts of PS, diluted them, and treated them with PSD as described before.
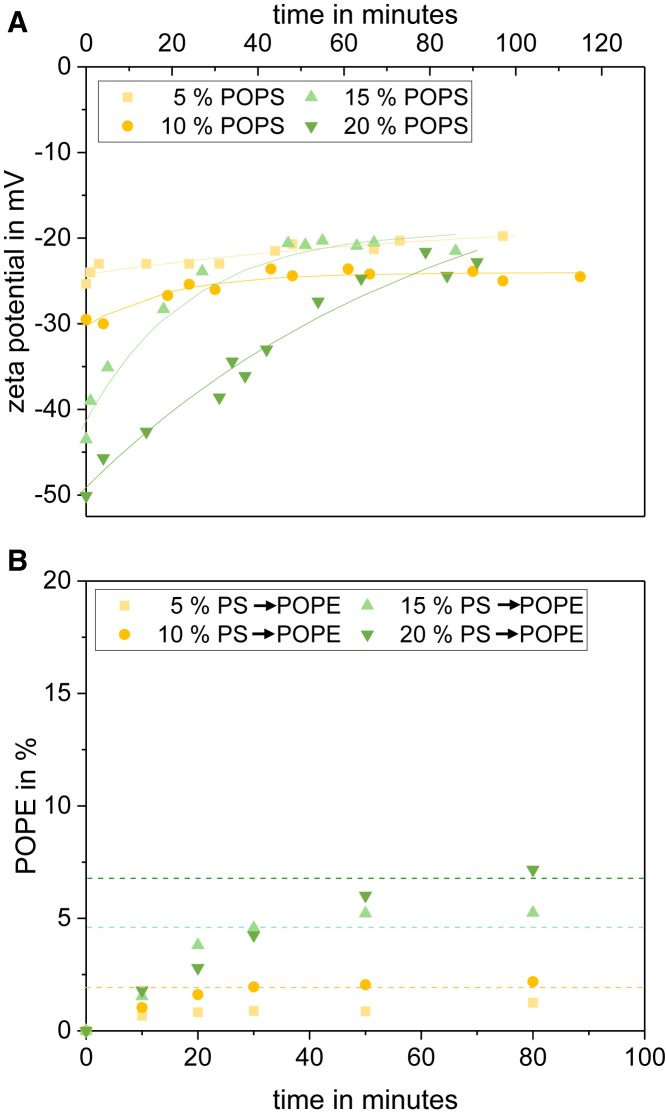
Fig. 6 shows the corresponding ζ potentials and HPTLC results. Naturally, increasing PS content in the liposomes correlated with increasingly negative starting values of ζ potential (Fig. 6 A). Upon PSD treatment, all curves approached a common ζ potential of the order of −25 to −20 mV, which corresponded to a residual PS content of ≈ 5–6 mol % in the outer leaflet. Decarboxylating PS from 20, 15, 10, and 5 mol % to a common 6 mol % should create a local of 14, 9, 4, and 0 mol %, respectively, which should yield average values of = 7, 4.5, 2, and 0 mol %. These values predicted from ζ potential measurements are represented by grid lines in Fig. 6 B and found to agree with the actual HPTLC results within ±1 mol %. Given that the final outside content of PS remained unchanged despite increasing starting values of , which agreed with the final , it is obvious that the asymmetry parameter increased with increasing starting PS content (Table 1).
Figure 6.
Influence of the initial amount of PS on kinetics and extent of decarboxylation by PSD. (A) ζ -potential measurements during the application of PSD with increasing initial PS contents (5–20 mol %, see legend in plot). Lines are to guide the eye only. (B) HPTLC results for rising PE concentration during the PSD assay for liposomes with different starting PS amounts are shown. Grid lines represent the PE prediction from ζ-potential measurements. To see this figure in color, go online.
Table 1.
Asymmetry Parameters, a, and Other Parameters Quantifying the Asymmetry after PSD Treatment
| / % | a | /mV | / % | / % |
|---|---|---|---|---|
| 5 | 0.04 | −20 | 5.3 | 3.7 |
| 10 | −0.21 | −24 | 6.7 | 7.8 |
| 15 | −0.49 | −21 | 5.5 | 9.7 |
| 20 | −0.54 | −23 | 6.2 | 13 |
Asymmetry parameters, a, are defined as in Eq. 6; recall that a = −1 for asymmetrical inside-only localization and a = 0 for symmetric lipid distribution, whereas for asymmetrical outside only, a = 1.
Variation of lipid composition
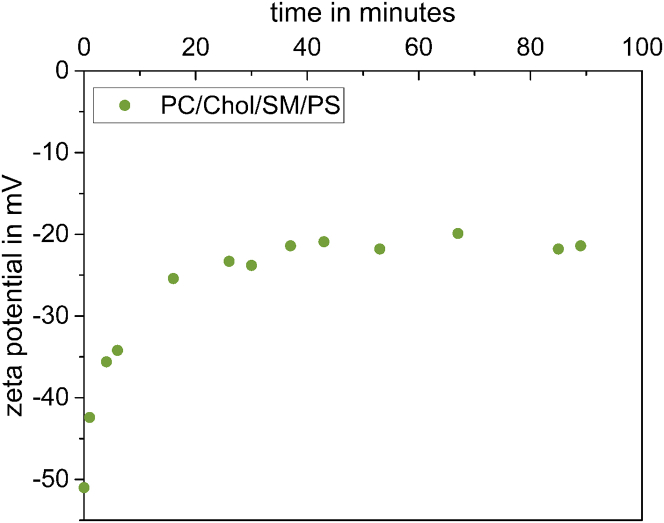
To determine whether other membrane components interfere with the decarboxylation process, liposomes consisting of several lipids were prepared. Because an enzyme was used, high specificity for the decarboxylation was expected meaning that the addition of other membrane components should not influence the outcome of the assay. In Fig. 7, a PSD assay was performed with liposomes containing 35 mol % EPC, 30 mol % cholesterol, 21 mol % SM, and 14 mol % POPS (20 mol % POPS of total phospholipid, without cholesterol), which served as a simple model membrane for mammalian cell membranes.
Figure 7.
PS decarboxylation by PSD in a more complex lipid mixture. Symmetric liposomes consisting of 35 mol % PC, 30 mol % cholesterol, 21 mol % SM, and 14 mol % PS were diluted to 1 mM total lipid, and PSD was added to a final protein concentration of 16 μg/mL. The assay mixture was incubated at 28°C with slight agitation. ζ potential was measured in regular intervals to observe the decarboxylation from PS to PE (green circles). To see this figure in color, go online.
As the ζ potential measurement of the assay indicated, the presence of cholesterol and SM did not interfere with the assay. Decarboxylation of PS in these liposomes approached the same residual PS content on the outside, ≈ 6 mol %, as obtained for our standard system.
Addition of anionic PG to increase PSD efficiency
As seen before, the PSD does not further decarboxylate PS below 6 mol % or −23 mV, independently of the lipid mixtures tested so far and the concentrations of PS or PSD used. To distinguish whether the key parameter was the 6 mol % of, specifically, PS or simply the ζ potential of −23 mV, we combined PS with another anionic lipid, PG, that is not a substrate of PSD.
Vesicles consisting of 75 mol % PC, 20 mol % PS, and 5 mol % PG were prepared. Additional liposomes, consisting of 75 mol % PC, 20 mol % PE, and 5 mol % PG were prepared as a control, showing the lipid mixture of the outer leaflet after complete decarboxylation. The PSD was added to the PC/PS/PG-containing liposomes, and a PSD assay was conducted. Table 2 shows the ζ potential of the vesicles before and after PSD treatment for 120 min.
Table 2.
PSD Efficiency in PS and PG Containing Liposomes
| Liposomes |
ζ potential |
||
|---|---|---|---|
| Lipids | Initial molar ratio | Ø PSD | + PSD |
| PC/PS | 80:20 | −50 | −23 |
| PC/PS/PG | 75:20:5 | −53 | −30 |
| PC/PE/PG | 75:20:5 | −19 | NA |
ζ potential of liposomes with different lipid composition, before and after PSD treatment. NA, not applicable.
The results of the test are in conflict with the prediction based on the electrostatic-attraction hypothesis. Instead of proceeding to −23 mV, decarboxylation now stops already at −30 mV.
Discussion
Suitability of the PSD assay to prepare PS-inside liposomes
We presented a new, to our knowledge, strategy and protocol for the preparation of liposomes with PS preferentially in the inner leaflet, for example with 20 mol % PS in the inner and around 6 mol % in the outer leaflet. Literature data on the amount of PS in outer membrane leaflets are scarce and suggest values of as low as 0–3.2% depending on the organism and cell type (2). That means, the minimal XPSout of 6 mol % might limit the applicability of the protocol in certain cases (see next section).
The preparation is fast (taking no more than 1–2 h), reliable, and does not require unusual instrumentation. The outcome can very conveniently be monitored by ζ-potential measurements.
The specific conversion of one lipid species has the advantage that a minor component can be changed in an exact 1:1 fashion without the need to exchange the other lipids and virtually independently of what the other lipids are.
The asymmetry of PS distribution between the leaflets of these liposomes remains stable over at least several days, implying that spontaneous flop rates of PS are low and that the preparation does not impose significant stress or damage to the membrane.
Limitations of the assay
A limitation may arise from the fact that we have not been able to eliminate outer-leaflet PS below ∼6 mol % PS. A better understanding of the physical basis of this threshold (see next section for some discussion) may permit adjusting the protocol to reach even lower outside PS.
However, the liposomes produced here can be expected to serve many purposes in asymmetry studies as they are. The question is whether or to which extent a 20 vs. 6 mol % asymmetry shows the behavior and functions of a natural, asymmetric membrane. There is no conclusive, general answer at this point, but one of the most prominent examples of asymmetry effects (the binding of annexin to a cell surface after partial scrambling of the PS content) may, for example, be modeled by these liposomes. Majd et al. (33) showed in 2006 that under suitable conditions, membranes with 6 mol % PS would bind very little annexin, whereas those with 13 mol % PS (as achievable by scrambling with 20 mol % inside) would cause substantial annexin binding. It remains to be shown whether recognition systems such as of phagocytes or anticancer peptides as well as asymmetry effects on intrinsic or peripheral membrane proteins rate 6 mol % as “little PS” or not.
Given that there may be cases requiring models with even lower outside PS, we should recall that the PSD construct used here is not the natural form. Instead, it is engineered to allow for a certain balance between aqueous solubility and activity (which requires membrane binding). A systematic variation of the design of the PSD construct might allow for a reduction or even elimination of the decarboxylation limit.
A second limitation is that PSD produces PE in the outer leaflet, whereas eukaryotic plasma membranes appear to localize PE in the inner leaflet preferentially. The natural PE asymmetry is not as strong as the PS asymmetry, and because PE is zwitterionic, it probably does not function as a signaling molecule if externalized like PS does. So maybe the low amounts of PE outside can be neglected. But to address this problem, one could use starting liposomes with a low, symmetric PE content. Alternatively, one could consider further converting outer PE to PC.
Finally, both limitations could, if needed, be addressed by combining the PSD assay described here with a subsequent cyclodextrin exchange assay (13, 15) to replace part of the remaining PS and the produced PE in the outer leaflet by, e.g., PC. Such a combination protocol may be faster and more gentle than achieving the same high asymmetry by extensive exchange alone.
Residual PS after PSD treatment
The PSD does not further decarboxylate PS below 6 mol % or around −20 mV, independently of the lipid mixtures tested here and the concentrations of PS or PSD used.
One hypothesis why the decarboxylation of PS stops at a ζ potential of about −23 mV was that this negative potential would be required for the membrane binding of the PSD construct used here. Below this value, a putative electrostatic attraction would become too weak to prevent the PSD from dissociating from the membrane, going back into solution, and stopping decarboxylation. This hypothesis was challenged by adding 5 mol % of another negatively charged lipid, PG. PG is not a substrate of the PSD but should maintain PSD binding to the membrane also when the PS content is reduced below 6 mol %. If the hypothesis was correct, decarboxylation should again stop at a putative binding threshold of about −23 mV, now resulting from the 5 mol % PG and only ∼1 mol % PS being left.
In conflict with this “electrostatic attraction hypothesis,” addition of 5 mol % PG made decarboxylation stop at −30 mV already. This suggests that the threshold is not defined by the potential (i.e., ∼6 mol % of any anionic lipid) alone. However, we cannot exclude electrostatics to have, at least, some effect. A rough estimate (ab)using the PS calibration curve for PG-PS mixtures would predict ζ = −35 mV for 11 mol % anionic lipid (5 mol % PG + 6 mol % PS). That means, the presence of PG might have reduced the residual PS at least somewhat, but the precision of the available data and interpretation routines do not permit a true quantification.
What else might govern the decarboxylation limit? Generally, incomplete action of an enzyme can also be explained by product inhibition. However, recall that the same limit was found independently of the starting PS content ranging from 5 to 20 mol % PS. That means that in the 5 mol % PS vesicles, nearly no decarboxylation happened in the first place, despite the lack of any product. This rules out product inhibition to explain the decarboxylation limit, too.
The failure of rather simple and straightforward explanations invites more complex and daring speculation. PSD might, for example, have more than one binding site for PS, and occupation of a second “activating” site is required to decarboxylate the PS in the first “acting” site. It should be noted that a reversible binding of lipid-modifying enzymes depending on a certain membrane property can have biological functions. One example is cytidyl transferase, apparently regulating the spontaneous curvature of the outer leaflet by tuning the PC/PE ratio (34). If there was an active mechanism of regulating PSD activity, it would be likely to apply not only to the construct used here but to the wild-type enzyme as well.
Conclusions
The assay presented here serves to produce liposomes mimicking the natural PS asymmetry of a eukaryotic plasma membrane in an easy, straightforward, and versatile fashion.
The specific, enzymatic conversion of PS allows for a minimal invasive protocol, leaving other lipids of, as it seems, whatever kind in place and unaffected. The asymmetry persists, virtually unchanged, for at least 4 days at 20°C but decays with a half-life of the order of 14 h at 65°C.
Decarboxylation by the PSD construct used here stops at 6 mol % and −23 mV. This limit does not seem to be solely due to a minimal electrostatic attraction needed nor to product inhibition or a limited stability or capacity of the enzyme. The discussion here provides a starting point for studies elucidating this unexpected behavior in more detail. This would be valuable for further improving the protocol and, possibly, also for gaining a better understanding of the biological function of PSD.
Using ζ potential measurements and HPTLC analysis, the degree and stability of the asymmetry can be determined.
Author Contributions
The method was established by C.D. PSD was purified by J.-Y.C. Additional experiments were performed by N.F. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Acknowledgments
The authors thank Eva Dengler and Nicole Specht for excellent technical assistance and Iulia Carabadjac for performing important additional experiments. We are indebted to Lipoid GmbH for providing phospholipids.
This work was supported by the Deutsche Forschungsgemeinschaft by grant RTG 2202 and a postdoctoral fellowship for S.F. (FI 2005/1-1).
Editor: Georg Pabst.
References
- 1.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zachowski A. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem. J. 1993;294:1–14. doi: 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sessions A., Horwitz A.F. Differentiation-related differences in the plasma membrane phospholipid asymmetry of myogenic and fibrogenic cells. Biochim. Biophys. Acta. 1983;728:103–111. doi: 10.1016/0005-2736(83)90442-x. [DOI] [PubMed] [Google Scholar]
- 4.Bretscher M.S. Asymmetrical lipid bilayer structure for biological membranes. Nat. New Biol. 1972;236:11–12. doi: 10.1038/newbio236011a0. [DOI] [PubMed] [Google Scholar]
- 5.Marquardt D., Geier B., Pabst G. Asymmetric lipid membranes: towards more realistic model systems. Membranes (Basel) 2015;5:180–196. doi: 10.3390/membranes5020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devaux P.F. Static and dynamic lipid asymmetry in cell membranes. Biochemistry. 1991;30:1163–1173. doi: 10.1021/bi00219a001. [DOI] [PubMed] [Google Scholar]
- 7.Fadok V.A., Bratton D.L., Henson P.M. The role of phosphatidylserine in recognition of apoptotic cells by phagocytes. Cell Death Differ. 1998;5:551–562. doi: 10.1038/sj.cdd.4400404. [DOI] [PubMed] [Google Scholar]
- 8.Riedl S., Rinner B., Zweytick D. In search of a novel target - phosphatidylserine exposed by non-apoptotic tumor cells and metastases of malignancies with poor treatment efficacy. Biochim. Biophys. Acta. 2011;1808:2638–2645. doi: 10.1016/j.bbamem.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riedl S., Rinner B., Zweytick D. Killing of melanoma cells and their metastases by human lactoferricin derivatives requires interaction with the cancer marker phosphatidylserine. Biometals. 2014;27:981–997. doi: 10.1007/s10534-014-9749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Meer G. Dynamic transbilayer lipid asymmetry. Cold Spring Harb. Perspect. Biol. 2011;3:1–11. doi: 10.1101/cshperspect.a004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pautot S., Frisken B.J., Weitz D.A. Engineering asymmetric vesicles. Proc. Natl. Acad. Sci. USA. 2003;100:10718–10721. doi: 10.1073/pnas.1931005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karamdad K., Law R.V., Ces O. Preparation and mechanical characterisation of giant unilamellar vesicles by a microfluidic method. Lab Chip. 2015;15:557–562. doi: 10.1039/c4lc01277a. [DOI] [PubMed] [Google Scholar]
- 13.Cheng H.T., Megha, London E. Preparation and properties of asymmetric vesicles that mimic cell membranes: effect upon lipid raft formation and transmembrane helix orientation. J. Biol. Chem. 2009;284:6079–6092. doi: 10.1074/jbc.M806077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markones M., Drechsler C., Fiedler S. Engineering asymmetric lipid vesicles: accurate and convenient control of the outer leaflet lipid composition. Langmuir. 2018;34:1999–2005. doi: 10.1021/acs.langmuir.7b03189. [DOI] [PubMed] [Google Scholar]
- 15.Heberle F.A., Marquardt D., Pabst G. Subnanometer structure of an asymmetric model membrane: interleaflet coupling influences domain properties. Langmuir. 2016;32:5195–5200. doi: 10.1021/acs.langmuir.5b04562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holzer M., Momm J., Schubert R. Lipid transfer mediated by a recombinant pro-sterol carrier protein 2 for the accurate preparation of asymmetrical membrane vesicles requires a narrow vesicle size distribution: a free-flow electrophoresis study. Langmuir. 2010;26:4142–4151. doi: 10.1021/la903386d. [DOI] [PubMed] [Google Scholar]
- 17.Huang Z., London E. Effect of cyclodextrin and membrane lipid structure upon cyclodextrin-lipid interaction. Langmuir. 2013;29:14631–14638. doi: 10.1021/la4031427. [DOI] [PubMed] [Google Scholar]
- 18.Kilsdonk E.P., Yancey P.G., Rothblat G.H. Cellular cholesterol efflux mediated by cyclodextrins. J. Biol. Chem. 1995;270:17250–17256. doi: 10.1074/jbc.270.29.17250. [DOI] [PubMed] [Google Scholar]
- 19.Ohvo H., Slotte J.P. Cyclodextrin-mediated removal of sterols from monolayers: effects of sterol structure and phospholipids on desorption rate. Biochemistry. 1996;35:8018–8024. doi: 10.1021/bi9528816. [DOI] [PubMed] [Google Scholar]
- 20.Tsamaloukas A., Szadkowska H., Heerklotz H. Interactions of cholesterol with lipid membranes and cyclodextrin characterized by calorimetry. Biophys. J. 2005;89:1109–1119. doi: 10.1529/biophysj.105.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsamaloukas A., Szadkowska H., Heerklotz H. Thermodynamic comparison of the interactions of cholesterol with unsaturated phospholipid and sphingomyelins. Biophys. J. 2006;90:4479–4487. doi: 10.1529/biophysj.105.080127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothman J.E., Kennedy E.P. Asymmetrical distribution of phospholipids in the membrane of Bacillus megaterium. J. Mol. Biol. 1977;110:603–618. doi: 10.1016/s0022-2836(77)80114-9. [DOI] [PubMed] [Google Scholar]
- 23.Denkins Y.M., Schroit A.J. Phosphatidylserine decarboxylase: generation of asymmetric vesicles and determination of the transbilayer distribution of fluorescent phosphatidylserine in model membrane systems. Biochim. Biophys. Acta. 1986;862:343–351. doi: 10.1016/0005-2736(86)90237-3. [DOI] [PubMed] [Google Scholar]
- 24.Choi J.Y., Duraisingh M.T., Voelker D.R. From protease to decarboxylase: the molecular metamorphosis of phosphatidylserine decarboxylase. J. Biol. Chem. 2015;290:10972–10980. doi: 10.1074/jbc.M115.642413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Bartolomeo F., Wagner A., Daum G. Cell biology, physiology and enzymology of phosphatidylserine decarboxylase. Biochim. Biophys. Acta. 2017;1862:25–38. doi: 10.1016/j.bbalip.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Dowhan W., Wickner W.T., Kennedy E.P. Purification and properties of phosphatidylserine decarboxylase from Escherichia coli. J. Biol. Chem. 1974;249:3079–3084. [PubMed] [Google Scholar]
- 27.Verma J.N., Goldfine H. Phosphatidylserine decarboxylase from Clostridium butyricum. J. Lipid Res. 1985;26:610–616. [PubMed] [Google Scholar]
- 28.Heerklotz H. Interactions of surfactants with lipid membranes. Q. Rev. Biophys. 2008;41:205–264. doi: 10.1017/S0033583508004721. [DOI] [PubMed] [Google Scholar]
- 29.Choi J.Y., Augagneur Y., Voelker D.R. Identification of gene encoding Plasmodium knowlesi phosphatidylserine decarboxylase by genetic complementation in yeast and characterization of in vitro maturation of encoded enzyme. J. Biol. Chem. 2012;287:222–232. doi: 10.1074/jbc.M111.313676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torchilin V.P., Weissig V. Second Edition. Oxford University Press; Oxford: 2003. Liposomes. A Practical Approach. [Google Scholar]
- 31.Bartlett G.R. Phosphorus assay in column chromatography. J. Biol. Chem. 1959;234:466–468. [PubMed] [Google Scholar]
- 32.Holzer M., Burghardt A., Schubert R. Quantitative high-performance thin-layer chromatography determination of common liposome components and critical parameters influencing the analysis results. J. Liposome Res. 2010;20:124–133. doi: 10.3109/08982100903218884. [DOI] [PubMed] [Google Scholar]
- 33.Majd S., Estes D.J., Mayer M. Assays for studying annexin binding to artificial bilayers. Calcium Bind. Proteins. 2006;1:26–29. [Google Scholar]
- 34.Johnson J.E., Kalmar G.B., Cornell R.B. Comparison of the lipid regulation of yeast and rat CTP: phosphocholine cytidylyltransferase expressed in COS cells. Biochem. J. 1992;285:815–820. doi: 10.1042/bj2850815. [DOI] [PMC free article] [PubMed] [Google Scholar]