Significance
Type 1 diabetes (T1D) is an incurable autoimmune disease caused by T cell-mediated destruction of insulin-producing beta cells. The beta-cell antigens recognized by human CD4+ T cells in T1D are poorly defined. Here, we show that C-peptide, derived from proinsulin, is recognized by CD4+ T cells from the blood of >60% of people with recent onset T1D. The majority of C-peptide–derived epitopes were recognized when presented by high-risk T1D HLA alleles. For some T cells, full-length C-peptide was a more potent stimulator than shorter peptides. Hence, our data suggests that full-length C-peptide may be uniquely antigenic in human T1D. C-peptide may be useful in assays to monitor changes in T cell autoimmunity and antigen-specific therapies for T1D.
Keywords: type 1 diabetes, proinsulin, CD4+ T cells, epitope, HLA
Abstract
Type 1 diabetes (T1D) is an autoimmune disease in which insulin-producing beta cells, found within the islets of Langerhans in the pancreas, are destroyed by islet-infiltrating T cells. Identifying the antigenic targets of beta-cell reactive T cells is critical to gain insight into the pathogenesis of T1D and develop antigen-specific immunotherapies. Several lines of evidence indicate that insulin is an important target of T cells in T1D. Because many human islet-infiltrating CD4+ T cells recognize C-peptide–derived epitopes, we hypothesized that full-length C-peptide (PI33–63), the peptide excised from proinsulin as it is converted to insulin, is a target of CD4+ T cells in people with T1D. CD4+ T cell responses to full-length C-peptide were detected in the blood of: 14 of 23 (>60%) people with recent-onset T1D, 2 of 15 (>13%) people with long-standing T1D, and 1 of 13 (<8%) HLA-matched people without T1D. C-peptide–specific CD4+ T cell clones, isolated from six people with T1D, recognized epitopes from the entire 31 amino acids of C-peptide. Eighty-six percent (19 of 22) of the C-peptide–specific clones were restricted by HLA-DQ8, HLA-DQ2, HLA-DQ8trans, or HLA-DQ2trans, HLA alleles strongly associated with risk of T1D. We also found that full-length C-peptide was a much more potent agonist of some CD4+ T cell clones than an 18mer peptide encompassing the cognate epitope. Collectively, our findings indicate that proinsulin C-peptide is a key target of autoreactive CD4+ T cells in T1D. Hence, full-length C-peptide is a promising candidate for antigen-specific immunotherapy in T1D.
Type 1 diabetes (T1D) is a chronic, incurable, autoimmune disease caused by T cell-mediated pancreatic beta-cell destruction (1) that leads to insulin deficiency and dysregulation of glucose metabolism (2). The International Diabetes Federation estimated that globally more than 0.5 million children, 14 y and younger, are living with T1D. Even with optimal glycemic control, vascular complications including ischemic heart disease, retinopathy, and nephropathy are rarely avoidable (3). Despite many significant advances in insulin-delivery technology and developments in synthetic insulins, the life expectancy of an individual with T1D remains over a decade less than the overall population (4).
Antigen-specific therapies that attenuate the pathogenic autoimmune response are an attractive approach to preventing and/or curing this disease. However, to date, attempts to develop antigen-specific therapies for T1D have been unsuccessful (5). Identification of the epitopes recognized by pathogenic CD4+ T cells is essential for developing both antigen-specific therapies for T1D and T cell assays to monitor changes in pathogenic T cell function. T cell assays are required for monitoring changes in T cell number and function following antigen-specific therapy to serve as a surrogate endpoint in clinical trials of new therapies for T1D (6).
Insulin is a prime candidate autoantigen in T1D (1). Insulin-specific autoantibodies are detectable before the symptomatic onset of T1D (7) and continue to be used to identify individuals at a high risk of developing T1D (8). Genetic association studies have also implicated insulin in the immune pathogenesis of human T1D. The T1D susceptibility locus, IDDM2, maps to a variable number of tandem repeats (VNTR) upstream of the insulin gene (9). This polymorphism is believed to modulate thymic proinsulin expression that, in turn, impacts upon T cell central tolerance (9, 10). Furthermore, T cell responses to insulin B9-23 are essential for the development of T1D in the NOD mouse (11).
The HLA region, on chromosome 6p21, is the locus most strongly associated with the risk of T1D (12). Within the HLA region, HLA-DQ8 (HLA-DQA1*03:01; HLA-DQB1*03:02) and HLA-DQ2 (HLA-DQA1*05:01; HLA-DQB1*02:01) confer the greatest risk of an individual developing T1D. Interestingly, individuals who are heterozygous for HLA-DQ2 and HLA-DQ8 are at greatest risk of developing T1D. This increased susceptibility to T1D is attributed to transdimers (trans), which form when the alpha chain of HLA-DQ8 pairs with the beta chain of HLA-DQ2 (HLA-DQ2trans: HLA-DQA1*03:01; HLA-DQB1*02:01) and, conversely, when the alpha chain of HLA-DQ2 pairs with the beta chain of DQ8 (HLA-DQ8trans: HLA-DQA1*05:01; HLA-DQB1*03:02) (13). The HLA class II heterodimers, HLA-DR, HLA-DQ, and HLA-DP, are expressed on professional antigen-presenting cells and present antigen-derived peptides to CD4+ T cells. Peptide-HLA binding studies have identified proinsulin-derived epitopes that bind to HLA-DQ2 and HLA-DQ8trans (14). However, despite the pivotal role HLA-DQ2 and HLA-DQ8 play in susceptibility to T1D, the epitopes presented by these HLA molecules recognized by CD4+ T cells are largely unknown. For example, to our knowledge, no blood-derived HLA-DQ2 (or HLA-DQ2trans) restricted, and only one HLA-DQ8 (or HLA-DQ8trans) restricted, insulin-specific CD4+ T cell clone has been reported (15).
In contrast to peripheral blood, HLA-DQ2/DQ8 restricted CD4+ T cells have been isolated from the residual pancreatic islets of deceased organ donors who suffered from T1D (16, 17). We found that many islet-infiltrating CD4+ T cell clones recognized epitopes derived from proinsulin C-peptide presented by HLA-DQ8 or HLA-DQ8trans (16). More recently, others have identified similar HLA-DQ8 restricted, islet-derived, T cell receptors (17). Although the numbers of donors are still small at this time, 10 of the 12 proinsulin-specific clones analyzed to date recognize epitopes derived from C-peptide (18).
C-peptide is a 31-aa peptide that is excised from proinsulin during the biosynthesis of insulin (19). In light of the evidence pointing to C-peptide–specific T cells playing a role in the immune pathogenesis of human T1D, we asked if C-peptide–specific CD4+ T cells could be detected in the blood of people with and without T1D. If present, we sought to characterize these cells. Our findings suggest that C-peptide is an important, but previously overlooked, antigen in T1D. This information will further guide the development of effective antigen-specific therapies for T1D and T cell assays for monitoring disease progression.
Results
CD4+ T Cell Responses to C-Peptide Are Detectable in the Blood of People with T1D.
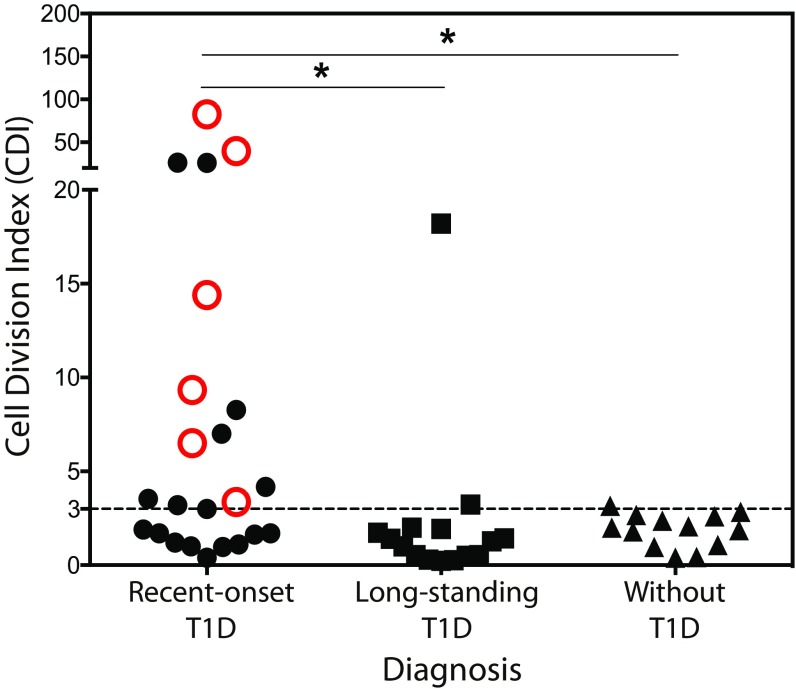
Previously, we identified six C-peptide–derived epitopes were recognized by human islet-infiltrating CD4+ T cells (16). To allow responses to all possible C-peptide–derived epitopes to be detected, with a small volume of blood, we used full-length C-peptide (PI33–63) to stimulate peripheral blood mononuclear cells (PBMCs) and detected CD4+ T cell responses using the sensitive CFSE-based proliferation assay (20). CD4+ T cell responses [cell division index (CDI) ≥ 3.0] to PI33–63 were detected in 14 of 23 subjects (60.8%) with recent-onset diabetes (within 100 d of diagnosis, stage III; ref. 21), 2 of 15 (13.3%) subjects with long-standing diabetes (greater than 100 d of diagnosis), and 1 of 13 (7.7%) healthy subjects (Fig. 1 and SI Appendix, Fig. S1 and Tables S1–S4, recent onset vs. healthy P = 0.005, recent onset vs. long-standing P = 0.016). The magnitude of the CD4+ T cell responses to PI33–63 was significantly greater in PBMCs from individuals with recent-onset T1D compared with healthy controls (10.8 ± 3.9 vs. 1.9 ± 0.25; P = 0.0024) and compared with long-standing subjects (10.8 ± 3.9 vs. 2.3 P = 0.0196). A CDI cutoff of ≥3.0 gave the greatest disease specificity (92.3%) and sensitivity (60.9%) (SI Appendix, Table S5). Insulin VNTR genotype showed no association with CD4+ T cell responses PI33–63 measured by CFSE-based proliferation assay (SI Appendix, Fig. S1). We found no correlation between CFSE responses and insulin dose-adjusted HbA1c (SI Appendix, Fig. S2).
Fig. 1.
CD4+ T cell responses to C-peptide in PBMC. A CDI ≥ 3.0, dotted line, is a positive response. The means of triplicate measurements for individuals with recent-onset T1D (n = 23), long-standing T1D (n = 15), and without T1D (n = 13) are plotted. Open red symbols indicate responses from which clones were isolated. Statistical significance was determined using unpaired Student’s t test, *P < 0.05 after log transformation of the CDIs.
C-Peptide Harbors Many CD4+ T Cell Epitopes.
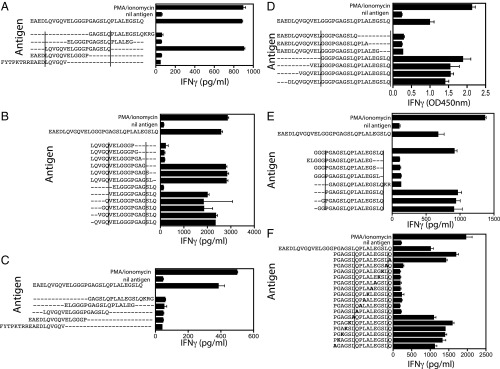
To further characterize the C-peptide–derived epitopes recognized by peripheral blood CD4+ T cells, clones were isolated using our CFSE-based T cell cloning protocol (22). A total of 32 CD4+ T cell clones that responded to PI33–63 were isolated from the peripheral blood of six subjects with recent-onset T1D. Of the 32, 7 could not be grown for analysis. The remaining 25 clones’ epitope specificities were determined using a panel of five overlapping 18mer peptides spanning C-peptide. The epitope mapping for two clones, H11.5 and K9.5, is shown in Fig. 2. Data for all of the other clones is shown in SI Appendix, Fig. S3. Fifteen of the 25 clones (60%), including H11.5 (Fig. 2 A and B), responded to one of the five 18mer peptides encompassing the entire C-peptide (Fig. 2A and SI Appendix, Fig. S3). The minimum epitopes required to stimulate these clones were then determined using a panel of peptides sequentially truncated by one amino acid from either the N or C terminus (Fig. 2B and SI Appendix, Fig. S3). Ten C-peptide–specific clones did not respond to any of the five 18mer peptides, including clone K9.5 (Fig. 2 C–F). For these clones, the minimum epitope was then determined by first testing against a panel of deletion variants of the full-length C-peptide (Fig. 2 D and E and SI Appendix, Fig. S4). The minimum epitope was identified by testing the clones against a panel of peptides with a single amino acid substitution expected to impair T cell recognition by disrupting either HLA binding or TCR recognition (Fig. 2F and SI Appendix, Fig. S4). Using this approach, the epitope specificities of 7 of these 10 clones were determined, but the specificity of the remaining 3 clones could not be determined. We concluded that clone H11.5 recognized PI42–51 (Fig. 2B) and clone K9.5 recognized PI54–62 (Fig. 2F). In all, we mapped epitopes recognized by 22 CD4+ T cell clones, from six subjects with recent-onset T1D (Table 1).
Fig. 2.
Analysis of epitope specificity of C-peptide–specific CD4+ T cell clones. CD4+ T cell clone responses to antigen were measured by the secretion of IFN-γ measured by ELISA, mean ± SEM of triplicate IFN-γ measurements are shown. (A) Clone H11.5 was tested against 18mer peptides (10 μM) spanning C-peptide. (B) Peptides truncated by a single amino acid. (C) Clone K9.5 doesn’t have a detectable response to the overlapping 18-mer peptides (10 μM). Clone K9.5 epitope mapping using truncated peptides (D and E) and a series of substituted peptides (F). Experiments were performed at least twice with similar results. In all figures, the parallel lines delineate the sequence of the minimum epitope determined.
Table 1.
Epitope mapping and HLA restriction analysis of blood-derived C-peptide–specific CD4+ T cells
| C-peptide | ||||||||||||||||||||||||||||||||
| Clone | E | A | E | D | L | Q | V | G | Q | V | E | L | G | G | G | P | G | A | G | S | L | Q | P | L | A | L | E | G | S | L | Q | HLA restriction* |
| B3.1 | X | X | X | X | X | X | X | X | X | X | X | X | DQ8 | |||||||||||||||||||
| B3.3 | X | X | X | X | X | X | X | X | X | X | X | DR4 | ||||||||||||||||||||
| K3.2 | X | X | X | X | X | X | X | X | X | DQ2 | ||||||||||||||||||||||
| K4.4 | X | X | X | X | X | X | X | X | X | X | DR4 | |||||||||||||||||||||
| K6.2 | X | X | X | X | X | X | X | X | X | X | DQ8trans | |||||||||||||||||||||
| K6.4 | X | X | X | X | X | X | X | X | X | X | DR4 | |||||||||||||||||||||
| K9.5 | X | X | X | X | X | X | X | X | X | DQ2 | ||||||||||||||||||||||
| K9.6 | X | X | X | X | X | X | X | X | X | X | X | DQ8 | ||||||||||||||||||||
| D1.1 | X | X | X | X | X | X | X | X | X | X | DQ8 | |||||||||||||||||||||
| D1.4 | X | X | X | X | X | X | X | X | X | X | DQ8 | |||||||||||||||||||||
| T6.1 | X | X | X | X | X | X | X | X | X | X | X | X | DQ2/DQ2trans | |||||||||||||||||||
| T6.6 | X | X | X | X | X | X | X | DQ2 | ||||||||||||||||||||||||
| T17.1 | X | X | X | X | X | X | X | DQ2/DQ2trans | ||||||||||||||||||||||||
| H3.3 | X | X | X | X | X | X | X | X | X | X | DQ8trans | |||||||||||||||||||||
| H3.7 | X | X | X | X | X | X | X | X | X | DQ8 | ||||||||||||||||||||||
| H6.4 | X | X | X | X | X | X | X | X | X | X | DQ8trans | |||||||||||||||||||||
| H7.4 | X | X | X | X | X | X | X | X | X | DQ8 | ||||||||||||||||||||||
| H8.5 | X | X | X | X | X | X | X | X | X | DQ8 | ||||||||||||||||||||||
| H11.5 | X | X | X | X | X | X | X | X | X | X | DQ8 | |||||||||||||||||||||
| H12.2 | X | X | X | X | X | X | X | X | X | DQ8/DQ8trans | ||||||||||||||||||||||
| E2.3 | X | X | X | X | X | X | X | X | X | DQ2 | ||||||||||||||||||||||
| E2.5 | X | X | X | X | X | X | X | X | X | X | X | X | DQ8 | |||||||||||||||||||
Nomenclature referenced by abbreviated HLA type is as follows (indicated in parentheses): HLA-DQ8 (A1*03:01, B1*03:02), HLA-DQ2 (A1*05:01, B1*02:01), HLA-DQ8trans (A1*05:01, B1*03:02), HLA-DQ2trans (A1*03:01, B1*02:01), HLA-DR4 (B1*04:01).
Most C-Peptide–Specific Clones Are HLA-DQ2, HLA-DQ8, HLA-DQ2trans, and HLA-DQ8trans Restricted.
The HLA restriction of the CD4+ T cell responses to proinsulin was determined by inhibiting responses by blocking HLA-DP, HLA-DQ, and HLA-DR, then testing against HLA-transfected cell lines (SI Appendix, Fig. S5). For the C-peptide–specific clones, 19 of 22 (86.3%) clones were HLA-DQ restricted and the remaining 3 (13.6%) were HLA-DR restricted (SI Appendix, Fig. S5). To define the restricting HLA alleles, panels of T2 or BLS lines transduced with individual HLA-DQ or HLA-DR genes, respectively, were used as antigen-presenting cells. Of the 22 clones analyzed: 9 (40.9%) were HLA-DQ8 restricted, 4 (18.2%) were HLA-DQ2 restricted, 3 (13.6%) were HLA-DR4 restricted, and 3 (13.6%) were HLA-DQ8trans restricted (SI Appendix, Figs. S5 and S6). The three remaining clones exhibited promiscuous recognition; one clone (4.5%) responded equally to both HLA-DQ8-expressing and HLA-DQ8trans-expressing antigen-presenting cells (APC) (SI Appendix, Fig. S5I) and two clones (9.1%) responded to both HLA-DQ2 and HLA-DQ2trans-expressing APC (SI Appendix, Figs. S5 M and N and S6). All three (13.6%) HLA-DR restricted clones were restricted by HLA-DR4; no HLA-DR3 restricted clones were isolated. Hence, with the exception of HLA-DR3, C-peptide–specific clones were restricted by all of the HLA alleles strongly associated with risk of T1D; notably 86% (19 of 22) of the clones were restricted by HLA-DQ2, HLA-DQ8 and/or their transdimers (Table 1). A comparison of the epitope specificity of islet-infiltrating and PBMC-derived C-peptide–specific clones is shown in SI Appendix, Table S6.
Private TCRs Encode C-Peptide Recognition.
To examine the clonal diversity of the peripherally derived C-peptide–specific CD4+ T cells, TCR genes expressed by the clones were sequenced. The clones used a range of TRAV and TRBV genes (SI Appendix, Table S7). TRAV and TRBV genes were sequenced from 21 of the 22 clones for which an epitope and HLA restriction could be determined. Fifteen distinct TRA/TRB combinations were found (SI Appendix, Table S7). There was no evidence of a “public” TCR. Clones from three of the six (50%) subjects expressed identical TCRs (SI Appendix, Table S8). Clones with identical TCRs had matching epitope specificity and HLA restriction, except T6.6 and T17.1. T6.6 was insufficiently sensitive to respond to HLA-DQ2trans, whereas T17.1 responded to both HLA-DQ2 and HLA-DQ2trans.
Full-Length C-Peptide Is a More Potent Antigen for Some T Cells.
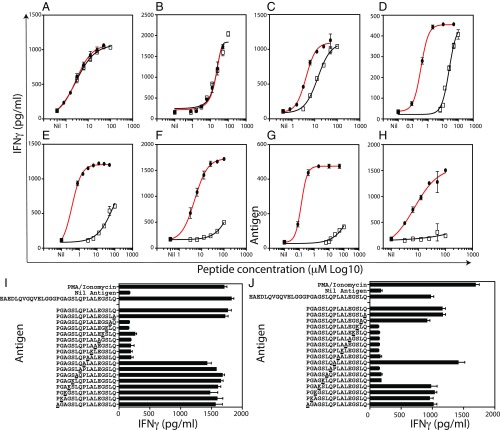
To determine the relative potency of PI33–63 as an agonist for CD4+ T cells, we compared the potency of full-length (PI33–63, 31 aa) C-peptide to 18mer peptides incorporating the cognate epitope for eight C-peptide–specific clones. In each case, the minimum epitope was flanked by at least three amino acids at both the N and C termini (SI Appendix, Table S9). Six of eight clones were more sensitive to the full-length C-peptide. Remarkably, for five of eight clones, full-length C-peptide was >100- to 1,000-fold more potent than the 18mer peptide incorporating the cognate epitope (Fig. 3 and SI Appendix, Table S9). In contrast, two of the eight clones tested responded equally to both full-length C-peptide and an overlapping 18mer. The clones that were more sensitive to the full-length C-peptide recognized epitopes toward the C terminus of the C-peptide (SI Appendix, Table S10). The enhanced response to full-length C-peptide is not due to protein splicing because amino acid substitution experiments, using a high concentration (50 μM) of 16mer peptides, show that 7–10 contiguous amino acids are required to stimulate the clones (Figs. 2F and 3 I and J and SI Appendix, Fig. S4G). We conclude, that for many C-peptide–specific CD4+ T cells, full-length C-peptide is a much stronger agonist than 18mer peptides incorporating the cognate epitope.
Fig. 3.
Comparison of the stimulatory capacity of full-length C-peptide and 18mer peptides. Titration of full-length C-peptide (closed circles) and 18mer peptides (open squares) incorporating the cognate epitope. KJ EBV or HLA-DR4 transfected BLS cells (both at 2 × 104 cells/well), were used as APC. T cell responses were measured in triplicate by IFN-γ ELISA (pg/mL). Mean (±SEM) of triplicate IFN-γ measurements are shown. Dose–response curves for the following clones are shown: K9.6 (A), H11.5 (B), D1.1 (C), E2.3 (D), T17.1 (E), K9.5 (F), B3.3 (G), H3.3 (H). Amino acid substitution experiments (50 μM peptide), where each amino acid is changed to alanine or lysine, reveal that clones T17.1 (I) and H3.3 (J) recognize contiguous 7- to 10-aa-long epitopes. One representative of at least two experiments is shown.
To confirm that the C-peptide–specific clones responded to beta cells, six clones were tested for responses against human islet lysates. Five of six clones responded to islet lysates (SI Appendix, Fig. S7) and, in some cases, weakly to acinar tissue.
Discussion
Here, we show that CD4+ T cells specific for proinsulin C-peptide can be detected in the peripheral blood of individuals with recent-onset T1D. This concurs with the detection of C-peptide–specific CD4+ T cells in the islets of the small number of organ donors with T1D who have been studied (16–18, 23). C-peptide–specific CD4+ T cell clones isolated from peripheral blood recognized epitopes from throughout the C-peptide with many in its C-terminal half. Eighty-six percent of T cell clones were restricted by HLA-DQ8, HLA-DQ2, HLA-DQ2trans, or HLA-DQ8trans. Dose–response experiments revealed that full-length C-peptide was a 100- to 1,000-fold more potent stimulator than 18mer peptides encompassing the cognate epitopes for some CD4+ T cell clones. Together, these data confirm that C-peptide–specific CD4+ T cells are present in both the islets and the peripheral blood of individuals with T1D. Furthermore, our data suggest that PI33–63 may be an important, but largely overlooked, target of pathogenic CD4+ T cells in T1D.
CD4+ T cell responses to C-peptide were detected in the peripheral blood of more than 60% (14 of 23) of people within 100 d of diagnosis of T1D, compared with 8% (1 of 13) of healthy controls. We note that many recent-onset T1D subjects will have residual C-peptide, which may impact upon CD4+ T cell responses. The comparison is also complicated by genetic differences between the T1D and without T1D groups. Nonetheless, the sensitivity and specificity of our assay was greater than in assays that used shorter peptides (24) or recombinant proinsulin (25). For example, CD4+ T cell responses to PI51–68 eluted from HLA-DR4 (26), which were seen in only ∼13% of individuals with recent-onset T1D, as detected by ELISpot (27). Our ability to detect responses in more than 60% of individuals with T1D may be attributable to the use of PI33–63. This provides more potential epitopes than a shorter peptide, increasing the pool of potential responding T cells. In addition, we found that many of the T cell clones that recognize epitopes in the C-terminal end of the C-peptide were 100- to 1,000-fold more sensitive to full-length C-peptide than to 18mer peptides incorporating the cognate epitope (Fig. 3). At this point, it is unclear why clones specific for epitopes in the C terminus of C-peptide are more sensitive to full-length C-peptide than shorter epitopes. However, our data suggest that hybrid insulin peptides (HIPs) (28), or an epitope formed by an altered conformation (29) of C-peptide, are not responsible for the greater potency of the full-length C-peptide. Instead, we suggest that the shorter peptides may be more rapidly degraded compared with full-length C-peptide, or the latter may be more efficiently processed and presented by antigen-presenting cells than shorter peptides.
In contrast to previous studies (30–33), 86% (19 of 22) of the C-peptide–specific clones we isolated were HLA-DQ restricted. It is unclear why it has been difficult to isolate HLA-DQ restricted CD4+ T cells from peripheral blood and why our approach, using CFSE-based proliferation assays and full-length C-peptide, stimulated a largely HLA-DQ2/8 restricted response. The detection of many HLA-DQ restricted CD4+ T cells fits neatly with the well-documented genetic association between these HLA alleles and risk of developing T1D (12), providing support for the clinical relevance of CD4+ T cell responses to full-length C-peptide. We did not seek to analyze C-peptide–specific CD4+ T cells restricted by HLA alleles other than HLA-DR3, HLA-DR4, HLA-DQ2, HLA-DQ8, HLA-DQ2trans, and HLA-DQ8trans. We did not attempt to clone T cells from subjects without T1D. We have found that clones cannot be isolated when the CDI is <3.0. However, it remains possible that there are C-peptide–specific CD4+ T cell clones restricted by other HLA alleles that we have not analyzed, or from subjects without T1D. Peptide HLA class II tetramers have been used to identify beta-cell antigen-specific T cells (34, 35). However, our results suggest that an HLA class II tetramer-based approach may not be feasible to comprehensively analyze C-peptide–specific responses because there are at least 13 different peptide/HLA combinations that can be derived from human C-peptide. Instead, we would advocate for functional assays (20, 36) that do not require a specific reagent for each peptide/HLA combination.
HLA-DQ2 and HLA-DQ8 are strongly associated with risk of T1D (12). While the mechanism by which HLA-DQ2 and HLA-DQ8 contribute to risk of T1D remains unclear, it is now clear that many islet-infiltrating CD4+ T cell clones are restricted by HLA-DQ2/DQ8 (16–18, 23). Broadly, there is overlap between the HLA-DQ2 and HLA-DQ8 restricted clones isolated from islets and peripheral blood (SI Appendix, Table S6) (16, 17), although fine mapping reveals that very few are identical. HLA-DQ8 restricted HIPs also infiltrate human islets (28). Interestingly, some CD4+ T cells that recognize a hybrid insulin peptide epitope formed by the fusion of the central region of C-peptide with a fragment of IAPP2, cross-react with unmodified C-peptide (28). Although the unmodified C-peptide is less potent than the HIP for these clones, this suggests that responses to HIPs and full-length C-peptide may both be HLA-DQ8 restricted and potentially reenforce each other.
The C-terminal half of C-peptide (PI51–68) has been tested in a phase I trial as a candidate for antigen-specific therapy to curb autoimmune CD4+ T cell responses in T1D (37, 38). This peptide, PI51–68, incorporates epitopes recognized by half (11 of 22) of the full-length C-peptide–specific clones described here. Although the PI51–68 peptide was identified in an HLA-DR4 restricted response (26), our data shows that this peptide contains epitopes presented by HLA-DQ2, HLA-DQ2trans, and HLA-DQ8 in addition to HLA-DR4 when T cells are stimulated with full-length C-peptide. Compared with PI51–68, full-length C-peptide comprises multiple epitopes, stimulates predominantly HLA-DQ2/8 restricted responses, and is a much more potent antigen.
The primary limitation of this work, like all clinical studies, is that we cannot demonstrate that C-peptide–specific CD4+ T cells cause T1D in humans. A direct demonstration of a pathogenic role for CD4+ T cell responses to C-peptide will require a mouse model of the human T cell responses to C-peptide, which is yet to be developed.
There are two clinical applications of our findings: T cell biomarker assays and antigen-specific therapy protocols. Developing T cell biomarker assays to monitor changes in beta cell-specific T cells in the blood of people with T1D has been an enduring challenge. Based on the results presented here, we suggest that C-peptide could usefully be included in functional T cell assays, such as ELIspot or CFSE-based proliferation assays. C-peptide can readily be applied to assays, using larger cohorts, to further delineate the temporal dynamics of CD4+ T cell responses to C-peptide in the blood of people developing T1D. Full-length C-peptide warrants further investigation as an antigen in antigen-specific therapies. As noted above, a shorter section of C-peptide has already been evaluated as an antigen-specific therapy, in a phase Ib clinical trial with HLA-DRB1*04:01 subjects (38). PI33–63 could readily be tested for prevention of T1D because it is relatively stable, nontoxic, and water soluble, and has already been used in clinical trials for diabetes-associated complications (39).
Materials and Methods
Subjects.
Studies received local ethical approval (St. Vincent’s Hospital, HREC-A 135/08), (Royal Melbourne Hospital, 2009.026), and (Monash Health, 12185B). All participants provided written informed consent. Participants (aged 3–45 y) without T1D, diagnosed with T1D within 100 d (recent-onset), or greater than 100 d (long-standing) were recruited. T1D was diagnosed according to American Diabetes Association criteria.
CFSE-Based Proliferation Assay and CD4+ T Cell Cloning.
Blood was obtained by venepuncture and PBMC isolated. CFSE-labeled PBMC were cultured with no antigen, PI33–63 (10 μM), or tetanus toxoid (20, 22). The results are presented as a CDI (20). C-peptide specific CD4+ T cell clones were isolated as described (22).
Screening Antigen-Specific Clones.
Clones were tested for antigen specificity using the 3H-thymidine assay (22). APC were either autologous PBMC or KJ-EBV. Each clone was tested in duplicate with and without antigen. Responses were measured by 3H-thymidine incorporation. Clones that had a stimulation index (>3.0) were considered to be antigen specific.
Functional Analysis of CD4+ T Cell Clones.
Cloned CD4+ T cells were incubated with synthetic peptides and the KJ cell line (22). Cloned CD4+ T cells (50,000 per well) were cultured with 20,000 APC with and without peptides. Responses to antigen were measured in triplicate as IFN-gamma (IFN-γ) secretion into the culture media, determined by ELISA (BioLegend).
Determining the HLA Restriction of T Cell Clones.
The HLA restriction of the C-peptide–specific CD4+ T cell clones was determined by mAb blocking and transfected cell assays (22, 31).
Sequencing TCR Genes Expressed by C-Peptide–Specific Clones.
Reverse transcription mix, SuperScript VILO Reverse Transcriptase (Invitrogen), was dispensed directly into wells containing five cells. TRA and TRB genes were amplified using pools of forward primers and Taq polymerase (Qiagen) as described by ref. 40.
Supplementary Material
Acknowledgments
We thank Prof. Vijaya Sundararajan for statistical advice. We thank Erin Hill, Gowri Selvaraj, Elham Mohammed-Nur, and Ashvin Nursing for clinical assistance. This work was supported by Juvenile Diabetes Research Foundation Grant 5-CDA-2014-210-A-N (to S.I.M.), National Health and Medical Research Council Grant GNT123586 (to S.I.M.), Diabetes Australia Research Trust Millennium Award Y17M1-MANS (to S.I.M.), and the Operational Infrastructure Support Program of the Victorian Government (S.I.M., H.E.T., and T.W.H.K.). M.S. is supported by NHMRC Postgraduate Scholarship APP1094337 and JDRF PhD Top-up Scholarship.
Footnotes
Conflict of interest statement: A provisional patent has been filed by St. Vincent’s Institute to protect the use of full-length C-peptide in T cell assays and antigen-specific therapies (2017904853).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809208115/-/DCSupplemental.
References
- 1.Mannering SI, Pathiraja V, Kay TW. The case for an autoimmune aetiology of type 1 diabetes. Clin Exp Immunol. 2016;183:8–15. doi: 10.1111/cei.12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziegler AG, et al. Type 1 diabetes prevention: A goal dependent on accepting a diagnosis of an asymptomatic disease. Diabetes. 2016;65:3233–3239. doi: 10.2337/db16-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lind M, et al. Glycemic control and excess mortality in type 1 diabetes. N Engl J Med. 2014;371:1972–1982. doi: 10.1056/NEJMoa1408214. [DOI] [PubMed] [Google Scholar]
- 4.Livingstone SJ, et al. Scottish Diabetes Research Network epidemiology group; Scottish Renal Registry Estimated life expectancy in a Scottish cohort with type 1 diabetes, 2008-2010. JAMA. 2015;313:37–44. doi: 10.1001/jama.2014.16425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staeva TP, Chatenoud L, Insel R, Atkinson MA. Recent lessons learned from prevention and recent-onset type 1 diabetes immunotherapy trials. Diabetes. 2013;62:9–17. doi: 10.2337/db12-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roep BO, Peakman M. Surrogate end points in the design of immunotherapy trials: Emerging lessons from type 1 diabetes. Nat Rev Immunol. 2010;10:145–152. doi: 10.1038/nri2705. [DOI] [PubMed] [Google Scholar]
- 7.Palmer JP, et al. Insulin antibodies in insulin-dependent diabetics before insulin treatment. Science. 1983;222:1337–1339. doi: 10.1126/science.6362005. [DOI] [PubMed] [Google Scholar]
- 8.Ziegler AG, et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA. 2013;309:2473–2479. doi: 10.1001/jama.2013.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pugliese A, et al. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet. 1997;15:293–297. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- 10.Durinovic-Belló I, et al. Insulin gene VNTR genotype associates with frequency and phenotype of the autoimmune response to proinsulin. Genes Immun. 2010;11:188–193. doi: 10.1038/gene.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakayama M, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005;435:220–223. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noble JA. Immunogenetics of type 1 diabetes: A comprehensive review. J Autoimmun. 2015;64:101–112. doi: 10.1016/j.jaut.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Nepom BS, Schwarz D, Palmer JP, Nepom GT. Transcomplementation of HLA genes in IDDM. HLA-DQ alpha- and beta-chains produce hybrid molecules in DR3/4 heterozygotes. Diabetes. 1987;36:114–117. doi: 10.2337/diab.36.1.114. [DOI] [PubMed] [Google Scholar]
- 14.van Lummel M, et al. Type 1 diabetes-associated HLA-DQ8 transdimer accommodates a unique peptide repertoire. J Biol Chem. 2012;287:9514–9524. doi: 10.1074/jbc.M111.313940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eerligh P, et al. Functional consequences of HLA-DQ8 homozygosity versus heterozygosity for islet autoimmunity in type 1 diabetes. Genes Immun. 2011;12:415–427. doi: 10.1038/gene.2011.24. [DOI] [PubMed] [Google Scholar]
- 16.Pathiraja V, et al. Proinsulin-specific, HLA-DQ8, and HLA-DQ8-transdimer-restricted CD4+ T cells infiltrate islets in type 1 diabetes. Diabetes. 2015;64:172–182. doi: 10.2337/db14-0858. [DOI] [PubMed] [Google Scholar]
- 17.Michels AW, et al. Islet-derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes. 2017;66:722–734. doi: 10.2337/db16-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent SC, Mannering SI, Michels AW, Babon JAB. Deciphering the pathogenesis of human type 1 diabetes (T1D) by interrogating T cells from the “scene of the crime”. Curr Diab Rep. 2017;17:95. doi: 10.1007/s11892-017-0915-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodson G, Steiner D. The role of assembly in insulin’s biosynthesis. Curr Opin Struct Biol. 1998;8:189–194. doi: 10.1016/s0959-440x(98)80037-7. [DOI] [PubMed] [Google Scholar]
- 20.Mannering SI, et al. A sensitive method for detecting proliferation of rare autoantigen-specific human T cells. J Immunol Methods. 2003;283:173–183. doi: 10.1016/j.jim.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Insel RA, et al. Staging presymptomatic type 1 diabetes: A scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care. 2015;38:1964–1974. doi: 10.2337/dc15-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mannering SI, et al. An efficient method for cloning human autoantigen-specific T cells. J Immunol Methods. 2005;298:83–92. doi: 10.1016/j.jim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Babon JA, et al. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat Med. 2016;22:1482–1487. doi: 10.1038/nm.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Semana G, Gausling R, Jackson RA, Hafler DA. T cell autoreactivity to proinsulin epitopes in diabetic patients and healthy subjects. J Autoimmun. 1999;12:259–267. doi: 10.1006/jaut.1999.0282. [DOI] [PubMed] [Google Scholar]
- 25.Dubois-LaForgue D, Carel JC, Bougnères PF, Guillet JG, Boitard C. T-cell response to proinsulin and insulin in type 1 and pretype 1 diabetes. J Clin Immunol. 1999;19:127–134. doi: 10.1023/a:1020558601175. [DOI] [PubMed] [Google Scholar]
- 26.Arif S, et al. Autoreactive T cell responses show proinflammatory polarization in diabetes but a regulatory phenotype in health. J Clin Invest. 2004;113:451–463. doi: 10.1172/JCI19585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arif S, et al. Peripheral and islet interleukin-17 pathway activation characterizes human autoimmune diabetes and promotes cytokine-mediated β-cell death. Diabetes. 2011;60:2112–2119. doi: 10.2337/db10-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delong T, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351:711–714. doi: 10.1126/science.aad2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zavala-Ruiz Z, Strug I, Walker BD, Norris PJ, Stern LJ. A hairpin turn in a class II MHC-bound peptide orients residues outside the binding groove for T cell recognition. Proc Natl Acad Sci USA. 2004;101:13279–13284. doi: 10.1073/pnas.0403371101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mannering SI, et al. The insulin A-chain epitope recognized by human T cells is posttranslationally modified. J Exp Med. 2005;202:1191–1197. doi: 10.1084/jem.20051251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mannering SI, et al. The A-chain of insulin is a hot-spot for CD4+ T cell epitopes in human type 1 diabetes. Clin Exp Immunol. 2009;156:226–231. doi: 10.1111/j.1365-2249.2009.03907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Lorenzo TP, Peakman M, Roep BO. Translational mini-review series on type 1 diabetes: Systematic analysis of T cell epitopes in autoimmune diabetes. Clin Exp Immunol. 2007;148:1–16. doi: 10.1111/j.1365-2249.2006.03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, et al. Antigen-specific T cell analysis reveals that active immune responses to β cell antigens are focused on a unique set of epitopes. J Immunol. 2017;199:91–96. doi: 10.4049/jimmunol.1601570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buckner JH, et al. Defining antigen-specific responses with human MHC class II tetramers. J Allergy Clin Immunol. 2002;110:199–208. doi: 10.1067/mai.2002.125976. [DOI] [PubMed] [Google Scholar]
- 35.Oling V, et al. GAD65- and proinsulin-specific CD4+ T-cells detected by MHC class II tetramers in peripheral blood of type 1 diabetes patients and at-risk subjects. J Autoimmun. 2005;25:235–243. doi: 10.1016/j.jaut.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 36.Mannering SI, et al. Immunology of Diabetes Society T-Cell Workshop Committee Current approaches to measuring human islet-antigen specific T cell function in type 1 diabetes. Clin Exp Immunol. 2010;162:197–209. doi: 10.1111/j.1365-2249.2010.04237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thrower SL, et al. Proinsulin peptide immunotherapy in type 1 diabetes: Report of a first-in-man phase I safety study. Clin Exp Immunol. 2009;155:156–165. doi: 10.1111/j.1365-2249.2008.03814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alhadj Ali M, et al. Metabolic and immune effects of immunotherapy with proinsulin peptide in human new-onset type 1 diabetes. Sci Transl Med. 2017;9:eaaf7779. doi: 10.1126/scitranslmed.aaf7779. [DOI] [PubMed] [Google Scholar]
- 39.Ekberg K, et al. C-peptide replacement therapy and sensory nerve function in type 1 diabetic neuropathy. Diabetes Care. 2007;30:71–76. doi: 10.2337/dc06-1274. [DOI] [PubMed] [Google Scholar]
- 40.Wang GC, Dash P, McCullers JA, Doherty PC, Thomas PG. T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci Transl Med. 2012;4:128ra42. doi: 10.1126/scitranslmed.3003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.