Significance
Bacteria live in aggregates at sites of chronic infection, and aggregated growth is thought to be important in producing key infection phenotypes such as antibiotic tolerance. We found that entropic forces generated by polymers abundant at chronic infection sites can cause bacteria to aggregate by a mechanism known as “depletion aggregation.” This aggregation mechanism does not require biofilm formation functions, but it does cause bacteria to become much less susceptible to killing by antibiotics. These findings indicate that aggregation could be a default growth mode at infection sites. It might be useful to target mechanisms of depletion-mediated antibiotic tolerance for the treatment of chronic infections.
Keywords: biofilm, Pseudomonas aeruginosa, chronic infection, antibiotic tolerance, depletion aggregation
Abstract
Bacteria causing chronic infections are generally observed living in cell aggregates suspended in polymer-rich host secretions, and bacterial phenotypes induced by aggregated growth may be key factors in chronic infection pathogenesis. Bacterial aggregation is commonly thought of as a consequence of biofilm formation; however the mechanisms producing aggregation in vivo remain unclear. Here we show that polymers that are abundant at chronic infection sites cause bacteria to aggregate by the depletion aggregation mechanism, which does not require biofilm formation functions. Depletion aggregation is mediated by entropic forces between uncharged or like-charged polymers and particles (e.g., bacteria). Our experiments also indicate that depletion aggregation of bacteria induces marked antibiotic tolerance that was dependent on the SOS response, a stress response activated by genotoxic stress. These findings raise the possibility that targeting conditions that promote depletion aggregation or mechanisms of depletion-mediated tolerance could lead to new therapeutic approaches to combat chronic bacterial infections.
Chronic infections are extremely difficult to eradicate once established. Recent work raises the possibility that treatment failure may be caused primarily by the growth mode of bacteria in vivo rather than by fixed antibiotic resistance caused by genetic changes (1, 2). This idea is based largely on the fact that chronic infections are treatment resistant even when ex vivo tests indicate that the infecting bacteria are antibiotic sensitive (3–5). Furthermore, bacteria at chronic infection sites are often found in multicellular aggregates, and aggregated growth can markedly increase bacterial antibiotic tolerance (6, 7). The opportunistic pathogen Pseudomonas aeruginosa is a prime example. This organism is frequently observed in aggregates in wounds and in the airways of people with cystic fibrosis (CF) (7, 8), and laboratory models that produce aggregated growth markedly increase bacterial resistance to killing (9).
The laboratory systems most commonly used to model aggregated bacterial growth generate surface-attached and matrix-encased communities known as “biofilms.” Biofilm formation in laboratory reactors is an active process that occurs when environmental cues activate bacterial functions such as surface adherence, motility, cell-to-cell communication, and extracellular matrix synthesis (10–12). Biofilm-forming bacteria use these functions to assemble themselves into complex multicellular structures. Significant progress has been made in understanding biofilm formation, and therapeutic strategies targeting biofilm functions are currently being developed (13).
However, several observations raise the possibility that biofilm formation functions may not be operative in some in vivo chronic infection conditions. For example, clinical isolates of P. aeruginosa, Staphylococcus aureus, and Stenotrophpmonas maltophilia cultured from people with chronic CF infections are often impaired in forming biofilms (6, 14–20). Furthermore, bacteria evolve genetically during CF and wound infections (3, 16, 21), and some genes mediating biofilm formation are consistently inactivated during within-host evolution. Examples include genes mediating flagellar motility, pili biosynthesis, and quorum sensing (11, 16, 22–25). Last, surface-associated growth is a key stimulus for biofilm formation (26), and most bacteria in wounds and CF airways are found suspended in secretions rather than being surface-attached (27, 28). Together, these findings raise the possibility that biofilm formation may not mediate bacterial aggregation in some in vivo settings.
An alternative model to explain bacterial aggregation during infection involves mechanisms mediated by the polymers and macromolecules present at chronic infection sites. High concentrations of glycosaminoglycans, glycoproteins, collagen, and other polymers are present in wounds (29). Likewise, CF airway secretions contain abundant mucin (30), DNA (31), and F-actin (32). Polymers can cause particles (e.g., bacteria) to aggregate by two general mechanisms: bridging (33) and depletion aggregation (34) (see below). Notably, both processes are passive from the standpoint of the bacteria, as bacterial activity is not required. This point is critical, because if these polymer-driven mechanisms operated in vivo, targeting biofilm assembly functions might not have therapeutic value.
Here we studied interactions between synthetic and host-derived polymers and P. aeruginosa to test the hypothesis that polymers present at infection sites cause bacterial aggregation independent of active biofilm formation functions. We also investigated the mechanism producing aggregation and its consequences for antibiotic tolerance, a critical factor in chronic infection persistence.
Results
Host Polymers Aggregate Bacteria via the Depletion Mechanism.
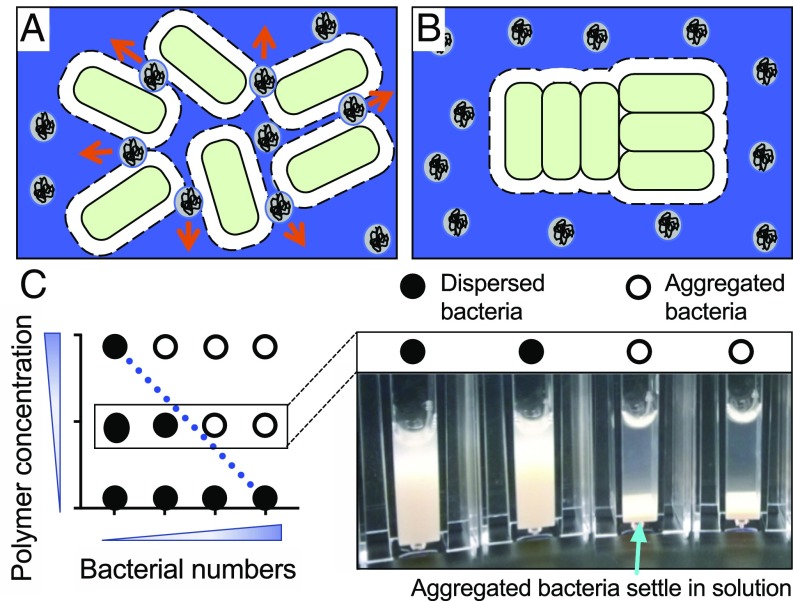
As noted above, polymers can aggregate particles by two general mechanisms: bridging (33) and depletion aggregation (34). Bridging occurs when polymers adsorb simultaneously on more than one particle, bringing them together. In contrast, depletion aggregation operates in environments in which nonadsorbing polymers and particles are present at high concentrations. In such conditions, the polymers present in the area between adjacent particles become constrained. Since polymers are generally more numerous and dynamic than particles, this constraint reduces the total entropy of the system (Fig. 1). However, spontaneous motion will produce configurations in which particles become closely aligned. Particle alignment is favored as it produces more space for polymer movement and thereby maximizes the entropy of the system. Furthermore, the resulting difference in polymer concentration across the particles establishes an unbalanced osmotic force that holds particles together in aggregates (34).
Fig. 1.
Schematic showing how the depletion mechanism aggregates bacteria. In polymer-rich conditions, entropy is maximized when bacterial cells aggregate. Dashed lines represent an exclusion volume around each cell, which is proportional to the radius (molecular weight) of the polymer coils. (A) As two cells approach one another, polymers spontaneously move out and away from exclusion volumes (arrows). (B) When exclusion volumes overlap, the volume available to the polymers increases, maximizing system entropy. The polymer concentration between cells is essentially zero, and the difference in polymer concentration across the cell generates an osmotic pressure that physically holds aggregates together. (C) Phase diagrams can be used to identify depletion aggregation. In the conceptual phase diagram shown here, bacteria and polymers are mixed at different concentrations in cuvettes. Bacteria remain dispersed when bacterial numbers and/or polymer concentrations are low (filled circles). When bacterial numbers and/or polymer concentrations are sufficiently high, aggregation occurs, causing bacteria to sediment (open circles). The blue line indicates the phase boundary separating the dispersed and aggregated phases. The negative slope of this line is indicative of depletion aggregation; see text and refs. 34, 53, and 54 for more details.
Bridging and depletion aggregation can be distinguished by two approaches. The first examines the relationship between the number of particles present and the concentration of polymer needed to produce aggregation. In bridging, more polymer is needed as particle concentration increases, because more bridging interactions are required. Thus, a phase diagram showing the polymer and particle concentrations at which particles transition from the dispersed to aggregated phase (i.e., the phase transition) shows a positive slope.
In contrast, depletion aggregation is enhanced by polymer and particle crowding. Thus, less polymer is needed to aggregate higher concentrations of particles, and phase transitions will show negative slopes (35). Fig. 1C shows a conceptual representation of a phase diagram that indicates depletion aggregation.
The second approach is to examine the morphology of aggregates. Depletion aggregation is driven by entropy gains realized when the space available for polymer movement is maximized. Thus, rod-shaped particles will align laterally to maximize space and entropy gains in depletion aggregates. Bridging does not generally produce lateral particle alignment, as polymers attach to multiple particles based on the locations of binding sites. Thus, bridging generally produces a less-ordered aggregate morphology.
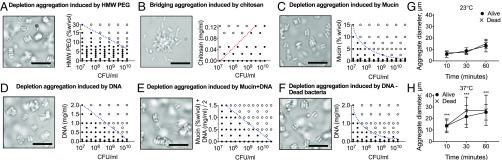
We began studying polymer-induced aggregation of P. aeruginosa (which carries a net negative charge) using two well-characterized polymers: positively charged chitosan and neutral polyethylene glycol (PEG, 10 kDa). While both polymers caused P. aeruginosa to aggregate, PEG produced a negatively sloped phase transition and laterally aligned aggregates characteristic of depletion aggregation (Fig. 2A). In contrast, chitosan produced a positively sloped phase transition and disordered aggregates (Fig. 2B), indicating bridging aggregation, as expected due to chitosan’s positive charge.
Fig. 2.
Diverse nonadsorbing polymers promote depletion aggregation of P. aeruginosa. (A–F, Left) Representative images were collected 30 min after mixing P. aeruginosa and polymers. (Scale bars, 10 µm.) (Right) Phase diagrams were generated by mixing bacteria and polymers in cuvettes at the indicated concentrations and visually scoring the cultures as either dispersed (filled circles) or aggregated (open circles) after 6 h. Dashed lines indicate phase boundaries; a positively sloped phase boundary (red line) indicates bridging; a negatively sloped phase boundary (blue line) indicates depletion aggregation. See Fig. 1C for a conceptual phase diagram. (G and H) Viable or dead P. aeruginosa were suspended in 30% (wt/vol) PEG 10k at 5 × 109 cfu/mL. Samples were incubated 23 °C (G) or 37 °C (H). Aggregate diameter was measured by microscopy at the indicated times. Results are the mean ± SD of three experiments, n = 50 aggregates for each experiment. ***P < 0.001, comparing 23 °C and 37 °C time points for both live and dead bacteria.
To determine if host-derived polymers that are abundant at infection sites could aggregate P. aeruginosa, we examined the effect of mucin, DNA, F-actin, and hyaluronan. All these polymers were highly effective in aggregating P. aeruginosa, and all generated negatively sloped phase transitions and laterally aligned aggregates (Fig. 2 C and D and SI Appendix, Fig. S1 A and B). The negatively charged bacteria-derived polymers alginate and filamentous Pf bacteriophage produced a similar effect (SI Appendix, Fig. S1 C and D). It is possible that the commercially available polymers used in these experiments might exhibit different properties from the polymers present in vivo. However, the general capacity of all tested negatively charged polymers to aggregate bacteria suggests that polymers abundant in chronic infection sites are also likely to cause depletion aggregation of P. aeruginosa.
Host Polymers Enhance Aggregation in Combination.
Infection sites contain multiple polymers, and we hypothesized that their aggregating effects would be enhanced in combination, as physical (entropic) rather than specific biological mechanisms are responsible. To test this, we mixed mucin and DNA together at varying concentrations that included levels present in CF airway secretions and found that polymers in combination aggregated bacteria at lower concentrations than when present alone (Fig. 2E). For example, used alone, 5% (wt/vol) of mucin or 1.0 mg/mL of DNA was required to aggregate ∼3.3 × 108 cfu/mL bacteria. In combination, similar numbers of bacteria could be aggregated by 1% (wt/vol) mucin and 0.5 mg/mL DNA. This finding indicates that physiologically relevant polymer mixtures could act to aggregate bacteria even if the concentration of individual polymers was insufficient (Discussion).
To determine if disease-relevant polymer concentrations cause aggregation of low numbers of bacteria that may be present early in disease, we mixed decreasing numbers of bacterial cells with a CF-relevant mixture of DNA (4 mg/mL), mucin [8% (wt/vol)], and F-actin (2.5 mg/mL). We observed small laterally aligned aggregates at bacterial concentrations as low as 105 cfu/mL (SI Appendix, Fig. S2).
Temperatures Encountered at Infection Sites Promote Depletion Aggregation.
Infection sites are generally warmer than the environmental niches occupied by P. aeruginosa. Higher temperatures should increase particle movement and thus the frequency at which particles spontaneously align. This might promote depletion aggregation, as chance alignments are a key step in aggregate formation (Fig. 1) (36). To test this idea, we measured aggregate formation as a function of temperature. Aggregates formed more quickly and developed larger sizes at 37 °C than at 23 °C (Fig. 2 G and H). Similar results were observed using viable and dead bacteria, indicating that active bacterial functions were not responsible for increased aggregate formation at the higher temperature. These results suggest that depletion aggregation of bacteria may be enhanced by the warmer temperatures likely encountered at infection sites.
DNase I Inhibits Mucin/DNA-Induced Depletion Aggregation.
Mucin and DNA are major components of airway sputum from CF patients, and DNase I is used therapeutically to cleave sputum DNA to enhance sputum clearance. We hypothesized that DNA cleavage would also inhibit depletion aggregation as low molecular weight (LMW) polymers produce smaller entropy gains when particles aggregate and generate lower aggregating forces (35). The addition of DNase I abolished the aggregating effects of DNA solutions and markedly reduced aggregation by DNA/mucin mixtures (SI Appendix, Fig. S1 H and I).
Depletion Aggregation Does Not Require Biofilm Functions.
While our findings that host-derived polymers promote the formation of laterally aligned bacterial aggregates and negatively sloped phase transitions implicate the depletion mechanism, it is possible that bacterial activity contributes somehow to aggregate formation or maintenance. To test this, we mixed polymers with biofilm-deficient strains of P. aeruginosa, including strains lacking flagellar motility (ΔfliC) and exopolysaccharide production (ΔpelA/pslBCD/algD) (SI Appendix, Fig. S3) (10, 12). Host polymers were as effective or more effective at producing depletion aggregation in biofilm-deficient strains as in isogenic wild-type bacteria (SI Appendix, Fig. S1 E and F). Similar results were obtained using formalin- and UV-killed bacteria (Fig. 2F and SI Appendix, Fig. S1J), indicating that aggregation caused by the depletion mechanism can be completely driven by host polymers and does not require bacterial activity.
Depletion Aggregation Promotes Antibiotic Tolerance.
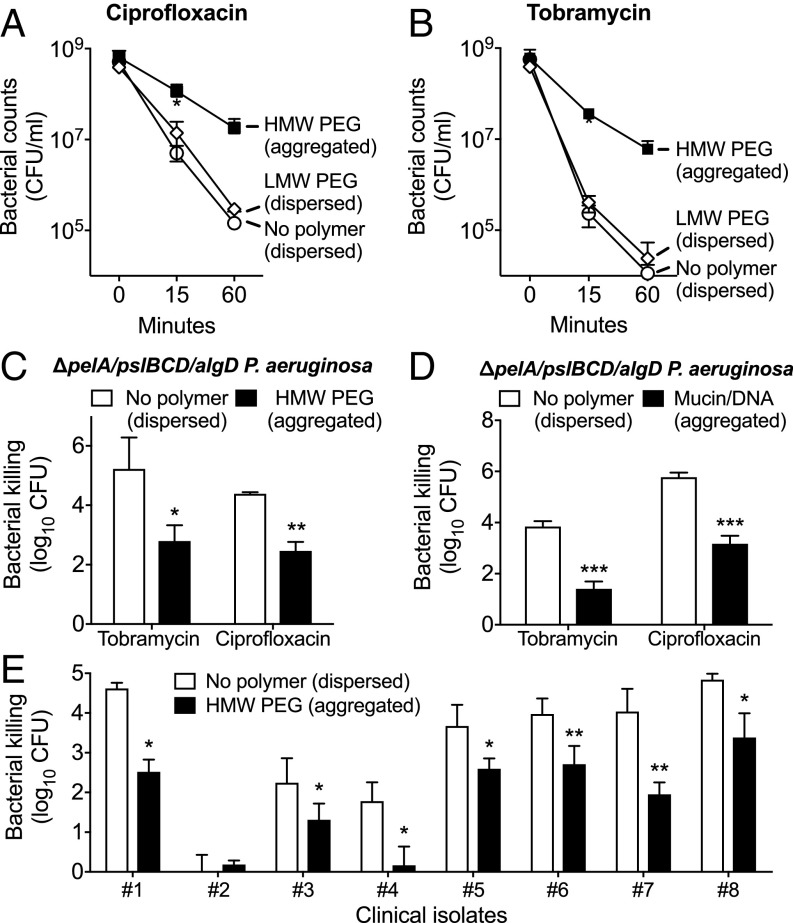
Laboratory biofilms exhibit marked resistance to antibiotic killing, and this phenotype has been attributed in part to tolerance mechanisms induced by biofilm formation. To determine if depletion aggregation produced antibiotic tolerance, we exposed P. aeruginosa to two conditions that induced depletion aggregation: high molecular weight (HMW) PEG [30% (wt/vol); 10 kDa] and a disease-relevant mixture of mucin (8% wt/vol) and DNA (4 mg/mL). Fifteen minutes later, we measured killing by ciprofloxacin and tobramycin. Relative to dispersed bacteria in polymer-free conditions, killing of polymer-induced aggregates was reduced by 10- to 100-fold (Fig. 3 A and B and SI Appendix, Fig. S4 A and B). Similar results were obtained using the biofilm-deficient strain ΔpelA/pslBCD/algD (Fig. 3 C and D).
Fig. 3.
Depletion aggregation promotes antibiotic tolerance in P. aeruginosa. (A and B) P. aeruginosa grown in conditions that produced dispersed bacteria [open symbols, no polymer or 30% (wt/vol) LMW PEG] or bacterial aggregates [filled squares, 30% (wt/vol) HMW PEG] were exposed to ciprofloxacin (A) or tobramycin (B) for 1 h, and viable bacteria were enumerated. Results are the mean ± SD of three experiments; *P < 0.05 relative to LMW PEG (dispersed) cultures. (C and D) HMW PEG (C) or a mixture of mucin (4% wt/vol) and DNA (2 mg/mL) (D) was used to induce depletion aggregation of biofilm-deficient ΔpelA/pslBCD/algD P. aeruginosa, and cultures were tested as above. Killing is represented as the log10 reduction of viable cells recovered after antibiotic treatment compared with untreated controls. Results are the mean ± SD of three experiments; *P < 0.05 relative to no polymer control. (E) Dispersed (no polymer) and aggregated (HMW PEG) cultures of eight P. aeruginosa CF clinical isolates were treated with 0.5 µg/mL ciprofloxacin for 1 h. Killing is represented as the log10 reduction of viable cells recovered after antibiotic treatment compared with untreated controls. In one case (#2) the clinical isolate was inherently resistant to ciprofloxacin. Results are the mean ± SD of at least three experiments; *P < 0.05, **P < 0.01 relative to no polymer control.
We also studied depletion-induced antibiotic tolerance in P. aeruginosa clinical isolates from people with CF that were either capable or deficient in biofilm formation (16) (SI Appendix, Fig. S3). Depletion aggregation promoted tolerance to ciprofloxacin in all clinical isolates tested, regardless of their ability to form in vitro biofilms (Fig. 3E). Collectively, these results indicate that biofilm assembly functions are not required for the antibiotic tolerance of P. aeruginosa aggregates formed by the depletion mechanism.
One explanation for the tolerance observed in the experiments above was that polymers somehow inactivated the antibiotics. We investigated this with three control experiments. First, we exploited the fact that it is possible to make solutions with identical weight per volume polymer concentrations that do (in the case of HMW polymers) and do not (in the case of LMW polymers) produce aggregation. This is possible because aggregation induced by short and LMW polymers produces smaller net entropy gains than those induced by identical concentrations of HMW polymers, so aggregating forces are smaller (35). For example, 30% (wt/vol) LMW PEG (2 kDa) did not cause P. aeruginosa aggregation until bacterial concentrations exceeded >5 × 1010 cfu/mL (SI Appendix, Fig. S1G), whereas 30% (wt/vol) HMW PEG (10 kDa) caused aggregation of 5 × 107 cfu/mL P. aeruginosa (Fig. 2A). Using these conditions, we found that 30% (wt/vol) HMW PEG produced aggregation and antibiotic tolerance (of 108 cfu/mL bacteria), whereas an identical (wt/vol) concentration of LMW PEG did not (Fig. 3 A and B).
Second, we changed the timing of drug addition, as an antibiotic inactivation mechanism should be enhanced (or at least maintained) if polymers and antibiotics are mixed together before the bacteria are added. However, antibiotic activity against dispersed bacteria was not diminished by premixing drugs with aggregate-inducing concentrations of mucin and DNA or HMW PEG (SI Appendix, Fig. S4 C and D). These experiments indicate that aggregates must form before antibiotic exposure to develop tolerance, as bacteria remain susceptible if exposed to antibiotics and polymers simultaneously. Third, we reduced bacterial density so that aggregation did not occur in the presence of HMW PEG (Fig. 2A). Nonaggregated bacteria in HMW PEG were equally susceptible to antibiotic killing as dispersed cells (SI Appendix, Fig. S4E). Together, these findings indicate that antibiotic inactivation is not responsible for polymer-induced antibiotic tolerance.
SOS Mediates Polymer-Induced Antibiotic Tolerance.
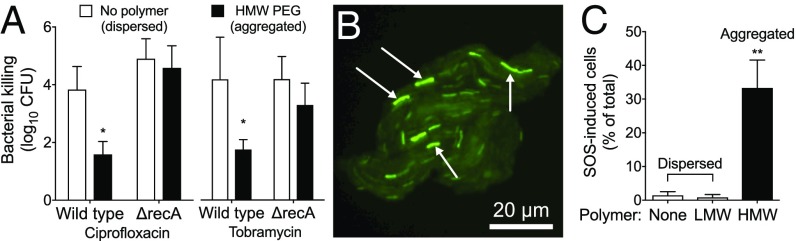
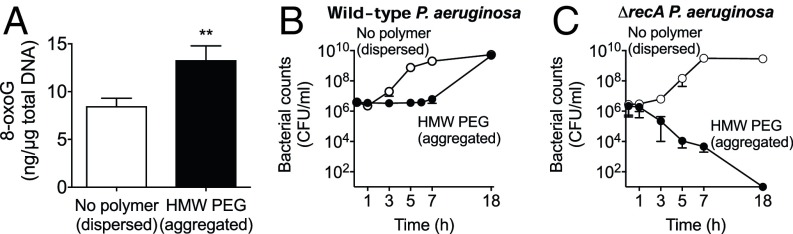
Our finding that polymer-induced aggregation produced tolerance to antibiotics that work by different mechanisms raised the possibility that a general stress response could be involved. Induction of the SOS response produces tolerance to several antibiotics (37, 38) and is initiated when the RecA protein binds to damaged DNA. This causes autoproteolytic cleavage of SOS transcriptional repressors and transcription of genes that mediate stress survival. We disabled the SOS response by inactivating the recA gene and found that while polymers aggregated ΔrecA P. aeruginosa in a manner indistinguishable from wild type, aggregated ΔrecA P. aeruginosa developed significantly lower tolerance to ciprofloxacin and tobramycin (Fig. 4A).
Fig. 4.
The SOS response is required for depletion aggregation-mediated tolerance. (A) Antibiotic tolerance of wild-type and ΔrecA P. aeruginosa was measured in dispersed (no polymer) or aggregated (HMW PEG) conditions. Bacteria were treated with the indicated antibiotic for 1 h. Killing is represented as the log10 reduction of viable cells treated with antibiotics compared with untreated controls. Results are the mean ± SD of three experiments; *P < 0.05 relative to no polymer control. (B) Representative image showing the fluorescent SOS reporter (recAp::gfp) 1 h after depletion aggregation was induced by HMW PEG. (C) Proportion of SOS-induced bacteria was measured by enumerating total and fluorescent cells. Results are the mean ± SD of three experiments with >300 cells per experiment; **P < 0.01.
The involvement of the SOS response and our finding that tolerance did not occur unless polymer-induced aggregation preceded antibiotic addition (SI Appendix, Fig. S4 C and D) led us to hypothesize that polymer exposure induced the SOS response in the absence of antibiotics. To test this, we aggregated P. aeruginosa carrying a recA transcriptional reporter (recAP::GFP) by a 1-h exposure to HMW PEG and counted the number of cells reporting SOS activation. The recA reporter was activated in 33% of cells that had been aggregated but in only 2% of cells that were not aggregated (i.e., bacteria exposed to polymer-free or LMW PEG solutions) (Fig. 4 B and C).
To determine if preinduction of the SOS response by a different mechanism could produce antibiotic tolerance, we exposed P. aeruginosa to a sublethal dose of the DNA-damaging agent mitomycin C. As shown in SI Appendix, Fig. S5A, mitomycin C activated the fluorescent recA transcriptional reporter in wild-type P. aeruginosa, indicating SOS activation. Pretreatment with mitomycin C also induced ciprofloxacin and tobramycin tolerance (SI Appendix, Fig. S5 B and C), consistent with previous observations in E. coli (39). However, pretreatment with mitomycin C did not promote tolerance in ΔrecA P. aeruginosa (SI Appendix, Fig. S5 D and E). Taken together, these findings suggest a mechanism whereby polymer exposure induces the SOS response and SOS-induced bacteria are protected from antibiotic-mediated killing.
Depletion Aggregation Induces Oxidative DNA Damage.
We investigated the possibility that aggregation-inducing polymers might activate SOS by producing oxidative DNA damage, as bacterial aggregation occurring via biofilm formation is known to produce endogenous oxidative stress (40). We measured the abundance of the oxidative DNA lesion 8-oxoguanine (8-oxoG) and found that P. aeruginosa that had been aggregated by HMW PEG for 1 h contained higher levels of 8-oxoG than bacteria that had not been aggregated (Fig. 5A). In addition, inactivation of recA, which is required for repair of some oxidative lesions, markedly reduced the survival of P. aeruginosa subjected to prolonged polymer aggregation (Fig. 5B). These findings suggest that depletion aggregation-inducing polymers can activate the SOS response, that SOS activation may occur as a consequence of oxidative DNA damage, and that SOS activation is required for bacterial survival in aggregates.
Fig. 5.
Depletion aggregation induces oxidative DNA damage. (A) 8-oxoG DNA lesions were measured by ELISA before and 1 h after exposure to conditions that produced dispersed bacteria (no polymer) or bacterial aggregates (HMW PEG). Results are the mean ± SD of three experiments; *P < 0.05 relative to no polymer. (B and C) Growth curves of wild-type (B) and ΔrecA (C) P. aeruginosa in dispersed (no polymer) or aggregated (HMW PEG) conditions. Results are the mean ± SD of duplicate experiments.
The LexA Regulon Mediates Antibiotic Tolerance.
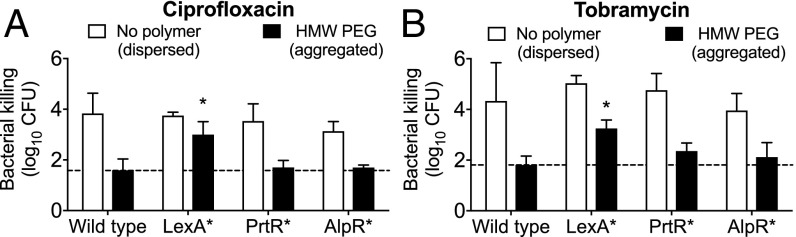
In P. aeruginosa, the binding of RecA to damaged DNA causes the autoproteolysis and inactivation of three SOS regulon repressors—LexA, PrtR, and AlpR—and each controls a regulon with distinct characterized functions. The LexA regulon contains DNA-repair genes, the PrtR regulon contains antimicrobial R2 pyocin genes (41), and the AlpR regulon has been reported to contain genes mediating a bacterial programmed cell death mechanism (42). We used P. aeruginosa harboring point mutations that produced proteolytic resistance in each SOS transcriptional repressor [including LexAG86V, PrtRS162A, and AlpRS153A (41)] to determine which SOS regulon(s) mediated polymer-induced tolerance. In the absence of polymers, the antibiotic sensitivity of all three SOS repressor mutants was similar to wild type (Fig. 6 A and B), as previously reported (38). Furthermore, HMW PEG-mediated depletion aggregation of the PrtRS162A and AlpRS153A mutants induced a level of antibiotic tolerance similar to that in wild-type bacteria. However, depletion-induced aggregates formed by the LexAG86V mutant exhibited 10- to 100-fold less tolerance than wild type, implicating genes in the LexA regulon in polymer-induced antibiotic tolerance.
Fig. 6.
The LexA regulon mediates depletion-mediated antibiotic tolerance. (A and B) Wild-type P. aeruginosa and the indicated SOS repressor autoproteolytic-resistant mutants were exposed to conditions that produced dispersed bacteria (no polymer) or bacterial aggregates (HMW PEG) for 1 h and then were treated with ciprofloxacin (A) or tobramycin (B) for an additional hour. Killing is represented as the log10 reduction of viable cells treated with antibiotics compared with untreated controls. Results are the mean ± SD of three experiments; *P < 0.05 compared with aggregated wild-type P. aeruginosa.
Aggregation Induces SulA-Dependent Growth Arrest.
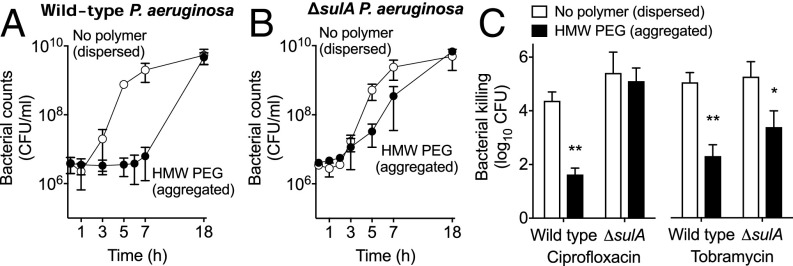
The LexA regulon includes SulA, a rapid and potent inhibitor of cell division. The speed at which depletion-mediated antibiotic tolerance develops (15 min) (Fig. 3 A and B) and the known association between growth and antibiotic susceptibility led us to hypothesize that sulA-dependent growth arrest contributed to depletion-mediated tolerance. To test this, we measured the growth rates and antibiotic susceptibility of wild-type and ΔsulA P. aeruginosa after aggregation by HMW PEG. As shown in Fig. 7 A and B, HMW PEG markedly reduced the growth of wild-type but not ΔsulA P. aeruginosa. Furthermore, inactivation of sulA eliminated polymer-induced tolerance to ciprofloxacin but not to tobramycin (Fig. 7C). These findings indicate that depletion aggregation causes growth arrest mediated by the SOS response and sulA and that this arrest is critical to recovery from ciprofloxacin toxicity. One mechanism that could explain the ciprofloxacin sensitivity of the sulA mutant is that inhibited cell division maintains multiple chromosomes within cells, which could enable recombinational repair of ciprofloxacin-induced DNA damage (43).
Fig. 7.
Depletion aggregation promotes sulA-mediated growth arrest. (A and B) Growth of wild-type (A) and ΔsulA (B) P. aeruginosa was measured in dispersed (no polymer, open symbols) or aggregated (HMW PEG, filled symbols) conditions. Results are the mean ± SD of triplicate experiments. (C) After 1 h in the dispersed or aggregated state, antibiotic susceptibility was measured. Killing is represented as the log10 reduction of viable cells treated with antibiotics compared with untreated controls. Results are the mean ± SD of three experiments; *P < 0.05, **P < 0.01 relative to growth medium.
Discussion
The persistence and treatment resistance of bacteria causing chronic infections are thought to be consequences, in large part, of phenotypes induced by conditions at infection sites. A leading model postulates that in vivo environmental cues activate biofilm-formation functions that cause bacteria to self-assemble into complex multicellular biofilm communities (44). Our findings raise an alternative possibility, as we found that host polymers abundant at chronic infection sites cause P. aeruginosa to aggregate by the depletion mechanism, which does not require biofilm assembly functions or even bacterial viability. Importantly, depletion aggregation markedly and rapidly increases the antibiotic tolerance of bacteria via mechanisms mediated by the LexA SOS regulon.
Because it is a consequence of physical forces, depletion aggregation will operate if the concentrations of bacteria and polymers are sufficient. We tested concentrations of DNA, mucin, F-actin, and bacteria found in CF airways with established infections and found these sufficient to produce aggregation ex vivo. If depletion aggregation maintained bacteria in established infections in an aggregated state, biofilm formation functions could become superfluous. This could explain why biofilm deficiency frequently evolves in CF and wound isolates.
Our finding that physiologically relevant polymer mixtures can also aggregate low concentrations of bacteria raises the possibility that depletion aggregation could operate in nascent infections. However, this is more speculative because in vivo concentrations of bacteria and polymers are difficult to measure early in infection. One scenario is that replication of organisms that initiate infection produces sufficiently high local bacterial densities in areas that contain sufficient host polymer concentrations. If this occurred, depletion aggregation could reduce the susceptibility of bacteria to killing, enabling the organisms to gain a foothold. However, it is also possible that the polymer and bacterial concentrations needed for depletion aggregation are not present until disease is more advanced.
Our finding that antibiotic tolerance caused by polymer exposure is a consequence of the SOS response, which may be induced by oxidative DNA damage, is reminiscent of other work linking in vivo stress conditions to infection pathogenesis. For example, starvation stress responses have been shown to protect E. coli and P. aeruginosa from antibiotic killing (45). Acid stress responses have been shown to have similar effects for Salmonella (46) and Listeria species (47). Thus, some host responses may induce physiological changes in bacteria that produce cross-protection against antibiotic killing and thereby enhance rather than counter infection persistence. It would be interesting to investigate the normal (nonpathogenic) microbiota to determine if polymers present at healthy mucosal surfaces such as the mouth, intestine, and vagina cause depletion aggregation, induce bacterial stress responses, and increase antimicrobial tolerance.
Our study had several limitations. First, we used commercially available host polymers that are not identical to polymers present at chronic infection sites. For example, the porcine gastric mucin and salmon sperm DNA we used might aggregate bacteria differently from the analogous human polymers. However, we think this unlikely, as the depletion mechanism does not require any specific biological activity. Instead, it is driven by the space-occupying characteristics of the polymers and their concentrations. Second, we used laboratory strains of P. aeruginosa, a small number of clinical isolates (n = 8), and bacterial cells with identical (rod) shapes. It is possible that other clinical isolates or bacterial species could produce different results. Furthermore, as cell shape is known to affect the spatial organization of cells in a biofilm (48), it would be interesting to study how the depletion mechanism affects interactions between bacterial species with different shapes.
Third, we do not yet know how depletion aggregation induces oxidative stress in P. aeruginosa. One possibility is that osmotic pressure generated by the polymers (Fig. 1) induces bacterial envelope stress. Envelope stress may generate endogenous oxidants by causing the release of iron from membrane proteins, thus promoting Fenton reactions (49). Another possibility involves oxygen limitation, which readily occurs within aggregates (50). To maintain redox balance under oxygen-limiting conditions, P. aeruginosa produces phenazines as alternate electron acceptors, and nonspecific electron transfer from phenazines to intracellular iron (or other molecules) could result in oxidative stress (51). Last, it is possible that depletion aggregation reduces the expression of antioxidant defense mechanisms, as observed in P. aeruginosa biofilms (52). Our observation that depletion aggregation increases 8-oxoG DNA lesions is consistent with all these ideas, as these lesions are primarily caused by hydroxyl radicals, which can be produced by a variety of endogenous oxidative stresses (49).
Finally, future work will be needed to determine if the depletion mechanism causes bacterial aggregation in chronic animal or human infections. If it does, current ideas about how to target bacteria causing chronic infection may need to be reconsidered. If host conditions cause bacterial aggregation as a default growth mode at chronic infection sites, targeting biofilm assembly functions might not be therapeutically useful. Instead, it may be more fruitful to devise strategies to block the physiologic responses that cause depletion aggregation-mediated antibiotic tolerance or to alter the host environment in a manner that decreases aggregating forces generated by host polymers.
Methods
Detailed methods are found in SI Appendix. Unless indicated, bacteria were grown in lysogeny broth (LB). Phase diagrams were constructed as described (34). Bacterial viability was measured by enumerating cfus after serial dilution. Levels of 8-oxoG were quantified using the OxiSelect Oxidative DNA Damage ELISA kit (Cell Biolabs) following the manufacturer’s instructions.
Supplementary Material
Acknowledgments
P.R.S. was supported by a Cystic Fibrosis Foundation Postdoctoral Fellowship, NIH Grants K22AI125282 and P20GM103546, and Cystic Fibrosis Foundation Grant SINGH15R0. P.K.S. was supported by NIH Grants R01AI101307 and K24HL102246, Defense Threat Reduction Agency Grant HDTRA1-14-1-0018, the Cystic Fibrosis Foundation, and the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806005115/-/DCSupplemental.
References
- 1.Tramper-Stranders GA, et al. Initial Pseudomonas aeruginosa infection in patients with cystic fibrosis: Characteristics of eradicated and persistent isolates. Clin Microbiol Infect. 2012;18:567–574. doi: 10.1111/j.1469-0691.2011.03627.x. [DOI] [PubMed] [Google Scholar]
- 2.Worlitzsch D, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorth P, et al. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe. 2015;18:307–319. doi: 10.1016/j.chom.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prince AS. Biofilms, antimicrobial resistance, and airway infection. N Engl J Med. 2002;347:1110–1111. doi: 10.1056/NEJMcibr021776. [DOI] [PubMed] [Google Scholar]
- 5.Thomassen MJ, Demko CA, Boxerbaum B, Stern RC, Kuchenbrod PJ. Multiple of isolates of Pseudomonas aeruginosa with differing antimicrobial susceptibility patterns from patients with cystic fibrosis. J Infect Dis. 1979;140:873–880. doi: 10.1093/infdis/140.6.873. [DOI] [PubMed] [Google Scholar]
- 6.Staudinger BJ, et al. Conditions associated with the cystic fibrosis defect promote chronic Pseudomonas aeruginosa infection. Am J Respir Crit Care Med. 2014;189:812–824. doi: 10.1164/rccm.201312-2142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjarnsholt T, et al. The in vivo biofilm. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Singh PK, et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 9.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 10.Colvin KM, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2012;14:1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilder CN, Allada G, Schuster M. Instantaneous within-patient diversity of Pseudomonas aeruginosa quorum-sensing populations from cystic fibrosis lung infections. Infect Immun. 2009;77:5631–5639. doi: 10.1128/IAI.00755-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 13.Sharma G, et al. Pseudomonas aeruginosa biofilm: Potential therapeutic targets. Biologicals. 2014;42:1–7. doi: 10.1016/j.biologicals.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Moskowitz SM, Foster JM, Emerson J, Burns JL. Clinically feasible biofilm susceptibility assay for isolates of Pseudomonas aeruginosa from patients with cystic fibrosis. J Clin Microbiol. 2004;42:1915–1922. doi: 10.1128/JCM.42.5.1915-1922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee B, et al. Heterogeneity of biofilms formed by nonmucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. J Clin Microbiol. 2005;43:5247–5255. doi: 10.1128/JCM.43.10.5247-5255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cassat JE, et al. Comparative genomics of Staphylococcus aureus musculoskeletal isolates. J Bacteriol. 2005;187:576–592. doi: 10.1128/JB.187.2.576-592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pompilio A, et al. Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: Genome diversity, biofilm formation, and virulence. BMC Microbiol. 2011;11:159. doi: 10.1186/1471-2180-11-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deligianni E, et al. Pseudomonas aeruginosa cystic fibrosis isolates of similar RAPD genotype exhibit diversity in biofilm forming ability in vitro. BMC Microbiol. 2010;10:38. doi: 10.1186/1471-2180-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Head NE, Yu H. Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: Biofilm formation, virulence, and genome diversity. Infect Immun. 2004;72:133–144. doi: 10.1128/IAI.72.1.133-144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Hal SJ, et al. In vivo evolution of antimicrobial resistance in a series of Staphylococcus aureus patient isolates: The entire picture or a cautionary tale? J Antimicrob Chemother. 2014;69:363–367. doi: 10.1093/jac/dkt354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock RE, et al. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: A class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect Immun. 1983;42:170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaber JA, et al. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J Med Microbiol. 2004;53:841–853. doi: 10.1099/jmm.0.45617-0. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez CJ, Jr, et al. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect Dis. 2013;13:47. doi: 10.1186/1471-2334-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donlan RM. Biofilms: Microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjarnsholt T, et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol. 2009;44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- 28.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol. 2016;14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark RAF, Henson PM. 1988. The Molecular and Cellular Biology of Wound Repair (Plenum, New York), p xxii, 597 p.
- 30.Fahy JV, Dickey BF. Airway mucus function and dysfunction. N Engl J Med. 2010;363:2233–2247. doi: 10.1056/NEJMra0910061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandt T, Breitenstein S, von der Hardt H, Tümmler B. DNA concentration and length in sputum of patients with cystic fibrosis during inhalation with recombinant human DNase. Thorax. 1995;50:880–882. doi: 10.1136/thx.50.8.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasconcellos CA, et al. Reduction in viscosity of cystic fibrosis sputum in vitro by gelsolin. Science. 1994;263:969–971. doi: 10.1126/science.8310295. [DOI] [PubMed] [Google Scholar]
- 33.Strand SP, Varum KM, Ostgaard K. Interactions between chitosans and bacterial suspensions: Adsorption and flocculation. Colloid Surface B. 2003;27:71–81. [Google Scholar]
- 34.Schwarz-Linek J, et al. Polymer-induced phase separation in suspensions of bacteria. Europhys Lett. 2010;89:68003. [Google Scholar]
- 35.Schwarz-Linek J, et al. Polymer-induced phase separation in Escherichia coli suspensions. Soft Matter. 2010;6:4540–4549. [Google Scholar]
- 36.Poon WCK. The physics of a model colloid-polymer mixture. J Phys Condens Matter. 2002;14:R859–R880. [Google Scholar]
- 37.Miller C, et al. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2004;305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- 38.Cirz RT, O’Neill BM, Hammond JA, Head SR, Romesberg FE. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol. 2006;188:7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dörr T, Lewis K, Vulić M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 2009;5:e1000760. doi: 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boles BR, Singh PK. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci USA. 2008;105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penterman J, Singh PK, Walker GC. Biological cost of pyocin production during the SOS response in Pseudomonas aeruginosa. J Bacteriol. 2014;196:3351–3359. doi: 10.1128/JB.01889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McFarland KA, et al. A self-lysis pathway that enhances the virulence of a pathogenic bacterium. Proc Natl Acad Sci USA. 2015;112:8433–8438. doi: 10.1073/pnas.1506299112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bos J, et al. Emergence of antibiotic resistance from multinucleated bacterial filaments. Proc Natl Acad Sci USA. 2015;112:178–183. doi: 10.1073/pnas.1420702111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen D, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leyer GJ, Johnson EA. Acid adaptation induces cross-protection against environmental stresses in Salmonella typhimurium. Appl Environ Microbiol. 1993;59:1842–1847. doi: 10.1128/aem.59.6.1842-1847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gandhi M, Chikindas ML. Listeria: A foodborne pathogen that knows how to survive. Int J Food Microbiol. 2007;113:1–15. doi: 10.1016/j.ijfoodmicro.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 48.Smith WP, et al. Cell morphology drives spatial patterning in microbial communities. Proc Natl Acad Sci USA. 2017;114:E280–E286. doi: 10.1073/pnas.1613007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 50.Wessel AK, et al. Oxygen limitation within a bacterial aggregate. MBio. 2014;5:e00992. doi: 10.1128/mBio.00992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price-Whelan A, Dietrich LE, Newman DK. Rethinking ‘secondary’ metabolism: Physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 52.Hassett DJ, et al. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 53.Dorken G, Ferguson GP, French CE, Poon WCK. Aggregation by depletion attraction in cultures of bacteria producing exopolysaccharide. J R Soc Interface. 2012;9:3490–3502. doi: 10.1098/rsif.2012.0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marenduzzo D, Finan K, Cook PR. The depletion attraction: An underappreciated force driving cellular organization. J Cell Biol. 2006;175:681–686. doi: 10.1083/jcb.200609066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.