Significance
Defensins are antimicrobial peptides exhibiting antibacterial, antifungal, and antiviral activity. They are expressed by epithelial cells at the intestinal mucosal surface where they play a crucial role in the host intestinal homeostasis. Therefore, approaches aiming to boost their expression represent a promising therapeutic strategy to treat infections and dysbiosis-driven diseases in humans at a time of increasing incidence of antibiotic resistance.
Keywords: epithelial cell, β-defensin, gene regulation, natural inducer, synergistic effect
Abstract
Antimicrobial peptides (AMPs) are mucosal defense effectors of the human innate immune response. In the intestine, AMPs are produced and secreted by epithelial cells to protect the host against pathogens and to support homeostasis with commensals. The inducible nature of AMPs suggests that potent inducers could be used to increase their endogenous expression for the prevention or treatment of diseases. Here we aimed at identifying molecules from the natural pharmacopoeia that induce expression of human β-defensin-3 (HBD3), one of the most efficient AMPs, without modifying the production of proinflammatory cytokines. By screening, we identified three molecules isolated from medicinal plants, andrographolide, oridonin, and isoliquiritigenin, which induced HBD3 production in human colonic epithelial cells. This effect was observed without activation of the NF-κB pathway or the expression of associated proinflammatory cytokines. We identified the EGF receptor as the target of these compounds and characterized the downstream-activated MAPK pathways. At the chromatin level, molecules increased phosphorylation of histone H3 on serine S10 and recruitment of the c-Fos, c-Jun, and Elk1 or c-Myc transcription factors at the HBD3 promoter. Interestingly, stimulating cells with a combination of andrographolide and isoliquiritigenin synergistically enhanced HBD3 induction 10-fold more than observed with each molecule alone. Finally, we investigated the molecular basis governing the synergistic effect, confirmed our findings in human colonic primary cells, and demonstrated that synergism increased cellular antimicrobial activity. This work shows the capability of small molecules to achieve induction of epithelial antimicrobial defenses while simultaneously avoiding the deleterious risks of an inflammatory response.
Antimicrobial defenses from the human intestinal tract rely on the ability of the mucosal immune system to recognize and neutralize pathogenic microbes. In this context of innate immunity, sensing of bacteria occurs mainly through the engagement of host cell Toll-like receptors (TLRs), nucleotide oligomerization domain (NOD) proteins, and Nod-like receptors (NLRs) by pathogen-associated molecular patterns (PAMPs) (1–3). After recognition of the pathogen, the antimicrobial response is achieved by immediate secretion of epithelial antimicrobial peptides (AMPs), such as defensins and lectins, and subsequent recruitment of immune cells (4–6).
In recent decades, several defensins able to rapidly kill microorganisms have been identified. Although varying in their structure and length, these peptides show common features. Defensins are mainly cationic and amphipathic, synthetized as propeptides and released in a mature form following hydrolysis by specific proteases. Defensins, like cytokines, are considered functional effectors of the innate immune system, also liaising with cells of the adaptive immune system, including monocytes, T cells, or immature dentritic cells, through their direct interactions with receptors such as CCR2, CCR6, and FPLR1 (7, 8).
In humans, defensins include two families: α- and β-defensins. Human α-defensins are stored in granules of polymorphonuclear leukocytes (HNP 1–4) or Paneth cells (HD-5 and HD-6) localized at the bottom of intestinal crypts. In contrast, β-defensins are produced by epithelial cells along the entirety of the intestinal tract, and their synthesis is regulated at the gene-expression level. Expression of the best-known β-defensins, HBD1–4, by epithelial cells, is either constitutive or inducible in response to proinflammatory stimuli. Expression of the HBD1 gene is essentially constitutive, whereas expression of HBD2–4 genes is inducible in response to various stimuli, including bacteria, PAMPs, and proinflammatory cytokines (5).
β-Defensins have a broad spectrum of antimicrobial efficacy and act on Gram-positive and Gram-negative bacteria as well as on fungi and enveloped viruses (9). They exert their action by interacting with the surface of microorganisms. In Gram-negative bacteria they bind the anionic portion of the LPS molecule; in Gram-positive bacteria they interact with teichoic acids or with anionic groups present in the peptidoglycan molecule. The interaction of β-defensins with the different structures of the bacterial cell envelope permeabilizes the membrane either through a detergent effect resulting in the leakage of cytoplasmic components or through the formation of pores subsequent to peptide aggregation (9).
The growing threat of antibiotic resistance in pathogenic bacteria and the need for new antibiotics stimulate interest in the use of β-defensins as candidate therapeutic molecules due to their significant antimicrobial activities and their additional protective properties, such as in wound healing, modulation of the immune response, angiogenesis, tissue remodeling, and ability to bind LPS in septic shock models (10). Moreover, diseases associated with defects in the expression of β-defensins continue to be identified by clinical studies (11). However, the limited knowledge accumulated about the circuits involved in the regulation of AMP expression has hampered efforts to develop innovative therapies aimed at stimulating their expression.
The idea of using natural AMPs as antibiotics has been explored in recent years and continues to be investigated. However, current AMPs-based antiinfectious strategies show that local or systemic administration of modified or synthetic AMPs provides mixed results that are largely due to the intrinsic biochemical and complex bioavailability characteristics of these peptides (12, 13). Several factors, including cost, toxicity, and susceptibility to several parameters (salt and proteases concentrations, oxido-reduction status, spontaneous aggregation, and difficulty in mimicking the optimal local condition of action) have limited their use as therapeutic agents (12, 14). In addition is the need for AMPs to be expressed and secreted at high levels, along with other antimicrobial molecules and possibly cells, in relevant sites. The possibility of boosting transcription of the host endogenous AMPs for greater antimicrobial protection and immuno-modulation thus appeared to us to be an alternative option. The inducible nature of AMPs such as β-defensins suggested that the use of biological screens was a promising option for identifying original pharmacological inducers stimulating endogenous AMPs for the prevention and treatment of infectious diseases or dysbiosis-related pathological conditions.
Insufficient investments in the development of novel compounds with original scaffolds and targets largely account for the poor pipeline of new antimicrobial molecules available to date, and alternative discovery strategies are urgently warranted. The environment has proven to be a rich source of diverse natural products with significant antibacterial, antiviral, antifungal, antiparasitic, antiinflammatory, antitumor, antioxidant, and immuno-modulatory activities (15). Many natural products, such as neoechinulin B, have been found to be promising drug candidates to alleviate the mortality and morbidity rates caused by drug-resistant infections, and several natural antiinfectious molecules have already entered phase 1, 2, and 3 clinical trials (16–18).
Following unsuccessful screening of an extended spectrum of molecules contained in edited and nonedited libraries from the pharmaceutical industry, we decided to turn our screening approach to molecules from the natural pharmacopoeia, particularly from plants used in Chinese traditional medicine (19, 20). We thereby successfully identified three molecules that selectively stimulated the inducible expression of the human β-defensin-3 (HBD3) in a dose-dependent manner at the gene and protein levels in the TC7 colonic epithelial cell line. These compounds are andrographolide, a labdane diterpenoid and the main bioactive component of Andrographis paniculata (21); oridonin, a diterpenoid purified from Rabdosia rubescens (22); and isoliquiritigenin, a flavonoid present in the root of Glycyrrhiza glabra (23). Induction of HBD3 was observed without activation of the NF-κB signaling pathway or expression of associated proinflammatory cytokines such as IL-1B, IL-8, and TNF. Using specific inhibitors, we identified the EGF receptor (EGFR) as a target of these compounds and showed that these molecules signal through different MAPK-dependent pathways. Whereas andrographolide triggered Erk and SAPK/JNK phosphorylation, oridonin and isoliquiritigenin induced Erk and p38 phosphorylation. At the chromatin level, we demonstrated that the three molecules increased phosphorylation of histone H3 on serine S10 (H3S10), leading to recruitment of the c-Fos, c-Jun, and Elk1 or c-Myc transcription factors at the HBD3 promoter. Interestingly, we found that stimulating cells with a combination of andrographolide and isoliquiritigenin dramatically enhanced HBD3 induction, much more than observed with either compound alone, thus showing a synergistic effect of the two molecules on HBD3 expression. We provided molecular insights supporting the synergistic effect of the molecule combination and confirmed our findings in primary human colonic cells using an ex vivo minigut organoid model. Finally, we showed that andrographolide and isoliquiritigenin synergism increased the antimicrobial activity of cells against pathogenic bacteria. This work opens the way to using molecular inducers of AMPs as an innovative therapeutic strategy to treat infections and dysbiosis-driven diseases at a time of increasing incidence of antibiotic resistance.
Results
Andrographolide, Oridonin, and Isoliquiritigenin Induce Expression of the Human AMP β-Defensin-3.
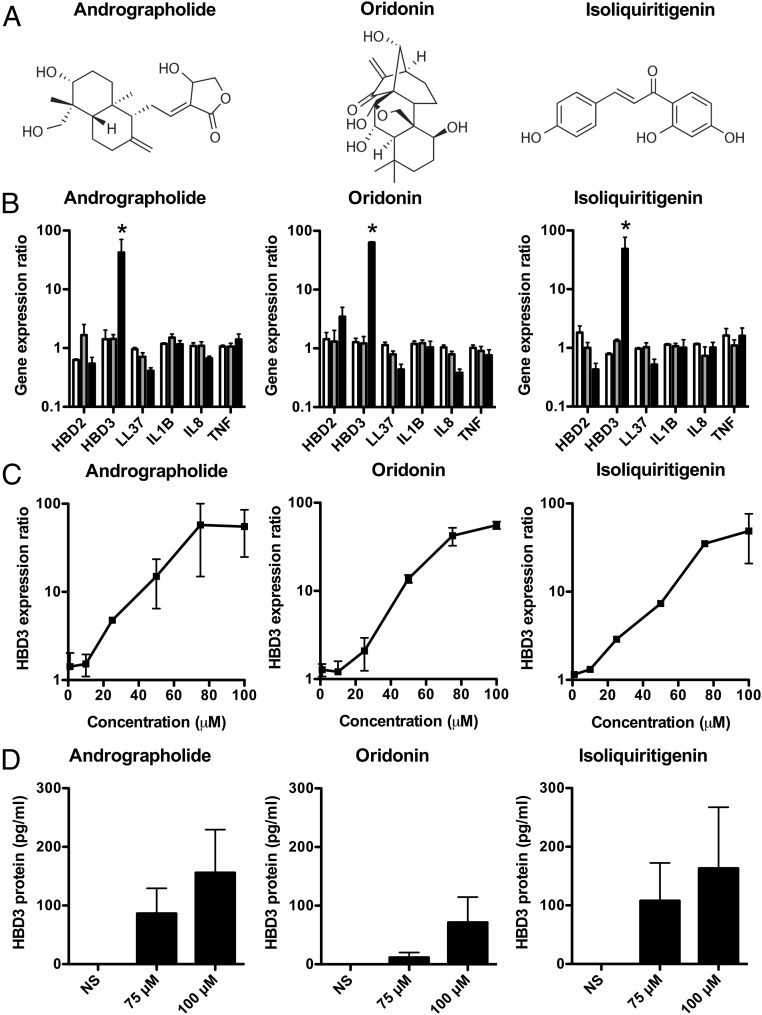
To identify molecules from the pharmacopeia inducing the expression of genes encoding antimicrobial peptides (β-defensins HBD2 and -3 and cathelicidin LL-37), we treated human colonic epithelial cells [Caco-2, subclone TC7 (24)] with a library of 170 natural compounds isolated from plants used in traditional Chinese medicine, which were structurally diverse, bioactive, and cell permeable. Treatments were performed on confluent cell monolayers using three different concentrations (1, 10, and 100 μM). RNA was extracted 24 h after treatment and was analyzed by qRT-PCR. We identified three molecules that induced expression of the human β-defensin HBD3 gene at 100 μM final concentration (Fig. 1 A and B and SI Appendix, Fig. S1A). These molecules were andrographolide, a labdane diterpenoid that is the main bioactive component of the medicinal plant A. paniculata; oridonin, a diterpenoid purified from R. rubescens; and isoliquiritigenin, a flavonoid from the root of G. glabra. Quantitatively, induction of HBD3 transcription was increased by almost 100-fold in cells treated with molecules as compared with nontreated cells. EC50s for HBD3 induction were measured and evaluated at 38, 49, and 42 μM for andrographolide, oridonin, and isoliquiritigenin, respectively (Fig. 1C). In contrast, the expression of other AMPs such as HBD2 and LL-37 was not significantly induced after treatment (Fig. 1B and SI Appendix, Fig. S1A). As a control, cellular viability as assessed by lactate dehydrogenase assay was not modified by molecule treatment (SI Appendix, Fig. S1B).
Fig. 1.
Andrographolide, oridonin, and isoliquiritigenin induce the expression of the human antimicrobial peptide β-defensin-3. (A) Chemical structure of andrographolide, oridonin, and isoliquiritigenin. (B) Transcription of genes encoding HBD2, HBD3, LL-37, IL-1B, IL-8, and TNF in TC7 cells treated for 24 h with increasing concentrations of andrographolide, oridonin, or isoliquiritigenin. Values are presented on a logarithmic scale as the ratio of gene expression in treated cells to that in nontreated cells: white bars, 1 μM; gray bars, 10 μM; black bars, 100 μM. Data are presented as the mean ± SD (n = 6 biological replicates). *P < 0.05 evaluated by two-tailed Mann–Whitney U test. (C) Dose–response analysis of HBD3 gene expression after treatment of cells for 24 h with andrographolide, oridonin, or isoliquiritigenin (0–100 μM). Values are presented on a logarithmic scale as the ratio of gene expression in treated cells to that in nontreated cells. Data are presented as mean ± SD (n = 4 biological replicates). (D) ELISA dosage of HBD3 in supernatants of cells treated for 48 h with increasing concentrations (0–100 μM) of andrographolide, oridonin, or isoliquiritigenin. Values are presented on a linear scale in picograms of peptide per milliliter. NS, nonstimulated cells. Data are presented as mean ± SD (n = 4 biological replicates).
To investigate secretion of the HBD3 peptide, cells were treated for 48 h with increasing concentrations (0–100 μM) of andrographolide, oridonin, or isoliquiritigenin, and ELISAs were performed on culture supernatants. The HBD3 peptide was not detected in supernatants of nontreated cells (Fig. 1D). In contrast, supernatants of treated cells revealed increasing concentrations of HBD3 peptide that depended on the concentration of molecules used for stimulation. For example, concentration of the HBD3 peptide was close to 80 and 160 pg/mL upon treatment with 75 and 100 μM andrographolide, respectively (Fig. 1D). Collectively, these results demonstrate the existence of natural compounds isolated from medicinal plants that are able to specifically induce expression of the human β-defensin HBD3.
We then investigated the expression of several genes encoding bona fide proinflammatory mediators (IL-1B, IL-8, and TNF), a set of genes that is generally considered to respond together with antimicrobial peptide genes in the course of an innate immune response. Confluent cells were treated with 1, 10, or 100 μM andrographolide, oridonin, or isoliquiritigenin. RNA was extracted at 24 h and analyzed by qRT-PCR. Expression of IL-1B, IL-8, and TNF was not induced throughout the time course (Fig. 1B). Whatever the concentration of molecules used, their expression at the transcriptional and translational levels was similar in treated cells and in nontreated cells (Fig. 1B and SI Appendix, Fig. S1C). As a control, the NF-κB inhibitor IκBα was analyzed by immunoblot, using an antibody detecting the IκBα protein (SI Appendix, Fig. S1D). IκBα inhibits NF-κB activation by keeping it sequestered in an inactive state in the cytoplasm, thus preventing expression of its targets, such as proinflammatory genes (25). The amount of IκBα was constant during the time course of the experiment, indicating the absence of NF-κB activation, in agreement with the absence of cytokine induction (Fig. 1B and SI Appendix, Fig. S1 C and D). Together, these data demonstrate the existence of molecules from the pharmacopoeia that are able to disconnect the expression of antimicrobial peptides from the proinflammatory NF-κB signaling pathway.
Andrographolide, Oridonin, and Isoliquiritigenin Activate the EGFR.
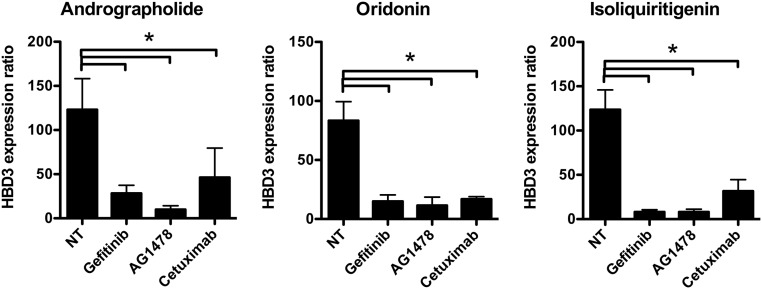
Based on these observations, we sought to determine the molecular mechanisms that led to the induction of HBD3 expression upon treatment with andrographolide, oridonin, and isoliquiritigenin. In a previous study carried out in keratinocytes, expression of HBD3 was induced by ligands for EGFR, such as TGF-α, EGF, amphiregulin, or HB-EGF, and was inhibited by antibodies against the EGFR (26). We therefore investigated the consequences of the treatment of colonic TC7 cells with andrographolide, oridonin, and isoliquiritigenin on EGFR activation and the effect of EGFR blocking by the use of the specific inhibitors gefitinib and AG1478 or the anti-EGFR humanized monoclonal antibody cetuximab. The inhibitors block the EGFR tyrosine kinase activity, whereas cetuximab prevents EGFR dimerization, both resulting in a loss of activation of the downstream signaling pathways. Upon treatment with 10 μM gefitinib or AG1478 or 100 nM cetuximab and 100 μM andrographolide, oridonin, or isoliquiritigenin, induction of HBD3 was decreased at least 75% regardless of the natural compound used to stimulate the cells, compared with nontreated cells (Fig. 2).
Fig. 2.
Andrographolide, oridonin, and isoliquiritigenin activate the EGF receptor. Transcription of the gene encoding HBD3 in cells pretreated with 10 μM gefitinib or AG1478 EGFR inhibitors or with 100 nM of the anti-EGFR humanized monoclonal antibody cetuximab and stimulated for 24 h with 100 μM andrographolide, oridonin, or isoliquiritigenin. Values are presented on a linear scale as the ratio of gene expression in inhibited and treated cells to that in inhibited and nontreated cells. NT, nontreated cells. Data are presented as mean ± SD (n = 4 biological replicates). *P < 0.05 evaluated by two-tailed Mann–Whitney U test.
During transactivation, the EGFR is phosphorylated at multiple tyrosine residues, such as Y1068, a residue which binds directly to the growth factor receptor-binding protein 2 (GRB2) for activation of the downstream signaling cascades (27, 28). We therefore analyzed its phosphorylation status during treatment of cells with 100 μM andrographolide, oridonin, or isoliquiritigenin. By immunoblot, we observed an increased phosphorylation of Y1068 that could not be attributed to increased EGFR protein levels as early as 15 min after treatment of cells with natural compounds, (SI Appendix, Fig. S2 A and B). This posttranslational modification of the EGFR Y1068 residue was highest with andrographolide and isoliquiritigenin and less so with oridonin. Collectively, these data show that EGFR is activated and transduces the signal for HBD3 induction upon treatment with natural molecules.
Andrographolide, Oridonin, and Isoliquiritigenin Signal Through Different MAPK Pathways.
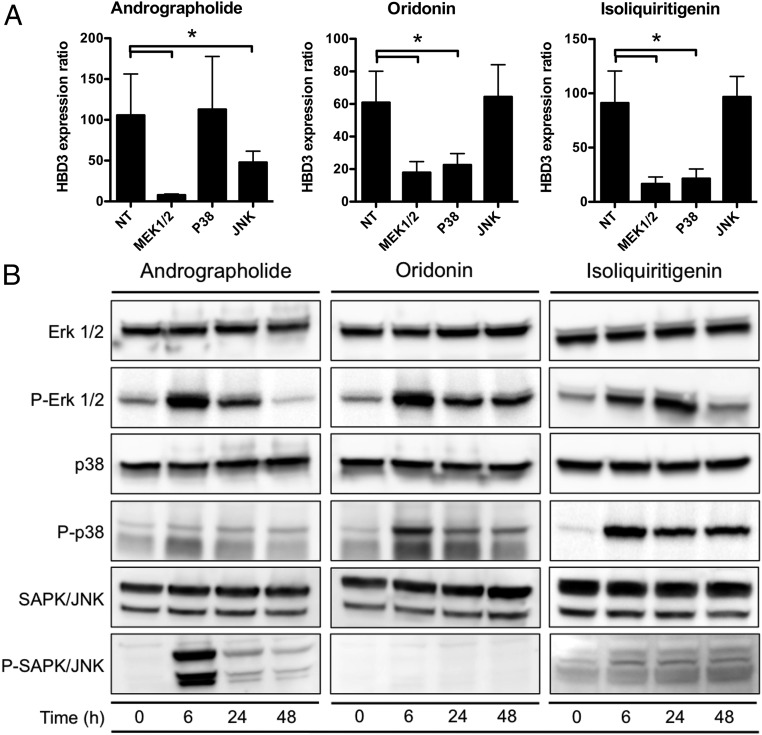
We next investigated the signaling pathways downstream of the EGFR by which andrographolide, oridonin, and isoliquiritigenin induce HBD3 expression. We analyzed the involvement of the MEK1/2, p38, and JNK MAPK cascades using the specific inhibitors U0126, SB203580, and SP600125, respectively. Confluent cell monolayers were pretreated for 3 h with inhibitors and were stimulated for 24 h with 100 μM of each natural molecule (Fig. 3A). Inhibition of MEK1/2 dramatically reduced HBD3 expression in cells stimulated with each of the three molecules. In contrast, inhibition of p38 and JNK led to a differential pattern of HBD3 expression depending on the molecule used to stimulate the cells. Inhibition of p38 decreased HBD3 expression upon oridonin and isoliquiritigenin treatment but not upon andrographolide treatment, whereas inhibition of JNK reduced HBD3 induction upon andrographolide treatment but not upon oridonin and isoliquiritigenin treatment.
Fig. 3.
Andrographolide, oridonin, and isoliquiritigenin signal through different MAPK pathways. (A) Transcription of the gene encoding HBD3 in cells pretreated with 10 μM inhibitor of MEK1/2 (U0126), p38 (SB203580), or JNK (SP600125) and stimulated for 24 h with 100 μM andrographolide, oridonin, or isoliquiritigenin. Values are presented on a linear scale as the ratio of gene expression in inhibited and treated cells to that in inhibited and nontreated cells. NT, nontreated cells. Data are presented as mean ± SD (n = 4 biological replicate). *P < 0.05 evaluated by two-tailed Mann–Whitney U test. (B) Immunoblot analysis of Erk1/2, phosphorylated Erk1/2, p38, phosphorylated p38, SAPK/JNK, and phosphorylated SAPK/JNK in cells treated or not treated with 100 μM andrographolide, oridonin, or isoliquiritigenin. After lysis of cells at the indicated time points, Western blots were performed using specific antibodies directed against proteins or phosphorylated proteins. Results are representative of three biological replicates. The prefix “P” indicates phosphorylation.
To confirm these results, we studied activation of the MAPK signaling pathways upon stimulation with the natural molecules. By immunoblot, we observed an increased phosphorylation of Erk1/2 as early as 6 h following treatment of cells with each of the three molecules (Fig. 3B). Moreover, stimulating cells with andrographolide led to phosphorylation of SAPK/JNK but not p38, whereas treating cells with oridonin or isoliquiritigenin induced phosphorylation of p38 but not SAPK/JNK. Together, these results show that compounds induce HBD3 expression by targeting different signaling cascades: Andrographolide activates the EGFR–Erk–JNK pathway, whereas oridonin and isoliquiritigenin signal through the EGFR–Erk–p38 pathway.
Andrographolide, Oridonin, and Isoliquiritigenin Induce H3S10 Phosphorylation and Promote Differential c-Fos, c-Jun, c-Myc, and ELK1 Recruitment at the HBD3 Promoter.
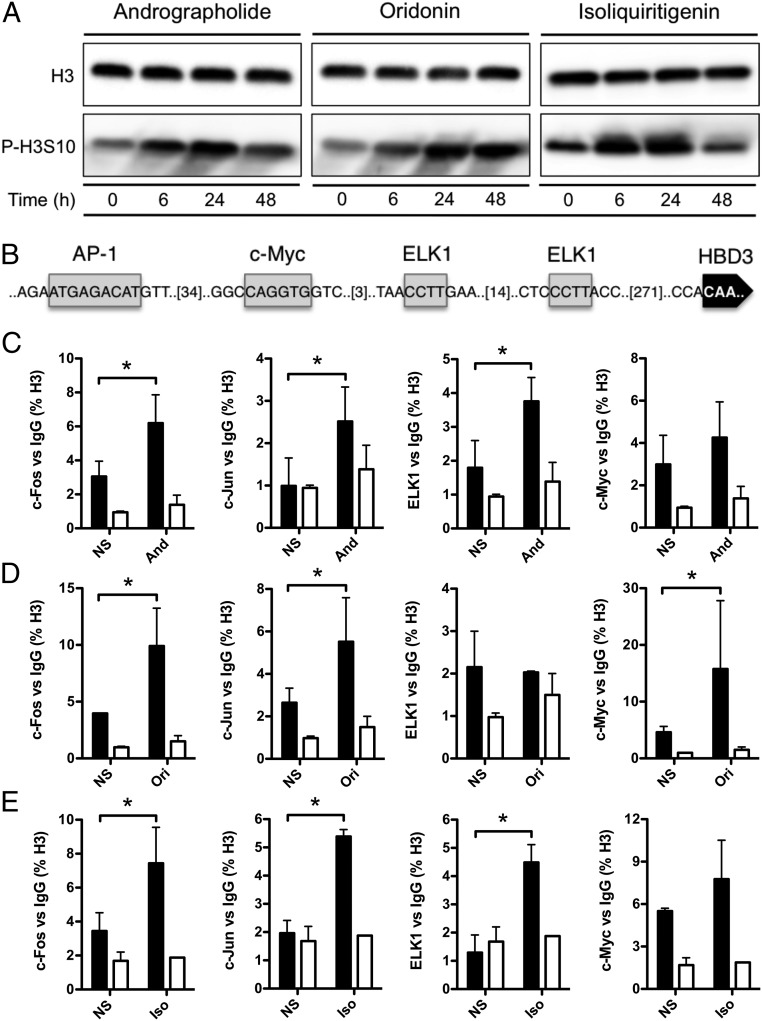
Accessibility of transcription factors to chromatin requires the acetylation of histones H3 and H4 on several lysine residues and the phosphorylation of H3S10 (29, 30). We therefore investigated the impact of natural molecules on the occurrence of these histone posttranslational modifications. By immunoblot, using antibodies against acetylated H3, H3K4, H3K9, H3K18, H3K27, and H4, we failed to detect variation of acetylation in cells stimulated with andrographolide, oridonin, or isoliquiritigenin and the basal state observed in nonstimulated cells (SI Appendix, Fig. S3). In contrast, using an antibody detecting phosphorylated H3S10, we observed a strong increase of this epigenetic mark following treatment of cells with each of the three molecules as early as 6 h poststimulation (Fig. 4A).
Fig. 4.
Andrographolide, oridonin, and isoliquiritigenin induce H3S10 phosphorylation and promote differential c-Fos, c-Jun, c-Myc, and ELK1 recruitment at the HBD3 promoter. (A) Immunoblot analysis of H3S10 phosphorylation in cells treated with 100 μM andrographolide, oridonin, or isoliquiritigenin. After lysis of cells at the indicated time points, Western blots were performed using antibodies directed against histone posttranslational modifications. Results are representative of three biological replicates. The prefix “P” indicates phosphorylation. (B) Promoter sequence of the human HBD3 gene. Putative DNA-binding motifs are indicated by gray boxes for AP-1 (c-Fos/c-Jun), c-Myc, and ELK1 transcription factors. (C–E) ChIP analysis of c-Fos, c-Jun, ELK1, and c-Myc protein recruitment at the HBD3 promoter in cells treated with 100 μM andrographolide (C), oridonin (D), or isoliquiritigenin (E). Enrichment in chromatin was detected using c-Fos-, c-Jun-, ELK1-, and c-Myc–specific antibodies, or rabbit IgG as control and was quantified by qRT-PCR using specific primers of the HBD3 promoter. Values are presented as the percentage of signal relative to the histone H3 protein. And, andrographolide; Iso, isoliquiritigenin; NS, nonstimulated cells; Ori, oridonin. Data are presented as mean ± SD (n = 4 biological replicates). *P < 0.05 evaluated by two-tailed Mann–Whitney U test.
A current model suggests that H3S10 phosphorylation accounts for a histone structure that favors the binding of chromatin-remodeling enzymes, thereby increasing the accessibility of transcription factors to promoter of specific genes from the innate immune response (29, 31). Because the EGFR pathway activates regulators such as c-Fos, c-Jun, c-Myc, or ELK1 and the promoter of HBD3 harbors putative binding sites for each of these transcription factors (Fig. 4B), we investigated the specific recruitment of these four proteins at this promoter upon andrographolide (Fig. 4C), oridonin (Fig. 4D), and isoliquiritigenin (Fig. 4E) treatment. ChIP experiments were carried out using antibodies directed against the transcription factors or an irrelevant IgG of the same isotype as a control. Because of the time existing between early regulatory events occurring at promoters, such as recruitment of transcription factors, and the subsequent gene transcription, all experiments were performed 3 h poststimulation. The size of the sonicated chromatin fragment was 150–900 bp, corresponding to one to five nucleosomes. Immunoprecipitated materials were analyzed by qRT-PCR using the ribosomal protein RPL30 housekeeping gene as control. Signals corresponding to c-Fos and c-Jun, forming the AP-1 complex, were increased regardless of the molecule used to stimulate the cells, with increases ranging from 2.5-fold to 10-fold compared with nonstimulated cells. Increased recruitment of ELK1 was observed upon andrographolide and isoliquiritigenin treatment but not upon oridonin treatment, whereas increased recruitment of c-Myc was detected upon oridonin treatment but not upon andrographolide or isoliquiritigenin treatment. These results indicate that natural molecules induce phosphorylation of the H3S10 residue and favor the recruitment of the transcription factors c-Fos, c-Jun, c-Myc, and ELK1 at the HBD3 promoter.
Andrographolide and Isoliquiritigenin Synergize to Boost HBD3 Expression in Vitro and ex Vivo.
Andrographolide, oridonin, and isoliquiritigenin induced HBD3 expression through different signaling pathways (EGFR–Erk–JNK for andrographolide and EGFR–Erk–p38 for oridonin and isoliquiritigenin). We therefore examined whether a combination of andrographolide and oridonin or isoliquiritigenin could elicit greater induction of HBD3 gene expression than the individual molecules. Monolayers of confluent cells were treated with 100 μM andrographolide, oridonin, or isoliquiritigenin alone or with combinations of andrographolide and oridonin or isoliquiritigenin. RNA was extracted at 6, 24, and 48 h after stimulation and was analyzed by qRT-PCR. In cells treated with andrographolide and oridonin, we detected HBD3 expression at the transcriptional and translational levels quite similar to that observed with the single natural compounds, even at later time points (SI Appendix, Fig. S4). In contrast, at 48 h, cells treated with the andrographolide and isoliquiritigenin combination induced HBD3 mRNA at significantly higher levels (3,000-fold) than seen with the addition of either compound alone (300-fold over basal levels) (Fig. 5A). This synergistic effect of the andrographolide and isoliquiritigenin combination on HBD3 transcription was observed at lower concentrations, e.g., 10 μM (Fig. 5B). The effect of the combinatorial treatment was confirmed at the translational level, with a significantly greater secretion of the HBD3 peptide upon andrographolide and isoliquiritigenin treatment (400 pg/mL) compared with molecules alone (100–130 pg/mL) (Fig. 5C). As controls, the production of the proinflammatory IL-8 cytokine was not modified in any of the tested conditions (SI Appendix, Figs. S4 and S5 A–C), and no synergism was observed between the three natural molecules and proinflammatory stimuli such as the IL-1B cytokine (SI Appendix, Fig. S5D). Taken together, these results show that andrographolide and isoliquiritigenin molecules synergize to boost the expression of HBD3.
Fig. 5.
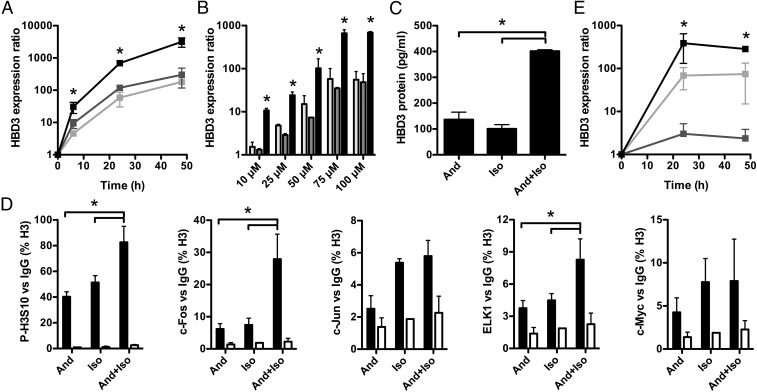
Andrographolide and isoliquiritigenin synergize to boost HBD3 expression in vitro and ex vivo. (A) Kinetics of HBD3 gene expression in cells treated with 100 μM andrographolide (light gray line), isoliquiritigenin (dark gray line), or andrographolide and isoliquiritigenin (black line). Values are presented on a logarithmic scale as the ratio of gene expression in treated cells to that in nontreated cells. Data are presented as mean ± SD (n = 6 biological replicates). *P < 0.05 evaluated by two-tailed Mann–Whitney U test. (B) Transcription of the HBD3 gene in cells treated for 24 h with increasing concentrations (10–100 μM) of andrographolide (light gray bars), isoliquiritigenin (dark gray bars), or andrographolide and isoliquiritigenin (black bars). Values are presented on a logarithmic scale as the ratio of gene expression in treated cells to that in nontreated cells. Data are presented as mean ± SD (n = 4 biological replicates). *P < 0.05 evaluated by two-tailed Mann–Whitney U test. (C) ELISA dosage of the HBD3 peptide in supernatants of cells treated for 48 h with 100 μM andrographolide (And), isoliquiritigenin (Iso), or andrographolide and isoliquiritigenin (And+Iso). Values are presented on a linear scale in picograms of peptide per milliliter. Data are presented as mean ± SD (n = 4 biological replicates). *P < 0.05 evaluated by two-tailed Mann–Whitney U test. (D) ChIP analysis of phosphorylated H3S10, c-Fos, c-Jun, ELK1, and c-Myc at the HBD3 promoter in cells treated with 100 μM andrographolide (And), isoliquiritigenin (Iso), or the andrographolide and isoliquiritigenin combination (And+Iso). Enrichment in chromatin was detected using anti–P-H3S10, anti–c-Fos, anti–c-Jun, anti-ELK1, and anti–c-Myc antibodies or rabbit IgG as control and was quantified by qRT-PCR using specific primers matching the HBD3 promoter. Values are presented as the percentage of signal relative to the H3 protein. Data are presented as mean ± SD (n = 4 biological replicates). *P < 0.05 evaluated by two-tailed Mann–Whitney U test. (E) Kinetics of HBD3 gene expression in 5-d-old human colonic organoids treated with 100 μM andrographolide (light gray line), isoliquiritigenin (dark gray line), or andrographolide and isoliquiritigenin (black line). Values are presented on a logarithmic scale as the ratio of gene expression in treated organoids to that in nontreated organoids. Data are presented as mean ± SD (n = 6 biological replicates). *P < 0.05 evaluated by two-tailed Mann–Whitney U test.
At the HBD3 promoter, the synergistic effect of andrographolide and isoliquiritigenin was supported by stronger phosphorylation of H3S10 than seen with each molecule alone (Fig. 5D). Quantitatively, at 3 h posttreatment, we detected an 80-fold enrichment of phosphorylated H3S10 upon treatment with andrographolide and isoliquiritigenin together, compared with 40-fold and 50-fold enrichment, respectively, upon treatment with andrographolide or isoliquiritigenin alone (Fig. 5D). Higher levels of c-Fos and ELK1 transcription factor recruitment were similarly detected following stimulation of cells with the combination of the two molecules as compared with each molecule alone (Fig. 5D).
We investigated the impact of the synergistic effect of the andrographolide and isoliquiritigenin combination in primary human colonic cells using ex vivo cultured organoids derived from healthy colon tissues. We analyzed the expression of HBD3 in 5-d-old organoids treated with 100 μM andrographolide, isoliquiritigenin, or the andrographolide and isoliquiritigenin combination (Fig. 5E). Briefly, crypts were isolated from normal human colons and embedded in Matrigel in the presence of growth factors to induce the formation of tridimensional structures termed “organoids.” Organoids recapitulate an intact architecture, harboring an internal lumen, stem cells, which are located in surface protrusions that correspond to novel crypts, and the different epithelial lineages, including colonocytes (32). RNA was extracted at 24 and 48 h and analyzed by qRT-PCR. Quantitatively, induction of HBD3 with the combination treatment was ∼400-fold at 24 h, as compared with 70-fold with andrographolide treatment alone or threefold with isoliquiritigenin treatment alone (Fig. 5E). As control, expression of IL-8 was similar for all tested conditions (SI Appendix, Fig. S5C). These data demonstrate the existence of small molecules that, alone or in combination, allow strong induction of the β-defensin HBD3 gene, but not those encoding proinflammatory mediators, in human colonic primary cells.
Andrographolide and Isoliquiritigenin Synergism Increases Cellular Antimicrobial Activity.
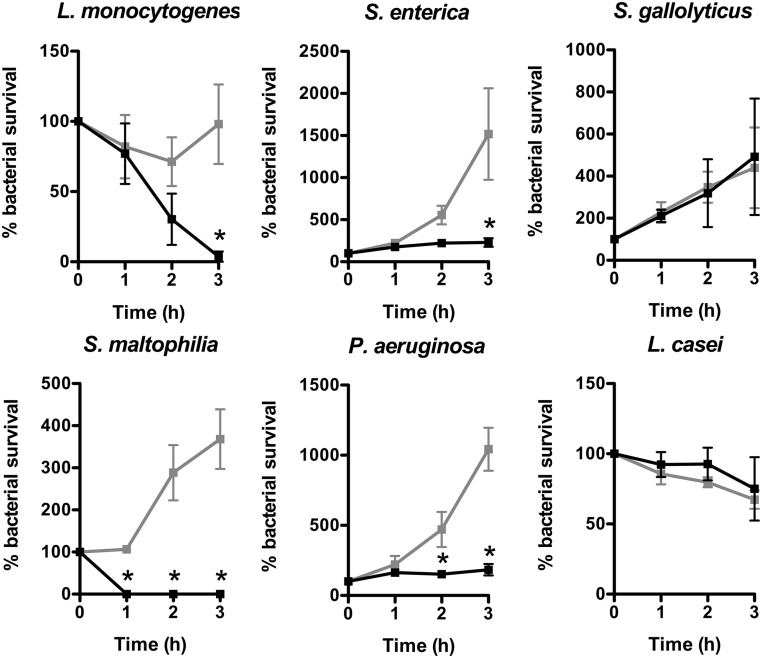
Finally, we tested the in vitro antimicrobial activity of supernatant from cells stimulated with andrographolide and isoliquiritigenin against commensal, pathobiontic, and pathogenic bacteria. For this purpose, the Gram-negative strains Pseudomonas aeruginosa, Salmonella enterica, and Stenotrophomonas maltophilia and the Gram-positive strains Lactobacillus casei, Streptococcus gallolyticus, and Listeria monocytogenes were sampled in the exponential phase, washed, and resuspended into the supernatant of cells collected 48 h after stimulation with 100 μM andrographolide and isoliquiritigenin. Bacteria were plated at different time points and counted (Fig. 6). In these conditions, the supernatant was bactericidal against the L. monocytogenes pathogen and the S. maltophilia pathobiont and was bacteriostatic against the P. aeruginosa and S. enterica pathogens. In contrast, supernatant did not affect the growth of the S. gallolyticus pathobiont and the L. casei commensal. As control, andrographolide and isoliquiritigenin molecules, by themselves, did not affect the growth of these bacterial strains (SI Appendix, Fig. S6). Collectively, these results show a change in the antimicrobial activity of cells treated with andrographolide and isoliquiritigenin, which differentially impact bacteria depending on whether the bacterium is a Gram-positive or a Gram-negative commensal, pathobiont, or pathogen.
Fig. 6.
Andrographolide and isoliquiritigenin synergism increases cellular antimicrobial activity. Antimicrobial activity of supernatant from cells treated for 48 h with 100 μM andrographolide and isoliquiritigenin on the growth of L. monocytogenes, S. gallolyticus, L. casei, P. aeruginosa, S. enterica, and S. maltophilia. Values are presented on a linear scale as the percentage of bacterial survival. Data are presented as mean ± SD (n = 4 biological replicates). Black lines, supernatant from treated cells; gray lines, supernatant from nontreated cells. *P < 0.05 evaluated by two-tailed Mann–Whitney U test.
Discussion
Most genes involved in the innate immune response are inducible genes whose expression needs to be tightly regulated and rapidly and specifically activated in response to diverse inducers (33). Intestinal epithelial cells, being the first line of interaction with microbes, are endowed with innate immune functions encompassing the balanced expression of an array of genes, including those encoding antimicrobial peptides and proinflammatory cytokines and chemokines. These two groups of genes are generally considered to be synchronously and coordinately expressed under the necessity to protect the epithelium against pathogenic microbes and to keep commensal bacteria away from the epithelial surface. We hypothesized, however, that these two groups of genes might obey differential regulatory rules that do not necessarily imply their synchronous expression. We previously showed the possibility of a disconnection between expression of the β-defensin HBD2 and bona fide proinflammatory genes, based upon epigenetic imprints that would modulate the differential expression of genes that are otherwise part of the innate immunity network (31). Here, we identified a regulatory circuit disconnecting expression of the β-defensin HBD3 from proinflammatory mediators, including chemokines IL-1B, IL-8, and TNF, when epithelial cells are treated with andrographolide, oridonin, or isoliquiritigenin. This circuit induces HBD3 through the activation of the EGFR and MAPK signaling cascades without activation of the NF-κB proinflammatory pathway. Other circuits have been described previously, such as the one used by the two rare sugars mannoheptulose and perseitol from avocado, which can pharmacologically stimulate HBD2 and HBD3 expression in human keratinocytes, in the absence of a proinflammatory response, through activation of the TLR2 receptor and the Erk MAPK (34–36). In addition to plant-derived compounds, other molecules from microbes have been reported to induce the expression of antimicrobial peptides without inflammation by inducing other circuits, such as short-chain fatty acids produced by most of commensals species activating GPR43-mTOR-STAT3 or the lipopeptide LP01 from Staphylococcus epidermis activating TLR2/CD36-p38 MAPK (37, 38). The existence of such regulatory circuits disconnecting the expression of these two groups of genes shows how epithelial cells can adapt their response when they are engaged by a pathogenic microbe that necessitates the mobilization of both arms of the innate immune system or in controlling and tolerating commensals that instead mobilize the expression of antimicrobial factors.
Why natural compounds such as andrographolide, oridonin, and isoliquiritigenin should serve as recognizable markers of the presence of commensal bacteria remains an open question. It is intriguing that molecules from plants act as β-defensin inducers. Because these natural compounds cannot be synthesized by the host, they must originate from an external source. One possible explanation for the emergence of these molecules as a marker of commensals is that nonpathogenic bacterial hydrolases, in the process of degrading plant components in the host diet, may release free andrographolide, oridonin, or isoliquiritigenin in amounts that are substantially in excess of those found upon infection with pathogenic bacteria. The release of these molecules would reflect the degradative activity of the microbiota and therefore its density, thus requiring tight shaping by the action of antimicrobial peptides without the need of inflammation beyond a certain level of bacterial overgrowth. It has been similarly reported that other classes of natural molecules, including branched-chain amino acids and short-chain fatty acids, are inducers of the β-defensin HBD2 and the cathelicidin LL-37 expression and may be used by the host as markers of the microbial presence (39, 40).
Although the nature of their receptor remains unknown, we have demonstrated that one feature of the cellular signals produced by andrographolide, oridonin, and isoliquiritigenin is activation of the EGFR-MAPK pathways. EGFR is known to be activated by membrane-bound ligands released in close vicinity to the receptor. This leads to its rapid and localized activation (28, 41). Subsequent to its activation, the EGFR is known to play a critical role in mucosal repair and wound-healing processes during infection and inflammation (42). This occurs through the engagement of the downstream MAPK pathways. High- and low-affinity interactions between EGFR and its ligands activate different signaling pathways. While high-affinity ligand binding is sufficient for the activation of most canonical signaling pathways, low-affinity binding is required for activation of the others. The existence of receptors with distinct signaling properties provides a way for EGFR to respond to different concentrations of the same ligand in qualitatively different ways (43). All studies show a dramatic increase in HBD2 expression during inflammation, suggesting that its antimicrobial and chemotactic properties provide a first line of defense; however, during the repair phase of tissues, inflammation recedes with declining HBD2 level, and HBD3 comes into play (44, 45). This suggests that MAPKs must be finely tuned according to time. This also suggests that MAPKs must synergize or cross-talk with other pathways, such as NF-κB, to differentially modulate the expression of antimicrobial and proinflammatory genes depending on whether the epithelium is engaged by pathogenic or nonpathogenic bacteria. In that sense, proinflammatory cytokines such as IL-1B and IL-6 modulate the EGFR activation leading to increased expression of HBD3 during inflammation, even though these cytokines did not induce HBD3 expression directly (26).
Few studies have examined the possible synergistic effect of inducers on antimicrobial peptide expression. Previously, we showed that the concomitant treatment of human intestinal epithelial cells in vitro and ex vivo with the histone deacetylase inhibitor trichostatin A and Escherichia coli K12 commensal bacteria enhanced the expression of the β-defensin HBD2 more strongly than treatment with each inducer alone (31). This effect resulted from activation of the IκB kinase complex, leading to an increased phosphorylation of H3S10 and an enriched recruitment of acetylated p65 subunits of NF-κB at the HBD2 promoter. Here, using the same models, we show that the combined treatment of cells with the two natural molecules andrographolide and isoliquiritigenin promoted higher expression of the β-defensin HBD3 than observed with single-molecule treatment. The mechanism supports the concept that these two compounds are synergized through the concomitant activation of the Erk, p38, and JNK MAPK pathways, leading to the strongest detected phosphorylation of H3S10 and the highest recruitment levels of c-Fos and ELK1 at the HBD3 promoter. The common thread that emerges from both studies is that the level of H3S10 phosphorylation at the promoter of β-defensin genes is crucial to their transcriptional activation state. Therefore, drug compounds that specifically promote this histone modification represent an effective strategy to boost host endogenous innate immune defenses.
Given the extent, spread, evolution, and impact of antibiotic resistance in pathogenic bacteria, new molecules and innovative strategies are urgently needed in the antiinfective drug-discovery pipeline. This has stimulated interest in the use of antimicrobial peptides as therapeutic targets. The inducible nature of antimicrobial peptides and the existence of regulatory circuits disconnecting their expression from proinflammatory mediators show the possibility of developing innovative approaches to increase their endogenous expression for the prevention or treatment of diseases. Moreover, beyond their own antibacterial activity, antimicrobial peptides have been shown to enhance the potency of several well-known antibiotics in vivo by facilitating their access to bacterial cells (46). This has been demonstrated for HBD3 in combination with the antibiotics amoxicillin, chlorhexidine, and metronidazole (47). Thus, therapeutic strategies based on the delivery of immunomodulatory molecules such as andrographolide and isoliquiritigenin, combined with antibiotics or not, would provide effective prophylaxis and therapy for drug-resistant infections by playing on both host and microbe sides. Major clinical situations that require strong boosting of mucosal innate immune defenses, such as recurrent enteric infections in pediatric populations in low-income countries (48, 49), chronic inflammatory bowel diseases (50), or myelosuppressive and immunosuppressive therapies (51), with the aim, respectively, of avoiding luminal bacterial overgrowth causing malnutrition, chronic intestinal inflammation, or deadly gut-derived bacterial translocation events, may benefit from such an approach.
Materials and Methods
Natural Molecules.
The natural product library from Selleck Chemicals (L1400; Selleck Chemicals) containing more than 170 bioactive molecules was used for the initial screening. The compounds andrographolide (S2261; Selleck Chemicals), oridonin (S2335; Selleck Chemicals) and isoliquiritigenin (S2404; Selleck Chemicals) were selected for further characterization.
Antibodies.
Antibodies used in this work include cetuximab [Erbitux; Research Resource Identifier (RRID) AB_2489605; Creative Diagnostics catalog no. AIT-19034]; anti-Erk1/2 (RRID AB_390779; Cell Signaling Technology catalog no. 4695), anti–phospho-Erk1/2 (T202/Y204) (RRID AB_2315112; Cell Signaling Technology catalog no. 4370); anti-p38 (RRID AB_10999090; Cell Signaling Technology catalog no. 8690); anti–phospho-p38 (T180/Y182) (RRID AB_2139682; Cell Signaling Technology catalog no. 4511); anti-SAPK/JNK (RRID AB_2250373; Cell Signaling Technology catalog no. 9252); anti–phospho-SAPK/JNK (T183/Y185) (RRID AB_2307320; Cell Signaling Technology catalog no. 4668); anti-EGFR (RRID AB_331707; Cell Signaling Technology catalog no. 2232); anti–phospho-EGFR Y1068 (RRID AB_305012; Abcam catalog no. ab5644); anti-IκBα (RRID AB_2235952; Santa Cruz Biotechnology catalog no. sc-371); anti-histone H3 (RRID AB_302613; Abcam catalog no. ab1791); anti–pan-acetylated histone H3 (RRID AB_8738; Abcam catalog no. ab47915, 60); anti–acetyl-histone H3K4 (RRID AB_673133; Millipore catalog no. 07-539); anti–acetyl-histone H3K9 (RRID AB_2118292; Abcam catalog no. ab4441); anti–acetyl-histone H3K18 (RRID AB_298692; Abcam catalog no. ab1191); anti–acetyl-histone H3K27 (RRID AB_2118291; Abcam catalog no. ab4729); anti–phospho-histone H3S10 (RRID AB_304763; Abcam catalog no. ab5176); anti–c-Myc (RRID AB_2631168; Cell Signaling Technology catalog no. 13987); anti-ELK1 (RRID AB_732141; Abcam catalog no. ab32106); anti–c-Fos (RRID AB_2247211; Cell Signaling Technology catalog no. 2250); anti–c-Jun (RRID AB_2130165; Cell Signaling Technology catalog no. 9165); anti-histone H4 (RRID AB_305837; Abcam catalog no. ab7311); anti–pan-acetylated histone H4 (RRID AB_310270; Millipore catalog no. 06-866); anti-actin (RRID AB_476693; Sigma-Aldrich catalog no. A2066); anti-mouse IgG-POX (RRID AB_772209; GE Healthcare catalog no. NXA931-1 mL); and anti-rabbit IgG-POX [GAR/IgG(H+L)/PO; Nordic Immunology].
Cell Culture.
The human colonic epithelial cell line Caco-2, subclone TC7 (24), was cultured with DMEM (Thermo Fisher) supplemented with 10% (vol/vol) decomplemented FBS (Thermo Fisher), 1% nonessential amino acids (Thermo Fisher), 100 U/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher) at 37 °C in 10% CO2. Cells were split two times/wk using Versene solution (Thermo Fisher).
Inhibitors.
We used the pharmacological inhibitors gefitinib (S1025; Selleck Chemicals), SB203580 (S1076; Selleck Chemicals), U0126 (S1102; Selleck Chemicals), LY294002 (S1105; Selleck Chemicals), ruxolitinib (S1378; Selleck Chemicals), SP600125 (S1460; Selleck Chemicals), and AG-1478 (S2728; Selleck Chemicals).
qRT-PCR.
RNA was isolated using the RNeasy Mini kit and the RNase-Free DNase kit (Qiagen). RT-PCR reactions were performed overnight using SuperScript II Reverse Transcriptase (Thermo Fisher) and the oligo(dT)18 primers (Thermo Fisher) as recommended by the supplier. Gene-specific primers were designed and purchased from Sigma: HBD2, GCCATGAGGGTCTTGTATCTC/TTAAGGCAGGTAACAGGATCG; HBD3, TTTGGTGCCTGTTCCAGGTCAT/GCCGCCTCTGACTCTGCAATAATA; LL-37, AAGGAAGCTGTGCTTCGTGCTA/AATCCTCTGGTGACTGCTGTGT; IL-1B, TACGATCACTGAACTGCACGCT/TCTTTCAACACGCAGGACAGGT; IL-8, AAGAAACCACCGGAAGGAACCA/ATTTCTGTGTTGGCGCAGTGTG; and TNF, AAACAACCCTCAGACGCCACAT/AGTGCTCATGGTGTCCTTTCCA. The qRT-PCR reactions were carried out in a 20-μL final volume containing 8 μL of cDNA (diluted at 1/100), 2 μL of primers (0.2 μM each), and 10 μL of Power SYBR Green PCR Master Mix (Thermo Fisher). Reactions were run on a QuantStudio 7 PCR system (Thermo Fisher) with the recommended universal thermal cycling parameters. Each sample reaction was run in duplicate on the same plate. Relative gene-expression quantification was performed using the comparative cycle threshold (Ct) method. Data were normalized to the β-2-microglobulin (B2M) housekeeping gene expression.
ELISA.
We used the ELISA kits for HBD3 (EK-072-38; Phoenix Pharmaceuticals) and IL-8 (900-K18; PeproTech), as recommended by the suppliers. Absorbance was measured on a M200PRO fluorimeter (Tecan).
Cytotoxicity Measurement.
The cytotoxic effect of molecules was evaluated by measurement of lactate dehydrogenase release using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega), as recommended by the supplier.
Immunoblotting.
Total cell lysates were harvested by removing growth medium and adding Nonidet P-40 lysis buffer [25 mM Tris⋅HCl (pH 7.5), 1 mM EDTA, 0.1 mM EGTA, 5 mM MgCl2, 1% Nonidet P-40, 10% (vol/vol) glycerol, 150 mM NaCl] supplemented by a mixture of protease inhibitors [sodium orthovanadate (Sigma) and Roche cOmplete Protease Inhibitor]. Samples were diluted with sample buffer [1 M Tris⋅HCl, 20% (vol/vol) glycerol, 6% (vol/vol) SDS, 0.02% bromophenol blue, 10% (vol/vol) β-mercaptoethanol] and were boiled for 5 min. Denatured proteins were loaded on 7.5%, 10%, or 12% acrylamide Mini-PROTEAN TGX Precast Gel (Bio-Rad). Separated proteins were transferred onto PVDF membranes using the Trans-Blot Turbo transfer system (Bio-Rad). Membranes were blocked with 3% (wt/vol) BSA (Sigma) or 5% (wt/vol) milk (Régilait) at room temperature before incubation with primary antibodies overnight at 4 °C in 1% BSA or 5% (wt/vol) milk. Incubation with the secondary HRP-conjugated IgG antibody was performed at room temperature. Blots were developed using the SuperSignal West Dura Extended Duration Substrate solution (Thermo Fisher) and an Amersham Imager 600 (GE). The results presented are representative of at least two independent experiments.
ChIP.
ChIP was performed using the SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling) using magnetic beads as recommended by the supplier. Chromatin inputs corresponded to 5–10 μg DNA for each individual ChIP assay. The ChIP DNA fractions were quantified by qRT-PCR on a QuantStudio 7 PCR system (Thermo Fisher), using the comparative Ct method. Gene-specific primers were designed and purchased from Sigma: HBD3, AGCTGTTGTGAGCTGTAATC/GTAGTGAGGTAAGGGAGGAA; IL-8, AGGACAAGAGCCAGGAAGAAACCA/AGAGCTGCAGAAATCAGGAAGGCT.
Human Colonic Organoid Culture.
This study was approved by the Institut Pasteur’s ethical and medical committee under agreement no. 2012-37. Surgically resected human colonic tissues were obtained from the Henri Mondor Hospital. All samples were obtained from patients who provided informed consent before surgery. Normal epithelia were isolated and cultured according to the protocol described by Sato et al. (32), with minor modifications. Organoids were cultured with Advanced DMEM/F12 (Thermo Fisher) supplemented with Hepes (Thermo Fisher), 2 mM GlutaMAX (Thermo Fisher), 100 U/mL penicillin, 100 μg/mL streptomycin (Thermo Fisher), 1× N2 and B27 supplements (Thermo Fisher), 1 mM N-acetyl-l-cysteine (Sigma), 10 μM Y-27632 (Sigma), 500 nM A83-01 (Tocris), 10 μM SB202190 (Sigma), 10 mM nicotinamide (Sigma), 10 nM gastrin I (Sigma), 100 ng/mL recombinant human Noggin (R&D Systems), 50 ng/mL recombinant human EGF (R&D Systems), 1 μg/mL recombinant human R-Spondin-1 (PeproTech), 100 ng/mL recombinant human Wnt-3A (R&D Systems), and 10% (vol/vol) FBS (Thermo Fisher) at 37 °C in 5% CO2. Organoids were cultured in 48-well plates with 100 crypts per well. RNA was isolated from 5-d-old organoids treated or not treated with 100 μM natural molecule for 24 or 48 h, using the RNeasy Mini Kit and the RNase-Free DNase Kit (Qiagen). Gene expression was analyzed using TaqMan probes (Thermo Fisher): HBD3 (Hs00218678_m1) and IL-8 (Hs00174103_m1). Data were normalized to the B2M (Hs00984230_m1) housekeeping gene expression.
Antibacterial Assay.
The method used for the antibacterial assay was described previously (52). Briefly, exponential cultures of bacterial strains (Listeria monocytogenes EGD-e, Streptococcus gallolyticus UCN34, Lactobacillus casei ATCC 334, Pseudomonas aeruginosa PAO1, Salmonella enterica subspecies Typhi, and Stenotrophomonas maltophilia BR12) were harvested and washed with PBS. The bacterial suspension was diluted with PBS to 5.108 cells/mL; 10 μL of suspension was inoculated into 1 mL of supernatant collected 48 h after stimulation of cells with 100 μM andrographolide and isoliquiritigenin and was incubated at 37 °C with shaking. At different time points, 100 μL of serial dilutions of the culture were plated on appropriate agar plates [LB (Thermo Fisher) for L. monocytogenes, P. aeruginosa, S. enterica, and S. maltophilia; Brain Heart Infusion (BHI) (Thermo Fisher) broth for S. gallolyticus; de Man, Rogosa, and Sharpe (MRS) (Thermo Fisher) for L. casei] and incubated at 37 °C overnight. Inoculum density (cfu/mL) was calculated from the number of colonies on each plate. The percentage of bacterial survival was determined as the number of cells surviving versus the total number of cells used.
Statistics.
Statistical analysis was performed on GraphPad Prism 5 (GraphPad software). Results are presented as a mean of at least four biological replicates. Error bars represent the SD. Statistical comparisons were performed using two-tailed Mann–Whitney U test. A P value < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Claude Parsot and Mark Anderson for critical reading of the manuscript, Aurélien Amiot from the Henri Mondor Hospital for providing human colon tissues, and Ludovic Reichard for helpful discussions. This study received funding from the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” Grant ANR-10-LABX-62-IBEID, and from L'Alliance pour les Sciences de la Vie et de la Santé (AVIESAN), Institut Thématique Multi-Organisme Immunologie, Inflammation, Infectiologie et Microbiologie (ITMO I3M). P.J.S. is supported by European Research Council DECRYPT Project Advanced Grant 339579 and the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: C.B. is a recipient of a Sanofi-funded PhD fellowship in the context of the French private–public partnership Convention Industrielle de Formation par la Recherche (CIFRE) program.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805298115/-/DCSupplemental.
References
- 1.Sansonetti PJ. The innate signaling of dangers and the dangers of innate signaling. Nat Immunol. 2006;7:1237–1242. doi: 10.1038/ni1420. [DOI] [PubMed] [Google Scholar]
- 2.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 4.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 5.Ganz T. Defensins: Antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 6.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radek K, Gallo R. Antimicrobial peptides: Natural effectors of the innate immune system. Semin Immunopathol. 2007;29:27–43. doi: 10.1007/s00281-007-0064-5. [DOI] [PubMed] [Google Scholar]
- 8.Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu Rev Immunol. 2004;22:181–215. doi: 10.1146/annurev.immunol.22.012703.104603. [DOI] [PubMed] [Google Scholar]
- 9.Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 10.Giacometti A, et al. Potential therapeutic role of cationic peptides in three experimental models of septic shock. Antimicrob Agents Chemother. 2002;46:2132–2136. doi: 10.1128/AAC.46.7.2132-2136.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivas-Santiago B, Serrano CJ, Enciso-Moreno JA. Susceptibility to infectious diseases based on antimicrobial peptide production. Infect Immun. 2009;77:4690–4695. doi: 10.1128/IAI.01515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hancock REW, Sahl H-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 13.Fox JL. Antimicrobial peptides stage a comeback. Nat Biotechnol. 2013;31:379–382. doi: 10.1038/nbt.2572. [DOI] [PubMed] [Google Scholar]
- 14.Marr AK, Gooderham WJ, Hancock RE. Antibacterial peptides for therapeutic use: Obstacles and realistic outlook. Curr Opin Pharmacol. 2006;6:468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Abdelmohsen UR, et al. Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect Dis. 2017;17:e30–e41. doi: 10.1016/S1473-3099(16)30323-1. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima S, et al. Fungus-derived neoechinulin B as a novel antagonist of liver X receptor, identified by chemical genetics using a hepatitis C virus cell culture system. J Virol. 2016;90:9058–9074. doi: 10.1128/JVI.00856-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, et al. Neoechinulin B and its analogues as potential entry inhibitors of influenza viruses, targeting viral hemagglutinin. Eur J Med Chem. 2015;93:182–195. doi: 10.1016/j.ejmech.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Kim K-S, et al. Anti-inflammatory effect of neoechinulin a from the marine fungus Eurotium sp. SF-5989 through the suppression of NF-кB and p38 MAPK pathways in lipopolysaccharide-stimulated RAW264.7 macrophages. Molecules. 2013;18:13245–13259. doi: 10.3390/molecules181113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu S, Pei J. Innovating chinese herbal medicine: From traditional health practice to scientific drug discovery. Front Pharmacol. 2017;8:381. doi: 10.3389/fphar.2017.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan H, Ma Q, Ye L, Piao G. The traditional medicine and modern medicine from natural products. Molecules. 2016;21:E559. doi: 10.3390/molecules21050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan WSD, Liao W, Zhou S, Wong WSF. Is there a future for andrographolide to be an anti-inflammatory drug? Deciphering its major mechanisms of action. Biochem Pharmacol. 2017;139:71–81. doi: 10.1016/j.bcp.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Owona BA, Schluesener HJ. Molecular insight in the multifunctional effects of oridonin. Drugs R D. 2015;15:233–244. doi: 10.1007/s40268-015-0102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng F, et al. A review: The pharmacology of isoliquiritigenin. Phytother Res. 2015;29:969–977. doi: 10.1002/ptr.5348. [DOI] [PubMed] [Google Scholar]
- 24.Chantret I, et al. Differential expression of sucrase-isomaltase in clones isolated from early and late passages of the cell line Caco-2: Evidence for glucose-dependent negative regulation. J Cell Sci. 1994;107:213–225. doi: 10.1242/jcs.107.1.213. [DOI] [PubMed] [Google Scholar]
- 25.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 26.Sørensen OE, et al. Differential regulation of beta-defensin expression in human skin by microbial stimuli. J Immunol. 2005;174:4870–4879. doi: 10.4049/jimmunol.174.8.4870. [DOI] [PubMed] [Google Scholar]
- 27.Hubbard SR, Miller WT. Receptor tyrosine kinases: Mechanisms of activation and signaling. Curr Opin Cell Biol. 2007;19:117–123. doi: 10.1016/j.ceb.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel) 2017;9:E52. doi: 10.3390/cancers9050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saccani S, Pantano S, Natoli G. p38-dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- 30.Arbibe L, et al. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat Immunol. 2007;8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- 31.Fischer N, et al. Histone deacetylase inhibition enhances antimicrobial peptide but not inflammatory cytokine expression upon bacterial challenge. Proc Natl Acad Sci USA. 2016;113:E2993–E3001. doi: 10.1073/pnas.1605997113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 33.Weake VM, Workman JL. Inducible gene expression: Diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 34.Donnarumma G, et al. Effects of AV119, a natural sugar from avocado, on Malassezia furfur invasiveness and on the expression of HBD-2 and cytokines in human keratinocytes. Exp Dermatol. 2007;16:912–919. doi: 10.1111/j.1600-0625.2007.00613.x. [DOI] [PubMed] [Google Scholar]
- 35.Sass V, et al. Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci. Infect Immun. 2010;78:2793–2800. doi: 10.1128/IAI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paoletti I, et al. Patented natural avocado sugar modulates the HBD-2 and HBD-3 expression in human keratinocytes through toll-like receptor-2 and ERK/MAPK activation. Arch Dermatol Res. 2012;304:619–625. doi: 10.1007/s00403-012-1237-1. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018;11:752–762. doi: 10.1038/mi.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li D, et al. A novel lipopeptide from skin commensal activates TLR2/CD36-p38 MAPK signaling to increase antibacterial defense against bacterial infection. PLoS One. 2013;8:e58288. doi: 10.1371/journal.pone.0058288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fehlbaum P, Rao M, Zasloff M, Anderson GM. An essential amino acid induces epithelial beta -defensin expression. Proc Natl Acad Sci USA. 2000;97:12723–12728. doi: 10.1073/pnas.220424597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raqib R, et al. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. Proc Natl Acad Sci USA. 2006;103:9178–9183. doi: 10.1073/pnas.0602888103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prenzel N, et al. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402:884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- 42.Leahy DJ. Structure and function of the epidermal growth factor (EGF/ErbB) family of receptors. Adv Protein Chem. 2004;68:1–27. doi: 10.1016/S0065-3233(04)68001-6. [DOI] [PubMed] [Google Scholar]
- 43.Krall JA, Beyer EM, MacBeath G. High- and low-affinity epidermal growth factor receptor-ligand interactions activate distinct signaling pathways. PLoS One. 2011;6:e15945. doi: 10.1371/journal.pone.0015945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boughan PK, et al. Nucleotide-binding oligomerization domain-1 and epidermal growth factor receptor: Critical regulators of beta-defensins during Helicobacter pylori infection. J Biol Chem. 2006;281:11637–11648. doi: 10.1074/jbc.M510275200. [DOI] [PubMed] [Google Scholar]
- 45.Dhople V, Krukemeyer A, Ramamoorthy A. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochim Biophys Acta. 2006;1758:1499–1512. doi: 10.1016/j.bbamem.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Darveau RP, et al. Beta-lactam antibiotics potentiate magainin 2 antimicrobial activity in vitro and in vivo. Antimicrob Agents Chemother. 1991;35:1153–1159. doi: 10.1128/aac.35.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maisetta G, et al. Activity of human beta-defensin 3 alone or combined with other antimicrobial agents against oral bacteria. Antimicrob Agents Chemother. 2003;47:3349–3351. doi: 10.1128/AAC.47.10.3349-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarleton JL, et al. Cognitive effects of diarrhea, malnutrition, and Entamoeba histolytica infection on school age children in Dhaka, Bangladesh. Am J Trop Med Hyg. 2006;74:475–481. [PubMed] [Google Scholar]
- 49.Wierzba TF, et al. The interrelationship of malnutrition and diarrhea in a periurban area outside Alexandria, Egypt. J Pediatr Gastroenterol Nutr. 2001;32:189–196. doi: 10.1097/00005176-200102000-00018. [DOI] [PubMed] [Google Scholar]
- 50.Fellermann K, Wehkamp J, Herrlinger KR, Stange EF. Crohn’s disease: A defensin deficiency syndrome? Eur J Gastroenterol Hepatol. 2003;15:627–634. doi: 10.1097/00042737-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 51.Wiest R, Rath HC. Gastrointestinal disorders of the critically ill. Bacterial translocation in the gut. Best Pract Res Clin Gastroenterol. 2003;17:397–425. doi: 10.1016/s1521-6918(03)00024-6. [DOI] [PubMed] [Google Scholar]
- 52.Midorikawa K, et al. Staphylococcus aureus susceptibility to innate antimicrobial peptides, beta-defensins and CAP18, expressed by human keratinocytes. Infect Immun. 2003;71:3730–3739. doi: 10.1128/IAI.71.7.3730-3739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.