Significance
While numerous long noncoding RNAs (lncRNAs) have been identified in muscle, most of their roles in myogenesis remain unclear, and many more lncRNAs have yet to be identified. In this study, we identified an intronic lncRNA, SYISL (SYNPO2 intron sense-overlapping lncRNA), which is highly expressed in muscle and interacts directly with polycomb repressive complex 2 (PRC2) to repress muscle development. The results reveal that SYISL acts as a regulatory RNA in PRC2-mediated myogenesis.
Keywords: lncRNA, SYISL, PRC2, H3K27 trimethylation, myogenesis
Abstract
Although many long noncoding RNAs (lncRNAs) have been identified in muscle, their physiological function and regulatory mechanisms remain largely unexplored. In this study, we systematically characterized the expression profiles of lncRNAs during C2C12 myoblast differentiation and identified an intronic lncRNA, SYISL (SYNPO2 intron sense-overlapping lncRNA), that is highly expressed in muscle. Functionally, SYISL promotes myoblast proliferation and fusion but inhibits myogenic differentiation. SYISL knockout in mice results in significantly increased muscle fiber density and muscle mass. Mechanistically, SYISL recruits the enhancer of zeste homolog 2 (EZH2) protein, the core component of polycomb repressive complex 2 (PRC2), to the promoters of the cell-cycle inhibitor gene p21 and muscle-specific genes such as myogenin (MyoG), muscle creatine kinase (MCK), and myosin heavy chain 4 (Myh4), leading to H3K27 trimethylation and epigenetic silencing of target genes. Taken together, our results reveal that SYISL is a repressor of muscle development and plays a vital role in PRC2-mediated myogenesis.
Myogenesis is a highly ordered process during which muscle stem cell proliferation, migration, differentiation, and fusion are activated to form myofibers (1). This process is controlled by a series of muscle-specific transcription factors, including myogenic differentiation 1 (MyoD), myogenin (MyoG), myogenic factor 5 (Myf5), muscle-specific regulatory factor 4 (MRF4, also known as “Myf6”), and myocyte enhancer factor 2 (MEF2) (2–4). In addition, epigenetic regulators, such as polycomb repressive complex 2 (PRC2), YIN-YANG-1 (YY1), histone deacetylase complexes (HDACs), histone methyltransferase complexes (HMTs), DNA methyltransferase complexes (DNMTs), the SWItch/Sucrose Non-Fermentable complex (SWI/SNF), and lysine methyltransferase 2A (KMT2A), also play crucial roles in muscle development (2, 5–7). Previous studies have shown that myogenic differentiation is associated with global chromatin landscape changes, especially for the well-known repressive marker histone H3 lysine 27 trimethylation (H3K27me3), which participates in the regulation of myogenic differentiation by silencing muscle-specific genes and cell-cycle genes (8, 9). Enhancer of zeste homolog 2 (EZH2), the core component of PRC2, is capable of H3K27me3 and is specifically required for early mouse muscle development (8, 10). Increased EZH2 expression inhibits skeletal muscle cell differentiation by silencing muscle-specific genes (10, 11), and EZH2 is associated with self-renewal of muscle satellite cells and muscle regeneration (12, 13).
Genome-wide analysis of human and mouse genomes has revealed that noncoding RNAs (ncRNAs), including miRNAs, long noncoding RNAs (lncRNAs), Piwi-interacting RNAs, and circular RNAs, are transcribed from most genomes. lncRNAs are defined as RNA molecules greater than 200 nt in length that have negligible protein-coding capacity; they play important roles in the epigenetic regulation of numerous biological processes, including X-chromosome inactivation (14), genome imprinting (15, 16), alternative splicing (17), stem cell maintenance and differentiation (18, 19), and human disease (20–22). During myogenesis, functional lncRNAs, such as SRA, Gtl2/Meg3, H19, linc-MD1, SINE-containing lncRNAs, Yam1, lncRNA-YY1, LncMyoD, MALAT1, Dum, MUNC, Linc-RAM, and Lnc-mg, have been found to regulate muscle development and regeneration through diverse mechanisms (23–29). For example, the skeletal muscle-specific overexpression of lnc-mg promotes muscle hypertrophy and increases muscle mass (29), and mutation in H19 leads to significant muscle mass increase in mice (30). Compared with WT mice, Linc-RAM and MALAT1-KO mice display impaired and enhanced muscle regeneration, respectively (28, 31). While numerous lncRNAs have been identified in muscle using microarray, high-throughput sequencing, and single-nucleus RNA sequencing (snRNA-seq) analysis (32, 33), most of their roles in myogenesis remain unclear, and many more lncRNAs have yet to be identified.
To systematically identify the lncRNAs that are potentially involved in myogenesis, we characterized the expression profiles of lncRNAs during C2C12 myoblast differentiation using microarray analysis and identified 1,913 differentially expressed lncRNAs. Based on these results, the lncRNA AK004418, located in the fourth intron of the SYNPO2 gene, was identified and named “SYISL” (for “SYNPO2 intron sense-overlapping lncRNA”). The SYISL gene is highly expressed in skeletal muscle, and its expression increases with C2C12 cell differentiation. Gain- and loss-of-function analysis revealed that SYISL promotes myoblast proliferation and inhibits myogenic differentiation. Significantly, SYISL-KO mice have more muscle mass and greater muscle fiber density than WT mice. Mechanistically, SYISL acts as a regulatory RNA that interacts directly with PRC2 to repress the expression of target genes.
Results
lncRNA and mRNA Expression Profiles During C2C12 Myoblast Differentiation.
To systematically identify lncRNAs involved in muscle cell differentiation, we used mRNA and lncRNA microarrays to identify differentially expressed lncRNAs during C2C12 cell differentiation (proliferating myoblasts and myoblasts at 2, 5, and 8 d after cell differentiation, represented as “D0,” “D2,” “D5,” and “D8,” respectively). This microarray contained 13,868 lncRNA probes and 19,465 mRNA probes; an overview of the lncRNA and mRNA expression profiles is summarized in Dataset S1. In total, 5,684 lncRNAs and 9,069 mRNA probes with signals in at least 6 of 12 samples had flags in the present or marginal categories (SI Appendix, Fig. S1 A and B and Dataset S1). Among 5,684 lncRNA probes with signals, 4,205 (73.98%) were annotated; intron sense-overlapping lncRNAs accounted for the largest proportion, followed by intergenic and exon sense-overlapping lncRNAs (SI Appendix, Fig. S1C). The chromosomal distribution of lncRNAs was not uniform; most of the lncRNAs with signals were transcribed from chromosomes 2 and 11 (SI Appendix, Fig. S1D). Previous studies showed that the expression levels of lncRNAs are highly correlated with those of their adjacent protein-coding genes (34, 35). In this study, we analyzed the expression correlation coefficient (r) of 896 annotated lncRNAs, including natural antisense, intron sense-overlapping, intronic antisense, exon sense-overlapping, and bidirectional lncRNAs, with their adjacent genes and found that 32 lncRNAs showed high expression correlation with their adjacent genes (|r| > 0.9; P < 0.05) (Dataset S2). Moreover, the expression levels of 12 intergenic lncRNAs were highly correlated with those of their flanking genes (|r| > 0.6; P < 0.05) (SI Appendix, Table S1), suggesting that they possibly exert their functions in cis. Next, the expression correlations of four randomly selected lncRNAs (NR_002864, NR_003647, AK046046, and AK012160) and their adjacent genes from the microarray data were confirmed by real-time quantitative PCR (qPCR), and the results showed that the r values from both methods were consistent (SI Appendix, Fig. S1 E–I).
In total, 1,931 lncRNAs and 4,526 mRNAs were identified as being differentially expressed during cell differentiation (fold-change ≥2; P < 0.05) (SI Appendix, Fig. S1 J and K and Datasets S3 and S4). Among the differentially expressed lncRNAs, 356 showed gradually increasing expression trends with myoblast differentiation, and 266 were persistently down-regulated during cell differentiation (Fig. 1A and Dataset S5). Among the gradually up-regulated lncRNAs, 96 lncRNAs, of which 58 were annotated, were up-regulated more than fivefold between D0 and D8 (Dataset S6). An expression heatmap of the 58 annotated lncRNAs showed that the intron sense-overlapping lncRNA AK004418 exhibited significantly higher expression than the others and was significantly up-regulated during myogenic differentiation (SI Appendix, Fig. S1L). AK004418 is transcribed from the fourth intron of the SYNPO2 gene (SI Appendix, Fig. S2A). Consistent with the microarray data, the SYISL expression level was increased with C2C12 cell differentiation, as determined by qPCR (Fig. 1B and SI Appendix, Fig. S2 B and C). Interestingly, the expression profiles of the SYISL gene in multiple adult tissues showed that SYISL was highly expressed in muscle tissues such as longissimus dorsi, leg muscle, and tongue when β-actin, GAPDH, and 18s RNA were used as reference genes, respectively (Fig. 1C and SI Appendix, Fig. S2 D and E). Gene Ontology (GO) analysis showed that SYISL-associated genes are clustered mainly into biological adhesion, biological regulation, developmental, metabolic, growth, and immune system processes (SI Appendix, Fig. S2 F and G).
Fig. 1.
Characterization of the lncRNA SYISL gene. (A) Heatmap showing the expression profiles of 356 up-regulated and 266 down-regulated lncRNAs during myogenic differentiation. (B) qPCR results showing that SYISL and MyHC were significantly up-regulated during myogenic differentiation. MyHC is the myogenic differentiation marker gene. (C) qPCR results showing that SYISL was highly expressed in muscle tissues including the longissimus dorsi, leg muscle, and tongue. (D) The distribution of SYISL in the cytoplasm and nuclei of proliferating C2C12 cells (D0) and C2C12 cells differentiated for 3 d (D3) was determined by qPCR. NEAT is a known nuclear lncRNA, and tRNA-lle is a cytoplasmic-enriched RNA. The relative RNA levels were normalized to those of the control β-actin. The data represent the means ± SD of three independent experiments.
Characterization of the lncRNA SYISL and Identification of Its Downstream Target Genes.
We first evaluated full-length SYISL cDNA in C2C12 cells using RACE and Northern blot assays, confirming that SYISL is a 1,181-nt polyadenylated lncRNA deposited in the National Center for Biotechnology Information database (SI Appendix, Fig. S2 H–J). The coding potential calculator (CPC) software indicated that SYISL is a noncoding RNA similar to the known lncRNAs HOTAIR, Braveheart, and LncMyoD (SI Appendix, Fig. S2K). Cell-fractionation assays demonstrated that SYISL is present in both the nuclei and cytoplasm of proliferating and differentiated C2C12 myoblasts (Fig. 1D). Furthermore, we examined the expression profiles of SYISL during postnatal muscle development, finding up-regulated SYISL expression in the first two postnatal weeks and down-regulated SYISL expression thereafter (SI Appendix, Fig. S2L). Because SYISL is localized to the fourth intron of the SYNPO2 gene, we tested the correlation between the spatiotemporal expression patterns of SYISL and SYNPO2 by qPCR. SYNPO2 was increased during C2C12 cell differentiation before decreasing after 8 d (SI Appendix, Fig. S2M), and it was mainly expressed in the stomach, small intestine, and heart (SI Appendix, Fig. S2N), indicating that the spatiotemporal expression patterns of SYISL and SYNPO2 were not consistent. To explore the regulatory mechanisms by which SYISL is regulated at the transcriptional level, we conducted luciferase assays with four reporter constructs containing different fragments of SYISL promoter (the region between −2000 bp and +200 bp) (SI Appendix, Fig. S3A). Deletion of the region between −200 bp and +200 bp led to a significant decrease in luciferase activity (SI Appendix, Fig. S3A). One potential MyoD-binding site (E-box) exists in this region (SI Appendix, Fig. S3B), and overexpression of MyoD increased the luciferase activity of reporter construct D1 containing this E-box (SI Appendix, Fig. S3C). Meanwhile, knockdown and overexpression of MyoD significantly reduced and increased the endogenous SYISL expression level, respectively (SI Appendix, Fig. S3 D and E). ChIP results in proliferating and differentiated C2C12 cells showed that MyoD could bind specifically to the E-box between −200 bp and +200 bp (SI Appendix, Fig. S3F). Therefore, we concluded that SYISL is a direct target of the MyoD gene.
To screen the target genes regulated by SYISL, we designed three RNAi oligonucleotides to knock down the SYISL gene specifically; the siRNA-2 fragments had the highest interference efficiency (SI Appendix, Fig. S4A). We further transfected siRNA-2 to knock down the SYISL gene and induced cell differentiation for 2 d. SYISL gene expression was significantly reduced when β-actin, GAPDH, and 18s RNA were used as reference genes, as indicated by qPCR (SI Appendix, Fig. S4B). We next used microarrays to analyze genome-wide gene-expression changes after SYISL knockdown in C2C12 cells differentiated for 2 d. In total, 399 genes, including MyoG, Myh1, Myh2, Myh4, Myh7, Tnni1, and Mybpc2, were significantly up-regulated, and 635 genes were down-regulated after SYISL knockdown (fold change >2.0; P < 0.05) (SI Appendix, Fig. S4C and Dataset S7). According to GO enrichment analysis, the differentially expressed genes were mainly related to cellular processes, metabolic processes, biological regulation, and responses to stimuli (SI Appendix, Fig. S4 D and E). Pathway analysis indicated that the up-regulated genes were mainly enriched for muscle differentiation and disease-associated pathways, such as the calcineurin-signaling and chemokine-signaling pathways, dilated cardiomyopathy, and viral myocarditis (SI Appendix, Fig. S4F). The down-regulated genes, including CDKs, N-Ras, ZEB2, IGF2BP3, Ki67, and PCNA, were mainly involved in axon guidance, the cell cycle, and MAPK-signaling pathways (SI Appendix, Fig. S4G). To validate the microarray results, we used qPCR to confirm the expression changes of some differentially expressed genes after SYISL overexpression and knockdown in C2C12 cells; the results from both techniques were consistent (SI Appendix, Fig. S4 H–K). From the above results, we deduced that SYISL might be involved in the regulation of myoblast proliferation and differentiation.
SYISL Promotes Myoblast Proliferation and Fusion but Inhibits Myogenic Differentiation.
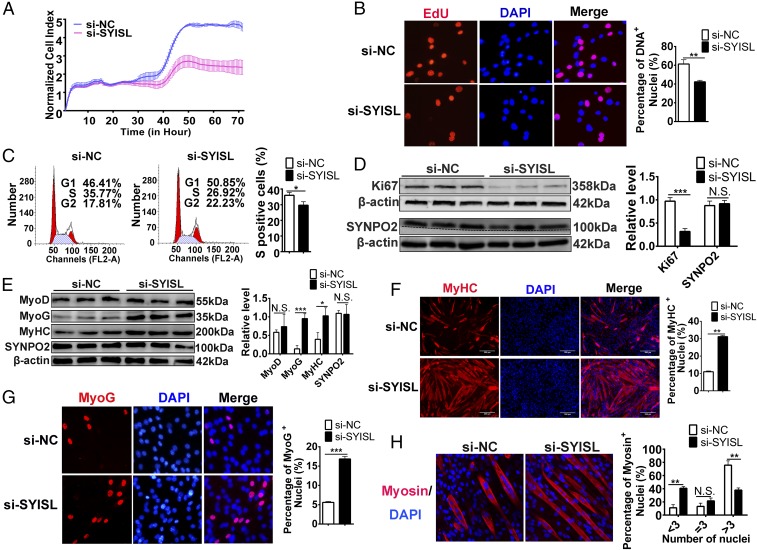
To determine whether the SYISL gene is involved in myogenesis, we used functional gain and loss to study the effects of SYISL on myoblast proliferation and differentiation in C2C12 cells. First, we used RTCA xCELLigence, 5-ethynyl-2′-deoxyuridine (EdU) staining, and flow cytometry assays to verify the potential roles of SYISL in C2C12 cell proliferation. The RTCA xCELLigence assay showed that SYISL knockdown significantly reduced the cell-proliferation capacity after 30 h of transfection, whereas SYISL overexpression markedly improved the cell-proliferation capacity after 40 h of transfection (Fig. 2A and SI Appendix, Fig. S5A). The EdU staining assay showed that SYISL knockdown significantly decreased EdU incorporation, and SYISL overexpression resulted in increased EdU positivity compared with that of the control (Fig. 2B and SI Appendix, Fig. S5B). Flow cytometry analysis indicated a significant reduction in the percentage of S-phase cells after SYISL knockdown, and opposite effects were observed after the overexpression of SYISL (Fig. 2C and SI Appendix, Fig. S5C). These results suggested that SYISL accelerates myoblast proliferation. In addition, we detected the expression of the key proliferation marker gene Ki67 after overexpression and knockdown of SYISL. The Ki67 protein was significantly down-regulated after SYISL knockdown, and SYISL overexpression promoted its protein expression, while no significant difference at the protein level was observed for SYNPO2 gene expression (Fig. 2D and SI Appendix, Fig. S5D).
Fig. 2.
SYISL promotes myoblast proliferation and fusion but inhibits myogenic differentiation. (A) RTCA xCELLigence analysis showed that C2C12 cell-proliferation capacity was significantly reduced after SYISL knockdown. (B) Representative photographs of EdU staining showing that SYISL knockdown significantly decreased EdU incorporation. Nuclei were stained with DAPI, magnification 40×. (C) Flow cytometry analysis showing that SYISL knockdown significantly decreased the percentage of S-phase cells. (D) Western blot results showing that SYISL knockdown significantly decreased the protein expression level of Ki67, while no significant difference for SYNPO2 protein expression was observed in proliferating C2C12 cells. The dotted line represents a dividing line. Below the dividing line is white space. (E) Western blot results showing that SYISL knockdown significantly increased the protein expression levels of MyoG and MyHC in C2C12 cells differentiated for 2 d but did not affect MyoD and SYNPO2 protein expression. (F) Representative photographs of MyHC immunofluorescence staining in C2C12 cells differentiated for 4 d showing that SYISL knockdown promoted myoblast differentiation. Positively stained cells were quantified, magnification 10×. (G) Representative photographs of MyoG immunofluorescence staining in C2C12 cells differentiated for 2 d showing that SYISL knockdown significantly promoted the MyoG protein expression level. Positively stained cells were quantified, magnification 40×. (H) Representative photographs of myosin immunofluorescence staining in C2C12 cells differentiated for 5 d showing that SYISL knockdown decreased the fusion rate of myoblasts, magnification 40×. The relative protein levels were normalized to those of the control β-actin. The data represent the means ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. NC, negative control; N.S. indicates statistical nonsignificance.
Next, we investigated the role of SYISL in myogenic differentiation. Real-time PCR, Western blot, and immunofluorescence staining were performed to detect the expression of myogenic marker genes including MyoD, MyoG, and MyHC. SYISL was successfully knocked down in C2C12 cells at D0, D2, D4, and D6 (SI Appendix, Fig. S6A). SYISL knockdown significantly promoted the myogenic differentiation of C2C12 cells, as demonstrated by the increased mRNA and protein expression of the myogenic marker genes MyoG and MyHC (Fig. 2 E–G and SI Appendix, Fig. S6 B and C). However, no significant change in protein expression of the MyoD gene was found after SYISL knockdown (Fig. 2E). Likewise, SYNPO2 expression was detected after SYISL knockdown; no significant differences were observed at either the mRNA or protein level (Fig. 2E and SI Appendix, Fig. S6D). To further confirm the knockdown results, we overexpressed the SYISL gene in C2C12 cells and then induced their differentiation. As expected, SYISL overexpression significantly decreased the expression of MyoG and MyHC at the mRNA and protein levels, but no significant changes in the expression of the MyoD and SYNPO2 genes were observed after SYISL overexpression (SI Appendix, Figs. S6 E–H and S7 A–C).
Finally, myosin immunofluorescence staining was used to analyze the function of SYISL in the regulation of myotube fusion. SYISL knockdown increased the proportion of myotubes with three or fewer nuclei (Fig. 2H) and decreased the mRNA expression of the fusion marker genes Myomaker and β-1integrin (SI Appendix, Fig. S7D), while SYISL overexpression increased the proportion of myotubes with more than three cell nuclei (SI Appendix, Fig. S7E) and promoted the mRNA expression levels of Myomaker and β-1 integrin (SI Appendix, Fig. S7F), suggesting that SYISL facilitates more myoblast fusion into one myotube. Moreover, we used the wound-healing test to determine the cell migration capacity at 12, 24, and 48 h after the knockdown or overexpression of SYISL in C2C12 cells. SYISL knockdown significantly promoted myoblast migration, and SYISL overexpression inhibited cell migration (SI Appendix, Fig. S8 A and B).
SYISL KO in Mice Significantly Increases Muscle Fiber Density, Muscle Mass, and Regeneration.
To determine the role of SYISL in muscle development at the individual animal level, we used CRISPR/Cas9-mediated genome editing to generate SYISL-KO mice. A 1,133-bp genomic region that contains most of the SYISL transcript was deleted, and different genotypes were identified by PCR (SI Appendix, Fig. S9A). SYISL-KO mice were healthy and manifested no significant differences in weight or growth rate compared with WT mice (SI Appendix, Fig. S9B). qPCR results showed that SYISL expression was barely detectable in the skeletal muscles of the SYISL-KO mice (SI Appendix, Fig. S9C). To validate the effects of SYISL depletion on muscle development, we examined the expression changes of the proliferation and myogenic marker genes in the skeletal muscles of 2-mo-old WT and SYISL-KO mice. Compared with WT mice, SYISL-KO mice displayed higher protein expression levels of MyoG and MyHC but lower protein expression levels of N-Ras (SI Appendix, Fig. S9D). Moreover, the protein expression of the SYNPO2 gene was not significantly altered (SI Appendix, Fig. S9D), a finding that was consistent with the results in C2C12 cells.
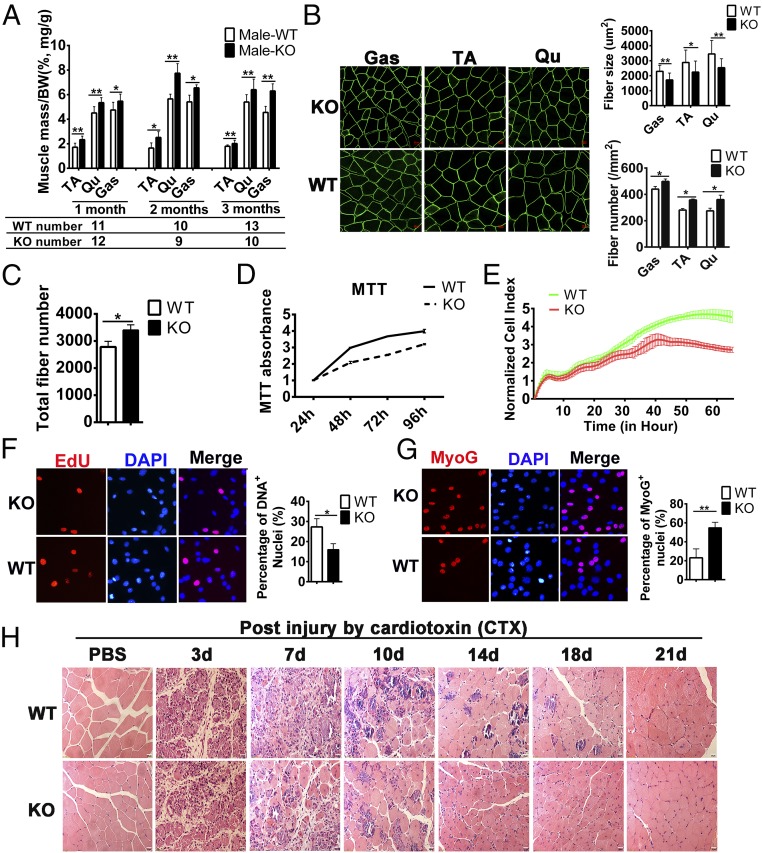
Although the SYISL-KO mice showed normal growth and body weight, the weights of their gastrocnemius (Gas), tibialis anterior (TA), and quadriceps (Qu) muscles in both males and females were significantly higher than those of WT mice (Fig. 3A and SI Appendix, Fig. S9E). Next, we used H&E staining and immunohistochemistry staining to assess changes in the number and size of myofibers. Compared with WT mice, SYISL-KO mice showed a significantly lower mean cross-sectional area of individual myofibers but a significantly higher number of myofibers/mm2 and a higher proportion of smaller myofibers (Fig. 3B and SI Appendix, Figs. S9 F–I and S10 A–E). Next, we scanned the whole cross-section of the TA muscle from SYISL-KO and WT mice and found that the TA muscle from SYISL-KO mice had a higher total fiber number than that of WT mice (Fig. 3C). To further confirm the roles of SYISL in cell proliferation, differentiation, and fusion, we isolated muscle primary myoblasts from the leg muscles of WT and SYISL-KO mice. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and RTCA xCELLigence assays showed that the proliferation capacity of SYISL-KO muscle primary myoblasts was significantly lower than that of WT cells (Fig. 3 D and E). Similarly, the EdU staining assay showed that SYISL KO in myoblasts significantly decreased EdU staining (Fig. 3F). Moreover, SYISL KO significantly decreased Ki67 and N-Ras gene mRNA expression but did not affect Pax7 gene expression (SI Appendix, Fig. S11A). Next, we analyzed the effects of SYISL deletion on the differentiation of primary myoblasts; the results showed that deletion of SYISL in primary myoblasts significantly promoted MyoG and MyHC mRNA expression, but the expression of MyoD was not significantly altered, a finding that was consistent with the results in C2C12 cells (SI Appendix, Fig. S11B). MyHC, MyoG, and myosin immunofluorescence staining further showed that SYISL KO in primary myoblasts significantly increased myogenic differentiation and the number of myotubes but decreased the myoblast fusion rate as judged by fewer cell nuclei in each myotube (Fig. 3G and SI Appendix, Fig. S11 C and D). These data suggested that SYISL promotes myoblast proliferation, increases myoblast fusion, and inhibits differentiation in vivo and in vitro.
Fig. 3.
SYISL KO in mice results in increased muscle fiber density, muscle mass, and regeneration. (A) The weights of the Gas, TA, and Qu muscles of male SYISL-KO mice were significantly higher than those of male WT mice. All the data were normalized to the body weight (mg/g). (B) Representative photographs of immunohistochemistry staining of dystrophin for Gas, TA, and Qu muscles from 2-mo-old SYISL-KO and WT mice. Compared with WT mice, SYISL-KO mice had lower average cross-sectional areas and more muscle fibers/mm2, magnification 40×. (C) SYISL-KO mice had more total muscle fibers than WT mice. The TA muscle was isolated from SYISL-KO and WT mice, and immunohistochemistry was performed using the anti-dystrophin antibody. The number of muscle fibers was measured by scanning the whole cross-section. (D–F) SYISL KO significantly decreased primary myoblast proliferation capacity as determined by MTT (D), RTCA xCELLigence analysis (E), and EdU staining (F), magnification 40×. (G) Representative photographs of MyoG immunofluorescence staining showing that SYISL KO in primary myoblasts significantly increased the myogenic differentiation, magnification 40×. (H) Representative photographs of H&E staining of GAS muscle at days 3, 7, 10, 14, 18, and 21 after injury showing that SYISL-KO mice with smaller myofibers complete muscle damage repair earlier than WT mice with larger myofibers. PBS was used as the control, magnification 40×. The data represent the means ± SD of at least three independent experiments. *P < 0.05, **P < 0.01.
To investigate the role of SYISL in postnatal muscle regeneration, we performed a cardiotoxin (CTX)-induced muscle-injury experiment in WT and SYISL-KO mice. H&E staining of muscle sections at different times of injury showed that at 14 and 18 d following CTX injection most of the inflammatory myofibers in SYISL-KO mice had been replaced by newly formed ones with centralized nuclei, but more inflammatory cells and necrotic myofibers still existed in WT mice. At day 21 after CTX injection, muscle regeneration and repair were completed in SYISL-KO mice, but newly formed myofibers identified by the presence of central nuclei were still present in WT mice. Moreover, the regenerated myofibers in SYISL-KO mice were denser and smaller than those in WT mice, which were similar to the uninjured state (Fig. 3H). To confirm these findings, we examined the expression of embryonic MyHC (eMyHC), a marker of muscle regeneration, at day 14 and day 21 after CTX injection. At these two stages, regenerating myofibers of WT mice exhibited higher eMyHC expression than those of SYISL-KO mice (SI Appendix, Fig. S12A). These results indicated that muscle regeneration in WT mice lags behind that of SYISL-KO mice. Muscle regeneration involves the activation and proliferation of satellite cells, followed by their terminal differentiation. To investigate how SYISL regulates muscle regeneration, we conducted Pax7 (a specific maker of muscle satellite cells), EdU, and MyoG staining to compare proliferation and differentiation capacities of satellite cells in the regenerating myofibers from WT and SYISL-KO mice at day 3 after injury. Consistent with the above results in cells, the percentage of proliferating muscle satellite cells (Pax7+/EdU+) decreased by about 27% in SYISL-KO mice compared with that in WT mice while no significant change was found in the percentage of satellite (Pax7+) cells (SI Appendix, Fig. S12B). In addition, the results of Pax7 and dystrophin staining in normal muscle tissues indicated that there was no significant difference in the total number of satellite cells in WT and SYISL-KO mice (SI Appendix, Fig. S12C). These results suggested that SYISL KO does not affect the Pax7 level but induces cell-cycle exit and prevents the expansion of satellite cells. Meanwhile, differentiated (MyoG+) cells increased by about 72% in SYISL-KO mice compared with WT mice (SI Appendix, Fig. S12D), suggesting an earlier myogenic differentiation potential of SYISL-KO satellite cells. Considering the opposing effects of SYISL on satellite cell proliferation and differentiation during muscle regeneration, we infer that the faster muscle-damage repair in SYISL-KO mice may be mainly attributable to the more efficient differentiation of SYISL-KO satellite cells. Additionally, it is also possible that the smaller myofibers in SYISL-KO mice require less time to regenerate than the larger myofibers in WT mice.
SYISL Interacts Directly with PRC2.
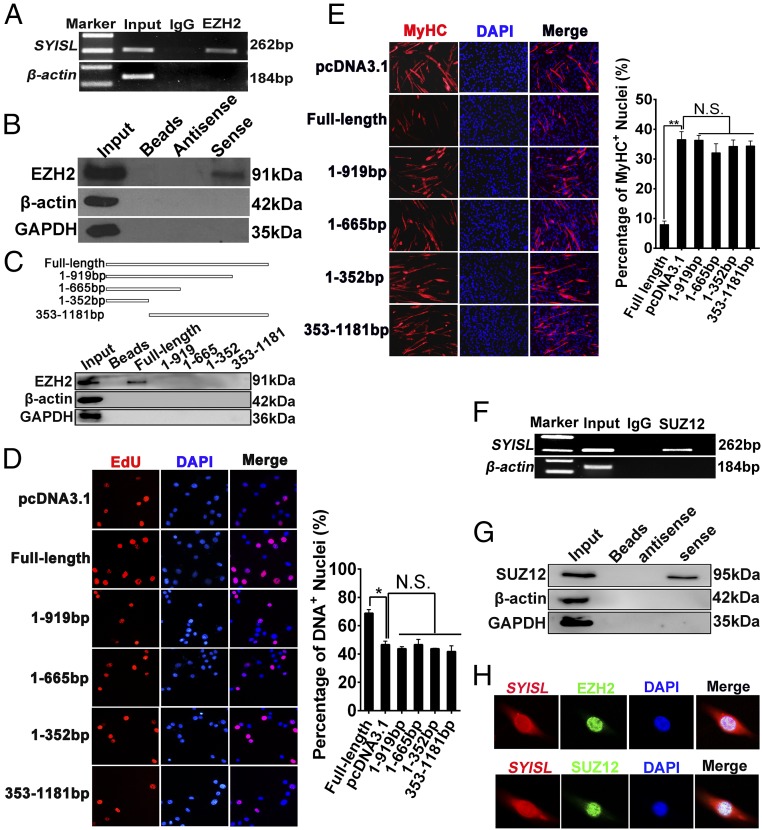
The nuclear localization of SYISL suggested that this lncRNA may recruit RNA-binding proteins to control the transcription of target genes. Thus, we attempted to identify the protein partners of SYISL. First, the potential SYISL-binding proteins were predicted using the catRAPID algorithm, and EZH2 was found to interact with SYISL (SI Appendix, Fig. S13A), a finding that was also confirmed by RNA–protein interaction prediction (RPISeq) (SI Appendix, Fig. S13B). Previous studies have shown that EZH2 interacts with various regulatory lncRNAs and plays important roles in myogenesis (25, 36). To confirm the roles of the EZH2 gene in myoblast proliferation and myogenic differentiation, we examined EZH2 protein expression during C2C12 cell differentiation and found that the EZH2 expression level was decreased with C2C12 cell differentiation (SI Appendix, Fig. S13C). Next, we overexpressed and knocked down the EZH2 gene in C2C12 cells. As expected, overexpression of the EZH2 gene significantly promoted cell proliferation and decreased myogenic differentiation (SI Appendix, Fig. S13 D–F), which were confirmed by the results of EZH2 gene knockdown (SI Appendix, Fig. S13G). To further elucidate the possible association of SYISL with EZH2, we compared the target genes of SYISL with EZH2 and H3K27me3 ChIP-seq data from C2C12 myoblasts. In total, 88 target genes regulated by SYISL overlapped with H3K27me3 target genes, and 36 genes overlapped with EZH2 target genes (SI Appendix, Fig. S13H and Dataset S8). Also, MyoG, Myh4, and MCK promoters were enriched by endogenous EZH2 and H3K27me3, as determined by ChIP in proliferating and differentiated C2C12 cells (SI Appendix, Fig. S13 I and J). To validate this interaction between SYISL and EZH2, we performed RNA immunoprecipitation (RIP) of EZH2. As expected, EZH2 pulled down SYISL in C2C12 cell lysates, suggesting that EZH2 interacts with SYISL in vivo (Fig. 4A). To further determine this specific interaction in vitro, we performed a biotinylated RNA-pulldown assay using biotinylated SYISL with biotinylated single-stranded antisense RNA as the control. Indeed, SYISL RNA could bind EZH2, but the antisense RNA strand pulled down no EZH2 protein (Fig. 4B). In conclusion, SYISL interacts specifically with EZH2 in C2C12 cells in vitro and in vivo.
Fig. 4.
SYISL interacts directly with PRC2. (A and B) Interaction between SYISL and EZH2 as determined by RIP (A) and biotin-labeled RNA pulldown (B). (C) The interaction of full-length and truncated SYISL (base pairs 1–352, 1–665, 1–919, and 353–1181) with EZH2 was determined by RNA pulldown. (D) Representative photographs of EdU staining in proliferating C2C12 cells and quantification showing that only full-length SYISL significantly promoted myoblast proliferation, magnification 40×. (E) Representative photographs of MyHC immunofluorescence staining in C2C12 cells differentiated for 2 d and quantification showing that only full-length SYISL significantly repressed myoblast differentiation, magnification 10×. (F and G) Interaction between SYISL and SUZ12 as determined by RIP (F) and biotin-labeled RNA pulldown (G). (H) RNA FISH and immunofluorescence staining showed that SYISL colocalized with EZH2 and SUZ12 in C2C12 cell nuclei, magnification 63×. GAPDH and β-actin were used as negative controls in RIP and RNA pulldown, respectively. The values are shown as means ± SD of three independent experiments. *P < 0.05, **P < 0.01. N.S. indicates statistical nonsignificance.
To determine the core protein-binding domain of SYISL, we constructed a series of truncated SYISL fragments. Unlike full-length SYISL, none of the truncated fragments could physically bind EZH2 (Fig. 4C). We next investigated the role of truncated fragments and full-length SYISL in myoblast proliferation and differentiation. Only the full-length SYISL significantly promoted cell proliferation, as shown by EdU staining (Fig. 4D), and significantly increased the expression of Ki67, N-Ras, and CDK6 genes (SI Appendix, Fig. S13K). Similarly, only full-length SYISL significantly repressed myogenic differentiation, as shown by MyHC immunofluorescence staining (Fig. 4E), and MyoG and MyHC gene expression (SI Appendix, Fig. S13L). These results supported the notion that full-length SYISL is required to exert its function in its physical interaction with EZH2. PRC2 is a multisubunit histone methyltransferase complex mainly including EZH2, SUZ12, EED, and RBBP4 (37, 38). To verify whether SYISL could interact with other members of the PRC complex, such as SUZ12, we performed RNA-pulldown and RIP experiments. The full-length SYISL interacted with SUZ12 (Fig. 4 F and G). This interaction of PRC2 and SYISL was also confirmed by their nuclear colocalization as shown by SYISL RNA FISH and EZH2/SUZ12 immunofluorescence staining (Fig. 4H). These results suggested that SYISL specifically recruits PRC2 to perform its epigenetic regulation roles.
SYISL Regulates Myogenesis by Recruiting PRC2 to the Promoters of Target Genes.
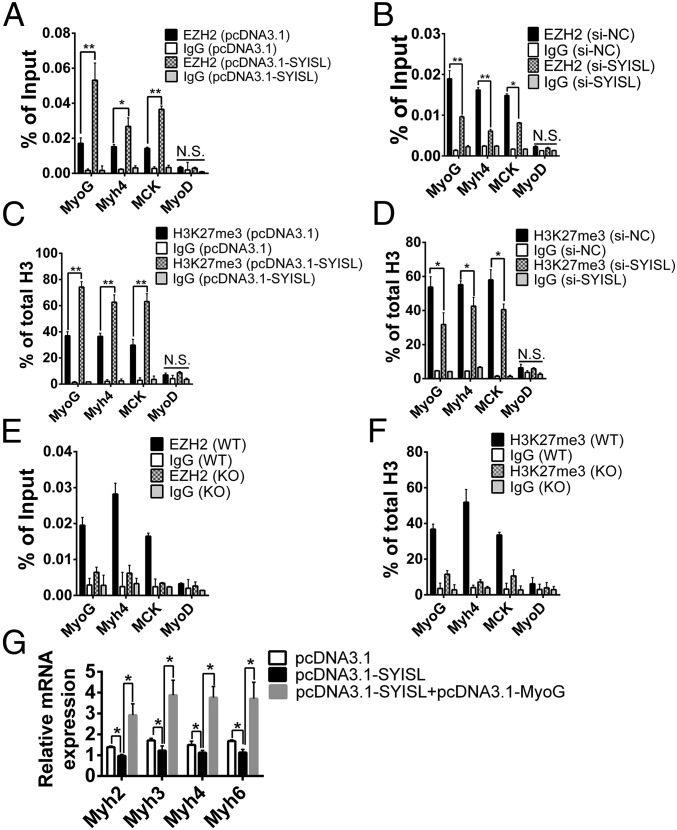
Because SYISL specifically interacts with PRC2, we investigated whether there is a mutual regulation relationship between SYISL and EZH2 in C2C12 cells. The results showed that SYISL knockdown or overexpression did not significantly influence EZH2 gene expression (SI Appendix, Fig. S14 A–D), nor was SYISL gene expression significantly altered by EZH2 knockdown or overexpression (SI Appendix, Fig. S14 E and F). Similarly, knockdown of SYISL did not influence SUZ12 gene expression (SI Appendix, Fig. S14G). These results suggested that SYISL regulates myogenesis through its interaction with PRC2 rather than by regulating EZH2 gene expression. Because EZH2 functions by binding to target promoters, we performed ChIP to elucidate whether SYISL affects the capacity of EZH2 to bind the promoters of its target myogenic genes such as MyoG, Myh4, and MCK in C2C12 cells. SYISL overexpression significantly increased and SYISL knockdown significantly decreased the enrichment of EZH2 at MyoG, Myh4, and MCK gene promoters (Fig. 5 A and B). Previous studies revealed that EZH2 repressed gene expression by binding chromatin and establishing H3K27me3 (10). Thus, we performed ChIP analysis for H3K27me3 after SYISL overexpression and knockdown in C2C12 cells, finding that SYISL overexpression significantly increased and SYISL knockdown significantly decreased H3K27me3 enrichment at the MyoG, Myh4, and MCK promoters (Fig. 5 C and D). To substantiate these observations, we conducted EZH2 and H3K27me3 ChIP assays in the primary myoblasts from SYISL-KO and WT mice and found that there was no significant enrichment of EZH2 and H3K27me3 at MyoG, Myh4, and MCK promoters in SYISL-KO primary myoblasts compared with levels in WT primary myoblasts (Fig. 5 E and F), indicating that SYISL KO results in the removal of EZH2 and H3K27me3 from target promoters. Because overexpression of SYISL led to the down-regulation of the MyoG expression level, we tested whether MyoG could efficiently rescue the expression of myogenic genes after overexpression of SYISL. Remarkably, MyoG overexpression rescued the inhibitory effects of myogenic gene expression mediated by SYISL overexpression (Fig. 5G and SI Appendix, Fig. S14 H and I). The SYISL regulation of myogenic differentiation through MyoG function was also confirmed by the bioinformatics analysis of the SYISL target genes with MyoG and MyoD ChIP-seq data from C2C12 myoblasts. In total, 422 target genes regulated by SYISL overlapped with both MyoG and MyoD target genes, 108 genes regulated by SYISL specifically overlapped with MyoG target genes, and only 26 genes that may be regulated by SYISL through other mechanisms or in an indirect manner overlapped with MyoD target genes (SI Appendix, Fig. S14J and Dataset S9). In conclusion, SYISL inhibits myogenic differentiation by recruiting PRC2 to the promoters of myogenic genes.
Fig. 5.
SYISL inhibits myogenic differentiation by recruiting EZH2 and H3K27me3 to the promoters of myogenic genes. (A) ChIP-qPCR results showing that SYISL overexpression significantly increased EZH2 enrichment at the MyoG, Myh4, and MCK promoters in C2C12 cells differentiated for 3 d. (B) ChIP-qPCR results showing that SYISL knockdown significantly decreased EZH2 enrichment at the MyoG, Myh4, and MCK promoters in C2C12 cells differentiated for 3 d. (C) ChIP-qPCR results showing that SYISL overexpression significantly increased H3K27me3 enrichment at the MyoG, Myh4, and MCK promoters in C2C12 cells differentiated for 3 d. (D) ChIP-qPCR results showing that SYISL knockdown significantly decreased H3K27me3 enrichment at the MyoG, Myh4, and MCK promoters in C2C12 cells differentiated for 3 d. (E and F) ChIP-qPCR results in primary myoblasts showing that there was no significant enrichment of EZH2 (E) or H3K27me3 (F) at MyoG, Myh4, and MCK promoters in SYISL-KO primary myoblasts differentiated for 3 d compared with levels in WT primary myoblasts. (G) qPCR results showing that MyoG overexpression rescued the inhibitory effects of myogenic gene expression mediated by SYISL overexpression. The relative RNA levels were normalized to those of the control β-actin. The values are shown as means ± SD of three independent experiments. *P < 0.05, **P < 0.01. N.S. indicates statistical nonsignificance.
Recent studies have reported that some lncRNAs, including Xist, APTR, and ANCR, can recruit EZH2 to epigenetically silence the cdkn1a (cyclin-dependent kinase inhibitor 1A, p21) gene, thereby promoting cell proliferation (39–41). p21 is an important cell-cycle regulator that restrains the cell cycle by inhibiting CDKs in mouse embryonic fibroblasts (42, 43). In this study, the inhibitory effects of p21 on cell proliferation were confirmed by EdU staining in C2C12 cells (SI Appendix, Fig. S14K). Thus, we deduced that SYISL may recruit EZH2 to inhibit p21 gene expression, thereby promoting myoblast proliferation. First, we investigated the effects of SYISL on the expression of p21 in C2C12 cells; the results indicated that SYISL significantly inhibited p21 gene expression and increased the expression of CDK6, its downstream target gene, at either the mRNA or the protein level (Fig. 6 A and B and SI Appendix, Fig. S14 L and M). Consistent with these results, deletion of SYISL in mice resulted in significantly increased p21 protein expression in muscle (Fig. 6C). Additionally, we analyzed the effects of the EZH2 gene on p21 gene expression in myoblasts. Similar to SYISL, EZH2 inhibited p21 gene expression and increased CDK6 gene expression (SI Appendix, Fig. S14N). Next, we analyzed the effects of SYISL on the capacity of EZH2 and H3K27me3 to bind the p21 gene promoter in C2C12 cells. As expected, SYISL overexpression significantly promoted EZH2 and H3K27me3 enrichment at the p21 promoter (Fig. 6 D and E), while SYISL knockdown reduced EZH2 and H3K27me3 enrichment at the p21 promoter (Fig. 6 F and G). Consistently, no enrichment of EZH2 and H3K27me3 at the p21 promoter was found in SYISL-KO primary myoblasts, while the p21 promoter was significantly enriched by EZH2 and H3K27me3 in WT primary myoblasts (Fig. 6 H and I). Taken together, these results indicate that SYISL inhibits p21 gene expression by recruiting EZH2 to the p21 promoter, ultimately resulting in failure to exit the cell cycle. To further confirm whether SYISL promotes the cell cycle through the p21 pathway, we cotransfected SYISL and p21 into C2C12 cells. The results showed that overexpression of SYISL induced the expression of CDK, N-Ras, and Ki67 genes. However, p21 overexpression significantly attenuated SYISL activity (Fig. 6J). These results demonstrated that the p21 gene is the main downstream target gene in the regulation of cell proliferation by SYISL.
Fig. 6.
SYISL promotes cell proliferation by epigenetically repressing the p21 gene. (A) Western blot results showing that SYISL knockdown significantly increased p21 protein expression and decreased CDK6 protein expression in proliferating C2C12 cells. The CDK6 gene is a known p21 target gene. (B) Western blot results showing that SYISL overexpression inhibited p21 protein expression and increased CDK6 protein expression in proliferating C2C12 cells. (C) Western blot results showing that SYISL KO in mice significantly increased p21 protein expression in muscle. The dotted line represents a dividing line. Above the dividing line is white space. (D and E) ChIP-qPCR results showing that SYISL overexpression significantly increased EZH2 (D) and H3K27me3 (E) enrichment at the p21 promoter in proliferating C2C12 cells. (F and G) ChIP-qPCR results showing that SYISL knockdown significantly decreased EZH2 (F) and H3K27me3 (G) enrichment at the p21 promoter in proliferating C2C12 cells. (H and I) ChIP-qPCR results showing that there was no significant enrichment of EZH2 (H) or H3K27me3 (I) at the p21 promoter in SYISL-KO primary myoblasts compared with levels in WT primary myoblasts. (J) qPCR results showing that p21 overexpression significantly attenuated SYISL activity. (K and L) ChIRP-qPCR results showing that SYISL could physically bind to the MyoG, Myh4, MCK, and p21 promoters, but not to the MyoD promoter, in C2C12 cells (K) and primary myoblasts (L). Probes against lacZ RNA were used as a negative control. Twelve probes tiling the full-length of SYISL RNA were separated into odd and even pools. (M and N) qPCR results showing that EZH2 gene overexpression significantly inhibited p21, MyoG, and Myh4 gene expression and increased CDK6 expression in WT primary myoblasts (M), but no significant changes were found in SYISL-KO primary myoblasts (N). The relative RNA and protein levels were normalized to those of the control β-actin. The values are shown as the means ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. NC, negative control; N.S. indicates statistical nonsignificance.
To confirm whether SYISL binds directly to the promoters of its target genes, we examined SYISL enrichment at the MyoG, p21, Myh4, MCK, and MyoD promoters by chromatin isolation by RNA purification (ChIRP) in C2C12 cells and primary myoblasts. The promoters of MyoG, p21, Myh4, and MCK, but not MyoD, were significantly enriched in SYISL ChIRP compared with the lacZ RNA controls (Fig. 6 K and L). These data confirmed that SYISL physically binds to the promoters of target genes and inhibits their transcription by recruiting PRC2. Finally, to test whether SYISL is required for the EZH2-mediated epigenetic silencing of muscle-specific genes and cell-cycle genes, we performed EZH2 gene overexpression experiments in primary myoblasts from WT and SYISL-KO mice. As expected, EZH2 gene overexpression significantly inhibited p21, MyoG, and Myh4 gene expression and increased CDK6 expression in WT primary myoblasts (Fig. 6M), while no significant differences in the expression levels of these genes were detected in SYISL-KO primary myoblasts (Fig. 6N).
Discussion
Recent studies have highlighted the importance of lncRNAs in diverse biological processes, most specifically in regulating cell proliferation and differentiation (44, 45). The expression levels of lncRNA are found to be more tissue- and cell type-specific than those of protein-coding genes, and lncRNAs have been shown to be differentially expressed across various stages of differentiation, indicating that they may be fine-tuners of cell fate (44). Current hotspots of lncRNA research focus mainly on tumorigenesis and stem cell pluripotency, while little is known about the role of lncRNAs in myogenesis, especially regarding their regulation of muscle mass. Using microarrays, Zhu et al. (29) identified 82 lncRNAs that are differentially expressed in original and differentiated muscle stem cells and revealed that the lncRNA lnc-mg regulates muscle mass at the individual animal level. In this study, we used microarray analysis to systematically analyze the expression profiles of lncRNAs during C2C12 cell differentiation and identified 356 persistently up-regulated and 266 persistently down-regulated lncRNAs. Our results provided a database to further screen functional lncRNAs that are involved in muscle fiber formation. Based on this database, we identified the lncRNA SYISL as a repressor of muscle development and found that SYISL KO significantly increased the number of smaller muscle fibers and muscle mass in mice. In general, muscle mass is determined by the number and size of muscle fibers. An increase in the size or cross-sectional area of muscle cells is deemed muscle hypertrophy, whereas hyperplasia involves an increase in the number of muscle cells (3, 46). Thus, we concluded that the increased muscle mass observed in SYISL-KO mice results mainly from muscle hyperplasia rather than from hypertrophy.
PRC2 is involved in various biological processes, including cell proliferation, differentiation, stem cell maintenance, and embryonic development, and myogenic differentiation is controlled by PRC2-mediated pathways (47–49). In undifferentiated myoblasts, PRC2 and HDAC1 are detected in the genomic regions of silent muscle-specific genes. By contrast, PRC2 dissociates from muscle-specific gene loci, and then MyoD and SRF are recruited to chromatin in differentiated myotubes (10). PRC-mediated gene silencing is mainly dependent on the regulation of EZH2-mediated H3K27me3 (50–52). In addition, the lncRNAs Xist, HOTAIR, H19, Chaer, Fendrr, Braveheart, and linc-YY1 are involved in the recruitment of PRC2 to the specific regulatory regions (25, 38, 53, 54). For example, the long intergenic noncoding RNA HOTAIR recruits PRC2 to specific target genes, leading to the trimethylation of H3K27 and epigenetic silencing of metastasis-suppressor genes (55). Linc-YY1 interacts with YY1/PRC2, evicts YY1/PRC2 from target promoters, and activates the expression of target genes (25). Here, we located EZH2-binding peaks within the promoters of 36 genes regulated by SYISL. Functional analysis further confirmed that SYISL recruits PRC2 to the promoters of its target genes and inhibits their expression through EZH2-mediated H3K27me3. Meanwhile, our results indicated that SYISL is indispensable for the repression of myogenic gene expression mediated by PRC2. Previous studies showed that mice with conditional knockout (cKO) of EZH2 in satellite cells have reduced muscle mass with smaller myofibers and compromised muscle regeneration (12), the opposite of the muscle phenotype of SYISL-KO mice. EZH2 cKO in mice results in impaired satellite-cell proliferation characterized by a reduction of Pax7+ cells and derepression of genes expressed in nonmuscle cell lineages (12), which may account for the reduction of muscle mass and regeneration. Interestingly, SYISL-KO mice show significantly increased muscle fiber density and muscle mass although myoblast proliferation is partly arrested. We speculate that the increased muscle density and mass of SYISL-KO mice are mainly caused by the elimination of SYISL’s inhibition of the expression of myogenic genes such as MyoG. Indeed, because the SYISL expression level increases significantly with myogenic differentiation in normal conditions, SYISL may play more important roles in the inhibition of myogenic differentiation than in the promotion of myoblast proliferation by recruiting EZH2 to target gene promoters or other mechanisms. Moreover, full-length SYISL is required for its physical interaction with EZH2 to repress the expression of target genes. Recent studies have reported that secondary and tertiary lncRNA structures might be correlated with their biological functions and protein-binding potential (56). Thus, we assume that full-length SYISL may be necessary for the formation of secondary or higher-order structures to interact with its binding proteins. Our results supply evidence that lncRNAs are involved in the PRC2-mediated epigenetic regulation of myoblast proliferation and differentiation.
Myoblast fusion is also a critical process that contributes to muscle growth and regeneration and that is controlled by multiple molecular and signaling pathways, such as the Myomaker, Kif2, Cav2, Trio, Cdh18, Itgb1, MAPK, Wnt, and calcineurin-NFATc2 pathways (57). In this study, the microarray data and qPCR results showed that SYISL knockdown reduces the expression of some myocyte fusion-associated genes, including Myomaker, β-1 integrin, Kif2, Cav2, Trio, Cdh18, and Itgb1. However, no EZH2-binding peaks exist within the promoters of these genes, according to the EZH2 and H3K27me3 ChIP-seq data. Additionally, only a subset of genes is coregulated by the SYISL and EZH2 genes. Thus, we hypothesize that during myogenesis SYISL may regulate the expression of some target genes, particularly genes related to myoblast fusion, through other mechanisms. lncRNAs were found to interact with multiple proteins (58). For example, Xist interacts with 81 proteins from the chromatin-modification, nuclear-matrix, and RNA-remodeling pathways (59). Similarly, like PRC2, other unknown proteins may participate in SYISL-mediated myogenesis. Beyond that, lncRNAs have been shown to function as miRNA molecular sponges to affect the expression of miRNA target genes (34). For instance, MALAT1 has been identified as a miR-133 molecular sponge (60), and lnc-mg functions as a competing endogenous RNA for miR-125b (29). Bioinformatics analysis showed the SYISL sequence also contains multiple potential miRNA-binding sites, such as sites for miR-1, miR-125, miR-214, miR-133, and miR-124, which are involved in the regulation of myogenic differentiation (61). Therefore, SYISL could feasibly exert its function by acting as miRNA sponge.
Compared with protein-coding genes, lncRNAs show lower conservation across species, especially in their nucleotide sequences. However, recent studies have indicated that lncRNAs exhibit cross-species conservation of their genomic position (62). For example, LncMyoD is located ∼30 kb upstream of the mouse MyoD gene, and in the human genome a conserved lncRNA (termed “hLncMyoD”) was identified ∼20 kb upstream of the human MyoD gene (26). Likewise, both human and mouse linc-YY1 are transcribed from the upstream genomic regions of YY1 (25). Bioinformatics analysis indicated that the human lncRNA AK021986 is transcribed from the fourth intron of the SYNPO2 gene (SI Appendix, Fig. S15A). We next analyzed the expression pattern of AK021986 in human normal tissues using the AnnoLnc web server and found that AK021986 is highly expressed in skeletal muscle (SI Appendix, Fig. S15B), a pattern similar to that of mouse SYISL. In addition, human AK021986 is mainly expressed in the nuclei of human ES cells and SK.N.SH cells, as determined using the lncATLAS annotation database (SI Appendix, Fig. S15 C and D). Recent studies have revealed that PRC2-binding lncRNAs have distinctive and evolutionarily conserved sequence features across species (63). Thus, we analyzed the binding capacity of AK021986 and PRC2 using the lncPro calculation method (64). Interestingly, human AK021986 shows a higher interaction score with PRC2 than the known PRC2-binding lncRNA HOTAIR (SI Appendix, Fig. S15E). Considering the important roles of SYISL in muscle development, further elucidation of the function and mechanism of AK021986 in human muscle development will be worthwhile.
In summary, we identify an lncRNA, SYISL, and propose a mechanistic model to elucidate its role in the regulation of muscle fiber formation through PRC2-mediated epigenetic silencing (SI Appendix, Fig. S16). At the individual animal level, SYISL cKO in mice leads to increased muscle fiber density and muscle mass. Mechanistically, on the one hand, SYISL recruits PRC2 to repress p21 gene expression, resulting in failure to exit the cell cycle and promotion of myoblast proliferation. On the other hand, SYISL guides PRC2 to repress the expression of muscle-specific genes, such as MyoG and Myh4, and restrains myoblast differentiation. Meanwhile, SYISL increases the rate of myocyte fusion by other unknown mechanisms.
Materials and Methods
The lncRNA SYISL was screened using microarray analysis, and its function was identified by gene overexpression and interference experiments in myoblasts. SYISL-KO mice were generated using the CRISPR/Cas9 system. Muscle fiber and regeneration phenotypes were measured to verify the effects of SYISL on myogenesis. Animal experiments were conducted based on the National Research Council Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee at Huazhong Agricultural University. The molecular mechanism by which SYISL regulates target gene expression was examined by RIP, RNA pulldown, ChIRP, ChIP, and other methods. See SI Appendix for detailed information.
Supplementary Material
Acknowledgments
We thank Dr. Wang Fengchao for helping in the generation of SYISL-KO mice and Dr. Wang Heng for helping in muscle regeneration experiments. This work was supported by National Project for Breeding of Transgenic Pig Grants 2016ZX08006-002 and 2018ZX080102B-002, Fundamental Research Funds for Central Universities Grant 2662018PY045, the National Natural Science Foundation of China Grant 91440114, and the Agricultural Innovation Fund of Hubei Province (B.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801471115/-/DCSupplemental.
References
- 1.Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev. 2006;16:525–532. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 3.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12:349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 4.Liu QC, et al. Comparative expression profiling identifies differential roles for myogenin and p38α MAPK signaling in myogenesis. J Mol Cell Biol. 2012;4:386–397. doi: 10.1093/jmcb/mjs045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albini S, et al. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Rep. 2013;3:661–670. doi: 10.1016/j.celrep.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perdiguero E, Sousa-Victor P, Ballestar E, Muñoz-Cánoves P. Epigenetic regulation of myogenesis. Epigenetics. 2009;4:541–550. doi: 10.4161/epi.4.8.10258. [DOI] [PubMed] [Google Scholar]
- 7.Bharathy N, Ling BM, Taneja R. Epigenetic regulation of skeletal muscle development and differentiation. Subcell Biochem. 2013;61:139–150. doi: 10.1007/978-94-007-4525-4_7. [DOI] [PubMed] [Google Scholar]
- 8.Asp P, et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci USA. 2011;108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol. 2007;179:1399–1412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stojic L, et al. Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenetics Chromatin. 2011;4:16. doi: 10.1186/1756-8935-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juan AH, et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodhouse S, Pugazhendhi D, Brien P, Pell JM. Ezh2 maintains a key phase of muscle satellite cell expansion but does not regulate terminal differentiation. J Cell Sci. 2013;126:565–579. doi: 10.1242/jcs.114843. [DOI] [PubMed] [Google Scholar]
- 14.Brockdorff N, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, et al. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J Cell Biol. 2014;204:61–75. doi: 10.1083/jcb.201304152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabory A, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 17.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu S, Shan G. LncRNAs in stem cells. Stem Cells Int. 2016;2016:2681925. doi: 10.1155/2016/2681925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Zhang S. Long noncoding RNAs in cell differentiation and pluripotency. Cell Tissue Res. 2016;366:509–521. doi: 10.1007/s00441-016-2451-5. [DOI] [PubMed] [Google Scholar]
- 20.Bhan A, Mandal SS. Long noncoding RNAs: Emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. 2014;9:1932–1956. doi: 10.1002/cmdc.201300534. [DOI] [PubMed] [Google Scholar]
- 21.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neguembor MV, Jothi M, Gabellini D. Long noncoding RNAs, emerging players in muscle differentiation and disease. Skelet Muscle. 2014;4:8. doi: 10.1186/2044-5040-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015;25:335–350. doi: 10.1038/cr.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou L, et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun. 2015;6:10026. doi: 10.1038/ncomms10026. [DOI] [PubMed] [Google Scholar]
- 26.Gong C, et al. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev Cell. 2015;34:181–191. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Mueller AC, et al. MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol Cell Biol. 2015;35:498–513. doi: 10.1128/MCB.01079-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X, et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat Commun. 2017;8:14016. doi: 10.1038/ncomms14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu M, et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat Commun. 2017;8:14718. doi: 10.1038/ncomms14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinet C, et al. H19 controls reactivation of the imprinted gene network during muscle regeneration. Development. 2016;143:962–971. doi: 10.1242/dev.131771. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, et al. Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov. 2017;3:17002. doi: 10.1038/celldisc.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weirick T, John D, Dimmeler S, Uchida S. C-It-Loci: A knowledge database for tissue-enriched loci. Bioinformatics. 2015;31:3537–3543. doi: 10.1093/bioinformatics/btv410. [DOI] [PubMed] [Google Scholar]
- 33.Zeng W, et al. Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucleic Acids Res. 2016;44:e158. doi: 10.1093/nar/gkw739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Engreitz JM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin CY, Cai H, Qing HR, Li L, Zhang HP. Recent advances on the role of long non-coding RNA H19 in regulating mammalian muscle growth and development. Yi Chuan. 2017;39:1150–1157. doi: 10.16288/j.yczz.17-193. [DOI] [PubMed] [Google Scholar]
- 37.Pereira JD, et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci USA. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA. 2015;21:2007–2022. doi: 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu T, Jiang W, Fan L, Gao Q, Li G. Upregulation of long noncoding RNA Xist promotes proliferation of osteosarcoma by epigenetic silencing of P21. Oncotarget. 2017;8:101406–101417. doi: 10.18632/oncotarget.20738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang F, Peng H. LncRNA-ANCR regulates the cell growth of osteosarcoma by interacting with EZH2 and affecting the expression of p21 and p27. J Orthop Surg Res. 2017;12:103. doi: 10.1186/s13018-017-0599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Negishi M, et al. A new lncRNA, APTR, associates with and represses the CDKN1A/p21 promoter by recruiting polycomb proteins. PLoS One. 2014;9:e95216. doi: 10.1371/journal.pone.0095216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Latres E, et al. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 2000;19:3496–3506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grange M, et al. Control of CD8 T cell proliferation and terminal differentiation by active STAT5 and CDKN2A/CDKN2B. Immunology. 2015;145:543–557. doi: 10.1111/imm.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Tian H, Yang J, Gong Z. Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. DNA Cell Biol. 2016;35:459–470. doi: 10.1089/dna.2015.3187. [DOI] [PubMed] [Google Scholar]
- 46.Otto A, Patel K. Signalling and the control of skeletal muscle size. Exp Cell Res. 2010;316:3059–3066. doi: 10.1016/j.yexcr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 47.Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- 48.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 49.Khan AA, Lee AJ, Roh TY. Polycomb group protein-mediated histone modifications during cell differentiation. Epigenomics. 2015;7:75–84. doi: 10.2217/epi.14.61. [DOI] [PubMed] [Google Scholar]
- 50.Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 52.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z, et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grote P, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li R, Zhu H, Luo Y. Understanding the functions of long non-coding RNAs through their higher-order structures. Int J Mol Sci. 2016;17:E702. doi: 10.3390/ijms17050702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hindi SM, Tajrishi MM, Kumar A. Signaling mechanisms in mammalian myoblast fusion. Sci Signal. 2013;6:re2. doi: 10.1126/scisignal.2003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chu C, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han X, Yang F, Cao H, Liang Z. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015;29:3054–3064. doi: 10.1096/fj.14-259952. [DOI] [PubMed] [Google Scholar]
- 61.Luo W, Nie Q, Zhang X. MicroRNAs involved in skeletal muscle differentiation. J Genet Genomics. 2013;40:107–116. doi: 10.1016/j.jgg.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tu S, Yuan GC, Shao Z. The PRC2-binding long non-coding RNAs in human and mouse genomes are associated with predictive sequence features. Sci Rep. 2017;7:41669. doi: 10.1038/srep41669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu Q, et al. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genomics. 2013;14:651. doi: 10.1186/1471-2164-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.