Significance
The Hippo pathway is an important regulator of animal growth, and its effector, Yorkie (in flies) or YAP/TAZ (in mammals), drives the expression of genes needed for cell proliferation and survival. In an effort to identify new regulators of Yorkie, we performed a genetic modifier screen in Drosophila. In this screen, we identified the RNA-binding protein Hrb27C as a positive regulator of Yorkie activity that modulates its phosphorylation status. Additional experiments identified the Hrb27C interacting proteins Glorund, Halfpint, Squid, and Pabp2 as negative modulators of Yorkie activity. Our results identify a link between the Hippo pathway and RNA binding factors and deepen the knowledge of this important growth control pathway.
Keywords: Hippo pathway, Hrb27C, hnRNP, aPKC, RNA-binding proteins
Abstract
The Hippo tumor-suppressor pathway regulates organ growth, cell proliferation, and stem cell biology. Defects in Hippo signaling and hyperactivation of its downstream effectors—Yorkie (Yki) in Drosophila and YAP/TAZ in mammals—result in progenitor cell expansion and overgrowth of multiple organs and contribute to cancer development. Deciphering the mechanisms that regulate the activity of the Hippo pathway is key to understanding its function and for therapeutic targeting. However, although the Hippo kinase cascade and several other upstream inputs have been identified, the mechanisms that regulate Yki/YAP/TAZ activity are still incompletely understood. To identify new regulators of Yki activity, we screened in Drosophila for suppressors of tissue overgrowth and Yki activation caused by overexpression of atypical protein kinase C (aPKC), a member of the apical cell polarity complex. In this screen, we identified mutations in the heterogeneous nuclear ribonucleoprotein Hrb27C that strongly suppressed the tissue defects induced by ectopic expression of aPKC. Hrb27C was required for aPKC-induced tissue growth and Yki target gene expression but did not affect general gene expression. Genetic and biochemical experiments showed that Hrb27C affects Yki phosphorylation. Other RNA-binding proteins known to interact with Hrb27C for mRNA transport in oocytes were also required for normal Yki activity, although they suppressed Yki output. Based on the known functions of Hrb27C, we conclude that Hrb27C-mediated control of mRNA splicing, localization, or translation is essential for coordinated activity of the Hippo pathway.
The conserved Hippo tumor suppressor pathway is an important regulator of cell proliferation and apoptosis and a key component of organ growth control (1). The Hippo pathway comprises a kinase cascade at its core. In Drosophila, these include the Hippo (Hpo; MST1/2 in mammals) and Warts (Wts; LATS1/2 in mammals) kinases that phosphorylate and thereby inactivate the transcriptional cofactor Yorkie (Yki; YAP/TAZ in mammals) (1). When the Hippo pathway kinases are inactive and Yki is not phosphorylated, it localizes in nuclei and binds to the transcription factor Scalloped (Sd; TEAD1–4 in mammals) to drive expression of target genes such as expanded (ex), Drosophila inhibitor of apoptosis 1 (diap1), and myc. Upstream, the Hippo pathway is regulated by multiple external inputs, including signals from cell polarity, mechanical forces, and metabolic conditions (1).
The Hippo pathway is involved in the control of organ size and cell proliferation, and hyperactivation of Yki or YAP/TAZ causes overgrowth of multiple organs such as imaginal discs in Drosophila, and liver and heart in mice (1). In the mouse and fly intestine and in mammalian skin, Yki/YAP hyperactivation drives stem cell hyperproliferation and expansion of progenitor cell compartments (1). Coordinated regulation of Hippo pathway activity is thus essential for normal development and homeostasis (1, 2). Furthermore, activation of YAP contributes to the development of cancer: YAP and TAZ protein levels and nuclear localization are elevated in a variety of human cancers, including liver, lung, breast, skin, colon, and ovarian cancers, and knockdown of YAP can reverse cancer cell phenotypes in vitro and in vivo in murine and human cancer models (2). However, genetic aberrations that directly affect known Hippo pathway components are rare in most cancers (2). Thus, deregulation of the Hippo pathway in cancer is likely mediated by unknown defects in regulatory mechanisms of the pathway.
Numerous upstream inputs that regulate Hippo kinase cascade activity have been identified, including integral membrane proteins, modulation of mechanical forces, the actin cytoskeleton, and protein complexes that establish apical-basal cell polarity, such as the Crumbs and atypical protein kinase C (aPKC) complex (1). However, how the actin cytoskeleton and cell polarity complexes regulate Hippo signaling is poorly understood (1, 2). Thus, unknown mechanisms likely exist that impact Yki/YAP/TAZ activity and output.
To identify new regulators of Hippo signaling, we used Drosophila to screen for mutations that dominantly suppress eye phenotypes caused by overexpression of aPKC during eye development. aPKC is a component of the apically localized Par complex, which is important for establishing and maintaining apical-basal polarity in epithelial cells (3). Loss of epithelial polarity is a common feature of cancer progression, and up-regulation of aPKC is observed in non-small-cell lung cancers (4) and in ovarian cancers, where it contributes to up-regulation of Cyclin E and poor prognoses (5–7). Overexpression of aPKC can promote Yki activity and tissue overgrowth in epithelial cells of Drosophila imaginal discs (8, 9), the precursors of many adult fly structures, as well as nuclear accumulation of YAP in mammalian cells by disrupting LATS activation (7, 10). In our screen for suppressors of aPKC-overexpression phenotypes, we identified mutations in the heterogeneous nuclear ribonucleoprotein (hnRNP) Hrb27C gene that strongly suppressed the aPKC-induced phenotypes. We found that Hrb27C regulates Yki activity and is required for Yki target gene expression and proper growth of imaginal discs. Thus, Hrb27C is a regulator of the Hippo pathway.
Results
Mutations in Hrb27C Dominantly Suppress the aPKC-Overexpression Phenotype in the Drosophila Eye.
We previously reported that hyperactivation of aPKC signaling by overexpression of an activated version of aPKC with a five-amino acid deletion in the autoregulatory pseudosubstrate domain (aPKCζ*) induced ectopic Cyclin E expression, entry into S-phase, and tissue overgrowth in larval eye and wing imaginal discs (5). These larval overgrowth phenotypes resemble those of Hippo pathway mutants that hyperactivate Yki and, indeed, the canonical Yki target gene ex was induced in aPKCζ*-overexpressing cells (Fig. 1 A and B) (8, 9). Similarly, overexpression of a constitutively active, membrane-targeted form of aPKC (aPKC-CAAX-WT) also induced Yki activity and tissue overgrowth in wing and eye discs (8, 9). Therefore, hyperactivation of aPKC promotes Yki activity in imaginal discs.
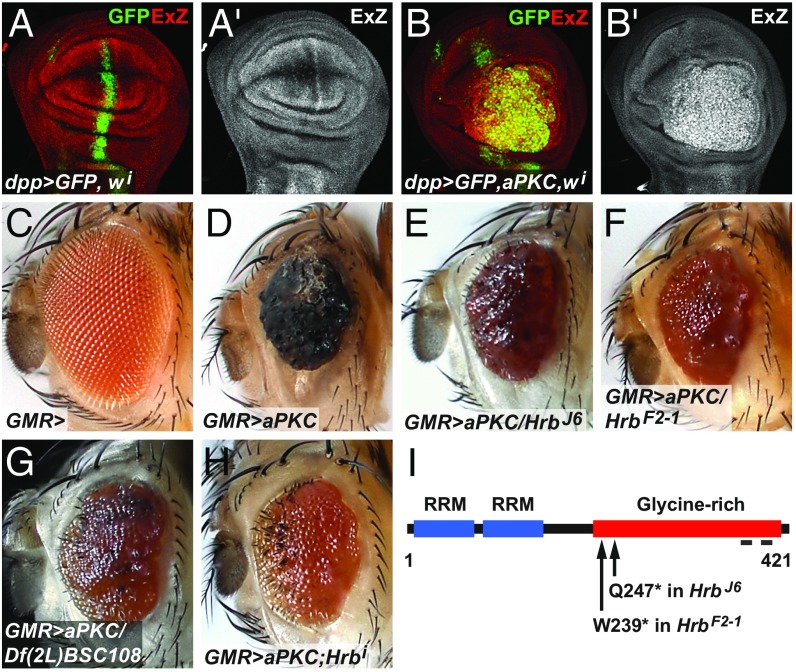
Fig. 1.
Hrb27C mutations are dominant suppressors of the aPKC overexpression phenotype in the Drosophila eye. (A and B) Confocal images of wing imaginal discs from third instar larvae expressing GFP (green or gray) in a stripe of cells along the anterior-posterior compartment boundary driven by dpp-Gal4 and white-RNAi (A) or aPKCζ* and white-RNAi (B). (C) Eye of a fly with one copy of the GMR-Gal4 transgene. (D) Eye of a fly with GMR-Gal4–driven overexpression of aPKCζ*. (E–G) GMR>aPKCζ* flies also carrying one copy of the allele Hrb27CJ6, Hrb27CF2-1, or Df(2L)BSC108 (which deletes Hrb27C). (H) GMR>aPKCζ* fly coexpressing Hrb27C-RNAi (v16041). (I) Diagram of the Hrb27C protein depicting its functional domains and the location of the Hrb27CF2-1 and Hrb27CJ6 mutations. The gray bars at the C-terminal end mark the locations of epitopes recognized by the Hrb27C polyclonal antibodies. RRM, RNA-recognition motif.
Overexpression of aPKCζ* during the late stages of eye development using the GMR-Gal4 driver produced viable adults with small, rough eyes, a consequence of deregulated cell proliferation and defects in differentiation and apical-basal cell polarity (5) (Fig. 1 C and D). This phenotype was sensitive to loss of Sd, the transcription factor partner of Yki (SI Appendix, Fig. S1 A and B), and we thus used it for a genetic modifier screen to identify new regulators of Yki activity (Fig. 1 E and F and SI Appendix, Fig. S1 for screen details). With this strategy, we identified a complementation group of two mutations (J6 and F2-1) that are new alleles of Hrb27C, also known as hrp48 (11). The Hrb27C gene encodes an RNA-binding protein of the hnRNP family and contains two RNA-recognition motifs and a glycine-rich motif that mediates protein–protein interactions (11). Sequencing of the Hrb27C gene from hemizygous J6 and F2-1 mutant animals revealed nonsense mutations at Gln247 and Trp239, respectively, predicting truncated proteins that lack the majority of the C-terminal glycine-rich region (Fig. 1I). The subcellular localization of Hrb27C was mainly cytoplasmic, but mitotic clones of the newly isolated mutants had strongly reduced Hrb27C levels (SI Appendix, Fig. S2 A–C). Notably, a deficiency that uncovers Hrb27C and Hrb27C knockdown by RNAi also suppressed the GMR>aPKCζ* phenotype (Fig. 1 G and H). Thus, the GMR>aPKCζ* phenotype depends on Hrb27C gene dose.
Hrb27C is conserved in humans, with DAZAP1 being the closest homolog (12), and is known to control diverse aspects of mRNA processing such as splicing, localization, and translation (12–17). Its function has been mostly studied in Drosophila oocytes, where it regulates osk and grk mRNA transport (13, 14, 17), and it is required for mushroom body formation (18) and Notch signaling (19, 20) in wing discs. However, Hrb27C has not previously been linked to Hippo signaling and growth control. Thus, we sought to identify its function in the Hippo pathway.
Hrb27C Is Required for Yki Target Gene Expression.
Because Hrb27C mutations dominantly suppressed the phenotypes caused by aPKCζ* overexpression, and because aPKCζ* activates Yki, we tested whether Hrb27C plays a role in aPKCζ*-driven overgrowth and in Yki target gene expression. We used the dpp-Gal4 and ptc-Gal4 drivers to drive ectopic expression of aPKCζ* and shRNAs in a stripe of cells along the anterior-posterior compartment boundary marked by coexpression of GFP (Fig. 2A) and used the hh-Gal4 driver to drive ectopic gene expression in the posterior compartment of imaginal discs (Fig. 2C). We depleted Hrb27C in aPKCζ*-overexpressing cells by coexpressing either of two different nonoverlapping genome-encoded shRNAs targeting the Hrb27C mRNA (Hrb27C-RNAi) and used ex-lacZ as a readout for Yki activity. Notably, expression of each Hrb27C-RNAi construct caused loss of Hrb27C protein (SI Appendix, Fig. S2 B and C), confirming that the RNAi constructs affect the Hrb27C gene. The expression of aPKCζ* induced an overgrowth of the GFP region and an increase in ex-lacZ expression (Fig. 1 A and B). Coexpression of Hrb27C-RNAi with aPKCζ* completely suppressed the aPKCζ*-driven overgrowth phenotype and the ectopic induction of ex-lacZ expression (Fig. 2 A and B). Notably, Hrb27C knockdown in wild-type cells using multiple drivers caused a strong reduction of ex-lacZ expression below normal expression levels in a cell autonomous manner (Fig. 2 A–D and SI Appendix, Fig. S2F). In addition, Hrb27C knockdown reduced the expression of other Yki target genes and readouts, including lacZ enhancer trap reporters for the diap1 and myc genes (1) (Fig. 2 F–I). Conversely, hh-Gal4-driven Hrb27C overexpression caused an increase in the compartment size and increased levels of ex-lacZ expression (Fig. 2E). Thus, Hrb27C is necessary and sufficient for Yki target gene induction in imaginal disc cells and is required for the elevated Yki output caused by aPKCζ* overexpression.
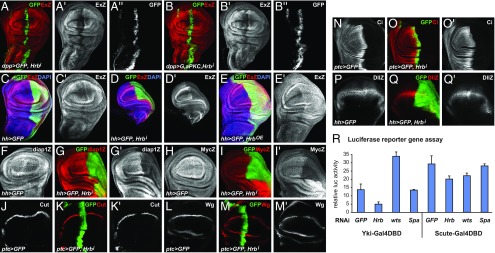
Fig. 2.
Loss of Hrb27C fucntion suppresses Yki activity. (A–Q) Confocal images of wing imaginal discs from third instar larvae expressing GFP (green or gray) in a stripe of cells along the anterior-posterior compartment boundary driven by dpp-Gal4 (A and B) or ptc-Gal4 (J–O), or in the posterior compartment driven by hh-Gal4 (C–I, P, and Q). In addition, discs expressed Hrb27C-RNAi (A, B, D, G, I, K, M, O, Q), Hrb27C-FLAG (E), and aPKCζ* and Hrb27C-RNAi (B). Discs were stained for β-galactosidase to detect ex-lacZ (ExZ), diap1-lacZ (diap1Z), myc-lacZ (MycZ), or Dll-lacZ (DllZ) expression, and Cut, Wg, or Cubitus interruptus (Ci), as indicated (red or gray). (R) Firefly luciferase expression levels of S2 cells transiently expressing Yki-Gal4DBD or Sc-Gal4DBD together with UAS-luc. Firefly expression levels were normalized against a constitutive Renilla luciferase. Cells were transfected with dsRNAs targeting GFP, Hrb27C, wts, or Spase25.
Hrb27C is involved in several aspects of mRNA production and regulation and could have general effects on gene expression. We thus tested effects of loss of Hrb27C function on the activity of transcriptional readouts of other signaling pathways. We found that knockdown of Hrb27C reduced the expression of the Notch target genes Cut and Wg at the presumptive wing margin (Fig. 2 J–M), consistent with previous reports (19, 20). However, the expression of Distal-less (Dll), Dll-lacZ, or Cubitus interruptus (a target of the Hedgehog pathway) and the expression of coexpressed GFP, lacZ, or white-RNAi were not affected by Hrb27C knockdown (Fig. 2 N–Q and SI Appendix, Fig. S2 H and I). Therefore, loss of Hrb27C does not cause a general defect in gene expression or protein production and its effect on the Hippo and other pathways must be due to specific functions of Hrb27C.
To further explore the requirement of Hrb27C for Yki activity, we tested the effect of Hrb27C knockdown on a Yki-dependent luciferase reporter in cultured cells. In this assay, a fusion protein between Yki and the Gal4 DNA binding domain (Yki-GDBD) drives expression of a UAS-luciferase reporter in cultured Drosophila Kc cells (21). Cotransfection of cells with dsRNA targeting Hrb27C resulted in a significant reduction of Yki-GDBD–driven luciferase expression, while dsRNA targeting GFP or an unrelated gene (Spase25) had no effect and knockdown of wts resulted in increased reporter expression as expected (Fig. 2R). To further test whether knockdown of Hrb27C generally affected transcription, we used an alternative GDBD construct in which Yki was replaced by Scute. Scute-GDBD is not regulated by the Hippo pathway but induces UAS-luciferase to about the same levels as Yki-GDBD (Fig. 2R) (22). Knockdown of Hrb27C had little effect on Scute-GDBD–induced reporter expression, similar to GFP, Spase25, or wts knockdown (Fig. 2R). Together, these results and the in vivo analysis indicate a specific role of Hrb27C in Yki-dependent gene expression, although Hrb27C likely affects other pathways in addition to the Hippo pathway.
Hrb27C Is Required for Tissue Growth.
Animals homozygous for either one of our Hrb27C alleles or heterozygous over a deficiency covering the Hrb27C region died as severely undersized third instars with small discs (Fig. 3A). Clones of Hrb27C mutant cells in imaginal discs were much smaller than their wild-type twin clones, which are born in the same cell division and serve as normally growing counterparts (Fig. 3 B and C). Hrb27C mutant cells in genetic mosaics were also underrepresented in adult eyes, and mosaic discs that were composed mainly of Hrb27C mutant cells produced eyes that were small and rough (Fig. 3 D and E). Similarly, Hrb27C knockdown in entire eye discs by eyeless-Gal4 or in the dorsal eye by DE-Gal4 resulted in adults that had markedly reduced eyes or small dorsal eye regions, respectively (Fig. 3 F and G and SI Appendix, Fig. S2 D and E), while knockdown with hh-Gal4 was pupal lethal and caused severe reduction of the posterior compartment in third instar discs (Fig. 2 C and D). These defects could be attributed to defects in cell proliferation, viability, or both. To determine whether Hrb27C is required for cell proliferation, we assayed BrdU/Edu (5-ethynyl-2′-deoxyuridine) incorporation and Cyclin E expression. Clones of Hrb27C mutant cells and posterior compartments of wing discs with knockdown of Hrb27C had reduced, but not abolished, BrdU incorporation and Cyclin E levels compared with wild-type cells (Fig. 3 H and I and SI Appendix, Fig. S3 A and B), most conspicuously observed in the second mitotic wave where cell cycles are synchronized (Fig. 3 H and I arrowheads). To assess whether Hrb27C is required for cell viability, we stained for apoptotic cells by anti-cleaved caspase-3 detection. Hrb27C mutant clones or knockdown of Hrb27C resulted in small clusters of apoptotic cells that were not present in controls (SI Appendix, Fig. S3 C and D) and in reduced cell density (SI Appendix, Fig. S4). In conclusion, cells lacking Hrb27C can still proliferate, although at a slower rate, and have reduced fitness, resulting in smaller clones and imaginal discs.
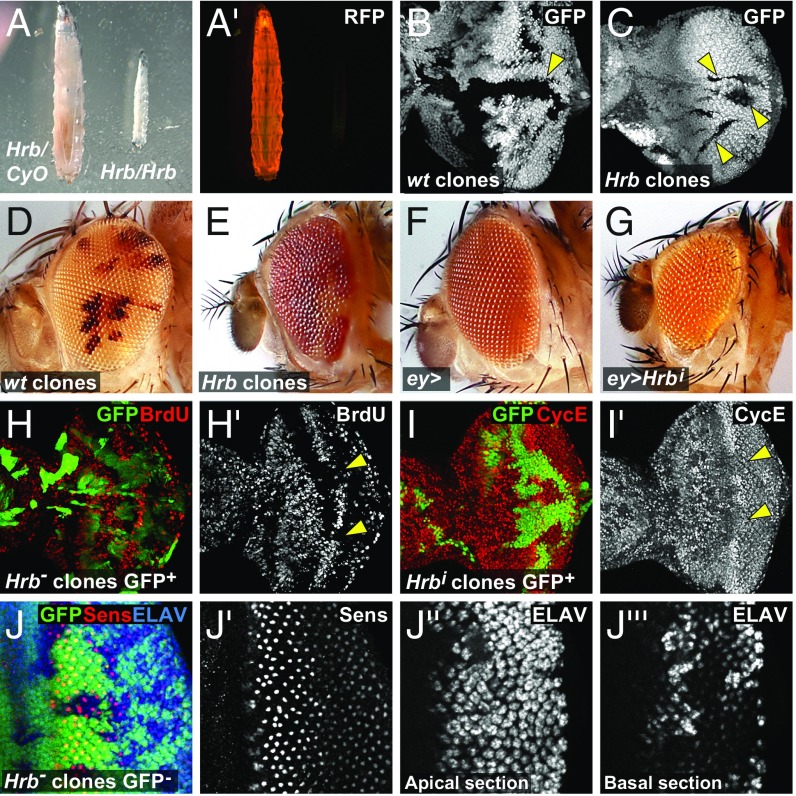
Fig. 3.
Hrb27C is required for tissue growth. (A) Hrb27CF2-1/Df(2L)Exel17029 (Right) and a Df(2L)Exel17029/CyO-RFP sibling (Left) third instar larva of the same age. Bright field picture (A) and RFP expression from the CyO-RFP balancer chromosome (A′). (B and C) Third instar eye discs with wild-type (wt) clones (B) and homozygous Hrb27CJ6 mutant clones (C) (black, indicated with arrowheads) flipped against ubi-GFP. (D and E) Eyes of flies in which a wild-type chromosome (D) or a Hrb27CJ6 mutant chromosome (E) was flipped against a w+ marked chromosome with a cell lethal mutation. Homozygous wild-type or Hrb27CJ6 homozygous cells are light orange, and heterozygous cells are red. While wild-type cells make big patches, Hrb27CJ6 homozygous cells form only small patches and most of the eye is composed of heterozygous red cells. (F) Eye of a fly with an ey-Gal4 driver (control). (G) Eye of a fly with ey-Gal4–driven Hrb27C-RNAi expression. (H and I) Eye discs with positively GFP-marked clones expressing Hrb27C-RNAi generated by MARCM stained for BrdU incorporation (red or gray, indicated with arrowheads) (H) or Cyclin E (CycE; red or gray, indicated with arrowheads) (I) . (J) Third instar eye disc with Hrb27CJ6 mutant clones marked by the lack of GFP expression and stained for Sens (red or gray) and the neuronal marker ELAV (blue or gray).
To investigate whether Hrb27C influences cellular differentiation, we monitored expression of markers for photoreceptor R8 specification (Sens) and neuronal differentiation (ELAV and 24B10, which recognizes the terminal differentiation marker Chaoptin) in eye discs with Hrb27C mutant clones. In wild-type discs, Sens expression commences in small clusters of cells in the morphogenetic furrow that then resolve into single cells destined to become R8 photoreceptors, while ELAV and 24B10 are expressed in differentiating neurons after their specification posterior to the morphogenetic furrow (Fig. 3J and SI Appendix, Fig. S3 E and F). R8 specification and neuronal patterning was typical in Hrb27C mutant cells (Fig. 3J), although ELAV-positive nuclei were displaced basally in the epithelia (Fig. 3J) and more-posterior clones had normal levels of ELAV and 24B10 expression (Fig. 3J and SI Appendix, Fig. S3 E and F). This indicates that Hrb27C mutant cells can survive and undergo differentiation but display mild defects in morphogenesis. Altogether, these data indicate that Hrb27C is essential for cell fitness and that its loss decreases cell proliferation rate and viability and causes morphological defects in differentiating cells.
Hrb27C Acts Genetically Downstream of Hpo and Wts but Upstream of Yki.
Our epistasis experiments place Hrb27C downstream of aPKC to control the expression of Yki target genes. To determine where Hrb27C intersects the Hippo pathway, we performed genetic epistasis experiments between Hrb27C and different Hippo pathway components. First, we tested whether Hrb27C was required for the up-regulation of ex-lacZ in cells with hpo or wts knockdown. Knockdown of hpo or wts led to an increase in ex-lacZ expression (Fig. 4 A, C, and E), but combined knockdown of hpo or wts with Hrb27C resulted in ex-lacZ levels that were lower than those in wild-type cells and resembled the phenotype caused by Hrb27C-RNAi alone (Fig. 4 A–F). Thus, Hrb27C intersects the Hippo pathway genetically downstream of Hpo and Wts.
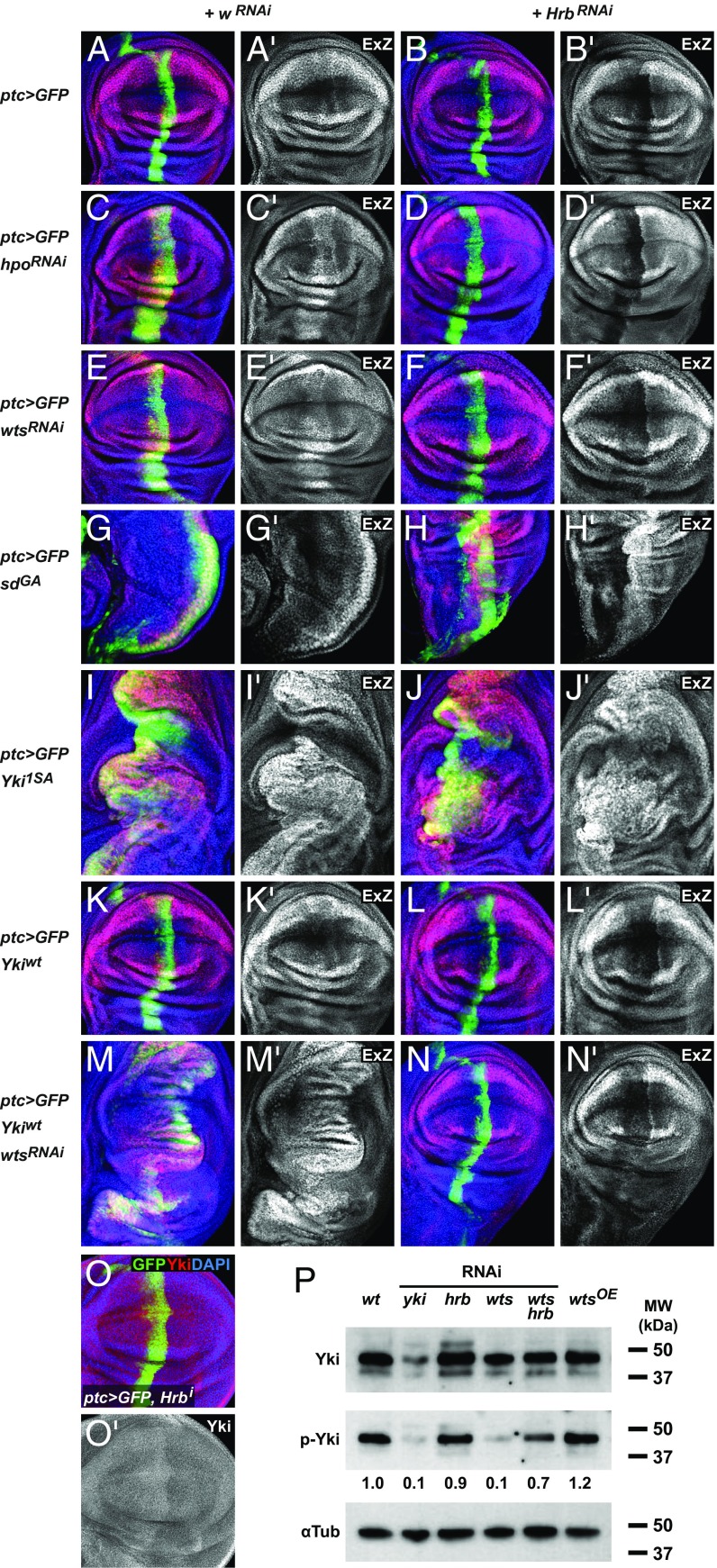
Fig. 4.
Hrb27C intersects the Hippo pathway between Wts and Yki. (A–N) Wing imaginal discs expressing ptc-Gal4, UAS-GFP (green) plus transgenes as indicated on the left, together with white-RNAi (A, C, E, G, I, K, and M) or Hrb27C-RNAi (B, D, F, H, J, L, and N). Discs were stained to detect the expression of the ex-lacZ reporter (ExZ; red or gray) and nuclei (DAPI; blue). Presumptive pouch regions are shown except for G and H, which show the notum regions, because SdGA impaired pouch development, likely by disrupting normal Sd function that is required for wing development independent of Yki. (O) Wing imaginal discs expressing ptc-Gal4, UAS-GFP, Hrb27C-RNAi stained to detect Yki (red or gray) and nuclei (DAPI; blue). (P) Western blot showing the effect of Hrb27C knockdown on Yki phosphorylation in combination with other genetic manipulations, as indicated.
Next, we explored interaction between Hrb27C and the Yki–Sd transcription factor complex, which acts downstream of Wts. By default, Sd acts as a repressor, but binding of Yki switches it to an activator (23). However, expression of constitutively active forms of Sd, where Sd was fused with the transcriptional activation of domain of Gal4 (SdGA), is sufficient to drive and rescue target gene expression in the absence of Yki (24). SdGA expression alone stimulated overgrowth and up-regulation of ex-lacZ as expected (Fig. 4G), and SdGA expression together with Hrb27C knockdown was still sufficient to induce ex-lacZ expression and tissue overgrowth (Fig. 4H, quantified in SI Appendix, Fig. S5). Thus, artificial Sd activation can rescue the growth and gene expression defects of Hrb27C knockdown, indicating that Hrb27C intersects the Hippo pathway upstream of Sd.
Next, we tested for interaction between Hrb27C and Yki, because Yki acts in-between Wts and Sd. Yki is phosphorylated by Wts, which causes its nuclear export and degradation and blocks Yki–Sd complex formation. Thus, overexpression of a constitutively active Yki mutant (Yki1SA), where the major Wts phosphorylation site has been mutated to alanine, cause increasingly severe overgrowth phenotypes and up-regulation of ex-lacZ expression in wing and eye discs (Fig. 4I) (25, 26). Notably, overexpression of Yki1SA rescued the growth phenotype and loss of ex-lacZ expression caused by Hrb27C knockdown (Fig. 4J and SI Appendix, Fig. S5). This was surprising because knockdown of wts, which also causes overgrowth and up-regulation of ex-lacZ, did not rescue the Hrb27C knockdown phenotype (Fig. 4F and SI Appendix, Fig. S5). This difference could be explained if Hrb27C regulated endogenous yki expression, because wts mutant cells express Yki from the endogenous locus while Yki1SA is expressed as an intronless cDNA from the artificial UAS promoter. However, Hrb27C knockdown did not reduce the amount of endogenous Yki protein (Fig. 4O and SI Appendix, Fig. S2G). Alternatively, Hrb27C may affect the phosphorylation status of Yki. To test this possibility, we monitored Yki phosphorylation in eye discs with knockdown of Hrb27C by Western blot (Fig. 4P). We found that Hrb27C knockdown did not significantly increase the fraction of phosphorylated Yki in otherwise wild-type discs. However, most Yki molecules may already be phosphorylated in wild-type cells, because even overexpression of Wts, which causes significant tissue reduction like Hrb27C knockdown (27), only led to a slight increase in Yki phosphorylation (Fig. 4P). To be able to better observe effects on Yki phosphorylation, we then depleted Hrb27C in eye discs with reduced levels of phosphorylated Yki. We thus used discs with wts knockdown as the baseline. We found that simultaneous knockdown of Hrb27C and wts caused a strong increase in Yki phosphorylation compared with wts knockdown alone (Fig. 4P).
We then wanted to genetically distinguish effects on yki gene expression from effects on Yki phosphorylation. We thus expressed Hrb27C-RNAi together with wild-type Yki using the same Gal4/UAS system and yki cDNA backbone as for the Yki1SA experiment (25). We found that knockdown of Hrb27C caused a decrease in ex-lacZ expression, even when wild-type Yki was overexpressed (Fig. 4 K and L). Next, to mimic the overgrowth triggered by Yki1SA, we added wts RNAi to activate the overexpressed wild-type Yki. Expression of wild-type Yki together with wts RNAi indeed paralleled the massive overgrowth and ex-lacZ induction caused by Yki1SA expression (Fig. 4M). Strikingly, however, knockdown of Hrb27C in this background fully suppressed the overgrowth and induction of ex-lacZ, in stark contrast to the overgrowth phenotype of Yki1SA expression with Hrb27C knockdown (Fig. 4N and SI Appendix, Fig. S5). This result shows that Hrb27C affects Yki activity, but only when it has intact Wts phosphorylation sites. Thus, Hrb27C affects the phosphorylation status of Yki.
Multiple hnRNPs Regulate Yki Activity.
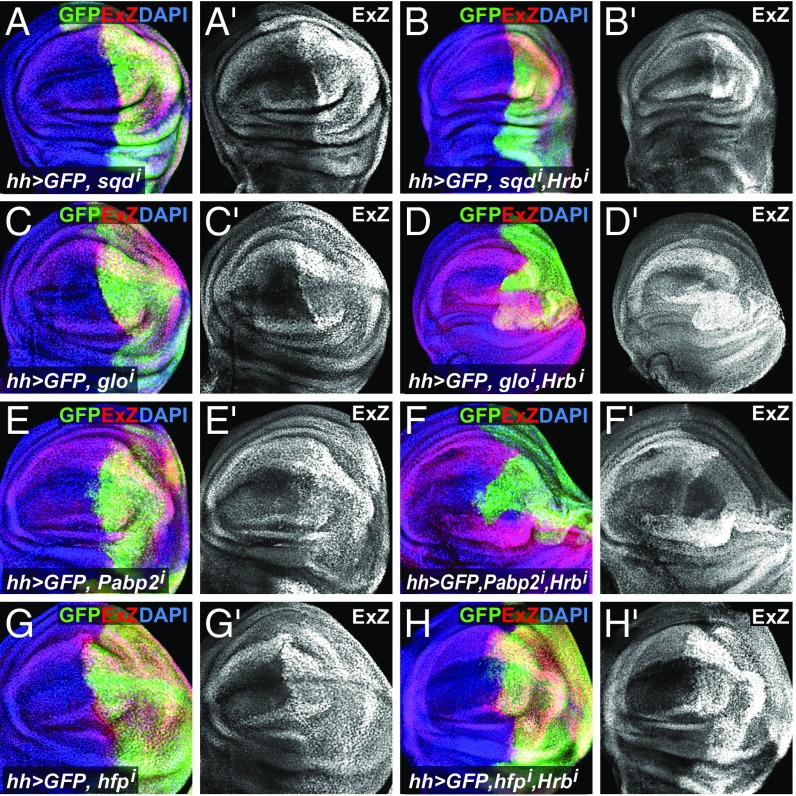
Hrb27C is known to regulate diverse aspects of mRNA biology in different protein complexes. Its known binding partners include Glorund (Glo), Squid (Sqd), Syncrip, IGF-II mRNA-binding protein, the translation initiation factor 4E-binding protein Cup, PolyA-binding protein (PABP), and many others (12, 16). To test whether any of these (or a subset of other RNA-binding proteins) regulate Yki function, we assayed ex-lacZ expression in wing discs with knockdown of members of different complexes and RNA-binding proteins that are related to Hrb27C using hh-Gal4 (SI Appendix, Fig. S6). While knockdown of most of these proteins had no effect, knockdown of sqd, glo, hfp, and the pAbp-related gene Pabp2 resulted in an increase of ex-lacZ expression similar to that observed with wts knockdown (Fig. 5 A, C, E, and G and SI Appendix, Fig. S6). Like Hrb27C knockdown, glo, hfp, and Pabp2, but not sqd, knockdown affected Cut levels in wing discs (SI Appendix, Fig. S7). To gain insight into the relationships of these proteins for the regulation of Yki activity, we performed an epistasis experiment between Hrb27C and Sqd, Glo, Pabp2, or Hfp. Loss of either Glo or Pabp2 required Hrb27C expression for the up-regulation of Yki activity, while sqd and hfp knockdown still up-regulated Yki activity in the absence of Hrb27C (Fig. 5 B, D, F, and H). We conclude that multiple RNA-binding proteins affect Yki activity and that Hrb27C acts genetically downstream of Glo and Pabp2 but upstream of Sqd and Hfp in the pathway that regulates Yki.
Fig. 5.
Yki regulation by RNA-binding proteins. Wing imaginal discs expressing hh-Gal4, UAS-GFP (green) plus sqd-RNAi (A), glo-RNAi (C), Pabp2-RNAi (E), hfp-RNAi (G), and in combination with Hrb27C-RNAi (B, D, F, and H, respectively). Discs were stained to detect the expression of the ex-lacZ reporter (ExZ; red or gray) and nuclei (DAPI; blue).
Discussion
Our data support a model in which the RNA-binding protein Hrb27C modulates the activity of the Hippo pathway effector Yki. First, Hrb27C loss of function suppressed Yki target gene expression and reduced tissue growth. Second, Hrb27C acts genetically downstream of Wts but upstream of Yki and affects Yki phosphorylation. Third, loss of Hrb27C function does not disable gene expression in general, although Hrb27C does affect the output of other signaling pathways such as Notch, Wingless, and Dpp signaling. Fourth, the Hrb27C-interacting proteins—hnRNPs Glo, Hfp, Pabp2, and Sqd—also regulate Yki target gene expression, either downstream or upstream of Hrb27C. Therefore, Hrb27C has a specific function required for normal Hippo pathway output.
How does Hrb27C affect Yki activity? Hrb27C has diverse functions: It functions as a splicing factor in the nucleus and is required for the polarized localization and translation of specific RNAs during oocyte maturation in Drosophila (12). Hrb27C is mainly cytoplasmic in imaginal disc cells, suggesting a function downstream of transcription and splicing, such as in mRNA translation or localization. Our data indicate that Hrb27C regulates the phosphorylation state of Yki; however, this must be indirect, as Hrb27C is neither a kinase nor a phosphatase. Notably, depletion of Hrb27C caused a strong increase in phospho-Yki levels, particularly in cells that had wts knockdown. Thus, Hrb27C may promote Yki dephosphorylation or modulate another kinase that can phosphorylate Yki, such as Tricornered, although we cannot exclude that it modulates Wts, with residual levels potentially present in wts knockdown cells. Systematic hnRNP purification schemes were recently used to identify mRNAs that are bound to Hrb27C in Drosophila S2 tissue culture cells and in vivo (28). These experiments identified about 3,000 mRNAs as targets of Hrb27C, including more than half of the known components of the Hippo pathway (SI Appendix, Fig. S8). However, how Hrb27C binding affects the function of these mRNAs and which of them mediate effects of Hrb27C on Yki activity is not known.
In Drosophila oocytes, Hrb27C interacts with the hnRNPs Sqd and Glo, among others. Knockdown of sqd, glo, hfp, or Pabp2 resulted in increased ex-lacZ expression, and knockdown of glo, hfp, and Pabp2, but not sqd, diminished expression of the Notch target gene Cut. Epistasis experiments revealed that Hrb27C is required for the up-regulation of Yki activity by glo and Pabp2 knockdown, but not by hfp or sqd knockdown. Thus, Hrb27C appears to act through different mechanisms in imaginal discs and oocytes, although it shared phenotypic specificity and acted downstream of glo and Pabp2, indicating that in discs, these factors interact in modulating mRNA biology to regulate the activity of the Hippo and Notch signaling pathways.
Hrb27C has several human homologs, including DAZAP1, which is important for growth during mouse development (29); members of the hnRNP A/B family that control cell proliferation in cancer cell lines (30); and Musashi proteins that play key functions in stem/progenitor cells in various murine tissues (31). Notably, several of these proteins have been linked to cancer phenotypes. The hnRNPs A1, A2, and B1 are frequently overexpressed in multiple tumor types (including breast and lung cancers) and are associated with poor prognosis (30, 32, 33). Furthermore, knockdown of hnRNP A2/B1 induces apoptosis in cancer cells, but not in normal cells (34), indicating that members of this hnRNP family might be novel and promising therapeutic targets. Further studies of the mechanisms by which Hrb27C regulates target gene expression will thus likely reveal novel mechanisms for the regulation of YAP/TAZ target genes in developmental and tumor contexts.
Materials and Methods
Methods for Drosophila culture and imaginal disc immunostaining were performed as described in ref. 22. Antibodies, shRNA, siRNA, and quantitative RT-PCR information is given in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank D. Rio, D. St. Johnston, K. Moberg, N. Dyson, N. Tapon, T. Yano, J. Jiang, D. Pan, H. Bellen, K. Basler, R. Durino, the Bloomington Drosophila Stock Center (NIH P40OD018537), the Vienna Drosophila Resource Center (stockcenter.vdrc.at/), the Transgenic RNAi Project at Harvard Medical School (NIH/NIGMS R01-GM084947), and the Developmental Studies Hybridoma Bank (University of Iowa) for fly stocks and antibodies. This work is funded by a Research Foundation–Flanders (www.foe.be/) Odysseus Group I Grant and Grant G.0640.13; NIH Grant R01 GM067997-06; and Cancer Prevention Research Institute of Texas Grant RP100773 (all to G.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807325115/-/DCSupplemental.
References
- 1.Johnson R, Halder G. The two faces of Hippo: Targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki A, Ohno S. The PAR-aPKC system: Lessons in polarity. J Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 4.Regala RP, et al. Atypical protein kinase C ι is an oncogene in human non-small cell lung cancer. Cancer Res. 2005;65:8905–8911. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- 5.Eder AM, et al. Atypical PKCiota contributes to poor prognosis through loss of apical-basal polarity and cyclin E overexpression in ovarian cancer. Proc Natl Acad Sci USA. 2005;102:12519–12524. doi: 10.1073/pnas.0505641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nanos-Webb A, et al. PKCiota promotes ovarian tumor progression through deregulation of cyclin E. Oncogene. 2016;35:2428–2440. doi: 10.1038/onc.2015.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar S, et al. PRKCI promotes immune suppression in ovarian cancer. Genes Dev. 2017;31:1109–1121. doi: 10.1101/gad.296640.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 9.Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev Biol. 2011;350:139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Archibald A, Al-Masri M, Liew-Spilger A, McCaffrey L. Atypical protein kinase C induces cell transformation by disrupting Hippo/Yap signaling. Mol Biol Cell. 2015;26:3578–3595. doi: 10.1091/mbc.E15-05-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matunis EL, Matunis MJ, Dreyfuss G. Characterization of the major hnRNP proteins from Drosophila melanogaster. J Cell Biol. 1992;116:257–269. doi: 10.1083/jcb.116.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piccolo LL, Corona D, Onorati MC. Emerging roles for hnRNPs in post-transcriptional regulation: What can we learn from flies? Chromosoma. 2014;123:515–527. doi: 10.1007/s00412-014-0470-0. [DOI] [PubMed] [Google Scholar]
- 13.Huynh J-R, Munro TP, Smith-Litière K, Lepesant J-A, St Johnston D. The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev Cell. 2004;6:625–635. doi: 10.1016/s1534-5807(04)00130-3. [DOI] [PubMed] [Google Scholar]
- 14.Yano T, et al. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev Cell. 2004;6:637–648. doi: 10.1016/s1534-5807(04)00132-7. [DOI] [PubMed] [Google Scholar]
- 15.Blanchette M, et al. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Mol Cell. 2009;33:438–449. doi: 10.1016/j.molcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson MR, et al. A multiprotein complex that mediates translational enhancement in Drosophila. J Biol Chem. 2007;282:34031–34038. doi: 10.1074/jbc.M706363200. [DOI] [PubMed] [Google Scholar]
- 17.Goodrich JS, Clouse KN, Schüpbach T. Hrb27C, Sqd and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development. 2004;131:1949–1958. doi: 10.1242/dev.01078. [DOI] [PubMed] [Google Scholar]
- 18.Bruckert H, Marchetti G, Ramialison M, Besse F. Drosophila Hrp48 is required for mushroom body axon growth, branching and guidance. PLoS One. 2015;10:e0136610. doi: 10.1371/journal.pone.0136610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suissa Y, et al. Hrp48 attenuates Sxl expression to allow for proper notch expression and signaling in wing development. Proc Natl Acad Sci USA. 2010;107:6930–6935. doi: 10.1073/pnas.0910570107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutta D, Paul MS, Singh A, Mutsuddi M, Mukherjee A. Regulation of notch signaling by the heterogeneous nuclear ribonucleoprotein Hrp48 and deltex in Drosophila melanogaster. Genetics. 2017;206:905–918. doi: 10.1534/genetics.116.198879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Sansores-Garcia L, et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koontz LM, et al. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev Cell. 2013;25:388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, et al. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin F, et al. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMahon AC, et al. TRIBE: Hijacking an RNA-editing enzyme to identify cell-specific targets of RNA-binding proteins. Cell. 2016;165:742–753. doi: 10.1016/j.cell.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu LC-L, et al. DAZAP1, an hnRNP protein, is required for normal growth and spermatogenesis in mice. RNA. 2008;14:1814–1822. doi: 10.1261/rna.1152808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han N, Li W, Zhang M. The function of the RNA-binding protein hnRNP in cancer metastasis. J Cancer Res Ther. 2013;9(Suppl):S129–S134. doi: 10.4103/0973-1482.122506. [DOI] [PubMed] [Google Scholar]
- 31.Horisawa K, Imai T, Okano H, Yanagawa H. The Musashi family RNA-binding proteins in stem cells. Biomol Concepts. 2010;1:59–66. doi: 10.1515/bmc.2010.005. [DOI] [PubMed] [Google Scholar]
- 32.Pino I, et al. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer. 2003;41:131–143. doi: 10.1016/s0169-5002(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 33.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu W-J, Liu H-L. Induction of pancreatic cancer cell apoptosis, invasion, migration, and enhancement of chemotherapy sensitivity of gemcitabine, 5-FU, and oxaliplatin by hnRNP A2/B1 siRNA. Anticancer Drugs. 2013;24:566–576. doi: 10.1097/CAD.0b013e3283608bc5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.