Significance
Chronic infection with the opportunistic pathogen Pseudomonas aeruginosa is the leading cause of lung transplant or death in cystic fibrosis (CF) patients. P. aeruginosa diversifies in the CF lung, although why this happens remains a mystery. We allowed P. aeruginosa to evolve in the laboratory under a range of conditions approximating the CF lung. The diversity of evolved populations was highest, and most closely resembled the range of phenotypes among clinical isolates, in environments resembling the spectrum of nutritional resources available in the CF lung. Our results point to the nutritional complexity of the CF lung as a major driver of diversification, and they suggest that diversity could be important in the development of chronic infections.
Keywords: experimental evolution, adaptation, diversity, pathogen evolution, chronic infection
Abstract
Chronic infection of the cystic fibrosis (CF) airway by the opportunistic pathogen Pseudomonas aeruginosa is the leading cause of morbidity and mortality for adult CF patients. Prolonged infections are accompanied by adaptation of P. aeruginosa to the unique conditions of the CF lung environment, as well as marked diversification of the pathogen into phenotypically and genetically distinct strains that can coexist for years within a patient. Little is known, however, about the causes of this diversification and its impact on patient health. Here, we show experimentally that, consistent with ecological theory of diversification, the nutritional conditions of the CF airway can cause rapid and extensive diversification of P. aeruginosa. Mucin, the substance responsible for the increased viscosity associated with the thick mucus layer in the CF airway, had little impact on within-population diversification but did promote divergence among populations. Furthermore, in vitro evolution recapitulated traits thought to be hallmarks of chronic infection, including reduced motility and increased biofilm formation, and the range of phenotypes observed in a collection of clinical isolates. Our results suggest that nutritional complexity and reduced dispersal can drive evolutionary diversification of P. aeruginosa independent of other features of the CF lung such as an active immune system or the presence of competing microbial species. We suggest that diversification, by generating extensive phenotypic and genetic variation on which selection can act, may be a key first step in the development of chronic infections.
The bacterium Pseudomonas aeruginosa is a globally ubiquitous, metabolically flexible, gram-negative opportunistic pathogen. While it is an important nosocomial pathogen causing a range of acute infections, P. aeruginosa is also commonly recovered from the airways of adult cystic fibrosis (CF) patients, where it causes chronic endobronchial infections in the majority of adult patients and is the leading cause of morbidity and mortality in this population (1–3). The majority of chronic CF lung infections are thought to be the result of colonization by P. aeruginosa from environmental sources, although highly transmissible epidemic strains are responsible for ∼25% of infections in Canadian patients (4). Once established, chronic infections can remain persistently associated with a host for decades, being virtually impossible to eradicate with standard antibiotic therapy (5).
The transition from environmental strain to one that causes chronic infection is characterized by a few repeatable phenotypic changes underlain by a much larger suite of genetic changes. Free-living environmental strains are typically motile, virulent, and nonmucoid, whereas strains isolated from chronically infected CF patients are often nonmotile, avirulent, mucoid, and highly antibiotic-resistant (6–9). These phenotypes represent parallel adaptations to a range of CF lung-specific stressors, including increased viscosity, osmotic stress, low oxygen, high concentrations of antibiotics, and immune system attack. This parallelism notwithstanding, the other striking feature common to P. aeruginosa isolates from CF patients is their diversity. Phenotypes resembling both environmental and chronically infected strains can persist in the same patient for long periods of time, and isolates from within (10–14) and among (15, 16) patients can be highly diverse, both phenotypically and genetically. This diversity makes it difficult to identify reliable markers for the onset of chronic infection (17) and predict clinical outcomes on the basis of single colony isolates alone. Indeed, the lack of correlation between the prevalence of a particular phenotype and clinical outcomes (13) hints that the diversity of the population, rather than the abundance of any particular phenotype, could be what makes CF lung infections by P. aeruginosa so recalcitrant to treatment (10).
Diversity in the CF lung typically evolves rapidly and de novo following colonization (10, 13, 18), with genetically and phenotypically diverse clones often persisting for years within the same host (8, 19, 20). This dynamic has many of the hallmarks of an adaptive radiation, the rapid diversification of a lineage into a range of niche specialist types. Both theory (21–23) and experiment (reviewed in refs. 24–27) suggest that adaptive diversification is most likely to occur when divergent selection, imposed by ecological opportunities in the form of vacant niche space or underutilized resources, is strong relative to the rate of dispersal. The extent of phenotypic divergence may be further exaggerated, or its rate accelerated, through ecological interactions such as resource competition or predation (28–31).
We suspect that the complex ecological conditions of the CF lung likely promote diversification in colonizing P. aeruginosa. The CF airway contains a rich spectrum of resources and nutrients that could provide ample ecological opportunity to drive specialization (13, 32, 33), and the thick mucus layer, a by-product of the impaired ability to transport chloride ions resulting from mutations in the CF transmembrane conductance regulator (CFTR) gene, serves to reduce dispersal among subpopulations. Dispersal will be further reduced due to the highly compartmentalized nature of the lung itself, with distinct patches or habitats, such as right and left lobes, upper and lower respiratory tract, and the alveoli and bronchioles, on different branches of the respiratory tree. The resource profile and spatial structure of the CF lung thus provide ideal conditions for strong divergent selection to drive diversification. Additional aspects of life in the CF lung, such as the emergence of hypermutator lineages (20, 34), high levels of antibiotic use (35), and interactions between P. aeruginosa and coinfecting species (36, 37), bacteriophages (38, 39), and the host immune system (40, 41), may further contribute to the genetic and phenotypic variability seen among isolates from chronically infected patients.
Here, we evaluate the contributions of resource complexity and reduced dispersal to P. aeruginosa diversification by tracking the extent of phenotypic diversification within and among independently evolved populations descended from P. aeruginosa strain Pa14 after ∼220 generations of selection in environments that varied in how closely they resemble the resource profile and viscosity of the CF lung. The most CF-like environment consisted of the nutritionally complex synthetic CF medium (SCFM), a defined medium based on the free amino acid, anion, cation, and carbon-source profiles from CF sputum (32), supplemented with mucin, the major protein component of mucus, to mimic the thick mucus layer in the lumen of CF patients. Several studies have shown that the presence of mucin increases the viscosity of the medium and reduces the motility of P. aeruginosa (42, 43) and is therefore very likely to reduce dispersal. The least CF-like environment was composed of a minimal medium supplemented with glucose as the sole carbon source in the absence of mucin. The experimental conditions consisted of factorial combinations of two levels of nutrient complexity (minimal medium vs. SCFM) and viscosity (with or without mucin). Phenotypic divergence among independently evolved replicate populations provided an estimate of the contribution of spatial compartmentalization to diversification. We scored both colony morphology and 10 traits thought to be associated with pathoadaptation tied to chronic infection of the CF lung (growth rate in LB, pyocyanin and pyoverdine production, biofilm formation, swim and twitch motility, and resistance to four classes of antibiotics: ciprofloxacin, ceftazidime, colistin, and tobramycin) for 12 randomly sampled isolates from each of 12 independently evolved populations within each treatment (for a total of 576 isolates) as well as a collection of 24 P. aeruginosa isolates from the lungs of CF patients from across Ontario (15) to gauge the extent to which our experimental conditions recapitulate the markers of phenotypic divergence associated with clinical strains.
Results and Discussion
Markers of Chronic Infection Evolve in the Most Lung-Like Environments.
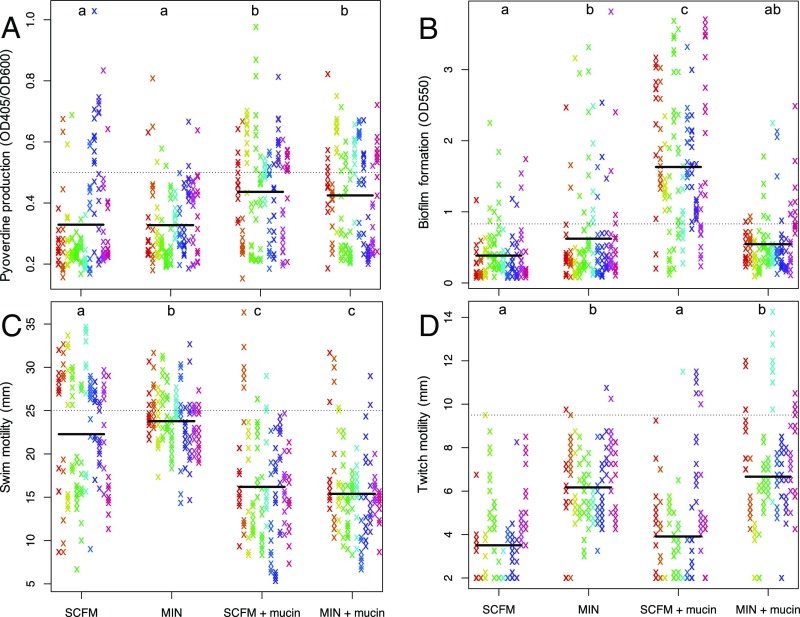
Adaptation to the CF lung is often accompanied by a suite of phenotypic changes that include loss of virulence factors, loss of motility, and increased biofilm formation. We determined the extent to which similar phenotypic changes evolved in our in vitro experiment by scoring a range of traits associated with CF lung adaptation and using a linear mixed-effects model with medium and mucin as fixed effects and population as a random effect to examine the impact of different treatment combinations on trait evolution. Our results are summarized for all traits in Table 1 (and SI Appendix, Table S1) and for four traits commonly associated with CF lung adaptation in Fig. 1.
Table 1.
Summary of phenotypic evolution
| Trait | SCFM | MIN | SCFM+mucin | MIN+mucin |
| Growth rate | — | — | — | — |
| Pyoverdine | ↓ | ↓ | ↓ | ↓ |
| Pyocyanin | — | — | ↑ | — |
| Biofilm formation | ↓ | ↓ | ↑ | ↓ |
| Swim motility | ↓ | ↓ | ↓ | ↓ |
| Twitch motility | ↓ | ↓ | ↓ | ↓ |
| MIC ciprofloxacin | ↑ | — | ↑ | ↑ |
| MIC ceftazidime | — | — | ↑ | ↑ |
| MIC colistin | — | — | — | ↓ |
| MIC tobramycin | — | — | — | — |
Arrows represent significant deviations from the ancestral value in each treatment group, Bonferroni-corrected for multiple comparisons. Antibiotic resistance was measured by minimum inhibitory concentration (MIC), the lowest concentration of drug at which growth was inhibited. See SI Appendix, Table S1 for test statistics and P values.
Fig. 1.
Phenotypic adaptation of evolved isolates for pyoverdine production (A), biofilm formation (B), swim motility (C), and twitch motility (D). Within each treatment, each column/color represents a replicate evolved population (n = 12 populations for each treatment). Solid lines represent treatment means, and dashed lines represent the value of that phenotype in the ancestral strain (Pa14).
We observed marked trait evolution, including a reduction in pyoverdine production (the main siderophore and a measure of iron-scavenging ability; Fig. 1A), swim motility (Fig. 1C), and twitch motility (Fig. 1D), and an increase in biofilm production (Fig. 1B), in the most CF-like conditions, all changes that mirror those seen during adaptation to the CF lung. These phenotypic changes persist when colonies are subcultured, implying that they have a genetic basis. Given that our experiment was conducted in vitro without competing microflora or an active immune system, these results suggest that selection driven by the combined effects of resource complexity and viscosity is sufficient to explain changes in these putatively pathoadaptive traits characteristic of chronic P. aeruginosa infections of the CF lung.
This interpretation might be questioned on the grounds that many of these trait changes, especially those involving a decrease in trait value relative to the common ancestor (pyoverdine production, swim motility, and twitch motility), reflect general adaptation to in vitro culture conditions rather than specific adaptation to CF-like environments. This explanation cannot, however, explain the treatment-specific changes in these and other traits (SI Appendix, Table S1). Pyoverdine production, for example, declined less in the presence of mucin than in its absence, while swim motility showed the reverse trend. Changes in twitch motility, on the other hand, were independent of mucin and decreased more in the nutrient-rich conditions of SCFM than in minimal medium. Pyocyanin and biofilm production both increased in the most CF lung-like conditions (SCFM+mucin), with either no change (pyocyanin) or decreases (biofilm) in the other treatments. The increase in pyocyanin (SI Appendix, Fig. S3), a virulence factor, was unexpected in light of the commonly observed loss of virulence factors among chronic CF isolates. Nonetheless, it has been noted that pyocyanin production is increased in isolates from early stage infections and decreases as disease severity progresses (44), suggesting that reduced pyocyanin production in isolates from chronic infections evolves as a by-product of prolonged infection or in response to an aspect of ecology that we did not investigate, with immune system attack or competition from other species being the most obvious candidates. Taken together, such treatment-specific responses to selection suggest that our results represent trait changes specifically associated with CF lung-like conditions. We note that while this may not suggest that the laboratory environment recapitulates the conditions of the human lung, it does demonstrate that the selection pressures driving these parallel phenotypic changes are recapitulated in the laboratory.
Intriguingly, our results also suggest that antibiotic resistance can evolve in these populations as a pleiotropic effect of adaptation, a phenomenon observed in several bacterial species (45). While a lack of evolutionary change in resistance to antibiotics such as colistin and tobramycin was unsurprising, as no drugs were used in the selection experiment, it was surprising to see a marked increase in ciprofloxacin resistance across all conditions except the simplest environment [M9 minimal salts medium supplemented with 0.7% glucose (MIN)] and an increase in ceftazidime resistance in environments containing mucin (Table 1). Although resistance is commonly observed following antibiotic treatment in clinical settings, our results suggest that both ciprofloxacin and ceftazidime resistance can evolve as a pleiotropic by-product of adaptation to CF lung-like conditions. Spontaneous resistance to antibiotics has been shown to arise via mutations to the MexGHI-OpmD efflux pump that are also associated with biofilm production (46). This mechanism seems unlikely to explain the increases in resistance in our experiment because we did not observe the expected positive relationship between biofilm formation and resistance for either drug class, with the correlation being significantly negative for ciprofloxacin and indistinguishable from zero for ceftazidime (SI Appendix, Fig. S1). Interestingly, a recent study found that resistance to ceftazidime arose in the absence of this antibiotic as a result of an increase in beta-lactamase production (47), a mechanism that warrants further exploration.
We did not observe any mucoid individuals in our evolved populations, despite this being a common phenotype of lung-evolved isolates that can often be associated with aggregation and biofilm formation. If mucoidy represents an adaptation to prolonged infection, it is possible that we would have observed it in a longer experiment. An alternative explanation is that mucoidy evolves in response to selection for some other factor not captured in our experiment. The mucoid colony morphology is often linked to the overproduction of alginate (48), a compound that can also effectively scavenge reactive oxygen species (49, 50). If mucoidy is a pleiotropic result of adaptation for tolerance to oxidative burst from macrophages (15) or neutrophils (51), for example, we would not expect to see it evolving in our experiment.
Within-Population Phenotypic Diversity Is Driven by Resource Complexity.
We observed extensive phenotypic diversity in all evolved populations. To capture the general trends associated with multivariate phenotypic diversification within and among populations, we used all 10 traits described above to calculate the Euclidean distance between pairs of isolates within a population (Eq. 1) and between the means of pairs of populations within each treatment (Eq. 2) as follows:
| [1] |
| [2] |
Here, xi and yi are standardized z scores for trait i of isolates x and y, respectively, and xa,i and yb,i are standardized z scores for trait i of isolates in populations a and b, respectively. Note that calculating among-population distance by using the population means controls for any effect of variation within populations on the measure of among-population distance.
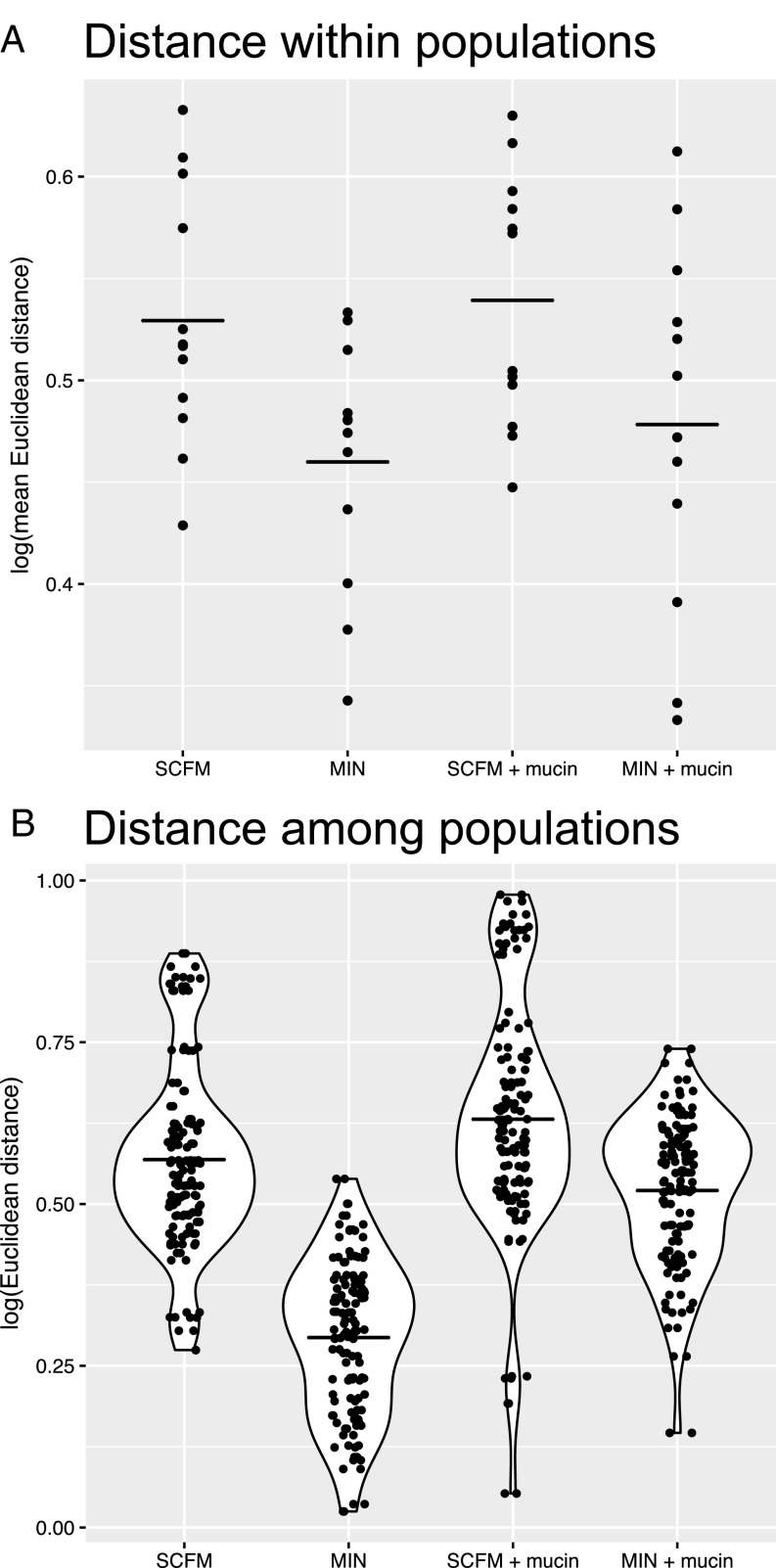
Our results, shown in Fig. 2, are striking. Populations evolved in the complex nutritional environment of SCFM supported higher amounts of within-population diversity than those evolved in the simpler nutritional environment of minimal medium with glucose as the sole carbon and energy source (Fig. 2A; two-factor ANOVA, F1,44 = 10.21, P = 0.003), independent of the presence of mucin (F1,44 = 0.65, P = 0.423). Moreover, we found no significant interaction between resource complexity and mucin (F1,44 = 0.79, P = 0.786). Our results contradict the prevailing view that the within-host diversification by P. aeruginosa accompanying chronic infection is driven primarily by restricted dispersal due to the thick mucus layer in the lung lumen. In evolutionary terms, the nutritional complexity of the lung likely represents abundant ecological opportunity for colonizing P. aeruginosa that generates strong divergent selection, leading to phenotypic divergence. Competition for resources among incipient niche specialists in the evolving population could further accelerate phenotypic diversification and contribute to coexistence, perhaps through negative frequency-dependent selection (reviewed in refs. 27 and 52), leading to long-term persistence of phenotypically divergent P. aeruginosa variants within a single host.
Fig. 2.
Euclidean distance within (A) and among (B) populations. (A) Each point represents a population with the distance within that population determined by the average of all pairwise comparisons of individuals within that population. (B) Each point represents the distance between the mean values of two populations. Solid lines represent treatment means. All distance calculations are Euclidean distance based on all 10 z-score-transformed phenotypic traits.
Nutritional Complexity and Spatial Structure Cause Populations to Diverge.
P. aeruginosa isolates from distinct, spatially separated compartments within the CF airway are often genetically and phenotypically distinct, a result that has been taken to imply niche-specific adaptive diversification following colonization (53). An alternative interpretation is that spatial separation of subpopulations in distinct lung compartments leads to genetic and phenotypic divergence, independent of niche-specific adaptation due to stochastic effects in the timing and order of mutations contributing to adaptation. Our results allow us to quantify the extent of phenotypic divergence among replicate-evolved populations within each treatment in our experiment as a proxy for the extent of among-compartment divergence. Both nutritional complexity and mucin had statistically significant effects on among-population divergence (randomization test where treatment labels were resampled and a null distribution of F statistics calculated; P values were <0.0001, <0.0001, and 0.0048 for medium, mucin, and the interaction, respectively, from a two-factor ANOVA), with the SCFM+mucin treatment displaying the most extensive between-population diversity, followed by the SCFM and MIN+mucin treatments and the MIN treatment (Fig. 2B). Since replicate populations within a treatment evolved, by design, under the same selective pressure, these results lend support to the idea that mutation-order effects in the most CF-like conditions can support substantial among-population divergence of multivariate phenotypes.
Comparison of Evolved Populations and Clinical Isolates from CF Patients Across Ontario.
To what extent do the conditions in our experiment recapitulate the range of variation associated with P. aeruginosa isolates from CF patients? To answer this question, we measured the same 10 phenotypic traits in each of 24 P. aeruginosa strains isolated from the lungs of CF patients from across the Canadian province of Ontario, a collection that included both nonepidemic and epidemic strains (4). We then calculated the Mahalanobis distance between each clinical strain and the multivariate distribution of all evolved isolates from each treatment group, separately, to obtain a measure of phenotypic similarity between the clinical strains and the isolates evolved under different conditions. MIN-evolved isolates were the most different (largest Mahalanobis distance) from the clinical strains (Fig. 3A; ANOVA, F3, 92 = 17.9, P < 0.001). The clinical strains were most similar to those evolved in SCFM, but not statistically more similar than those evolved in SCFM+mucin or MIN+mucin, evaluated by post hoc pairwise t tests, Holm-adjusted for multiple comparisons. Interestingly, strains classified as epidemic were consistently less similar to laboratory-evolved isolates than those classified as nonepidemic (two-factor ANOVA, effect of epidemic/nonepidemic, F1,88 = 6.21, P = 0.015; Fig. 3A). This difference is largely attributable to levels of antibiotic resistance, as excluding antibiotic resistance traits eliminates the difference in Mahalanobis distance between epidemic and nonepidemic strains from our in vitro isolates (Fig. 3B; two-factor ANOVA, effect of epidemic/nonepidemic, F1,88 = 1.69, P = 0.197). This result further supports those noted by ref. 15 that the key distinguishing feature of epidemic strains is that they are more resistant to antibiotics than nonepidemic strains. Importantly, epidemic and nonepidemic strains are indistinguishable in the other phenotypic dimensions measured here, making it difficult to distinguish them on the basis of nonantibiotic phenotypes alone.
Fig. 3.
Comparison of clinical isolates to laboratory-evolved isolates. Each point represents the multivariate (Mahalanobis) distance between a clinical strain and the distribution of all individual isolates from replicate populations in each treatment. Mahalanobis distances are calculated for all traits (A) and excluding antibiotic resistance (AR) traits (B). Black circles are nonepidemic clinical isolates, and gray circles are epidemic isolates. Solid lines represent treatment means.
Interestingly, the Mahalanobis distance between the clinical isolates and those from even the most CF-like conditions in our experiment was always significantly greater than zero, whether or not we included antibiotic resistance traits in the analysis (Hotelling’s T2 test, P < 10−7 for all comparisons). There are two, not mutually exclusive, interpretations of this result. The first is that a positive Mahalanobis distance reflects the absence in our experiment of additional selective forces commonly experienced by P. aeruginosa in the lungs of CF patients such as an active immune system or competition from a diverse microbial community. The second stems from the observation that most chronic CF infections arise from de novo colonization by genetically unique strains of P. aeruginosa. If so, then some portion of the phenotypic divergence among isolates from different patients could be associated with phylogenetic diversity among colonizing strains. The relative contribution of phylogenetic vs. selective forces in driving CF lung-associated adaptation and divergence is unclear and constitutes a subject for future investigation.
Variation in Colony Morphology Is also Linked to Nutritional Complexity.
Colony morphologies are often used as indicators for progression to chronic infection, so we assayed colonies from each evolved population for eight morphological characteristics (pigmentation, opacity, iridescence, surface texture, margin, halo, autolysis, and small colony variant). We identified a total of 18 distinct morphotypes across all 120 populations and an average of ∼2 morphotypes per population (range from 1 to 4; SI Appendix, Fig. S2). Notably, and consistent with the results presented above, populations evolved in SCFM contained more distinct colony morphs on average than those evolved in MIN (ANOVA, medium, F1,116 = 7.36, P = 0.008), while the presence of mucin had no significant effect on the number of colony morphs (ANOVA, presence of mucin, F1,116 = 0.64, P = 0.427). Moreover, there was little correspondence between colony morphology and the changes in the suite of putatively pathoadaptive traits we measured, as revealed by inspection of Spearman-rank correlations for all pairs of traits (SI Appendix, Fig. S1). These results lend further support to the growing consensus that colony morphotypes are unreliable markers of adaptation and the onset of chronic infection.
Clinical Significance and Implications.
Our results provide direct experimental evidence that phenotypic diversification of P. aeruginosa in the airways of CF patients can be driven by divergent selection imposed by the ecological opportunity associated with the nutritionally complex lung environment. Divergence among subpopulations can be further exaggerated by reduced dispersal resulting from the thick mucus layer associated with the CFTR defect and colonization of different airway compartments. Together, these results suggest that the combination of nutritional complexity and reduced dispersal alone is sufficient to drive rapid and repeatable diversification, independently of other sources of selection associated with adaptation such as immune evasion, resource competition from other microbial species, or redox stress.
This interpretation must be qualified, of course, by the fact that our experiments were done in vitro under conditions that were a far cry from the more complex and dynamic environment of the CF lung itself. The advantage of our approach is that it affords us the opportunity to construct focused, highly replicated tests of the role of nutrient complexity and mucin in driving diversification on scales that would not otherwise be possible. The disadvantage, of course, is that our environments can only represent a crude simulacrum of the conditions actually experienced over the course of an infection. That said, it is worth noting that many of the phenotypes thought to be hallmarks of chronic infection, especially loss of motility and biofilm formation, were also observed in the more CF-like conditions in our experiment, suggesting that we have done a reasonable job of recapitulating some of the selective conditions driving adaptation to the CF lung. These changes are likely due to loss-of-function mutations, a result commonly observed in the initial stages of adaptation to novel environments in microbial selection experiments (27). While the detailed genetic changes underlying these phenotypes will have to await whole-genome sequencing (currently underway), the striking phenotypic parallelism between adaptation in vitro and in vivo suggests that the initial stages of colonization of the CF lung can be understood as a specific instance of the more general phenomenon of adaptation and diversification to a novel, nutrient-rich environment.
There are two important implications of our results for clinical practice. First, our results provide direct evidence that CF lung-like conditions promote rapid phenotypic and colony morph diversification, a result that is in line with both longitudinal (14, 53) and cross-sectional (11, 13) studies at the level of both phenotype and genotype. Such rapid diversification, together with the observation that a number of hallmark phenotypes evolved rapidly in vitro, imply that colony morphology or other phenotypic biomarkers are not reliable diagnostic traits of the transition to chronic infection. Second, rapid and extensive diversification both within and among independently evolved populations suggests that the transition to chronic infection can occur by many different phenotypic and genetic routes. The presence of genetically distinct subpopulations within the lung or among different patients complicates treatment because it means that no single therapy targeting P. aeruginosa is likely to be effective at clearing or managing infection for all patients. Rather, a more tailored, patient-specific approach that focuses therapy on the phenotypic and genomic properties of the strain infecting a given host may prove more useful.
The long-term fate of diversity and its consequences for patient health remain unclear. Longitudinal studies suggest that P. aeruginosa diversification can occur rapidly following colonization and can persist for decades within a single host (53), likely due to specialization of subpopulations to different conditions of growth in distinct lung compartments. The comparatively short duration of our experiment does not allow us to distinguish whether the diversity we observed was transient, being the product of high mutation supply rates generating competition among independently arising genotypes, or stable, being supported by negative frequency-dependent selection linked to resource specialization. Nevertheless, our results do lend support to the idea that the nutritional conditions of the CF lung provide a substrate that spurs strain diversification and supports higher levels of genetic variation than would be otherwise available in a more nutritionally homogenous environment. It may be that this diversity provides the raw material for colonization of different airway compartments, leading to compartment-based specialization. Under this view, the transition to chronic infection is intimately tied to, and is in fact the result of, diversification.
Materials and Methods
Sources of strains and growth conditions are presented in SI Appendix. Following isolation of individual colonies, 30 replicate populations in each of four selection environments (SCFM, MIN, SCFM+mucin, and MIN+mucin; conditions are described in SI Appendix) were propagated by daily serial transfer for ∼220 generations. Following this period of selection, morphological diversity was quantified by classifying a minimum of 100 colonies from each population based on eight characteristics: pigmentation, opacity, iridescence, surface texture, margin, halo, autolysis, and small colony variant (details are in SI Appendix). To quantify diversity, 10 additional phenotypes were measured for each of 12 random colonies from 12 random populations of each treatment, for a total of 576 isolates. Phenotyping included growth rate, swimming and twitching motility, pyocyanin and pyoverdine production, biofilm formation, and resistance to one of four classes of antibiotic: ciprofloxacin (fluoroquinolone), ceftazidime (beta-lactam), tobramycin (aminoglycoside), and colistin (polymyxin). Methods for assaying all phenotypes above are described in SI Appendix. Statistics for individual traits, quantifying diversity, Mahalanobis distances, and correlations between traits are also described in SI Appendix.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All data and R scripts for data analysis have been deposited in the Dryad Digital Repository database, datadryad.org/ (doi:10.5061/dryad.5c38fv8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721270115/-/DCSupplemental.
References
- 1.Rajan S, Saiman L. Pulmonary infections in patients with cystic fibrosis. Semin Respir Infect. 2002;17:47–56. doi: 10.1053/srin.2002.31690. [DOI] [PubMed] [Google Scholar]
- 2.Schaedel C, et al. Predictors of deterioration of lung function in cystic fibrosis. Pediatr Pulmonol. 2002;33:483–491. doi: 10.1002/ppul.10100. [DOI] [PubMed] [Google Scholar]
- 3.Hoiby N, Pressler T. Emerging pathogens in cystic fibrosis. Eur Respir Monogr. 2006;35:66–78. [Google Scholar]
- 4.Aaron SD, et al. Infection with transmissible strains of Pseudomonas aeruginosa and clinical outcomes in adults with cystic fibrosis. JAMA. 2010;304:2145–2153. doi: 10.1001/jama.2010.1665. [DOI] [PubMed] [Google Scholar]
- 5.Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med. 2003;168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 6.Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 7.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mowat E, et al. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Respir Crit Care Med. 2011;183:1674–1679. doi: 10.1164/rccm.201009-1430OC. [DOI] [PubMed] [Google Scholar]
- 9.Sousa AM, Pereira MO. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs—A review. Pathogens. 2014;3:680–703. doi: 10.3390/pathogens3030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jorth P, et al. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe. 2015;18:307–319. doi: 10.1016/j.chom.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fothergill JL, Mowat E, Ledson MJ, Walshaw MJ, Winstanley C. Fluctuations in phenotypes and genotypes within populations of Pseudomonas aeruginosa in the cystic fibrosis lung during pulmonary exacerbations. J Med Microbiol. 2010;59:472–481. doi: 10.1099/jmm.0.015875-0. [DOI] [PubMed] [Google Scholar]
- 12.Ashish A, et al. Extensive diversification is a common feature of Pseudomonas aeruginosa populations during respiratory infections in cystic fibrosis. J Cyst Fibros. 2013;12:790–793. doi: 10.1016/j.jcf.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Workentine ML, et al. Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS One. 2013;8:e60225. doi: 10.1371/journal.pone.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark ST, et al. Phenotypic diversity within a Pseudomonas aeruginosa population infecting an adult with cystic fibrosis. Sci Rep. 2015;5:10932. doi: 10.1038/srep10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dettman JR, Rodrigue N, Aaron SD, Kassen R. Evolutionary genomics of epidemic and nonepidemic strains of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2013;110:21065–21070. doi: 10.1073/pnas.1307862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams D, et al. Divergent, coexisting Pseudomonas aeruginosa lineages in chronic cystic fibrosis lung infections. Am J Respir Crit Care Med. 2015;191:775–785. doi: 10.1164/rccm.201409-1646OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winstanley C, O’Brien S, Brockhurst MA. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 2016;24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folkesson A, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat Rev Microbiol. 2012;10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 19.Ciofu O, Mandsberg LF, Wang H, Høiby N. Phenotypes selected during chronic lung infection in cystic fibrosis patients: Implications for the treatment of Pseudomonas aeruginosa biofilm infections. FEMS Immunol Med Microbiol. 2012;65:215–225. doi: 10.1111/j.1574-695X.2012.00983.x. [DOI] [PubMed] [Google Scholar]
- 20.Feliziani S, et al. Coexistence and within-host evolution of diversified lineages of hypermutable Pseudomonas aeruginosa in long-term cystic fibrosis infections. PLoS Genet. 2014;10:e1004651. doi: 10.1371/journal.pgen.1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewontin RC. The Genetic Basis of Evolutionary Change. Vol 560 Columbia Univ Press; New York: 1974. [Google Scholar]
- 22.Whittaker RH, Levin SA. Niche: Theory and Application. Vol 3 Halsted Press; New York: 1975. [Google Scholar]
- 23.Nevo E. Genetic variation in natural populations: Patterns and theory. Theor Popul Biol. 1978;13:121–177. doi: 10.1016/0040-5809(78)90039-4. [DOI] [PubMed] [Google Scholar]
- 24.Futuyma DJ, Moreno G. The evolution of ecological specialization. Annu Rev Ecol Syst. 1988;19:207–233. [Google Scholar]
- 25.Schluter D. The Ecology of Adaptive Radiation. Oxford Univ Press; Oxford: 2000. [Google Scholar]
- 26.Kassen R. Toward a general theory of adaptive radiation: Insights from microbial experimental evolution. Ann N Y Acad Sci. 2009;1168:3–22. doi: 10.1111/j.1749-6632.2009.04574.x. [DOI] [PubMed] [Google Scholar]
- 27.Kassen R. Experimental Evolution and the Nature of Biodiversity. Roberts; Greenwood Village, CO: 2014. [Google Scholar]
- 28.Felsenstein J. Skepticism towards Santa Rosalia, or why are there so few kinds of animals? Evolution. 1981;35:124–138. doi: 10.1111/j.1558-5646.1981.tb04864.x. [DOI] [PubMed] [Google Scholar]
- 29.Rozen DE, Lenski RE. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am Nat. 2000;155:24–35. doi: 10.1086/303299. [DOI] [PubMed] [Google Scholar]
- 30.Doebeli M, Dieckmann U. Evolutionary branching and sympatric speciation caused by different types of ecological interactions. Am Nat. 2000;156:S77–S101. doi: 10.1086/303417. [DOI] [PubMed] [Google Scholar]
- 31.Friesen ML, Saxer G, Travisano M, Doebeli M. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution. 2004;58:245–260. [PubMed] [Google Scholar]
- 32.Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marvig RL, et al. Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. mBio. 2014;5:e00966-14. doi: 10.1128/mBio.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 35.Wright EA, Fothergill JL, Paterson S, Brockhurst MA, Winstanley C. Sub-inhibitory concentrations of some antibiotics can drive diversification of Pseudomonas aeruginosa populations in artificial sputum medium. BMC Microbiol. 2013;13:170. doi: 10.1186/1471-2180-13-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sibley CD, et al. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci USA. 2013;110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brockhurst MA, Buckling A, Rainey PB. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc Biol Sci. 2005;272:1385–1391. doi: 10.1098/rspb.2005.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.James CE, et al. Lytic activity by temperate phages of Pseudomonas aeruginosa in long-term cystic fibrosis chronic lung infections. ISME J. 2015;9:1391–1398. doi: 10.1038/ismej.2014.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jensen PØ, Givskov M, Bjarnsholt T, Moser C. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol. 2010;59:292–305. doi: 10.1111/j.1574-695X.2010.00706.x. [DOI] [PubMed] [Google Scholar]
- 41.McKeon DJ, et al. Cystic fibrosis neutrophils have normal intrinsic reactive oxygen species generation. Eur Respir J. 2010;35:1264–1272. doi: 10.1183/09031936.00089709. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Lory S, Ramphal R, Jin S. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol Microbiol. 1996;22:1005–1012. doi: 10.1046/j.1365-2958.1996.01533.x. [DOI] [PubMed] [Google Scholar]
- 43.Landry RM, An D, Hupp JT, Singh PK, Parsek MR. Mucin-Pseudomonas aeruginosa interactions promote biofilm formation and antibiotic resistance. Mol Microbiol. 2006;59:142–151. doi: 10.1111/j.1365-2958.2005.04941.x. [DOI] [PubMed] [Google Scholar]
- 44.Fung C, et al. Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J Med Microbiol. 2010;59:1089–1100. doi: 10.1099/jmm.0.019984-0. [DOI] [PubMed] [Google Scholar]
- 45.Hershberg R. Antibiotic-independent adaptive effects of antibiotic resistance mutations. Trends Genet. 2017;33:521–528. doi: 10.1016/j.tig.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Sakhtah H, et al. The Pseudomonas aeruginosa efflux pump MexGHI-OpmD transports a natural phenazine that controls gene expression and biofilm development. Proc Natl Acad Sci USA. 2016;113:E3538–E3547. doi: 10.1073/pnas.1600424113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies EV, James CE, Brockhurst MA, Winstanley C. Evolutionary diversification of Pseudomonas aeruginosa in an artificial sputum model. BMC Microbiol. 2017;17:3. doi: 10.1186/s12866-016-0916-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marvig RL, Sommer LM, Molin S, Johansen HK. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet. 2015;47:57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 49.Simpson JA, Smith SE, Dean RT. Scavenging by alginate of free radicals released by macrophages. Free Radic Biol Med. 1989;6:347–353. doi: 10.1016/0891-5849(89)90078-6. [DOI] [PubMed] [Google Scholar]
- 50.Hassett DJ, et al. Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways. Trends Microbiol. 2009;17:130–138. doi: 10.1016/j.tim.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 51.Makam M, et al. Activation of critical, host-induced, metabolic and stress pathways marks neutrophil entry into cystic fibrosis lungs. Proc Natl Acad Sci USA. 2009;106:5779–5783. doi: 10.1073/pnas.0813410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rainey PB, Cooper TF. Evolution of bacterial diversity and the origins of modularity. Res Microbiol. 2004;155:370–375. doi: 10.1016/j.resmic.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 53.Markussen T, et al. Environmental heterogeneity drives within-host diversification and evolution of Pseudomonas aeruginosa. mBio. 2014;5:e01592-14. doi: 10.1128/mBio.01592-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.