Significance
Animal–microbial mutualistic symbioses are important examples of evolutionary adaptation in which symbionts shape diverse traits of their hosts, from physiology to morphology; yet few examples are known of multiple convergences on identical phenotypes within a clade of closely related hosts. Fungus-farming ants possess complex cuticular structures for maintaining Pseudonocardia symbionts. By reconstructing evolutionary history, we find ant–Pseudonocardia associations originated early in evolution of fungus-farming ants and multiple subsequent losses occurred over evolutionary timescales. Complex, strikingly similar structures for maintaining Pseudonocardia have arisen at least three times in fungus-farming ants. The anatomical convergence for maintaining symbionts reveals repeated selection for mutualistic adaptation, likely driven by the necessity of controlling a coevolved fungus-garden parasite.
Keywords: Formicidae, Attina, mutualistic adaptation, Actinobacteria, phylogenomics
Abstract
Evolutionary adaptations for maintaining beneficial microbes are hallmarks of mutualistic evolution. Fungus-farming “attine” ant species have complex cuticular modifications and specialized glands that house and nourish antibiotic-producing Actinobacteria symbionts, which in turn protect their hosts’ fungus gardens from pathogens. Here we reconstruct ant–Actinobacteria evolutionary history across the full range of variation within subtribe Attina by combining dated phylogenomic and ultramorphological analyses. Ancestral-state analyses indicate the ant–Actinobacteria symbiosis arose early in attine-ant evolution, a conclusion consistent with direct observations of Actinobacteria on fossil ants in Oligo-Miocene amber. qPCR indicates that the dominant ant-associated Actinobacteria belong to the genus Pseudonocardia. Tracing the evolutionary trajectories of Pseudonocardia-maintaining mechanisms across attine ants reveals a continuum of adaptations. In Myrmicocrypta species, which retain many ancestral morphological and behavioral traits, Pseudonocardia occur in specific locations on the legs and antennae, unassociated with any specialized structures. In contrast, specialized cuticular structures, including crypts and tubercles, evolved at least three times in derived attine-ant lineages. Conspicuous caste differences in Pseudonocardia-maintaining structures, in which specialized structures are present in worker ants and queens but reduced or lost in males, are consistent with vertical Pseudonocardia transmission. Although the majority of attine ants are associated with Pseudonocardia, there have been multiple losses of bacterial symbionts and bacteria-maintaining structures in different lineages over evolutionary time. The early origin of ant–Pseudonocardia mutualism and the multiple evolutionary convergences on strikingly similar anatomical adaptations for maintaining bacterial symbionts indicate that Pseudonocardia have played a critical role in the evolution of ant fungiculture.
Evolutionary adaptations associated with mutualistic associations are ubiquitous in nature and mutualistic evolution is an important driver of phenotypic complexity (1–4). In animals, internal specialized cells, tissues, and/or organs often evolve in host organisms to accommodate and maintain mutualists, such as bacteriocytes in the aphid–Buchnera symbiosis (5, 6) and light organs in the squid–Vibrio symbiosis (7). Likewise, in many insect groups, external cuticular modifications have arisen to house microbial symbionts, such as antennal gland reservoirs in the beewolf wasp–Streptomyces symbiosis (8) and mycangia in beetle–fungus symbioses (9). Such symbiont-associated traits are often regarded as inherently contingent evolutionary outcomes, the results of complex sequences of unique historical events (10).
Attine ants (subfamily Myrmicinae, tribe Attini, subtribe Attina), an exclusively New World monophyletic group of 17 genera that comprise ∼250 described species, have cultivated fungi for some 55–65 My (11–13). Primitively, attine ants forage on insect frass, seeds, flower parts, and other organic detritus as substrates for their fungus gardens, but higher-attine leaf-cutting ants cut fresh vegetation, making them dominant herbivores in Neotropical ecosystems. As part of the fungus-farming life history, attine ants participate in elaborate symbiotic associations with multiple microbial lineages, spanning fungi and bacteria (14–17). Species in the genus Escovopsis (Ascomycota, Hypocreales), specialized parasitic fungi that are only known to occur in attine fungus gardens, constitute a “crop disease” of attine agriculture, competing with the ants to use their fungal cultivars for food. As a defense against Escovopsis, attine ants participate in mutualistic associations with Actinobacteria that produce antibiotics with potent antagonistic properties against Escovopsis parasites (16, 18, 19). The attine ant–cultivar–parasite–bacterium association is therefore, minimally, a quadripartite symbiosis and so far one of the most complex symbiotic interactions discovered in nature (20).
The exoskeletons of numerous attine-ant species are anatomically modified to house Actinobacteria symbionts and nourish them with glandular secretions through pores lining the specialized cuticle (21), as indicated by the trophic position of the symbionts one level higher than the trophic position of the ants (22). This key evolutionary innovation likely favors the resident Actinobacteria, thereby stabilizing the attine ant–Actinobacteria mutualistic association. An earlier survey of specialized structures that maintain Actinobacteria in attine ants revealed high diversity (21). In the early-diverging paleoattine genus Apterostigma, filamentous Actinobacteria occur on the propleura (in between the forelegs) and the mesopleura (between the forelegs and middle legs), where they grow directly on the ventral surface of the cuticle, which is lined with gland cell-associated pores. In the “higher” attine-ant genus Trachymyrmex, the Actinobacteria symbiont occurs on the propleura, where they grow on tubercles within crypts, in which each tubercle is connected to an internal glandular cell. In the leaf-cutting genus Acromyrmex (Fig. 1A), the bacteria grow on tubercles directly on the exoskeleton rather than in crypts (Fig. 1 B and C). When interpreted with reference to the ant phylogeny, this diversity in cuticular structures conforms to broad evolutionary patterns, with important implications for understanding evolutionary adaptations associated with ant–bacteria mutualism.
Fig. 1.
Cuticular structures associated with bacterial symbionts of extant and extinct attine ants. (A) Extant A. octospinosus garden-tending ant with Actinobacteria (white) covering the entire body. Image courtesy of Alexander L. Wild (photographer). (B) Actinobacteria growing directly on tubercles (arrows) on the propleural plate of A. octospinosus and (C) A. octospinosus tubercles from which Actinobacteria have been removed, revealing details. (D) Extant T. bugnioni ant with crypts over the entire body. (E) Actinobacteria growing directly on tubercles within crypts (arrows) on the propleural plate of T. bugnioni, and (F) T. bugnioni crypts from which Actinobacteria have been removed, revealing details in which each crypt contains a tubercle. (G) T. primaevus ant fossil embedded in Oligo-Miocene amber from the Dominican Republic, ventral view. (H) Detail of T. primaevus head showing foveae (i.e., pits or crypts). (I) Detail of T. primaevus propleural plate and (J) enlargement, showing dense foveae (arrows indicate pits or crypts). (K) Extant Apterostigma dentigerum, ventral view. Arrows indicate Actinobacteria on mesopleura. (L) A. eowilsoni ant fossil embedded in Oligo-Miocene amber from the Dominican Republic, ventral view. Arrows indicate plumes of bubbles on the mesopleura.
Despite the apparent advantages of mutualism with Actinobacteria, some extant higher-attine ants have nearly or completely lost the cuticular structures associated with the maintenance of bacterial symbionts (e.g., Atta spp.) (21, 23). Such secondary losses of complex structural traits and, presumably, of the symbiosis with Actinobacteria implies alternative evolutionary interpretations, including (i) a single principal origin followed by multiple losses, (ii) multiple independent origins and no losses, or (iii) some combination of multiple origins and losses. Each scenario, in turn, has implications for the evolution of the ant–bacteria symbiosis. To date, these scenarios have not been tested phylogenetically due both to the lack of ultramorphological studies of ants sampled from all major attine-ant clades and the lack of a well-supported phylogeny for those sampled ants. To address these issues, we perform a phylogenomic analysis that includes 69 taxa, covering 15 of the 17 fungus-farming attine-ant genera as well as closely related, nonfungus-growing ants. Only two monotypic genera Paramycetophylax and Pseudoatta were excluded. In addition, we conduct scanning electron microscopy (SEM), quantitative PCR (qPCR), divergence dating, and ancestral-state character reconstruction analyses to reconstruct the origin and subsequent evolution of specialized cuticular structures for maintaining and nourishing bacteria in fungus-farming ants.
Results and Discussion
Ancient Ant–Pseudonocardia Symbiosis.
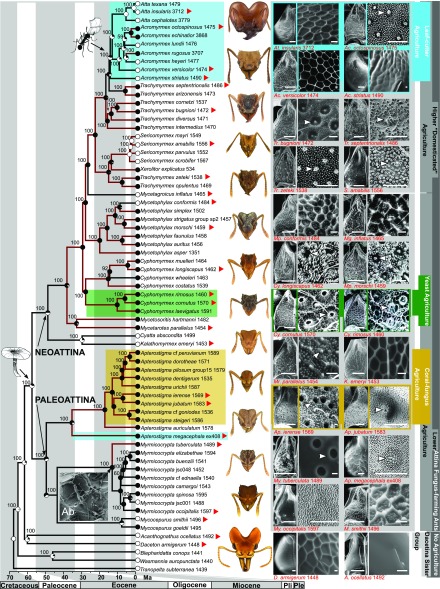
To reconstruct the phylogenetic relationships of attine fungus-farming ants, we obtained phylogenomic data for 69 taxa (five outgroup and 64 ingroup taxa) consisting of 672 ultraconserved-element (UCE) loci, resulting in a concatenated alignment containing 416,786 bp with 13% missing data and 131,357 parsimony-informative sites (PISs). Maximum-likelihood analyses resulted in a resolved phylogeny of the fungus-farming ants in which nearly all nodes are maximally well supported (Fig. 2, details provided in SI Appendix, Fig. S1 and Table S1). The recovered phylogenetic relationships are fully congruent with those proposed in a recent study with more outgroup taxa by Branstetter et al. (11).
Fig. 2.
A time-calibrated phylogeny of the fungus-farming attine ants, indicating the ancestral-state reconstruction probabilities (circles at nodes and tips) for the presence (black) or absence (white) of associated Actinobacteria. Branch colors indicate the status of specialized structures for housing and maintaining mutualistic Actinobacteria: brown branches indicate the presence of cuticular structures on the propleural plate and black branches indicate the absence of such structures. Red arrows adjacent to taxon names refer to the images on the Right (additional images are in SI Appendix, Fig. S5). Numbers above branches are maximum-likelihood bootstrap proportions. [Scale bars: 100 μm for propleural plate; 10 μm for other images (except A. jubatum, for which scale bar: 5 μm).] Ab, antennal Actinobacteria (Myrmicocrypta spp.).
To gauge the timescale over which the ant–Actinobacteria symbiosis emerged and diversified, we conducted divergence-dating analysis in which four nodes were calibrated using information from four Dominican amber fossils. The posterior dates of three of our four fossil-calibrated nodes (N1, N3, and N4) (SI Appendix, Fig. S2) are significantly older (59 Ma, 27 Ma, and 22 Ma, respectively) than our prior dates based on the minimum age of Dominican amber (15 Ma), indicating that the UCE data are highly decisive. Our results indicate that fungus-farming ants arose at the end of the Paleocene around 57 Ma [95% highest posterior density (HPD) interval: 48–66 Ma] (details provided in SI Appendix, Fig. S2). The crown clades of the “Paleoattina” and “Neoattina,” the two major sister clades within fungus-farming ants, originated 49 (41–58) Ma and 48 (40–57) Ma, respectively. Although our inferred dates are largely congruent with those of other molecular studies, they imply a slightly younger date for the origin of fungus-farming ants (11, 12), possibly due to our sparser taxon sampling.
Our electron microscopy survey established the presence/absence of filamentous bacterial symbionts across a broad, representative sample of fungus-farming ants (Fig. 2, details provided in SI Appendix, Fig. S3). Combined evidence from maximum-likelihood ancestral-state reconstruction, divergence-dated phylogenetic analysis, and electron-microscopic ultramorphological study indicates that the ant–Actinobacteria association originated coincident with or shortly after the origin of fungus-farming ants, no later than 49 Ma (Fig. 2).
Ants embedded in amber (i.e., fossilized tree resin) are often extremely well preserved, providing the opportunity to study microscopic morphological structures. We examined two attine-ant fossils in Oligo-Miocene amber from the Dominican Republic, which has been dated variously at 15–20 million years old (24, 25), Trachymyrmex primaevus and Apterostigma eowilsoni (26, 27). We found crypts housing filamentous bacteria in a modern Trachymyrmex species (Fig. 1 D–F) and we likewise observed crypts in the fossil T. primaevus (Fig. 1G) in the same locations on the head (Fig. 1H) and propleural plates (Fig. 1 I and J). For extant Apterostigma species, dense filamentous Actinobacteria are found in the area of the mesopleura under the forelegs (Fig. 1K), corresponding to the location on A. eowilsoni where we found conspicuous bubbles that are likely due to bacterial respiration (Fig. 1L). The presence of Actinobacteria on ants fossilized in 15–20 million-year-old amber is consistent with an early origin of the ant–Actinobacteria symbiosis.
To reconstruct the evolution of specialized cuticular structures for maintaining Actinobacteria, we conducted micromorphological examinations of 69 ant species, sampled from across the full range of the tribe Attina, with electron microscopy (Fig. 2, details provided in SI Appendix, Figs. S4 and S5). This led to the unexpected discovery of numerous conspicuous crypts distributed across large parts of the integuments of Myrmicocrypta tuberculata and Apterostigma ierense and of diverse specialized structures in other Apterostigma species. The presence of such structures in both the Paleoattina and the Neoattina, sister clades resulting from the basalmost split in the attine ant phylogeny, adds further support to an early origin of the ant–Actinobacteria mutualism.
To assess the presence and relative abundance of filamentous bacterial symbionts across attine ants, we screened 25 species representing all attine-ant genera for the abundance of Pseudonocardia with a combination of qPCR and electron microscopy (Fig. 3 and SI Appendix, Table S2). Ant genera closely related to attine ants, including Blepharidatta conops, Wasmannia auropunctata, Tranopelta subterranea, Acanthognathus ocellatus, and Daceton armigerum (the latter two in the subtribe Dacetina, the sister group of the Attina), do not have Pseudonocardia bacteria nor any filamentous bacteria on their integuments. Pseudonocardia bacteria were found to be present in the paleoattine genera Mycocepurus, Myrmicocrypta, and Apterostigma by both qPCR and eSEM. In most species of early-diverging Neoattina, qPCR found relatively low abundances of Pseudonocardia (e.g., in Cyphomyrmex costatus, C. longiscapus, C. muelleri, Mycetarotes parallelus, Mycetosoritis hartmanni, and Mycetophylax asper), which is in agreement with our microscopic observations of lower densities on workers. Relatively lower abundances in these groups are likely due to the substantially smaller bodies of these workers (28). Both qPCR and microscopy found relatively high concentrations of Pseudonocardia in Mycetophylax morschi. Similarly, qPCR detected high concentrations of Pseudonocardia in most higher attine ants, such as Trachymyrmex cf. bugnioni and Trachymyrmex septentrionalis but not in Sericomyrmex mayri or Xerolitor explicatus (microscopic study found visible filamentous bacteria absent in Sericomyrmex and present in low density in X. explicatus). Also, no Pseudonocardia is detectable in the fungus gardens of S. mayri (SI Appendix, Fig. S6). In the leaf-cutting genus Atta, Pseudonocardia is completely undetectable via qPCR, consistent with microscopic observations. In leaf-cutting Acromyrmex species, the highest concentration of Pseudonocardia was found in A. echinatior and A. laticeps, but Pseudonocardia was absent in A. versicolor. Our direct qPCR evidence, considered together with past long-term isolation experiments (29) as well as culture-independent studies by us and others (30, 31), indicate that the genus Pseudonocardia is the dominant filamentous Actinobacteria symbiont found on attine ant exoskeletons.
Fig. 3.
qPCR for measurement of abundance of Pseudonocardia. Sample sizes (maximum number of biological replicates) are provided next to their respective data points.
Evolutionary Trajectory of Pseudonocardia Maintenance in Fungus-Farming Ants.
To reconstruct the evolutionary history of the ant–Pseudonocardia symbiosis, we examined the locations of bacteria on the ant cuticle and the specialized cuticular structures for maintaining bacteria across the phylogeny of attine ants (Fig. 2, details provided in SI Appendix, Fig. S3). Species in the genera closely related to attine ants, Tranopelta, Wasmannia, Blepharidatta, Acanthognathus, and Daceton (the latter two representing the Dacetina, the sister group of the fungus-farming ants) do not have any specialized structures for maintaining Pseudonocardia bacteria. Similarly, species in the paleoattine genera Mycocepurus and Myrmicocrypta have no Pseudonocardia-associated morphological structures present on the external exoskeleton. The only exception is the species M. tuberculata, which has been recovered as the sister group of all other Myrmicocrypta species and has obvious crypts both on the propleural plate and distributed over the rest of the exoskeleton (Fig. 2) (32). Despite the absence of specialized structures in other Myrmicocrypta species, in 24 of 26 species examined, dense concentrations of filamentous Pseudonocardia occur on the antennae and legs, specifically on the antennal scape and on the femora and tibiae (SI Appendix, Fig. S7 and Table S3). In the paleoattine genus Apterostigma, diverse specialized cuticular structures were found on both the propleural and mesopleural plates, including, on the former, crypts and tubercles. In species with tubercles only (e.g., Apterostigma megacephala and Apterostigma jubatum, Fig. 2), the tubercles have pores in the locations where the filamentous Pseudonocardia are cultured. In species in which crypts are present (e.g., A. ierense, Fig. 2), each crypt contains dense concentrations of filamentous Pseudonocardia. In the neoattine species, tubercles are absent in Kalathomyrmex emeryi and Cyatta abscondita, but present in the clade containing M. hartmanni and Mycetarotes parallelus. Crypts are also present in Cyphomyrmex and Mycetophylax species. In these species, each crypt contains a tubercle with a duct connected to a glandular cell in the ant body (in Fig. 2) (21). In the paraphyletic higher-attine genus Trachymyrmex, tubercles were found in the clade that is the sister group of Sericomyrmex. Tubercles are also present in Sericomyrmex species and X. explicatus but were highly reduced in terms of size and abundance. In the clade containing Trachymyrmex cornetzi, Trachymyrmex bugnioni, and Trachymyrmex diversus, crypts are present, with the exception of Trachymyrmex intermedius, in which tubercles are present. Significantly, T. intermedius is the sister to all other species in that clade. Interestingly, tubercles are present in the clade containing T. septentrionalis and Trachymyrmex arizonensis, which is the sister clade to the leaf-cutter ants. Similarly, tubercles are present in the leaf-cutting species A. echinatior, Acromyrmex octospinosus, and Acromyrmex rugosus, whereas specialized cuticular structures are absent in the leaf-cutter species Acromyrmex striatus, Acromyrmex versicolor, Acromyrmex heyeri, Acromyrmex lundii, and Atta spp. Interestingly, the inquiline social parasite Acromyrmex charruanus, which is considered a close relative of its host A. heyeri (33), is also lacking specialized cuticular modifications to house Pseudonocardia symbionts. The specialized cuticular modifications for maintaining Pseudonocardia symbionts across the attine ants represent an apparent continuum of adaptive traits, from antennal gland reservoirs to gland-associated crypts and tubercles. Maximum-likelihood ancestral-state reconstruction indicates that specialized cuticular structures for maintaining Pseudonocardia originated separately three times (Fig. 2). Repeated anatomical convergent evolution is consistent with the defensive role of Pseudonocardia against the garden pathogen Escovopsis, a ubiquitous and ancient crop disease of fungus-farming ant agriculture (14, 16).
Caste Differences in Maintenance of Pseudonocardia Symbionts.
Comparisons of the worker castes (minor and major workers) in A. echinatior revealed significant differences in the structure and height of tubercles (SI Appendix, Fig. S8). These differences are correlated with the abundance of visible filamentous Pseudonocardia, which achieve their highest densities in major workers, the caste most likely to apply Pseudonocardia-produced antibiotics. We further compared workers, males, and alate queens of five attine-ant species, A. echinatior, Mycetophylax faunulus, M. asper, M. hartmanni, and T. septentrionalis. Specialized Pseudonocardia-associated cuticular structures, including crypts and tubercles, were consistently present in workers and queens, but absent or highly reduced in males (SI Appendix, Fig. S9). These differences across castes in the presence/absence of cuticular structures, together with observed differences in the abundance of bacterial symbionts, indicate that Pseudonocardia symbionts are transferred from worker to worker within colonies (34) and vertically by daughter queens when they found new colonies. In contrast to some marine and terrestrial symbioses with consistent horizontal acquisition, the vertical transmission of bacterial symbionts from mother to daughter colonies in fungus-farming ants, while not precluding occasional horizontal transmission, can be expected to reinforce the stability of particular pairs of mutualistic partners.
Repeated Loss of Symbiosis.
Despite the apparent benefits of ant–Pseudonocardia symbiosis in attine ants, our results indicate the occurrence of multiple losses of ant–Pseudonocardia associations and of Pseudonocardia-maintaining structures during the course of attine ant evolution. Specialized cuticular structures are absent in Mycetophylax conformis, Mycetagroicus inflatus, A. striatus, A. versicolor, A. heyeri, A. lundii, and the genus Atta. Reduced tubercles are present in Sericomyrmex, even though Pseudonocardia are absent. Our maximum-likelihood ancestral-state reconstruction indicates that Pseudonocardia symbionts have been lost at least six times and that specialized Pseudonocardia-maintaining cuticular structures have been lost at least seven times. Thus, ant–Pseudonocardia mutualistic associations have been lost repeatedly, possibly due to environmental factors such as migration into dry or colder environments where parasite pressure might be reduced or where the growth of the exosymbionts may not be possible. Given the high metabolic cost associated with maintaining the Pseudonocardia (35), the repeated loss of Pseudonocardia symbionts when pressure is removed is perhaps not surprising.
Conclusion
Many animals and plants have important morphological, physiological, and behavioral traits that help establish and maintain beneficial microbes (3, 36). In this study, we demonstrate the convergent origin of elaborate cuticular structures for maintaining Pseudonocardia in multiple fungus-farming ant lineages. These multiple convergences, each producing strikingly similar yet complex morphological structures, indicate the presence of strong selection pressures for maintaining Pseudonocardia to help control the ancient garden parasite Escovopsis. The vertical transmission of Pseudonocardia across ant colony generations, coupled with the provision by the ants of glandular nutrients to the bacteria, is expected to reinforce the stability of partner associations with Pseudonocardia. Considered together, these results indicate that the mutualism between fungus-farming ants and antibiotic-producing Pseudonocardia is an ancient defensive symbiosis.
The medicinal use of antibiotics by humans dates only from 1945, yet the rapid evolution of antibiotic resistance in human pathogens has rendered the original antibiotics largely ineffective. In contrast, our results indicate that fungus-farming ants have effectively used antibiotics for millions of years. Understanding the mechanisms associated with the long-term use of antibiotics in this ancient symbiosis has the potential to inform our own attempts to counter antibiotic resistance in human pathogens. Likewise, the small molecules produced by ant-associated Pseudonocardia for controlling the Escovopsis crop disease represent a promising resource for antibiotic drug discovery (18, 37, 38).
Experimental Procedures
Taxon Sampling and UCE Data Preparation.
We sampled a total of 69 ant species for phylogenetic analysis, representing the phylogenetic diversity of the tribe Attini for both fungus-farming and nonfungus-farming ant species (SI Appendix, Table S1). Within the fungus-farming ants (subtribe Attina), we included 64 species, representing ∼26% of the 245 currently known species and covering 15 of the 17 currently recognized genera. We did not have material for the genera Paramycetophylax or Pseudoatta, each of which contains a single species, and the latter of which is a derived social parasite of Acromyrmex known to be nested within that genus. The monophyly/nonmonophyly of the included genera has been previously tested in multiple studies (SI Appendix, Table S4). Five outgroup taxa were included, two from the sister group Dacetina and three from other distantly related clades in the tribe Attini. Our analyses are based on a modified version of the alignment used in Branstetter et al. (11), into which we incorporated sequences for seven species belonging to the fungus-farming ants (ingroup). The output from the demultiplexed FASTQ data were trimmed for adapter contamination and low-quality bases using Illumiprocessor (39), which contains the package Trimmomatic (40, 41). Further data processing followed a series of scripts available in the Phyluce package v1.5 (42) to process the reads and extract targeted UCE loci, and is similar to that employed in Branstetter et al. (11) and Ješovnik et al. (43) (SI Appendix, Tables S5 and S6).
Alignment of each UCE locus was performed using MAFFT v7.310 (44) and the resulting alignments were trimmed with GBLOCKS v0.91b (45) using relaxed settings (-b1 = 0.5 -b2 = 0.5 -b3 = 12 -b4 = 7).
Phylogenetic Inference.
After removing loci with poor taxon representation and gap-rich regions, our data consisted of 672 UCE loci, which were on average 620 bp long. We employed IQ-TREE (46) to infer the best substitution model under the corrected Akaike Information Criterion (AICc) for each UCE locus, performed 2,000 ultrafast bootstrap approximations (47), and increased the number of unsuccessful attempted iterations to 200 (from the default 100) using the command (iqtree-omp -s $f -nt 2 -bb 2000 -merit AICc -wbt -nstop 200).
We then constructed a 70% complete (data from ≥48 of the 69 taxa for each locus) concatenated alignment (416,786 bp long, including 13.1% of missing data and gaps) using the program AMAS (48) and created a by-locus partition file with the appropriate model selected for each UCE locus based on the IQ-TREE analysis described above. We used this concatenated alignment to infer a maximum-likelihood tree using IQ-TREE. Node support was obtained by performing 2,000 ultrafast bootstrap approximations (47).
Divergence-Dating Analyses.
Estimation of species divergence times was conducted using the approximate-likelihood approach implemented in the program MCMCTREE as part of the PAML package (49). To calibrate the analysis, information from four Dominican amber fossils (Acanthognathus poinari, Apterostigma electropilosum, Cyphomyrmex spp., and T. primaevus) were employed as independent constraints to calibrate our analysis (SI Appendix, Fig. S2 and Tables S7 and S80, black box symbols indicate constraints, N1–N4). We employed lower bounds (minimum-age bounds) for all four calibrations following Branstetter et al. (11). Except for the root age, all four fossil calibration points were specified as a truncated Cauchy distribution indicated by L(tL, p, c), where tL = minimum-age bound (set as 15 Ma), P = offset value (default value of 0.1), and c = scale parameter value (default value of 1) representing a heavy-tailed density (50). The minimum (lower) bounds here specified represent “soft” bounds, allowing molecular data to correct for conflicting fossil information (51), with a 2.5% probability that the bounds may be violated. The 97.5% upper limit of the probability distribution is at 366 Ma; there is no mean and variance is infinite (49). Since the dating of Dominican amber is ambiguous, ranging from 15 to 20 Ma (24, 25), we chose a conservative minimum age of 15 Ma and employed relatively flat priors to accommodate a wide range of posterior dates. Because of the lack of a fossil for directly calibrating the root node, we employed an admittedly more problematic secondary calibration based on the inferred age of the corresponding internal node of the phylogeny of Branstetter et al. (11). For the root of the tree, we employed soft minimum and maximum bounds [as B (0.56, 0.76), representing the secondary calibration range of 56–76 Ma], to incorporate the 95% HPD value range estimated by Branstetter et al. (11) for the node corresponding to our root, with lower (pL) and upper (pU) tail probabilities set at pL = pU = 0.025 (default values). In this case the prior density distribution is a flat uniform density between 56 and 76 Ma, with 2.5% of density mass lying outside this range (52). To decrease computation time, we performed the analyses using an unpartitioned dataset (ndata = 1) using the HKY+G4 substitution model (model = 4 and alpha = 0.5). We conducted two independent MCMCTREE runs using the following settings: sampfreq = 5,000, nsample = 10,000, and burnin = 5,000,000. We assessed run convergence and performance by examining mcmc.txt files in Tracer v1.6 (53) and convergence plots in either Excel or R v.3.4.3 (R Development Core Team, 2014).
Fossil Ants.
Two amber fossils were available for morphological study: (i) A. eowilsoni (Holotype); worker caste; Oligo-Miocene; Dominican Republic and (ii) T. primaevus; worker caste; Oligo-Miocene; Dominican Republic (details provided in SI Appendix, SI Materials and Methods).
Electron Microscopy.
Besides the 69 taxa in the phylogeny, 21 additional taxa were chosen for electron microscopic study, 17 species of the genus Myrmicocrypta (SI Appendix, Table S3), Mycocepurus obsoletus, and Acromyrmex silvestrii. In addition, we included the two inquiline social parasites Mycocepurus castrator and A. charruanus, which exploit colonies of Mycocepurus goeldii and A. heyeri, respectively (33) (SI Appendix, Table S9). See details in SI Appendix, SI Materials and Methods.
qPCR.
A total of 25 species were screened for the abundance of Pseudonocardia using qPCR, representing all major taxa in the phylogeny (SI Appendix, Table S2). Material from four fungus gardens was also included. See SI Appendix, SI Materials and Methods for details of DNA extraction, primer sets, and gene amplification.
Ancestral-State Reconstruction.
We carried out maximum-likelihood–based ancestral-state reconstruction analyses in the software program Mesquite (54) (details provided in SI Appendix, SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank R. Noll, S. Swanson, and J. G. Pennington for expert help conducting SEM work; D. Grimaldi (American Museum of Natural History) for the loan of amber fossil specimens; E. Okonski for fossil amber ant imaging and laboratory assistance; and M. Borowiec and M. Lloyd for assistance with UCE statistics. This study was funded by National Institutes of Health (NIH) Grant U19 TW009872-05 (to C.R.C., M.T.P., and J.C.), NIH Grant U19 AI109673 (to C.R.C.), National Science Foundation (NSF) Grants Division of Environmental Biology 1456964 and 1654829 (to C.R. and T.R.S.), São Paulo Research Foundation-Fundação de Amparo à Pesquisa de Estado de São Paulo Grant 2013/50954-0 (to M.T.P.), and partially supported by the University of Wisconsin (UW)-Madison College of Engineering Shared Research Facilities and the NSF through the Materials Science Research and Engineering Center (DMR-1720415) using instrumentation provided at the UW-Madison Materials Science Center. Collections of Brazilian samples are approved according to Sistema de Autorização e Informação em Biodiversidade 46555-5 and Conselho Nacional de Desenvolvimento Científico e Tecnológico 010936/2014-9 authorizations.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in National Center for Biotechnology Information database through Sequence Read Archive under BioProject (accession no. PRJNA454746).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809332115/-/DCSupplemental.
References
- 1.Joy JB. Symbiosis catalyses niche expansion and diversification. Proc Biol Sci. 2013;280:20122820. doi: 10.1098/rspb.2012.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci USA. 2007;104:8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douglas AE. Symbiotic Interactions. Oxford Univ Press; Oxford: 1994. [Google Scholar]
- 5.Baumann P, Moran NA, Baumann LC. Bacteriocyte-associated endosymbionts of insects. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Springer; Berlin: 2013. pp. 465–496. [Google Scholar]
- 6.Braendle C, et al. Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 2003;1:E21. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nyholm SV, McFall-Ngai MJ. The winnowing: Establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 8.Kaltenpoth M, et al. Partner choice and fidelity stabilize coevolution in a Cretaceous-age defensive symbiosis. Proc Natl Acad Sci USA. 2014;111:6359–6364. doi: 10.1073/pnas.1400457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulcr J, Stelinski LL. The ambrosia symbiosis: From evolutionary ecology to practical management. Annu Rev Entomol. 2017;62:285–303. doi: 10.1146/annurev-ento-031616-035105. [DOI] [PubMed] [Google Scholar]
- 10.Gerardo NM. Harnessing evolution to elucidate the consequences of symbiosis. PLoS Biol. 2015;13:e1002066. doi: 10.1371/journal.pbio.1002066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Branstetter MG, et al. Dry habitats were crucibles of domestication in the evolution of agriculture in ants. Proc Biol Sci. 2017;284:20170095. doi: 10.1098/rspb.2017.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultz TR, Brady SG. Major evolutionary transitions in ant agriculture. Proc Natl Acad Sci USA. 2008;105:5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sosa-Calvo J, Schultz TR, Ješovnik A, Dahan RA, Rabeling C. Evolution, systematics, and natural history of a new genus of cryptobiotic fungus-growing ants. Syst Entomol. 2018;43:549–567. [Google Scholar]
- 14.Currie CR, et al. Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science. 2003;299:386–388. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
- 15.Pinto-Tomás AA, et al. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science. 2009;326:1120–1123. doi: 10.1126/science.1173036. [DOI] [PubMed] [Google Scholar]
- 16.Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398:701–704. [Google Scholar]
- 17.Currie CR, Mueller UG, Malloch D. The agricultural pathology of ant fungus gardens. Proc Natl Acad Sci USA. 1999;96:7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Arnam EB, et al. Selvamicin, an atypical antifungal polyene from two alternative genomic contexts. Proc Natl Acad Sci USA. 2016;113:12940–12945. doi: 10.1073/pnas.1613285113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Arnam EB, Currie CR, Clardy J. Defense contracts: Molecular protection in insect-microbe symbioses. Chem Soc Rev. 2018;47:1638–1651. doi: 10.1039/c7cs00340d. [DOI] [PubMed] [Google Scholar]
- 20.Youngsteadt E. Community ecology. All that makes fungus gardens grow. Science. 2008;320:1006–1007. doi: 10.1126/science.320.5879.1006. [DOI] [PubMed] [Google Scholar]
- 21.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science. 2006;311:81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 22.Steffan SA, et al. Microbes are trophic analogs of animals. Proc Natl Acad Sci USA. 2015;112:15119–15124. doi: 10.1073/pnas.1508782112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boomsma JJ, Aanen DK. Rethinking crop-disease management in fungus-growing ants. Proc Natl Acad Sci USA. 2009;106:17611–17612. doi: 10.1073/pnas.0910004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iturralde-Vinent MA, MacPhee DE. Age and paleogeographical origin of Dominican amber. Science. 1996;273:1850–1852. [Google Scholar]
- 25.Grimaldi DA, Agosti D. A formicine in New Jersey Cretaceous amber (Hymenoptera: Formicidae) and early evolution of ants. Proc Natl Acad Sci USA. 2000;97:13678–13683. doi: 10.1073/pnas.240452097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimaldi D, Engel MS. Evolution of Insects. Cambridge Univ Press; Cambridge, UK: 2005. [Google Scholar]
- 27.Schultz T. The fungus-growing ant genus Apterostigma in Dominican amber. Mem Am Entomol Inst. 2007;80:425–436. [Google Scholar]
- 28.Andersen SB, Hansen LH, Sapountzis P, Sørensen SJ, Boomsma JJ. Specificity and stability of the Acromyrmex-Pseudonocardia symbiosis. Mol Ecol. 2013;22:4307–4321. doi: 10.1111/mec.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cafaro MJ, et al. Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc Biol Sci. 2011;278:1814–1822. doi: 10.1098/rspb.2010.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersen SB, Yek SH, Nash DR, Boomsma JJ. Interaction specificity between leaf-cutting ants and vertically transmitted Pseudonocardia bacteria. BMC Evol Biol. 2015;15:27. doi: 10.1186/s12862-015-0308-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poulsen M, Cafaro M, Boomsma JJ, Currie CR. Specificity of the mutualistic association between actinomycete bacteria and two sympatric species of Acromyrmex leaf-cutting ants. Mol Ecol. 2005;14:3597–3604. doi: 10.1111/j.1365-294X.2005.02695.x. [DOI] [PubMed] [Google Scholar]
- 32.Sosa-Calvo J, Fernández F, Schultz TR. Phylogeny and evolution of the cryptic fungus-farming ant genus Myrmicocrypta F. Smith (Hymenoptera: Formicidae) inferred from multilocus data. Syst Entomol. 2018 doi: 10.1111/syen.12313. [DOI] [Google Scholar]
- 33.Rabeling C, Schultz TR, Bacci M, Bollazzi M. Acromyrmex charruanus: A new inquiline social parasite species of leaf-cutting ants. Insectes Soc. 2015;62:335–349. [Google Scholar]
- 34.Marsh SE, Poulsen M, Pinto-Tomás A, Currie CR. Interaction between workers during a short time window is required for bacterial symbiont transmission in Acromyrmex leaf-cutting ants. PLoS One. 2014;9:e103269. doi: 10.1371/journal.pone.0103269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poulsen M, Bot ANM, Currie CR, Nielsent MG. Within-colony transmission and the cost of transmission in the leaf-cutting bacterium a mutualistic ant Acromyrmex octospinosus. Funct Ecol. 2003;17:260–269. [Google Scholar]
- 36.Parniske M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 37.Sit CS, et al. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc Natl Acad Sci USA. 2015;112:13150–13154. doi: 10.1073/pnas.1515348112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh DC, Poulsen M, Currie CR, Clardy J. Dentigerumycin: A bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faircloth BC, Sorenson L, Santini F, Alfaro ME. A phylogenomic perspective on the radiation of ray-finned fishes based upon targeted sequencing of ultraconserved elements (UCEs) PLoS One. 2013;8:e65923. doi: 10.1371/journal.pone.0065923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One. 2013;8:e85024. doi: 10.1371/journal.pone.0085024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faircloth BC. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics. 2016;32:786–788. doi: 10.1093/bioinformatics/btv646. [DOI] [PubMed] [Google Scholar]
- 43.Ješovnik A, et al. Phylogenomic species delimitation and host-symbiont coevolution in the fungus-farming ant genus Sericomyrmex Mayr (Hymenoptera: Formicidae): Ultraconserved elements (UCEs) resolve a recent radiation. Syst Entomol. 2017;42:523–542. [Google Scholar]
- 44.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borowiec ML. AMAS: A fast tool for alignment manipulation and computing of summary statistics. PeerJ. 2016;4:e1660. doi: 10.7717/peerj.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 50.Inoue J, Donoghue PCJ, Yang Z. The impact of the representation of fossil calibrations on Bayesian estimation of species divergence times. Syst Biol. 2010;59:74–89. doi: 10.1093/sysbio/syp078. [DOI] [PubMed] [Google Scholar]
- 51.Barba-Montoya J, Dos Reis M, Yang Z. Comparison of different strategies for using fossil calibrations to generate the time prior in Bayesian molecular clock dating. Mol Phylogenet Evol. 2017;114:386–400. doi: 10.1016/j.ympev.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Z, Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol Biol Evol. 2006;23:212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
- 53.Rambaut A, Suchard M, Xie W, Drummond AJ. 2014. Tracer (Univ Edinburgh, Edinburgh), Version 1.6.0.
- 54.Maddison WP, Maddison DR. 2017 Mesquite: A modular system for evolutionary analysis. Version 3.2. Available at https://mesquiteproject.org. Accessed May 30, 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.