Significance
Autophagosomes are double-membraned compartments that serve a central role in maintaining cellular metabolism and homeostasis. These compartments form de novo adjacent to the endoplasmic reticulum (ER), engulfing cytoplasmic macromolecules and organelles for delivery to lysosomes for recycling. The ER has been thought to supply lipids into the precursor membrane phagophore via the PI3P lipid-enriched membrane unit omegasome, enabling the phagophore to expand for engulfment of a bulk of cytoplasmic materials. We have structurally and biochemically characterized the ATG2A-WIPI4 complex, a factor localized at the omegasome, and show that it forms a rod-shaped structure that can tether PI3P-containing membranes to non–PI3P-containing membranes. Our data suggest that the ATG2A-WIPI4 complex is the long-sought factor that tethers the omegasome to the ER and/or the phagophore.
Keywords: autophagy, ATG2, membrane tethering, single-particle analysis, chemical cross-linking coupled with mass spectrometry
Abstract
Autophagy is an enigmatic cellular process in which double-membrane compartments, called “autophagosomes, form de novo adjacent to the endoplasmic reticulum (ER) and package cytoplasmic contents for delivery to lysosomes. Expansion of the precursor membrane phagophore requires autophagy-related 2 (ATG2), which localizes to the PI3P-enriched ER–phagophore junction. We combined single-particle electron microscopy, chemical cross-linking coupled with mass spectrometry, and biochemical analyses to characterize human ATG2A in complex with the PI3P effector WIPI4. ATG2A is a rod-shaped protein that can bridge neighboring vesicles through interactions at each of its tips. WIPI4 binds to one of the tips, enabling the ATG2A-WIPI4 complex to tether a PI3P-containing vesicle to another PI3P-free vesicle. These data suggest that the ATG2A-WIPI4 complex mediates ER–phagophore association and/or tethers vesicles to the ER–phagophore junction, establishing the required organization for phagophore expansion via the transfer of lipid membranes from the ER and/or the vesicles to the phagophore.
Macroautophagy (hereafter autophagy) is a catabolic process essential for the maintenance of nutrition homeostasis and the elimination of cytotoxins, such as damaged organelles, invading bacteria, and aberrant protein aggregates (1, 2). During autophagy, cytoplasmic contents, including cytotoxins, are engulfed in the autophagosome and are broken down by lysosomal hydrolases upon autophagosome–lysosome fusion (3). The materials resulting from the degradation, such as amino acids, are recycled. The engulfment of cytoplasmic contents is enabled by the de novo formation of the autophagosome. This process involves three dynamic membrane reorganization steps: (i) nucleation of the precursor membrane, called the “isolation membrane” or “phagophore,” (ii) expansion of the phagophore into a cup-shaped structure, and (iii) closure of the open end of the cup-shaped membrane. Despite extensive study, the molecular mechanisms associated with these steps remain elusive due to lack of information regarding the functional roles of the autophagy-related (ATG) proteins (4).
Upon autophagy induction, early ATG factors such as ATG1/ULK kinase, ATG9 membrane protein, and VPS34 PI3 kinase mediate nucleation of both the early phagophore and the omegasome (5–8). The latter is a PI3-enriched membrane unit observed as an omega/ring-shaped subdomain of the endoplasmic reticulum (ER) by fluorescence microscopy (5) or a cluster of ER-associated thin tubular membranes by EM (9). Subsequently, the omegasome recruits ATG18/WD-repeat proteins interacting with phosphoinositides (WIPIs) (ATG18 in yeast and WIPI1-4 in mammals) (10–15), the PI3P effector that belongs to the PROPPIN (β-propellers that bind polyphosphoinositides) family (16, 17), and the binding protein ATG2, the largest member of the ATG family. Recruitment of these cofactors leads to the expansion of the early phagophore into a cup-shaped membranous sac. During expansion, the omegasome remains associated with the edge of the open end of the cup-shaped phagophore (5), and ATG18/WIPI and ATG2 also remain concentrated at this ER–phagophore junction (18, 19). When the phagophore has expanded sufficiently, the omegasome starts to shrink and finally disappears as the phagophore is sealed (5). Neither ATG2 nor ATG18/WIPI remains associated with the mature autophagosome (10). The precise spatiotemporal correlation between the localization of ATG2 and ATG18/WIPI and the phagophore/omegasome membrane dynamics suggests that these proteins play direct roles in phagophore expansion at the ER–phagophore interface. Indeed, depletion of ATG2A/B in mammalian cells, which does not affect omegasome formation, leads to an accumulation of small immature phagophores and small autophagosome-like vesicles that are distant from the ER (20).
Although the precise function of ATG2 is unknown, previous studies in yeast suggest that ATG2 may be a peripheral membrane protein (21, 22) that binds directly to membranes (10, 23). In accordance with this described affinity for membranes, mammalian ATG2A/B also localizes to lipid droplets (LDs) and thereby regulates their size (13, 24) as well as localizing at the ER–phagophore junction. These observations collectively suggest that ATG2 may directly mediate a membrane reorganization process, although functional studies have not yielded results to support such claims.
The sequences of ATG2 proteins span ∼1,600–2,300 residues across eukaryotes and contain evolutionarily conserved regions at the N and C termini as well as in the middle of the polypeptide. These domains have been assigned in the Pfam database (25) to the Chorein_N (ID: PF12624), ATG_C (ID: PF09333), and ATG2_CAD (ID: PF13329) families, respectively (Fig. 1A). Chorein_N and ATG_C share sequence similarity with the N and C termini of VPS13 (24, 26), a paralog of VPS13A/Chorein (27). The 200 N-terminal residues containing Chorein_N and the ATG_C region of ATG2A have been reported to be required for the localization of ATG2A to autophagosome-forming sites and LDs, respectively (28). ATG2_CAD contains a highly conserved cysteine-alanine-aspartic acid triad, but its role in autophagosome formation is unknown. In addition, a short region (residues 1,723–1,829) preceding ATG_C is also conserved but is not registered in the Pfam database. This short region is required for the localization of ATG2A to both phagophores and LDs and in isolation localizes to LDs (13, 28). We hereafter refer to this region as the “C-terminal localization region” (CLR). The CLR has been predicted to contain an amphipathic α-helix, indicative of association with membranes (28). Apart from these domains, the regions flanked by Chorein_N and ATG2_CAD and by ATG2_CAD and ATG2_C in yeast ATG2 were reported to share similarities with the mitochondrial protein FMP27, whose function is unknown, and with the Golgi-localized protein of maize APT1, which has been suggested to be involved in membrane trafficking, respectively (Fig. 1A) (23, 29). A fragment containing the APT1 region of yeast ATG2 was shown to interact with membranes containing phosphatidylinositol phosphates, including PI3P (23). However, whether these similarities also apply to higher eukaryotic species is unclear.
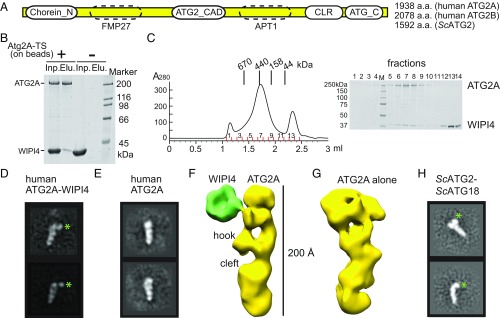
Fig. 1.
Structural analyses of the human ATG2A-WIPI4 complex and the ScATG2-ATG18 complex by negative-stain EM. (A) Diagram of the primary structure of ATG2. The lengths of human ATG2A/B and ScATG2 are indicated. The regions conserved among all species are indicated by ovals with solid outlines. The similarities suggested in ScATG2 to FMP27 and APT1 proteins are indicated as ovals with dashed outlines. (B) Affinity capture experiment with ATG2A immobilized on the beads and WIPI4 in solution. (C) Superose 6 size-exclusion chromatography profile of the mixture of ATG2A and an excess amount of WIPI4. (D and E) 2D class averages of the ATG2A-WIPI4 complex (D) and ATG2A alone (E). (F and G) Reconstructed 3D structures of the ATG2A-WIPI4 complex (F) and ATG2A alone (G). (H) 2D class averages of the ScATG2-ATG18 complex. Green asterisks in 2D class averages in D and H indicate the locations of WIPI4 and ATG18, respectively.
To better understand the role of ATG2, we characterized human ATG2A in complex with WIPI4 using structural and biochemical methods. Using EM and chemical cross-linking coupled with mass spectrometry (CXL-MS), we show that ATG2A has an elongated structure with a WIPI4-binding site at one tip (end). We determined that ATG2A is a bipartite membrane-binding protein that bridges two membranes through interactions at each tip of its elongated structure. Furthermore, we demonstrate that the ATG2A-WIPI4 complex can mediate asymmetric tethering between liposomes with and without PI3P. Placed in the context of the current literature, our findings indicate that the ATG2-WIPI4 complex tethers the PI3P-enriched omegasome to a neighboring membrane(s), such as the ER, phagophore, and other vesicles that may be recruited as a membrane source.
Results
Reconstitution and Overall Structure of the Human ATG2A-WIPI4 Complex.
To enable the structural characterization of ATG2 and investigate its interactions with WIPI/ATG18, we expressed and purified human ATG2A and WIPI4, a pair of proteins that have been reported to interact strongly (12–14), from baculovirus-infected insect cells. The binding was confirmed by an affinity capture experiment in which WIPI4 bound to beads preimmobilized with ATG2A but not to beads lacking ATG2A (Fig. 1B). Furthermore, during size-exclusion chromatography the mixture of ATG2A and WIPI4 comigrated as a single peak (Fig. 1C), thus supporting their ability to form a stable complex. Negative-stain EM studies with the purified ATG2A-WIPI4 complex showed that the particles were monodisperse and homogeneous in size and shape (SI Appendix, Fig. S1). The 2D class averages of the stained particles revealed that the ATG2A-WIPI4 complex is composed of a rod-shaped protein associated with a small, distinct, bead-like feature at one end of the molecule (Fig. 1D). The structural details visible in the 2D averages suggest that the rod-shaped portion of the images corresponds to the multidomain protein ATG2A. The bead-shaped feature can be provisionally attributed to WIPI4 since the overall shape and size is consistent with its predicted β-propeller fold (30–32). Comparison of these class averages with 2D averages of ATG2A alone supports this proposed organization (Fig. 1E) and establishes that ATG2A and WIPI4 form a 1:1 stoichiometric complex upon reconstitution.
3D reconstructions of the ATG2A-WIPI4 complex and free ATG2A further support the 2D analyses, resolving a rod-shaped ATG2A about ∼200 Å in length with a width of ∼30 Å. One end of the rod is hook-shaped with a cleft in the middle (Fig. 1 F and G). The WIPI4 density exhibited characteristics consistent with a β-propeller and directly contacts ATG2A through a thin density (Fig. 1F). This contact likely serves as a hinge through which WIPI4 can adopt a range of orientations relative to ATG2A, as observed in both 2D analyses (Movie S1) and 3D reconstructions (SI Appendix, Fig. S1). Collectively, these results establish the overall structure of ATG2A in complex with WIPI4. WIPI4 is flexibly associated with ATG2A, inducing no significant conformational change in ATG2A.
The Overall Shape Is Conserved in the Yeast ATG2-ATG18 Complex.
The significance of the interactions between mammalian WIPIs and ATG2A/B has not been thoroughly studied. Much of our knowledge regarding this interaction comes from studies of the Saccharomyces cerevisiae (Sc) ATG2-ATG18 complex. Thus, we investigated whether the structural organization of the human ATG2A-WIPI4 complex described above is conserved in the yeast complex. ScATG2 is smaller than mammalian ATG2A (Fig. 1A) and appears to bind ATG18 weakly (SI Appendix, Fig. S1), which makes EM studies of the yeast complex more challenging than studies of its human counterpart. Nevertheless, we obtained 2D class averages of the ATG2-ATG18 complex (Fig. 1H), which show an elongated object with a bead-like density at one end, very similar to the human complex. These results confirm that the overall structure of the ATG2-ATG18 complex is evolutionarily conserved from yeast to human and indicate that functional studies in yeast are relevant in the context of structural work with the human version.
Identification of the WIPI4-Binding Site and Insights into the Chain Topology of ATG2A.
We used an integrative approach to gain further structural information about the ATG2A-WIPI4 complex. First, we sought to establish a coarse-grained chain trace of ATG2A and identify the sites of interaction between WIPI4 and ATG2A by CXL-MS. However, there were some foreseeable technical obstacles in performing a CXL-MS analysis of the ATG2-WIPI4 complex. For example, the overall 3D organization of the ATG2A-WIPI4 complex, comprising an elongated ATG2A and the small binding interface between ATG2A and WIPI4, would limit the number of residue pairs that can be cross-linked. Furthermore, the protein complex was prone to aggregation at higher concentrations, limiting the highest protein concentration achievable without introducing aggregation to a moderate level for a CXL-MS analysis. Therefore, to maximize the number of the cross-linked pairs, we performed two cross-linking reactions: a standard amine-coupling reaction with disuccinimidyl suberate (DSS), which cross-links pairs of lysines up to ∼30 Å apart (33), and another reaction with the coupling reagent 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMTMM) and a linker, pimelic acid dihydrazide (PDH), which cross-links pairs of acidic and lysine residues with zero-length (ZL) or two acidic residues via PDH (34). We used relatively low concentrations of the cross-linkers to suppress nonspecific intermolecular cross-linking that would cause protein aggregation (Methods). Despite these technical challenges, mass spectrometry of these samples successfully identified 20 cross-linked peptide fragments overall (Fig. 2A and SI Appendix, Table S1).
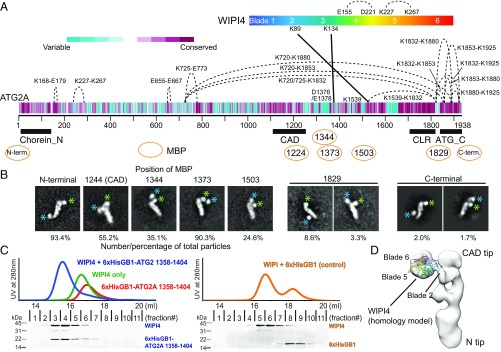
Fig. 2.
Structural characterizations of the ATG2A-WIPI4 complex. (A) CXL-MS analysis of the ATG2A-WIPI4 complex. Cross-links within ATG2A and between ATG2A and WIPI4 are indicated by dashed and solid lines, respectively. The cross-linked residues are labeled. The diagram of the primary structure of ATG2A is colored per the conservation score of each residue as indicated. The conservation score was calculated on the ConSeq server (56). Pfam conserved domains (Chorein_N, ATG2_CAD, and ATG_C) and the CLR are indicated. (B) 2D class averages of MBP-fused or inserted ATG2A in complex with WIPI4. For each construct, a representative 2D class in which the MBP is consistently located adjacent to a tip of ATG2A is shown; the number of particles (in percentage) that constitute all the similar 2D classes in each dataset is indicated below. The remaining particles for each dataset in which the MBP density is missing in the 2D classes are not shown. The green and blue asterisks indicate the locations of WIPI4 and MBP, respectively. (C) Superdex 200 size-exclusion chromatography profiles of 6xHis-GB1-ATG2A (residues 1358–1404), WIPI4, 6xHis-GB1, and their mixtures as indicated. SDS/PAGE shows the size-exclusion fraction contents of the sample mixtures containing stoichiometric amounts of WIPI4 and 6xHis-GB1-ATG2A (1358–1404) (Left) and of WIPI4 and 6xHis-GB1 (Right), respectively. (D) WIPI4-docked electron density map of the ATG2A-WIPI4 complex.
One cross-link of DSS and one cross-link of PDH were identified within WIPI4, and both are consistent with the WIPI4 homology structure model (SI Appendix, Fig. S2), validating our experiments. Three cross-links were intermolecular between ATG2A and WIPI4: one DSS cross-link between Lys1539 of ATG2A and Lys89 on blade 2 of WIPI4 and two ZL cross-links between Asp1376 or Glu1378 of ATG2A and Lys134 on blade 3 of WIPI4 (SI Appendix, Fig. S2). These data agree with previous work reporting that yeast Atg18 interacts with ScATG2 through blade 2 and loop 2, which connects blades 2 and 3 (30, 35), and with another study showing that a truncation construct (residues 1–1561) of ATG2A containing the WIPI4 cross-linked residues is able to bind to ATG18 in a yeast two-hybrid assay (36), suggesting evolutionary conservation of this interaction mode. Furthermore, these data also suggest that the WIPI4-bound tip comprises amino acids that are located in a central region of the primary structure. Fifteen cross-links were collected within ATG2A: 11 short- to midrange (11–93 residues) DSS/PDH/ZL cross-links, which are indicative of locally folded subdomains (Fig. 2A), and five long-range (293–1,160 residues) DSS cross-links between the residues in ATG_C and Lys720/725/1539. SDS/PAGE analysis of the DSS cross-linking reveals an intense band at ∼250 kDa (SI Appendix, Fig. S3). Because the mass of the ATG2A-WIPI4 complex is ∼250 kDa, and the ∼250 kDa band is the single major band, it seems likely that the five long-range DSS cross-links are intramolecular (SI Appendix, Fig. S3), indicating that the folded ATG2A polypeptide adopts a nonlinear chain topology. However, a faint smear in the ∼500- to 600-kDa range is also observable, which may result from two cross-linked ATG2A-WIPI4 complexes, raising the possibility that the long-range cross-links could be inter-ATG2A molecules. Attempts to identify cross-links from in-gel digestion of the monomeric band were unsuccessful, probably due to limited recovery of cross-linked peptides from the gel, preventing us from drawing an unambiguous conclusion.
To map the conserved regions of ATG2A as well as the WIPI4-interacting site, a 42-kDa maltose-binding protein (MBP) was fused to ATG2A at the N terminus and separately inserted into the ATG2_CAD (after residue 1224) (Fig. 2A). 2D image analyses of negatively stained samples revealed that the MBP fused to the N terminus localized to the tip opposite the WIPI4-bound tip of the elongated ATG2A (Fig. 2B). The MBP inserted into the ATG2_CAD localized to the same tip that binds WIPI4 (Fig. 2B). These data suggest that the previously described conserved regions are located at opposite ends of the ATG2A rod-like complex and thus have different functional roles. Hereafter, we refer to the tips of the ATG2A complex as the “N tip” and the “CAD tip.” Our identification of the CAD region is consistent with our CXL-MS experiments, as the CAD region is in close proximity to residues that cross-linked with WIPI4. To further confirm this localization, we also inserted MBP into three positions adjacent to the residues cross-linked to WIPI4 (after residues 1344, 1373, and 1503). As expected, all these MBP inserts were found to be in close proximity to WIPI4 in negative-stain 2D class averages (Fig. 2B).
To identify the C-terminal region, we fused an MBP to the C terminus and also inserted an MBP between the CLR and ATG_C (after residue 1829). In contrast to our previous labeling experiments, the MBP molecules in these C-terminal regions were not identifiable in most 2D classes. The remaining subset of particles (∼12% particles with MBP at the position 1829–1830 and ∼4% particles with MBP at the C terminus) (Fig. 2B) show an additional globular density attached to ATG2A-WIPI4 that we attribute to MBP. However, in both constructs this MBP density was observed adjacent to both tips of ATG2A-WIPI4 (Fig. 2B). These data do not allow unambiguous localization of the C-terminal regions but rather suggest that the C terminus of ATG2A is flexible with respect to the rest of the molecule. In the CXL-MS experiments, three lysine residues in ATG2_C cross-linked to two residues in the middle of the ATG2A sequence (720/725 and 1539) (Fig. 2A). If these cross-links are indeed intramolecular, then this also supports the notion that the C terminus of ATG2 is flexible and can reach the WIPI4-bound tip.
Given that our EM structural analysis revealed that WIPI4 was flexibly attached to ATG2A and that β-propellers often bind to peptides, we hypothesized that the WIPI4 binding site is in a flexible linear region of ATG2A. To test this hypothesis, we generated a fragment of ATG2A (residues 1358–1404) containing two of the intermolecularly cross-linked residues (Asp1375 and Glu1378) as a fusion to the B1 domain of streptococcal protein G (GB1) protein. The GB1-fused peptide comigrated with WIPI4 in a size-exclusion chromatography column and eluted earlier than WIPI4 alone (Fig. 2C). A control using only GB1 and WIPI4 shows no comigration, demonstrating that this linear region is indeed capable of binding to WIPI4.
Based on these results, we fit a WIPI4 homology model into the 3D EM density with blade 2 facing ATG2A (30, 35) and the membrane-binding surface of WIPI4, including the two PI3P recognition sites (blades 5 and 6) (30–32), on the opposite side of ATG2A (Fig. 2D).
ATG2A Associates with Membranes Through Its Tips.
Next, we sought to characterize membrane binding by ATG2A by performing a liposome flotation assay using a Nycodenz density gradient (37). Small unilamellar vesicles (SUVs) and large unilamellar vesicles (LUVs) were prepared by sonication and extrusion methods, respectively. These vesicles were mixed with ATG2A in the presence of Nycodenz, floated to the top of a gradient by centrifugation, and subsequently collected and further analyzed. ATG2A rose to the top of the gradient only in the presence of liposomes (Fig. 3A), confirming direct membrane binding. The recovery of ATG2A proteins was substantially higher with SUVs than with LUVs [14- to 32-fold with 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) vesicles], suggesting that ATG2A prefers binding to highly curved membranes. Additionally, incorporation of negatively charged lipid (1,2-dioleoyl-sn-glycero-3-phospho-l-serine; DOPS) into the liposomes increased ATG2A–liposome association by 1.3- to ninefold, suggesting some electrostatic contribution to this interaction.
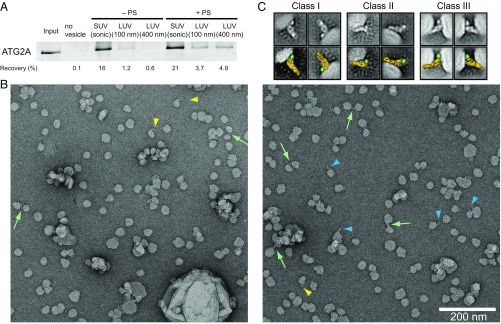
Fig. 3.
Interaction between ATG2A and liposomes and its visualization by EM. (A) Liposome flotation assay with 50 nM ATG2A. The liposomes composed of 99% DOPC and 1% 1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate (DiD) (indicated as “−PS”) or 74% DOPC, 25% DOPS, and 1% DiD (+PS) were prepared by sonication (SUVs) or extrusion (LUVs) using 100- or 400-nm filters. The inputs (4%) and the top layers after centrifugation (24%) were loaded onto SDS/PAGE. The percentage of ATG2A recovered in each of the top fractions was quantified and is shown below the gel image. (B) Micrographs of the negative-stained ATG2A-WIPI4-SUV complex. Colored arrowheads and arrows denote an elongated object that emanates perpendicularly (blue arrowheads) or tangentially (yellow arrowheads) from an SUV or is tethering two SUVs (green arrows). (C) 2D class averages of the ATG2A-WIPI4-SUV complex shown without (Upper) and with (Lower) a manually placed 3D model of ATG2A (shown in yellow). The green dot marks the WIPI4 density.
To better understand how ATG2A associates with membranes, we performed negative-stain single-particle EM analysis on the ATG2A-WIPI4 complex bound to SUVs. Because ATG2A is a thin, somewhat featureless rod, we foresaw difficulties in clearly visualizing such proteins on large membrane surfaces. Therefore, we added WIPI4 to serve as a molecular marker, allowing us to determine unambiguously the orientation of ATG2A bound to the liposomes. We used SUVs composed of DOPC and DOPS, but not PI3P, to avoid any effects introduced by the WIPI4–PI3P interaction. Because SUVs produced by sonication were highly heterogeneous in size, we generated more homogenous SUVs using a dialysis methodology (38). We performed a flotation assay with WIPI4 and these SUVs and confirmed that WIPI4 does not bind to these SUVs (SI Appendix, Fig. S4). In the raw micrographs of the ATG2A-WIPI4-SUV complex (Fig. 3B), we observed elongated objects resembling ATG2A associated with either one or two liposomes as well as clustered liposomes. 2D analyses focused on the elongated object produced averages containing features consistent with the previously observed ATG2A-WIPI4 complex, including the characteristic hook and cleft (Fig. 3C). The 2D classes could be categorized into three major structural classes of the protein–SUV complexes. In the first class, WIPI4 and the CAD tip of ATG2A are bound to the membrane, with the long axis of ATG2A aligned roughly orthogonal to the membrane, positioning the N tip away from the membrane. In the second class, ATG2A is bound to the membrane through the N tip, with the WIPI4 directed away from the membrane. These data indicate that both tips of ATG2A can interact with membranes independently of each other. Strikingly, particles belonging to the third class can be described as a combination of the first and second classes: ATG2A is bound to one liposome through the CAD tip and to another through the N tip, spacing neighboring liposomes ∼10–15 nm apart. These EM data thus suggest that ATG2A is a bipartite membrane-binding protein that can bridge two membranes.
ATG2A Tethers SUVs.
To confirm the membrane-tethering activity of ATG2A, we examined the effect of the presence of ATG2A on the size distribution of liposomes using dynamic light scattering (DLS). The DLS profile of the SUVs shifted markedly to larger sizes upon incubation with ATG2A (Fig. 4A), whereas those of the LUVs (100 and 400 nm) did not change (Fig. 4 B and C). To confirm that the increase in the SUV particle size was due to liposome clustering mediated by the protein, we added proteinase K to the final sample of the ATG2A-SUV mixture and monitored its effect. Upon incubation, the observed particle size decreased to its original dimensions (Fig. 4 D and E), demonstrating that homotypic tethering mediated by ATG2A resulted in the clustering.
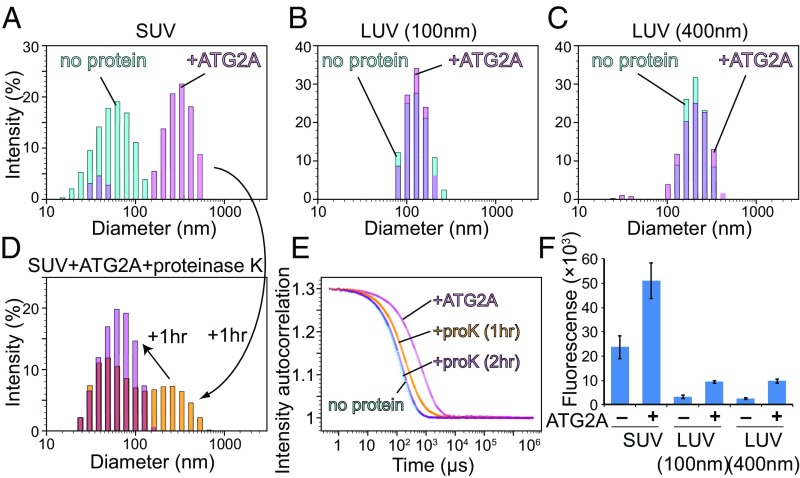
Fig. 4.
Membrane tethering by ATG2A. (A–C) The DLS profiles of SUVs (A), LUVs (100 nm) (B), and LUVs (400 nm) (C) in the absence (cyan) or presence (magenta) of 200 nM ATG2A. All liposomes consisted of 75% DOPC and 25% DOPS. The samples were incubated for 1 h before the measurements. (D) The final sample of A was mixed with proteinase K and remeasured after 1 (yellow) and 2 h (purple) incubation. (E) Auto-scaled autocorrelation functions of the four DLS measurements with SUVs (B and D) are plotted. (F) Fluorescence liposome-tethering assay. Liposomes consisting of 73.3% DOPC, 25% DOPS 0.2% biotinylated lipids, and 1.5% rhodamine-PE were mixed with liposomes of the same size consisting of 73% DOPC, 25% DOPS, and 2% 1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate (DiD) in the presence of 200 nM ATG2A. The fluorescence reports the number of DiD-containing liposomes associated with the biotinylated liposomes. For each experiment, the average of three repeats is shown; whiskers indicate the SD.
We also performed a fluorescence-based liposome-tethering assay, in which biotin-incorporated liposomes were mixed with liposomes containing fluorescent lipids and separated by streptavidin beads (39). The fluorescence intensity of the beads reports the degree of tethering occurring between these two types of liposomes. The results show that the fluorescence signals of the liposomes, regardless of size, were increased by the presence of ATG2A (Fig. 4F), but the difference between the signals in the presence or absence of ATG2A was approximately five times larger in the SUVs than in the LUVs. These data suggest that ATG2A is capable of tethering liposomes, with a preference for SUVs. Taken together, the results from flotation assays, DLS, and fluorescence measurements establish that ATG2A can tether small liposomes (i.e., membranes with high curvature).
The ATG2A-WIPI4 Complex Tethers PI3P-Containing and Non–PI3P-Containing Membranes.
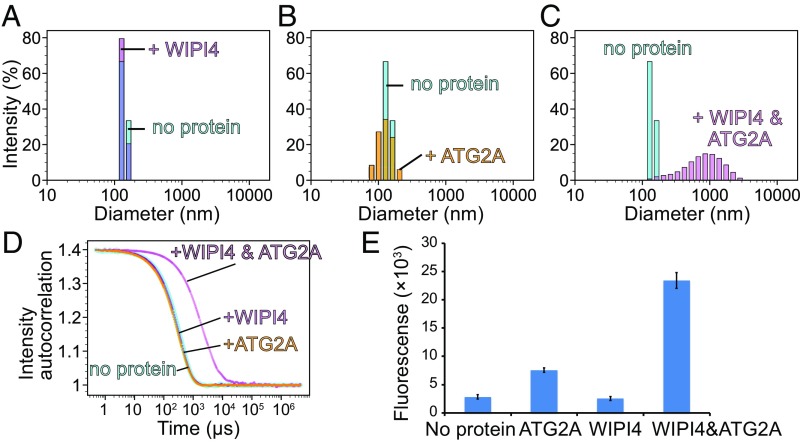
We reasoned that in the homotypic tethering experiments described above ATG2A was unable to tether LUVs (Fig. 4 B, C, and F) due to its weak affinity to LUVs (Fig. 3A). Notably, the liposomes used in these experiments lacked PI3P. Because the PI3P effector WIPI4 robustly binds to PI3P-containing LUVs (31), we hypothesized that the ATG2A-WIPI4 complex could also efficiently bind PI3P-containing LUVs, thereby facilitating tethering of these liposomes. To test this hypothesis, we used DLS. As shown in Fig. 5 A and B, neither WIPI4 nor ATG2A individually changed the size distribution of the LUVs. Thus, the presence of PI3P does not appear to increase the affinity between ATG2A and LUVs. In contrast, the LUV particle size increased markedly in the presence of both proteins (Fig. 5 C and D), demonstrating that the WIPI4–PI3P interaction triggers the clustering of PI3P-containing LUVs.
Fig. 5.
Tethering of PI3P-containing LUVs by the ATG2-WIPI4 complex. (A–C) DLS profiles of LUVs (100 nm) consisting of 75% DOPC, 15% DOPS, and 10% PI3P in the absence (A–C: cyan) or the presence of 200 nM WIPI4 (A), 200 nM ATG2A (B), or both proteins (C). (D) Auto-scaled autocorrelation functions of the four DLS measurements. (E) Fluorescence-based liposome tethering assay. The higher the fluorescence, more associations there are between the liposomes with and without PI3P. LUVs consisting of 73.3% DOPC, 15% DOPS, 10% PI3P, 0.2% biotinylated lipids, and 1.5% rhodamine-PE were mixed with LUVs consisting of 73% DOPC, 25% DOPS, and 2% 1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate in the presence of the indicated proteins. For each experiment, the average of three repeats is shown; whiskers indicate the SD.
Our structure of the ATG2A-WIPI4 complex in which WIPI4 is located adjacent to only one of the two membrane-binding sites (the CAD tip) suggests that the ATG2A-WIPI4 complex could asymmetrically bridge two membranes, one containing PI3P to one without PI3P, via WIPI4 and the N tip, respectively. To test the capacity for such heterotypic tethering directly, we performed fluorescence tethering assays with LUVs containing PI3P and biotinylated lipids and with LUVs lacking both. WIPI4 on its own did not change the fluorescence signal compared with the control (Fig. 5E). Addition of ATG2A showed only a slight increase in signal, similar to that observed using non–PI3P-containing LUVs (Fig. 4F), demonstrating that the presence of PI3P does not improve ATG2A’s poor ability to tether LUVs. In stark contrast, the signal increased markedly in the presence of the ATG2A-WIPI4 complex, with rises in fluorescence that were on par with those observed for the homotypic tethering between non–PI3P-containing SUVs (Fig. 4F). Taken together, these data demonstrate heterotypic membrane tethering by the ATG2A-WIPI4 complex.
The CLR Fragment Binds to Membranes in an Amphipathic α-Helical Conformation.
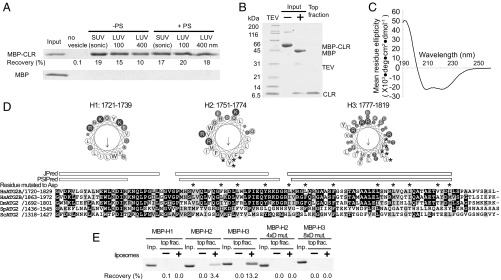
The observation that the CLR in isolation localizes to LDs (13) raises the possibility that the CLR is a membrane-binding domain, as many proteins localize to LDs through direct interaction with the lipid monolayer surface (40). To further characterize the role of CLR in membrane binding, we generated a CLR fragment as an MBP fusion construct, since MBP was required to maintain the solubility of the CLR fragment. In a liposome flotation assay, MBP-CLR was recoverable in the top fraction only in the presence of liposomes, whereas MBP alone was not detected in the top fraction, demonstrating direct membrane binding by the CLR (Fig. 6A). MBP-CLR did not exhibit any preferences for membrane curvature, as it associated with both SUVs and LUVs.
Fig. 6.
Characterization of the CLR. (A) Liposome flotation assay with 100 nM MBP-fused ATG2A CLR (residues1723–1819) or MBP alone (control). The liposomes used are indicated as in Fig. 3A. The inputs (4%) and the top layers after the centrifugation (25%) were loaded onto SDS/PAGE. The percentage of MBP-CLR recovered in each of the top fractions was quantified and is shown below the gel image. (B) Preparation of the CLR-SUV complex by liposome flotation. The input containing MBP-CLR and SUVs (75% DOPC and 25% DOPS) was mixed with tobacco etch virus (TEV) to cleave off MBP, and the resulting CLR-SUV complex was isolated in the top fraction after centrifugation. (C) The CD spectrum of the CLR-SUV complex. (D) Predicated α-helical regions in the CLR. A multiple sequence alignment (MSA) of the CLR of Homo sapiens (Hs) ATG2A, HsATG2B, Drosophila melanogaster (Dm) ATG2, Schizosaccharomyces pombe (Sp) ATG2, and ScATG2 was generated by ClustalW. Secondary structure predictions were obtained using JPred (57) and PSIPred (58) servers. The three fragments generated for flotation assays (H1: residues 1721–1739; H2: residues 1751–1774; H3: residues 1777–1819) are shown as a cartwheel drawing generated by the HeliQuest server (59). The asterisks shown above the MSA and in the cartwheel drawings indicate the residues mutated to aspartic acid. (E) Liposome flotation assay with 1 µM MBP-fused CLR fragments and LUVs prepared by extruding a lipid mixture consisting of 74% POPC, 25% POPS, and 1% 1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate through a 100-nm membrane.
Because LD-localized proteins often interact with the LD membrane through their amphipathic α-helices (40), we sought to examine the secondary structure of the CLR. We purified a CLR-SUV complex by removing the MBP tag from the SUV-bound MBP-CLR by proteolytic cleavage, followed by liposome flotation (Fig. 6B). The CD spectrum of the purified CLR-SUV complex shows a profile typical of an α-helix, with local minima at 208 and 220 nm (Fig. 6C). The helical content predicted from the CD spectrum is 61%. A secondary structure prediction suggests that the CLR may contain three α-helical regions, and helical wheel drawings of these regions show that all three (referred to as “H1,” “H2,” and “H3”) may form amphipathic α-helices (Fig. 6D). To determine which region is responsible for membrane binding, we generated each region as an MBP fusion and tested its membrane-binding ability. The results of the flotation assay revealed that H2 and H3, but not H1, bind to LUVs consisting of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS). H3 appears to have a higher affinity for membranes than H2, based on its higher recovery. We then replaced four residues in the hydrophobic side of H2 and eight residues in the hydrophobic side of H3 with aspartic acids. These mutations abolished the membrane-binding capability of each fragment (Fig. 6E), supporting the likelihood that H2 and H3 bind to membranes via an amphipathic α-helix.
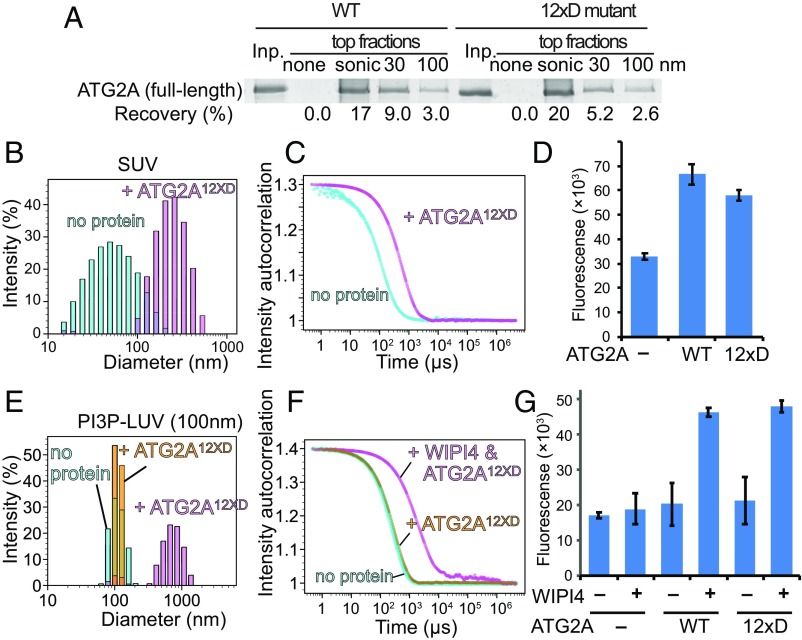
The CLR Is Not Responsible for Membrane Tethering by ATG2A.
To determine whether the membrane-binding property of the CLR plays a role in membrane tethering, we incorporated all the mutations described above (a total 12 mutations to aspartic acid) into the full-length ATG2A protein and characterized this mutant (ATG2A12xD). We tested membrane binding by flotation assay using liposomes containing PO lipids (POPC and POPS), which were prepared by sonication or extrusion with a 30- or 100-nm filter to eliminate potential artifacts (nonspecific binding) caused by DO lipids and sonication. ATG2A bound to all types of liposomes tested but exhibited higher affinity for smaller liposomes (Fig. 7A). These results with DO lipids are consistent with those observed for wild-type ATG2A. The higher preference for sonicated liposomes over the 30-nm liposomes suggests that ATG2A binds to membrane surfaces with local defects rather than sensing overall membrane curvature, as sonication introduces surface defects (37). Our results with ATG2A12xD were very similar to those with the wild type, indicating that these mutated residues are not essential for membrane binding. We then performed membrane-tethering assays with ATG2A12xD and found that these mutations do not affect tethering activity. That ATG2A12xD clusters SUVs and also mediates the PI3P- and WIPI4-dependent homotypic and heterotypic tethering as efficiently as the wild type (Fig. 7 B–G) leads us to conclude that the CLR is not involved in membrane tethering.
Fig. 7.
The CLR is not involved in membrane tethering. (A) Flotation assay with the ATG2A12xD mutant and liposomes composed of 74% POPC, 25% POPS, and 1% 1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate. The liposomes were prepared by sonication or extrusion using 30- or 100-nm filters. The result of SDS/PAGE is shown as in Figs. 3A and 6A. (B and C) DLS homotypic membrane-tethering assay with ATG2A12xD. The DLS profiles (B) and the autocorrelation functions (C) of the 75%DOPC/25%DOPS SUVs in the absence and presence of ATG2A12xD are shown. (D) Fluorescence homotypic membrane assay with ATG2A wild-type and ATG2A12xD performed and presented as in Fig. 4F. (E and F) DLS homotypic membrane-tethering assay with PI3P-incorporated LUVs (100 nm). The DLS profiles (E) and the autocorrelation functions (F) are shown. (G) Fluorescence heterotypic tethering assay with ATG2A12xD performed as shown in Fig. 5E. The experiments with the wild-type ATG2A repeat the data shown in Fig. 5E but were performed at the same time as the experiments with mutants. Although the fluorescence values are different from those in Fig. 5E, the results from both experiments with the wild type are consistent with each other.
Discussion
Recent related studies by Zheng et al. (41) on the rat ATG2B-WIPI4 complex described the overall shape of the ATG2B-WIPI4 complex by negative-stain EM as well as PI3P-independent membrane binding by ATG2B. They also identified the WIPI4-binding site of ATG2B by a combination of CXL-MS and mutagenesis. Our findings are in agreement with this published work, and their mutational studies reinforce our identification of the WIPI4-binding site on ATG2A. Here, however, we structurally and biochemically demonstrate that ATG2A is capable of tethering membranes, which provides valuable insights into autophagosome biogenesis. Gómez-Sánchez et al. (42) also recently characterized the ScATG2 protein and its interaction with ATG18 and ScATG9. Their discovery that ScATG2 binds to membranes by recognizing surface defects is in agreement with our observation that ATG2A binds most strongly with sonicated liposomes. Additionally, their conclusion that ScATG2 is a mediator of ER–phagophore association complements our structural and biochemical data that demonstrate membrane tethering by the ATG2A-WIPI4 complex.
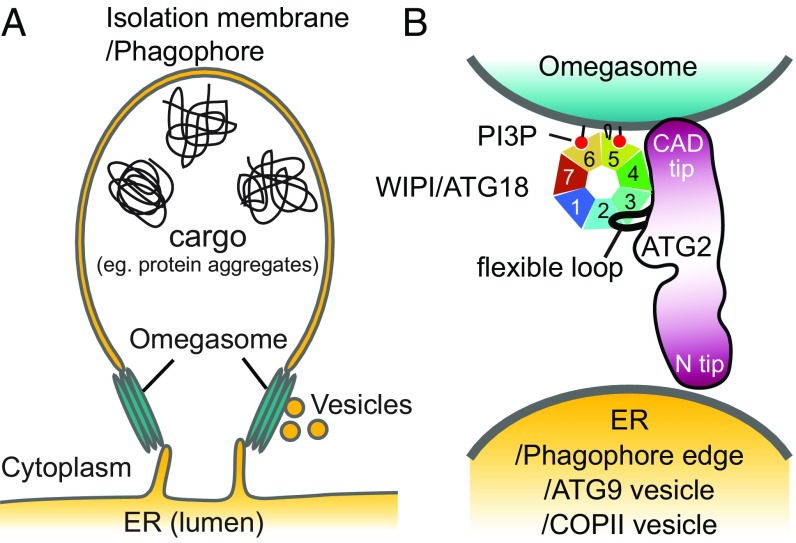
The membrane organization of the ER–phagophore junction, a site intricately tied to the omegasome, is poorly understood due to its highly complex and dynamic nature. Thus, predicting the precise location of ATG2A within this junction is a challenge. In Fig. 8 we illustrate possible pairings of membranes that may be tethered by the ATG2A-WIPI4 complex at the ER–phagophore junction. The PI3P-enriched omegasome has been suggested to be a cluster of tubular membranes with a diameter of ∼30 nm (9), similar to the diameters of the SUVs used in this study (Fig. 8A). Our 2D averages of the ATG2A-WIPI4-SUV complex show that the CAD tip and WIPI4 can simultaneously contact the same membrane (Fig. 3B). Thus, it is most logical to assume that the ATG2A-WIPI4 complex associates directly with the omegasome through the CAD tip as well as through WIPI4 (Fig. 8B). With the CAD tip attached to the omegasome, the N tip could bind to either the ER or the phagophore edge, resulting in a tethering of the omegasome to the ER and/or the phagophore. Alternatively, the N tip may bind to membrane vesicles, such as ATG9 vesicles or COPII vesicles, since ATG2 has been reported to interact with ATG9 and SEC23 (a component of COPII vesicles) (18, 21, 42). These vesicles have been proposed to transform into early phagophores and also may serve as membrane sources for phagophore expansion (6, 43–47). Therefore, tethering of these membrane vesicles to the omegasome by ATG2 would be consistent with the requirement for ATG2 for phagophore expansion (13, 18, 19, 28).
Fig. 8.
Proposed models of the ER-phagophore/isolation membrane association mediated by the ATG2-WIPI/ATG18 complex. (A) Illustration of the ER–phagophore junction based on current knowledge from cell biological studies. Each gray line represents a lipid bilayer. (B) Structural model of the ATG2-WIPI/ATG18 complex tethering the omegasome to its neighboring membranes (ER, phagophore edge, ATG9 vesicle, or COPII vesicle). The dark red color of ATG2 represents conserved regions as in Fig. 2A. The WIPI/ATG18-binding region of ATG2 is represented as a black line emanating from the middle region of ATG2 to indicate the flexible attachment of WIPI/ATG18.
Although the precise manner by which ATG2 interacts with this multitude of vesicular components is largely unknown, Gómez-Sánchez et al. (42) report that a short region of ScATG2 mediates ATG9 interaction. This region, located in the reported APT1 domain (23), is partially conserved in ATG2A and starts at residue 1589 (Figs. 1A and 2A). The closest MBP insertion site among our MBP-tagged constructs was at the position 1503–1504, and this MBP was observed adjacent to WIPI4 and the CAD tip in the 2D class averages (Fig. 2B). It is difficult to determine how far this ATG9-binding region is located from the CAD tip, as the 85 residues between the MBP insertion site and the start of the ATG9-binding region could span a long distance if it is flexible. If the ATG9-binding site is located closer to the CAD tip, then the CAD tip, ATG18/WIPI, and ATG9 could all be localized to the omegasome. Indeed, under conventional fluorescence microscopy, ATG9 colocalizes with ATG2 and ATG18/WIPI at the ER–phagophore junction (18, 19). However, according to the superresolution fluorescence microscopy data, the ATG9 compartment and the omegasome are independent units located in close proximity (6). If the ATG9-binding site is rather closer to the N tip, the ATG2–ATG9 interaction could help stabilize the omegasome–ATG9 compartment association. In either case, there is currently insufficient information to produce a meaningful and complete structural model describing how ATG2, ATG9, and ATG18/WIPI are involved in membrane tethering at the ER–phagophore junction. Higher-resolution structural information about these protein complexes and their interactions with membranes, along with more precise localization of these proteins at the ER–phagophore junction, will be required to elucidate such a model (48).
Our negative-stain EM analysis of the SUV-bound ATG2A-WIPI4 complex suggests that the primary regions in or adjacent to the N terminus and ATG2_CAD are responsible for membrane binding and tethering (Fig. 3B). Demonstrating membrane interaction of each tip using isolated fragments is particularly challenging, as these fragments are exceptionally recalcitrant to purification. Similarly, obtaining ATG2A proteins containing deletions at these regions has also proven extremely difficult. Such issues have prevented further investigation into the role that these regions play in the membrane-tethering process. On a related note, we also attempted to characterize two other previously studied ATG2A constructs having a deletion of the CLR or ATG_C (13, 28) but again failed to obtain sufficient amounts of protein to perform in vitro assays. Although the challenges that have been met during the preparation of these truncated proteins may be attributed to suboptimal constructs or expression and purification conditions, we cannot rule out the possibility that the regions targeted for deletion are integral to the structural stability of the protein. Therefore, site-directed mutagenesis, rather than truncations, would serve as a better strategy for probing the molecular mechanics of membrane tethering, but such studies would require an accurate atomic model of the complexes. While we are unable to provide direct demonstrations of membrane interaction by each tip in isolation, the following evidence supports our conclusions.
First, the N terminus of VPS13 has been shown to interact with membranes containing negatively charged lipids (49). Thus, the N terminus of ATG2, which shares sequence similarity with VPS13’s N terminus, could also be a membrane-interacting domain. Second, the APT1 domain located between ATG2_CAD and the CLR in ScATG2 has been shown to interact with PI3P-containing membranes (23). Although recent reports (41, 42) and the data we presented here show that ATG2(A/B) bind membranes irrespective of the presence of PI3P, it is still possible that the same region in human ATG2A is responsible for membrane interaction. In that case, the residues of this region, rather than those of ATG2_CAD, may form the CAD tip.
In this work, we focused on characterizing another conserved region, the CLR. Our data show that the CLR fragment binds to membranes through its two amphipathic α-helices, which is consistent with the LD localization of the CLR fragment (13). However, the mutations in this region that disrupt membrane interaction of the fragment did not affect the membrane-tethering activity of the full-length protein (Fig. 7). Recently, a similar set of mutations has been shown to reduce cellular autophagic activity (28). We confirmed that this mutant protein, referred to as “AH-E” (28), is also active in membrane tethering in vitro (SI Appendix, Fig. S5). Thus, the CLR is likely to possess another role essential for phagophore expansion. The membrane-tethering activity of ATG2A described in our work cannot alone explain how the phagophore expansion, a process that must involve either lipid transfer or new lipid synthesis, would occur. We speculate that the biochemical function of the CLR may hold the key to this long-standing mystery in autophagosome biogenesis.
Some ATG factors, such as the ATG1 kinase complex, the ATG12-ATG5-ATG16 complex, and ATG8-phosphatidylethanolamine (PE) conjugate, have been reported to mediate membrane tethering in vitro (39, 50, 51). While these factors are distinct in their molecular organization, they all use protein self-oligomerization to tether neighboring membranes: The ATG1 and ATG12-ATG5-ATG16 complexes both self-dimerize so that two molecules of their membrane-interacting subunits (ATG1 and ATG5, respectively) can associate independently with two separate membranes to tether two vesicles (39, 50, 52). ATG8 is associated with a membrane via its covalent linkage to a PE molecule in the membrane, and multimerization of ATG8 molecules on separate vesicles leads to clustering of the vesicles (51). In contrast, ATG2A has a capacity of bridging two membranes without the requirement for self-oligomerization. ATG2A is rather similar to multisubunit tethering complexes, such as the Dsl1, HOPS, COG, and TRAPPIII complexes, all of which tether two membranes through the tips of their elongated shapes (53). The roles of the variety of autophagic membrane tethers in autophagosome biogenesis must be different from one other, as they function at different steps. In the earliest step of biogenesis, the ATG1 kinase complex multimerizes ATG9 vesicles, triggering nucleation of a phagophore. The membrane tethering by the ATG12-ATG5-ATG16 complex, which is the E3-ligase–like factor for ATG8-PE conjugation, and tethering by ATG8-PE have also been suggested to occur during the nucleation step (44, 50, 51). There is also evidence that ATG8-PE is involved in the final step of phagophore membrane closure (54, 55). ATG2A, which is required for phagophore expansion during the intermediate steps of the autophagosome formation, may be an important collaborator of these other membrane tethers and requires future study.
Materials and Methods
Experimental procedures for protein expression and purification, affinity capture binding assays of the ATG2-WIPI4 and ATG2-ATG18 complexes, reconstitution of the ATG2A-WIPI4-SUV complex for negative-stain EM analysis, negative-stain EM, liposome flotation assay, CXL-MS analysis, DLS analysis, fluorescence liposome tethering assays, and CD spectroscopy are described in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Imre Berger and Maria Martinez-Yamout for sharing the MultiBac system and cDNA of GB1, respectively; Drs. Elizabeth M. Wilson-Kubalek and Zoltan Metlagel for assistance in preliminary EM studies; and Drs. Peter E. Wright and Qinghai Zhang for access to CD and DLS instruments, respectively. This research was supported by NIH Grants GM092740 (to T.O.) and DP2EB020402 (to G.C.L.) and by European Research Council Advanced Grant 670821: Proteomics4D (to R.A.). G.C.L. is a Searle Scholar and a Pew Scholar.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The EM density maps of ATG2A and the ATG2A-WIPI4 complex have been deposited in the Electron Microscopy Data Bank [accession codes 8900 (ATG2A) and 8899 (ATG2A-WIPI4)].
See Commentary on page 10540.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811874115/-/DCSupplemental.
References
- 1.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 2.Reggiori F, Klionsky DJ. Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics. 2013;194:341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: Origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018;20:521–527. doi: 10.1038/s41556-018-0092-5. [DOI] [PubMed] [Google Scholar]
- 5.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karanasios E, et al. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun. 2016;7:12420. doi: 10.1038/ncomms12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karanasios E, et al. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci. 2013;126:5224–5238. doi: 10.1242/jcs.132415. [DOI] [PubMed] [Google Scholar]
- 8.Hurley JH, Young LN. Mechanisms of autophagy initiation. Annu Rev Biochem. 2017;86:225–244. doi: 10.1146/annurev-biochem-061516-044820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uemura T, et al. A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Mol Cell Biol. 2014;34:1695–1706. doi: 10.1128/MCB.01327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 12.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell. 2012;23:896–909. doi: 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Q, et al. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell. 2011;21:343–357. doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 15.Bakula D, et al. WIPI3 and WIPI4 β-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat Commun. 2017;8:15637. doi: 10.1038/ncomms15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dove SK, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Proikas-Cezanne T, Takacs Z, Dönnes P, Kohlbacher O. WIPI proteins: Essential PtdIns3P effectors at the nascent autophagosome. J Cell Sci. 2015;128:207–217. doi: 10.1242/jcs.146258. [DOI] [PubMed] [Google Scholar]
- 18.Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24:2918–2931. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- 20.Kishi-Itakura C, Koyama-Honda I, Itakura E, Mizushima N. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J Cell Sci. 2014;127:4089–4102. doi: 10.1242/jcs.156034. [DOI] [PubMed] [Google Scholar]
- 21.Wang CW, et al. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J Biol Chem. 2001;276:30442–30451. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shintani T, Suzuki K, Kamada Y, Noda T, Ohsumi Y. Apg2p functions in autophagosome formation on the perivacuolar structure. J Biol Chem. 2001;276:30452–30460. doi: 10.1074/jbc.M102346200. [DOI] [PubMed] [Google Scholar]
- 23.Kaminska J, et al. Phosphatidylinositol-3-phosphate regulates response of cells to proteotoxic stress. Int J Biochem Cell Biol. 2016;79:494–504. doi: 10.1016/j.biocel.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Pfisterer SG, et al. Lipid droplet and early autophagosomal membrane targeting of Atg2A and Atg14L in human tumor cells. J Lipid Res. 2014;55:1267–1278. doi: 10.1194/jlr.M046359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang RY, et al. Identification of VPS13C as a galectin-12-binding protein that regulates galectin-12 protein stability and adipogenesis. PLoS One. 2016;11:e0153534. doi: 10.1371/journal.pone.0153534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Velayos-Baeza A, Vettori A, Copley RR, Dobson-Stone C, Monaco AP. Analysis of the human VPS13 gene family. Genomics. 2004;84:536–549. doi: 10.1016/j.ygeno.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Tamura N, et al. Differential requirement for ATG2A domains for localization to autophagic membranes and lipid droplets. FEBS Lett. 2017;591:3819–3830. doi: 10.1002/1873-3468.12901. [DOI] [PubMed] [Google Scholar]
- 29.Xu Z, Dooner HK. The maize aberrant pollen transmission 1 gene is a SABRE/KIP homolog required for pollen tube growth. Genetics. 2006;172:1251–1261. doi: 10.1534/genetics.105.050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe Y, et al. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem. 2012;287:31681–31690. doi: 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol Cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krick R, et al. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a β-propeller protein family. Proc Natl Acad Sci USA. 2012;109:E2042–E2049. doi: 10.1073/pnas.1205128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leitner A, Walzthoeni T, Aebersold R. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat Protoc. 2014;9:120–137. doi: 10.1038/nprot.2013.168. [DOI] [PubMed] [Google Scholar]
- 34.Leitner A, et al. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc Natl Acad Sci USA. 2014;111:9455–9460. doi: 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rieter E, et al. Atg18 function in autophagy is regulated by specific sites within its β-propeller. J Cell Sci. 2013;126:593–604. doi: 10.1242/jcs.115725. [DOI] [PubMed] [Google Scholar]
- 36.Romanyuk D, et al. Human hAtg2A protein expressed in yeast is recruited to preautophagosomal structure but does not complement autophagy defects of atg2Δ strain. Acta Biochim Pol. 2011;58:365–374. [PubMed] [Google Scholar]
- 37.Nath S, et al. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat Cell Biol. 2014;16:415–424. doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhoden V, Goldin SM. Formation of unilamellar lipid vesicles of controllable dimensions by detergent dialysis. Biochemistry. 1979;18:4173–4176. doi: 10.1021/bi00586a020. [DOI] [PubMed] [Google Scholar]
- 39.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiam AR, Farese RV, Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol. 2013;14:775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng JX, et al. Architecture of the ATG2B-WDR45 complex and an aromatic Y/HF motif crucial for complex formation. Autophagy. 2017;13:1870–1883. doi: 10.1080/15548627.2017.1359381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gómez-Sánchez R, et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J Cell Biol. 2018;217:2743–2763. doi: 10.1083/jcb.201710116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. eLife. 2014;3:e04135. doi: 10.7554/eLife.04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan D, et al. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc Natl Acad Sci USA. 2013;110:19432–19437. doi: 10.1073/pnas.1316356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto H, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orsi A, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell. 2012;23:1860–1873. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hurley JH, Nogales E. Next-generation electron microscopy in autophagy research. Curr Opin Struct Biol. 2016;41:211–216. doi: 10.1016/j.sbi.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De M, et al. The Vps13p-Cdc31p complex is directly required for TGN late endosome transport and TGN homotypic fusion. J Cell Biol. 2017;216:425–439. doi: 10.1083/jcb.201606078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romanov J, et al. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 52.Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem. 2010;285:1508–1515. doi: 10.1074/jbc.M109.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker RW, Hughson FM. Chaperoning SNARE assembly and disassembly. Nat Rev Mol Cell Biol. 2016;17:465–479. doi: 10.1038/nrm.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuboyama K, et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036–1041. doi: 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- 55.Fujita N, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berezin C, et al. ConSeq: The identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- 57.Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013;41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.