Significance
The antimicrobial effect of copper has been known since ancient times and it is well known that copper alloy surfaces kill various disease-causing bacteria, fungi, and viruses, preventing the spread of antibiotic resistance through horizontal gene transfer between pathogens. However, there is little knowledge about specific targets of copper toxicity in bacteria. Here we show that copper inhibits peptidoglycan LD-transpeptidases, causing higher permeability in the outermembrane and precluding strains of Escherichia coli and Enterococcus faecium to utilize these enzymes in a bypass mechanism to achieve β-lactam resistance. Hence, copper affects bacterial cell envelope biogenesis at low concentration that does not affect growth rate.
Keywords: Escherichia coli, Enterococcus faecium, copper, LD-transpeptidase, peptidoglycan
Abstract
The peptidoglycan (PG) layer stabilizes the bacterial cell envelope to maintain the integrity and shape of the cell. Penicillin-binding proteins (PBPs) synthesize essential 4–3 cross-links in PG and are inhibited by β-lactam antibiotics. Some clinical isolates and laboratory strains of Enterococcus faecium and Escherichia coli achieve high-level β-lactam resistance by utilizing β-lactam–insensitive LD-transpeptidases (LDTs) to produce exclusively 3–3 cross-links in PG, bypassing the PBPs. In E. coli, other LDTs covalently attach the lipoprotein Lpp to PG to stabilize the envelope and maintain the permeability barrier function of the outermembrane. Here we show that subminimal inhibitory concentration of copper chloride sensitizes E. coli cells to sodium dodecyl sulfate and impair survival upon LPS transport stress, indicating reduced cell envelope robustness. Cells grown in the presence of copper chloride lacked 3–3 cross-links in PG and displayed reduced covalent attachment of Braun’s lipoprotein and reduced incorporation of a fluorescent d-amino acid, suggesting inhibition of LDTs. Copper dramatically decreased the minimal inhibitory concentration of ampicillin in E. coli and E. faecium strains with a resistance mechanism relying on LDTs and inhibited purified LDTs at submillimolar concentrations. Hence, our work reveals how copper affects bacterial cell envelope stability and counteracts LDT-mediated β-lactam resistance.
Copper is an essential trace metal and cofactor of several enzymes in bacteria but is toxic at high concentrations. The antimicrobial effect of copper has been known since ancient times, when copper was used to sterilize drinking water and chest wounds (1). Multiple studies show that copper alloy surfaces kill efficiently and rapidly various disease-causing bacteria, such as Escherichia coli (2, 3), Staphylococcus aureus (4), Clostridium difficile (5), Salmonella enterica and Camphylobacter jejuni (6), and Enterococcus faecalis and Enterococcus faecium (7, 8). Currently, copper is used as a self-sanitizing material in high-risk areas in hospitals and care units to reduce the spread of infections.
How copper ions affect bacteria is poorly understood. Once taken up, copper ions cycle between cupric (Cu2+) and cuprous (Cu+) states, potentially disturbing the intracellular redox potential. Additionally, copper generates superoxide and other reactive oxygen species in the presence of molecular oxygen, causing damage to the cell membrane through lipid peroxidation (9, 10). Copper can outcompete and replace other metals from their binding sites in metallo-proteins, such as the iron-sulfur protein fumarase A, isopropylmalate isomerase, and 6-phosphogluconate dehydratase in E. coli (11, 12). It is likely that copper has other yet unknown targets, for example in the bacterial cell envelope.
The bacterial cell envelope is composed of several layers and its integrity is essential for viability. The cell wall peptidoglycan (PG) layer is made of glycan chains that are connected by short peptides and encases the cytoplasmic membrane to provide mechanical stability to the cell (13). PG transpeptidases (TPases) cross-link peptides during PG synthesis. Many bacteria, for example E. coli, mainly contain 4–3 cross-links in their PG, made by the classic DD-TPases, the penicillin-binding proteins (PBPs), which are essential and targeted by β-lactam antibiotics. However, ∼5–15% of the peptide cross-links are unusual 3–3 cross-links (14) formed by LD-TPases (LDTs), which are not essential (15), have an active-site cysteine residue (16), and are not inhibited by most β-lactams, with the notable exception of carbapenems (17). DD- and LD-TPases are structurally and evolutionary unrelated and use different peptide donors for the TPase reaction (pentapeptides and tetrapeptides, respectively) (17–20).
E. coli has six LDTs (LdtA-F) with a YkuD-like domain (PFAM 03744), which all locate in the periplasm. Of these, only LtdD and LtdE form 3–3 cross-links, while LdtA, LdtB, and LdtC attach the outermembrane (OM)-anchored lipoprotein Lpp (Braun’s lipoprotein) to PG, stabilizing the cell envelope (15, 21). The enzymatic function of a sixth homolog, LdtF (YafK), which is involved in biofilm formation in pathogenic E. coli (22), is unknown. Interestingly, several strains (e.g., E. coli M1 and E. faecium M512) are able to bypass the essential DD-TPase activity of PBPs by utilizing an LDT (E. coli LdtD or E. faecium Ldtfm), resulting in β-lactam resistance (23, 24). In these strains, LDTs work together with PG glycosyltransferases (glycan chain polymerases) and DD-carboxypeptidases, which are required to produce the tetrapeptide donor peptides.
Here we show that subminimal inhibitory concentration (sub-MIC) of copper ions inhibit the LDTs of E. coli, causing cell envelope defects due to the reduced attachment of Braun’s Lpp and the loss of 3–3 cross-links. Inhibition of LdtD and Ldtfm prevented the bypass of PBPs in E. coli and E. faecium, respectively, resensitizing them to β-lactams.
Results
Copper Impairs the Robustness of the Cell Envelope.
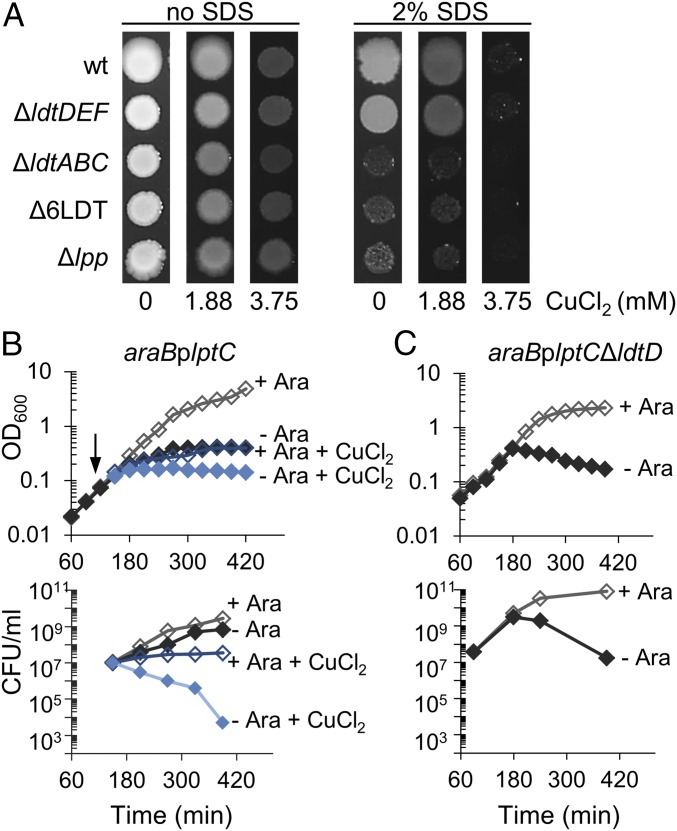
The OM of E. coli is intimately linked to the PG layer via covalent attachment of an abundant lipoprotein, called Lpp. This tight connection contributes to the function of the OM as permeability barrier, protecting the cell from otherwise lethal doses of toxic compounds. Here we aimed to characterize the growth of an E. coli strain lacking all known LDTs, BW25113Δ6LDT, in the presence of the anionic detergent sodium dodecyl sulfate (SDS) and the toxic metal salt, copper chloride. BW25113Δ6LDT cannot produce 3–3 cross-links in PG and cannot attach Lpp to PG, although it produces Lpp. We observed that BW25113Δ6LDT was sensitive to SDS (Fig. 1A and SI Appendix, Fig. S1A), suggesting that SDS resistance requires not only the presence of Lpp, but its covalent attachment to PG by LDTs. Using different mutant strains, we verified that SDS resistance required Lpp and LDTs for attachment of Lpp (strain BW25113ΔldtABC in Fig. 1A and SI Appendix, Fig. S1A) and not LDTs for the formation of 3–3 cross-links (strain BW25113ΔldtDEF in Fig. 1A and SI Appendix, Fig. S1A). Mutant strains with a single deletion of ldtA, ldtB, or ldtC were resistant to SDS, indicating that the absence of each LDT can be compensated for by the two others (SI Appendix, Fig. S1A).
Fig. 1.
Copper impairs the robustness of the cell envelope. (A) Growth in presence of SDS and copper. Overnight cultures of E. coli BW25113 (wt), BW25113ΔldtDΔldtEΔldtF (ΔldtDEF), BW25113ΔldtAΔldtBΔldtC (ΔldtABC), BW25113Δ6LDT, and BW25113Δlpp were adjusted to an equal OD and serial dilutions were spotted on plates with or without 2% SDS, containing no CuCl2, 1.88 mM or 3.75 mM CuCl2 (0, 0.25, and 0.5× MIC, respectively). Plates were incubated at 37 °C for 48 h. Representative results of three independent experiments are shown. Spots of the 10−3 dilution are shown; the complete spot plate assay is shown in SI Appendix, Fig. S1B. (B) Copper induces cell killing when the transport of LPS to the OM is compromised. Cells of the strain araBplptC were grown to an OD600 of 0.2 under permissive conditions (LD medium in the presence of 0.2% arabinose). Cells were harvested, washed three times in LD and diluted 1/100 in LD medium + 0.2% Ara and LD medium without Ara. When cells reached an OD600 of 0.1 the cultures were split and 3.75 mM CuCl2 was added (arrow). Cell growth was monitored by OD600 measurements (Upper) and viability was assessed by determining CFU (Lower). Results of the wild-type strain are shown in SI Appendix, Fig. S2. (C) An ldtD mutant loses viability upon lptC depletion. Growing cells of the araBplptCΔldtD were shifted into media with or without arabinose and the OD (Upper) and viability (Lower) was followed. The ldtD mutant lysed and lost viability in the absence of arabinose, similar to the LdtD-containing wild-type cells grown in the presence of copper chloride (B), suggesting that LdtD is inhibited by copper chloride.
The MIC of all strains for CuCl2 in tryptic soy broth (TSB) was 7.5 mM. Interestingly, in the course of these experiments we noticed that the addition of 0.5× MIC (3.75 mM) of copper chloride sensitized the wild-type and ΔldtDEF cells to SDS, phenocopying Δlpp, ΔldtABC, and Δ6LDT cells (Fig. 1A and SI Appendix, Fig. S1B). These results suggest that copper inhibits LDTs involved with the attachment of Lpp, although the inhibition of other targets by copper could contribute to SDS sensitivity.
We next aimed to test if copper chloride impairs cell envelope integrity in a different situation. For this we induced severe OM assembly stress by depleting LptC, an essential component of the lipopolysaccharide (LPS) export machinery. It is known that E. coli arrests growth but is capable to survive the depletion of lptC (25). We grew cells of BW25113araBplptC in the presence of arabinose and then inoculated them into growth media with or without arabinose, and in the presence or absence of copper chloride, and followed cell growth (optical density, OD) and viability (colony forming units, CFU) (Fig. 1B). Wild-type cells and cells of BW25113araBplptC repleted of lptC (with arabinose) stopped growing but survived in the presence of copper chloride (Fig. 1B and SI Appendix, Fig. S2B). In contrast, the presence of copper chloride reduced the OD and CFUs of BW25113araBplptC cells grown in the absence of arabinose (i.e., upon depletion of lptC) (Fig. 1B). This effect was remarkably similar to our observation that depletion of lptC caused lysis in mutant cells that lack one of the LDTs for the formation of 3–3 cross-links, LdtD (Fig. 1C), and suggested that copper chloride might inhibit LdtD.
In summary, our cellular data suggest that copper inhibits some or all LDTs at a concentration below the MIC, affecting the robustness of the cell envelope under two conditions of envelope stress, the presence of an exogenous detergent, and severe LPS transport stress.
Copper Alters the PG Composition.
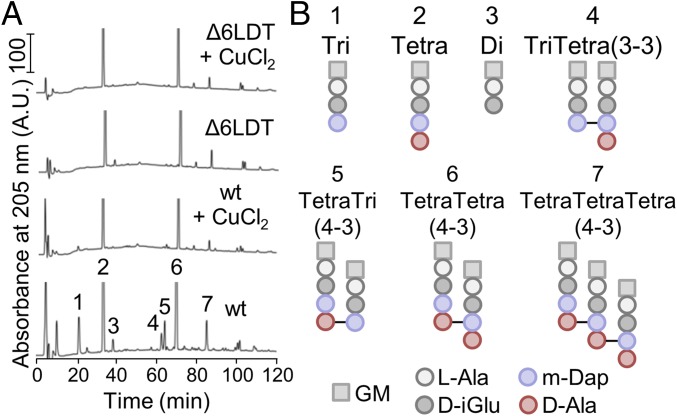
To test whether the presumed inhibition of LDTs by copper alters the PG composition, we isolated PG from wild-type BW25113 and the corresponding strain lacking all LDTs, BW25113Δ6LDT, grown in the presence or absence of 0.5× MIC of copper chloride, and determined the muropeptide (disaccharide peptide subunit) composition. We initially analyzed the PG of stationary cells to better detect attachment of Lpp to PG, which is recognizable as LysArg-modified muropeptides and is enriched in stationary cells (14). Indeed, we identified the monomeric TriLysArg muropeptide in PG isolated from stationary cells (SI Appendix, Fig. S3). When cells were grown in the presence of copper chloride, the relative percentage of TriLysArg decreased by 45 ± 11% (mean ± SD; n = 3) compared with cells grown without copper chloride. Interestingly, we noticed that muropeptides with 3–3 cross-links or tripeptides [Tri, TriTetrai(3–3) and TetraTri(4–3)] were not detectable in PG from cells grown in the presence of copper chloride (SI Appendix, Fig. S4). This observation prompted us to perform detailed analysis of PG from exponentially growing cells (Fig. 2).
Fig. 2.
Copper changes the muropeptide composition of E. coli. (A) Muropeptide profiles of E. coli BW25113 (wt) and BW25113Δ6LDT grown in the presence of 0.5× MIC of CuCl2 or without CuCl2. Main muropeptides are numbered. SI Appendix, Fig. S4 shows the full chromatograms with the full height of all peaks. (B) Structures of the muropeptides numbered in A. d-Ala, d-alanine; d-iGlu, d-isoglutamate; GM, N-acetylglucosamine-N-acetylmuramitol; l-Ala, l-alanine; m-Dap, meso-diaminopimelic acid.
E. coli strains, including BW25113, produce a characteristic muropeptide profile containing ∼5 major and ∼40 minor muropeptides. The latter arise from variations in peptide length (di- to pentapeptide), oligomeric state of cross-linked peptides (monomer to tetramer), different types of cross-links (3–3 or 4–3), and the presence of 1,6-anhydroMurNAc–containing muropeptides. The presence of copper chloride during exponential growth altered the muropeptide composition (Fig. 2 and SI Appendix, Table S2), causing a drastic and specific reduction in all muropeptides with tripeptides and 3–3 cross-links: monomeric disaccharide tripeptide (Tri) dropped from 7.3% (without copper) to 0.7% (with copper), and dimeric TetraTri (4–3 cross-link) from 4.1 to 0.5%. In addition, all muropeptides with 3–3 cross-links, such as TriTetra(3–3) disappeared (Fig. 2 and SI Appendix, Table S2). The affected muropeptides are all generated by LDTs. Intriguingly, the muropeptide composition of BW25113 grown in the presence of copper chloride was similar to that of BW25113Δ6LDT grown with or without copper, strongly supporting the conclusion that copper inhibited the LDTs responsible for the formation of 3–3 cross-links in the PG. This effect was specific for copper, as the presence of 0.5× MIC of NiCl2 or CoCl2 did not reduce the portion of muropeptides with tripeptides or 3–3 cross-links, suggesting that nickel and cobalt ions do not inhibit LDTs in E. coli (SI Appendix, Fig. S4 and Table S3). Overall, the results of this muropeptide analysis indicate that copper mainly targets LDTs that generate 3–3 cross-links and tripeptides (LdtD, LdtE, and LdtF).
Copper Reduces the Incorporation of a Fluorescent d-Amino Acid in the Sidewall.
PBPs and LDTs perform d-amino acid exchange reactions and are therefore capable of incorporating fluorescent d-amino acids (FDAAs) into PG, which allows visualizing PG in live bacteria (26). We used this methodology to test if the inhibition of the LDTs by copper leads to a detectable change in the incorporation of the FDAA 7-hydroxycoumarin-carbonylamino-d-alanine (HADA).
When using a published protocol for in situ labeling of E. coli cells with HADA (27, 28), we noticed that, unexpectedly, constricting cells displayed a label-free zone at midcell (SI Appendix, Fig. S5 A, 1). We reasoned that this zone of intense PG synthesis (PBP activity) was free of labels because the incorporated HADA is quickly removed by the cellular DD-carboxypeptidases (DD-CPases) during harvesting and washing of cells at neutral pH. We therefore optimized the HADA-labeling procedure (SI Appendix, Supplemental Methods and Fig. S5) by harvesting the cells and performing the first wash at a low pH of 3.0, at which DD-CPases are inactive (29, 30). Using this protocol, the cells retained a strong HADA signal at midcell (SI Appendix, Fig. S5 A, 2).
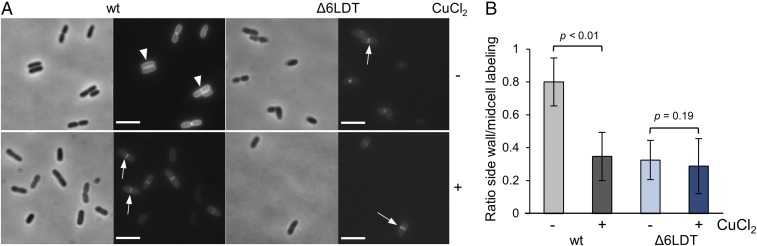
Using the optimized protocol BW25113 cells showed robust HADA labeling at the sidewall and had the expected strong signal at the septum of constricting cells (SI Appendix, Fig. S5 A, 2). The same labeling pattern was obtained with purified PG sacculi (SI Appendix, Fig. S5 B, 2), confirming that the signal is indeed in PG. Interestingly, in BW25113Δ6LDT cells the signal at the sidewall was significantly reduced but the septa of constricting cells were still labeled (SI Appendix, Fig. S5 A, 2), and this pattern was also seen with purified sacculi (SI Appendix, Fig. S5 B, 2). This is consistent with a scenario according to which most of the sidewall label is incorporated by LDTs, and most of the septal label by PBPs.
We next optimized the HADA label of cells grown in the presence of copper, which required the removal of excessive copper by the chelating agent triethylenetetramine dihdrochloride (TETA). When grown in the presence of 0.5× MIC of CuCl2, BW25113 cells lost most of the label at the sidewall, presumably due to inactivation of the LDTs, but septal labeling due to the high activity of PBPs was retained (Fig. 3A), resulting in a significantly lower ratio of sidewall to midcell fluorescence density compared with cells grown without copper (Fig. 3B). As expected from the previous results, the presence of copper did not change the HADA localization profile in BW25113Δ6LDT and the ratio of sidewall to midcell fluorescence density was low with or without copper (Fig. 3). These findings provide further support for the inhibition of LDTs by copper.
Fig. 3.
Copper reduces the incorporation of HADA into the sidewall PG. (A) Cells of BW25113 (wt) and BW25113Δ6LDT were grown exponentially in the presence of 0.5× MIC CuCl2 or without CuCl2, and labeled with HADA for 30 min. Cells were collected and the excess of HADA was removed (SI Appendix, Fig. S5 A, 4). Cells were fixed and imaged by phase-contrast microscopy and HADA was detected by fluorescence microscopy. White arrows, septal localization; white triangles, membrane localization. (Scale bar, 2 μm.) (B) Quantification of the sidewall fluorescence label of cells grown in the presence or absence of copper. The density of the fluorescence signals at the sidewall and midcell in BW25113 (wt) and BW25113Δ6LDT cells grown in the presence of 0.5× MIC CuCl2 (+) or without CuCl2 (−) were quantified, and are shown as ratio of the sidewall over midcell signal density. The values are mean ± SD of 44–57 cells.
Copper Inhibits LDTs from E. coli and E. faecium.
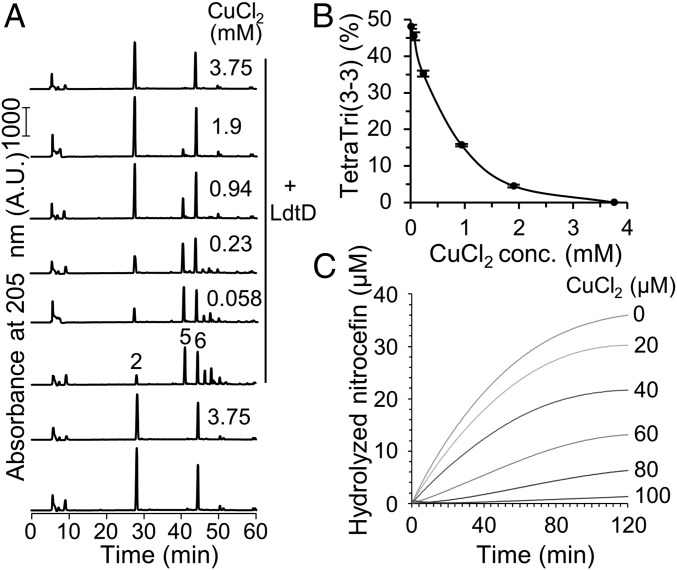
To test a possible direct inhibition by copper we purified two LDTs that were previously shown to bypass PBPs and confer β-lactam resistance, LdtD from E. coli (23) and Ldtfm from E. faecium (31). LdtD was assayed with purified sacculi from BW25113Δ6LDT without copper or in the presence of CuCl2 (different concentrations), followed by digestion with the muramidase cellosyl and analysis of the muropeptide profile (Fig. 4). As expected, LdtD was highly active in the absence of copper, causing a decrease in monomeric Tetra from 61 to 14% (compared with the control without LdtD) and producing a total of 48% LDT product, TriTetra (3–3). Copper chloride inhibited LdtD in a concentration-dependent manner; half-inhibition was reached at ∼0.6 mM CuCl2, and LdtD was virtually inactive in the presence of 3.75 mM CuCl2 (Fig. 4B).
Fig. 4.
Copper inhibits LdtD from E. coli and Ldtfm from E. faecium. (A) PG sacculi from BW25113Δ6LDT were incubated with LdtD from E. coli. CuCl2 was added at different concentrations. Control samples contained no LdtD or no CuCl2. The numbers of the main muropeptides correspond to the structures shown in Fig. 2B. (B) The percentage of TetraTri(3–3) produced by LdtD was plotted against the concentration of CuCl2. The values are mean ± variation of two independent experiments. (C) E. faecium Ldtfm was incubated with nitrocefin (50 µM) in the presence of CuCl2 at a different concentration.
We previously showed that Ldtfm slowly hydrolyzes the chromogenic cephalosporin nitrocefin providing a sensitive assay to determine inhibition of the enzyme (32). Here we show that the hydrolytic activity of Ldtfm was inhibited by CuCl2 in a concentration-dependent manner in the 20- to 100-µM range, with nearly complete inhibition (>98%) at the highest concentration tested (Fig. 4C). The EC50 value for CuCl2 was estimated to be 43 µM. Ldtfm-independent hydrolysis of nitrocefin was not observed in the presence of CuCl2 at 100 µM. Hence, copper inhibits two key LDTs from different species at submillimolar concentrations.
Sub-MIC Copper Resensitizes β-Lactam–Resistant Strains by Inhibition of LDTs.
Several β-lactam–resistant strains, for example E. coli M1 and E. faecium M512, rely on LDTs to bypass the otherwise essential PBPs, replacing the canonical 4–3 cross-links by 3–3 cross-links (18, 23, 24). If copper inhibits the LD-TPases, these strains should be unable to express β-lactam resistance. To test this possibility, we determined the MIC of ampicillin against M1 and M512 in the presence and absence of copper chloride (Table 1).
Table 1.
Effect of copper on the MIC of ampicillin (µg/mL) against E. coli and E. faecium strains
| CuCl2 (mM) | E. coli BW25113* | E. faecium* | |||
| WT (none) | M1 (LDT) | pTRC99aΩ blaTEM116 (β-Lactamase) | M512 (LDT) | D344R (PBP5) | |
| 0 | 8 | 128 | >512 | 512 | 32 |
| 0.25 | 8 | 64 | >512 | 512 | 32 |
| 0.5 | 8 | 64 | >512 | 512 | 32 |
| 1 | 16 | 8 | >512 | 512 | 32 |
| 2 | 8 | 8 | >512 | 4 | 32 |
| 4 | 16 | 16 | >512 | 8 | 64 |
| 8 | 32 | 32 | >512 | 8 | 512 |
The values are medians of five independent experiments.
The resistance mechanism of strains is in parenthesis.
The MICs to CuCl2 of all E. coli strains in Mueller–Hinton medium and E. faecium strains in brain–heart infusion broth were 16 mM. Consistent with earlier results (23), E. coli strain M1 had a 16-fold higher MIC against ampicillin (128 µg/mL) compared with the sensitive wild-type strain due to the overproduction of LdtD and an increased synthesis of the alarmone (p)ppGpp. In the presence of 2 mM CuCl2 the MIC of ampicillin against M1 decreases to 8 µg/mL, which is the MIC of the wild-type strain (Table 1). The MIC of E. coli BW25133 pTRC99aΩblaTEM116, which is resistant to ampicillin, by production of the TEM116 β-lactamase remained high (>512 μg/mL) in the presence of copper chloride, showing that copper specifically prevents LDT-mediated resistance.
Similar results were obtained with E. faecium. Copper strongly reduced β-lactam resistance in strain M512, which relies on LDTs, reducing the MIC from 512 to 4 µg/mL (Table 1). No decrease in MIC was observed for the control strain D344R, which shows moderate intrinsic ampicillin resistance due to the production of a low-affinity PBP5fm (33, 34). Hence, copper prevented the expression of β-lactam resistance in both strains of E. coli and E. faecium, respectively, that rely on LDT activity to bypass PBPs.
Discussion
Copper is an essential cofactor in many organisms, driving processes such as respiration and photosynthesis. However, aquate (or free) copper ions are extremely toxic even at low concentration (in the micromolar range) due to their high affinity for the active site of iron-cluster–containing metallo-enzymes (11, 12). When misloaded with copper ions, these enzymes produce highly toxic reactive oxygen species. Hence, bacteria and other organisms employ specific copper storage proteins and export systems to maintain nontoxic levels of intracellular copper.
At higher concentrations (millimolar range), copper ions can bind to many biomolecules, for example enzymes with active site cysteine residues, and cause protein misfolding and degradation (35). To the best of our knowledge, the impact of copper ions on cell envelope biogenesis has not been previously investigated. Here we show that the LDT cell envelope enzymes are inhibited by copper ions at a concentration that does not prevent cell growth. Presumably, cupric (Cu2+) ions inhibit LDTs by binding to the thiol group of the active site cysteine residue, preventing catalysis. The inhibition of LDTs is not lethal, but results in E. coli in a lower amount of PG-attached Braun’s lipoprotein and abolishes 3–3 cross-link formation, reducing the robustness of cells against extracellular detergent (SDS) and LPS export defects. Interestingly, the inhibition of 3–3 cross-link formation also prevents the LDT-mediated PBP bypass mechanism for β-lactam resistance in both E. coli and E. faecium, illustrating how the targeting of nonessential enzymes can dramatically alter the physiology of a bacterial cell, and how sub-MIC copper ion levels can resensitize resistant strains to β-lactam antibiotics. It is known that copper ions can chemically degrade β-lactams in microbiological growth media (36, 37), which likely explains the increase in MIC values to ampicillin at the higher copper concentrations (Table 1).
Remarkably, copper ions were recently reported to inhibit the metallo-β-lactamase NDM-1 in E. coli MS6192, resulting in a significant increase in susceptibility to ertapenem and meropenem (38). In addition to the inhibition of LDTs, copper ions likely target other periplasmic enzymes with cysteine residues. In the presence of molecular oxygen, copper can also cause damage to the cell membrane through lipid peroxidation (9, 10). Hence, copper weakens the bacterial cell envelope in multiple ways, which might all contribute to the intrinsic, highly efficient antibacterial properties of copper surfaces and agents. The clinical perspectives of our findings are not clear at this time and it remains to be determined if and how copper ions affect pathogens with a high percentage of 3–3 cross-links in their cell wall, such as C. difficile and Mycobacterium tuberculosis (39–42). The latter species has five LDTs, of which one, LdtMt2, is required for virulence (43).
In summary, our study revealed that copper ions affect bacterial cell envelope biogenesis and stability and identified LDTs as targets of copper ions. The physiological consequence of this inhibition is the inability of E. coli and E. faecium strains to utilize LDTs in a bypass mechanism to achieve β-lactam resistance.
Materials and Methods
Bacterial Strains and Growth Conditions.
Strains used in this study are listed in SI Appendix, Table S1. Bacteria were grown on TSB plates or in liquid TSB or LD medium (LB-Lennox medium: 10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl; Difco) at 37 °C.
Spot Plate Assay.
For each strain, 5 mL liquid TSB media were inoculated with a single colony and incubated overnight at 37 °C. The OD578 of the cultures was measured and the cultures were diluted to obtain the same OD578. A 10-fold serial dilution series was prepared using a sterile 96-well microtiter plate (Greiner Bio-One) and multichannel pipettes. The serial dilution was spotted on TSB plates with or without 0.25 or 0.5× MIC of CuCl2 (1.88 or 3.75 mM) and with or without 2% SDS using a replica plater (Sigma Aldrich). The plates were incubated at 37 °C. Pictures were taken after 24 and 48 h with the InGenius Syngene Bio Imaging system.
Antibiotic Susceptibility Testing.
MICs of ampicillin were determined by the microdilution method according to the Clinical and Laboratory Standards Institute recommendations (31), except that cation-adjusted Mueller–Hinton medium was replaced by brain–heart infusion broth for E. faecium strains.
PG Isolation and Analysis.
PG was isolated from E. coli strains and analyzed by HPLC, as previously described (44). The muropeptide fraction of TriLysArg was collected and analyzed at the Newcastle University Pinnacle facility, as described previously (45). SI Appendix, Supplemental Methods contains the details of these methods.
Optimized Protocol for the Incorporation of FDAA (HADA Labeling).
The optimization of in situ labeling of PG with HADA is described in the SI Appendix. Strains were streaked on TSB plates. Single colonies were used to inoculate 25 mL of TSB media and were incubated overnight at 30 °C. Overnight cultures were diluted to an OD578 of 0.1 in TSB media with or without 0.5× MIC of CuCl2 (3.75 mM) and grown at 37 °C until an OD578 of 0.4 was reached. Cells were diluted to an OD578 of 0.1 in a final volume of 500 µL. The cells were incubated with 250 µM of the fluorescent derivative of d-Ala (HADA) for 30 min at 37 °C. The samples were put on ice, and copper ions and the excess of HADA were removed by short (2 min) centrifugation steps at 16,200 × g at 4 °C. The samples were washed with 1.5 mL of 7.5 mM TETA, pH of 3.0 and twice with 1× PBS (1.7 mM KH2PO4, 5 mM Na2HPO4, 150 mM NaCl, pH 7.4) on ice. The initial wash at pH 3.0 and the rapid processing of samples on ice prevented potential removal of HADA by DD-carboxypeptidases and reactivation of LD-TPases for further HADA incorporation. The cells were either fixed for immediate cell imaging or processed to purify the PG sacculi.
Cell Fixation.
The final cell pellets were resuspended in 12.5 µL 1× PBS and 12.5 µL of 3% paraformaldehyde (diluted in 1× PBS) were added for cell fixation. A sample of 3 µL was pipetted on a 1% agarose slide and analyzed by microscopy.
Fast Sacculi Purification for Validation.
The final cell pellets were resuspended in 500 µL H2O and 500 µL of 8% SDS were added. After boiling the samples for 30 min at 100 °C, the cells were centrifuged for 30 min at 16,200 × g. The cells were washed twice by removing the supernatant, resuspension in 1.5 mL H2O and centrifugation at 16,200 × g for 30 min. Samples were resuspended in 20 µL H2O. A 5-µL sample was pipetted on a polylysine-coated glass slide and analyzed by microscopy.
Microscopic Imaging.
Labeled cell samples and isolated sacculi were analyzed using a Nikon Eclipse Ti microscope (Nikon Plan Fluor × 100/1.30 Oil Ph3 DLL objective) equipped with a photometrics/Cool SNAP HQ2 CCD camera using the phase contrast and DAPI channel (filter set: Chroma 49000, excitation at 350/50 nm, emission 460/50 nm). Exposure times were 100 ms for phase contrast and 1 s for fluorescence images. ImageJ was used to crop the images to prepare figures and MetaMorph (v7) was used to quantify the fluorescence signals from 55 (BW25113), 57 (BW25113 with CuCl2), 44 (BW25113Δ6LDT), or 50 (BW25113Δ6LDT with CuCl2) cells. All analyzed cells were from at least two independent experiments and showed a clear HADA labeling at the septum. The fluorescence intensity per unit of area at septal and sidewall positions were quantified, the background subtracted, and the ratio of the sidewall over the midcell signal density was calculated. The values are given as mean ± SD; the Student’s t-test was used to determine the significance of the results.
LdtD Activity Assay.
E. coli LdtD was purified as a soluble protein without the putative membrane anchor using an established and published procedure (23). The purified LdtD protein (2 µM) was incubated with 15 µL of PG sacculi (∼38 µg) from E. coli Δ6LDT strain in absence and presence of different concentrations of CuCl2 (0.058, 0.23, 0.94, 1.9, 3.75 mM). The reactions were performed overnight in a Thermomixer at 37 °C and 700 rpm in a total volume of 50 µL and buffered at pH 7.5 (25 mM Tris, 10 mM MgCl2, and 0.1% Triton X-100). The enzyme was inactivated by boiling for 10 min at 100 °C. The samples were digested with cellosyl at 37 °C and 700 rpm overnight, followed by heat inactivation for 10 min at 100 °C. After a centrifugation step for 15 min at 17,000 × g, the muropeptides present in the supernatant were reduced with sodium borohydride and separated by HPLC according to a previously published procedure (14). The muropeptide profiles were quantified using the Laura software (Lab Logic Systems).
Inhibition Assay of Ldtfm.
The catalytic domain of E. faecium Ldtfm was produced and purified as previously described except that the final size-exclusion chromatography step was performed in ammonium acetate (50 mM; pH 6.4) (32). Ldtfm (10 µM) was incubated with nitrocefin (50 µM) and various concentrations of CuCl2 (0–100 µM) in ammonium acetate (50 mM; pH 6.4). The absorbance at 486 nm was monitored to determine nitrocefin hydrolyzed by Ldtfm (Δε = 15,200 M−1 cm−1).
Supplementary Material
Acknowledgments
We thank Christian Otten (Newcastle University) for purified LdtD from Escherichia coli; Daniela Vollmer (Newcastle University) for help with peptidoglycan purification; and Joe Gray (Newcastle University) for performing mass spectrometry analysis. This work was funded by Wellcome Trust Grant 101824/Z/13/Z; the United Kingdom Medical Research Council within the Antimicrobial Resistance Cross-Council Initiative Collaborative Grant MR/N002679/1 (to W.V.); the European Commission via the International Training Network Train2Target (721484) (to A.P. and W.V.); the Fondation pour la Recherche Médicale Grant ECO20160736080 (to Z.E.); and NIH Grant GM113172 (to M.S.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809285115/-/DCSupplemental.
References
- 1.Dollwet HHA, Sorenson JRJ. Historic uses of copper compounds in medicine. Trace Elem Med. 1985;2:80–87. [Google Scholar]
- 2.Wilks SA, Michels H, Keevil CW. The survival of Escherichia coli O157 on a range of metal surfaces. Int J Food Microbiol. 2005;105:445–454. doi: 10.1016/j.ijfoodmicro.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 3.Warnes SL, Caves V, Keevil CW. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ Microbiol. 2012;14:1730–1743. doi: 10.1111/j.1462-2920.2011.02677.x. [DOI] [PubMed] [Google Scholar]
- 4.Noyce JO, Michels H, Keevil CW. Potential use of copper surfaces to reduce survival of epidemic meticillin-resistant Staphylococcus aureus in the healthcare environment. J Hosp Infect. 2006;63:289–297. doi: 10.1016/j.jhin.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Weaver L, Michels HT, Keevil CW. Survival of Clostridium difficile on copper and steel: Futuristic options for hospital hygiene. J Hosp Infect. 2008;68:145–151. doi: 10.1016/j.jhin.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Faúndez G, Troncoso M, Navarrete P, Figueroa G. Antimicrobial activity of copper surfaces against suspensions of Salmonella enterica and Campylobacter jejuni. BMC Microbiol. 2004;4:19. doi: 10.1186/1471-2180-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warnes SL, Keevil CW. Mechanism of copper surface toxicity in vancomycin-resistant enterococci following wet or dry surface contact. Appl Environ Microbiol. 2011;77:6049–6059. doi: 10.1128/AEM.00597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warnes SL, Green SM, Michels HT, Keevil CW. Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNAs. Appl Environ Microbiol. 2010;76:5390–5401. doi: 10.1128/AEM.03050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grass G, Rensing C, Solioz M. Metallic copper as an antimicrobial surface. Appl Environ Microbiol. 2011;77:1541–1547. doi: 10.1128/AEM.02766-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong R, Kang TY, Michels CA, Gadura N. Membrane lipid peroxidation in copper alloy-mediated contact killing of Escherichia coli. Appl Environ Microbiol. 2012;78:1776–1784. doi: 10.1128/AEM.07068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan G, et al. Anaerobic copper toxicity and iron-sulfur cluster biogenesis in Escherichia coli. Appl Environ Microbiol. 2017;83:e00867-17. doi: 10.1128/AEM.00867-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci USA. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2011;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glauner B, Höltje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- 15.Magnet S, Dubost L, Marie A, Arthur M, Gutmann L. Identification of the L,D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J Bacteriol. 2008;190:4782–4785. doi: 10.1128/JB.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mainardi JL, et al. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J Biol Chem. 2005;280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- 17.Mainardi JL, et al. Unexpected inhibition of peptidoglycan LD-transpeptidase from Enterococcus faecium by the beta-lactam imipenem. J Biol Chem. 2007;282:30414–30422. doi: 10.1074/jbc.M704286200. [DOI] [PubMed] [Google Scholar]
- 18.Biarrotte-Sorin S, et al. Crystal structure of a novel beta-lactam-insensitive peptidoglycan transpeptidase. J Mol Biol. 2006;359:533–538. doi: 10.1016/j.jmb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Triboulet S, et al. Inactivation kinetics of a new target of beta-lactam antibiotics. J Biol Chem. 2011;286:22777–22784. doi: 10.1074/jbc.M111.239988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Triboulet S, et al. Kinetic features of L,D-transpeptidase inactivation critical for β-lactam antibacterial activity. PLoS One. 2013;8:e67831. doi: 10.1371/journal.pone.0067831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnet S, et al. Identification of the L,D-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J Bacteriol. 2007;189:3927–3931. doi: 10.1128/JB.00084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheikh J, Hicks S, Dall’Agnol M, Phillips AD, Nataro JP. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol Microbiol. 2001;41:983–997. doi: 10.1046/j.1365-2958.2001.02512.x. [DOI] [PubMed] [Google Scholar]
- 23.Hugonnet JE, et al. Factors essential for L,D-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli. eLife. 2016;5:e19469. doi: 10.7554/eLife.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mainardi JL, et al. Balance between two transpeptidation mechanisms determines the expression of beta-lactam resistance in Enterococcus faecium. J Biol Chem. 2002;277:35801–35807. doi: 10.1074/jbc.M204319200. [DOI] [PubMed] [Google Scholar]
- 25.Martorana AM, et al. Dissecting Escherichia coli outer membrane biogenesis using differential proteomics. PLoS One. 2014;9:e100941. doi: 10.1371/journal.pone.0100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radkov AD, Hsu YP, Booher G, VanNieuwenhze MS. Imaging bacterial cell wall biosynthesis. Annu Rev Biochem. 2018;87:991–1014. doi: 10.1146/annurev-biochem-062917-012921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuru E, et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuru E, Tekkam S, Hall E, Brun YV, Van Nieuwenhze MS. Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat Protoc. 2015;10:33–52. doi: 10.1038/nprot.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stefanova ME, Davies C, Nicholas RA, Gutheil WG. pH, inhibitor, and substrate specificity studies on Escherichia coli penicillin-binding protein 5. Biochim Biophys Acta. 2002;1597:292–300. doi: 10.1016/s0167-4838(02)00311-4. [DOI] [PubMed] [Google Scholar]
- 30.Amanuma H, Strominger JL. Purification and properties of penicillin-binding proteins 5 and 6 from Escherichia coli membranes. J Biol Chem. 1980;255:11173–11180. [PubMed] [Google Scholar]
- 31.Sacco E, et al. Activation of the L,D-transpeptidation peptidoglycan cross-linking pathway by a metallo-D,D-carboxypeptidase in Enterococcus faecium. Mol Microbiol. 2010;75:874–885. doi: 10.1111/j.1365-2958.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- 32.Edoo Z, Arthur M, Hugonnet JE. Reversible inactivation of a peptidoglycan transpeptidase by a β-lactam antibiotic mediated by β-lactam-ring recyclization in the enzyme active site. Sci Rep. 2017;7:9136. doi: 10.1038/s41598-017-09341-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fontana R, Cerini R, Longoni P, Grossato A, Canepari P. Identification of a streptococcal penicillin-binding protein that reacts very slowly with penicillin. J Bacteriol. 1983;155:1343–1350. doi: 10.1128/jb.155.3.1343-1350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson R, le Bouguénec C, Gutmann L, Horaud T. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J Gen Microbiol. 1985;131:1933–1940. doi: 10.1099/00221287-131-8-1933. [DOI] [PubMed] [Google Scholar]
- 35.Dupont CL, Grass G, Rensing C. Copper toxicity and the origin of bacterial resistance—New insights and applications. Metallomics. 2011;3:1109–1118. doi: 10.1039/c1mt00107h. [DOI] [PubMed] [Google Scholar]
- 36.Fernández-González A, Badía R, Díaz-García ME. Insights into the reaction of beta-lactam antibiotics with copper(II) ions in aqueous and micellar media: Kinetic and spectrometric studies. Anal Biochem. 2005;341:113–121. doi: 10.1016/j.ab.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 37.Beard SJ, Ciccognani DT, Hughes MN, Poole RK. Metal ion-catalysed hydrolysis of ampicillin in microbiological growth media. FEMS Microbiol Lett. 1992;75:207–211. doi: 10.1016/0378-1097(92)90405-d. [DOI] [PubMed] [Google Scholar]
- 38.Djoko KY, et al. Copper ions and coordination complexes as novel carbapenem adjuvants. Antimicrob Agents Chemother. 2018;62:e02280-17. doi: 10.1128/AAC.02280-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peltier J, et al. Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3-3 cross-links. J Biol Chem. 2011;286:29053–29062. doi: 10.1074/jbc.M111.259150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lavollay M, et al. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J Bacteriol. 2008;190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavollay M, et al. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by L,D-transpeptidases. J Bacteriol. 2011;193:778–782. doi: 10.1128/JB.00606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sütterlin L, Edoo Z, Hugonnet JE, Mainardi JL, Arthur M. Peptidoglycan cross-linking activity of l,d-transpeptidases from Clostridium difficile and inactivation of these enzymes by β-lactams. Antimicrob Agents Chemother. 2017;62:e01607-17. doi: 10.1128/AAC.01607-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta R, et al. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med. 2010;16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 45.Bui NK, et al. The peptidoglycan sacculus of Myxococcus xanthus has unusual structural features and is degraded during glycerol-induced myxospore development. J Bacteriol. 2009;191:494–505. doi: 10.1128/JB.00608-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.