Significance
It has been appreciated for decades that translation can strongly impact transcription in bacteria. A popular view is that RNA polymerase and the lead ribosome tend to be physically coupled, explaining how translation can influence transcription. Here, we investigate this model by comparing single- versus multiple-round translation of genes in six representative operons of Escherichia coli. Our data lend no support for a mechanism to ensure or promote coupling between RNA polymerase and the lead ribosome, implying that in these cases any physical coupling occurs stochastically. This work provides important insight on a fundamental aspect of gene expression in bacteria.
Keywords: ribosome, RNA polymerase, NusG, NusE, Rho
Abstract
In prokaryotes, the synthesis of RNA and protein occurs simultaneously in the cytoplasm. A number of studies indicate that translation can strongly impact transcription, a phenomenon often attributed to physical coupling between RNA polymerase (RNAP) and the lead ribosome on the nascent mRNA. Whether there generally exists a mechanism to ensure or promote RNAP–ribosome coupling remains unclear. Here, we used an efficient hammerhead ribozyme and developed a reporter system to measure single- versus multiple-round translation in Escherichia coli. Six pairs of cotranscribed and differentially translated genes were analyzed. For five of them, the stoichiometry of the two protein products came no closer to unity (1:1) when the rounds of translation were severely reduced in wild-type cells. Introduction of mutation rpoB(I572N), which slows RNAP elongation, could promote coupling, as indicated by stoichiometric SspA and SspB products in the single-round assay. These data are consistent with models of stochastic coupling in which the probability of coupling depends on the relative rates of transcription and translation and suggest that RNAP often transcribes without a linked ribosome.
In Bacteria and Archaea, transcription and translation occur together in time and space. Ribosomes can load onto mRNA as soon as the translation initiation region (TIR) emerges from the elongating RNA polymerase (RNAP) (1). Consequently, translation can have large effects on transcription. One well-known example is transcriptional polarity, when a block in translation leads to Rho-dependent transcription termination (2). Another example is transcription attenuation, whereby a slow-moving ribosome promotes the formation of an RNA antiterminator structure to allow continuation of transcription (3).
Several studies have suggested a direct physical link between RNAP and the lead ribosome. Vogel and Jensen (4) showed a tight correlation between transcription and translation elongation rates of lacZ under different growth conditions. Later work by Nudler and coworkers showed that the transcription rate of lacZ is dependent on the translation rate in vivo. Namely, a reduction in the translation elongation rate due to either an antibiotic or a ribosomal mutation resulted in a corresponding reduction in the transcription elongation rate (5). They also provided evidence that the ribosome can prevent RNAP backtracking or stalling and proposed that this tight relationship between transcription and translation helps maintain genome stability (6).
Structural studies have suggested several possible ways that RNAP and the ribosome might interact. The transcriptional factor NusG (or its paralog RfaH) can act as a linker for physical coupling (7–9). The N-terminal domain of NusG interacts with RNAP, while the C-terminal domain binds to protein S10 of the ribosome (also known as “NusE”). The interaction between NusG and the ribosome was suggested to prevent NusG from binding or activating the termination factor Rho. Other studies indicate that RNAP can interact directly with the ribosome. An “expressome” complex, determined by cryo-EM, revealed extensive interaction between RNAP and the 70S ribosome (10). Four of the five subunits of RNAP contact the 30S subunit of the ribosome, with the RNA exit region of RNAP docked onto the mRNA entry tunnel of the ribosome, resulting in seamless protection of the bound mRNA. Another cryo-EM structure showed RNAP bound to the isolated 30S subunit in a different way (11). In this complex, which lacks mRNA, the RNA exit region of RNAP lies near the mRNA exit tunnel of the ribosome, close to the anti-Shine–Dalgarno sequence of the 16S rRNA. In vitro binding studies have shown that core RNAP binds the ribosome or either subunit with a similar affinity (Kd ∼0.9 μM) in the absence of mRNA (12). While these studies are generally consistent with a physical link between RNAP and the ribosome, it remains unclear which, if any, of these interactions are involved in coupling RNAP and the ribosome in the cell.
Whether a mechanism typically exists to ensure or promote the coupling of transcription and translation remains an open question. One can imagine, for example, that RNAP is programmed to pause downstream of the TIR to wait for the lead ribosome to catch up and connect with it (13, 14). Alternatively, RNAP could actively recruit the ribosome to the site of translation initiation, for example with the help of other factors (8, 15). In this study, we developed a reporter system to compare single-round translation and multiple-round translation. Our results provide no evidence for a mechanism to ensure coupling. They suggest instead that coupling is stochastic and that RNAP often transcribes without a linked ribosome.
Results
Experimental Rationale.
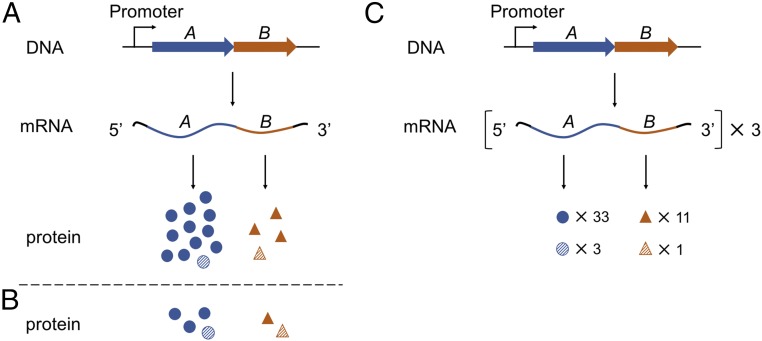
Coupling of transcription and translation requires an interaction between the elongating RNAP and the first ribosome to translate the nascent mRNA. As only this lead ribosome can potentially engage RNAP, we reasoned that a means to distinguish first-round and multiple-round translation would inform on the phenomenon of coupling. Consider, for example, the case of an operon in which genes A and B are transcribed at a ratio of 1:1 and are translated at a ratio of 3:1 (Fig. 1A). If a mechanism exists to ensure RNAP–ribosome interaction throughout transcription, one would expect the ratio of first-round translation products to match that of transcription, 1:1. On the other hand, if there is no such mechanism, any coupling would be stochastic, and the ratio of cotranscriptional translation products would match that of multiple-round translation, 3:1 (Fig. 1C).
Fig. 1.
Models of strict versus stochastic coupling. (A and B) Scenarios in which transcription and translation are strictly coupled. (A) Genes A and B are cotranscribed, resulting in stoichiometric levels of A and B mRNA (1:1). First-round translation generates equivalent amounts of proteins A and B (1:1, striped symbols), whereas multiple-round translation yields different levels of total proteins (3:1, all symbols). (B) A hypothetical case: Partial reduction of translation rounds yields a product ratio of 2:1 clearly distinct from the original 3:1 ratio. (C) A scenario in which transcription and translation are uncoupled or stochastically coupled. In this case, RNAP has effectively no impact on the lead ribosome, and hence the ratio of protein products made during transcription (3:1, striped symbols) matches that of multiple-round translation (3:1, all symbols).
A Reporter System to Measure Single- Versus Multiple- Round Translation.
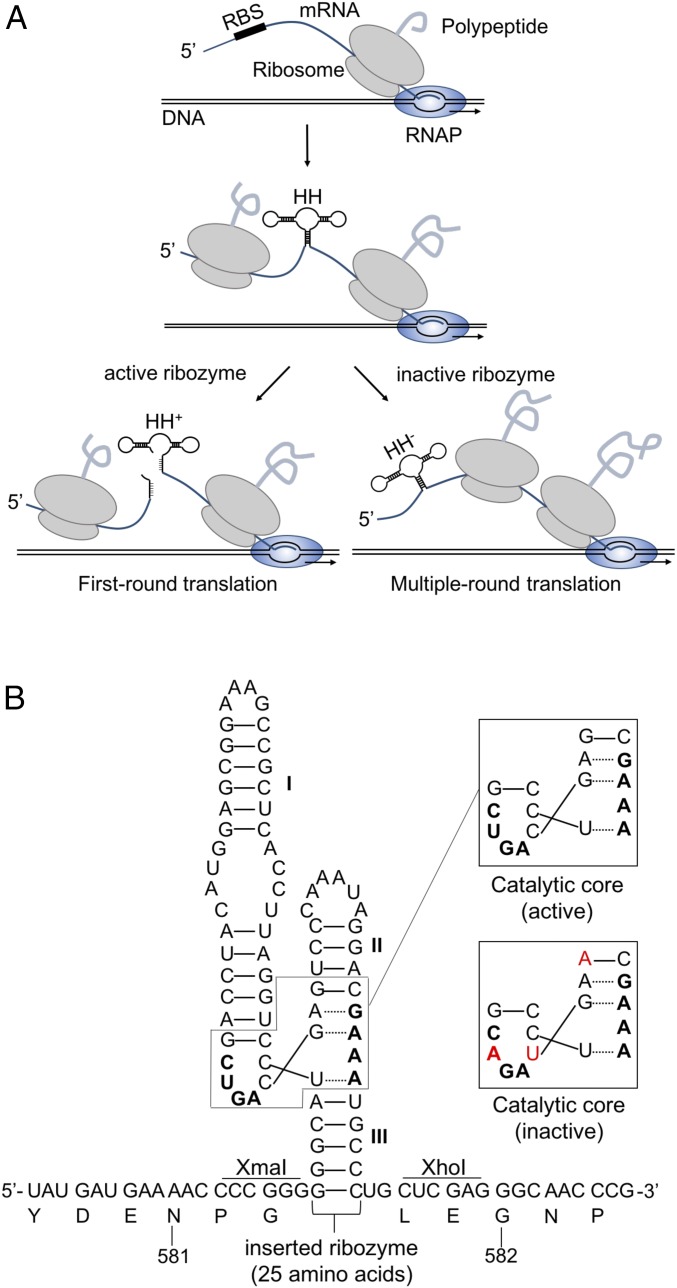
A reporter system was developed to compare cotranscriptional (single-round) translation and multiple-round translation (Fig. 2). DNA encoding a self-cleaving hammerhead ribozyme derived from Schistosoma mansoni (16, 17) was inserted into a translational lacZ reporter after codon 581, a position corresponding to a solvent-exposed loop (residues 578–584) of the β-gal protein (Fig. 2B). During expression of this construct, the coupled ribosome should, in theory, be able to complete synthesis of LacZ, but trailing ribosomes will not due to rapid folding and cleavage of the ribozyme (Fig. 2A). In other words, the hammerhead ribozyme effectively limits translation to one round. To measure multiple-round translation, three mutations were introduced in the catalytic core of the ribozyme without altering the polypeptide sequence encoded (Fig. 2).
Fig. 2.
A reporter system to measure single-round versus multiple-round translation. (A) DNA encoding the hammerhead ribozyme was inserted into lacZ. Due to rapid cotranscriptional folding and cleavage of the ribozyme, only the lead (coupled) ribosome should be able to translate the entire mRNA, providing a measure of single-round translation (left pathway). An analogous reporter with an inactive hammerhead provides a measure of multiple-round translation (right pathway). (B) Sequence of the inserted hammerhead ribozyme between codons 581 and 582 of lacZ mRNA. The catalytic core of the ribozyme is boxed, with critical nucleotides indicated in bold. Mutated bases are labeled in red.
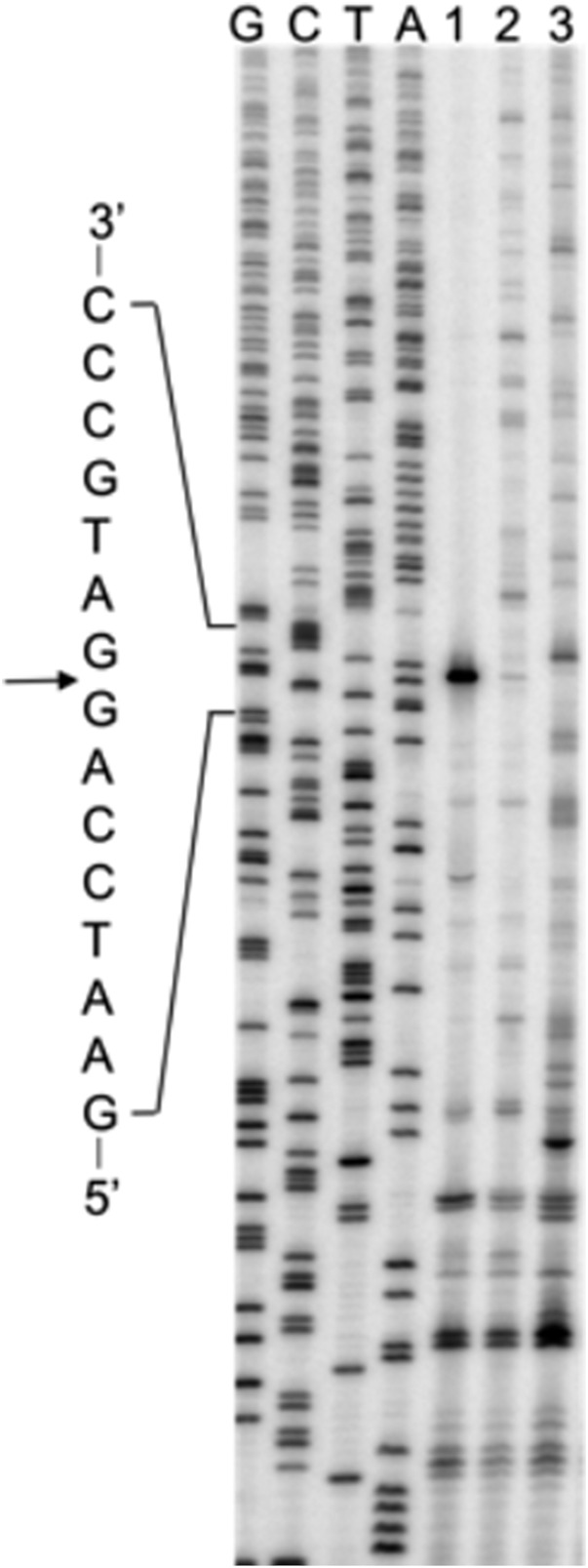
To assess the feasibility of the system, we first moved the hammerhead sequence into lacZ on a high-copy plasmid, pJC27 (18), generating pMC55 (with the inactive hammerhead sequence, lacZ-HH−) and pMC56 (with the active hammerhead sequence, lacZ-HH+). Transformants carrying pMC55 had β-gal activity (1.5 × 104 units) similar to those carrying pJC27 (1.9 × 104 units), consistent with earlier evidence that loop 578–584 can tolerate various insertions (19) and showing that the hammerhead secondary structure has no appreciable effect on LacZ production. Transformants carrying pMC56 had about 40-fold lower β-gal activity (4.3 × 102 units), consistent with rapid ribozyme cleavage. Total RNA was isolated from cells harboring pJC27, pMC55, or pMC56 and was subjected to primer extension analysis (Fig. 3). When the active ribozyme was present (lacZ-HH+), a predominant cDNA product corresponding to the ribozyme cleavage site was observed, and virtually no larger cDNA products were seen. By contrast, in the lacZ-HH− and the wild-type lacZ cases, multiple longer cDNA products were seen throughout the lanes. These data are consistent with rapid cleavage of lacZ-HH+ mRNA, which substantially limits the rounds of translation.
Fig. 3.
Ribozyme cleavage is efficient in vivo. Total RNA was isolated from strains expressing lacZ-HH+ mRNA (lane 1), lacZ-HH− mRNA (lane 2), or wild-type lacZ mRNA (lane 3) and was subjected to primer extension analysis. DNA sequencing of pMC55 (lacZ-HH−) was performed, and reactions were run in adjacent lanes (G, C, T, and A). A portion of the cDNA complementary to the hammerhead sequence is shown, with the cleavage site indicated by the arrow. This experiment was performed twice with virtually identical results.
Operon Choice and Strain Construction.
The first two genes of six operons were chosen for analysis (SI Appendix, Fig. S1). For each operon (i) the mRNA levels of the genes are similar, based on RNA-sequencing coverage (20); (ii) the translation rates of the genes are substantially different, based on ribosome profiling (ribo-seq) coverage (20); and (iii) the protein products of the genes are cytosolic (21), avoiding potential problems caused by cotranslational membrane insertion. Two reporter fusions were made for each gene. A DNA fragment extending from the operon promoter to about the 50th codon of the particular gene was cloned upstream of the engineered lacZ in pMC95 (lacZ-HH−) and pMC120 (lacZ-HH+), derivatives of the suicide vector pNPTS-lacZ (22). Each resulting plasmid was integrated into the chromosome of strain CSH142 (23) by single cross-over recombination. This yielded a set of strains, each having an intact copy of the operon downstream of the engineered translational fusion (SI Appendix, Fig. S2). None of the strains exhibited a growth defect.
The Stoichiometry of Protein Products from Differentially Translated Genes Is Typically Unaltered by Limiting the Rounds of Translation.
β-Gal activity was measured in exponentially growing cells, with lacZ-HH− and lacZ-HH+ fusions reporting on multiple- and single-round translation, respectively (Table 1). For operon sspAB, multiple-round translation of sspA was over three times higher than that of sspB (3.2:1), consistent with the ribo-seq data (4:1). When measuring single-round translation, ∼30-fold drops in β-gal activity were seen, consistent with ribozyme cleavage limiting translation rounds substantially. However, the ratio of protein products remained basically the same (3.8:1), a result contrary to that predicted by the strict coupling model (Fig. 1A). Similar results were seen for four other operons. In the case of purMN, the ratio of products from multiple-round translation (2.7:1) was in line with that predicted by ribo-seq (3:1). The presence of the active hammerhead ribozyme reduced translation by ∼50-fold for both fusions, leaving the product ratio virtually unchanged (3:1). In the case of moeAB, the product ratio from multiple-round translation was 3.2:1. This ratio is somewhat smaller than that expected from ribo-seq coverage (6:1), which could be a consequence of the particular fusion junctions. Substantial drops (∼40 fold) in protein production were observed in corresponding strains reporting on single-round translation, which increased the product ratio slightly (3.8:1). For operon purHD, the multiple-round translation reporters gave a product ratio of 3.4:1. The corresponding single-round translation reporters yielded a somewhat higher product ratio (5.5:1) due to a 49-fold and 80-fold decrease in purH′-lacZ and purD′-lacZ translation, respectively. In the case of aceBA, the multiple-round translation reporters indicated a product ratio of 1:4, in line with the ribo-seq data. Translation of aceB′-lacZ and aceA′-lacZ decreased by 150-fold and 55-fold, respectively, in the presence of the active hammerhead, leading to a somewhat larger difference in protein production (1:11). Thus, a consistent trend was observed for these five operons. Limiting the rounds of translation failed to change the product ratio toward unity (1:1). These data argue against the strict coupling model.
Table 1.
Protein production from multiple- and single-round translation reporters for six representative operons in wild-type (rpoB+) E. coli
| Operon | Reporter design | Fusions* | β-Gal activity† | Product ratio‡ |
| sspAB | Multiple-round | sspA′-lacZ-HH− | 403 ± 6.0 | 3.2:1 |
| sspB′-lacZ-HH− | 125 ± 8.9 | |||
| Single-round | sspA′-lacZ-HH+ | 15 ± 0.7 | 3.8:1 | |
| sspB′-lacZ-HH+ | 3.9 ± 0.3 | |||
| purMN | Multiple-round | purM′-lacZ-HH− | 59 ± 3.8 | 2.7:1 |
| purN′-lacZ-HH− | 22 ± 0.6 | |||
| Single-round | purM′-lacZ-HH+ | 1.2 ± 0.1 | 3.0:1 | |
| purN′-lacZ-HH+ | 0.4 ± 0.1 | |||
| moeAB | Multiple-round | moeA′-lacZ-HH− | 58 ± 0.9 | 3.2:1 |
| moeB′-lacZ-HH− | 18 ± 0.4 | |||
| Single-round | moeA′-lacZ-HH+ | 1.5 ± 0.1 | 3.8:1 | |
| moeB′-lacZ-HH+ | 0.4 ± 0.1 | |||
| purHD | Multiple-round | purH′-lacZ-HH− | 54 ± 5.4 | 3.4:1 |
| purD′-lacZ-HH− | 16 ± 0.3 | |||
| Single-round | purH′-lacZ-HH+ | 1.1 ± 0.1 | 5.5:1 | |
| purD′-lacZ-HH+ | 0.2 ± 0.1 | |||
| aceBA | Multiple-round | aceB′-lacZ-HH− | 30 ± 3.7 | 1:4.0 |
| aceA′-lacZ-HH− | 121 ± 6.2 | |||
| Single-round | aceB′-lacZ-HH+ | 0.2 ± 0.1 | 1:11 | |
| aceA′-lacZ-HH+ | 2.2 ± 0.1 | |||
| frmRAB | Multiple-round | frmR′-lacZ-HH− | 4.5 ± 0.1 | 1:11 |
| frmA′-lacZ-HH− | 50 ± 1.6 | |||
| Single-round | frmR′-lacZ-HH+ | 0.5 ± 0.1 | 1:2.8 | |
| frmA′-lacZ-HH+ | 1.4 ± 0.2 |
HH−, inactive hammerhead ribozyme; HH+, active hammerhead ribozyme.
β-Galactosidase activity based on cleavage of CPRG. Data represent mean ± SEM (n ≥ 3). Background ≤0.1, based on measurements of CSH142 cells.
First gene:second gene.
One gene pair analyzed, frmRA of the frmRAB operon, behaved differently. The product ratio based on multiple-round translation reporters was 1:11, quite different from the 1:4 ratio predicted by ribo-seq. In the context of single-round translation, the product ratio given by the corresponding reporters changed to 1:2.8. While this change toward unity is consistent with a mechanism to promote coupling, there are caveats to this experiment. The translation of frmR′-lacZ-HH− was considerably lower than expected, based on ribo-seq coverage (SI Appendix, Fig. S3), and the difference in β-gal activity between frmR′-lacZ-HH+ and frmR′-lacZ-HH− was unusually small (ninefold).
A Mutation That Slows RNAP Can Promote Transcription–Translation Coupling.
The data above provide no direct evidence for physical coupling. The lacZ-HH+ activity detected may be the product of RNAP-coupled ribosomes and/or uncoupled ribosomes fast enough to translate the hammerhead RNA before cleavage occurs. In fact, our data are consistent with either stochastic coupling or no coupling and cannot distinguish these possibilities.
We reasoned that transcription–translation coupling might become evident if the elongation rate of RNAP was reduced. To test this, we moved reporters for the sspAB, purMN, and moeAB operons into strains containing rpoB(H526Y) (24) or rpoB(I572N) (25), mutations that increase or decrease the intrinsic speed of RNAP, respectively (Table 2). For all three operons, increasing the rate of transcription elongation had little bearing on the results. Product ratios of single-round translation were similar to those of multiple-round translation and were no closer to unity, data comparable to that seen in the wild-type (rpoB+) strain (Table 1). By contrast, slowing RNAP elongation had an obvious effect on sspAB (Table 2). In the presence of rpoB(I572N), multiple-round translation yielded SspA and SspB at a ratio of 2.9:1, whereas single-round translation yielded near-stoichiometric products (1:1.1). These data are consistent with tight coupling between transcription and first-round translation, as depicted in Fig. 1A. While slowing transcription clearly impacted sspAB translation, mutation rpoB(I572N) had no such effect on purMN or moeAB. While the basis of this idiosyncrasy remains unclear, we suspect that only in the case of sspAB is RNAP slowed sufficiently by rpoB(I572N) to allow the lead ribosome translating sspB to catch up and associate with RNAP.
Table 2.
Protein production from multiple- and single-round translation reporters in rpoB mutant strains
| Operon | Reporter design | rpoB(H526Y)* | rpoB(I572N)† | ||
| β-Gal activity‡ | Product ratio§ | β-Gal activity‡ | Product ratio§ | ||
| sspAB | Multiple-round | 347 ± 55 | 3.7:1 | 408 ± 50 | 2.9:1 |
| 95 ± 5.7 | 143 ± 7.3 | ||||
| Single-round | 12 ± 0.2 | 4.4:1 | 7.5 ± 0.8 | 1:1.1 | |
| 2.7 ± 0.3 | 8.4 ± 0.1 | ||||
| purMN | Multiple-round | 49 ± 0.4 | 2.6:1 | 55 ± 1.6 | 2:1 |
| 19 ± 0.4 | 28 ± 0.8 | ||||
| Single-round | 1.1 ± 0.1 | 3.7:1 | 1.2 ± 0.1 | 2.4:1 | |
| 0.3 ± 0.1 | 0.5 ± 0.1 | ||||
| moeAB | Multiple-round | 47 ± 1.7 | 3.1:1 | 87 ± 0.7 | 2.6:1 |
| 15 ± 0.4 | 33 ± 2.1 | ||||
| Single-round | 1.9 ± 0.1 | 6.3:1 | 3.2 ± 0.1 | 4:1 | |
| 0.3 ± 0.1 | 0.8 ± 0.1 | ||||
Causes increased transcription elongation rate.
Causes decreased transcription elongation rate.
β-Gal activity based on cleavage of CPRG. Data represent mean ± SEM (n ≥ 3). Background ≤0.1, based on measurements of CSH142 cells.
First gene:second gene.
Discussion
It is widely thought that ribosomes inhibit premature transcription termination by blocking potential rut (Rho utilization) sites, blocking Rho’s access to RNAP, and/or preventing the formation of intrinsic RNA terminator structures (2, 26, 27). Models depicting physical coupling between RNAP and the lead ribosome are attractive, as they can rationalize the effects of translation on transcription in a straightforward way. An implication of such models is that RNAP is usually linked to a ribosome to avoid premature termination. In this study, we examined cotranscribed gene pairs in six operons, testing the hypothesis that first-round translation is normally coupled to transcription. For five of them, we found that the difference in protein production between the two genes becomes no smaller when hammerhead-catalyzed mRNA cleavage substantially limits the rounds of translation. These data argue against the idea that there generally exists a mechanism to ensure (or even promote) coupling between RNAP and the lead ribosome. We infer that coupling is stochastic, rather than ensured, and depends on the relative rates of transcription and translation of the particular gene.
For our sixth gene pair (frmRA of the frmRAB operon), the product ratio did become closer to unity when translation rounds were limited. While this trend is consistent with a mechanism to promote coupling (Fig. 1A), there are caveats to this particular experiment. Namely, β-gal activity of the frmR′-lacZ-HH− strain was considerably lower than expected, based on ribo-seq data, and was only ninefold higher than that of the frmR′-lacZ-HH+ strain. FrmR functions as a repressor that regulates frmRAB transcription (28), a fact we initially overlooked. This could be related to the anomalous β-gal data and in any event complicates the experiment, so we are hesitant to draw conclusions regarding this gene pair.
One might be concerned about our lack of evidence confirming that translation of HH+ mRNA is limited to one round. However, our conclusions rely only on the assumption that translation rounds are substantially reduced by the active hammerhead. For example, even if several ribosomes manage to pass through the hammerhead before its cleavage, the ratio of products would still move closer to unity, according to the strict-coupling model (Fig. 1B). In other words, if RNAP rate-limits the lead ribosome, this effect should be detectable as long as translation rounds are substantially reduced. With the sole exception of frmR (discussed above), the active hammerhead reduces β-gal production by 27- to 150-fold in wild-type cells (Table 1). Based on bulk measurements of macromolecular synthesis, Bremer and Dennis deduced that, on average, mRNA is translated ∼60 times in rapidly growing Escherichia coli (29). Quantitative analyses of specific genes lacZ and trpE showed that the corresponding mRNA is translated ∼40 and ∼20 times, respectively (30, 31). These values are on par with the fold reductions in translation caused by the active hammerhead, suggesting that translation rounds are being limited to near 1 in our HH+ strains.
Recent studies on Rho-independent transcription termination suggest that coupling between the ribosome and RNAP is generally stochastic (26), consistent with our current findings. Li et al. (26) varied the position of an intrinsic terminator with respect to the stop codon. They found that the efficiency of transcription termination gradually increases with distance from the end of the coding region, and their data fit well to the stochastic-coupling model. They also showed that the position of a terminator within a gene matters—terminators embedded at the 5′ end are suppressed by translation to a lesser degree than those at the 3′ end. These data can also be explained by stochastic dynamics of the molecular machines involved. In the latter (3′-end terminator) case, ribosomes have more time to catch up with RNAP and inhibit transcription termination.
Nudler and coworkers (5) suggested that for most genes the lead ribosome would catch up to RNAP by the time ≤200 nt of nascent RNA are made. However, we see no evidence for coupling in wild-type cells even though the hammerhead lies more than 1,700 nt from the transcription start site. This indicates that the probability of coupling is generally lower than what Nudler and coworkers predicted and that RNAP often transcribes without a linked ribosome. In our study, the operons chosen are widely distributed across genome, and each measurement relied on a single-copy lacZ reporter integrated at the endogenous chromosomal locus of interest. Moreover, the translation rates of the genes analyzed fall within the normal range, based on ribosome-profiling data collected under analogous conditions (SI Appendix, Fig. S4). Hence we believe that our data are quite representative of gene expression in the cell. It is possible that translation initiation is particularly rapid in the experimental system of Nudler (5) and in the similar system of Kim and coworkers (32), which allows highly efficient coupling.
One limitation of our study is that it involved only a handful of genes, all of which lie in operons and encode cytoplasmic products. It is possible—even probable—that some other genes normally do exhibit tight transcription–translation coupling. Transcription of such genes may be inherently prone to Rho-dependent termination, an issue remedied by coupling, as often envisaged. Indeed, our current findings challenge only the view that tight coupling represents the norm in the cell.
Finally, it should be mentioned that questions regarding the basis of polarity remain. de Smit et al. (33, 34) varied the rate of translation initiation and did not observe intracistronic transcription termination unless the rate of ribosome loading was very small (≤0.006/s). Recent ChIP–chip analyses revealed that Rho associates with virtually all transcripts shortly after transcription begins but does not cause termination in most cases (35). These observations suggest that there must be other signals that allow Rho to distinguish normal from abnormal translation and promote termination only in response to the latter. Further studies will be needed to understand Rho activation and the ability of ribosomes to impact transcription processivity, as coupling seems to be only part of the story.
Methods
Plasmid and Strain Construction.
Two restriction sites (XhoI and XmaI) were introduced at codon 581 of the lacZ gene of pJC27 (18) to generate pMC09. Hammerhead ribozyme sequences were synthesized as described (36) using primers US-HH− and DS-HH− or US-HH+ and DS-HH+ (SI Appendix, Table S1) and Sequenase version 2.0 (Thermo Fisher Scientific). Fragments were cut with XhoI and XmaI and cloned into pMC09 to generate pMC55 (lacZ-HH−) and pMC56 (lacZ-HH+).
To make translational fusions, Bsu 36I-SacI restriction fragments from pMC55 or pMC56 were subcloned to the suicide vector pNPTS-lacZ (22) to generate pMC95 (lacZ-HH−) and pMC120 (lacZ-HH+), using the cloning strain DH5α-λpir. For each gene of interest, a fragment extending from the operon promoter to around codon 50 of the gene was amplified by PCR (SI Appendix, Table S1) and cloned into pMC95 and pMC120 via restriction sites NcoI and EcoRI, resulting in a translational lacZ fusion. For moeAB, compatible restriction enzyme BspHI was used instead of NcoI. For purHD and frmRA, BamHI was used instead of EcoRI.
Resulting plasmids were transformed into a diaminopimelic acid (DAP) auxotrophic E. coli donor strain WM3064 [thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::(erm pir)] and then were moved into CSH142 [F− ara Δ(gpt-lac)5 thi] (23) by conjugation (37, 38). Donor strains were mixed with CSH142 at a ratio of 1:2, spotted on LB + DAP (0.3 mM) plates, and incubated at 37 °C for 3–5 h. Cells were collected and plated on LB with kanamycin (50 μg/mL) and X-Gal (20 μg/mL) to select for transconjugants. Strains harboring a single integrated plasmid at the expected location were confirmed by PCR. Three or more independent isolates of each strain were assayed for β-gal activity.
Mutations rpoB(H526Y) and rpoB(I572N) were moved into CSH142 separately by P1 phage transduction, screening for rifampicin (50 μg/mL) resistance. Fusions were then moved by conjugation as described above. Three or more independent isolates of each strain were assayed for β-gal activity.
Primer Extension Analysis.
Cultures (10 mL) of CSH142 (pJC27), CSH142 (pMC55), and CSH142 (pMC56) were grown in LB medium with chloramphenicol (30 μg/mL) to midlog phase and were induced with 1 mM isopropyl β-d-thiogalactopyranoside for 2 h. Cells were collected, resuspended in TE buffer [10 mM Tris⋅HCl (pH 8.0), 1 mM EDTA], and extracted with TRIzol (Invitrogen). The aqueous phase was subsequently extracted with chloroform, and total RNA was ethanol precipitated. RNA pellets were washed with 70% ethanol and were dissolved in water. A radiolabeled reverse primer (5′-[32P]-GACCAGACCGTTCATACAG-3′) was annealed to codons 602–608 of lacZ mRNA in AMV reaction buffer (Life Sciences Advanced Technologies, Inc.). AMV reverse transcriptase (2 U; Life Sciences Advanced Technologies, Inc.) and dNTPs (0.25 mM each) were added, and reactions were incubated for 10 min at 37 °C to allow primer extension. Products were resolved using 6% denaturing (7 M urea) PAGE. In parallel, the same radiolabeled primer was used to sequence pMC55, and reactions were run in adjacent lanes. Gels were visualized by a Typhoon FLA 9000 phosphorimager (GE Healthcare).
β-Gal Assays.
Cells were grown in LB with kanamycin (50 μg/mL) and grown to midlog phase (OD600 ∼0.5). β-Gal assays were performed as described previously (39) using the substrate chlorophenol red-β-d-galactopyranoside (CPRG; Roche).
Supplementary Material
Acknowledgments
We thank D. Watkins for help with plasmid construction, K. Jung and J. Lassak for the vector pNPTS-lacZ and strains DH5α-λpir and WM3064, I. Artsimovitch for RNAP mutant strains, and C. Gross, A. Buskirk, M. Ibba, I. Artsimovitch, B. Roy, L. Ying, and Z. McNutt for comments on the manuscript. This work was supported by NIH Grant GM072528.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812940115/-/DCSupplemental.
References
- 1.McGary K, Nudler E. RNA polymerase and the ribosome: The close relationship. Curr Opin Microbiol. 2013;16:112–117. doi: 10.1016/j.mib.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson JP. Preventing the synthesis of unused transcripts by Rho factor. Cell. 1991;64:1047–1049. doi: 10.1016/0092-8674(91)90257-y. [DOI] [PubMed] [Google Scholar]
- 3.Yanofsky C. Transcription attenuation: Once viewed as a novel regulatory strategy. J Bacteriol. 2000;182:1–8. doi: 10.1128/jb.182.1.1-8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel U, Jensen KF. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J Bacteriol. 1994;176:2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burmann BM, et al. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 8.Burmann BM, et al. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxena S, et al. Escherichia coli transcription factor NusG binds to 70S ribosomes. Mol Microbiol. 2018;108:495–504. doi: 10.1111/mmi.13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohler R, Mooney RA, Mills DJ, Landick R, Cramer P. Architecture of a transcribing-translating expressome. Science. 2017;356:194–197. doi: 10.1126/science.aal3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demo G, et al. Structure of RNA polymerase bound to ribosomal 30S subunit. eLife. 2017;6:e28560. doi: 10.7554/eLife.28560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan H, et al. Transcription-translation coupling: Direct interactions of RNA polymerase with ribosomes and ribosomal subunits. Nucleic Acids Res. 2017;45:11043–11055. doi: 10.1093/nar/gkx719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts JW. Molecular biology. Syntheses that stay together. Science. 2010;328:436–437. doi: 10.1126/science.1189971. [DOI] [PubMed] [Google Scholar]
- 14.Burmann BM, Rösch P. The role of E. coli Nus-factors in transcription regulation and transcription:translation coupling: From structure to mechanism. Transcription. 2011;2:130–134. doi: 10.4161/trns.2.3.15671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knauer SH, Rösch P, Artsimovitch I. Transformation: The next level of regulation. RNA Biol. 2012;9:1418–1423. doi: 10.4161/rna.22724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol. 1998;18:3880–3888. doi: 10.1128/mcb.18.7.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canny MD, et al. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- 18.Curran JF, Yarus M. Base substitutions in the tRNA anticodon arm do not degrade the accuracy of reading frame maintenance. Proc Natl Acad Sci USA. 1986;83:6538–6542. doi: 10.1073/pnas.83.17.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feliu JX, Villaverde A. Engineering of solvent-exposed loops in Escherichia coli beta-galactosidase. FEBS Lett. 1998;434:23–27. doi: 10.1016/s0014-5793(98)00943-0. [DOI] [PubMed] [Google Scholar]
- 20.Balakrishnan R, Oman K, Shoji S, Bundschuh R, Fredrick K. The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 2014;42:13370–13383. doi: 10.1093/nar/gku1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misra RV, Horler RS, Reindl W, Goryanin II, Thomas GH. EchoBASE: An integrated post-genomic database for Escherichia coli. Nucleic Acids Res. 2005;33:D329–D333. doi: 10.1093/nar/gki028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fried L, Lassak J, Jung K. A comprehensive toolbox for the rapid construction of lacZ fusion reporters. J Microbiol Methods. 2012;91:537–543. doi: 10.1016/j.mimet.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Miller JH. Bacterial Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1992. A short course. [Google Scholar]
- 24.McDowell JC, Roberts JW, Jin DJ, Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- 25.Zhou YN, et al. Isolation and characterization of RNA polymerase rpoB mutations that alter transcription slippage during elongation in Escherichia coli. J Biol Chem. 2013;288:2700–2710. doi: 10.1074/jbc.M112.429464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R, Zhang Q, Li J, Shi H. Effects of cooperation between translating ribosome and RNA polymerase on termination efficiency of the Rho-independent terminator. Nucleic Acids Res. 2016;44:2554–2563. doi: 10.1093/nar/gkv1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roland KL, Liu CG, Turnbough CL., Jr Role of the ribosome in suppressing transcriptional termination at the pyrBI attenuator of Escherichia coli K-12. Proc Natl Acad Sci USA. 1988;85:7149–7153. doi: 10.1073/pnas.85.19.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herring CD, Blattner FR. Global transcriptional effects of a suppressor tRNA and the inactivation of the regulator frmR. J Bacteriol. 2004;186:6714–6720. doi: 10.1128/JB.186.20.6714-6720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. Ecosal Plus. 2008;3 doi: 10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- 30.Kennell D, Riezman H. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J Mol Biol. 1977;114:1–21. doi: 10.1016/0022-2836(77)90279-0. [DOI] [PubMed] [Google Scholar]
- 31.Baker R, Yanofsky C. Transcription initiation frequency and translational yield for the tryptophan operon of Escherichia coli. J Mol Biol. 1972;69:89–102. doi: 10.1016/0022-2836(72)90025-3. [DOI] [PubMed] [Google Scholar]
- 32.Iyer S, Le D, Park BR, Kim M. Distinct mechanisms coordinate transcription and translation under carbon and nitrogen starvation in Escherichia coli. Nat Microbiol. 2018;3:741–748. doi: 10.1038/s41564-018-0161-3. [DOI] [PubMed] [Google Scholar]
- 33.de Smit MH, Verlaan PW, van Duin J, Pleij CW. Intracistronic transcriptional polarity enhances translational repression: A new role for Rho. Mol Microbiol. 2008;69:1278–1289. doi: 10.1111/j.1365-2958.2008.06360.x. [DOI] [PubMed] [Google Scholar]
- 34.de Smit MH, Verlaan PW, van Duin J, Pleij CW. In vivo dynamics of intracistronic transcriptional polarity. J Mol Biol. 2009;385:733–747. doi: 10.1016/j.jmb.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Mooney RA, et al. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi JJ, Kierzek R, Huang T, Walker PA, Itakura K. An alternate method for synthesis of double-stranded DNA segments. J Biol Chem. 1982;257:9226–9229. [PubMed] [Google Scholar]
- 37.Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J Bacteriol. 1997;179:538–540. doi: 10.1128/jb.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang S, et al. Paradigm for industrial strain improvement identifies sodium acetate tolerance loci in Zymomonas mobilis and Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2010;107:10395–10400. doi: 10.1073/pnas.0914506107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin D, Abdi NM, Fredrick K. Characterization of 16S rRNA mutations that decrease the fidelity of translation initiation. RNA. 2007;13:2348–2355. doi: 10.1261/rna.715307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.