Significance
The ability of cells to migrate collectively underlies many biological processes. The parapineal is a small group of cells that requires Fgf8 to migrate from the midline to the left side of the zebrafish forebrain. Studying the dynamics of FGF pathway activation reveals that FGF activity is restricted to a few left-sided parapineal cells. Global activation of the FGF pathway interferes with parapineal migration in wild-type embryos, while focal activation in few parapineal cells can restore migration in fgf8−/− mutants, indicating that FGF pathway activation in leading cells is required for collective migration. We show that focal FGF activity is influenced by left-sided Nodal signaling. Our findings may apply to other contexts of FGF-dependent cell migration during development or metastasis.
Keywords: collective cell migration, FGF signaling pathway, left/right brain asymmetry, Nodal signaling pathway, zebrafish brain development
Abstract
The ability of cells to collectively interpret surrounding environmental signals underpins their capacity to coordinate their migration in various contexts, including embryonic development and cancer metastasis. One tractable model for studying collective migration is the parapineal, a left-sided group of neurons that arises from bilaterally positioned precursors that undergo a collective migration to the left side of the brain. In zebrafish, the migration of these cells requires Fgf8 and, in this study, we resolve how FGF signaling correlates with—and impacts the migratory dynamics of—the parapineal cell collective. The temporal and spatial dynamics of an FGF reporter transgene reveal that FGF signaling is activated in only few parapineal cells usually located at the leading edge of the parapineal during its migration. Overexpressing a constitutively active Fgf receptor compromises parapineal migration in wild-type embryos, while it partially restores both parapineal migration and mosaic expression of the FGF reporter transgene in fgf8−/− mutant embryos. Focal activation of FGF signaling in few parapineal cells is sufficient to promote the migration of the whole parapineal collective. Finally, we show that asymmetric Nodal signaling contributes to the restriction and leftwards bias of FGF pathway activation. Our data indicate that the first overt morphological asymmetry in the zebrafish brain is promoted by FGF pathway activation in cells that lead the collective migration of the parapineal to the left. This study shows that cell-state differences in FGF signaling in front versus rear cells is required to promote migration in a model of FGF-dependent collective migration.
The formation of tissue and organs during embryonic development relies on the ability of cells to coordinate their behavior through physical and chemical communication between each other and with their environment. Striking examples of collective cell behavior are directed cell migrations, which occur widely during development, tissue repair, regeneration, angiogenesis, and metastasis. In these different contexts, coherent actions of cells improve the robustness and efficiency of their collective migration (1–4). Collective migration also facilitates cell differentiation and morphogenesis through maintenance of cell–cell interactions and signaling during migration (5–7). Collective migration is thus the predominant mode of migration adopted by epithelial and mesenchymal cells (8, 9).
Cells can migrate in different size groups, over variable distances, and in mechanically different environments, and can adopt different multicellular arrangements, such as sheets, chains, or groups with variable cohesivity. Over the last decade, advances in genetic methods and imaging tools have considerably improved our ability to observe and study collective cell migration in vivo. For example, studies imaging the migration of border cells and tracheal cells in Drosophila, and the lateral line primordium (LLP) and vascular system in zebrafish, have revealed the behavior of individual cells within the migrating group and have highlighted some common features of collective cell migration, such as the importance of cell communication between leaders and followers (1, 3, 5, 6). These studies also indicate that many features of cell migration vary between models. For example, although FGF signaling is implicated in many cell migration events, its function varies from a role in promoting chemotaxis (10–13), cell motility (14), and cell adhesion (15) or in coupling migration to epithelial morphogenesis (16–20). Despite progress in our understanding of cell migration, we are still far from fully understanding how cells collectively interpret signals from the surrounding environment and, consequently, there remains a need for studies that correlate, in vivo, dynamic signaling events with individual cell behaviors.
In the present study, we analyze the unusual left-sided migration of the parapineal in the zebrafish brain as an optically and genetically tractable model for studying collective cell migration. The parapineal is a small group of 15–20 cells located in the epithalamic region of the forebrain. In zebrafish, the epithalamus has been intensively studied as a powerful model to understand how left–right (LR) asymmetry develops in the brain (21–24). It is composed of a pair of asymmetric nuclei called the habenulae and of the photoreceptive pineal complex, which itself consists of the medially located epiphysis and, to its left, the parapineal. Although it almost always resides on the left, the parapineal derives from a group of cells that span the midline, delaminate from the anterior epiphysis, and collectively migrate leftward to lie adjacent to the left habenula (25). The function of the mature parapineal is not clear but during development it has an instructive role in the development of LR differences between the habenulae (25–29).
We have previously shown that local Fgf8 signaling is required for parapineal cells to migrate away from the midline (30). Although Fgf8 is expressed bilaterally in the epithalamus, the parapineal migrates to the left in most wild-type embryos. This bias in the orientation/laterality of parapineal migration depends on the activity of Cyclops (25, 30), a secreted signal of the Nodal/TGF-β family that is transiently expressed in the left epithalamus before parapineal migration (31–33). In embryos lacking Cyclops/Nodal, asymmetry develops but the parapineal migrates to the left or to the right with equal probability (32). Thus, one signaling pathway promotes parapineal cell migration (FGF), while another (Nodal/TGF-β) imparts directionality to the migration (30).
In this study, we elucidate the mechanisms by which Fgf8 promotes parapineal migration. Using a well-established genetically encoded dynamic reporter of FGF signaling activity (34), we observe that just a few parapineal cells, most often located on the left posterior side, show FGF pathway reporter transgene activation. This mosaic and asymmetric expression of Tg(dusp6:d2EGFP) FGF reporter in the parapineal recapitulates the pattern of endogenous dusp6 gene expression and is dependent on Fgf8. Time-lapse confocal imaging in live embryos shows that the dynamics of FGF reporter activity correlates with the behavior of migrating parapineal cells and that transgene expression is enriched in leading parapineal cells throughout migration. Global expression of a constitutively active Fgf receptor (CA-FgfR1) is able to partially rescue parapineal migration in fgf8−/− mutants. However, despite the global expression of the activated receptor, FGF reporter transgene activity resolves to leading cells as in wild-type embryos. This suggests that focal activation of the FGF pathway promotes parapineal migration. Supporting this finding, the focal expression of CA-FgfR1 in few parapineal cells is sufficient to partially restore parapineal migration in fgf8−/− mutants. Finally, we show that left-sided Nodal activity is required for the lateralization and restriction of FGF pathway activation and that absent or bilateral Nodal signaling contexts differ in their impact on the pattern of FGF pathway activation. Altogether, our data indicate that Fgf8 triggers a focal activation of the FGF pathway in leading parapineal cells that is influenced by left-sided Nodal activity, and this in turn promotes the migration of the whole parapineal cell collective.
Results
Focal and Lateralized Activation of Tg(dusp6:d2EGFP) FGF Signaling Reporter Transgene in the Parapineal.
Although fgf8 is expressed bilaterally in the epithalamus before and during parapineal migration (30), whether Fgf8-dependent parapineal migration requires pathway activation in the parapineal or in surrounding cells is not known. To resolve the spatial and temporal dynamics of FGF signaling in the epithalamus, we used an FGF pathway reporter transgenic line, Tg(dusp6:d2EGFP)pt6, in which a destabilized version of green fluorescent protein (d2EGFP) is expressed under the control of the dusp6/mkp3 gene promoter (34). dusp6/mkp3 is a well-characterized direct and immediate FGF target gene involved in negative feedback inhibition of FGF signaling (35–37).
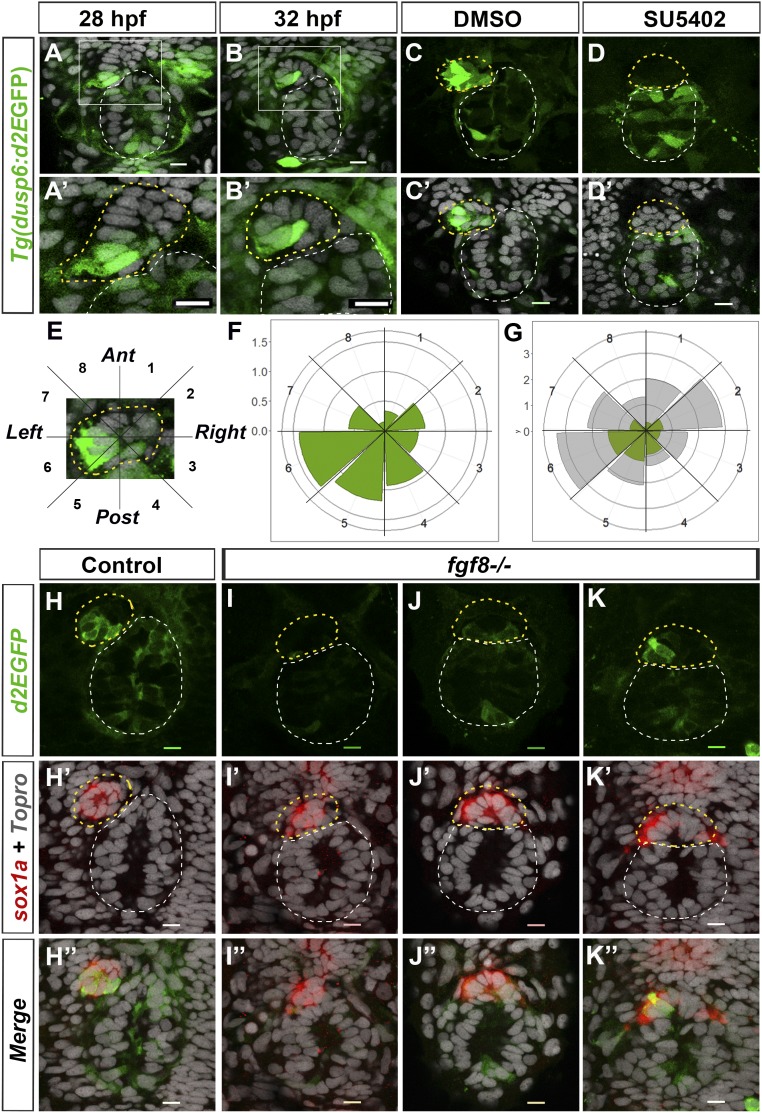
Confocal imaging of the epithalamus in Tg(dusp6:d2EGFP) embryos revealed robust transgene expression in a few parapineal cells that are usually found at the border between the parapineal and the epiphysis on the left side of the parapineal at the onset of migration (Fig. 1 A–B′). Because the pattern of d2EGFP expression was variable from one embryo to another, we quantified the number and position of expressing cells in the parapineals of 32-h postfertilization (hpf) embryos; at this stage, parapineal cells are organized in a rosette-like structure that is distinct from the epiphysis and can easily be delineated by staining nuclei. At 32 hpf, an average of 5.8 (±2.7) cells expressed Tg(dusp6:d2EGFP) with variable intensity of a total average of 16.8 (±5.6) parapineal cells per embryo. The d2EGFP+ cells were frequently found on the left posterior quadrants of the parapineal (Fig. 1 F and G) (an average of 1.2 d2EGFP+ cells per embryo in semiquadrant 5 and 1.4 in semiquadrant 6) as well as in the posterior semiquadrant 4 (Fig. 1 F and G) (0.9 cells d2EGFP+ cells per embryo). Thus, while total parapineal cells distribute equally along a clockface 2pm to 8pm axis (Fig. 1G), the distribution of d2EGFP+ cells is enriched in the posterior and left side of the parapineal. This localized expression of the FGF reporter transgene recapitulated the expression of the endogenous dusp6 gene in the epithalamus; although dusp6 mRNA was weakly detected by in situ hybridization, when visible, it overlapped with d2EGFP staining in the parapineal and elsewhere (SI Appendix, Fig. S1 A–C′). The spatial localization of Tg(dusp6:d2EGFP)pt6 expression was also confirmed with a second allele of the reporter transgene [Tg(dusp6:d2EGFP)pt8] (SI Appendix, Fig. S1 D–E′).
Fig. 1.
The Tg(dusp6:d2EGFP) FGF pathway reporter is focally activated in the parapineal by Fgf8. (A–B′) Confocal sections showing expression of Tg(dusp6:d2EGFP) (green) in the epithalami of 28-hpf (A) or 32-hpf (B) embryos with cell nuclei labeled (Topro-3, gray) visualizing the epiphysis (white circle) and parapineal (yellow circle). White boxes in A and B are magnified in A′ and B′. (C–D′) Confocal maximum projections (C and D) or sections (C′ and D′) of the epithalami of 35-hpf Tg(dusp6:d2EGFP) embryos treated with DMSO (C and C′) or SU5402 (D and D′) immunostained for GFP (green) and additionally for nuclei (C′ and D′; gray). In the control embryos (C and C′; n = 10), Tg(dusp6:d2EGFP) is expressed in both the epiphysis and the parapineal; in the SU5402 treated embryos (D and D′; n = 11), Tg(dusp6:d2EGFP) is absent in the parapineal. (E) Image of a 32-hpf parapineal defining eight 45 °C semiquadrants (1–8) along the antero-posterior and LR axes relative to the mean position of the parapineal (center). (F and G) Polar graphs showing the distribution and mean number of total (G, gray) or Tg(dusp6:d2EGFP)+ (F and G, green) parapineal cells in each 45 °C semiquadrant relative to the parapineal mean position (center) at 32 hpf; the distribution of total (gray) and Tg(dusp6:d2EGFP)+ cells (green) are shown at the same scale in G. The radial axis (vertical scale on the left side of polar graphs) represents the mean number of cells per semiquadrant (n = 27 embryos). (H–K″) Confocal sections showing the expression of Tg(dusp6:d2EGFP) (green) and sox1a (red) at 32 hpf in control embryos (H–H″; n = 34) and in three illustrative fgf8−/− mutant embryos (I–K″) displaying no expression (I and I′) weakly and barely lateralized expression (J–J″) or relatively normally patterned (K and K″). The distribution and mean number of Tg(dusp6:d2EGFP)- and sox1a-expressing cells are quantified in SI Appendix, Fig. S2. In all panels (A–D′ and H–K″), embryo view is dorsal, anterior is up. (Scale bars, 10 µm.)
Expression of the Tg(dusp6:d2EGFP) Transgene Depends on Fgf8.
Although Tg(dusp6:d2EGFP) transgene expression generally recapitulates dusp6 expression and depends on FGF pathway activity (34), in some contexts it has been shown to depend on Lef1, a transcriptional activator of the Wnt pathway (38). To determine whether Tg(dusp6:d2EGFP) expression in the parapineal reflects FGF pathway activation, we treated Tg(dusp6:d2EGFP) embryos with the SU5402 inhibitor of FGF signaling (39) between 25 and 35 hpf. Although d2EGFP was still detected in some tissues, it was abolished in the parapineal, suggesting that expression of the Tg(dusp6:d2EGFP) transgene reflects activation of the FGF pathway in parapineal cells (Fig. 1 C–D′). Consequently, the loss of Tg(dusp6:d2EGFP) expression in the parapineal correlates with compromised parapineal migration in comparably SU5402-treated embryos (30).
Several Fgf ligands are expressed in the epithalamus (40) but only Fgf8 has been shown to influence parapineal migration (30). To determine whether FGF pathway activation in parapineal cells depends on Fgf8, we analyzed the expression of Tg(dusp6:d2EGFP) in fgf8ti282a/acerebellar (ace) mutant embryos. Because the parapineal is not always easy to detect in fgf8−/− mutants, we used sox1a expression to delineate parapineal cells (40).
In most fgf8−/− embryos, Tg(dusp6:d2EGFP) expression was either absent or markedly reduced in the parapineal (Fig. 1 I–J″), indicating that Fgf8 is required to activate the FGF pathway reporter in the parapineal. The loss of d2EGFP expression was not completely penetrant as, in some mutant embryos (Fig. 1 K–K″), the expression of Tg(dusp6:d2EGFP) was similar to control embryos (Fig. 1 H–H″); this lack of penetrance has also been noted for the parapineal migration phenotype (30) and may be due to the hypomorphic nature of the fgf8ti282a mutation (41) or compensatory activity of other Fgf ligands. However, overall, the average number of Tg(dusp6:d2EGFP)+ cells was significantly decreased in fgf8−/− mutants (SI Appendix, Fig. S2 A and C), while the total number of sox1a-expressing parapineal cells was not affected [average of 10.2 (±4.2) sox1a+ cells in controls and of 10.2 (±4.7) in mutants] (Fig. 1 H′–K″ and SI Appendix, Fig. S2 B and D).
Altogether, our results show Fgf8-dependent FGF pathway activation in a few cells on the posterior and left side of the parapineal at the time of onset of migration.
Localized Tg(dusp6:d2EGFP) Expression Correlates with Time and Direction of Parapineal Migration.
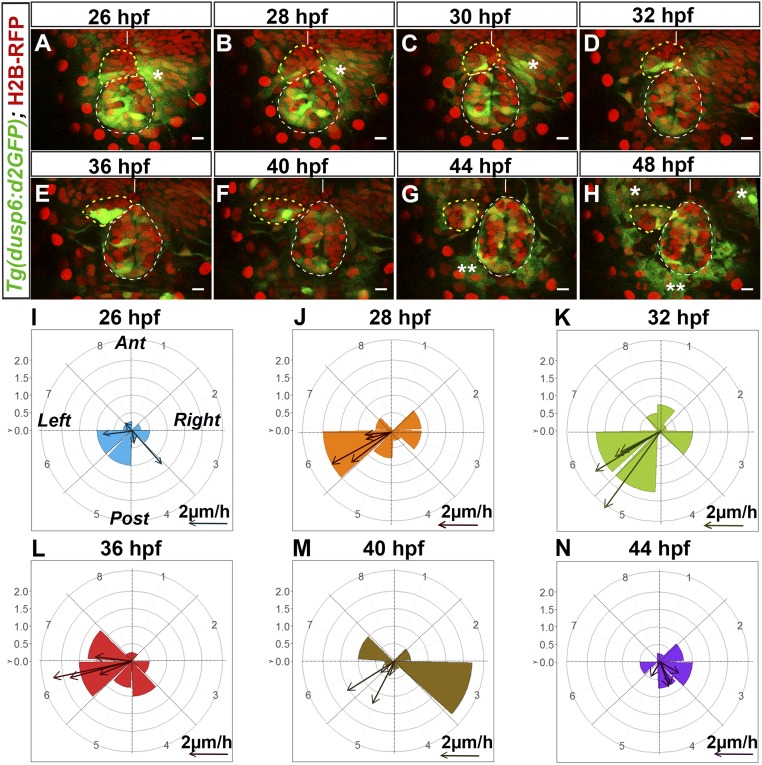
To assess how activation of the FGF pathway correlates with the temporal dynamics of parapineal migration, we performed time-lapse analysis of the distribution of Tg(dusp6:d2EGFP)-expressing cells before and during migration (Fig. 2 A–H, SI Appendix, Fig. S3, and Movies S1–S4). For 37 embryos, movies were for 10- to 14-h periods from the onset of migration (from 26–28 hpf to 36–40 hpf). In four cases, we analyzed a 22-h period from 26 to 48 hpf and averaged the number and position of Tg(dusp6:d2EGFP)-expressing cells for each time point (Fig. 2 I–N).
Fig. 2.
Tg(dusp6:d2EGFP) expression is enriched in cells at the leading edge of the migrating parapineal. (A–H) Time series of thin confocal maximum projection (3 µm) of the brain of a live Tg(dusp6:d2EGFP)pt6 (green) embryo (embryo no. 1 shown in Movie S1) expressing H2B-RFP protein (red) in cell nuclei at different stages of development (26, 28, 30, 32, 36, 40, 44, 48 hpf). The epiphysis and the parapineal are shown as white or yellow dotted circles. Tg(dusp6:d2EGFP) is also expressed in the presumptive habenulae, shown by an asterisk (*); two asterisks (**) indicate autofluorescence that appears from 44 hpf. Embryo view is dorsal, anterior is up. (Scale bars, 10 µm.) Position of Tg(dusp6:d2EGFP)+ cells relative to the mean position of all parapineal cells at different stages of parapineal migration are shown in SI Appendix, Fig. S3I. (I–N) Polar graph showing the distribution of Tg(dusp6:d2EGFP) cells per 45 °C semiquadrant (1–8) relative to the parapineal mean position (center) and the antero-posterior line at a given time point: 26 hpf (I), 28 hpf (J), 32 hpf (K), 36 hpf (L), 40 hpf (M), and 44 hpf (N). The radial axis (0–2, vertical scale, upper left) represents the mean number of Tg(dusp6:d2EGFP)+ parapineal cells per semiquadrant and per embryo (n = 4). At each specific time point, arrows show the orientation of migration for each of the four embryos [defined by the extrapolated line passing through the parapineal mean positions at T and at T + 2 h (for 26 hpf) or T + 4 h (28, 32, 36, 40, 44 hpf)]. The length of the arrow is proportional to the extrapolated distance migrated per hour. Anterior (Ant), posterior (Post), left and right orientations are shown in the I graph.
Before migration, localized expression of Tg(dusp6:d2EGFP) usually predicted the direction of subsequent parapineal migration. At 26–28 hpf, presumptive parapineal cells are detected at the midline in the most anterior part of the pineal complex and subsequently organize into a rosette-like structure. At this stage, in the majority of embryos, a few cells express weakly Tg(dusp6:d2EGFP) on the left, usually at the border between epiphysis and parapineal (n = 26 of 41) (Fig. 2A and SI Appendix, Fig. S4 A–A″). In other embryos, d2EGFP+ cells were also detected at the epiphysis–parapineal border but on both the left and right sides (n = 10 of 41) or on the right side only (n = 5 of 41) (SI Appendix, Figs. S3A and S4 B–C″); subsequently, right-sided Tg(dusp6:d2EGFP)+ cells either relocated to the left side (n = 5 of 10 and n = 3 of 5, respectively) (Movie S2) or stopped expressing the transgene. From 28 to 32 hpf (Fig. 2 B–D, J, and K and SI Appendix, Fig. S3I), expression of Tg(dusp6:d2EGFP) became more robust in the left-posterior quadrant at the border between the epiphysis and the parapineal. This robust lateralized d2EGFP expression correlated with an active phase of leftward and caudally directed migration; the distance of parapineal migration was usually highest between 32 and 36 hpf [about 14 µm (±5 µm) over the 4-h period, n = 4] (Fig. 2K).
Tg(dusp6:d2EGFP) expression continued to delineate cells at the leading edge of the parapineal throughout the period of migration. At 36 hfp, the expression of Tg(dusp6:d2EGFP) usually remained very strong on the left and posterior sides of the parapineal (Fig. 2 E and L). However, from 36 to 40 hpf, transgene expression progressively decreased in cells at the front while concommitantly arising in medially positioned parapineal cells. This change prefigured a caudal reorientation of parapineal migration from 40 to 44 hpf and medial/caudal reorientation from 44 to 48 hpf (Fig. 2 F, G, M, and N and SI Appendix, Fig. S3 F, G, I, and J). By 48 hpf, Tg(dusp6:d2EGFP) expression was only retained weakly in cells at the interface with the epiphysis (Fig. 2H, SI Appendix, Fig. S3H, and Movies S1 and S2).
At all stages of parapineal migration but most often during the active phase of migration between 30 and 36 hpf, protrusions characteristic of active migratory behavior were visible on Tg(dusp6:d2EGFP)-expressing parapineal cells (Movies S1, S2, and S4) (42). Our approach of visualizing protrusions using photoverted cytoplasmic Kaede in Tg(flhBAC:Kaede)vu376 embryos did not enable unequivocal allocation of protrusions to individual cells (and may miss fine processes) but we were nevertheless able to quantify processes on Tg(dusp6:d2EGFP)-expressing and Tg(dusp6:d2EGFP)-negative parapineal cells. Although Tg(dusp6:d2EGP)-negative parapineal cells did display protrusions (SI Appendix, Fig. S5 A–B″ and Movie S5), they were about half as frequent as on Tg(dusp6:d2EGFP)-expressing cells (SI Appendix, Fig. S5C). Because there were about half as many Tg(dusp6:d2EGFP)-expressing cells as Tg(dusp6:d2EGFP)-negative cells (Fig. 1), visible protrusions were consequently about four times more frequent in parapineal cells that express Tg(dusp6:d2EGFP) FGF reporter transgenes than those that do not. Moreover, we found that long protrusions were predominantly observed on Tg(dusp6:d2EGP)+ cells (SI Appendix, Fig. S5D).
Taken together, these data suggest that FGF signaling pathway activity is robustly enriched at the leading edge of the parapineal during its migration and raises the question of whether localized pathway activation is required for effective migration.
Global Ectopic Expression of CA-FgfR1 Compromises Parapineal Migration.
In 3 of 41 imaged embryos, we noticed that an increase in the number of Tg(dusp6:d2EGFP)-expressing cells correlated with delayed migration (Movie S3). To assess if restriction of pathway activation to a few parapineal cells is required for migration, we activated the FGF pathway broadly before and during the initiation of migration using a transgenic line in which CA-FgfR1 is under the control of a heat-shock inducible promoter (43). Heat shocking Tg(hsp70:ca-fgfr1) transgenic embryos resulted in strong ectopic expression of the endogenous dusp6 gene but only for up to 2 to 3 h after heat shock (SI Appendix, Fig. S6 A–E). Therefore, to ensure that the CA-FgfR1 transgene is expressed throughout the period of initiation of parapineal migration, we performed a first heat shock at 26 hpf, a second at 29 hpf and, in some cases, a third at 32 hpf.
Constitutive FgfR1 activation during the early stage of parapineal migration often led to reduced or occasionally absent migration (SI Appendix, Fig. S6 F–I). The parapineal migrated at least 25 µm away from the midline to the left in about 90% of heat-shocked control embryos (n = 32 of 35) (SI Appendix, Fig. S6H, light blue), whereas in embryos expressing CA-FgfR1, this frequency decreased to 60% (n = 21 of 35) SI Appendix, Fig. S6H, dark blue) (P = 0.023); in the remaining CA-FgfR1–expressing embryos, the parapineal either migrated partially (between −15 and −25 µm in n = 9 of 35) or did not migrate (within −15 µm and +15 µm of the midline in n = 4 of 35) (SI Appendix, Fig. S6H, dark blue). Although receptor activation compromised the extent of migration, when migration did occur, its direction to the left was not affected (SI Appendix, Figs. S3I and S6G).
These results suggest that widespread activation of FgfR signaling compromises parapineal migration, although less severely than when FGF signaling is absent (30).
Global Ectopic Expression of Fgf8 or CA-FgfR1 Rescues Parapineal Migration in fgf8−/− Mutants.
In the experiments above, exogenous FgfR1 activation occurs in the context of normal Fgf8-mediated signaling in the epithalamus and consequently it is not possible to disentangle the contribution of endogenous and exogenous pathway activation to migration. To more cleanly resolve the requirement of FGF pathway activation on parapineal migration, we expressed CA-FgfR1 or Fgf8 ligand itself in fgf8−/− mutant embryos and assessed effects upon migration.
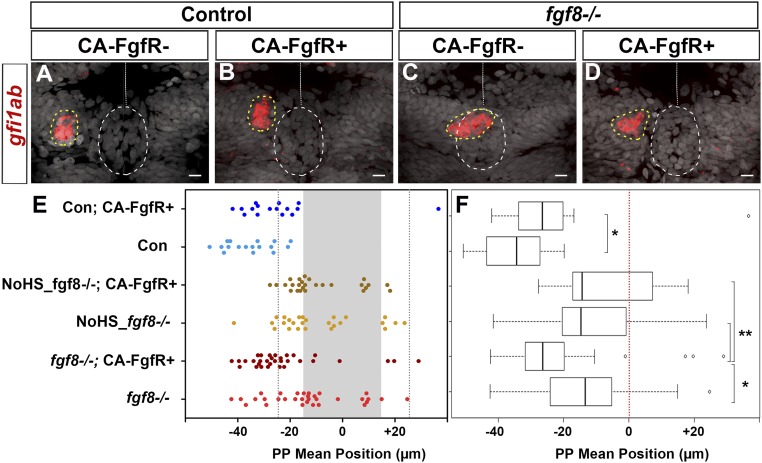
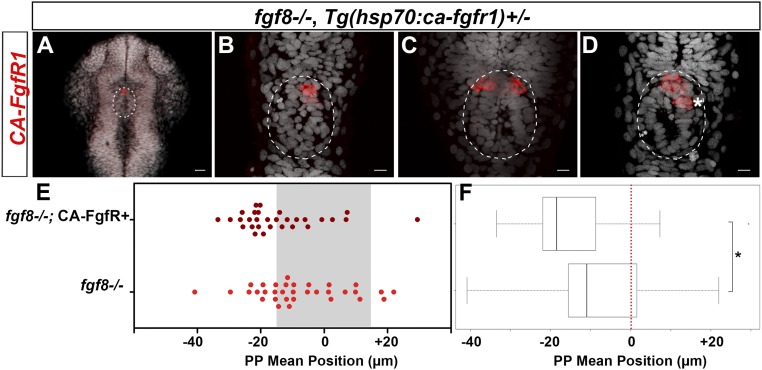
Widespread expression of CA-FgfR1 during the period when the parapineal initiates its migration reduced the penetrance and expressivity of the parapineal migration deficit in fgf8−/− embryos. In most control embryos (90%, n = 15 of 17), the mean position of parapineal cells is between −25 and −55 μm to the left of the midline (Fig. 3 A and E, light blue) and, as above, this frequency decreased upon expression of CA-FgfR1 (56%, n = 9 of 16) (Fig. 3 A and E, dark blue). Also as expected, in the absence of the Tg(hsp70:ca-fgfr1) transgene, the parapineal failed to migrate in about half of the fgf8−/− embryos (n = 17 of 32) while, in the other half, it migrated normally (n = 7 of 32) or at least partially toward the left (between −15 and −25 µm in n = 7 of 32); rarely, the parapineal was found to migrate on the right side (n = 1 of 32) (Fig. 3 C and E, light red). In contrast, in fgf8−/− mutant embryos with activated CA-FgfR1, only 3 of 32 embryos failed to show any migration, while the parapineal migrated normally or partially leftward in respectively 56% (n = 18 of 32) and 25% (n = 8 of 32) of the embryos or migrated rightward (n = 3 of 32) (Fig. 3 C and E, dark red). This leftward shift in the mean position of the parapineal in fgf8−/−;Tg(hsp70:ca-fgfr1)+ embryos after heat shock (Fig. 3 E and F) (P = 0.039, SI Appendix, Table S1) indicates that global activation of FgfR1 can partially rescue parapinal migration without a major effect on leftward orientation. There was no rescue of migration in mutant embryos that were not heat-shocked before parapineal migration (Fig. 3 E and F).
Fig. 3.
FgfR1 receptor activation partially restores parapineal migration in fgf8−/− mutants. (A–D) Confocal maximum projection (10 µm) showing gfi1ab expression (red) and cell nuclei (gray) in representative control embryos (A and B) and fgf8−/− mutants (C and D) that express CA-FgfR1 (B and D) or not (A and C); embryo view is dorsal, anterior is up. (Scale bars, 10 µm.) Control embryos are siblings of fgf8−/− mutants and thus correspond to both wild-type or fgf8+/− heterozygotes. gfi1ab expression marks the parapineal (yellow circle) while cell nuclei staining allows us to define the epiphysis (white circle) and the brain midline (straight dotted white line). (E) Dot plot showing, for each embryo, the mean parapineal position in micrometers distant to the brain midline (x = 0), at 52 hpf, in control embryos (Con, blue dots) and in fgf8−/− mutant embryos (fgf8−/−, red dots) that expressed (CA-FgfR+, dark color) or not (light color) CA-FgfR1 after a heat shock at 26 and 29 hpf; “NoHS_fgf8−/−;CA-FgfR+” and “NoHS_fgf8−/−” correspond to fgf8−/− mutants that do or do not carry the Tg(hsp70:ca-fgfr1) transgene but were not heat-shocked (dark and light yellow dots, respectively). Gray-shaded zone (−15 µm and +15 µm) define the “no migration” domain as corresponding to the average width of the epiphysis; gray dotted lines show −25 µm and +25 µm. (F) Boxplot showing the distribution of parapineal mean position relative to the brain midline (reference 0, red dotted line) in the same embryos. Parapineal mean position is shifted toward the midline in wild-type embryos expressing CA-FgfR1 (dark blue in E; n = 16) compared with control embryos that do not express CA-FgfR1 (light blue in E; n = 17); P = 0.023 (Wilcoxon test). The expression of CA-FgfR1 partially restores parapineal migration in fgf8−/− mutants (dark vs. light red in E; n = 32); P = 0.039. Parapineal mean position did not differ significantly between fgf8−/− mutants that do or do not carry the Tg(hsp70:ca-fgfr1) transgene but were not heat-shocked (dark versus light yellow in E; n = 26); P = 1 (see pairwise Wilcoxon test in SI Appendix, Table S1). Statistical significance is indicated in F, *P < 0.05, **P < 0.01.
Because Fgf8 can influence the specification of the parapineal (40), one possibility is that pathway activation might improve migration in fgf8 mutants indirectly by increasing the number of parapineal cells. However, the number of gfi1ab-expressing parapineal cells did not vary significantly between control and fgf8−/− mutant embryos not carrying the Tg(hsp70:ca-fgfr1) transgene [12.4 (±3.8) for controls vs. 11.5 (±3.6) cells for fgf8−/− mutants; adjusted P = 1 in pairwise Wilcoxon test] nor between heat-shocked embryos that do or do not carry the Tg(hsp70:ca-fgfr1) transgene [11.5 (±3.6) cells for fgf8−/−; compared with 12.4 (±3.3) cells for fgf8−/−; Tg(hsp70:ca-fgfr1)+/−; adjusted P = 1]. Therefore, the activation of FgfR signaling does not affect parapineal cell number but promotes its migration.
Supporting the conclusions above, global activation of Fgf8 ligand by heat shocking Tg(hsp70:fgf8) embryos at 26 and 29 hpf similarly partially restored parapineal migration without influencing parapineal size or its leftward orientation (SI Appendix, Fig. S7).
Rescue of Parapineal Migration by Ectopic Expression of CA-FgfR1 Correlates with Restoration of Localized Expression of Tg(dusp6:d2EGFP).
Because fgf8 is expressed broadly in the epithalamus and parapineal migration still usually occurs after widespread FgfR activation, one possibility is that if spatially localized activation of signaling is important for migration, then this may occur downstream of ligand and receptor. To ascertain if this may be the case, we assessed expression of Tg(dusp6:d2EGFP) reporter transgene in fgf8−/− mutants following widespread activation of the Tg(hsp70:ca-fgfr1) transgene.
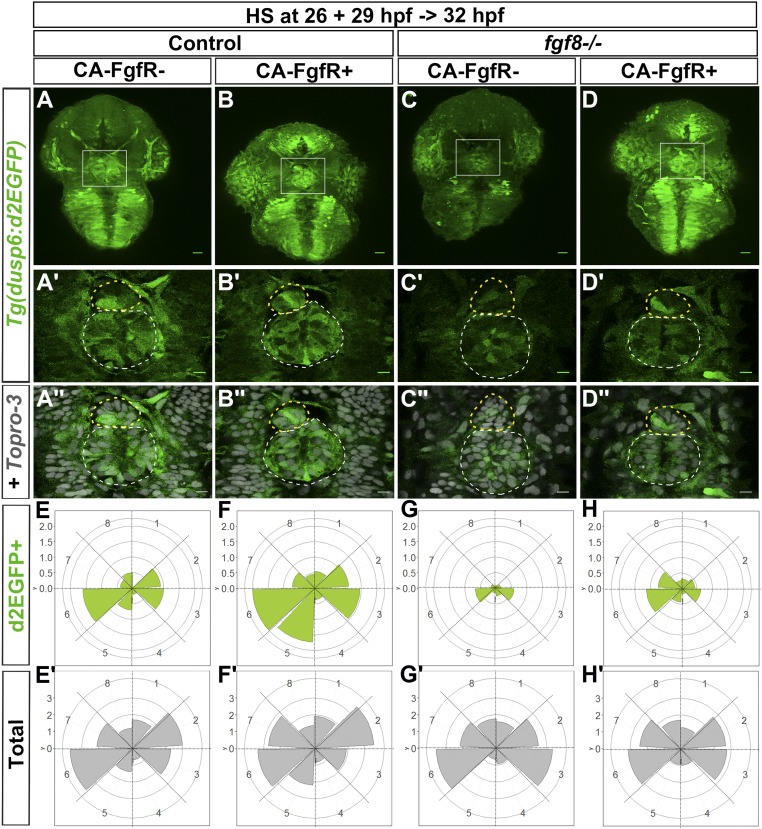
Global activation of FgfR1 restored Tg(dusp6:d2EGFP) expression within the parapineal and despite the nonlocalized expression of CA-FgfR1, within a few hours, the FGF pathway reporter was only activated mosaically as in the wild-type condition. As mentioned above (Fig. 1), the expression of Tg(dusp6:d2EGFP) was either absent or strongly decreased in the parapineal of fgf8−/− mutants (Fig. 4 C–C″) and, when it was detectable in parapineal cells, expression was most often not lateralized (Fig. 4G). However, following expression of CA-FgfR1, within 3–6 h, Tg(dusp6:d2EGFP) expression was robustly detected in the parapineal of fgf8−/− mutants (Fig. 4 D–D″ and H) with a pattern that is very similar to that of control embryos (Fig. 4 A–A″ and E).
Fig. 4.
Global overexpression of CA-FgfR1 in fgf8−/− mutants rescues focal left-sided expression of Tg(dusp6:d2EGFP). (A–D″) Confocal maximum projection (55 µm) showing the whole head (A–D) or confocal sections (A′–D″) showing the epithalamus of 32-hpf Tg(dusp6:d2EGFP) control embryos (A–B″) or Tg(dusp6:d2EGFP) fgf8−/− mutants (C–D″) that express (B–B″ and D–D″) or not (A–A″ and C–C″) Tg(hsp70:ca-fgfr1) transgene after heat shock at 26 and 29 hpf. (Scale bars, 25 µm in A–D and 10 µm in A′–D″.) The image in A″–D″ corresponds to the image in A′–D′ (green) superimposed on nuclear staining (gray) allowing visualization of the epiphysis (white circle) and parapineal (yellow circle); embryo view is dorsal, anterior is up. (E–H′) Polar graphs showing the distribution and mean number of total (E′–H′) or Tg(dusp6:d2EGFP)+ (E–H) parapineal cells in each 45 °C semiquadrant (1–8) relative to the parapineal mean position (center) at 32 hpf. Cell distribution and mean number are shown for control embryos that express (F and F′; n = 11) or not (E and E′; n = 10) CA-FgfR1 transgene and for fgf8−/− mutants that express (H and H′; n = 16) or not (G and G′; n = 12) CA-FgfR1. Radial axis (vertical scale on the left side) represent the mean number of Tg(dusp6:d2EGFP)+ parapineal cells (green area in E–H, scale 0–2) or all parapineal cells (gray area in E′–H′, scale 0–3) per semiquadrant and per embryo. The expression of Tg(dusp6:d2EGFP) is increased in the parapineal of embryos expressing CA-FgfR1 although it is still mosaic and enriched on the left/posterior side (B–B″ and F; n = 11) as in controls (A–A″ and E; n = 10). In fgf8−/− embryos, Tg(dusp6:d2EGFP) is either not expressed in the parapineal or weakly detected (C–C″ and G; n = 12) while its expression is rescued in the parapineal of fgf8−/− embryos expressing CA-FgfR1 transgene (D–D″ and H; n = 16).
These results show that in fgf8−/− mutants that express CA-FgfR1, FGF signaling is reactivated mosaically despite the constitutively activated receptor being expressed ubiquitously. This suggests that the intracellular pathways downstream of the receptor are tightly and rapidly regulated to spatially localize pathway activation. This is also consistent with the observation that although Tg(dusp6:d2EGFP) expression was globally increased upon ectopic activation of FgfR1 in control wild-type embryos, reporter transgene expression also rapidly resolved to being mosaic and enriched at the front of the migrating parapineal (Fig. 4 A–A″ and E and E′ vs. Fig. 4 B–B″ and F and F′).
Targeted Focal Activation of CA-FgfR1 Expression in Few Parapineal Cells Improves Parapineal Migration in fgf8−/− Mutants.
The results above are consistent with localized FGF pathway activation mediating parapineal migration. However, they do not exclude the possibility that it is the activation of FGF signaling in neighboring habenular cells that indirectly promotes parapineal cell migration. Additionally, although Tg(dusp6:d2EGFP) is only expressed in some parapineal cells, it is still possible that other intracellular branches of the FGF pathway need to be activated in all parapineal cells. To address whether the observed focal activation of the FGF pathway in parapineal cells is indeed sufficient for parapineal migration, we tested whether we could restore parapineal migration in fgf8−/− mutants by activating the FGF pathway in only few parapineal cells. To do so, we performed highly localized heat shock using an adapted infrared laser-evoked gene operator (IR-LEGO) optical system (44).
By irradiating two or three cells in the anterior part of the pineal complex where the future parapineal rosette forms, we could trigger the subsequent expression of CA-FgfR1 in ∼six cells (n = 10) (Fig. 5 A–D). In irradiated fgf8−/− embryos carrying the Tg(hsp70:ca-fgfr1) transgene (n = 30), the parapineal migrated leftward further than −15 µm in 60% of embryos (Fig. 5E, dark red) compared with only 33% of similarly treated fgf8−/− mutant embryos lacking the transgene (Fig. 5E, light red, and Fig. 5F) (P = 0.02 in Wilcoxon test). In these experiments, we never detected cells expressing CA-FgfR1 in the habenulae or other areas outside of the pineal complex, suggesting that activation of the FGF pathway is indeed required in parapineal cells. Therefore, we conclude that focal activation of CA-FgfR1 in a few parapineal cells is able to improve parapineal migration in fgf8−/− mutants.
Fig. 5.
Focal activation of FGF signaling in few parapineal cells partially restores parapineal migration in fgf8−/− mutants. (A–D) Confocal 40-µm maximum projection and confocal thin 2-µm projections (B–D) showing the expression of the ca-fgfr1 transgene by in situ hybridization (red) in four different representative fgf8−/−; Tg(hsp70:ca-fgfr1)+/− embryos, 1–2 h after two cells were irradiated in the anterior epiphysis. (Scale bars, 25 µm in A and 10 µm in B–D.) Superimposed nuclear staining (Topro-3, gray) visualizes the pineal complex (white circle) consisting of both the pineal and the parapineal at this stage (26–28 hpf). ca-fgfr1 mRNA is only detected in the anterior pineal complex, in three cells (A and B) or six cells (C and D), the average number being 6 CA-FgfR1–expressing cells (n = 10 embryos). Given their location in the anterior pineal complex, most of these CA-FgfR1–expressing cells are expected to become parapineal cells; some more posterior cells (*) might become epiphyseal cells; embryo view is dorsal, anterior is up. (E) Dot plot showing, for each embryo, the mean parapineal position in micrometers distant to the brain midline (x = 0), at 52 hpf, in fgf8−/− mutants that were locally heat-shocked (two to three irridiated cells) between 25 and 29 hpf; gray-shaded zone (–15 µm and +15 µm) defines the average width of the epiphysis. Parapineal mean position in fgf8−/− mutant embryos carrying the Tg(hsp70:ca-fgfr1) transgene is slightly shifted toward the left (dark red dots, n = 30) compared with fgf8−/− mutants that do not express CA-FgfR1 (light red dots, n = 33). (F) Boxplot showing the distribution of parapineal mean position relative to the brain midline (reference 0) in same embryos. Local expression of CA-FgfR1 in few parapineal cells partially rescues parapineal migration (P = 0.02; Wilcoxon test; *indicates statistical significance in F).
Nodal Signaling Restricts FGF Pathway Activation to the Left Posterior Side of the Parapineal.
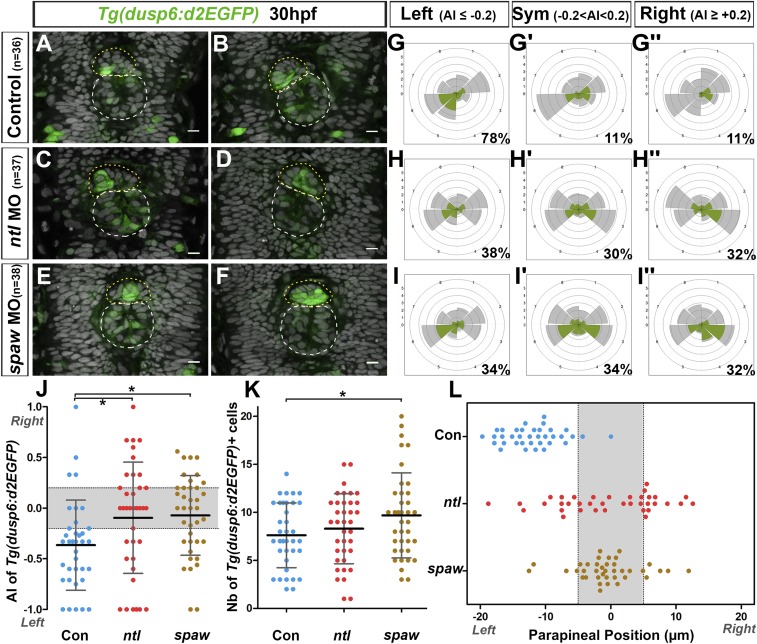
The orientation of parapineal migration depends on activation of Nodal signaling in the left epithalamus (32). Previous results described that absent or bilateral Nodal signaling in the brain lead to comparable outcomes on parapineal migration: that is, a randomization of directionality (25, 30, 32). To address whether the lateralization of FGF pathway activation is influenced by left-sided Nodal activity, we analyzed Tg(dusp6:d2EGFP) expression at the onset of parapineal migration in contexts where Nodal is either absent or bilateral following the injection of validated morpholino oligonucleotides against southpaw (spaw morphants; epithalamic Nodal signaling absent) (45) or no tail (ntl morphants; epithalamic Nodal signaling bilateral) (25), respectively (SI Appendix, Table S2).
As described in Figs. 1, 2, and 5, at 29–30 hpf, most parapineals showed a higher number of Tg(dusp6:d2EGFP)+ cells in the left than in the right posterior quadrant: for example, an asymmetry index (AI) in the number of Tg(dusp6:d2EGFP)-expressing cells was smaller than or equal to −0.2 in about 80% of control fish (Fig. 6 A, B, G, and J and SI Appendix, Table S3). In both contexts of absent or bilateral Nodal signaling, we observed a randomization of lateralized Tg(dusp6:d2EGFP) expression with one-third of embryos displaying a left bias (AI ≤ −0.2 in 38% of ntl morphants and in 34% of spaw morphants) (Fig. 6 C and H–J and SI Appendix, Table S3) and one-third displaying a right bias (AI ≥ +0.2 in 32% of both ntl and spaw morphants) (Fig. 6 D, H″, I″, and J and SI Appendix, Table S3); in the remaining third of embryos, Tg(dusp6:d2EGFP) expression either was not (AI = 0) or was weakly lateralized (AI comprised between −0.2 and +0.2) (Fig. 6 E, F, H′, I′, and J and SI Appendix, Table S3). While the distribution of AIs looks similar in ntl and spaw morphants, the variance of these AI was significantly higher in ntl morphants (AI more spread along the −1 +1 axis) than in spaw morphants (AI closer to the median 0) (variance ratio =1.9, P = 0.045 in F test) (Fig. 6J). This reflects Tg(dusp6:d2EGFP) expression showing overall less lateralization in spaw morphants than in ntl morphants (Fig. 6J).
Fig. 6.
Left biased lateralization of FGF pathway activation depends on lateralized Nodal signaling. (A–F) Confocal sections of the pineal complex at 30 hpf showing the expression of Tg(dusp6:d2EGFP) (green) in two illustrative control embryos (A and B; n = 36) and in two embryos injected with no tail morpholinos (ntl MO) (C and D; n = 37) or southpaw morpholinos (spaw MO) (E and F; n = 38); the superimposed nuclear staining (gray) allows visualization of the epiphysis (white circle) and parapineal (yellow circle). Embryo view is dorsal, anterior is up. (Scale bars, 10 µm.) (G–I′) Polar graphs showing the distribution and mean number of total (gray) or Tg(dusp6:d2EGFP)+ (green) parapineal cells in each 45 °C hemiquadrant relative to the parapineal mean position (graph center) at 30 hpf in control embryos (G–G″, n = 36), ntl morphants (H–H″; n = 37), or spaw morphants (I–I″; n = 38). For each context, cell distributions and mean numbers of cells are averaged within embryos that display an AI (see Materials and Methods and SI Appendix) in Tg(dusp6:d2EGFP) expression ≤ −0.2 (G, n = 28 of 36; H, n = 14 of 37; I, n = 13/38), AI ≥ +0.2 (G″, n = 4 of 36; H″, n = 11 of 37; I′, n = 13 of 38) or −0.2 ≤ AI ≥ +0.2 (G′, n = 4 of 36; H′, n = 12 of 37; G″, n = 12 of 38); percentages of embryos in each category are shown on the lower right side of the polar graph. Radial axis (scale 0–6) represents the mean number of Tg(dusp6:d2EGFP)+ parapineal cells (green area) or all parapineal cells (gray area) per hemiquadrant and per embryo. In both ntl and spaw morphants, the expression of Tg(dusp6:d2EGFP) is either not clearly lateralized or lateralized with random orientation (enriched on the left or on the right side). (J) Dot plot showing, for each embryo, the AI in the number of Tg(dusp6:d2EGFP)+ cells in the left posterior versus the right posterior quadrant in control embryos (n = 36), ntl morphants (n = 37) or spaw morphants (n = 38). Left-sided lateralization of Tg(dusp6:d2EGFP) expression observed in controls is lost in both ntl and spaw morphants. The distribution of AIs is less spread in spaw morphants than in ntl morphants (P = 0.045 in F test comparing variances) showing that Tg(dusp6:d2EGFP) expression is overall less lateralized in spaw morphants than in ntl morphants. Gray-shaded zone shows AI between −0.2 and +0.2. (K) Dot plot showing, for each embryo, the number of Tg(dusp6:d2EGFP)+ parapineal cells in control embryos, ntl morphants and spaw morphants at 30 hpf. The number of Tg(dusp6:d2EGFP)+ cells is overall increased in the parapineal of spaw morphants compared with controls (P = 0.026, Welsh t test). In J and K, means ± SD are indicated as a horizontal (mean) and vertical lines (SD); statistical significance is indicated, *P < 0.05. (L) Dot plot showing, for each embryo, the mean parapineal position in micrometers distant to the brain midline (x = 0) in control embryos, ntl morphants and spaw morphants at 30 hpf. Gray-shaded zone shows parapineal mean position between −5 µm and +5 µm. In controls and ntl morphants, the parapineal had usually started to migrate, respectively, toward the left or randomly by 30 hpf, while it had not initiated migration in spaw morphants at this stage.
The difference observed in the pattern of Tg(dusp6:d2EGFP) expression in contexts of absent or bilateral Nodal signaling correlated with a difference in the timing of initiation of parapineal migration. At 29–30 hpf, the parapineal rosette is formed and has usually initiated its migration toward the left side in control embryos: for example, in over 90% of control embryos, the parapineal mean position was displaced more than 5 µm to the left of the midline (Fig. 6L and SI Appendix, Table S4). At this stage, the parapineal in ntl morphants had usually started to migrate toward the left (30%, n = 37) or the right side (35%, n = 37) or was observed at/near the midline (35% of embryos with a parapineal mean position between −5 µm and +5 µm, n = 37) (Fig. 6L and SI Appendix, Table S4). In contrast, the parapineal was found at the midline in most spaw morphants at this stage (76%, n = 38), while it had initiated its migration toward the left or the right side in only 11% and 13% of the embryos, respectively (Fig. 6L and SI Appendix, Table S4). This delay in parapineal migration in spaw morphants was not due to a global delay of development as, at that time point, the parapineal rosette was clearly visible in the anterior epiphysis (Fig. 6 E and F). Moreover, as late as 36–38 hpf, the parapineal was still observed at the midline in about one-quarter of spaw morphants (25%, n = 32) (SI Appendix, Fig. S8 C and C′ and Table S5) while, at this stage, the parapineal had initiated migration in all controls (n = 17) (SI Appendix, Fig. S8 A and A′) and in all ntl morphants (54% with a left and 46% with a right parapineal, n = 26) (SI Appendix, Fig. S8 B and B′).
Finally, the total number of Tg(dusp6:d2EGFP)+ parapineal cells was increased in spaw morphants at 30 hpf (mean = 9.7; median = 9.5) compared with control embryos (mean = 7.6; median = 7.0) (Fig. 6K) (P = 0.026, Welch t test). The number of Tg(dusp6:d2EGFP)+ cells was also slightly increased in ntl morphants (mean = 8.3; median = 9.0), although this difference is not significant (P = 0.41, Welsh t test).
In summary, we show that left-sided Tg(dusp6:d2EGFP) activation is Nodal signaling-dependent. In the presence of bilateral Nodal signaling, Tg(dusp6:d2EGFP) expression is no longer consistently lateralized to the left, although expression is usually spatially restricted within the parapineal as in wild-type; this correlates with the parapineal initiating its migration toward the left or the right in most embryos from 29 to 30 hpf. Consistent lateralization of Tg(dusp6:d2EGFP) expression is also compromised in the absence of Nodal signaling but, in this condition, expression is generally less restricted and less lateralized within the parapineal and this correlates with delayed parapineal migration.
Discussion
In this study we show that during their leftward migration, parapineal cells respond to Fgf8 and that, despite it being likely that all parapineal cells are exposed to Fgf ligands, FGF signaling is activated focally in only few cells usually located at the leading edge. Activation of FGF reporter transgene expression in parapineal leading cells is lost in fgf8−/− mutant embryos in which parapineal migration is compromised. Widespread activation of the FGF pathway in fgf8−/− mutant embryos rescues migration, while concommitantly restoring the localized expression of Tg(dusp6:d2EGFP) at the leading edge of the parapineal and targeted activation of the FGF pathway in a small number of parapineal cells is sufficient to promote parapineal migration in fgf8−/− mutants. Finally we show that lateralized Nodal signaling influences the spatial localization and restriction of Tg(dusp6:d2EGFP) expression and subsequent timing and direction of parapineal migration. Our results show that parapineal cells respond as a collective rather than as individuals to environmental signals and that the capacity of parapineal cells to coordinate cellular responses to such signals impacts the ability of the parapineal to undergo efficient directed collective migration.
During border cell migration and tracheal sprouting in Drosophila or during endothelial cell migration in vertebrate angiogenesis, selection and guidance of cells at the migration front, so-called leading cells or tip cells, are dependent on receptor tyrosine kinase (RTK) signaling (3). Here, we establish the parapineal as an example of a freely migrating group of cells that depends on RTK signaling for the selection of leading cells and promotion of migration. Although Tg(dusp6:d2EGFP) expression is always detected in cells located at the migration front during the active phase of migration, in some embryos it can also be detected at the rear or on the lateral side of the parapineal rosette and the intensity of transgene activation can also vary among expressing cells. This variability in the number, position, and fluorescence intensity of Tg(dusp6:d2EGFP)+ cells might reflect the dynamic nature of the cell-to-cell communication events within the parapineal that define leading cells and that may mediate competition among parapineal cells for the leading position; indeed, during the active phase of migration, parapineal cells can exchange the leading position as cyclists do in the peleton (Movie S4). A similar dynamic in leader-exchange has also been described during Drosophila border cell migration (46) and vessel sprouting in zebrafish (47), and may provide a way for the migrating cluster to better adapt to the environment. Such variability in the number and in the position of Tg(dusp6:d2EGFP)-expressing cells could underlie the variability that exists in the timing of parapineal migration in wild-type embryos. Indeed, we noticed that parapineal migration is more frequently delayed in the rare embryos in which Tg(dusp6:d2EGFP)-expressing cells are initially found on both sides or on the right side only than when they locate to the left side at the onset of migration.
One interesting aspect of our study is the relative robustness of parapineal migration in response to global activation of FGF signaling. Various observations suggest that all parapineal cells are competent to activate the FGF pathway; for example, ablating the left or the right side of the presumptive parapineal results in the remaining half migrating normally, indicating that both left and right parapineal cells are competent to migrate toward the left (25). Moreover, we show that all parapineal cells can activate expression of the dusp6 gene rapidly after ectopic expression of Fgf8 (SI Appendix, Fig. S9). Although all parapineal cells seem able to respond to Fgf8, activation of FGF signaling is nonetheless restricted to few cells. This suggests the existence of a mechanism downstream of the activated receptor that permits pathway activation in a few cells while silencing it in others. The processes involved in restricting FGF pathway activation are likely functional in fgf8−/− mutants, as we can rescue the mosaic pattern of Tg(dusp6:d2EGFP) expression after global misexpression of CA-FgfR1. The mechanisms restricting FGF pathway activation are likely to depend on parapineal cells being able to communicate their state of FGF pathway activation. In both the Drosophila tracheal system and vertebrate vessel sprouting, Notch-Delta signaling contributes to tip cell selection by restricting the ability of follower cells to activate RTK signaling (48–50), and this pathway is an obvious candidate for a comparable role in the parapineal.
The mechanisms underlying the roles of the FGF signaling pathway in collective migration vary depending on the context in which they have been addressed and even within the same model of cell migration (51). In the zebrafish LLP for example, FGF signaling is required upstream of Notch signaling for the epithelialization and apical constriction that underlies neuromast rosette formation at the back of the primordium, thereby coupling morphogenesis with LLP migration (16–20, 52). Besides this Notch-mediated role in rosette self-organization (52), FGF signaling is also required in this system to trigger the coalescence of the LLP at the onset of migration (12), to maintain LLP polarity via restriction of Wnt/ß-catenin signaling to the leading zone (53), and to maintain cohesion among cells of the cluster during migration, as trailing cells are attracted toward Fgf signals produced by leading cells (54). However, despite progress in understanding the various specific functions of the FGF pathway in promoting cell migration, it is not clear yet how Fgf signals are interpreted by cell collectives. In Drosophila, expression of an active version of the PDGF and VEGF related receptor (PVR) receptor in a single border cell can rescue migration of the entire cluster in the absence of ligand (55). This study suggested that it is the difference in RTK signaling levels between cells in the cluster rather than, or in addition to, an asymmetry in RTK signaling at the level of the individual cells (along the leading to trailing axis of the migrating cell) that triggers migration. In contrast, expression of a constitutively active Breathless Fgf receptor in a single cell is not sufficient to rescue the migration of Drosophila tracheal cells (56), despite expression of a wild-type FgfR doing so (57). Therefore, in tracheal cells, the activation of FGF signaling needs to be asymmetrically distributed, not only between cells in the cluster as in border cells, but within the cell itself for it to become a leading cell and to drive the migration of the whole cluster.
Here, we show that parapineal migration can be restored by expressing constitutively activated CA-FgfR1 focally in few cells in a context of reduced ligand level. Therefore, our results differ from those described for Drosophila tracheal cells and suggest that, similar to border cells, differences in the levels of FGF pathway activation between parapineal cells can define leading cells and promote migration. The fact that global activation of FgfR1 delays migration in wild-type embryos further supports this idea. However, in contrast to what has been suggested by studies in border cells (58), we show that parapineal migration can also be restored in fgf8−/− mutants by expressing constitutively activated CA-FgfR1 in all cells as, in this context, the global activation of FGF signaling resolves to become spatially restricted downstream of the activated receptor. Therefore, our data highlight common features and point out important differences in the mechanisms underlying the interpretation of Fgf signals in different models of Fgf-dependent collective cell migration.
One unusual feature of parapineal migration is that it almost always is directed to the left side of the brain. This is a consequence of bilaterally expressed Fgf8 promoting cell migration while left-sided expression of Cyclops, a Nodal/TGF-β signal, determines directionality (25, 30). Our results show that consistent left-sided lateralization of Tg(dusp6:d2EGFP) expression is lost when unilateral Nodal pathway activation is abrogated, and this correlates with randomized parapineal cell migration. Our results also reveal Nodal-dependent influence upon restriction of Tg(dusp6:d2EGFP) expression to a few parapineal cells, which in turn influences the timing of onset of parapineal migration. Indeed, although absent or bilateral Nodal signaling both result in randomized parapineal migration (29, 30, 32), we find that these two contexts differ in their impact on the pattern of FGF activation. Bilateral Nodal signaling leads to a randomization of Tg(dusp6:d2EGFP) lateralization and a randomization of parapineal migration without significant delay. In contrast, in absence of Nodal pathway activation, lateralization and restriction of Tg(dusp6:d2EGFP) expression is reduced and this correlates with delayed parapineal cell migration. Therefore, our data suggest that Nodal signaling directionally biases and times parapineal migration by contributing to the restriction and lateralization of Fgf signaling.
This work sets the stage for studies aimed at understanding how the Nodal pathway could contribute to restrict the activation of the FGF pathway to a few leading cells and how it provides a leftwards bias to the focal FGF pathway activation.
Materials and Methods
Fish Lines.
Adults heterozygous for the fgf8 mutation (fgf8ti282a/acerebellar/ace) (59) were identified by PCR genotyping (60). Heterozygous embryos carrying the Tg(hsp70:fgf8a)x17 transgenic insertion were identified by PCR genotyping (61). Embryos heterozygous for Tg(hsp70:ca-fgfr1;cryaa:DsRed)pd3 (43) were identified by the presence of DsRed expression in the lens from 48 hpf or at 32 hpf by PCR, as described previously (62). Tg(dusp6:d2EGFP)pt6 and Tg(dusp6:d2EGFP)pt8 (34) lines were used as reporters for FGF pathway activity; Tg(flhBAC:Kaede)vu376 was used as marker of the pineal complex (40).
Quantification of the Number and Position of Tg(dusp6:d2EGFP)+ and gfi1ab+ Parapineal Cells.
The position and number of parapineal cells negative or positive for the Tg(dusp6:d2EGFP) transgene were analyzed using the ImageJ software (ROI Manager tool), the position of each cell being defined by the center of the cell nucleus detected with the Topro-3 staining. For each parapineal cell, we calculate its x and y position relative to the center of the parapineal to create the polar graph (R Studio). Parapineal cells positive for the gfi1ab marker were counted using the Multipoint tool on ImageJ software and the position of each parapineal cell was measured relative to the brain midline (reference origin = 0) to define parapineal mean position.
Calculation of the AI of Tg(dusp6:d2EGFP) Expression.
The AI of Tg(dusp6:d2EGFP) expression was calculated using the following equation: n(Rp) − n(Lp)/n(Rp) + n(Lp), where n(Rp) is the number of Tg(dusp6:d2EGFP)+ cells in the right posterior quadrant of the parapineal rosette and n(Lp), the number in the left posterior quadrant.
Ethics Statement.
All experiments were performed in accordance with both the guidelines from the European directive on the protection of animals used for scientific purposes (2010/63/UE) and national guidelines. In France, all animals were maintained in a facility certified by the French Ministry of Agriculture (approval no. A3155510). The work received the project no. APAFIS#3653-2016011512005922 on the 01/12/2016. M.R. received authorization to experiment on vertebrates models (311255556) from the Direction Départementale de la Protection des Populations de la Haute-Garonne. In Germany, all experimental procedures were performed according to the guidelines of the German animal welfare law and approved by the local government (Tierschutzgesetz §11, Abs. 1, Nr. 1, husbandry permit no. AZ 35-9185.64/BH). In the United Kingdom, all experiments were conducted with Project and Personal license approval. Anesthesia and euthanasia procedures were performed in Tricaine Methanesulfonate (MS-222) solutions as recommended for zebrafish (0.16 mg/mL for anesthesia, 0.30 mg/mL for euthanasia). Efforts were made to minimize the number of animals used and their suffering.
A detailed description of all materials and methods can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the P.B. and S.W.W. laboratories for insightful discussions; Michael Tsang for kindly providing Tg(dusp6:d2EGFP)pt6 and Tg(dusp6:d2EGFP)pt8 transgenic lines, as well as reagents and plasmids; Bruce Riley for the Tg(hsp70:fgf8a)x17 line; Qiling Xu for providing the Tg(hsp70:ca-fgfr1)pd3 line generated in Ken Poss’s group; Pauline Rataud for technical assistance; Brice Ronsin from the Toulouse RIO Imaging platform; Stephane Relexans, Richard Brimicombe, and Aurore Laire and the University College London Fish Facility team for fish care; and Bertrand Benazeraf, Damien Ramel, and Alain Vincent for critical reading of the manuscript. This work was supported by the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, Université de Toulouse III, Fondation pour la Recherche Médicale (DEQ20131029166), Fédération pour la Recherche sur le Cerveau, Association pour la Recherche sur le Cancer (PJA 20131200173), the European Community (254195), and the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812016115/-/DCSupplemental.
References
- 1.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 2.Rørth P. Fellow travellers: Emergent properties of collective cell migration. EMBO Rep. 2012;13:984–991. doi: 10.1038/embor.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pocha SM, Montell DJ. Cellular and molecular mechanisms of single and collective cell migrations in Drosophila: Themes and variations. Annu Rev Genet. 2014;48:295–318. doi: 10.1146/annurev-genet-120213-092218. [DOI] [PubMed] [Google Scholar]
- 4.Scarpa E, Mayor R. Collective cell migration in development. J Cell Biol. 2016;212:143–155. doi: 10.1083/jcb.201508047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aman A, Piotrowski T. Cell-cell signaling interactions coordinate multiple cell behaviors that drive morphogenesis of the lateral line. Cell Adhes Migr. 2011;5:499–508. doi: 10.4161/cam.5.6.19113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ochoa-Espinosa A, Affolter M. Branching morphogenesis: From cells to organs and back. Cold Spring Harb Perspect Biol. 2012;4:a008243. doi: 10.1101/cshperspect.a008243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilmour D, Rembold M, Leptin M. From morphogen to morphogenesis and back. Nature. 2017;541:311–320. doi: 10.1038/nature21348. [DOI] [PubMed] [Google Scholar]
- 8.Theveneau E, Mayor R. Collective cell migration of epithelial and mesenchymal cells. Cell Mol Life Sci. 2013;70:3481–3492. doi: 10.1007/s00018-012-1251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell K, Casanova J. A common framework for EMT and collective cell migration. Development. 2016;143:4291–4300. doi: 10.1242/dev.139071. [DOI] [PubMed] [Google Scholar]
- 10.Sutherland D, Samakovlis C, Krasnow MA. Branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- 11.Sato A, et al. FGF8 signaling is chemotactic for cardiac neural crest cells. Dev Biol. 2011;354:18–30, and erratum (2012) 370:164. doi: 10.1016/j.ydbio.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breau MA, Wilson D, Wilkinson DG, Xu Q. Chemokine and Fgf signalling act as opposing guidance cues in formation of the lateral line primordium. Development. 2012;139:2246–2253. doi: 10.1242/dev.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadam S, Ghosh S, Stathopoulos A. Synchronous and symmetric migration of Drosophila caudal visceral mesoderm cells requires dual input by two FGF ligands. Development. 2012;139:699–708. doi: 10.1242/dev.068791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bénazéraf B, et al. A random cell motility gradient downstream of FGF controls elongation of an amniote embryo. Nature. 2010;466:248–252. doi: 10.1038/nature09151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMahon A, Reeves GT, Supatto W, Stathopoulos A. Mesoderm migration in Drosophila is a multi-step process requiring FGF signaling and integrin activity. Development. 2010;137:2167–2175. doi: 10.1242/dev.051573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lecaudey V, Cakan-Akdogan G, Norton WHJ, Gilmour D. Dynamic Fgf signaling couples morphogenesis and migration in the zebrafish lateral line primordium. Development. 2008;135:2695–2705. doi: 10.1242/dev.025981. [DOI] [PubMed] [Google Scholar]
- 17.Nechiporuk A, Raible DW. FGF-dependent mechanosensory organ patterning in zebrafish. Science. 2008;320:1774–1777. doi: 10.1126/science.1156547. [DOI] [PubMed] [Google Scholar]
- 18.Ernst S, et al. Shroom3 is required downstream of FGF signalling to mediate proneuromast assembly in zebrafish. Development. 2012;139:4571–4581. doi: 10.1242/dev.083253. [DOI] [PubMed] [Google Scholar]
- 19.Harding MJ, Nechiporuk AV. Fgfr-Ras-MAPK signaling is required for apical constriction via apical positioning of Rho-associated kinase during mechanosensory organ formation. Development. 2012;139:3130–3135. doi: 10.1242/dev.082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durdu S, et al. Luminal signalling links cell communication to tissue architecture during organogenesis. Nature. 2014;515:120–124. doi: 10.1038/nature13852. [DOI] [PubMed] [Google Scholar]
- 21.Beretta CA, Dross N, Guiterrez-Triana JA, Ryu S, Carl M. Habenula circuit development: Past, present, and future. Front Neurosci. 2012;6:51. doi: 10.3389/fnins.2012.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Concha ML, Bianco IH, Wilson SW. Encoding asymmetry within neural circuits. Nat Rev Neurosci. 2012;13:832–843. doi: 10.1038/nrn3371. [DOI] [PubMed] [Google Scholar]
- 23.Duboc V, Dufourcq P, Blader P, Roussigné M. Asymmetry of the brain: Development and implications. Annu Rev Genet. 2015;49:647–672. doi: 10.1146/annurev-genet-112414-055322. [DOI] [PubMed] [Google Scholar]
- 24.Roussigne M, Blader P, Wilson SW. Breaking symmetry: The zebrafish as a model for understanding left-right asymmetry in the developing brain. Dev Neurobiol. 2012;72:269–281. doi: 10.1002/dneu.20885. [DOI] [PubMed] [Google Scholar]
- 25.Concha ML, et al. Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron. 2003;39:423–438. doi: 10.1016/s0896-6273(03)00437-9. [DOI] [PubMed] [Google Scholar]
- 26.Aizawa H, et al. Laterotopic representation of left-right information onto the dorso-ventral axis of a zebrafish midbrain target nucleus. Curr Biol. 2005;15:238–243. doi: 10.1016/j.cub.2005.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bianco IH, Carl M, Russell C, Clarke JD, Wilson SW. Brain asymmetry is encoded at the level of axon terminal morphology. Neural Dev. 2008;3:9. doi: 10.1186/1749-8104-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamse JT, Thisse C, Thisse B, Halpern ME. The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development. 2003;130:1059–1068. doi: 10.1242/dev.00270. [DOI] [PubMed] [Google Scholar]
- 29.Gamse JT, et al. Directional asymmetry of the zebrafish epithalamus guides dorsoventral innervation of the midbrain target. Development. 2005;132:4869–4881. doi: 10.1242/dev.02046. [DOI] [PubMed] [Google Scholar]
- 30.Regan JC, Concha ML, Roussigne M, Russell C, Wilson SW. An Fgf8-dependent bistable cell migratory event establishes CNS asymmetry. Neuron. 2009;61:27–34. doi: 10.1016/j.neuron.2008.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bisgrove BW, Essner JJ, Yost HJ. Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development. 2000;127:3567–3579. doi: 10.1242/dev.127.16.3567. [DOI] [PubMed] [Google Scholar]
- 32.Concha ML, Burdine RD, Russell C, Schier AF, Wilson SW. A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron. 2000;28:399–409. doi: 10.1016/s0896-6273(00)00120-3. [DOI] [PubMed] [Google Scholar]
- 33.Liang JO, et al. Asymmetric nodal signaling in the zebrafish diencephalon positions the pineal organ. Development. 2000;127:5101–5112. doi: 10.1242/dev.127.23.5101. [DOI] [PubMed] [Google Scholar]
- 34.Molina GA, Watkins SC, Tsang M. Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Dev Biol. 2007;7:62. doi: 10.1186/1471-213X-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eblaghie MC, et al. Negative feedback regulation of FGF signaling levels by Pyst1/MKP3 in chick embryos. Curr Biol. 2003;13:1009–1018. doi: 10.1016/s0960-9822(03)00381-6. [DOI] [PubMed] [Google Scholar]
- 36.Kawakami Y, et al. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat Cell Biol. 2003;5:513–519. doi: 10.1038/ncb989. [DOI] [PubMed] [Google Scholar]
- 37.Tsang M, et al. A role for MKP3 in axial patterning of the zebrafish embryo. Development. 2004;131:2769–2779. doi: 10.1242/dev.01157. [DOI] [PubMed] [Google Scholar]
- 38.Matsuda M, et al. Lef1 regulates Dusp6 to influence neuromast formation and spacing in the zebrafish posterior lateral line primordium. Development. 2013;140:2387–2397. doi: 10.1242/dev.091348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammadi M, et al. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- 40.Clanton JA, Hope KD, Gamse JT. Fgf signaling governs cell fate in the zebrafish pineal complex. Development. 2013;140:323–332. doi: 10.1242/dev.083709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: A quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- 42.Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–112. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marques SR, Lee Y, Poss KD, Yelon D. Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Dev Biol. 2008;321:397–406. doi: 10.1016/j.ydbio.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamei Y, et al. Infrared laser-mediated gene induction in targeted single cells in vivo. Nat Methods. 2009;6:79–81. doi: 10.1038/nmeth.1278. [DOI] [PubMed] [Google Scholar]
- 45.Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- 46.Prasad M, Montell DJ. Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev Cell. 2007;12:997–1005. doi: 10.1016/j.devcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 47.Jakobsson L, et al. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat Cell Biol. 2010;12:943–953. doi: 10.1038/ncb2103. [DOI] [PubMed] [Google Scholar]
- 48.Ghabrial AS, Krasnow MA. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature. 2006;441:746–749. doi: 10.1038/nature04829. [DOI] [PubMed] [Google Scholar]
- 49.Ikeya T, Hayashi S. Interplay of Notch and FGF signaling restricts cell fate and MAPK activation in the Drosophila trachea. Development. 1999;126:4455–4463. doi: 10.1242/dev.126.20.4455. [DOI] [PubMed] [Google Scholar]
- 50.Siekmann AF, Lawson ND. Notch signalling and the regulation of angiogenesis. Cell Adhes Migr. 2007;1:104–106. doi: 10.4161/cam.1.2.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bae Y-K, Trisnadi N, Kadam S, Stathopoulos A. The role of FGF signaling in guiding coordinate movement of cell groups: Guidance cue and cell adhesion regulator? Cell Adhes Migr. 2012;6:397–403. doi: 10.4161/cam.21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozlovskaja-Gumbrienė A, et al. Proliferation-independent regulation of organ size by Fgf/Notch signaling. eLife. 2017;6:e21049. doi: 10.7554/eLife.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aman A, Piotrowski T. Wnt/beta-catenin and Fgf signaling control collective cell migration by restricting chemokine receptor expression. Dev Cell. 2008;15:749–761. doi: 10.1016/j.devcel.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Dalle Nogare D, et al. Leading and trailing cells cooperate in collective migration of the zebrafish posterior lateral line primordium. Development. 2014;141:3188–3196. doi: 10.1242/dev.106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inaki M, Vishnu S, Cliffe A, Rørth P. Effective guidance of collective migration based on differences in cell states. Proc Natl Acad Sci USA. 2012;109:2027–2032. doi: 10.1073/pnas.1115260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lebreton G, Casanova J. Ligand-binding and constitutive FGF receptors in single Drosophila tracheal cells: Implications for the role of FGF in collective migration. Dev Dyn. 2016;245:372–378. doi: 10.1002/dvdy.24345. [DOI] [PubMed] [Google Scholar]
- 57.Lebreton G, Casanova J. Specification of leading and trailing cell features during collective migration in the Drosophila trachea. J Cell Sci. 2014;127:465–474. doi: 10.1242/jcs.142737. [DOI] [PubMed] [Google Scholar]
- 58.Poukkula M, Cliffe A, Changede R, Rørth P. Cell behaviors regulated by guidance cues in collective migration of border cells. J Cell Biol. 2011;192:513–524. doi: 10.1083/jcb.201010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reifers F, et al. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- 60.Albertson RC, Yelick PC. Fgf8 haploinsufficiency results in distinct craniofacial defects in adult zebrafish. Dev Biol. 2007;306:505–515. doi: 10.1016/j.ydbio.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwon H-J, Bhat N, Sweet EM, Cornell RA, Riley BB. Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet. 2010;6:e1001133. doi: 10.1371/journal.pgen.1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gonzalez-Quevedo R, Lee Y, Poss KD, Wilkinson DG. Neuronal regulation of the spatial patterning of neurogenesis. Dev Cell. 2010;18:136–147. doi: 10.1016/j.devcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.