Significance
Glial-guided neuronal migration is a key step in the histogenesis of cortical regions in the mammalian brain and requires the expression of adhesion proteins by the migrating neuron and the glial fiber. The neuronal receptor astrotactin (ASTN1) regulates glial-guided migration, but the glial ligand has long been unknown. Here we demonstrate that neuron–glia attachment and neuronal migration depend on glial expression of the neural cadherin (CDH2) and that ASTN1 promotes migration by a direct interaction with neuronal and glial CDH2. Thus, ASTN1 and CDH2 form a cis and trans asymmetric bridge complex in the migration junction that is essential for glial-guided neuronal migration.
Keywords: astrotactin, N-cadherin, cerebellum, glial-guided migration, migration junction
Abstract
Prior studies demonstrate that astrotactin (ASTN1) provides a neuronal receptor for glial-guided CNS migration. Here we report that ASTN1 binds N-cadherin (CDH2) and that the ASTN1:CDH2 interaction supports cell–cell adhesion. To test the function of ASTN1:CDH2 binding in glial-guided neuronal migration, we generated a conditional loss of Cdh2 in cerebellar granule cells and in glia. Granule cell migration was slowed in cerebellar slice cultures after a conditional loss of neuronal Cdh2, and more severe migration defects occurred after a conditional loss of glial Cdh2. Expression in granule cells of a mutant form of ASTN1 that does not bind CDH2 also slowed migration. Moreover, in vitro chimeras of granule cells and glia showed impaired neuron–glia attachment in the absence of glial, but not neuronal, Cdh2. Thus, cis and trans bindings of ASTN1 to neuronal and glial CDH2 form an asymmetric neuron–glial bridge complex that promotes glial-guided neuronal migration.
In cortical regions of mammalian brain, glial-guided neuronal migration directs postmitotic cells into neuronal layers, a process that underlies the formation of the cortical circuitry (1–3). The cerebellar cortex has long provided a key model for understanding the molecular basis of glial-guided migration, as granule cell precursors (GCPs) migrate from the external germinal layer (EGL) along the radial processes of Bergmann glia (BG) to a position deep to the Purkinje neuron, the sole output neuron of the cerebellar cortex (4). Correlated video and electron microscopy (EM) imaging of GCP migration along BG demonstrates that migrating neurons form a puncta adherens migration junction beneath the cell soma and extend a motile leading process in the direction of forward movement (5, 6). During migration, the neuron forms and releases the migration junction by a process that involves endocytosis of the receptor astrotactin (ASTN1), which is expressed in neurons but not in glia (7). Molecular experiments demonstrate that the conserved polarity complex mPar6 regulates the cadence of locomotion by controlling the forward movement of the centrosome (8) as well as microtubule dynamics and actomyosin motor function in the proximal aspect of the leading process (9), with the Rho GTPase Cdc42 controlling actin dynamics required for the polarity of the migrating GCP and for the formation of the migration junction with the glial fiber (10). While biochemical and genetic experiments have confirmed the key role of the neuronal guidance receptor ASTN1 in the migration junction (11–13), evidence is lacking for the glial ligand for ASTN1.
Cadherins are cell-surface proteins composed of an adhesive extracellular domain and a cytoplasmic tail that links to the actin cytoskeleton through a complex of catenins. The extracellular domain allows cadherins to form lateral (cis) homodimers or mediate cell adhesion through trans homodimers. A large body of evidence demonstrates a key role for homophilic trans cadherin interactions in the formation and maintenance of puncta adherens junctions in the developing heart and neural tube (14) and in synapse formation (15, 16). In addition, disruption of the neural cadherin, N-cadherin (CDH2), leads to defects in neuronal migration during development of the cerebral cortex (17–22). Here we show that an asymmetric cis and trans complex of ASTN1 and CDH2 functions in neuronal migration. Conditional loss of glial CDH2 in mice impaired GCP migration in vivo and ex vivo and perturbed the formation of a migration junction between GCPs and BG in cell-based assays. Moreover, CDH2-deficient GCPs expressing an ASTN1 variant that lacks the binding domain for CDH2 failed to migrate on CDH2-expressing glia. This suggests that ASTN1 in neurons and CDH2 in neurons and glial fibers form an asymmetric bridge complex that is required for glial-guided migration, and, more generally, that CDH2 might function as a heterophilic binding partner in the formation of other cell–cell junctions.
Results
CDH2 Is Expressed in the Migration Junction and Interacts with ASTN1.
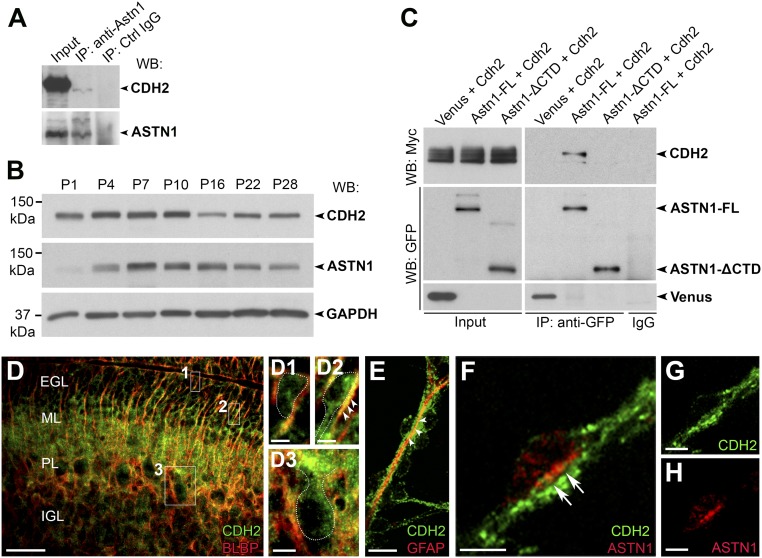
To investigate whether CDH2 interacts with ASTN1, we performed immunoprecipitation on protein lysates from postnatal day 7 (P7) mouse cerebella using an ASTN1 antibody. In this assay, we found that ASTN1 interacts with CDH2 (Fig. 1A). We then used Western blotting to examine the developmental expression of CDH2 in the cerebellum. Western blotting of whole-cerebellar lysates showed maximal levels of CDH2 in the early postnatal stages (P4–P10), when ASTN1 expression is high (Fig. 1B). In HEK 293T cells, we detected coimmunoprecipitation of CDH2 with full-length ASTN1 (ASTN1-FL) but not with a mutant variant of ASTN1 lacking a large portion of the C-terminal ectodomain (ASTN1-ΔCTD) that included the membrane attack complex/perforin (MACPF), fibronectin type III (FNIII), and annexin-like (ANX-like) domains (Fig. 1C). Flow cytometry showed that ASTN1-FL and ASTN1-ΔCTD localized to the cell surface at similar levels (58% and 49%, respectively) (SI Appendix, Fig. S1). Thus, the extracellular C terminus of ASTN1 forms a cis interaction with the ectodomain of CDH2.
Fig. 1.
ASTN1 and CDH2 form cis interactions and colocalize in the migration junction. (A) In vivo immunoprecipitation of ASTN1 in P7 whole-cerebellar lysates blotted with ASTN1 and CDH2 antibodies. ASTN1 is part of a protein complex with CDH2. (B) Developmental protein expression of CDH2 and ASTN1 in the cerebellum of postnatal mice (P1–P28) by Western blot. CDH2 expression was highest between P4–P10, decreased by P16, and reached a steady level at P22–P28. ASTN1 expression increased after P1 and was highest at P7–P10. Protein expression was compared with GAPDH levels. (C) Western blots showing coimmunoprecipitation of ASTN1-Venus and CDH2-Myc in HEK 293T cells. CDH2 interacted with ASTN1-FL but not with ASTN1-ΔCTD. (D and E) Endogenous protein expression of CDH2 at P7 in sagittal mouse cerebellar sections (D) and in GCP/BG in vitro cocultures (E). CDH2 was expressed in GCPs in the EGL (D1), in migrating GCPs in the ML (D2), and in Purkinje cells (D3) and colocalized with BLBP and GFAP in BG fibers (arrowheads in D2 and E). (F–H) GCP/BG cocultures labeled with antibodies against CDH2 (F and G) and ASTN1 (F and H). CDH2 localized to neuronal processes, glial fibers, and the migration junction beneath the neuronal soma. ASTN1 colocalized with CDH2 in the migration junction (arrows in F). PL, Purkinje cell layer. (Scale bars: 50 µm in D; 5 µm in D1, D2, and F–H; and 10 µm in D3 and E.)
In sections of early postnatal (P5–P7) mouse cerebellum, a CDH2 antibody labeled GCPs in the EGL, GCPs migrating across the molecular layer (ML), and mature granule cells (GCs) in the internal granule layer (IGL) as well as in the radial processes of BG stretching across the ML and in Purkinje cells (Fig. 1D). CDH2 also localized to the migration junction, a puncta adherens junction between migrating GCPs (6), identified by their elongated profile and close apposition with the glial fiber (5, 23), and BG fibers in cultures of purified neurons and glia (Fig. 1 E–G). Antibodies against CDH2 intensely labeled the neuronal soma at the junction with the glial fiber and also stained the underlying glial fiber. In agreement with prior confocal and immuno-EM localization studies (11), antibodies against ASTN1 labeled the neuronal aspect of the migration junction (Fig. 1 F and H). Thus, ASTN1 and CDH2 colocalize to the migration junction of GCPs migrating along BG fibers.
CDH2 and ASTN1 Form Heterophilic Trans Interactions.
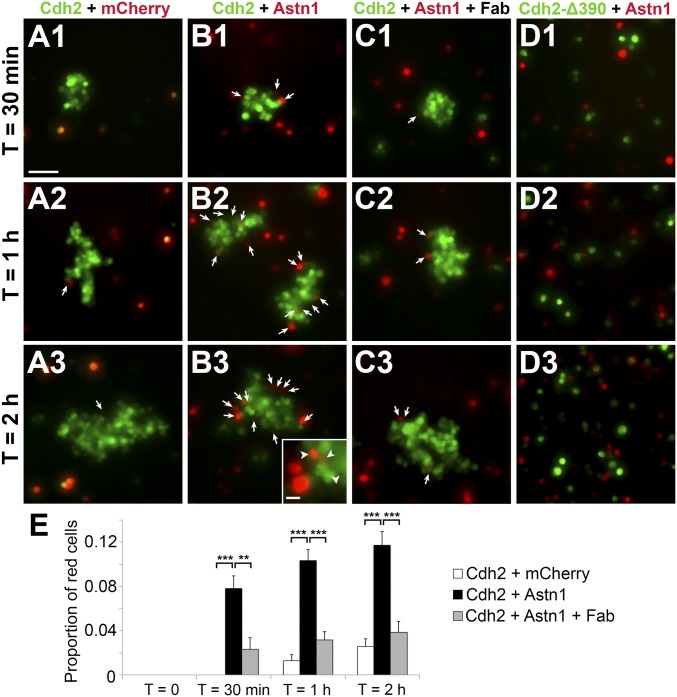
To analyze whether CDH2 interacts with ASTN1 in trans to promote cell adhesion, we used a classical Schneider 2 (S2) cell-adhesion assay (24). For this assay, we transfected S2 cells with bicistronic expression constructs (25) of Cdh2;GFP or Astn1;mCherry cDNA and measured cell aggregation rates over 2 h. Cells transfected with Cdh2;GFP formed aggregates within minutes, demonstrating a rapid homophilic trans binding of CDH2 (Fig. 2A). In contrast, ASTN1-positive cells did not form homophilic aggregates over the 2-h incubation period. ASTN1-positive cells did, however, form coaggregates with CDH2-positive cells, indicating heterophilic trans binding between ASTN1 and CDH2 (Fig. 2B). At 2 h, CDH2-positive aggregates contained 11.7 ± 1.2% ASTN1-positive cells compared with 2.6 ± 0.7% mCherry-expressing control cells (P < 0.0001). Moreover, in contrast to the control cells, ASTN1-positive cells frequently integrated into the core of the aggregates. Taken together, these results confirmed earlier findings that ASTN1 does not promote cell adhesion through homophilic binding (26) and showed that CDH2 provides a trans ligand for ASTN1 that functions in cell–cell adhesion.
Fig. 2.
Heterophilic trans interactions of ASTN1 and CDH2. Drosophila S2 cell-adhesion assays were prepared in four conditions: Cdh2;GFP + mCherry;GFP (A), Cdh2;GFP + Astn1;mCherry (B), Cdh2;GFP + Astn1;mCherry + ASTN1 Fab (C), and Cdh2-Δ390;GFP + Astn1;mCherry (D). ASTN1-positive cells were adhering to the CDH2-expressing aggregates after 30 min (arrows in B1), with more coaggregation seen after 1 h (arrows in B2) and 2 h (arrows in B3), indicating heterophilic trans interactions. Significantly lower proportions of cells were adhering to the aggregates in the conditions with cells expressing control vector (A) or Astn1;mCherry blocked with ASTN1 Fab fragments (C). (D) Expression of CDH2-Δ390;GFP did not result in cell aggregation within 2 h, demonstrating the importance of the cadherin ectodomain for homophilic and heterophilic interactions and cell adhesion. The proportion of mCherry-expressing cells in the CDH2;GFP-positive aggregates is quantified in E. **P < 0.01; ***P < 0.001. (Scale bars: 50 µm in A–D and 10 µm in Inset in B3.)
To test the specificity of the heterophilic CDH2:ASTN1 trans interaction, we measured the aggregation of ASTN1- and CDH2-positive S2 cells in the presence of Fab fragments of an ASTN1 antibody raised against the C terminus of ASTN1 (12). After addition of Fab fragments, heterophilic CDH2:ASTN1 trans cell adhesion was reduced to control levels (Fig. 2C), suggesting that the C terminus of ASTN1 is required for the interaction with CDH2. We then assessed the specificity of both homophilic CDH2:CDH2 and heterophilic CDH2:ASTN1 binding using a Cdh2-Δ390 construct with a deletion of the extracellular domain of CDH2. In S2 cells expressing CDH2-Δ390, both homophilic adhesion and heterophilic adhesion with ASTN1-positive cells failed (Fig. 2D), demonstrating a requirement for the ectodomain of CDH2 in trans cell adhesion.
Cell-Specific Deletion of Cdh2 in the Cerebellum.
To provide a genetic model for the function of CDH2 in GCP migration in the developing mouse cerebellum, we generated conditional knockout (cKO) of Cdh2 by crossing a floxed Cdh2 (Cdh2fl/fl) mouse line with a NeuroD1-Cre line to delete Cdh2 in GCPs. In addition, we crossed the Cdh2fl/fl mice with an mGFAP-Cre line to delete Cdh2 in BG or with an hGFAP-Cre line to delete Cdh2 in both GCPs and glia (27–29). Western blot analysis of lysates of GCPs and BG purified from each of the lines at P7 (30) confirmed the cell-specific deletion of Cdh2 (SI Appendix, Fig. S2A). To examine the development of the cerebellum in each of these cKO lines, we first analyzed fixed sections of P7 cerebellum by Nissl staining (SI Appendix, Fig. S2 B–E). Although the overall size of the cerebellum did not differ significantly in the three lines (n = 7 per genotype), defects in the foliation pattern of the cerebellum of Cdh2fl/fl;mGFAP-Cre and Cdh2fl/fl;hGFAP-Cre mice were observed compared with controls. These defects included additional fissures in the ventral (I–III) lobes, with fewer fissures and irregularly shaped lobes in mediodorsal (VI–VIII) areas.
Loss of Cdh2 in GCs and/or Glia Has a Differential Effect on Migration.
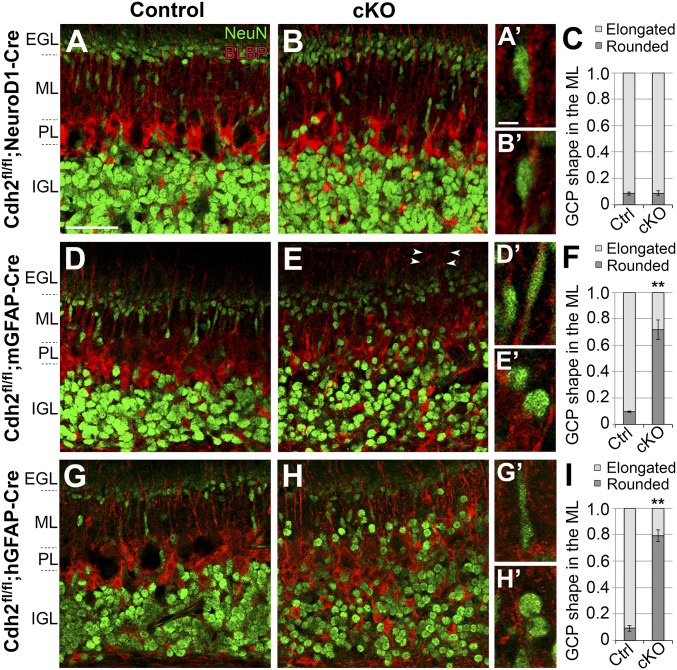
Immunostaining of cerebellar sections of the three cKO lines with NeuN, a marker for GCs, and with BLBP, a marker for BG, revealed striking differences in GCP migration and formation of the neuronal layers, especially in lines where BG or both BG and GCPs lacked Cdh2 (Fig. 3). In all three lines, the density and organization of NeuN-positive GCPs in the inner EGL were identical to that seen in control mice (n = 7 per genotype). In Cdh2fl/fl;NeuroD1-Cre mice, the elongated profile of GCPs migrating across the molecular layer was indistinguishable from controls, and the overall laminar organization of the cerebellum appeared to be normal (Fig. 3 A–C). In contrast, NeuN-positive GCPs had a significantly higher proportion of rounded soma relative to elongated soma in the ML of both Cdh2fl/fl;mGFAP-Cre (control: 9.6 ± 0.6%, cKO: 72 ± 13%; P = 0.0026) and Cdh2fl/fl;hGFAP-Cre (control: 9.5 ± 1.7%, cKO: 79 ± 6.5%; P = 0.0014) mice, and many GCPs were located in the ML, suggesting slowed or stalled GCP migration (Fig. 3 D–I) (31). Immunostaining for BLBP in these mice also revealed defects in positioning of BG cell bodies and radial patterning of BG fibers in some areas of the cerebellum (Fig. 3 E and H). Overall, the laminar patterning of the cerebellum was disorganized, as the classic boundaries of the ML and IGL were uneven relative to controls. In contrast, immunostaining with a calbindin antibody showed no changes in the gross morphology or positioning of Purkinje cells (SI Appendix, Fig. S3). Thus, a conditional loss of Cdh2 in BG perturbed the migration of GCPs and the formation of the GC layer.
Fig. 3.
Neuronal migration in Cdh2-cKO mice. Sagittal cerebellar sections of P7 Cdh2fl/fl control mice (A, D, and G) and Cdh2-cKO littermates expressing NeuroD1-Cre (B), mGFAP-Cre (E), or hGFAP-Cre (H) and labeled with NeuN and BLBP antibodies. In control mice, NeuN-positive GCPs displayed elongated somas along BG fibers in the ML, indicating migrating cells. A similar phenotype is seen in mice with GCPs lacking Cdh2 (B). In contrast, a loss of Cdh2 in BG (E) or in both GCPs and BG (H) resulted in rounded GCPs and a stalled migration in the ML. The proportion of elongated and rounded GCPs in the ML is shown in stacked bar charts (C, F, and I). Abnormal radial patterning of BG fibers was observed in some areas (arrowheads in E). Higher magnifications in A′, B′, D′, E′, G′, and H′ show representative GCPs from each genotype. PL, Purkinje cell layer. **P < 0.01. (Scale bars: 50 µm in A, B, D, E, G, and H and 5 µm in A′, B′, D′, E′, G′, and H′.)
To quantitate the rate of GCP migration along BG fibers, we performed BrdU birth-dating experiments, injecting BrdU at P5 and killing the animals at P7 (n = 4 per genotype; SI Appendix, Fig. S4 A–I). By BrdU labeling, the migration distance of GCs was reduced in Cdh2fl/fl;NeuroD1-Cre mice, in which 40 ± 3.7% of labeled GCs reached the IGL compared with 50 ± 3.9% in control mice (P = 0.035). However, GCP migration was dramatically reduced in the Cdh2fl/fl;mGFAP-Cre mice, in which 33 ± 4.4% reached the IGL compared with 60 ± 1.9% in control mice (P = 0.040). Similarly, in the Cdh2fl/fl;hGFAP-Cre mice, 29 ± 2.9% of GCs reached the IGL compared with 55 ± 4.4% in control mice (P = 0.0002). The latter two lines also had a significantly higher proportion of BrdU-labeled cells in the ML (22% and 21% higher than in control littermates, P = 0.02 and 0.0002, respectively). These findings suggest that the expression of CDH2 in BG fibers is required for GCP migration.
Since a decrease in the number of GCs reaching the IGL could also be due to changes in cell proliferation or cell death, we stained sections with antibodies to the mitosis marker phospho-histone H3 and the apoptosis marker caspase-3. We found no differences in proliferation or apoptosis in the three cKO lines (SI Appendix, Fig. S4 J and K), suggesting that the lower proportion of cells in the IGL resulted from migration defects.
Glial CDH2 Is Essential for GC Migration in Organotypic Slice Cultures.
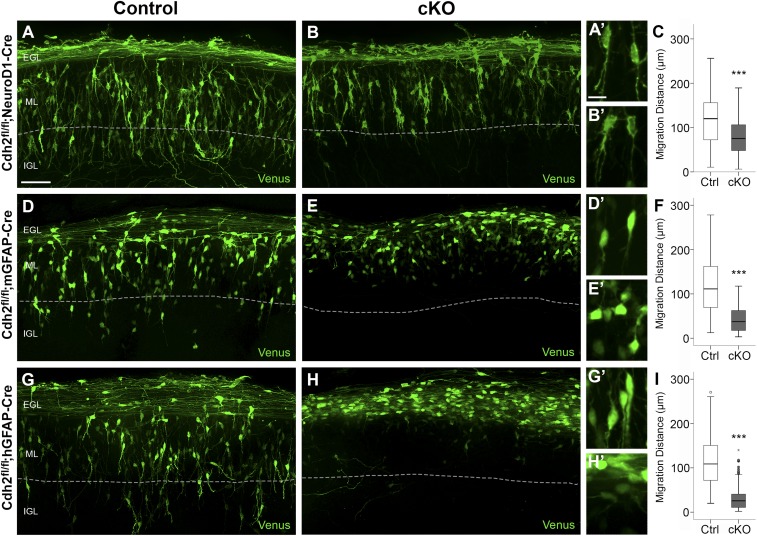
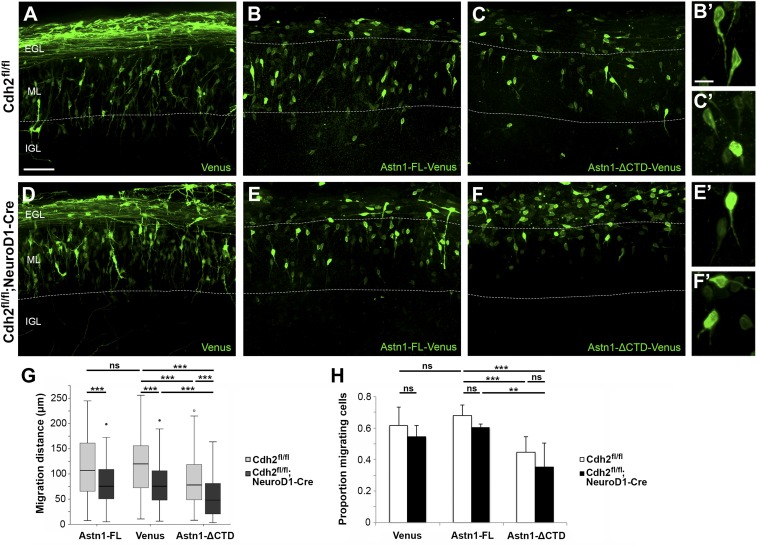
To analyze the features of migrating GCPs in the three cKO lines in more detail, we used electroporation to express the fluorophore Venus in GCPs in P8 organotypic slices of cerebellar cortex and imaged labeled cells by spinning-disk confocal microscopy (Fig. 4). In control slices, after 60 h, Venus-positive GCs were observed in the inner EGL, the ML, and the outer portion of the IGL. Venus-positive cells in the inner EGL extended long parallel fiber axons, with labeled cells in the ML showing the bipolar morphology typical of migrating neurons with a leading process in the radial plane (Fig. 4 A, D, and G). Although the polarity and overall morphology of labeled GCPs in ex vivo slices of Cdh2fl/fl;NeuroD1-Cre cerebellum were similar to controls (Fig. 4 A and B), the median distance of migration, calculated by measuring the distance of the cell soma to the parallel fiber axons, was reduced by 37% compared with controls (control: 120 µm, cKO: 75 µm; P < 0.001), indicating a slowed migration rate (Fig. 4C).
Fig. 4.
Glial CDH2 is essential for glial-guided neuronal migration. Organotypic ex vivo slice cultures prepared from the cerebellum of P8 Cdh2fl/fl control and Cdh2-cKO mice, electroporated with Venus, and fixed after 60 h. In slice cultures from control mice (A, D, and G) Venus-expressing GCPs migrated radially across the ML and extended a leading process in the direction of migration. Although most Venus-positive GCPs lacking Cdh2 extended a leading process (B), their median migration distance was reduced by 37% (C). In ex vivo slices where BG lacked Cdh2 (E), or where both GCPs and BG lacked Cdh2 (H), Venus-positive GCPs had a rounded or multipolar morphology, failed to extend a leading process, and migrated a shorter distance away from the field of labeled parallel fibers into the ML, indicating a stalled migration. Dotted lines indicate the ML/IGL boundary. (A′, B′, D′, E′, G′, and H′) Representative cell morphologies are shown at higher magnification. (F and I) The median migration distance was reduced by 66% (BG cKO) (F) and 76% (GCP + BG cKO) (I). ***P < 0.001. (Scale bars: 50 µm in A, B, D, E, G, and H and 10 µm in A′, B′, D′, E′, G′, and H′.)
While Venus-positive GCPs in ex vivo organotypic cultures of both Cdh2fl/fl;mGFAP-Cre and Cdh2fl/fl;hGFAP-Cre mice appeared to extend parallel fiber axons normally, the GCPs had dramatic morphological defects, as nearly all the cells were rounded or irregularly shaped, rather than elongated, and failed to extend a leading process in the direction of migration (Fig. 4 E and H). Consequently, the median migration distance was severely reduced in the Cdh2fl/fl;mGFAP-Cre (control: 111 µm, cKO: 38 µm; 66% reduction; P < 0.001) and Cdh2fl/fl;hGFAP-Cre (control: 109 µm, cKO: 26 µm; 76% reduction; P < 0.001) organotypic cultures (Fig. 4 F and I). Importantly, virtually all the Venus-positive cells were stalled in the EGL or the upper portion of the ML, indicating a failure of glial-guided GCP migration.
Functional Interaction of CDH2 and ASTN1 During GC Migration.
Since glial, but not neuronal, loss of Cdh2 stalled GCP migration, we hypothesized that ASTN1 may promote glial-guided migration in the absence of neuronal CDH2. To examine the function of ASTN1 in GCPs positive or negative for CDH2, we electroporated the Venus, Astn1-FL-Venus, or Astn1-ΔCTD-Venus plasmids into cerebella of P8 control and Cdh2fl/fl;NeuroD1-Cre mice before generating organotypic cultures. No significant differences in migration distance or proportion of migrating cells were observed between slice cultures with GCPs expressing Venus and Astn1-FL-Venus (Fig. 5 A, B, D, and E), indicating that ASTN1-FL overexpression did not alter glial-guided migration. However, expression of the Astn1-ΔCTD-Venus plasmid in GCPs in control slices reduced the median migration distance by 35% compared with control slices expressing Venus (78 µm and 120 µm, respectively; P < 0.001) (Fig. 5 C and G), which is similar to the reduction in the Cdh2fl/fl;NeuroD1-Cre slices expressing Venus (Fig. 5G). This indicates that the ASTN1-ΔCTD protein acted as a dominant-negative variant of ASTN1-FL. Expression of Astn1-ΔCTD-Venus in GCPs lacking Cdh2 (Cdh2fl/fl;NeuroD1-Cre) further reduced migration distance by 39% compared with the controls with Astn1-ΔCTD-Venus (48 µm and 78 µm, respectively; P < 0.001) and by 60% compared with the controls with Venus (48 µm and 120 µm, respectively; P < 0.001) (Fig. 5 F and G). Importantly, combined disruption of CDH2 and ASTN1 in GCPs resulted in a significant failure to migrate out of the EGL. In addition, expression of Astn1-ΔCTD-Venus in GCPs generated a lower proportion of migrating cells, as characterized by their morphology (rounded/multipolar vs. bipolar) (Fig. 5H). These experiments show that CDH2-deficient GCPs that expressed ASTN1 lacking the domains that bind CDH2 failed to extend a leading process and to migrate. Thus, although homophilic CDH2:CDH2 interactions may contribute to neuron–glial binding, these data demonstrate that heterophilic ASTN1:CDH2 binding is required for glial-guided neuronal migration.
Fig. 5.
ASTN1 and CDH2 interact functionally to regulate migration. Organotypic slice cultures from the cerebellum of P8 Cdh2fl/fl and Cdh2fl/fl;NeuroD1-Cre mice were electroporated with Venus (A and D), Astn1-FL-Venus (B and E), or Astn1-ΔCTD-Venus (C and F). ASTN1-Venus fluorescence labeled the cell soma and processes but not the parallel fibers. After 60 h, the distance migrated by GCPs expressing Astn1-FL-Venus was similar to that of GCPs expressing Venus (A, B, and G) both in the presence and absence of neuronal Cdh2 (D, E, and G). However, in slice cultures of control mice where GCPs expressed Astn1-ΔCTD-Venus, GCPs migrated a 35% shorter distance (78 µm median) than GCPs expressing Venus (120 µm median) (C and G). Loss of Cdh2 combined with overexpression of Astn1-ΔCTD-Venus in GCPs resulted in more severe migration defects, indicated by a 60% reduction in the median migration distance (48 µm) and a higher number of cells stalled in the EGL (F and G). (H) A significantly higher proportion of cells expressing Astn1-ΔCTD-Venus were rounded or multipolar. Dotted lines indicate EGL/ML and ML/IGL boundaries. **P < 0.01; ***P < 0.001; ns, not significant. (Scale bars: 50 µm in A–F; 10 µm in B′, C′, E′, and F′.)
Neuron–Glia Attachment Is Dependent on Glial Expression of CDH2.
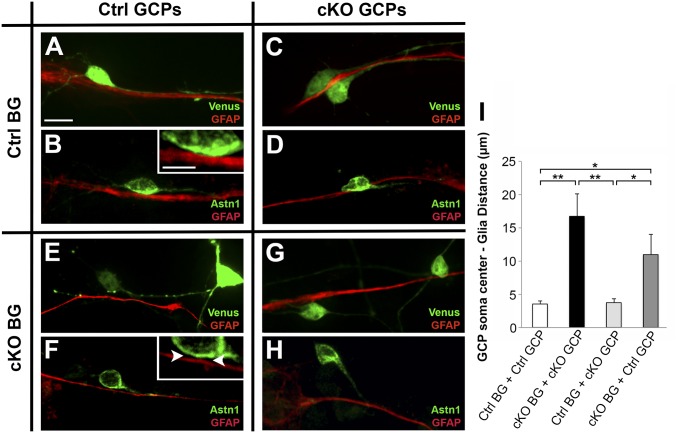
To directly analyze the formation of the migration junction in GCPs and BG lacking Cdh2, we generated in vitro chimeras (23, 31). For these experiments, we purified GCPs or BG using a step gradient of Percoll (30) and mixed and matched GCPs and BG from Cdh2fl/fl control and Cdh2fl/fl;hGFAP-Cre (GCP + BG-cKO) cerebella. Purified GCPs from each genotype were electroporated with either a Venus plasmid to visualize the GCP soma and processes or an Astn1-FL-Venus plasmid to examine the ASTN1 protein localization and were cocultured with purified glia from each genotype. In control cultures of wild-type GCPs and BG, Venus- or ASTN1-FL-Venus–expressing GCPs adhered to GFAP-labeled BG fibers, formed an elongated profile along the fiber, and extended a leading process in the direction of migration (Fig. 6 A and B). ASTN1-FL-Venus localized to the migration junction, and the overexpression did not disrupt neuron–glia attachment or migration. Similar results were observed in cocultures of GCPs lacking CDH2 with control BG (Fig. 6 C and D). A strikingly different result was seen when either control or CDH2-deficient GCPs were cultured with BG lacking CDH2. In both cases, although the ASTN1 protein localized to the basal portion of the soma, the neurons failed to form a migration junction (Fig. 6 E–H). Measurement of the distance between the GCP soma and BG fiber confirmed this observation (Fig. 6I). In addition, GCPs were rounded or multipolar, rather than elongated, as seen on control BG fibers, and failed to extend a leading process. Thus, formation of the migration junction between GCPs and BG fibers required glial CDH2.
Fig. 6.
Neuron–glia attachment is dependent on glial CDH2. In vitro chimera cocultures of GCPs and BG purified from Cdh2fl/fl (control) or Cdh2fl/fl;hGFAP-Cre (cKO) mice at P7. GCPs were electroporated with Venus (A, C, E, and G) or Astn1-Venus (B, D, F, and H). GCPs attached and migrated along BG fibers expressing CDH2 (A–D), irrespective of the GCP genotype. In contrast, GCPs cocultured with BG lacking Cdh2 did not form a migration junction with the glial fibers (E–H) and had an increased separation between the GCP somas and the glial fibers (I). The mean distance (± SEM) between control BG fibers and control or cKO GCPs was 3.53 ± 0.42 and 3.74 ± 0.54 µm, respectively. When cKO BG were used, the mean distance (± SEM) was 11.0 ± 3.03 µm (control GCPs) and 16.8 ± 3.36 µm (cKO GCPs). Note the gap between the neuronal soma and BG fiber even as the GCP process contacts the fiber (Inset in F, arrowheads). *P < 0.05; **P < 0.01. (Scale bars: 10 µm in A–H; 5 µm in Insets in B and F.)
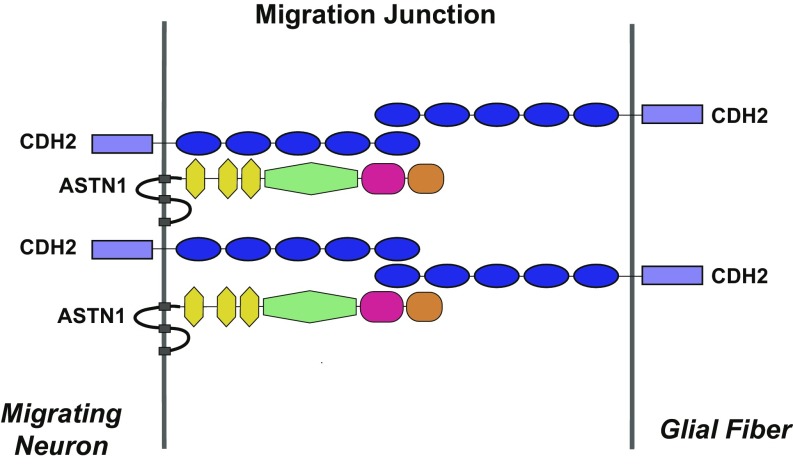
Taken together, our results propose a bridge model in which a cis complex of ASTN1 and CDH2 in the neuronal membrane interacts in trans with CDH2 on the glial fiber (Fig. 7) to promote glial-guided migration.
Fig. 7.
Proposed model of the ASTN1:CDH2 cis and trans asymmetric bridge complex in the neuron–glial migration junction. Blue ovals represent the cadherin domain; yellow hexagons represent the EGF-like domain; green hexagons represent the MACPF domain; magenta ovals represent the FNIII domain; and brown ovals represent the ANX-like domain.
Discussion
This study reports evidence for the formation of a cis and trans asymmetric bridge complex between two families of CNS guidance receptors, CDH2 and ASTN1, as well as the discovery that CDH2 is the glial ligand for ASTN1, the neuronal receptor for glial-guided migration. These findings are supported by biochemical evidence that ASTN1 binds CDH2 in cis, by S2 adhesion assays showing that ASTN1 binds CDH2 in trans, by genetic conditional loss-of-function studies showing defects in glial-guided migration in mice lacking Cdh2 in glia, and by in vitro chimeras showing a failure of GCPs to form a migration junction with glia lacking CDH2. Overexpression of an ASTN1 variant lacking the binding domain for CDH2 confirmed that homophilic CDH2 binding between GCPs and glia is not sufficient to support neuronal migration. Thus, heterophilic bridge complexes of ASTN1 and CDH2 are required for glial-guided neuronal migration in the developing cerebellum.
Support for cis interactions of ASTN1 and CDH2 was provided by immunoprecipitation assays, which further confirmed that three specific extracellular domains of ASTN1—MACPF, FNIII, and ANX-like—are required for ASTN1 binding to CDH2. This finding provided the molecular basis for studies on the overexpression of ASTN1 lacking these domains, which appeared to act as a dominant negative. The biochemical findings were also supported by S2 cell-adhesion assays showing that heterophilic ASTN1:CDH2 binding supports cell–cell adhesion in addition to homophilic CDH2-induced cell adhesion. Thus, CDH2 binds ASTN1 in both cis and trans and in intercellular adhesion. These findings are reminiscent of cis and trans binding between CDH2 and the AMPA receptor subunit GluR2 in dendritic spines (32).
Conditional loss-of-function experiments have been instrumental in defining the site of action of specific neuronal or glial receptors. The significance of glial CDH2, relative to neuronal CDH2, in glial-guided migration was evident from our finding that a glial loss of Cdh2 resulted in dramatic effects on GCP morphology and a failure to adhere to glial fibers, whereas a neuronal loss of Cdh2 did not affect neuron–glia binding or stall migration. We therefore propose that the binding of ASTN1 to glial CDH2 is sufficient to promote neuron–glia attachment and glial-guided migration. The finding that a loss of Cdh2 in GCPs slowed glial-guided migration suggests that the cis interaction of ASTN1 and CDH2 is also important for migration, likely by stabilizing the migration junction between the neuron and the glial fiber. One mechanism for stabilizing the migration junction would be through endosomal recycling of ASTN1 back to the plasma membrane, a role that has previously been ascribed for ASTN2, the second member of the astrotactin family (7). Indeed, CDH2 has been reported to function in AMPA receptor trafficking by increasing surface expression of AMPA receptors in neurons (33).
Other studies have proposed that homophilic CDH2:CDH2 binding regulates neuron–glia attachment and migration (19, 20). Our bridge model does not exclude this scenario; however, the discrepancy in migration defects caused by the cell-specific loss of Cdh2 in neurons or glia indicates that additional proteins as well as CDH2 are involved in the complex. Thus, we suggest that homophilic trans CDH2 interactions contribute to the formation of a migration junction but the CDH2:CDH2 complex is not sufficient for migration. The present study extends the general role for cadherins in homophilic cell–cell interactions by directly demonstrating that both cis and trans ASTN1:CDH2 interactions function in glial-guided migration.
Further support for the asymmetric bridge complex comes from the finding that a combined deletion of Cdh2 in both GCPs and BG produced migration defects similar to the glia-specific deletion. While the combined deletion also resulted in developmental defects in the cerebral cortex, notably a double cortex (SI Appendix, Fig. S2F), this was likely due to earlier expression of the hGFAP promoter at E13.5 (29) compared with the postnatal expression of the mGFAP promoter (28). Since the double-cortex phenotype indicates migration defects of cortical neurons, it is possible that the asymmetric ASTN1:CDH2 complex also regulates glial-guided migration in the cerebral cortex. However, detailed analyses will be required to address this issue. Importantly, all cerebellar migration assays in this study were comparable, as they assessed GCP migration at postnatal stages (P5–P8) where Cdh2 was deleted in all three lines (SI Appendix, Fig. S2A).
Heterophilic interactions between different cadherins have been reported (34, 35). However, no cerebellar defects have been described in mice with a targeted deletion of R-cadherin (36), M-cadherin (37), or cadherin-11 (38), which are expressed in the postnatal mouse cerebellum (39), suggesting that these cadherins do not function in GCP migration. Still, we cannot exclude the possibility that a partial compensation of other cadherins occurred in the neuron-specific Cdh2 cKO. Interestingly, cadherin-11 was recently shown to regulate neural crest migration via binding of its cleaved EC1–3 domains to ErbB2 (40). Moreover, CDH2 has been demonstrated to interact in trans with the AMPA receptor subunit GluR2 to regulate spine formation (32). This corroborates our findings that cell adhesion and migration can be regulated by cadherins independently of homophilic or compensatory cadherin bindings.
The heterophilic ASTN1:CDH2 interaction was shown to occur via the C-terminal ectodomain of ASTN1, which included the MACPF, FNIII, and ANX-like domains (Fig. 1C). We were unable to pinpoint whether a single domain binds CDH2, since ASTN1 constructs lacking only the MACPF or FNIII domains failed to localize to the cell surface. Expression of ASTN1-ΔCTD in control organotypic slices did not fully stall GCP migration but significantly slowed migration, similar to the neuronal loss of Cdh2. Slowed GCP migration is consistent with previous migration studies in Astn1 mutant mice (13). However, it is possible that endogenous ASTN1 might not have been fully competed out by ASTN1-ΔCTD and still might have contributed to GCP migration. In addition, we cannot exclude the possibility that other adhesion proteins are also involved in the ASTN1:CDH2 bridge complex. Nevertheless, the combined deletion of Cdh2 with ASTN1-ΔCTD expression in GCPs resulted in a migration failure similar to that seen with glial Cdh2 cKO, demonstrating that a cis interaction of ASTN1 and CDH2 in GCPs promotes migration. This is consistent with the observation that CDH2 acts in combination with nectin-based adhesion to regulate radial glia-independent somal translocation (41).
Although the intracellular aspect of ASTN1 does not contain any domains known to be involved in intracellular signaling pathways, it is possible that the complex of ASTN1:CDH2 functions in intracellular signaling during migration. The best-characterized signaling cascades involving CDH2 are β-catenin (42) and GTPases (22, 43). While recent studies support a key role for Rho, Rab, and Rap GTPases in the control of CDH2 function (18, 22, 43) and in GCP migration via actin-regulatory pathways (10), their signaling role in CDH2:ASTN1 complexes remains to be determined. In addition, since prior EM studies revealed the presence of microfilaments in the migration junction (6) where ASTN1 localizes (11), it will be important to assay the involvement of β-catenin and cytoskeletal elements, including actin-binding proteins, in the migration junction.
Our study provides a direct demonstration of cis and trans interactions of CDH2 with a CNS migration receptor and raises the possibility for a general function for heterophilic cadherin–receptor complexes in the formation of a wide range of cell–cell junctions.
Materials and Methods
Animals.
B6.129S6(SJL) Cdh2fl/fl mice (backcrossed to C57BL/6) carrying loxP sites flanking exon 1 of the Cdh2 gene (stock no. 007611; Jackson Laboratory) were crossed with Tg(NeuroD1-Cre) RZ24, Tg(hGFAP-Cre) PK90, or Tg(mGFAP-Cre) lines. Cdh2fl/+;NeuroD1-Cre, Cdh2fl/+;mGFAP-Cre, and Cdh2fl/+;hGFAP-Cre progeny were crossed with Cdh2fl/fl mice to generate Cdh2fl/fl;Cre+/− experimental mice and Cre-negative Cdh2fl/fl control littermates. Genotyping details are described in SI Appendix. All procedures were performed according to guidelines approved by the Rockefeller University Institutional Animal Care and Use Committee.
DNA Constructs.
See SI Appendix for details.
GC/BG Cocultures.
Cocultures of GCPs and BG from P7 cerebella were prepared as described previously (30). Briefly, a dissociated cerebellar cell suspension was applied to a two-step gradient of 35%/60% Percoll (Sigma-Aldrich) in Tyrode’s solution and was centrifuged at 2,000 × g for 10 min at 4 °C. The large cell fraction at the Tyrode’s solution/35% Percoll interface (BG) and the small cell fraction at the 35%/60% Percoll interface (GCPs) were subsequently washed and preplated in GC medium (SI Appendix) on untreated Petri dishes at 35 °C/5% CO2 for 20 min to remove fibroblasts. The BG fraction was thereafter cultured on 0.1 mg/mL poly-d-lysine–precoated 12-mm coverslips for 1 h at 35 °C/5% CO2. The unbound cell suspension was then removed, and the adhering glial cells were cultured in GC medium at 35 °C/5% CO2. The GCP fraction was transferred to a 60-mm tissue-culture dish and incubated for 30 min at 35 °C/5% CO2. The dish was then tapped to dislodge the GCPs. The cell suspension was transferred to a new tissue-culture dish, and the process was repeated for maximal purification of GCPs. Purified GCPs were then electroporated with an Amaxa Mouse Neuron Nucleofection kit (Lonza) as described in SI Appendix. GCPs were added to the BG cultures at a GCP:BG ratio of 5:1. The cocultures were incubated at 35 °C/5% CO2 for 48–72 h.
Organotypic Slice Cultures.
The method is described in previous work (10) and detailed in SI Appendix. Briefly, P8 cerebella from Cdh2fl/fl and Cdh2-cKO littermates were dissected out and electroporated with pCIG2-Venus (0.5 µg/µL), pCIG2-Astn1-FL-Venus (1 µg/µL), or pCIG2-Astn1-ΔCTD-Venus (1 µg/µL) plasmids dorsal-to-ventral for 50 ms at 80 V, for a total of five pulses with an interval of 500 ms between pulses, using an ECM 830 Electro Square Porator (BTX Genetronics). The cerebella were then embedded in 3% agarose in HBSS, and 250-µm horizontal slices were made using a Leica VT1000S vibratome. Slices were placed on Millicell-CM 0.4-µm culture plate inserts (Millipore) and cultured for 60 h at 35 °C/5% CO2.
Immunohistochemistry/Cytochemistry.
Brains were dissected out and fixed in 4% paraformaldehyde in PBS at 4 °C overnight and thereafter were cryoprotected in 20% sucrose in PBS at 4 °C overnight. Sagittal cryosections (25 µm), organotypic slice cultures, or GCP/BG cocultures were processed for immunohistochemistry as described in SI Appendix.
BrdU Labeling.
BrdU in PBS (BD Biosciences) (50 µg/g of body weight) was injected s.c. in the neck of P5 Cdh2fl/fl and Cdh2-cKO littermates. The brains were dissected out 48 h later and processed for immunohistochemistry as described in SI Appendix.
Immunoprecipitation and Western Blotting.
Briefly, transfected HEK 293T cells (clone 17; ATCC CRL-11268) or whole cerebella were extracted in ice-cold lysis buffer (SI Appendix) and precleared with 25 µL Protein G/A Agarose beads (Calbiochem). The lysates were incubated with 3 µg of a rabbit GFP antibody (Invitrogen), rabbit Astn1 antibody (12), or normal rabbit IgG (Santa Cruz Biotechnology) for 2 h at 4 °C. Immunoprecipitates were collected on 50 µL Protein G/A Agarose beads by overnight rotation at 4 °C, washed with lysis buffer, and resuspended in 50 µL 2× Laemmli buffer. Western blotting was performed as described in SI Appendix.
S2 Cell-Adhesion Assay.
Drosophila S2 cells (Life Technologies) were transfected for 24 h as described in SI Appendix, and 1.5 × 106 cells from each condition were mixed together at a density of 3 × 106 cells per well (1 × 106 cells/mL) and shaken gently at 28 °C for up to 2 h to allow aggregation. Cells were imaged immediately after the conditions were set up (t = 0) and after 30 min, 1 h, and 2 h. For full details, see SI Appendix.
Flow Cytometry.
Transfected HEK 293T cells were harvested in 1 mM EDTA in PBS. The surface fraction of Venus-linked ASTN1 variants was labeled with rabbit anti-GFP (1:5,000; Invitrogen) for 20 min at 4 °C followed by Alexa Fluor 647 donkey anti-rabbit (1:5,000; Life Technologies) for 25 min at 4 °C. Flow cytometry analysis was carried out on a BD Accuri C6 flow cytometer system (BD Biosciences) using 488-nm and 640-nm lasers as described in SI Appendix.
Statistical Analyses.
See SI Appendix for details. Differences between conditions were determined using unpaired t tests for equal or unequal variances, except for migration distance in the slice cultures where Kruskal–Wallis and Mann–Whitney U nonparametric tests were used. Significance was set at P < 0.05 (two-sided). In the bar diagrams, data are presented as means; error bars represent SDs in Figs. 2, 3, and 5H and SI Appendix, Fig. S4 and SEM in Fig. 6. The migration distance data from the slice cultures are presented in box plots (Figs. 4 and 5G).
Supplementary Material
Acknowledgments
We thank Dr. Eve-Ellen Govek for advice on carrying out organotypic slice culture assays and for comments on the manuscript; Andrew F. Iannone for technical assistance; Dr. Nathaniel Heintz and the Gene Expression Nervous System Atlas (GENSAT) Project for providing the NeuroD1-Cre and hGFAP-Cre lines; Dr. Alexandra Joyner for providing the mGFAP-Cre line and for helpful discussions; Dr. Richard Huganir for providing the pRK5-Cdh2 constructs; Dr. Leslie Vosshall for providing the pAc5-STABLE2 construct; Dr. Franck Polleux for the pCIG2 construct; Drs. David Solecki and Robert Gilbert for valuable comments on the manuscript; Drs. Svetlana Mazel and Stanka Semova for assistance with cell-surface protein measurements carried out in the Rockefeller University Flow Cytometry Resource Center; and Drs. Alison North, Christina Pyrgaki, and Tao Tong for assistance with bright-field and confocal microscopy, which was partly carried out in the Rockefeller University Bio-Imaging Resource Center. This study was supported by funding from the Swedish Research Council (Z.H.), the Nicholson Postdoctoral Exchange Program between the Rockefeller University and Karolinska Institutet (Z.H.), and the Hofmann Trust (M.E.H. and H.B.).
Footnotes
The authors declare no conflict of interest.
See Profile on page 10537.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811100115/-/DCSupplemental.
References
- 1.Solecki DJ. Sticky situations: Recent advances in control of cell adhesion during neuronal migration. Curr Opin Neurobiol. 2012;22:791–798. doi: 10.1016/j.conb.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evsyukova I, Plestant C, Anton ES. Integrative mechanisms of oriented neuronal migration in the developing brain. Annu Rev Cell Dev Biol. 2013;29:299–353. doi: 10.1146/annurev-cellbio-101512-122400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawauchi T. Cellullar insights into cerebral cortical development: Focusing on the locomotion mode of neuronal migration. Front Cell Neurosci. 2015;9:394. doi: 10.3389/fncel.2015.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rakic P. Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electronmicroscopic study in Macacus rhesus. J Comp Neurol. 1971;141:283–312. doi: 10.1002/cne.901410303. [DOI] [PubMed] [Google Scholar]
- 5.Edmondson JC, Hatten ME. Glial-guided granule neuron migration in vitro: A high-resolution time-lapse video microscopic study. J Neurosci. 1987;7:1928–1934. doi: 10.1523/JNEUROSCI.07-06-01928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory WA, Edmondson JC, Hatten ME, Mason CA. Cytology and neuron-glial apposition of migrating cerebellar granule cells in vitro. J Neurosci. 1988;8:1728–1738. doi: 10.1523/JNEUROSCI.08-05-01728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson PM, Fryer RH, Fang Y, Hatten ME. Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration. J Neurosci. 2010;30:8529–8540. doi: 10.1523/JNEUROSCI.0032-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- 9.Solecki DJ, et al. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Govek EE, et al. Cdc42 regulates neuronal polarity during cerebellar axon formation and glial-guided migration. iScience. 2018;1:35–48. doi: 10.1016/j.isci.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fishell G, Hatten ME. Astrotactin provides a receptor system for CNS neuronal migration. Development. 1991;113:755–765. doi: 10.1242/dev.113.3.755. [DOI] [PubMed] [Google Scholar]
- 12.Zheng C, Heintz N, Hatten ME. CNS gene encoding astrotactin, which supports neuronal migration along glial fibers. Science. 1996;272:417–419. doi: 10.1126/science.272.5260.417. [DOI] [PubMed] [Google Scholar]
- 13.Adams NC, Tomoda T, Cooper M, Dietz G, Hatten ME. Mice that lack astrotactin have slowed neuronal migration. Development. 2002;129:965–972. doi: 10.1242/dev.129.4.965. [DOI] [PubMed] [Google Scholar]
- 14.Radice GL, et al. Developmental defects in mouse embryos lacking N-cadherin. Dev Biol. 1997;181:64–78. doi: 10.1006/dbio.1996.8443. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka H, et al. Molecular modification of N-cadherin in response to synaptic activity. Neuron. 2000;25:93–107. doi: 10.1016/s0896-6273(00)80874-0. [DOI] [PubMed] [Google Scholar]
- 16.Reinés A, et al. N-cadherin prodomain processing regulates synaptogenesis. J Neurosci. 2012;32:6323–6334. doi: 10.1523/JNEUROSCI.0916-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadowaki M, et al. N-cadherin mediates cortical organization in the mouse brain. Dev Biol. 2007;304:22–33. doi: 10.1016/j.ydbio.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Kawauchi T, et al. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron. 2010;67:588–602. doi: 10.1016/j.neuron.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Shikanai M, Nakajima K, Kawauchi T. N-cadherin regulates radial glial fiber-dependent migration of cortical locomoting neurons. Commun Integr Biol. 2011;4:326–330. doi: 10.4161/cib.4.3.14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu C, et al. Radial glial cell-neuron interaction directs axon formation at the opposite side of the neuron from the contact site. J Neurosci. 2015;35:14517–14532. doi: 10.1523/JNEUROSCI.1266-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara Y, et al. ADP ribosylation factor 6 regulates neuronal migration in the developing cerebral cortex through FIP3/arfophilin-1-dependent endosomal trafficking of N-cadherin. eNeuro. 2016;3:ENEURO.0148-16.2016. doi: 10.1523/ENEURO.0148-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Müller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011;69:482–497. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao WQ, Liu XL, Hatten ME. The weaver gene encodes a nonautonomous signal for CNS neuronal differentiation. Cell. 1992;68:841–854. doi: 10.1016/0092-8674(92)90028-b. [DOI] [PubMed] [Google Scholar]
- 24.Felsenfeld DP, Hynes MA, Skoler KM, Furley AJ, Jessell TM. TAG-1 can mediate homophilic binding, but neurite outgrowth on TAG-1 requires an L1-like molecule and beta 1 integrins. Neuron. 1994;12:675–690. doi: 10.1016/0896-6273(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 25.González M, et al. Generation of stable Drosophila cell lines using multicistronic vectors. Sci Rep. 2011;1:75. doi: 10.1038/srep00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stitt TN, Hatten ME. Antibodies that recognize astrotactin block granule neuron binding to astroglia. Neuron. 1990;5:639–649. doi: 10.1016/0896-6273(90)90218-5. [DOI] [PubMed] [Google Scholar]
- 27.Lee JK, et al. Expression of neuroD/BETA2 in mitotic and postmitotic neuronal cells during the development of nervous system. Dev Dyn. 2000;217:361–367. doi: 10.1002/(SICI)1097-0177(200004)217:4<361::AID-DVDY3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Imura T, Sofroniew MV, Fushiki S. Loss of adenomatous polyposis coli in Bergmann glia disrupts their unique architecture and leads to cell nonautonomous neurodegeneration of cerebellar Purkinje neurons. Glia. 2011;59:857–868. doi: 10.1002/glia.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuang Y, et al. Dicer1 and MiR-9 are required for proper Notch1 signaling and the Bergmann glial phenotype in the developing mouse cerebellum. Glia. 2012;60:1734–1746. doi: 10.1002/glia.22392. [DOI] [PubMed] [Google Scholar]
- 30.Hatten ME. Neuronal regulation of astroglial morphology and proliferation in vitro. J Cell Biol. 1985;100:384–396. doi: 10.1083/jcb.100.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatten ME, Liem RK, Mason CA. Weaver mouse cerebellar granule neurons fail to migrate on wild-type astroglial processes in vitro. J Neurosci. 1986;6:2676–2683. doi: 10.1523/JNEUROSCI.06-09-02676.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saglietti L, et al. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron. 2007;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 33.Nuriya M, Huganir RL. Regulation of AMPA receptor trafficking by N-cadherin. J Neurochem. 2006;97:652–661. doi: 10.1111/j.1471-4159.2006.03740.x. [DOI] [PubMed] [Google Scholar]
- 34.Volk T, Cohen O, Geiger B. Formation of heterotypic adherens-type junctions between L-CAM-containing liver cells and A-CAM-containing lens cells. Cell. 1987;50:987–994. doi: 10.1016/0092-8674(87)90525-3. [DOI] [PubMed] [Google Scholar]
- 35.Prakasam AK, Maruthamuthu V, Leckband DE. Similarities between heterophilic and homophilic cadherin adhesion. Proc Natl Acad Sci USA. 2006;103:15434–15439. doi: 10.1073/pnas.0606701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dahl U, et al. Genetic dissection of cadherin function during nephrogenesis. Mol Cell Biol. 2002;22:1474–1487. doi: 10.1128/mcb.22.5.1474-1487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hollnagel A, Grund C, Franke WW, Arnold HH. The cell adhesion molecule M-cadherin is not essential for muscle development and regeneration. Mol Cell Biol. 2002;22:4760–4770. doi: 10.1128/MCB.22.13.4760-4770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manabe T, et al. Loss of cadherin-11 adhesion receptor enhances plastic changes in hippocampal synapses and modifies behavioral responses. Mol Cell Neurosci. 2000;15:534–546. doi: 10.1006/mcne.2000.0849. [DOI] [PubMed] [Google Scholar]
- 39.Gliem M, et al. Expression of classical cadherins in the cerebellar anlage: Quantitative and functional aspects. Mol Cell Neurosci. 2006;33:447–458. doi: 10.1016/j.mcn.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathavan K, Khedgikar V, Bartolo V, Alfandari D. The ectodomain of cadherin-11 binds to erbB2 and stimulates Akt phosphorylation to promote cranial neural crest cell migration. PLoS One. 2017;12:e0188963. doi: 10.1371/journal.pone.0188963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gil-Sanz C, et al. Cajal-Retzius cells instruct neuronal migration by coincidence signaling between secreted and contact-dependent guidance cues. Neuron. 2013;79:461–477. doi: 10.1016/j.neuron.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, et al. AKT activation by N-cadherin regulates beta-catenin signaling and neuronal differentiation during cortical development. Neural Dev. 2013;8:7. doi: 10.1186/1749-8104-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jossin Y, Cooper JA. Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat Neurosci. 2011;14:697–703. doi: 10.1038/nn.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.