Significance
Cyanobacteria were responsible for the origin of oxygenic photosynthesis, and have since come to colonize almost every environment on Earth. Here we show that their ecological range is not limited by the presence of sunlight, but also extends down to the deep terrestrial biosphere. We report the presence of microbial communities dominated by cyanobacteria in the continental subsurface using microscopy, metagenomics, and antibody microarrays. These cyanobacteria were related to surface rock-dwelling lineages known for their high tolerance to environmental and nutritional stress. We discuss how these adaptations allow cyanobacteria to thrive in the dark underground, a lifestyle that might trace back to their nonphotosynthetic ancestors.
Keywords: astrobiology, extreme environments, deep/dark biosphere, endolithic cyanobacteria, metagenomics
Abstract
Cyanobacteria are ecologically versatile microorganisms inhabiting most environments, ranging from marine systems to arid deserts. Although they possess several pathways for light-independent energy generation, until now their ecological range appeared to be restricted to environments with at least occasional exposure to sunlight. Here we present molecular, microscopic, and metagenomic evidence that cyanobacteria predominate in deep subsurface rock samples from the Iberian Pyrite Belt Mars analog (southwestern Spain). Metagenomics showed the potential for a hydrogen-based lithoautotrophic cyanobacterial metabolism. Collectively, our results suggest that they may play an important role as primary producers within the deep-Earth biosphere. Our description of this previously unknown ecological niche for cyanobacteria paves the way for models on their origin and evolution, as well as on their potential presence in current or primitive biospheres in other planetary bodies, and on the extant, primitive, and putative extraterrestrial biospheres.
The deep terrestrial biosphere is receiving increasing interest as it harbors a significant fraction of the total microbial biomass of the planet (1–3), yet also imposes severe energy and nutrient limitations on its inhabitants (4). Subsurface life is dependent on buried organic matter and lithogenically sourced compounds such as molecular hydrogen (5). Despite its inhospitality, the deep subsurface is a stable and sheltered environment, which makes it a good candidate habitat for the development of early life on Earth, as well as on potential extraterrestrial scenarios (6). Despite the increasing interest in deep subsurface ecosystems, obtaining reliable data on their microbiology is severely hampered by the high cost and technical difficulty of retrieving pristine samples, its heterogeneity and extent, and the vanishingly small microbial loads (7). The majority of deep subsurface habitats thus remain significantly underexplored (6).
The Iberian Pyrite Belt (IPB) in southwestern Spain hosts one of the largest sulphide deposits in the world, as well as the Río Tinto Mars analog (8–10). In previous work, we characterized pyrite-rich drill core samples down to 166-m depth with several molecular ecology techniques, and revealed a subsurface ecosystem with active iron and sulfur cycles (9). To investigate the microbiota and their activities in a deeper, more pristine location, we carried out a second drilling at a site ∼500 m from the previous one, where geophysical studies and tritium measurements showed the presence of an aquifer formed by ∼60-y-old groundwater at a depth of around 400 m (10). A 613-m-deep borehole was drilled, implementing procedures for aseptic sampling and for tracing potential contamination events during drilling and subsequent processing of the retrieved rock cores. These procedures minimized contamination of samples with extraneous microbes, enabled us to identify any samples that had been contaminated, and if so, to quantify any such contamination.
Results and Discussion
Biogeochemical Characterization of the Deep Subsurface of the Iberian Pyrite Belt.
Rock cores from a 613-m-deep borehole were retrieved and processed following protocols for aseptic sampling and the tracing of potential contaminations (SI Appendix, Fig. S1). The geochemistry of the borehole was characterized with several complementary techniques (SI Appendix, Fig. S2), and microbial profiles were obtained via MiSeq 16S rRNA gene amplicon sequencing (SI Appendix, Fig. S3) and an in situ antibody microarray immunoassay (11) (SI Appendix, Fig. S4).
Detection of Deep Subsurface Cyanobacteria.
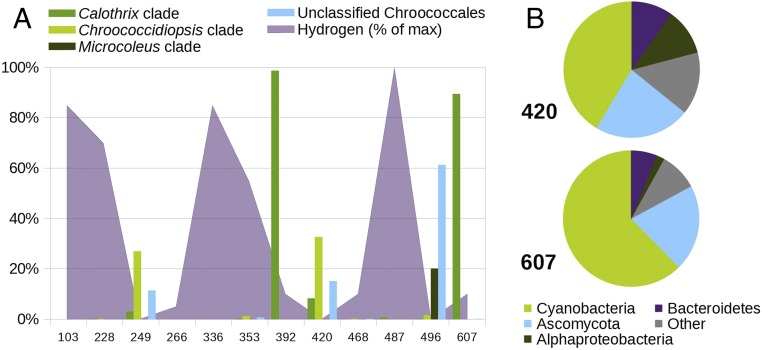
Strikingly, the on-site immunoassay detected, inter alia, cyanobacterial markers in samples from several depths, and 16S rRNA gene sequencing showed a predominance of cyanobacteria, whose exact sequence variants (ESVs) were related to endolithic and hypolithic representatives of the genera Calothrix, Chroococcidiopsis, and Microcoleus (SI Appendix, Fig. S4 and Dataset S1). The presence of cyanobacteria was associated with local decreases in hydrogen concentrations (Fig. 1A). The cyanobacterial ESVs present in the subsurface samples were absent from the drilling fluid and the internal laboratory controls (SI Appendix, Fig. S3), which confirms that their detection is not a consequence of contamination during sample retrieval and processing, and that they are indigenous to the retrieved cores.
Fig. 1.
Distribution of cyanobacteria and other microbial taxa in core samples from the Iberian Pyrite Belt deep subsurface. (A) Relative abundance of different cyanobacterial clades based on 16S rRNA amplicons (bars) and hydrogen concentrations (purple area) across the borehole samples. Hydrogen concentrations are shown relative to the sample with the highest hydrogen concentration. (B) Taxonomic composition of the metagenomic reads from samples 420 and 607.
We further focused on the samples from 420 and 607 m of depth (from now on referred to as samples 420 and 607, respectively), as they showed higher amounts of fatty acids (SI Appendix, Fig. S2). The rocks from 420 m below the surface floor (mbsf) are dominantly made up of quartz, with minor proportions of pyrite, carbonates (ankerite), and white mica, and show a conspicuous layering between some millimeters and 1 cm. Permeability is mainly controlled by widespread unoriented fracturing, with very variable openings between 0.01 and 0.1 mm. The rocks from 607 mbsf, on the other hand, consist of alternating dark shale (with abundant centimeter-sized nodules of pyrite) and sandstone. Fractures appear in the abundant contact zones between both minerals (SI Appendix, Fig. S2). Overall, the rocks at both depths have low porosity, but the presence of fractures provides space for microbial colonization and allows for a limited input of water and nutrients.
Cyanobacteria were the most abundant organisms in the metagenomes of both samples, followed by the Ascomycota, Alphaproteobacteria, and Bacteroidetes groups (Fig. 1B and Dataset S2). These organisms may form a microbial consortium similar to those found in cyanobacterial crusts (SI Appendix, Supplementary Text). In previous work, we described the presence of biofilms in the same borehole using fluorescence microscopy (12), but only universal probes were used. In this work, we confirmed the presence of viable cyanobacteria by catalyzed reported deposition fluorescent in situ hybridization (CARD-FISH) with specific probes against the cyanobacterial 16S ribosomal RNA. CARD-FISH is the best-practice method to search for viable cells—as defined by the presence of ribosomes—in deep subsurface settings (7, 13, 14). Ribosomal RNA has a half-life of days (15) and readily degrades upon cell starvation (16). Further, our samples contain pyrite, which is known to mediate the degradation of RNA via hydroxyl radicals under oxic and anoxic conditions (17). Sorption on certain mineral surfaces, such as clays, can increase the extracellular stability of ribosomal RNA, but complete degradation still occurs after a short time (18). Therefore, under the conditions of this study, positive CARD-FISH signals are a strong proof of extant viability.
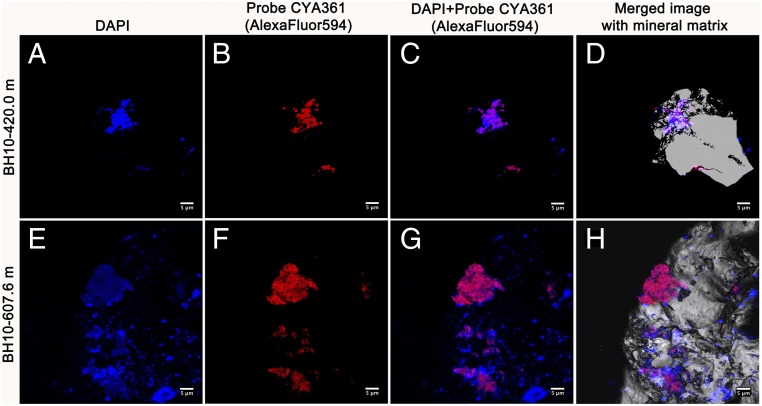
CARD-FISH with specific probes revealed clusters of cyanobacterial cells tightly attached to the mineral matrix and associated with other microorganisms (Fig. 2). These cells did not show photosystem II-related autofluorescence, indicating that they lacked active photosynthetic pigments. The inactivation of the photosynthetic apparatus when under environmental stress is a known trait of desert-dwelling cyanobacteria, such as Microcoleus sp (19), which helps them cope with both desiccation and photoinhibition.
Fig. 2.
Fluorescence micrographs (CARD-FISH) showing the presence of clusters of cyanobacterial cells attached to rock surfaces in deep subsurface samples. (A and E) Microbial DNA stained with DAPI (blue signal). (B and F) Hybridization signals with the specific cyanobacterial oligonucleotide probe CYA361 (red signal). (C and G) Merged image of DAPI and probe hybridization signals (blue and red, respectively). (D and H) Merged image of DAPI and probe hybridization together with the mineral matrix. The gray and white signal shows the host mineral. (Scale bars, 5 μm in all cases.)
Cyanobacteria have long been known to be ecologically versatile microorganisms (20) capable of light-independent energy generation (21), but until now, their ecological range appeared to be restricted to environments with at least occasional or prior exposure to sunlight (22). A few studies have reported the presence of cyanobacteria in deep subsurface environments (23–25), but to the best of our knowledge, only in ref. 25 have the authors attempted to discuss their origin. They proposed that a bloom of aquatic cyanobacteria had been trapped thousands of years ago into a groundwater aquifer with no further connection with the surface. That scenario strongly differs from the one described in this study: we analyze rock samples instead of groundwater, the IPB subsurface aquifer has recent connection to the surface (10), and the cyanobacterial lineages detected in this work are endolithic rather than aquatic. We thus believe that our results correspond to modern cyanobacteria with the ability to colonize deep subsurface environments.
Hydrogen as an Electron Donor for Cyanobacteria.
We found an apparent inverse correlation between the cyanobacteria predominance and hydrogen concentration in our samples (Fig. 1A). Hydrogen can be produced in the subsurface by several abiotic mechanisms, and its concentration in deep continental settings has recently been found to be controlled by biological sinks (26). To identify putative hydrogen consumers, we tested whether hydrogen concentration was dependent on taxa abundances using multiple linear regression. We considered the phylum, class, order, and family levels and tested models including all possible combinations of one to six taxa. Cyanobacteria was the only taxon that significantly explained hydrogen abundances when considered alone (negative correlation, P = 0.03, R2 = 0.33). The addition of more taxa to the model helped explain residual variance. The best model included the cyanobacterial families Rivulareaceae and Xenococcaceae, as well as the noncyanobacterial families Sphingomonadaceae and Bradyrhizobiaceae (negative correlation, P = 0.003, R2 = 0.80). These results, together with the presence of metabolic pathways for hydrogen utilization in the protein-coding metagenomic sequences affiliated with cyanobacteria (cyanobacterial pangenome), lead us to hypothesize that they obtain their energy by coupling the oxidation of hydrogen to the reduction of different electron acceptors (Fig. 3; see discussion below).
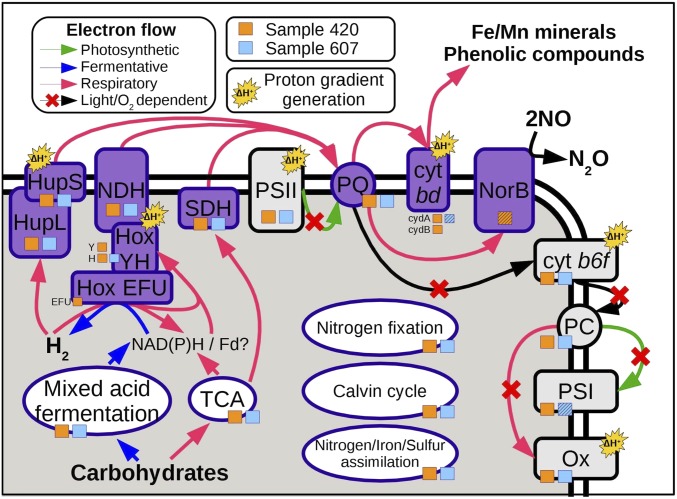
Fig. 3.
Schematic representation of the photosynthetic, respiratory, and fermentative pathways detected in the cyanobacterial pangenomes of two deep subsurface metagenomes. Orange and blue squares indicate whether an enzyme was detected in sample 420 or sample 607, respectively. Enzymes detected in the metagenomic reads but not in the assemblies have their square marked with a diagonal hatching. Reactions dependent on light or oxygen, and thus unlikely to be active in anoxic deep subsurface environments, are marked with a red cross. Abbreviations: cyt b6f, cytochrome b6f; cyt bd, cytochrome bd; Fd, ferredoxin; Hox, bidirectional hydrogenase; Hup, uptake hydrogenase; NDH, NDH-1 complex; NorB, quinol-dependent nitric oxide reductase; Ox, cytochrome c oxidase; PC, plastocyanin; PQ, plastoquinone; PSI, photosystem I; PSII, photosystem II; SDH, succinate dehydrogenase; TCA, tricarboxylic acid cycle.
Hydrogenases are widespread in cyanobacteria, which are believed to have originated from hydrogenotrophic ancestors (27). We detected both uptake and bidirectional hydrogenases in the two retrieved cyanobacterial pangenomes (Dataset S2). The uptake hydrogenase (Hup) transfers electrons from hydrogen to an unknown acceptor from the electron transport chain, most likely plastoquinone (28). Its main function is to minimize energy losses during nitrogen fixation and protect nitrogenase from oxygen toxicity by transferring electrons from hydrogen to oxygen via the electron transport chain (29). Cyanobacteria also have a bidirectional hydrogenase (Hox), which is hypothesized to function as an electron valve, providing a rapid way to balance the redox state of the cell. Hox can transfer electrons from/to either NAD(P) or plastoquinone via the NDH-I complex, contributing to both hydrogen uptake (providing reducing power for CO2 fixation) and hydrogen production (in dark-to-light transitions, and also coupled to fermentation) (21, 28, 30). Crucially, the cyanobacterial Hox has also been shown to be induced under dark anaerobic conditions (31), to participate in respiratory electron flow under prolonged darkness, and to be essential for growth when the photosynthetic and respiratory electron transport chains are overreduced (32).
Cyanobacterial Electron Transport Chains in Deep Subsurface Environments.
In several cyanobacterial genera, the overreduction of plastoquinone triggers the transfer of electrons to extracellular acceptors, via a cytochrome bd quinol oxidase (33). This has a protective effect in light-intense conditions, where cytochrome b6f is unable to accept electrons from plastoquinone at a sufficient rate. We note that growth under the dark, anoxic conditions of the deep subsurface would also lead to an overreduction of the plastoquinone pool, potentially triggering electron transfer from plastoquinone to cytochrome bd quinol oxidase. We thus propose that this protection mechanism would also provide the means for the anaerobic oxidation of hydrogen or other compounds using extracellular electron acceptors such as iron and manganese oxides, or phenolic compounds derived from the degradation of recalcitrant organic matter by other members of the microbial community (SI Appendix, Supplementary Text).
An additional potential electron acceptor could be nitric oxide, as we found a quinol-dependent nitric oxide reductase in the cyanobacterial pangenome from sample 420 (Dataset S2). Cyanobacterial nitric oxide reductases connected to the electron transport chain have been proposed to participate in nitric oxide detoxification and energy conversion (34). Interestingly, incomplete denitrification by noncyanobacterial partners is predominant in cyanobacteria-dominated biological crusts, leading to the emission of nitric and nitrous oxides (35, 36). Thus, cyanobacteria might profit from nitric oxide reductases by using them to exploit the nitric oxide produced by other members of the consortium as an alternative electron acceptor (34).
Evolution of Light-Independent Electron Transport Chains in Cyanobacteria.
Hoehler and Jørgensen (4), and more recently Starnawski et al. (37), have argued that the slow rates of biomass turnover in deep subsurface environments provide minimal opportunities for the introduction and propagation of beneficial mutations. Survivability will thus be determined by traits gained in other ecosystems showing similar (to some extent) restrictions, but with higher energy fluxes. In this context, endolithic cyanobacteria are perfect candidates for inhabiting the deep subsurface, as they are already adapted to living inside rocks and are able to withstand severe nutritional and environmental stresses and experience periodic anoxia during the diel cycle (38). Further, some cave-dwelling cyanobacteria survive for long periods in the near-total absence of light, where photosynthesis is no longer possible (39). Further, they possess several defense mechanisms that, having likely evolved to cope with light stress and desiccation in their original habitats (40), could also be triggered under the reducing conditions found in the deep subsurface and result in functional electron transport chains. This proposed mechanism relies on traits that are conserved across cyanobacterial lineages, and might thus reflect the lifestyle of the nonphotosynthetic ancestor of cyanobacteria (27). Under this second hypothesis, part of the energy transduction machinery of such an ancestor would have been coopted to serve as stress defense mechanisms in cyanobacteria, while still retaining its original capabilities in the absence of light.
Conclusions
We report the existence of cyanobacteria-dominated microbial communities in the deep continental subsurface, and discuss their potential metabolism based on geochemical and metagenomic data. Our proposal of cyanobacterial hydrogenotrophy is consistent with a large body of literature, as well as several parallel lines of evidence presented in this work. While the dark metabolism of cyanobacteria is still a matter of ongoing research, their unequivocal presence in samples from this and other studies calls for a reevaluation of their potential roles in deep subsurface ecosystems and increases their relevance in early life and astrobiological scenarios.
Materials and Methods
Drilling and Sampling.
Boreholes were continuously cored by rotary diamond-bit drilling using a Boart Longyear HQ wireline system producing 3 m of 60-mm-diameter cores. Well water was used as a drilling fluid to lubricate the bit and return cuttings to the surface. Fluids were recirculated. To detect potential contamination of the samples, sodium bromide (200 ppm) was added to the drilling fluid as a marker. Upon retrieval from the drilling rig, cores were divided into 60-cm-length pieces, inspected for signs of alteration, and stored in boxes for permanent storage and curation in the Instituto Geológico Minero de España lithoteque in Peñarroya. Selected cores were deposited in plastic bags, oxygen was displaced with N2, sealed, and transported to a field laboratory located at the Museo Minero de Riotinto village. Upon arrival at the field laboratory, cores were placed in an anaerobic chamber (5% H2, 95% N2), logged, and photographed. The anaerobic chamber and the airlock were decontaminated daily with Virkon S (Antec International Limited), a mixture of surfactants, organic acids, and strong oxidizers with the ability to disrupt bacterial membranes and degrade their nucleic acids. Furthermore, the chamber and the airlock were cleaned with ethanol and a 50:50 bleach:water solution before the introduction of a new core sample. Once in the anaerobic chamber, aseptic subsamples were obtained by splitting cores with a hydraulic core splitter and drilling out the central untouched portion with a rotary hammer with sterile bits. Bit temperature was strictly controlled (maximum 40 °C) with an infrared thermometer. Subsamples intended for CARD-FISH were instead obtained by chipping away bits of the central portion of the split core with a sterile chisel and grounding them with a sterile mortar and pestle.
Physicochemical Characterization of Rock Core Samples.
The concentrations of inorganic anions such as nitrite, nitrate, and sulfate, and small molecular weight organic acids such as acetate, formate, and oxalate, were estimated by ion chromatography as described elsewhere (11). pH was measured as described in ref. 41. The amounts of occluded hydrogen, methane, and CO2 in rock pores were measured as follows: 10 g of rock shards were placed into 100-mL vials, under sterile and anoxic conditions. The vials were in turn sealed with a gastight rubber septum and an aluminum cap, and their headspace was flushed with nitrogen gas. After a year of incubation at room temperature, it was assumed that the gases originally present in the rock pores had reached equilibrium with the headspace. The concentrations of hydrogen, methane, and CO2 were then measured in a Bruker 450GC gas chromatographer using a Hayesep 80/100 column (Valco Instruments). The presence of Fe3+, Fe2+, and NH4+ was assessed by using the Reflectoquant system (Merck Millipore), in accordance with the manufacturer’s instructions.
Total Sugars, Proteins, and Total Organic Carbon Determination.
Proteins, sugars, and total organic carbon were measured as described in ref. 11.
Lipid Extraction and Characterization.
Rock powders were extracted using a modified Bligh–Dyer method (42). Samples were placed in a 250-mL Teflon bottle, submerged in a monophasic solution of 4:10:5 water:methanol:dichloromethane, and disrupted with a sonicator wand (Branson Ultrasonics) for 1 h while maintained on ice. Following sonication, the bottles were shaken at 200 rpm for 1 h and centrifuged at 1,500 rpm for 15 min. The supernatant was removed and the extraction repeated twice. A total of 10 mL of dichloromethane and 10 mL of water were added to the pooled supernatant to induce phase separation, and the organic phase was collected. The aqueous phase was extracted with 10 mL of dichloromethane two additional times, and the pooled organic phases were dried under N2 and weighed. A total of 2.5 μg of pregnane diol was added to each sample as an internal standard. The extracts were acetylated by dissolving them in 50 μL of acetic anhydride and 50 μL of pyridine and heating them at 60 °C for an hour, after which they were dried and redissolved in 100 μL of dichloromethane. Samples were analyzed on a Trace 1310 GC coupled to an ISQ LT single quad mass spectrometer (Thermo Scientific). The programmable temperature vaporizing inlet was operated in constant temperature splitless mode at 300 °C. Separation was achieved on a Rxi-5HT fused silica column (Restek; 30 m, 0.25 mm inner diameter, 0.25 µm film) with a He flow rate of 1.5 mL/min using the following temperature program: 2 min hold at 40 °C; 25 °C/min to 120 °C; 6 °C/min to 320 °C; 30 min hold at 320 °C. The ISQ LT was operated in electron ionization mode with a 230 °C source temperature, scanning a mass range of 43–800 Da with a 0.2-s dwell time. Fatty acid methyl esters (formed from fatty acids during extraction in methanol-containing Bligh–Dyer solution) were quantified by comparing analyte peak areas to the area of the internal standard pregnane diacetate, assuming a 1:1 response factor. Finally, the calculated amount of fatty acid methyl esters in each sample was normalized to sample weight.
Sandwich Microarray Immunoassays with LDChip.
Sandwich-type microarray immunoassays (SMIs) were performed as described previously (11). Briefly, printed microscope slides with LDChip300 antibody microarray were blocked with 0.5 M Tris⋅HCl in 5% BSA for 5 min and then in 0.5 M Tris⋅HCl with 2% BSA for 30 min. After washing with TBSTRR buffer (0.4 M Tris⋅HCl pH 8, 0.3 M NaCl, 0.1% Tween 20) and drying the chip by quick centrifugation, the slides were mounted on a portable multiarray analysis module (MAAM) cassette for nine samples. Approximately 0.5 g of ground core samples were resuspended in 2 mL of TBSTRR and sonicated [3 × 1 min cycle with a handheld Ultrasonic Processor, UP50H (Hielscher Ultrasonics)]. Coarse material was removed by filtering through 10-μm nylon filter and 50 mL of the extracts were injected into each MAAM chamber and incubated for 1 h with the LDChip300 at ambient temperature. After a wash with TBSTRR, the chips were incubated with a fluorescently labeled antibody mixture for 1 h. The slides were then washed, dried, and scanned for fluorescence at 635 nm in a GenePix4100A scanner. Buffer was used as a blank control sample in parallel immunoassays. The scanned images were analyzed in the field with the GenePix Pro software (Genomic Solutions). The final fluorescence intensity was quantified as previously reported (11).
CARD-FISH.
For CARD-FISH analysis, rock powders and small chips were fixed in the field in 4% (vol/vol, final concentration) formaldehyde–1× PBS (137 mM NaCl; 2.7 mM KCl; 10 mM Na2HPO4; 1.8 mM KH2PO4) at 4 °C for 2 h. The samples were washed twice with 1× PBS and stored at −20 °C in 1:1 ethanol:1× PBS. Approximately 150 mg of small chips were subsequently subjected to analysis. Samples were embedded in 0.2% (wt/vol) agarose. Endogenous peroxidases were inactivated in 0.1% H2O2 in methanol for 30 min at room temperature. Hybridization was performed following the method described in ref. 43 with some modifications to facilitate the handling of small fragments of rock. Alexa Fluor 594-labeled tyramide was used as a fluorochrome. Samples were counterstained with 4′, 6′-diamidino-2-phenylindole (DAPI). The oligonucleotide probe used in this study for targeting rRNA genes was CYA361 (5′-CCCATTGCGGAAAATTCC-3′) (44) at 35% (vol/vol) formamide concentration. HRP-labeled probes were synthesized by Biomers.net GmbH. Negative controls were performed with the control probe NON338 (5′-ACTCCTACGGGAGGCAGC-3′) (45). Additional controls were carried out by subjecting the sample to the whole hybridization process without adding the probe, to evaluate whether the complex fluorophore-tyramide could interact with some mineral giving rise to a false positive. No signal was obtained from these controls. Further controls were conducted to evaluate whether the CARD-FISH signal could have been overlapping with autofluorescence from the cyanobacterial cells. CYA361 probe with both Alexa 488-labeled tyramide and Alexa 594-labeled tyramide were tested separately, and fluorescence was measured at 488 nm (green), 594 nm (red), and 633 nm (far red). In none of these controls could we achieve any autofluorescence signal from the cyanobacterial cells. Samples were mounted onto ibidi μ-slides 8 Well (ibidi GmbH) embedded in Citifluor:Vectashield (4:1) and examined with a Nikon AiR+ Resonant Scanning Confocal System (Nikon) at the Confocal Microscopy service at the Centro de Biología Molecular Severo Ochoa, CSIC-UAM.
DNA Extraction, Amplification, and Sequencing.
DNA extraction was performed in a UV- and ethanol-sterilized flow chamber according to ref. 46. Briefly, 0.5 g of powdered core sample were introduced into an Ultra-Clean Bead Tube (MoBio Laboratories), whose original buffer had been previously removed and substituted with 1 mL of phosphate buffer (1 M sodium phosphate, 15% ethanol). After adding 60 μL of MoBio Ultra-Clean Soil DNA solution S1, the tubes were subjected to two FastPrep (MP Biomedicals) cycles (30 s each, power setting of 5.5 m/s) separated by 1 min of ice cooling. Subsequently, the tubes were incubated in a thermomixer at 80 °C for 40 min, while shaking at 300 rpm. The MoBio Ultra-Clean Soil DNA extraction protocol was then followed from the addition of solution S2, according to the manufacturer’s instructions. All materials and stock solutions were UV sterilized for 5 min in either a GS Gene Linker UV Chamber (Bio-Rad Laboratories) or a Stratalinker 1800 UV crosslinker (Stratagene) to eliminate trace DNA contaminations. The isolated DNA was later subjected to multiple displacement amplification (MDA) using either the MagniPhi Phi29 polymerase (Genetrix, formerly X-Pol Biotech) or the REPLI-g Single Cell Kit (Qiagen). The nonenzymatic MDA reagents and the random hexamers were decontaminated following ref. 47. Briefly, they were aliquoted into 0.2-mL PCR tubes, which were laid down horizontally on the UV crosslinker chamber and subjected to a total UV dose of 0.04 mJ/cm2. The resulting MDA amplification products were finally purified using a MicroSpin G-50 column (GE Healthcare). Successful amplification was confirmed by PCR of the 16S rRNA gene using primers 16SF (5′-AGAGTTTGATCCTGGCTCAG-3′) and 16SR (5′-CACGAGCTGACGACAGCCG-3′). Negative controls were also run from MDA reagents without template DNA. Furthermore, in the cases where the MDA-amplified DNA gave no 16S PCR product, another nine individual rounds of DNA isolation were performed using the same extraction protocol (for a total of ∼5 g of powdered core sample). The 10 DNA isolations from the same rock core sample were immediately pooled, cleaned with phenol:chloroform:isoamyl alcohol (25:24:1), and precipitated with 70% ethanol (also UV sterilized) as described elsewhere. The DNA pellet was finally eluted in 50 μL of 10 mM Tris pH 8.0, amplified by MDA, and purified using the MicroSpin G-50 columns.
Additionally, an upscaled modification of the previous protocol was applied to a set of samples using the PowerMax Soil DNA Isolation Kit (MoBio Laboratories). A total of 15 mL of phosphate buffer was mixed with up to 10 g of powdered core sample into the provided PowerMax Bead Tube. After the addition of 5 mL of PowerMax Soil DNA solution C1, the tubes were vortexed vigorously for 30 s and subjected to two FastPrep cycles (40 s, power setting of 6.0 m/s). Samples were then incubated at 80 °C in a water bath during 40 min and centrifuged at 2,500 × g for 3 min at room temperature. The supernatant was recovered in a new collection tube and the protocol was followed from the addition of solution C2. Once again, the solutions employed were UV sterilized as described above, and the resulting DNA was also subjected to MDA amplification, purification, and PCR of the 16S rRNA gene.
The extraction of DNA from different depths with either one or several of the above-described methods is summarized in SI Appendix, Table S1. DNA was additionally isolated from a drilling water sample, to trace potential contamination events occurring during the core retrieval process. Two 250-mL drill cooling water samples, collected with a 15-d difference, were pooled and filtered through a 0.22-nm pore size filter (Millipore). The filter was introduced into an empty Ultra-Clean Bead Tube and the DNA was isolated using the first protocol described above. Additionally, during the preparation of the aliquots used for MiSeq sequencing (see below) DNA was also extracted from an empty PowerMax Bead Tube to account also for laboratory contaminations during extraction of the nucleic acids (DNA isolation control). DNA was finally eluted in 50 μL of 10 mM Tris⋅HCl buffer and quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific). The V5–V6 hypervariable regions of the bacterial 16S rRNA gene were PCR amplified using primers 807F and 1050R (48). The barcoding of the DNA amplicons, as well as the addition of Illumina adaptors, was carried out as described previously (49). The PCR-generated amplicon libraries were sent for 250-nt paired-end sequencing on a Illumina MiSeq platform (Illumina) at the Genome Analytics platform of the Helmholtz Centre for Infection Research. For the construction of metagenomic libraries, 0.8 μg of amplified DNA were mixed with 1× fragmentase reaction buffer in a final volume of 18 μL, vortexed thoroughly, and incubated on ice for 5 min. The fragmentation reaction was then started by mixing the samples with 2 μL of NEBNext dsDNA fragmentase (New England Biolabs, Inc.) and carried out for 25 min at 37 °C. After incubation, the fragmentation was halted by the addition of 5 μL 0.5 M EDTA. The ensuing DNA was purified with the QIAquick PCR Purification Kit (Qiagen) and eluted in a final volume of 35 μL before quantification with a Nanodrop (Thermo Scientific). Metagenomic libraries were prepared with the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs, Inc.) using ∼200 ng of fragmented DNA as initial input. Size selection of 400–500 bp DNA library fragments was carried out using the Agencourt AMPure XP magnetic beads (Beckman Coulter, Inc.) according to NEBNext Ultra DNA Library Prep Kit instructions. Each metagenomic DNA library was sequenced (100-nt paired-end sequencing) with the Illumina HiSEq. 2500 platform using the TruSeq SR Cluster Kit, v3-cBot-HS (Illumina).
16S Community Profiling.
Raw 16S MiSeq paired reads were assembled and quality filtered with moira (50) (v 1.3.2) with the -q posterior flag, and then preprocessed with mothur (51, 52) to remove chimeras and sequences with poor alignments to the SILVA reference database (53, 54). The preprocessed reads were analyzed with CFF (55) to achieve a subspecies resolution. The chimera-scanning step included in CFF’s standard pipeline was omitted, as chimeras had already been removed with mothur’s distribution of the VSEARCH software (56). The resulting subspecies ESVs were classified with SINA (57), and also by homology search against the nt and 16S ribosomal RNA NCBI databases and the SILVA nr128 database distributed with mothur.
Correlations between the abundances of different bacterial taxa and hydrogen concentrations were tested by multiple linear regression. For each taxonomic level from phylum to family, multiple linear models were fitted to explain hydrogen concentrations as a function of the abundances of all of the possible combinations of one to six taxa, considering only the taxa whose global abundance was over 5%. Statistical analyses were performed in R (v. 3.4.4).
Metagenomic Analysis.
Paired raw metagenomic reads were quality filtered with PRINSEQ (58) with the following parameters: -derep 14 -lc_method dust -lc_threshold 7 -ns_max_p 2 -trim_ns_left 2 -trim_ns_right 2 -trim_qual_left 20 -trim_qual_right 20 -min_qual_mean 25. Reads with more than one mismatch in the barcode sequence were removed. The filtered dataset was then subjected to digital normalization using khmer (59). Due to low coverage, a single-step normalization was performed as recommended in the author’s webpage (khmer.readthedocs.org/en/v1.1/guide.html). To achieve this, the normalize-by-median.py script included with khmer was run with the following parameters: -C 5 -k 20 -N 4 -x 2.5e8--paired. The filtered and normalized reads were annotated in the Metagenomic Rapid Annotations using Subsystems Technology (MG-RAST) server (60). Reads were classified based on the taxonomic classification reported by MG-RAST, and the four most abundant groups (Cyanobacteria, Ascomycota, Alphaproteobacteria, and Bacteroidetes) were analyzed independently to reconstruct their putative metabolisms, using the MG-RAST subsystem-based classification; if this led to missing enzymes for a pathway of interest, we manually searched for known synonyms in the annotation using the MG-RAST metagenome overview page.
Additionally, paired raw reads were analyzed with the SqueezeM pipeline (ref. 61; https://github.com/jtamames/squeezeM, commit 242644d1a1f6380b6df92b1c70924924cd1070c8) using the sequential mode. Briefly, the reads from each sample were assembled using metaSPAdes (62), and ORFs and rRNA coding sequences were predicted with Prodigal v2.6.2 (63) and barrnap v0.9-dev (https://github.com/tseemann/barrnap), respectively. ORFs were aligned against the Clusters of Orthologous Groups (64) and Kyoto Encyclopedia of Genes and Genomes (65) databases using diamond (66), and functionally annotated. Taxonomic annotation of ORFs was performed with a last common ancestor algorithm, based on the best hits against the NCBI nr database (downloaded on June 13, 2018). Binning was not attempted, as MDA introduces amplification biases which would affect contig coabundance patterns. Sequencing and assembly stats are summarized in SI Appendix, Table S2.
Supplementary Material
Acknowledgments
We thank Javier Tamames, Carlos Pedrós-Alió, Ramón Roselló-Mora, Barbara Sherwood-Lollar, and Chris McKay for useful feedback and discussions of the draft manuscript and all the members of the Iberian Pyrite Belt Subsurface Life (IPBSL) team and the personnel of the Museo Minero de Riotinto. This work was supported by the European Research Council Advanced Grant ERC250350-IPBSL, and the Spanish Ministerio de Economía y Competitividad/Fondo Europeo de Desarrollo Regional Grants AYA2011-24803 and ESP2015-69540-R. F.P.-S. is currently funded by the Spanish Ministerio de Economía y Competitividad/Fondo Europeo de Desarrollo Regional Grant CTM2016-80095-C2-1-R.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw sequence data and assemblies that support the findings of this study have been deposited in NCBI under the BioProject ID PRJNA476489. MG-RAST results are deposited under project ID mgp83581.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808176115/-/DCSupplemental.
References
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA. 2012;109:16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMahon S, Parnell J. Weighing the deep continental biosphere. FEMS Microbiol Ecol. 2014;87:113–120. doi: 10.1111/1574-6941.12196. [DOI] [PubMed] [Google Scholar]
- 4.Hoehler TM, Jørgensen BB. Microbial life under extreme energy limitation. Nat Rev Microbiol. 2013;11:83–94. doi: 10.1038/nrmicro2939. [DOI] [PubMed] [Google Scholar]
- 5.Colman DR, Poudel S, Stamps BW, Boyd ES, Spear JR. The deep, hot biosphere: Twenty-five years of retrospection. Proc Natl Acad Sci USA. 2017;114:6895–6903. doi: 10.1073/pnas.1701266114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards KJ, Becker K, Colwell F. The deep, dark energy biosphere: Intraterrestrial life on earth. Annu Rev Earth Planet Sci. 2012;40:551–568. [Google Scholar]
- 7.Wilkins MJ, et al. Trends and future challenges in sampling the deep terrestrial biosphere. Front Microbiol. 2014;5:481. doi: 10.3389/fmicb.2014.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tornos F. Environment of formation and styles of volcanogenic massive sulfides: The Iberian Pyrite Belt. Ore Geol Rev. 2006;28:259–307. [Google Scholar]
- 9.Puente-Sánchez F, et al. Deep subsurface sulfate reduction and methanogenesis in the Iberian Pyrite Belt revealed through geochemistry and molecular biomarkers. Geobiology. 2014;12:34–47. doi: 10.1111/gbi.12065. [DOI] [PubMed] [Google Scholar]
- 10.Gómez-Ortiz D, et al. Identification of the subsurface sulfide bodies responsible for acidity in Río Tinto source water, Spain. Earth Planet Sci Lett. 2014;391:36–41. [Google Scholar]
- 11.Parro V, et al. A microbial oasis in the hypersaline Atacama subsurface discovered by a life detector chip: Implications for the search for life on Mars. Astrobiology. 2011;11:969–996. doi: 10.1089/ast.2011.0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escudero C, Vera M, Oggerin M, Amils R. Active microbial biofilms in deep poor porous continental subsurface rocks. Sci Rep. 2018;8:1538. doi: 10.1038/s41598-018-19903-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schippers A, et al. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature. 2005;433:861–864. doi: 10.1038/nature03302. [DOI] [PubMed] [Google Scholar]
- 14.Lloyd KG, May MK, Kevorkian R, Steen AD. Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor. Appl Environ Microbiol. 2013;79:7790–7799. doi: 10.1128/AEM.02090-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karnahl U, Wasternack C. Half-life of cytoplasmic rRNA and tRNA, of plastid rRNA and of uridine nucleotides in heterotrophically and photoorganotrophically grown cells of Euglena gracilis and its apoplastic mutant W3BUL. Int J Biochem. 1992;24:493–497. doi: 10.1016/0020-711x(92)90044-2. [DOI] [PubMed] [Google Scholar]
- 16.Davis BD, Luger SM, Tai PC. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohn CA, et al. Pyrite-induced hydroxyl radical formation and its effect on nucleic acids. Geochem Trans. 2006;7:3. doi: 10.1186/1467-4866-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franchi M, Gallori E. A surface-mediated origin of the RNA world: Biogenic activities of clay-adsorbed RNA molecules. Gene. 2005;346:205–214. doi: 10.1016/j.gene.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Harel Y, Ohad I, Kaplan A. Activation of photosynthesis and resistance to photoinhibition in cyanobacteria within biological desert crust. Plant Physiol. 2004;136:3070–3079. doi: 10.1104/pp.104.047712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitton BA. Introduction to the cyanobacteria. In: Whitton BA, Potts M, editors. Ecology of Cyanobacteria II. Springer; Dordrecht, The Netherlands: 2012. pp. 1–11. [Google Scholar]
- 21.Stal LJ. Cyanobacterial mats and stromatolites. In: Whitton BA, Potts M, editors. Ecology of Cyanobacteria II. Springer; Dordrecht, The Netherlands: 2012. pp. 65–125. [Google Scholar]
- 22.Philippot L, et al. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol. 2010;8:523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- 23.Kormas KA, Smith DC, Edgcomb V, Teske A. Molecular analysis of deep subsurface microbial communities in Nankai Trough sediments (ODP Leg 190, Site 1176) FEMS Microbiol Ecol. 2003;45:115–125. doi: 10.1016/S0168-6496(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 24.Rastogi G, et al. Microbial and mineralogical characterizations of soils collected from the deep biosphere of the former Homestake gold mine, South Dakota. Microb Ecol. 2010;60:539–550. doi: 10.1007/s00248-010-9657-y. [DOI] [PubMed] [Google Scholar]
- 25.Hubalek V, et al. Connectivity to the surface determines diversity patterns in subsurface aquifers of the Fennoscandian shield. ISME J. 2016;10:2447–2458. doi: 10.1038/ismej.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagnoud A, et al. Reconstructing a hydrogen-driven microbial metabolic network in Opalinus Clay rock. Nat Commun. 2016;7:12770. doi: 10.1038/ncomms12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulkidjanian AY, et al. The cyanobacterial genome core and the origin of photosynthesis. Proc Natl Acad Sci USA. 2006;103:13126–13131. doi: 10.1073/pnas.0605709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamagnini P, et al. Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol Mol Biol Rev. 2002;66:1–20. doi: 10.1128/MMBR.66.1.1-20.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Sherman DM, Sherman LA. The uptake hydrogenase in the unicellular diazotrophic cyanobacterium Cyanothece sp. strain PCC 7822 protects nitrogenase from oxygen toxicity. J Bacteriol. 2014;196:840–849. doi: 10.1128/JB.01248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutta I, Vermaas WF. The electron transfer pathway upon H2 oxidation by the NiFe bidirectional hydrogenase of Synechocystis sp. PCC 6803 in the light shares components with the photosynthetic electron transfer chain in thylakoid membranes. Int J Hydrogen Energy. 2016;41:11949–11959. [Google Scholar]
- 31.Schwarz C, Poss Z, Hoffmann D, Appel J. Hydrogenases and hydrogen metabolism in photosynthetic prokaryotes. In: Hallenbeck PC, editor. Recent Advances in Phototrophic Prokaryotes. Springer; New York: 2010. pp. 305–348. [DOI] [PubMed] [Google Scholar]
- 32.De Rosa E, et al. [NiFe]-hydrogenase is essential for cyanobacterium Synechocystis sp. PCC 6803 aerobic growth in the dark. Sci Rep. 2015;5:12424. doi: 10.1038/srep12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pisciotta JM, Zou Y, Baskakov IV. Role of the photosynthetic electron transfer chain in electrogenic activity of cyanobacteria. Appl Microbiol Biotechnol. 2011;91:377–385. doi: 10.1007/s00253-011-3239-x. [DOI] [PubMed] [Google Scholar]
- 34.Büsch A, Friedrich B, Cramm R. Characterization of the norB gene, encoding nitric oxide reductase, in the nondenitrifying cyanobacterium Synechocystis sp. strain PCC6803. Appl Environ Microbiol. 2002;68:668–672. doi: 10.1128/AEM.68.2.668-672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abed RM, Lam P, de Beer D, Stief P. High rates of denitrification and nitrous oxide emission in arid biological soil crusts from the Sultanate of Oman. ISME J. 2013;7:1862–1875. doi: 10.1038/ismej.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber B, et al. Biological soil crusts accelerate the nitrogen cycle through large NO and HONO emissions in drylands. Proc Natl Acad Sci USA. 2015;112:15384–15389. doi: 10.1073/pnas.1515818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Starnawski P, et al. Microbial community assembly and evolution in subseafloor sediment. Proc Natl Acad Sci USA. 2017;114:2940–2945. doi: 10.1073/pnas.1614190114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajeev L, et al. Dynamic cyanobacterial response to hydration and dehydration in a desert biological soil crust. ISME J. 2013;7:2178–2191. doi: 10.1038/ismej.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giordano M, Mobili F, Pezzoni V, Hein MK, Davis JS. Photosynthesis in the caves of Frasassi (Italy) Phycologia. 2000;39:384–389. [Google Scholar]
- 40.Raanan H, et al. Towards clarifying what distinguishes cyanobacteria able to resurrect after desiccation from those that cannot: The photosynthetic aspect. Biochim Biophys Acta. 2016;1857:715–722. doi: 10.1016/j.bbabio.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 41.Fernández-Remolar DC, et al. Underground habitats in the Río Tinto basin: A model for subsurface life habitats on Mars. Astrobiology. 2008;8:1023–1047. doi: 10.1089/ast.2006.0104. [DOI] [PubMed] [Google Scholar]
- 42.Talbot HM, Rohmer M, Farrimond P. Rapid structural elucidation of composite bacterial hopanoids by atmospheric pressure chemical ionisation liquid chromatography/ion trap mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:880–892. doi: 10.1002/rcm.2911. [DOI] [PubMed] [Google Scholar]
- 43.Pernthaler A, Pernthaler J, Amann R. Sensitive multi-color fluorescence in situ hybridization for the identification of environmental microorganisms. In: Kowalchuk GA, de Bruijn FJ, Head IM, Akkermans AD, van Elsas JD, editors. Molecular Microbial Ecology Manual. Kluwer; Dordrecht, The Netherlands: 2004. pp. 711–726. [Google Scholar]
- 44.Schönhuber W, et al. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1999;65:1259–1267. doi: 10.1128/aem.65.3.1259-1267.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 46.Direito SO, Marees A, Röling WF. Sensitive life detection strategies for low-biomass environments: Optimizing extraction of nucleic acids adsorbing to terrestrial and Mars analogue minerals. FEMS Microbiol Ecol. 2012;81:111–123. doi: 10.1111/j.1574-6941.2012.01325.x. [DOI] [PubMed] [Google Scholar]
- 47.Woyke T, et al. Decontamination of MDA reagents for single cell whole genome amplification. PLoS One. 2011;6:e26161. doi: 10.1371/journal.pone.0026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bohorquez LC, et al. In-depth characterization via complementing culture-independent approaches of the microbial community in an acidic hot spring of the Colombian Andes. Microb Ecol. 2012;63:103–115, and erratum (2012) 63:238. doi: 10.1007/s00248-011-9943-3. [DOI] [PubMed] [Google Scholar]
- 49.Camarinha-Silva A, et al. Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ Microbiol. 2014;16:2939–2952. doi: 10.1111/1462-2920.12362. [DOI] [PubMed] [Google Scholar]
- 50.Puente-Sánchez F, Aguirre J, Parro V. A novel conceptual approach to read-filtering in high-throughput amplicon sequencing studies. Nucleic Acids Res. 2016;44:e40. doi: 10.1093/nar/gkv1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tikhonov M, Leach RW, Wingreen NS. Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J. 2015;9:68–80. doi: 10.1038/ismej.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pruesse E, Peplies J, Glöckner FO. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown CT, Howe A, Zhang Q, Pyrkosz AB, Brom TH. 2012. A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv:1203.4802. Preprint, posted May 21, 2012.
- 60.Meyer F, et al. The metagenomics RAST server–A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamames J, Puente-Sanchez F. 2018. SqueezeM, a fully automatic metagenomic analysis pipeline from reads to bins. bioRxiv:10.1101/347559. Preprint, posted June 14, 2018.
- 62.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hyatt D, et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tatusov RL, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101, discussion 101–103, 119–128, 244–252. [PubMed] [Google Scholar]
- 66.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.