Abstract
A mounting body of evidence has revealed that microRNAs (miRs) serve pivotal roles in various developmental processes, and in tumourigenesis, by binding to target genes and subsequently regulating gene expression. Continued activation of the Wnt/β-catenin signalling is positively associated with human malignancy. In addition, miR-194 dysregulation has been implicated in gastric cancer (GC); however, the molecular mechanisms underlying the effects of miR-194 on GC carcinogenesis remain to be elucidated. The present study demonstrated that miR-194 was upregulated in GC tissues and SUFU negative regulator of Ηedgehog signaling (SUFU) was downregulated in GC cell lines. Subsequently, inhibition of miR-194 attenuated nuclear accumulation of β-catenin, which consequently blocked Wnt/β-catenin signalling. In addition, the cytoplasmic translocation of β-catenin induced by miR-194 inhibition was mediated by SUFU. Furthermore, genes associated with the Wnt/β-catenin signalling pathway were revealed to be downregulated following inhibition of the Wnt signalling pathway by miR-194 suppression. Finally, the results indicated that cell apoptosis was markedly increased in response to miR-194 inhibition, strongly suggesting the carcinogenic effects of miR-194 in GC. Taken together, these findings demonstrated that miR-194 may promote gastric carcinogenesis through activation of the Wnt/β-catenin signalling pathway, making it a potential therapeutic target for GC.
Keywords: microRNA-194, SUFU negative regulator of Ηedgehog signaling, Wnt/β-catenin, apoptosis
Introduction
According to the World Health Organization, gastric cancer (GC) was the fifth most common malignancy and the third leading cause of cancer-associated mortality worldwide in 2012. In addition, it was estimated to cause 723,000 cases of mortality in 2012; notably, half of the reported cases occur in East Asia, particularly in China (1). The risk factors for GC include Helicobacter pylori infection, dietary factors, cigarette smoking, occupation, physical activity and obesity (2); however, the processes involved in the development of GC remain to be completely elucidated. It is therefore essential to explore mechanisms underlying gastric carcinogenesis, in order to identify molecular biomarkers for earlier detection and novel molecular targets for more effective therapy.
MicroRNAs (miRNAs/miRs) were initially discovered in Caenorhabditis elegans; lin-4 was revealed to inhibit lin-14 translation via the binding of lin-4 transcripts to the 3′ untranslated region (3′UTR) of lin-14 mRNA (3). Since then, thousands of miRNAs, which are non-coding RNA genes ~22 nt long, have been discovered in worms, flies, vertebrates and plants. By targeting the 3′UTR of target mRNAs, miRNAs either inhibit translation or induce mRNA degradation (4). Accumulating evidence has revealed that miRNAs are involved in the regulation of various cellular processes, including development, proliferation, differentiation, migration, apoptosis and cell cycle progression (5–7). In our previous study, it was reported that miR-194 is aberrantly overexpressed in GC tissues compared with in adjacent tissues. Furthermore, miR-194 has exhibited a strong ability to promote cell proliferation, migration and tumourigenicity (8).
The Wnt/β-catenin signalling pathway has been reported to serve a crucial role in embryonic development, tissue regeneration and numerous other biological processes. Mutations or dysregulated expression of components of the Wnt pathway are associated with various types of cancer (9). Numerous miRNAs have been demonstrated to directly regulate the Wnt signalling pathway in human malignancies, including pancreatic (10), colon (11) and breast cancer (12). Our previous study revealed that, in GC, by targeting a negative regulator of the Ηedgehog signalling pathway, SUFU negative regulator of Ηedgehog signaling (SUFU), overexpression of miR-194 resulted in significant nuclear accumulation of β-catenin, which subsequently activated the Wnt/β-catenin signalling pathway and promoted GC progression (8).
In this follow-up study, it was demonstrated that miR-194 was highly expressed in GC tissues, whereas SUFU was downregulated in GC tissues and cell lines, thus suggesting that miR-194 may be a carcinogenic miRNA, whereas SUFU may act as a tumour suppressor. The nuclear accumulation of β-catenin was significantly reduced following suppression of miR-194. Furthermore, the Wnt/β-catenin signalling transduction pathway was blocked, at least in part, when miR-194 was inhibited. Inhibition of miR-194 also increased cell apoptosis of GC cells. Taken together, these findings provided information on the mechanism underlying GC pathogenesis and may contribute to the development of novel targeted therapies.
Materials and methods
Tissue samples
The present study was approved by the Ethics Committee of The Shenzhen University School of Medicine (Shenzhen, China). All patients (n=62; age, 34–84 years; female to male ratio, 1:2.1, recruited between January 2016 and December 2017) provided written informed consent, according to the institutionally approved protocol. All GC tissues and adjacent normal tissues were obtained from Shenzhen Second People's Hospital (Shenzhen, China). The samples were pathologically and histologically confirmed to be GC tissues. Specimens were submerged in RNAlater solution and were stored in liquid nitrogen until use. Total RNA was subsequently isolated using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol.
Cell culture and cell transfection
Immortalised human normal gastric epithelial cells (HFE145) were obtained from Dr Hassan Ashktorab at Department of Medicine and Cancer Center, Howard University and Dr Duane Smoot at Meharry Medical Center (Nashville, TN, USA). BGC-823 and SGC7901 cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). AGS cells were obtained from the American Type Culture Collection (Manassas, VA, USA). MKN28 cells, which are known to be MKN74 derivatives (13), were obtained from China Infrastructure of Cell Line Resources (Beijing, China). All cells were cultured in Dulbecco's modified Eagle's medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% foetal bovine serum (HyClone; GE Healthcare Life Sciences), 100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator containing 5% CO2. The miR-194 hairpin inhibitor (cat. no. IH-300642-05-0002) and its corresponding non-specific control (cat. no. IN-001005-01-05) were purchased from GE Healthcare Dharmacon, Inc. (Lafayette, CO, USA). The non-specific control was designed based on cel-miR-67, mature sequence: 5′-UCACAACCUCCUAGAAAGAGUAGA-3′ (accession no. MIMAT0000039). Cel-miR-67 has been confirmed to have minimal sequence identity with miRNAs in human, mouse and rat (14). The miR-194 inhibitor and non-specific control were transfected using Lipofectamine® RNAimax (Invitrogen; Thermo Fisher Scientific, Inc.) at a final concentration of 60 nM, once BGC-823 cells had reached 30–50% confluence. Negative Control (NC) small interfering (si)RNA (cat. no. siN05815122147-1-5), SUFU siRNAs (5′-CGGCCTGAGTGATCTCTAT-3′, 5′-GATCCACACCTGCAAGAGA-3′ and 5′-GCAGCTTGAGAGCGTACAT-3′) and β-catenin siRNAs (5′-GCCACAAGATTACAAGAAA-3′, 5′-GACTACCAGTTGTGGTTAA-3′ and 5′-GATGGACAGTATGCAATGA-3′) were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The NC siRNA, or a mixture of the three SUFU or β-catenin siRNAs was transfected using Lipofectamine® RNAimax (Invitrogen; Thermo Fisher Scientific, Inc.) at a final concentration of 60 nM, once BGC-823 cells had reached 30–50% confluence. Cells were harvested 48 h post-transfection.
Cell immunofluorescence analysis
BGC-823 cells were transfected with a miR-194 inhibitor and cultured at 37°C for 48 h, or were incubated with 1 µM XAV-939 (cat. no. S1180; Selleck Chemicals, Houston, TX, USA) or dimethyl sulfoxide (cat. no. D2650; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 16 h at 37°C. The cells were then fixed in 4% formaldehyde (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) for 15 min and washed with PBS three times prior to permeabilisation with 0.2% Triton X-100 (Beijing Solarbio Science & Technology Co., Ltd.) (diluted with PBS) for 10 min at room temperature. Subsequently, the cells were stained with anti-β-catenin (L54E2) mouse monoclonal antibody (1:100; Cell Signaling Technology, Inc., Danvers, MA, USA) and anti-SUFU rabbit polyclonal antibody (1:100, cat. no. ab28083; Abcam, Cambridge, MA, USA) diluted in 1% bovine serum albumin (cat. no. SW3015; Beijing Solarbio Science & Technology Co., Ltd.) for 2 h at room temperature. After washing with PBS, the cells were incubated with anti-mouse immunoglobulin (Ig)G (H+L), F(ab')2 Fragment (Alexa Fluor® 594 Conjugate) (1:1,000, cat. no. 8890; Cell Signaling Technology, Inc.) and anti-rabbit IgG (H+L), F(ab')2 Fragment (Alexa Fluor® 488 Conjugate) (1:1,000, cat. no. 4412; Cell Signaling Technology, Inc.) for 1 h at room temperature in a dark box Nuclear staining was performed by mounting the cells in DAPI II (Abbott Laboratories, Abbott Park, IL, USA). The cells were imaged on a Leica SP5 AOBS confocal microscope (Leica Microsystems, Inc., Buffalo Grove, IL, USA) and were analysed using Leica Application Suite X software (Leica Microsystems, Inc.).
Western blotting
BGC-823 cells (30–50% confluent) were transfected with SUFU siRNA, β-catenin siRNA, non-specific inhibitor and NC siRNA, miR-194 inhibitor and NC siRNA, or miR-194 inhibitor and SUFU siRNA; after 48 h, the cells were harvested. For cytoplasmic and nuclear isolation, proteins were isolated using NE-PER™ nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Protein concentrations were estimated using a Bicinchoninic Acid Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). Cell lysates (30 µg) were electrophoresed on 10% polyacrylamide gels (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and were transferred to Immobilon-PSQ polyvinylidene difluoride membranes (EMD Millipore, Bedford, MA, USA). The membranes were blocked with Tris-buffered saline containing 5% skim milk and 0.1% Tween-20 and were then incubated with the primary antibodies overnight at 4°C. The primary antibodies used for western blotting were as follows: Anti-SUFU (1:1,000, cat. no. 2522; Cell Signaling Technology, Inc.), anti-β-catenin (1:1,000, cat. no. 8480; Cell Signaling Technology, Inc.), anti-histone deacetylase 1 antibodies (1:1,000, cat. no. A0238; ABclonal Biotech Co., Ltd., Wuhan, China) and anti-GAPDH monoclonal antibody (1:5,000, cat. no. 5174; Cell Signaling Technology, Inc.). Following secondary antibody incubation (cat. no. 7074, 1:3,000; Cell Signaling Technology, Inc.) and washing, the membranes were visualised using Pierce™ Enhanced Chemiluminescence Western Blotting Substrate (cat. no. 32106; Thermo Fisher Scientific, Inc.). Densitometric analyses of western blots from three independent subcellular fractionation experiments, with normalization to the control, were performed using ImageJ 1.50i software (National Institutes of Health, Bethesda, MD, USA). The relative expression levels of the control condition were designated as 1.
RNA extraction, TaqMan® Array 96-well Human WNT Pathway Plate assay and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Cells were transfected as aforementioned. Total cellular RNA was extracted from cultured cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, 1 ml TRIzol® reagent per 10 cm2 culture dish surface area was added directly to the cells in the culture dish, and the cells were lysed for 2 min on ice. The lysate was transferred to a tube and 0.2 ml chloroform per 1 ml TRIzol® reagent was added for homogenisation. After centrifugation at 12,000 × g for 15 min at 4°C, the aqueous phase was placed into a new tube. An equal volume of ethanol was added to the aqueous phase and mixed thoroughly. After incubation for 10 min at room temperature, the mixture was centrifuged at 12,000 × g for 10 min at 4°C. Finally, the RNA pellet was collected and washed twice with 75% ethanol, after which, the RNA was resuspended in RNase-free water. cDNA was synthesised using the Advantage RT-for-PCR kit (Takara Biotechnology Co., Ltd., Dalian, China) with random primers, according to the manufacturer's protocol. TaqMan® Array 96-well Human WNT Pathway Plates (Thermo Fisher Scientific, Inc.) were used to quantify the expression levels of Wnt signalling-associated genes, according to the manufacturer's protocols. 18S, GAPDH, hypoxanthine phosphoribosyltransferase 1 and glucuronidase β were used as internal controls. Primer sequences are listed in the instructions for the TaqMan® Array Human WNT Pathway kit. miR-194 TaqMan® RT-PCR amplicons (Thermo Fisher Scientific, Inc.) were used to confirm the expression of miR-194, according to the manufacturer's protocol. RNU6B TaqMan® RT-PCR amplicon (Thermo Fisher Scientific, Inc.) served as an internal control. The mRNA expression levels for other genes were determined by RT-qPCR performed on a CFX96 real-time PCR detection system (Bio-Rad Laboratories, Inc.) using the SYBR-Green Master Mix (Takara Biotechnology, Co., Ltd.). The thermocycling conditions were as follows: Initial denaturation at 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec, annealing at 60°C for 20 sec and extension at 72°C for 30 sec. The 2−ΔΔCq method was used for quantitative analysis (15). The specific primer sequences (Sangon Biotech Co., Ltd., Shanghai, China) used for PCR amplification were as follows: GAPDH, forward (F) 5′-AGCCTCAAGATCATCAGCAA-3′ and reverse (R) 5′-CCATCACGCCACAGTTTCC-3′; SUFU, F 5′-GACCCTGGTTACAAATTCTGTTGA and R 5′-AGGCAGGATGGAGACCTTCAG-3′; casein kinase 2 α2 (CSNK2A2), F 5′-AATGTTCGTGTAGCCTCAAGGTACTTC-3′ and R 5′-CAAGCTGGTCATAGTTGTCCTGTCC-3′; frizzled (FZD)-related protein (FRZB), F 5′-CGATCTGCTCTTCTTCCTCTGTGC-3′ and R 5′-CACGAGTGGCGGTACTTGATGAG-3′; FZD class receptor 7 (FZD7), F 5′-CGGCATCACCACTGGCTTCTG-3′ and R 5′-CCTTGCTGCTGTGGCTAAGTCTG-3′; F-box and WD repeat domain containing 11 (FBXW11), F 5′-TGAACGAATGGTACGCACTGATCC-3′ and R 5′-CGTCCACACCGCCAGTTAGATTC-3′; Wnt family member (WNT)7A, F 5′-CTGGAACTGCTCTGCACTGGGA-3′ and R 5′-GTACAGGCAGCTGTGATGGCGT-3′; and WNT7B, F 5′-CCCCCTCCCTGGATCATGCACA-3′ and R 5′-GCCACCACGGATGACAGTGCT-3′. GAPDH was used as the internal control.
Apoptosis analysis
Cells were transfected as aforementioned. After 48 h transfection, the cells were dissociated with Accutase® solution, centrifuged at ~200 × g for 5 min at room temperature and resuspended at 1×106 cells/ml in Annexin-binding buffer (cat. no. V13242; Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, 5 µl fluorescein isothiocyanate-conjugated Annexin V and 1 µl propidium iodide stock solution (diluted to a concentration of 100 µg/ml with Annexin-binding buffer) were added to a 100-µl cell suspension and incubated for 15 min at room temperature. Finally, 400 µl Annexin-binding buffer was added to each tube prior to flow cytometric analysis. Flow cytometry data were analysed by FlowJo 7.6.1 software (FlowJo, LLC, Ashland, OR, USA) and the experiments were repeated independently three times.
Statistical analysis
All statistical analyses were performed using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA). Data comparisons between two groups were made using Student's t-test. For comparisons in datasets containing multiple groups, one-way analysis of variances followed by Dunnett's test was used. Spearman's correlation analysis was used to determine the correlation between miR-194 and SUFU expression in paired GC samples. Data are presented as the means ± standard error of the mean derived from three independent experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-194 expression is elevated and SUFU expression is downregulated in GC
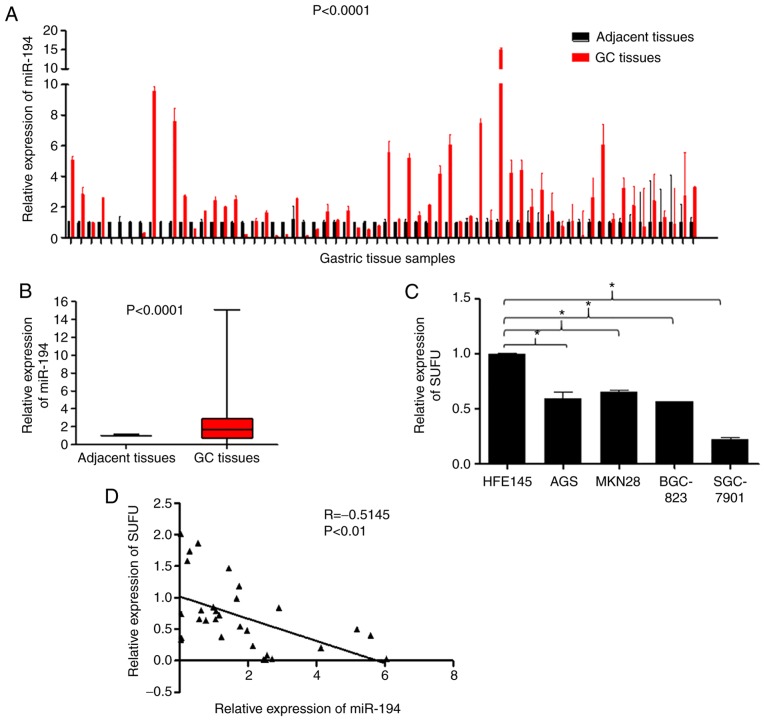
To explore the role of miR-194 in the carcinogenesis and development of GC, the present study detected the expression levels of miR-194 in 62 tumour tissues and paired adjacent non-cancerous tissues. RT-qPCR analysis indicated that miR-194 was markedly upregulated in the tumour tissues compared with in the adjacent non-cancerous tissues (Fig. 1A and B; P<0.0001). In addition, SUFU mRNA expression was detected in the HFE145 human normal gastric epithelial cell, and in the AGS, MKN28, BGC-823 and SGC-7901 human GC cell lines. As shown in Fig. 1C, SUFU expression was reduced in GC cell lines compared with in the HFE145 normal gastric epithelial cell line (P<0.05). Furthermore, Spearman's correlation analysis indicated a negative correlation between miR-194 and SUFU expression in 30 paired GC specimens (Fig. 1C; r=−0.5148, P<0.01), which was in line with our previous findings (8). These results suggested that miR-194 may be carcinogenic, whereas SUFU may act as a tumour suppressor. In addition, SUFU may be targeted by miR-194 in GC.
Figure 1.
miR-194 is upregulated, whereas SUFU is downregulated in GC. (A and B) Reverse transcription-quantitative polymerase chain reaction was performed using 62 GC tissues and corresponding adjacent normal tissues, in order to analyse the relative expression levels of miR-194. P<0.0001. (C) Relative expression levels of SUFU in the HFE145 human normal gastric epithelial cell line, and in the AGS, MKN28, BGC-823 and SGC-7901 human GC cell lines. One-way analysis of variance followed by Dunnett's test was used for statistical analysis. *P<0.05. (D) Spearman's correlation analysis between miR-194 and SUFU expression in 30 paired GC specimens. GC, gastric cancer; miR-194, microRNA-194; SUFU, SUFU negative regulator of Ηedgehog signaling.
Inhibition of miR-194 in BGC-823 cells induces cytoplasmic translocation of β-catenin and SUFU
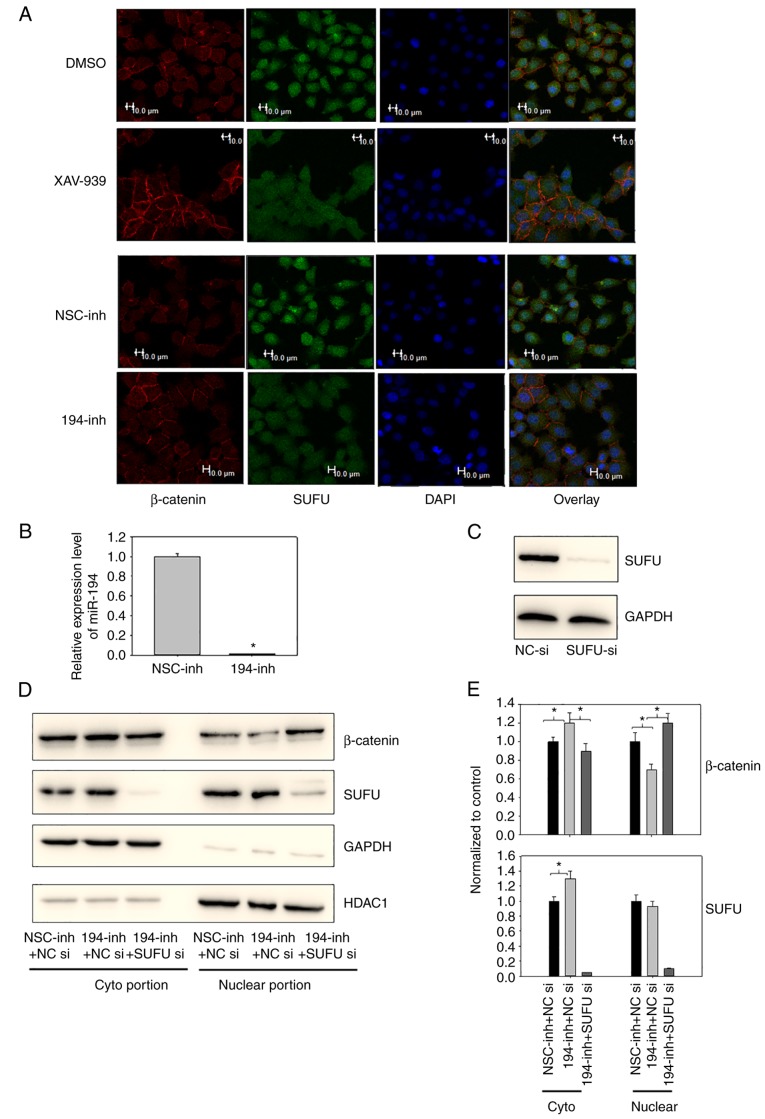
Extensive evidence has suggested that sustained activation of the Wnt/β-catenin signalling pathway is involved in the carcinogenesis of GC (16–18). It is well known that miR-194 is highly expressed and SUFU is expressed at low levels in BGC-823 cells (8). In addition, our previous study revealed that SUFU is the functional target of miR-194 and suppression of miR-194 in BGC-823 cells allows β-catenin to translocate to the cell membrane in GC (8). To further ascertain the effects of miR-194 on SUFU and β-catenin, immunofluorescence staining assays were conducted in BGC-823 cells (Fig. 2A). miR-194 was downregulated using a hairpin inhibitor (Fig. 2B). As shown in Fig. 2A, inhibition of miR-194 resulted in marked translocation of β-catenin to the cell membrane. In addition, the SUFU distribution pattern changed from predominantly nuclear to nuclear-cytoplasmic. This result was consistent with that observed in cells treated with XAV-939, a Wnt/β-catenin signalling inhibitor, thus suggesting that miR-194 may be involved in the localisation of not only β-catenin, but also SUFU.
Figure 2.
(A) Inhibition of miR-194 caused β-catenin to translocate to the cell membrane and caused SUFU to translocate to the cytoplasm. BGC-823 cells were transfected with a miR-194 inhibitor for 48 h or were incubated with 1 µM XAV-939 for 16 h. Subsequently, subcellular β-catenin and SUFU localisation was assessed by immunofluorescence staining. Scale bar, 10 µm. (B) Reverse transcription-quantitative polymerase chain reaction analysis of miR-194 was performed on BGC-823 cells 48 h following miR-194 inhibitor transfection to confirm successful transfection. Student's t-test was performed to analyse results. (C) SUFU expression was successfully knocked down using SUFU siRNA; BGC-823 cells were transfected with NC or SUFU siRNAs, and 48 h post-transfection, the cell lysates were immunoblotted with a SUFU antibody. (D) Suppression of SUFU alleviated the cytoplasmic translocation of β-catenin induced by miR-194 inhibition. BGC-823 cells were transfected with a miR-194 inhibitor or a miR-194 inhibitor and SUFU siRNA, and 48 h post-transfection, the cell lysates were fractionated into cytoplasmic and nuclear portions. Cell lysates were then immunoblotted with β-catenin and SUFU antibodies. (E) Densitometric analyses of western blots from three independent experiments, with normalization to the control, were performed with ImageJ software. The relative expression levels of the control condition were designated as 1. One-way analysis of variance followed by Dunnett's test was used for statistical analysis, *P<0.05. DMSO, dimethyl sulfoxide; GC, gastric cancer; HDAC1, histone deacetylase; miR-194, microRNA-194; NC, negative control; NSC, non-specific; si/siRNA, small interfering RNA; SUFU, SUFU negative regulator of Ηedgehog signaling.
To confirm the immunofluorescence results, and to determine if inhibition of miR-194-induced β-catenin translocation was mediated by SUFU, the subcellular localisation of β-catenin and SUFU were examined when miR-194 was inhibited alone or in combination with SUFU siRNAs. Western blotting was initially conducted to confirm that SUFU expression was successfully knocked down using SUFU siRNAs (Fig. 2C). In addition, SUFU expression was knocked down in cells transfected with SUFU siRNAs and a miR-194 inhibitor, indicating that SUFU siRNAs were effective under combined transfection conditions (Fig. 2D and E). In agreement with the immunofluorescence results, significantly more β-catenin was translocated to the cyptoplasmic region when cells were transfected with the miR-194 inhibitor alone. Notably, β-catenin expression was not significantly altered following miR-194 inhibitor transfection, as revealed in the western blot analysis presented in our previous study (8), and in the results of the TaqMan® Array 96-well Human WNT Pathway Plate assay (CTNNB1, Table I). These findings indicated that miR-194 may modulate the Wnt/β-catenin signalling principally by regulating β-catenin distribution, rather than its expression. In addition, as shown in Fig. 2D and E, suppression of SUFU alleviated the β-catenin cytoplasmic translocation induced by miR-194 inhibition. These findings suggested that miR-194 may modulate β-catenin subcellular localisation in a SUFU-dependent manner. Furthermore, consistent with the immunofluorescence analysis, more SUFU was translocated to the cytoplasmic region when miR-194 was inhibited. miR-194 directly targets SUFU at its 3′UTR; in our previous study, the expression levels of total SUFU were elevated when miR-194 was inhibited (8). Consistently, as shown in Fig. 2D and E, the increased expression of SUFU was mostly localized to the cytoplasmic region, whereas nuclear SUFU expression was comparable before and after miR-194 inhibitor transfection. These findings suggested that miR-194 may regulate the expression and localisation of SUFU.
Table I.
Relative expression levels of Wnt signaling pathway-associated genes in BGC-823 cells post-transfection with β-catenin siRNA or a miR-194 inhibitor.
| Gene | β-catenin siRNA | miR-194 inhibitor |
|---|---|---|
| CTNNB1 | 0.1010 | 0.8991 |
| TCF7 | 0.2024 | 0.2535 |
| KREMEN2 | 0.3353 | 0.6856 |
| FOXN1 | 0.3842 | 0.9433 |
| SFRP4 | 0.4322 | 0.2241 |
| PITX2 | 0.4323 | 0.4980 |
| SFRP1 | 0.4409 | 0.1021 |
| WNT4 | 0.4969 | 0.0816 |
| MYC | 0.5255 | 0.8198 |
| DKK1 | 0.5334 | 0.9630 |
| WNT11 | 0.5505 | 0.9622 |
| AXIN2 | 0.5596 | 0.7282 |
| FZD8 | 0.5795 | 0.7110 |
| NKD1 | 0.5951 | 1.0167 |
| LRP5 | 0.6482 | 0.7654 |
| SLC9A3R1 | 0.6521 | 0.7010 |
| WNT7B | 0.6646 | 0.2173 |
| CSNK1G1 | 0.6665 | 0.8205 |
| SENP2 | 0.6833 | 0.8449 |
| FRZB | 0.6926 | 0.3514 |
| PORCN | 0.6959 | 0.9331 |
| FZD1 | 0.7005 | 0.8605 |
| WNT5A | 0.7087 | 0.9652 |
| CSNK2A2 | 0.7173 | 0.4434 |
| WNT5B | 0.7258 | 0.9242 |
| FZD4 | 0.7523 | 0.7541 |
| PPP2R1A | 0.7633 | 0.7752 |
| LEF1 | 0.7701 | 0.8539 |
| CTNNBIP1 | 0.7743 | 1.8675 |
| TLE3 | 0.7831 | 0.8353 |
| PPP2CA | 0.8020 | 0.8453 |
| CSNK2B | 0.8089 | 0.8229 |
| CSNK1D | 0.8163 | 0.8483 |
| DKK3 | 0.8194 | 1.0270 |
| FZD7 | 0.8226 | 0.0020 |
| FZD10 | 0.8275 | 0.6828 |
| FZD6 | 0.8326 | 0.9536 |
| DVL1 | 0.8365 | 0.7208 |
| WNT6 | 0.8374 | 1.0771 |
| FZD9 | 0.8394 | 0.7183 |
| CSNK2A1 | 0.8427 | 0.7573 |
| EP300 | 0.8494 | 0.8390 |
| DVL3 | 0.8654 | 0.8581 |
| APC | 0.8745 | 1.0586 |
| DVL2 | 0.8824 | 0.8407 |
| AXIN1 | 0.8833 | 0.7570 |
| PYGO1 | 0.8903 | 0.9665 |
| KREMEN1 | 0.8926 | 0.5684 |
| WNT10B | 0.9044 | 0.7967 |
| WNT2B | 0.9231 | 1.2638 |
| GSK3A | 0.9456 | 1.0150 |
| TCF7L2 | 0.9504 | 1.0049 |
| CSNK1G3 | 0.9594 | 0.8100 |
| TLE2 | 0.9770 | 0.7009 |
| TLE4 | 0.9772 | 0.9552 |
| LRP6 | 0.9778 | 0.8770 |
| TCF7L1 | 1.0066 | 0.6388 |
| CBY1 | 1.0073 | 0.7859 |
| FRAT1 | 1.0132 | 0.6921 |
| CSNK1A1 | 1.0142 | 0.7752 |
| FZD2 | 1.0166 | 0.7472 |
| CSNK1G2 | 1.0258 | 0.8320 |
| RHOU | 1.0334 | 0.8648 |
| TLE1 | 1.0401 | 0.9018 |
| FBXW11 | 1.1036 | 0.4307 |
| WNT9A | 1.1052 | 0.6090 |
| WNT10A | 1.1198 | 3.0422 |
| WNT1 | 1.1331 | 1.4755 |
| GSK3B | 1.1437 | 0.8613 |
| FRAT2 | 1.1476 | 0.7411 |
| NLK | 1.1966 | 0.7700 |
| TLE6 | 1.1983 | 0.9940 |
| BTRC | 1.2124 | 0.8591 |
| PYGO2 | 1.2596 | 0.9025 |
| WNT8A | 1.2806 | 0.2067 |
| FZD3 | 1.5928 | 0.8283 |
| WNT7A | Not available | 0.5113 |
Relative expression levels compared with the control group are presented. miR-194, microRNA-194; siRNA, small interfering RNA.
miR-194 directly or indirectly regulates Wnt/β-catenin signalling and other associated genes involved in signal transduction
It is widely accepted that Wnt signalling is triggered when Wnt ligands bind to their membrane receptors, FZD and low-density-lipoprotein-related protein 5/6 (LRP5/6) co-receptor complex, thus resulting in the inhibition of β-catenin degradation (19,20). The accumulated β-catenin in the cytoplasm is translocated to the nucleus, where it interacts with the lymphoid enhancer factor/T cell factor (LEF/TCF) to transactivate downstream genes implicated in tumourigenesis.
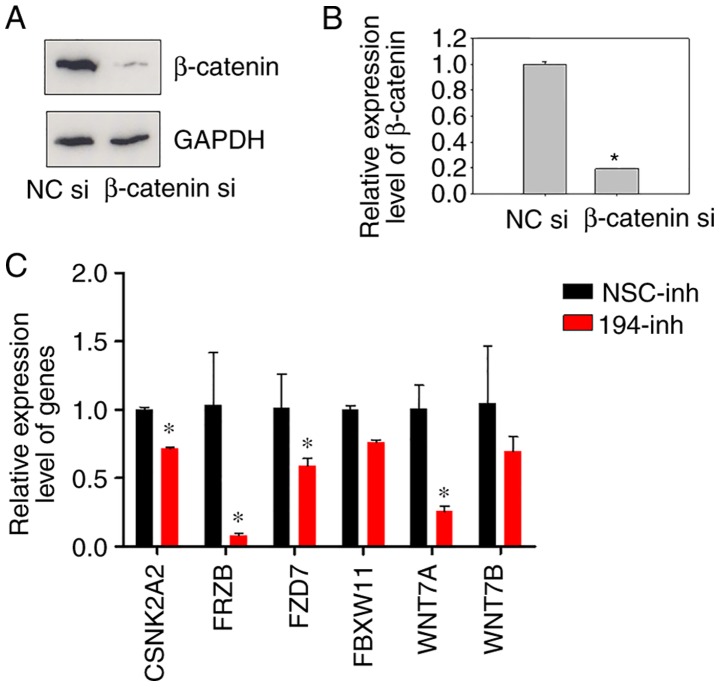
Our previous study revealed that miR-194 activates the Wnt/β-catenin signalling pathway by targeting SUFU (8); therefore, it was hypothesised that the expression of Wnt signalling-associated genes may be altered in response to inhibition of the Wnt signalling pathway by miR-194 suppression. Therefore, miR-194 was inhibited and β-catenin expression was knocked down (Fig. 3A and B), and their gene expression profiles were compared. A TaqMan® Array 96-well Human WNT Pathway Plate assay was used to quantify the expression levels of Wnt signalling-associated genes. The genes were listed from most downregulated to most upregulated in response to β-catenin siRNA. Notably, the expression levels of β-catenin were successfully suppressed in the β-catenin siRNA-transfected cells; however, its expression was relatively stable following miR-194 inhibition, which agreed with our previous findings (8). It may be hypothesized that miR-194 mainly regulates Wnt signalling through modulating β-catenin localisation, but not its expression.
Figure 3.
Inhibition of miR-194 results in downregulation of Wnt/β-catenin signalling-associated genes. (A) Transfection efficiency of β-catenin siRNA was evaluated prior to the TaqMan® Wnt plate assay. BGC-823 cells were transfected with NC siRNA or β-catenin siRNA, and 48 h post-transfection, the cell lysates were immunoblotted with a β-catenin antibody. (B) RT-qPCR of β-catenin was performed on BGC-823 cells 48 h post-transfection with β-catenin siRNA. Student's t-test was performed for statistical analysis. *P<0.05. (C) Relative expression levels of CSNK2A2, FRZB, FZD7, FBXW11, WNT7A and WNT7B. BGC-823 cells were transfected with NSC inhibitor or miR-194 inhibitor, and 48 h post-transfection, cells were harvested, and relative gene expression levels were determined using RT-qPCR. Data were normalised to GAPDH mRNA. Student's t-test was performed for statistical analysis. *P<0.05. CSNK2A2, casein kinase 2 α2; FBXW11, F-box and WD repeat domain containing 11; FRZB, frizzled-related protein; FZD7, frizzled class receptor 7; miR-194, microRNA-194; NC, negative control; NSC, non-specific; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA/si, small interfering RNA; WNT7, Wnt family member 7.
As shown in Table I, following inhibition of miR-194, various components of the Wnt/β-catenin signalling pathway, including members of the Wnt, FZD G-protein coupled receptor and Disheveled families, LRP5/6, glycogen synthase kinase 3β, Axin, casein kinase 1/2, β-transducin repeat containing E3 ubiquitin protein ligase, LEF and TCF, and the target genes MYC proto-oncogene, bHLH transcription factor and forkhead box N1 (21) were downregulated to different extents. Notably, among the seven genes in which the relative expression levels were <0.5 following transfection with β-catenin siRNAs, five genes, including TCF7, secreted FZD-related protein (SFRP)4, paired like homeodomain 2 (PITX2), SFRP1 and WNT4 were also downregulated by >2-fold in response to miR-194 inhibition. These findings suggested that similar to β-catenin suppression-mediated effects, inhibition of miR-194 suppressed the Wnt/β-catenin signalling pathway (Table I).
TCF7, which was the most downregulated gene in response to β-catenin siRNA transfection, was also downregulated to a similar degree by miR-194 inhibition. TCF7 (also known as TCF-1) serves a vital role in Wnt signalling; in the absence of β-catenin, TCF7 can interact with Groucho corepressors and act as a transcriptional repressor of Wnt target genes, whereas TCF7 can activate target gene expression when it binds to β-catenin (22,23). TCF7 is upregulated in breast cancer (24), osteosarcoma (25) and renal cell carcinoma (26). In the present study, inhibition of β-catenin and miR-194 led to the downregulation of TCF7, which further downregulated Wnt signalling.
Compared with the effects of β-catenin siRNA on β-catenin-interacting protein 1 (CTNNBIP1; also known as ICAT), which is a negative regulator of the Wnt signalling pathway that acts by blocking the interaction between β-catenin and TCF family members (27,28), CTNNBIP1 was upregulated in response to miR-194 inhibition. These findings suggested that miR-194 may regulate the Wnt pathway, similarly to β-catenin, but via different underlying mechanisms. Furthermore, the results revealed that inhibition of miR-194 not only downregulated molecules activating Wnt but also upregulated proteins blocking Wnt signalling activity. Overall, the Wnt signalling pathway may be inhibited by miR-194 suppression, potentially via SUFU. Wnt-associated genes SFRP (29,30), nemo-like kinase (31), pygopus family PHD finger 2 (32,33), FBXW11 (34), PITX2 (35,36), FRAT genes (37) and transducer-like enhancer of split genes (38); signal transducers Dickkopf Wnt signalling pathway inhibitors (39), chibby family member 1, β-catenin antagonist (40) and kringle containing transmembrane protein ½ (41); and embryogenesis-associated genes, such as SUMO-specific peptidase 2 (42), were also detected to better understand the association between miR-194 suppression and the Wnt/β-catenin pathway.
Since there is only one well for each gene in the TaqMan® Array 96-well Human WNT Pathway Plate, to verify the reliability of the plate assay, the mRNA expression levels of some genes were detected using RT-qPCR with specifically designed primers. Genes that were markedly downregulated in response to miR-194 inhibition were selected for analysis. As expected, the expression levels of CSNK2A2, FRZB, FZD7, FBXW11, WNT7A and WNT7B were markedly reduced in BGC-823 cells transfected with a miR-194 inhibitor compared with in those transfected with a non-specific control inhibitor (Fig. 3C). These results indicated that inhibition of the Wnt signalling pathway by miR-194 suppression may downregulate the expression of molecules involved in Wnt signalling, in agreement with our previous findings.
miR-194 inhibits cell apoptosis
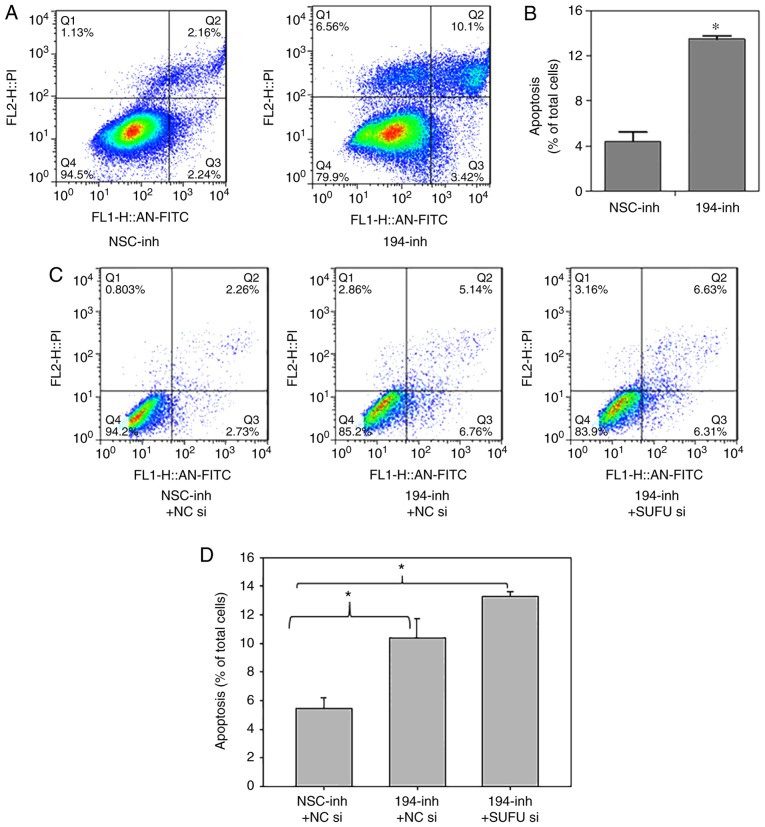
As Wnt signalling is involved in regulating cell apoptosis, to explore the effects of miR-194 on programmed cell death, an apoptosis assay was performed in BGC-823 cells transfected with miR-194 and non-specific control inhibitors. As shown in Fig. 4A and B, flow cytometric analysis revealed that transfection with a miR-194 inhibitor increased apoptosis threefold compared with in the control group, thus indicating that miR-194 may serve a crucial role in cell survival, and the pathological elevation of miR-194 may inhibit cell apoptosis. As shown in Fig. 4C and D, the effects of SUFU on apoptosis were determined. Notably, miR-194 inhibition combined with SUFU siRNA did not rescue the increased apoptosis; rather, it promoted apoptosis. These findings suggested that miR-194 may regulate apoptosis via another mechanism, which requires further investigation.
Figure 4.
miR-194 inhibition enhances cell apoptosis, but not via SUFU. (A and B) BGC-823 cells were transfected with NSC or miR-194 inhibitors. A total of 48 h post-transfection, cells were stained with Annexin V-FITC and apoptosis was analysed. Student's t-test was performed for statistical analysis. *P<0.05. (C and D) BGC-823 cells were transfected with miR-194 inhibitor plus NC siRNA or miR-194 inhibitor plus SUFU siRNA, and 48 h post-transfection, cells were stained with Annexin V-FITC and apoptosis was analysed. One-way analysis of variance followed by Dunnett's test was used for statistical analysis. *P<0.05. FITC, fluorescein isothiocyanate; miR-194, microRNA-194; NC, negative control; NSC, non-specific; si/siRNA, small interfering RNA; SUFU, SUFU negative regulator of Ηedgehog signaling.
Discussion
Our previous study reported that miR-194 activates the Wnt signalling pathway by targeting SUFU. In the present study, relatively high miR-194 expression and low SUFU expression was detected in paired GC tissues and GC cell lines; furthermore, miR-194 was revealed to exert regulatory effects on SUFU and β-catenin subcellular localisation, and inhibition of miR-194 resulted in downregulation of Wnt signalling pathway-associated genes and increased apoptosis.
SUFU has been reported to act as a negative regulator of the Ηedgehog signal transduction pathway, which mediates various fundamental processes, including cell growth, proliferation, survival during embryonic development, and adult tissue homoeostasis (43,44). SUFU is a major regulator of Gli/Ci transcription factors in vertebrates and invertebrates. By binding to the N- and C-terminal SUFU-interacting sites on the Gli/Ci family of transcription factors, SUFU tethers Gli/Ci in the cytoplasm and inhibits Gli/Ci activity in the nucleus, thus leading to inhibition of the transcription of downstream target genes (45–47). Compared with wild type SUFU, mutant SUFU is unable to decrease nuclear levels of β-catenin and cannot inhibit β-catenin/TCF-mediated transcription (48). In humans, children carrying germline and somatic mutations in the SUFU gene are predisposed to Gorlin's syndrome tumours and medulloblastomas (49,50). Downregulation of SUFU has also been reported in glioma (51), lung cancer (52) and other cancers. In GC, low SUFU expression has been detected in GC samples (53); however, the molecular mechanisms underlying SUFU dysregulation in gastric carcinogenesis are rarely reported. In our previous study, it was demonstrated that SUFU is a target of miR-194, and inhibition of miR-194 causes SUFU upregulation leading to β-catenin cytoplasmic translocation (8). The present study further investigated the regulation of β-catenin and SUFU by miR-194. The results indicated that more β-catenin was translocated to the cyptoplasmic region when miR-194 was inhibited alone. Furthermore, suppression of SUFU alleviated the β-catenin cytoplasmic translocation induced by miR-194 inhibition, thus suggesting that SUFU may mediate miR-194 inhibition-induced β-catenin cytoplasmic localisation. In addition, miR-194 inhibition resulted in elevated expression of SUFU and translocation of more SUFU to the cytoplasm. These findings suggested that miR-194 not only regulated subcellular localisation of β-catenin but also SUFU; however, the precise mechanisms underlying the distribution of SUFU regulated by miR-194 in GC require further investigation.
miR-194 expression is lost or downregulated in glioblastoma, hepatocellular carcinoma, and cervical, ovarian, colon and lung cancer. Conversely, miR-194 acts as a cancer promoter in endometrial cancer (54) and oesophageal cancer (55). The role of miR-194 in GC is controversial. Some independent groups have reported that miR-194 inhibits the proliferation, invasion and migration of GC cells (56–59), whereas others, including our group, revealed that miR-194 is overexpressed in GC (60–62). The present study also indicated that miR-194 may regulate the distribution of β-catenin to activate Wnt signalling by targeting SUFU.
In the present study, the expression levels of genes associated with Wnt signalling and modifiers of Wnt signalling were detected using a TaqMan® 96-well plate assay and RT-qPCR. Following miR-194 inhibition, the Wnt pathway was suppressed. Members of the Wnt family, particularly WNT4/7A/7B/8A, were markedly downregulated. Furthermore, the expression of an important negative regulator of β-catenin, CTNNBIP1, exhibited a twofold increase in miR-194 inhibitor-transfected cells. CTNNBIP1 contains two inhibitory regions, the N-terminal helical domain, which blocks CBP/p300 binding, and the extended C-terminal region, which inhibits the interaction of TCF/Lef with β-catenin, leading to the inhibition of Wnt signalling (28,63). Another downregulated gene upon miR-194 inhibition, PITX2, interacts with LEF-1 and β-catenin to increase LEF-1 expression, resulting in the activation of downstream target genes (35,36). Notably, the expression of some Wnt negative regulators also decreased under miR-194 inhibition. For example, FBXW11, which is a Wnt antagonist that encodes a protein responsible for the ubiquitylation and subsequent degradation of β-catenin (34). In the present study, FBXW11 expression was reduced following miR-194 inhibitor transfection. Identified as negative regulators of Wnt signalling, secreted FZD-related proteins (SFRPs), including SFRP1 and SFRP3 (FRZB), bind to WNT8 directly and block its activity (29). SFRP1 expression was also decreased in response to miR-194 inhibition. Therefore, it may be hypothesized that the overall Wnt signalling activity was inhibited by miR-194 suppression; however, negative feedback may balance this inhibition to a certain extent. These data demonstrated that following inhibition of the Wnt signalling pathway by miR-194 suppression through SUFU, genes relevant to Wnt/β-catenin signalling were inhibited, indicating a potential therapeutic effect of miR-194 in GC.
Since miR-194 has exhibited the ability to activate Wnt/β-catenin signalling, the present study performed further experiments to investigate cell apoptosis following miR-194 inhibition. The results indicated that suppression of miR-194 significantly induced apoptosis, strongly implying the carcinogenic effect of miR-194 in BGC-823 cells. Therefore, inhibition of miR-194 may reduce miR-194 to its normal levels in patients with GC and have a potential anticancer therapeutic effect. However, miR-194 inhibition combined with SUFU siRNA did not rescue the increased apoptosis; rather, it promoted apoptosis. This finding suggested that miR-194 may regulate apoptosis via other target genes, which require further investigation.
In conclusion, the present study revealed that miR-194 may contribute to carcinogenesis in GC by targeting SUFU. Notably, miR-194 inhibition resulted in significant translocation of β-catenin and SUFU from the nucleus to the nucleus-cytoplasm. Furthermore, this study revealed that miR-194 may regulate, either directly or indirectly, components of the Wnt/β-catenin signalling pathway and various related genes involved in signal transduction. In addition, miR-194 may inhibit cell apoptosis, suggesting it may be a therapeutic target for GC.
Acknowledgements
The authors would like to thank to Dr Hassan Ashktorab (Department of Medicine and Cancer Center, Howard University) and Dr Duane Smoot (Department of Medicine, Meharry Medical Center) for providing the HFE145 cells.
Funding
The present study was supported by the National Nature Science Foundation of China (grant no. 81772592) to ZJ; the National Natural Youth Science Foundation of China (grant no. 31601028) to YP; the Science Technology and Innovation Committee of Shenzhen (grant no. JCYJ20170818142852491) to ZJ; the Nature Science Foundation of Guangdong Province (grant no. 2017A030313144) and Startup Fund of Shenzhen University (grant no. 2018015) to YP; the Medical Science and Technology Research Foundation of Guangdong Province (grant no. A2016112), Nature Science Foundation of Guangdong Province (grant no. 2017A030313479) and The Planned Science and Technology Project of Shenzhen (grant no. JCYJ20160422170722474) to XZ; the Guangdong Provincial Department of Science and Technology (grant no. 2016B090918122), the Science Technology and Innovation Committee of Shenzhen (grant nos. JCYJ20160331190123578 and GJHZ20170314154722613) to YW; and NIH grants DK087454, CA146799, CA173390 and an American Cancer Society Clinical Research Professorship to SJM. Dr Meltzer is the Harry B. Myerberg-Thomas R. Hendrix Professor of Gastroenterology.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YP, XZ, ZZ, ZJ, SJM and SL designed the study. YP, XZ, HL, SD, YH, XFe, RY, YZ, YC, JW and WC performed the experiments. YQ communicated with patients, asked for written informed consent and collected GC samples. YW conducted the statistical analysis. HA and DS provided the HFE145 cell line and contributed to conception of the study. YP, HL, XFa and ZZ wrote the manuscript. XFa was also involved in the conception of the study. YW, SJM and SL reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Shenzhen University School of Medicine. All patients provided written informed consent according to the institutionally-approved protocol.
Patient consent for publication
Written informed consent was obtained from all patients prior to the study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Forman D, Burley VJ. Gastric cancer: Global pattern of the disease and an overview of environmental risk factors. Best Pract Res Clin Gastroenterol. 2006;20:633–649. doi: 10.1016/j.bpg.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambrost V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 4.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 5.Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 6.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng Y, Zhang X, Ma Q, Yan R, Qin Y, Zhao Y, Cheng Y, Yang M, Wang Q, Feng X, et al. MiRNA-194 activates the Wnt/β-catenin signaling pathway in gastric cancer by targeting the negative Wnt regulator, SUFU. Cancer Lett. 2017;385:117–127. doi: 10.1016/j.canlet.2016.10.035. [DOI] [PubMed] [Google Scholar]
- 9.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 10.Zhou W, Li Y, Gou S, Xiong J, Wu H, Wang C, Yan H, Liu T. MiR-744 increases tumorigenicity of pancreatic cancer by activating Wnt/β-catenin pathway. Oncotarget. 2015;6:37557–37569. doi: 10.18632/oncotarget.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T, Lai Q, Wang S, Cai J, Xiao Z, Deng D, He L, Jiao H, Ye Y, Liang L, et al. MicroRNA-224 sustains Wnt/β-catenin signaling and promotes aggressive phenotype of colorectal cancer. J Exp Clin Cancer Res. 2016;35:21. doi: 10.1186/s13046-016-0287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taipaleenmäki H, Farina NH, van Wijnen AJ, Stein JL, Hesse E, Stein GS, Lian JB. Antagonizing miR-218-5p attenuates Wnt signaling and reduces metastatic bone disease of triple negative breast cancer cells. Oncotarget. 2016;7:79032–79046. doi: 10.18632/oncotarget.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA, Reddel RR, et al. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- 14.Jin HY, Gonzalez-Martin A, Miletic AV, Lai M, Knight S, Sabouri-Ghomi M, Head SR, Macauley MS, Rickert RC, Xiao C. Transfection of microRNA mimics should be used with caution. Front Genet. 2015;6:340. doi: 10.3389/fgene.2015.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Song X, Xin N, Wang W, Zhao C. Wnt/β-catenin, an oncogenic pathway targeted by H. pylori in gastric carcinogenesis. Oncotarget. 2015;6:35579–35588. doi: 10.18632/oncotarget.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy TA, Wroblewski LE, Wang D, Piazuelo MB, Delgado A, Romero-Gallo J, Noto J, Israel DA, Ogden SR, Correa P, et al. β-Catenin and p120 mediate PPARδ-dependent proliferation induced by Helicobacter pylori in human and rodent epithelia. Gastroenterology. 2011;141:553–564. doi: 10.1053/j.gastro.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franco AT, Johnston E, Krishna U, Yamaoka Y, Israel DA, Nagy TA, Wroblewski LE, Piazuelo MB, Correa P, Peek RM., Jr Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68:379–387. doi: 10.1158/0008-5472.CAN-07-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, Mohammed S, Heck AJ, Maurice MM, Mahmoudi T, et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell. 2012;149:1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 20.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: Arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 21.Balciunaite G, Keller MP, Balciunaite E, Piali L, Zuklys S, Mathieu YD, Gill J, Boyd R, Sussman DJ, Holländer GA. Wnt glycoproteins regulate the expression of FoxN1, the gene defective in nude mice. Nat Immunol. 2002;3:1102–1108. doi: 10.1038/ni850. [DOI] [PubMed] [Google Scholar]
- 22.Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 23.Eastman Q, Grosschedl R. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr Opin Cell Biol. 1999;11:233–240. doi: 10.1016/S0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 24.Wang SH, Li N, Wei Y, Li QR, Yu ZP. β-catenin deacetylation is essential for WNT-induced proliferation of breast cancer cells. Mol Med Rep. 2014;9:973–978. doi: 10.3892/mmr.2014.1889. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Zhang S, Xu Y, Zhang Y, Guan H, Li X, Li Y, Wang Y. Upregulation of miR-192 inhibits cell growth and invasion and induces cell apoptosis by targeting TCF7 in human osteosarcoma. Tumour Biol. 2016;37:15211–15220. doi: 10.1007/s13277-016-5417-z. [DOI] [PubMed] [Google Scholar]
- 26.Nikuševa-Martić T, Serman L, Zeljko M, Vidas Z, Gašparov S, Zeljko HM, Kosović M, Pećina-Šlaus N. Expression of secreted frizzled-related protein 1 and 3, T-cell factor 1 and lymphoid enhancer factor 1 in clear cell renal cell carcinoma. Pathol Oncol Res. 2013;19:545–551. doi: 10.1007/s12253-013-9615-3. [DOI] [PubMed] [Google Scholar]
- 27.Zhang K, Zhu S, Liu Y, Dong X, Shi Z, Zhang A, Liu C, Chen L, Wei J, Pu P, et al. ICAT inhibits glioblastoma cell proliferation by suppressing Wnt/β-catenin activity. Cancer Lett. 2015;357:404–411. doi: 10.1016/j.canlet.2014.11.047. [DOI] [PubMed] [Google Scholar]
- 28.Tago K, Nakamura T, Nishita M, Hyodo J, Nagai S, Murata Y, Adachi S, Ohwada S, Morishita Y, Shibuya H, et al. Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev. 2000;14:1741–1749. [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Krinks M, Lin K, Luyten FP, Moos M., Jr Frzb, a secreted protein expressed in the Spemann organizer, binds and inhibits Wnt-8. Cell. 1997;88:757–766. doi: 10.1016/S0092-8674(00)81922-4. [DOI] [PubMed] [Google Scholar]
- 30.Kiper POS, Saito H, Gori F, Unger S, Hesse E, Yamana K, Kiviranta R, Solban N, Liu J, Brommage R, et al. Cortical-bone fragility - insights from sFRP4 deficiency in Pyle's disease. N Engl J Med. 2016;374:2553–2562. doi: 10.1056/NEJMoa1509342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ota S, Ishitani S, Shimizu N, Matsumoto K, Itoh M, Ishitani T. NLK positively regulates Wnt/β-catenin signalling by phosphorylating LEF1 in neural progenitor cells. EMBO J. 2012;31:1904–1915. doi: 10.1038/emboj.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Townsley FM, Cliffe A, Bienz M. Pygopus and Legless target Armadillo/beta-catenin to the nucleus to enable its transcriptional co-activator function. Nat Cell Biol. 2004;6:626–633. doi: 10.1038/ncb1141. [DOI] [PubMed] [Google Scholar]
- 33.Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Züllig S, Basler K. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell. 2002;109:47–60. doi: 10.1016/S0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- 34.Paluszczak J, Wiśniewska D, Kostrzewska-Poczekaj M, Kiwerska K, Grénman R, Mielcarek-Kuchta D, Jarmuż-Szymczak M. Prognostic significance of the methylation of Wnt pathway antagonists-CXXC4, DACT2, and the inhibitors of sonic hedgehog signaling-ZIC1, ZIC4, and HHIP in head and neck squamous cell carcinomas. Clin Oral Investig. 2017;21:1777–1788. doi: 10.1007/s00784-016-1946-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amen M, Liu X, Vadlamudi U, Elizondo G, Diamond E, Engelhardt JF, Amendt BA. PITX2 and beta-catenin interactions regulate Lef-1 isoform expression. Mol Cell Biol. 2007;27:7560–7573. doi: 10.1128/MCB.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vadlamudi U, Espinoza HM, Ganga M, Martin DM, Liu X, Engelhardt JF, Amendt BA. PITX2, beta-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J Cell Sci. 2005;118:1129–1137. doi: 10.1242/jcs.01706. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Liu S, Zhu H, Zhang W, Zhang G, Zhou X, Zhou C, Quan L, Bai J, Xue L, et al. FRAT1 overexpression leads to aberrant activation of beta-catenin/TCF pathway in esophageal squamous cell carcinoma. Int J Cancer. 2008;123:561–568. doi: 10.1002/ijc.23600. [DOI] [PubMed] [Google Scholar]
- 38.Chodaparambil JV, Pate KT, Hepler MR, Tsai BP, Muthurajan UM, Luger K, Waterman ML, Weis WI. Molecular functions of the TLE tetramerization domain in Wnt target gene repression. EMBO J. 2014;33:719–731. doi: 10.1002/embj.201387188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- 40.Li FQ, Mofunanya A, Harris K, Takemaru K. Chibby cooperates with 14-3-3 to regulate beta-catenin subcellular distribution and signaling activity. J Cell Biol. 2008;181:1141–1154. doi: 10.1083/jcb.200709091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 42.Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, Cheng J, Yeh ET. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell. 2010;38:191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingham PW, Placzek M. Orchestrating ontogenesis: Variations on a theme by sonic Ηedgehog. Nat Rev Genet. 2006;7:841–850. doi: 10.1038/nrg1969. [DOI] [PubMed] [Google Scholar]
- 44.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Ηedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 45.Merchant M, Vajdos FF, Ultsch M, Maun HR, Wendt U, Cannon J, Desmarais W, Lazarus RA, de Vos AM, de Sauvage FJ. Suppressor of fused regulates Gli activity through a dual binding mechanism. Mol Cell Biol. 2004;24:8627–8641. doi: 10.1128/MCB.24.19.8627-8641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hana Y, Shia Q, Jiang J. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc Natl Acad Sci USA. 2015;112:6383–6388. doi: 10.1073/pnas.1421628112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svärd J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergström A, Ericson J, Toftgård R, Teglund S. Genetic elimination of suppressor of fused reveals an essential repressor function in the mammalian Ηedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Taylor MD, Zhang X, Liu L, Hui CC, Mainprize TG, Scherer SW, Wainwright B, Hogg D, Rutka JT. Failure of a medulloblastoma-derived mutant of SUFU to suppress WNT signaling. Oncogene. 2004;23:4577–4583. doi: 10.1038/sj.onc.1207605. [DOI] [PubMed] [Google Scholar]
- 49.Smith MJ, Beetz C, Williams SG, Bhaskar SS, O'Sullivan J, Anderson B, Daly SB, Urquhart JE, Bholah Z, Oudit D, et al. Germline mutations in SUFU cause Gorlin syndrome-associated childhood medulloblastoma and redefine the risk associated with PTCH1 mutations. J Clin Oncol. 2014;32:4155–4161. doi: 10.1200/JCO.2014.58.2569. [DOI] [PubMed] [Google Scholar]
- 50.Brugières L, Remenieras A, Pierron G, Varlet P, Forget S, Byrde V, Bombled J, Puget S, Caron O, Dufour C, et al. High frequency of germline SUFU mutations in children with desmoplastic/nodular medulloblastoma younger than 3 years of age. J Clin Oncol. 2012;30:2087–2093. doi: 10.1200/JCO.2011.38.7258. [DOI] [PubMed] [Google Scholar]
- 51.Liu X, Wang X, Du W, Chen L, Wang G, Cui Y, Liu Y, Dou Z, Wang H, Zhang P, et al. Suppressor of fused (Sufu) represses Gli1 transcription and nuclear accumulation, inhibits glioma cell proliferation, invasion and vasculogenic mimicry, improving glioma chemo-sensitivity and prognosis. Oncotarget. 2014;5:11681–11694. doi: 10.18632/oncotarget.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee Y, Kawagoe R, Sasai K, Li Y, Russell HR, Curran T, McKinnon PJ. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26:6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 53.Yan R, Peng X, Yuan X, Huang D, Chen J, Lu Q, Lv N, Luo S. Suppression of growth and migration by blocking the Ηedgehog signaling pathway in gastric cancer cells. Cell Oncol. 2013;36:421–435. doi: 10.1007/s13402-013-0149-1. [DOI] [PubMed] [Google Scholar]
- 54.Chung TK, Lau TS, Cheung TH, Yim SF, Lo KW, Siu NS, Chan LK, Yu MY, Kwong J, Doran G, et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130:1036–1045. doi: 10.1002/ijc.26060. [DOI] [PubMed] [Google Scholar]
- 55.Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ. South Australian Oesophageal Research Group: MicroRNA profiling of Barrett's oesophagus and oesophageal adenocarcinoma. Br J Surg. 2010;97:853–861. doi: 10.1002/bjs.7000. [DOI] [PubMed] [Google Scholar]
- 56.Song Y, Zhao F, Wang Z, Liu Z, Chiang Y, Xu Y, Gao P, Xu H. Inverse association between miR-194 expression and tumor invasion in gastric cancer. Ann Surg Oncol. 2012;19(Suppl 3):S509–S517. doi: 10.1245/s10434-011-1999-2. [DOI] [PubMed] [Google Scholar]
- 57.Li Z, Ying X, Chen H, Ye P, Shen Y, Pan W, Zhang L. MicroRNA-194 inhibits the epithelial-mesenchymal transition in gastric cancer cells by targeting FoxM1. Dig Dis Sci. 2014;59:2145–2152. doi: 10.1007/s10620-014-3159-6. [DOI] [PubMed] [Google Scholar]
- 58.Chen X, Wang Y, Zang W, Du Y, Li M, Zhao G. miR-194 targets RBX1 gene to modulate proliferation and migration of gastric cancer cells. Tumour Biol. 2015;36:2393–2401. doi: 10.1007/s13277-014-2849-1. [DOI] [PubMed] [Google Scholar]
- 59.Bao J, Zou JH, Li CY, Zheng GQ. miR-194 inhibits gastric cancer cell proliferation and tumorigenesis by targeting KDM5B. Eur Rev Med Pharmacol Sci. 2016;20:4487–4493. [PubMed] [Google Scholar]
- 60.Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J, et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 61.Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 62.Shiotani A, Uedo N, Iishi H, Murao T, Kanzaki T, Kimura Y, Kamada T, Kusunoki H, Inoue K, Haruma K. H. pylori eradication did not improve dysregulation of specific oncogenic miRNAs in intestinal metaplastic glands. J Gastroenterol. 2012;47:988–998. doi: 10.1007/s00535-012-0562-7. [DOI] [PubMed] [Google Scholar]
- 63.Daniels DL, Weis WI. ICAT inhibits beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol Cell. 2002;10:573–584. doi: 10.1016/S1097-2765(02)00631-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.