Abstract
The function of the expression of microRNA (miR)-224-5p in prostate adenocarcinoma (PCa) remains to be elucidated, therefore, the present study aimed to investigate the clinical significance and potential molecular mechanism of miR-224-5p in PCa. Data on the expression of miR-224-5p in PCa were extracted from The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), ArrayExpress and previous literature, and meta-analyses with standardized mean difference (SMD) and summary receiver operating characteristic (sROC) methods were performed for statistical analyses. The prospective target genes of miR-224-5p were collected by overlapping the differentially expressed mRNAs in TCGA and GEO, and target genes predicted by miRWalk2.0. Subsequently, in silico analysis was performed to examine the associated pathways of miR-224-5p in PCa. The expression of miR-224-5p was markedly lower in PCa; the overall SMD was −0.562, and overall sROC area under the curve was 0.80. In addition, Kyoto Encyclopedia of Genes and Genomes analysis revealed that the prospective target genes of miR-224-5p were largely enriched in the amino sugar and nucleotide sugar metabolism signaling pathway, and three genes [UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1), hexokinase 2 (HK2) and chitinase 1 (CHIT1)] enriched in this pathway showed higher expression (P<0.05). In addition, key genes in the protein-protein interaction network analysis [DNA topoisomerase 2-α (TOP2A), ATP citrate lyase (ACLY) and ribonucleotide reductase regulatory subunit M2 (RRM2)] exhibited significantly increased expression (P<0.05). The results suggested that the downregulated expression of miR-224-5p may be associated with the clinical progression and prognosis of PCa. Furthermore, miR-224-5p likely exerts its effects by targeting genes, including UAP1, HK2, CHIT1, TOP2A, ACLY and RRM2. However, in vivo and in vitro experiments are required to confirm these findings.
Keywords: microRNA-224-5p, prostate cancer, molecular mechanism, The Cancer Genome Atlas, Gene Expression Omnibus, in silico
Introduction
According to cancer statistics from 2017, prostate adenocarcinoma (PCa) was ranked highest for newly diagnosed cases of cancer in men in the USA, and was considered the 3rd leading cause of cancer-associated mortality in men (1). Although the rates of incidence and mortality vary in different nations and regions, PCa poses a health risk to men due to its high incidence and mortality rates (2–4). PCa is a heterogeneous type of cancer; patients with PCa are prone to a relapse and metastasis, and the prognosis of PCa is associated with the age of patients (5,6). The majority of patients are diagnosed with PCa in its intermediate or terminal stage, which presents challenges to treatment and recovery of patients. Fortunately, the diagnosis and treatment of PCa have been modified owing to medical advances. For example, screening prostate-specific antigen in the early stage is considered an effective approach for early diagnosis and immediate treatment of PCa (7). Despite this, the incidence and mortality rates of PCa remain high, therefore, further investigations are urgently required to clarify the onset and mechanism of progression of PCa (8).
Based on previous studies, in addition to obesity, gene expression is closely associated with the onset and development of PCa (9,10). MicroRNAs (miRNAs) are small, non-coding RNA molecules of ~ 22 nucleotides, which perform functions in the post-transcriptional regulation of gene expression (11–15). miRNAs act as oncogenes or antioncogenes in tumorigenesis, thus influencing the onset and development of tumors (16–20). Previous studies have shown that the downregulated expression of miRNA may be essential in the onset and development of PCa (21–23). In addition, increased expressed of miRNA was likely to regulate the expression of target genes and exert its influences on the progression of PCa (24,25).
Currently, investigations on the role of miRNA-224-5p (miR-224-5p) in PCa have been limited. Mavridis et al performed a case-control study involving 73 cases of PCa and 66 cases of benign prostatic hyperplasia, demonstrating that the expression of miR-224-5p was downregulated in PCa tissues and, with disease progression, the expression of miR-224-5p was decreased further; patients with the lower expression of miR-224-5p were also more likely to have a poorer prognosis (26). Fu et al investigated the targeting association between miR-224-5p and calcium/calmodulin-dependent protein (CAMKK2) in tissues from 20 cases of PCa and non-cancerous counterpart tissues; it was found that the expression of miR-224-5p was markedly lower in PCa tissues, and patients with a lower expression of miR-224-5p tended to have a poorer survival rate; additionally, miR-224-5p inhibited the proliferation of tumor cells by targeting CAMKK2 (27). Having applied reverse transcription-quantitative polymerase chain reaction analysis to examine tissues from 36 cases of PCa and 14 non-cancerous tissues, Kristensen et al confirmed that the expression of miR-224-5p was decreased in PCa tissues and predicted an unsatisfactory outcome for patients (28). These previous studies offer important insights into the effects of miR-224-5p on PCa tissues, however, they were performed with a small sample size (n<80) and the results were not confirmed with a larger sample size, which may reduce the convincingness of the results. Furthermore, the studies mentioned above failed to perform bioinformatics analyses, which may assist in identifying more prospective target genes of miR-224-5p in PCa. Therefore, further verification is required of the expression of miR-224-5p and its clinical significance in PCa. In addition, the mining of multiple databases and bioinformatics analyses is required to examine the prospective molecular mechanism underlying the role of miR-224-5p in PCa.
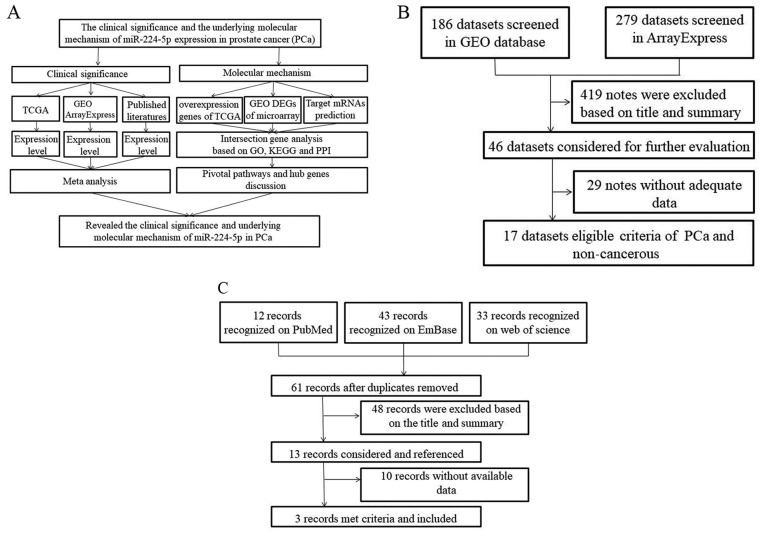
The present study aimed to verify the expression of miR-224-5p in PCa using data from different databases, including The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), ArrayExpress, and previous literature. In addition, prospective target genes of miR-224-5p were collected using online prediction tools and differentially expressed genes in TCGA and GEO. Bioinformatics analyses were also used to further examine the signaling pathways of miR-224-5p in PCa (Fig. 1A).
Figure 1.
Study design. (A) Flow chart of the study design. (B) Flow chart of microarray retrieval for differential expression of miR-224-5p in PCa. (C) Flow chart of literature retrieval for the differential expression of miR-224-5p in PCa. GO, Gene Ontology; KEGG, l TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; PPI, protein-protein interaction DEGs, differentially expressed genes; miR, microRNA; PCa, prostate adenocarcinoma.
Materials and methods
Collection of PCa data from TCGA
Data of Illumina HiSeq level 3 were acquired from the Launch ata portal of TCGA (https://cancergenome.nih.gov/). Following summarizing of the raw count data, raw data on miRNA expression in tissues of 500 cases of PCa and 52 normal prostatic tissues were obtained, from which reads per million data of pre-miR224 were extracted, including tissues from 498 cases of PCa and 51 normal PCa tissues. In these 500 cases, the age of the patients ranged between 41 and 78 years, with a mean age of 61 years. Additional clinicopathological parameters are listed in Table I.
Table I.
Association between the expression of miR-224-5p between prostate cancer tissue and non-cancerous tissue based on The Cancer Genome Atlas data.
| miR-224 expression | T-test | |||||
|---|---|---|---|---|---|---|
| Clinicopathological parameter | Cases | Mean | SD | FC | T-value | P-value |
| Group | ||||||
| Normal adjacent | 51 | 5.4773 | 0.86449 | 1.0000 | −3.413 | 0.001 |
| Cancer | 498 | 5.0255 | 1.19453 | 0.9175 | ||
| Age (years) | ||||||
| <60 | 204 | 4.9880 | 1.36730 | 1.0000 | −0.086 | 0.931 |
| ≥60 | 296 | 4.9982 | 1.26126 | 1.0020 | ||
| Pathological T stage | ||||||
| T1-T2 | 188 | 5.1306 | 1.21866 | 1.0000 | 1.880 | 0.061 |
| T3-T4 | 305 | 4.9052 | 1.33624 | 0.9561 | ||
| N stage | ||||||
| N0 | 348 | 5.0312 | 1.28350 | 1.0000 | 1.347 | 0.179 |
| N1 | 79 | 4.8182 | 1.19763 | 0.9577 | ||
| M stage | ||||||
| M0 | 456 | 4.9999 | 1.25252 | 1.0000 | 1.715 | 0.087 |
| M1 | 4 | 3.9179 | 1.76728 | 0.7836 | ||
| Gleason score | ||||||
| ≤7 | 291 | 5.1057 | 1.13197 | 1.0000 | 1.143 | 0.254 |
| 8≥ | 203 | 4.9815 | 1.26629 | 0.9757 | ||
| Gleason grade | ||||||
| 2 | 1 | 4.8074 | 1.048 | 0.371 | ||
| 3 | 197 | 5.1616 | 1.12858 | |||
| 4 | 248 | 5.0052 | 1.23926 | |||
| 5 | 48 | 4.8763 | 1.16617 | |||
| Recurrence | ||||||
| No | 373 | 5.0491 | 1.30796 | 1.0000 | 1.008 | 0.314 |
| Yes | 58 | 4.8636 | 1.27528 | 0.9633 | ||
| Clinical T stage | ||||||
| T1-T2 | 351 | 5.0312 | 1.31167 | 1.0000 | −0.338 | 0.735 |
| T3-T4 | 55 | 5.0945 | 1.14167 | 1.0126 | ||
miR, microRNA; SD, standard deviation; FC, fold-change.
Screening of differentially expressed microarrays of miRNAs in PCa
In the microarray GEO (https://www.ncbi.nlm.nih.gov/gds/) and ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) databases, a search was performed using the following key words: (parastata OR prostatic gland OR prostate gland OR prostat* AND (cancer OR carcinoma OR adenocarcinoma OR tumor OR malignan* OR neoplas*) AND (miR OR miRNA OR microRNA OR miR OR miRNA OR microRNA OR ‘miR’ OR ‘miRNA’ OR ‘microRNA’). Subsequently, the differentially expressed microarrays of miRNAs in PCa were screened and downloaded. Finally, studies were included which fulfilled the following criteria: i) compared the cancerous tissues with the controls; ii) contained microarrays of miRNA expression in PCa tissues, biofluids and cell lines; and iii) had more than 3 samples in each microarray. The procedures for the search are shown in Fig. 1B.
Literature search
From the PubMed (https://www.ncbi.nlm.nih.gov/pubmed), Embase (https://embase.com/) and Web of Science (http://apps.webofknowledge.com/UA_General Search_input.do?product=UA&search_mode=GeneralSearch&SID=8ErJvGamhqk4GR7zNPu&preferencesSaved=) databases, studies on differentially expressed miR-224-5p in PCa tissues and in non-cancerous controls were collected. The key words used for the search included the following: (parastata OR prostatic gland OR prostate gland OR prostat*) and (cancer OR carcinoma OR adenocarcinoma OR tumor OR malignan* OR neoplas*) and (miR-224 OR miRNA-224 OR microRNA-224 OR miR224 OR miRNA224 OR microRNA224 OR ‘miR 224’ OR ‘miRNA 224’ OR ‘microRNA 224’ OR miR-224-5p OR miRNA-224-5p OR microRNA-224-5p). The studies were included if they met the following standards: i) involved the comparison of PCa with non-cancerous controls in tissues, biofluids and cell lines; and ii) provided the mean ± standard deviation (SD) or diagrams from which data extraction was possible. The procedures for the literature screening are shown in Fig. 1C.
Collection of differentially expressed genes of PCa in TCGA
Gene expression profiling interactive analysis (GEPIA; http://gepia.cancer-pku.cn/), a visualized website based on TCGA database developed by Peking University (Beijing, China), contains various functional analyses, including the comparison of differentially expressed genes in cancerous and non-cancerous tissues (29). Data on the differential genes of PCa on GEPIA were retrieved, and differentially expressed genes calculated using the Linear Models for Microarray Data (LIMMA) package (http://gepia.cancer-pku.cn/detail.php?gene=) were downloaded. The genes were selected for further analysis if log2 fold change (FC)>1.
Selection of microarrays of differentially expressed genes of PCa
In the GEO and ArrayExpress databases, a search was performed using the aforementioned key words for prostate and cancer. The relevant microarrays were included in the study if they conformed to the following criteria: i) investigation of differentially expressed mRNA based on post-transcriptional miR-224-5p; and ii) comparison of PCa cell lines and normal cell lines. If there existed numerous similar samples in one microarray, the intersections were obtained. When dealing with different microarrays, the unions were obtained. Further analyses were performed on all results.
Prediction of potential target genes of miR-224-5p
The microRNA-mRNA prediction was performed with the miRWalk2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) online prediction tools involving 12 prediction tools, namely, DIANA microT v4, RNA22, Pictar2, miRWalk, miRNAMap, RNAhybrid, mirBridge, TargetScan, miRMap, miRanda, PITA and miRDB. Genes that were predicted by three tools qualified for the present study. In order to acquire the potential target genes with accuracy, the overexpressed genes in TCGA, mRNAs expressed at low levels following miR-224-5p transcription and the predicted microarrays were combined, and the unions were obtained. Bioinformatics analysis was performed on these results.
In silico analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed on the genes that appeared in TCGA, GEO and target genes prediction tools on Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/). Bingo on Cytoscape 3.5.0 (http://www.cytoscape.org/) was applied to construct network analysis of GO terms, and the ClueGO and CluePedia plugins were used to establish the KEGG network. For genes enriched in significant pathways, the data in TCGA were used to verify their expression levels in PCa. In addition, protein-protein interaction (PPI) analysis was performed, a PPI network was constructed and the interactive associations between proteins were confirmed on the Search Tool for the Retrieval of Interacting Genes 0.5 (https://string-db.org/cgi/input.pl?sessionId=Ce1Dx9pYDluc&input_page_show_search=on) (30–37) database. Based on TCGA data, the mRNA expression of key genes in the PPI network were also confirmed, and the mechanism of miR-224-5p in PCa was further examined.
Statistical analysis
SPSS 23.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis of the expression of miR-224-5p in PCa. An independent t-test was applied to evaluate the differentials of miR-224-5p between PCa tissues and non-cancerous tissues, and the results are presented as the mean ± SD. The standardized mean difference (SMD) was used to combine all the included studies on STATA 2.0 (StataCorp, College Station, TX, USA), and to calculate the expression trend of miR-224-5p in PCa. In addition, a receiver operating characteristic (ROC) was used to analyze the sensitivity and specificity of each study, and their cut-off value was calculated. Subsequently, the cut-off value was applied to identify the true positive, false positive, false negative and true negative of each study, following which a diagnostic test four-fold table was produced. STATA 12.0 was then used to confirm the expression of miR-224-5p in PCa, and the summary ROC (sROC) was used to measure its credibility. In addition, in order to examine the expression trend in each study, scatter diagrams were produced to show the expression of miR-224-5p in PCa tissues and adjacent tissues via GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate statistically significant difference.
Results
Expression of miR-224-5p in PCa
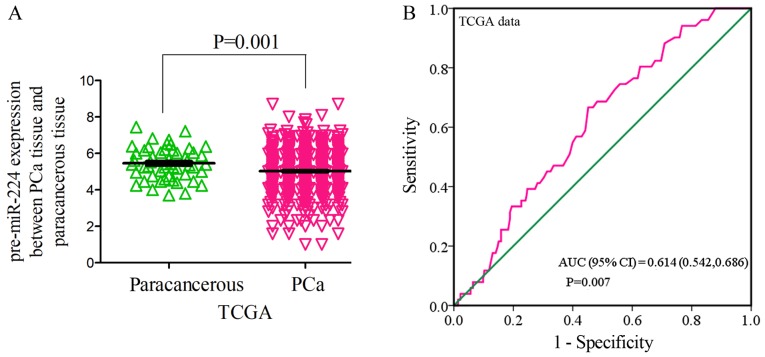
According to the data in TCGA, the expression of pre-miR-224 in PCa tissues was 5.0255±1.1945 (PCa, vs. normal, 498, vs. 52), which was significantly lower than that of normal adjacent tissues (5.4773±0.8645, FC=0.9175, P=0.001; Fig. 2A). The area under the curve (AUC) of the downregulated pre-miR-224 in PCa tissues was 0.614 (95% CI, 0.542, 0.686. P=0.007; Fig. 2B). It was found that pre-miR-224 tended to exhibit lower expression with the progression of clinical staging by comparing T3-4 and T1-2 (4.9052±1.3362 vs. 5.1306±1.2187, FC=0.9561, P=0.061), M1 and M0 (3.9179±1.7677 vs. 4.9999±1.2525, FC=0.7836, P=0.087; Table I). However, no clear associations were found between its expression and prognosis or other types of staging.
Figure 2.
Expression of miR-224. (A) Scatter diagram of miR-224 precursor expression between PCa tissue and normal prostate tissue based on TCGA data. (B) ROC curve of miR-224-5p expression in PCa tissue. miR, microRNA; PCa, prostate adenocarcinoma; AUC, area under the curve; TCGA, The Cancer Genome Atlas.
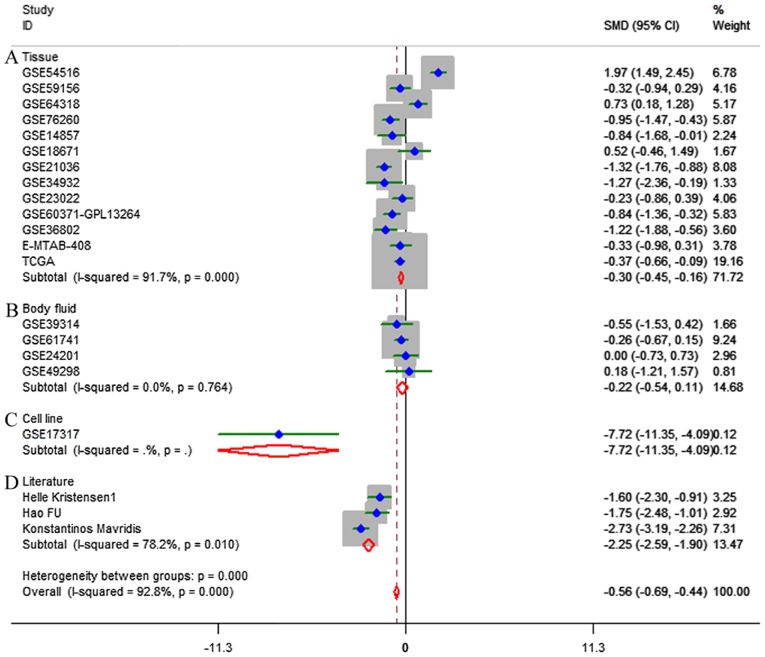
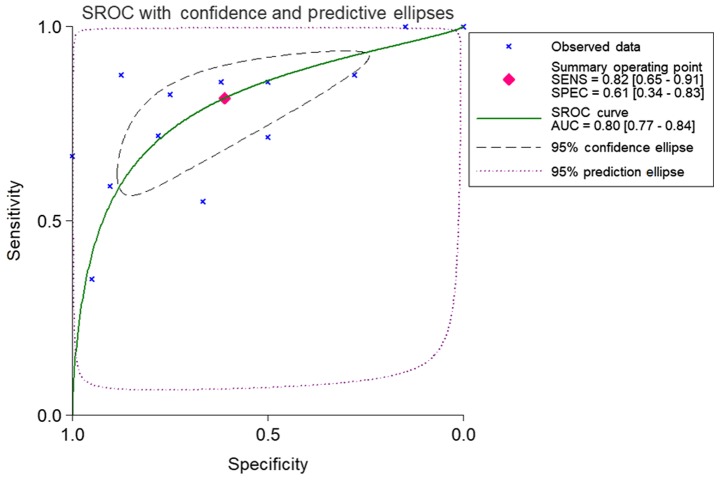
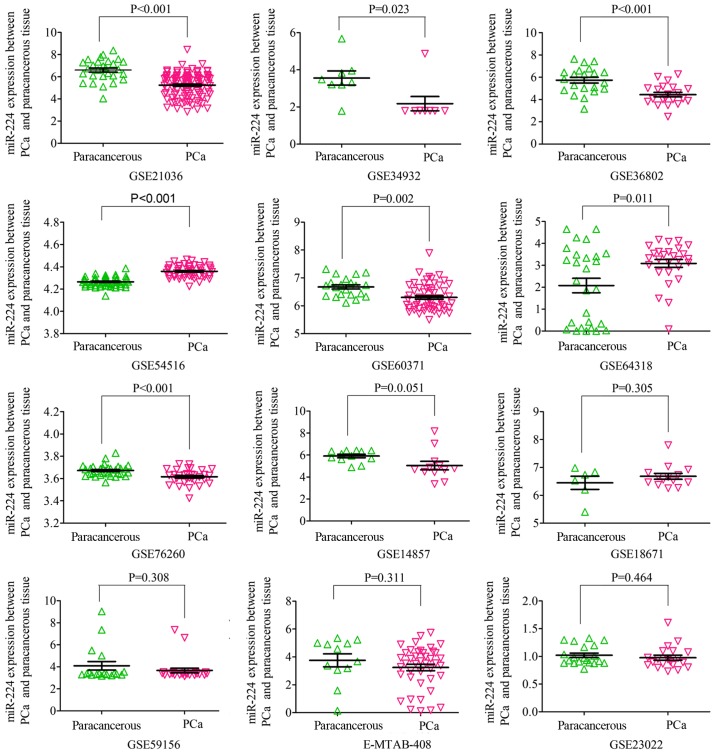
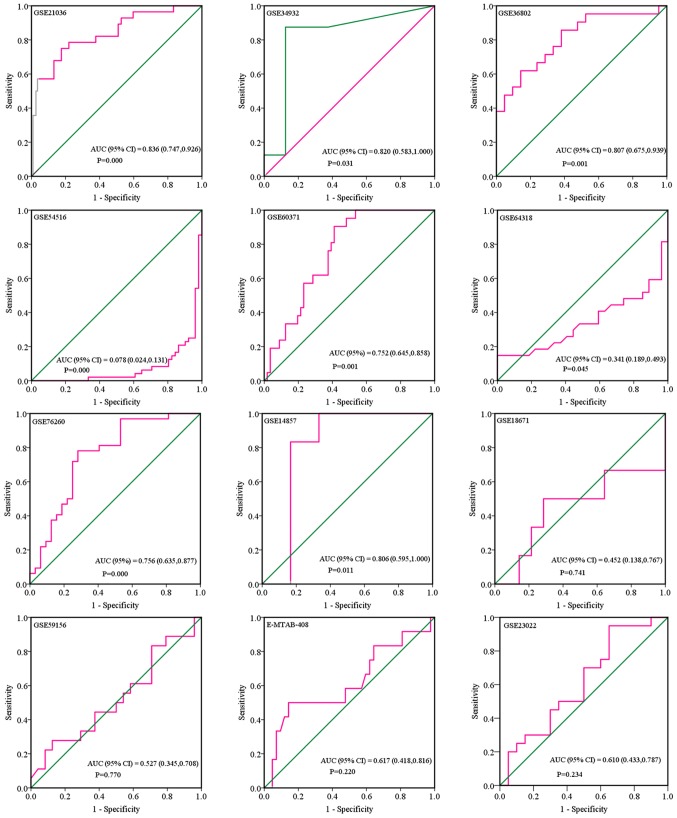
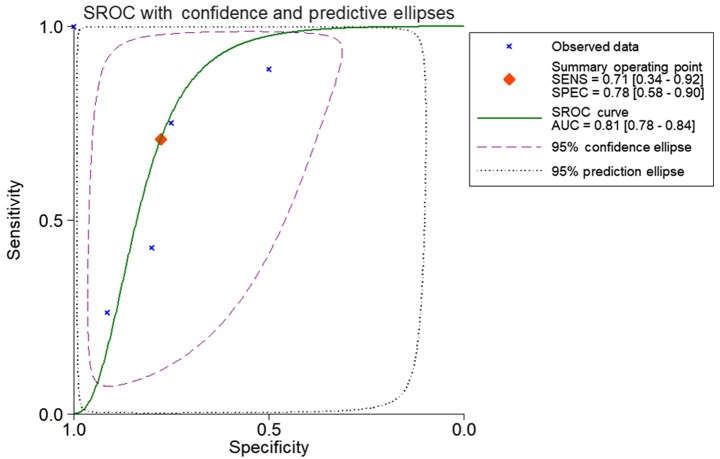
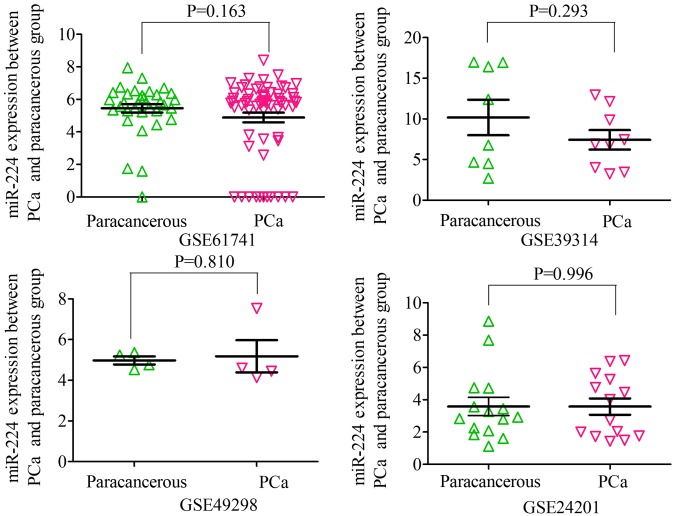
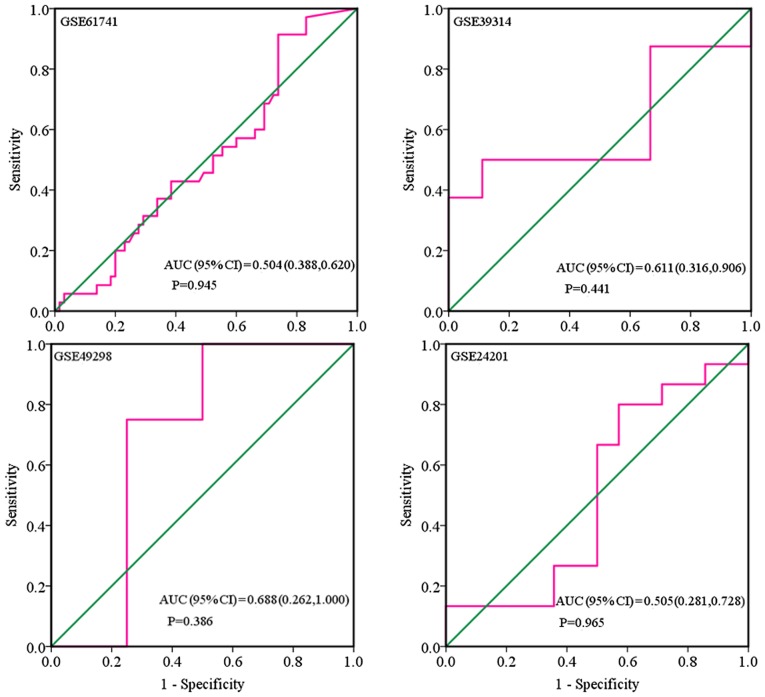
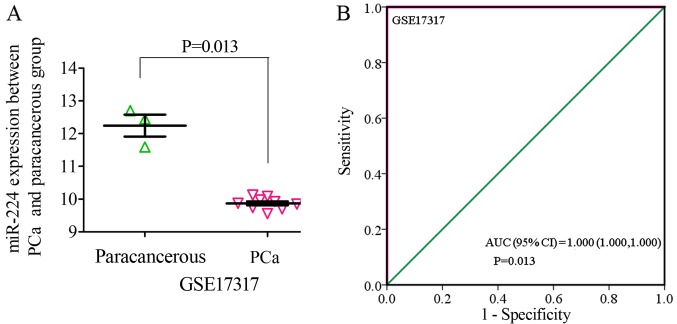
Based on the inclusion criteria, a total of 17 microarrays were eventually considered eligible for the present study, which were categorized into three subtypes: Tissues, biofluids and cell lines (Fig. 3). In the subgroup of tissues, 12 microarrays were included, among which the expression of miR-224-5p was notably lower in PCa tissues in GSE76260, GSE21036, GSE34932, GSE60371 and GSE36802. In GSE54516 and GSE64318, the expression of miR-224-5p was upregulated. In terms of SMD, a low expression of miR-224-5p was identified in PCa tissues: Sub-SMD (95% CI)=−0.304 (−0.452, −0.156) (Table II and Fig. 3A; P<0.001) PCa, vs. normal=421, vs. 253); sROC AUC (95% CI)=0.80 (0.77, 0.84) (Fig. 4). The optimum sensitivity and specificity were 0.82 (95% CI: 0.65, 0.91) and 0.61 (95% CI: 0.34, 0.83, Fig. 4), respectively. The scatter diagram and ROC curve of expression of each microarray are shown in Figs. 5 and 6. When analyzing the biofluids subgroup, four microarrays were included (GSE39314, GSE61741, GSE24201 and GSE49298). The SMD result indicated that Sub-SMD (95% CI)=−0.216 (−0.544, −0.111), P=0.195 (Table III and Fig. 3B) PCa, vs. normal, 92, vs 62. sROC analysis revealed the following: AUC (95% CI)=0.81 (0.78, 0.84), sensitivity (SENS; 95% CI)=0.71 (0.34, 0.92), specificity (SPEC; 95% CI)=0.78 (0.58, 0.90) (Fig. 7). The scatter diagrams and ROC curves are shown in Figs. 8 and 9. The expression of miR-224-5p was markedly lower in PCa biofluids despite no clear significance. In the subgroup of cell lines, only one microarray was included: Mean ± SD: 9.8705±0.1835 (P=0.013, PCa, vs. normal, 9, vs. 3; ROC AUC (95% CI)=1.000 (1.000, 1.000), P=0.013 (Fig. 10). The expression of miR-224-5p was significantly lower in the PCa cell lines, and exhibited higher specificity.
Figure 3.
Forest profile of SMD of the expression of microRNA-224-5P between prostate adenocarcinoma and non-cancerous (A) tissues, (B) body fluid, (C) cell line and (D) literature. SMD, standardized mean difference.
Table II.
Characteristics of the expression of microRNA-224-5p between studies of prostate adenocarcinoma and non-cancerous tissue based on Gene Expression Omnibus, ArrayExpress and TCGA datasets.
| Patients | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
| Study | N | Mean | SD | N | Mean | SD | T-value | P-value |
| GSE54516 | 51 | 4.3588 | 0.0525 | 48 | 4.2649 | 0.0420 | 9.858 | <0.001 |
| GSE59156 | 24 | 3.6686 | 1.0369 | 18 | 4.0963 | 1.6407 | −1.033 | 0.308 |
| GSE64318 | 27 | 3.0859 | 0.9412 | 27 | 2.0776 | 1.7120 | 2.682 | 0.011 |
| GSE76260 | 32 | 3.6162 | 0.0662 | 32 | 3.6723 | 0.0512 | −3.794 | <0.001 |
| GSE14857 | 12 | 5.0417 | 1.3522 | 12 | 5.9110 | 0.5401 | −2.068 | 0.051 |
| GSE18671 | 14 | 6.6809 | 0.3943 | 6 | 6.4473 | 0.5782 | 1.057 | 0.305 |
| GSE21036 | 114 | 5.2423 | 1.0451 | 28 | 6.6041 | 0.9873 | −6.243 | <0.001 |
| GSE34932 | 8 | 2.1788 | 1.0875 | 8 | 3.5600 | 1.0803 | −2.549 | 0.023 |
| GSE23022 | 20 | 0.9755 | 0.2065 | 20 | 1.0201 | 0.1732 | −0.740 | 0.464 |
| GSE60371 | 56 | 6.3003 | 0.4778 | 21 | 6.6752 | 0.3432 | −3.285 | 0.002 |
| GSE36802 | 21 | 4.4416 | 0.9294 | 21 | 5.7362 | 1.1766 | −3.957 | <0.001 |
| E-MTAB-408 | 42 | 3.2419 | 1.5142 | 12 | 3.7560 | 1.6093 | −1.023 | 0.311 |
| TCGA | 498 | 5.0164 | 1.2154 | 52 | 5.4589 | 0.8661 | −3.413 | 0.001 |
| Total | Standardized mean difference (95% CI)=−0.304 (−0.452, −0.156) P<0.001 | |||||||
TGCA, The Cancer Genome Atlas; SD, standard deviation.
Figure 4.
Combined sROC curve for the expression of miR-224-5P between prostate adenocarcinoma and normal prostate tissue. sROC, summary receiver operating characteristic; AUC, area under the curve; SENS, sensitivity; SPEC, specificity.
Figure 5.
Scatter plot of levels of miR-224-5p between PCa tissues and normal prostate tissues based on microarray. miR, microRNA; PCa, prostate adenocarcinoma.
Figure 6.
Receiver operating characteristic curves of microRNA-224-5p levels between prostate adenocarcinoma tissues based on microarray. AUC, area under the curve.
Table III.
Characteristics of the expression of microRNA-224-5p between prostate adenocarcinoma and non-cancerous body fluid studies based on the Gene Expression Omnibus dataset.
| Patients | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
| Study | N | Mean | SD | N | Mean | SD | T-value | P-value |
| GSE39314 | 9 | 7.4306 | 3.6046 | 8 | 10.1763 | 6.1584 | −1.104 | 0.293 |
| GSE61741 | 65 | 4.8860 | 2.4333 | 35 | 5.4507 | 1.5656 | −1.407 | 0.163 |
| GSE24201 | 14 | 3.5766 | 1.8929 | 15 | 3.5814 | 2.1733 | −0.006 | 0.996 |
| GSE49298 | 4 | 5.1792 | 1.5787 | 4 | 4.9743 | 0.3945 | 0.252 | 0.810 |
| Total | Standardized mean difference (95% CI)=−0.216 (−0.544, 0.111) P=0.195 | |||||||
SD, standard deviation.
Figure 7.
Combined sROC curve for the expression of microRNA-224-5p between prostate adenocarcinoma body fluids based on microarray. sROC, summary receiver operating characteristic; AUC, area under the curve; SENS, sensitivity; SPEC, specificity.
Figure 8.
Scatter plot of the expression of miR-224-5p between PCa body fluid and non-cancerous body fluid based on microarray. miR, microRNA; PCa, prostate adenocarcinoma.
Figure 9.
Receiver operating characteristic curves of the expression of microRNA-224-5p between prostate adenocarcinoma body fluid based on microarray. AUC, area under the curve.
Figure 10.
Expression of miR-224-5p between PCa cell line and normal cell line based on microarray. (A) Scatter diagram. (B) Receiver operating characteristic curve. miR, microRNA; PCa, prostate adenocarcinoma; AUC, area under the curve.
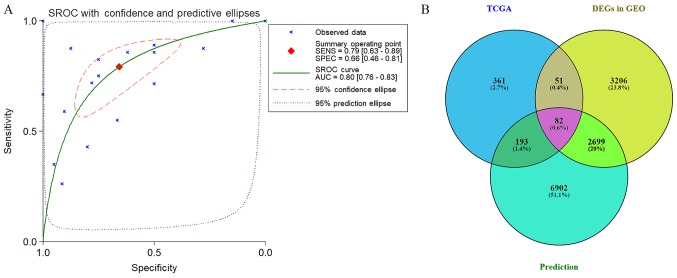
Furthermore, in the literature search, three studies were retrieved providing a mean ± SD (Fig. 1C) (15–17). The results also suggested that the expression of miR-224-5p was downregulated in PCa tissues: SMD (95% CI)=−2.245 (−2.587, −1.904), P<0.001 (Table IV and Fig. 3D; PCa, vs. normal, 129, vs. 100). The results of all studies provided the following results: Overall SMD (95% CI)=−0.562 (−0.687, −0.436; P<0.001); overall sROC AUC (95% CI)=0.80 (0.76, 0.83); SENS (95% CI) 0.79 (0.63, 0.89); SPEC (95% CI)=0.66 (0.46, 0.81) (Table V and Figs. 3 and 11A). This provided a more reliable conclusion that the expression of miR-224-5p was reduced in PCa.
Table IV.
Characteristics of three studies selected from previous literature.
| Control | Prostate adenocarcinoma | ||||||
|---|---|---|---|---|---|---|---|
| Author, date | Country | N | Mean ± SD | N | Mean ± SD | Detection method | Refs:PMID |
| Fu et al (27) | China | 20 | 2.171±0.564 | 20 | 1.344±0.358 | RT-qPCR | 25394900 |
| Mavridis et al (26) | Athens | 66 | 8.470±2.090 | 73 | 4.228±0.809 | RT-qPCR | 23136246 |
| Kristensen et al (28) | Denmark | 14 | 2.929±0.147 | 36 | 2.562±0.252 | RT-qPCR | 24737792 |
| Total | Standardized mean difference (95% CI)=−2.245 (−2.587, −1.904) P=0.195, P<0.001 | ||||||
RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SD, standard deviation.
Table V.
Characteristics of the expression of microRNA-224-5p between prostate adenocarcinoma and non-cancerous studies.
| Patients | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
| Study | N | Mean | SD | N | Mean | SD | T-value | P-value |
| GSE54516 | 51 | 4.3588 | 0.0525 | 48 | 4.2649 | 0.0420 | 9.858 | <0.001 |
| GSE59156 | 24 | 3.6686 | 1.0369 | 18 | 4.0963 | 1.6407 | −1.033 | 0.308 |
| GSE64318 | 27 | 3.0859 | 0.9412 | 27 | 2.0776 | 1.7120 | 2.682 | 0.011 |
| GSE76260 | 32 | 3.6162 | 0.0662 | 32 | 3.6723 | 0.0512 | −3.794 | <0.001 |
| GSE14857 | 12 | 5.0417 | 1.3522 | 12 | 5.9110 | 0.5401 | −2.068 | 0.051 |
| GSE18671 | 14 | 6.6809 | 0.3943 | 6 | 6.4473 | 0.5782 | 1.057 | 0.305 |
| GSE21036 | 114 | 5.2423 | 1.0451 | 28 | 6.6041 | 0.9873 | −6.243 | <0.001 |
| GSE34932 | 8 | 2.1788 | 1.0875 | 8 | 3.5600 | 1.0803 | −2.549 | 0.023 |
| GSE23022 | 20 | 0.9755 | 0.2065 | 20 | 1.0201 | 0.1732 | −0.740 | 0.464 |
| GSE60371 | 56 | 6.3003 | 0.4778 | 21 | 6.6752 | 0.3432 | −3.285 | 0.002 |
| GSE36802 | 21 | 4.4416 | 0.9294 | 21 | 5.7362 | 1.1766 | −3.957 | <0.001 |
| E-MTAB-408 | 42 | 3.2419 | 1.5142 | 12 | 3.7560 | 1.6093 | −1.023 | 0.311 |
| TCGA | 498 | 5.0164 | 1.2154 | 52 | 5.4589 | 0.8661 | −3.413 | 0.001 |
| GSE39314 | 9 | 7.4306 | 3.6046 | 8 | 10.1763 | 6.1584 | −1.104 | 0.293 |
| GSE61741 | 65 | 4.8860 | 2.4333 | 35 | 5.4507 | 1.5656 | −1.407 | 0.163 |
| GSE24201 | 14 | 3.5766 | 1.8929 | 15 | 3.5814 | 2.1733 | −0.006 | 0.996 |
| GSE49298 | 4 | 5.1792 | 1.5787 | 4 | 4.9743 | 0.3945 | 0.252 | 0.810 |
| GSE17317 | 9 | 9.8705 | 0.1835 | 3 | 12.2457 | 0.5820 | −7.559 | 0.013 |
| Kristensen et al (28) | 36 | 2.5623 | 0.2523 | 14 | 2.9291 | 0.1469 | <0.001 | |
| Fu et al (27) | 20 | 1.3444 | 0.3578 | 20 | 2.1705 | 0.5643 | <0.001 | |
| Mavridis et al (26) | 73 | 4.2280 | 0.8090 | 66 | 8.4700 | 2.0900 | <0.001 | |
| Total | Standardized mean difference (95% CI)=−0.562 (−0.687, −0.436) P<0.001 | |||||||
SD, standard deviation; TCGA, The Cancer Genome Atlas.
Figure 11.
(A) Combined sROC curve for the expression of miR-224-5p between PCa based on the overall microarray. (B) Venn diagram for the crossing potential target genes of miR-224-5p in PCa. sROC, summary receiver operating characteristic; AUC, area under the curve; SENS, sensitivity; SPEC, specificity; miR, microRNA; PCa, prostate adenocarcinoma; TCGA, The Cancer Genome Atlas; DEGs, differentially expressed genes; GEO, Gene Expression Omnibus.
Prospective target genes of miR-224-5p in PCa
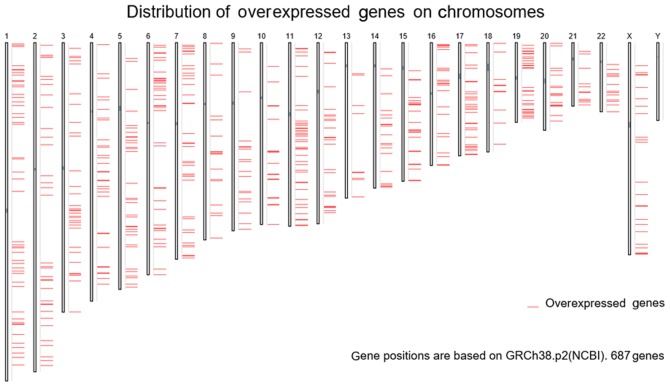
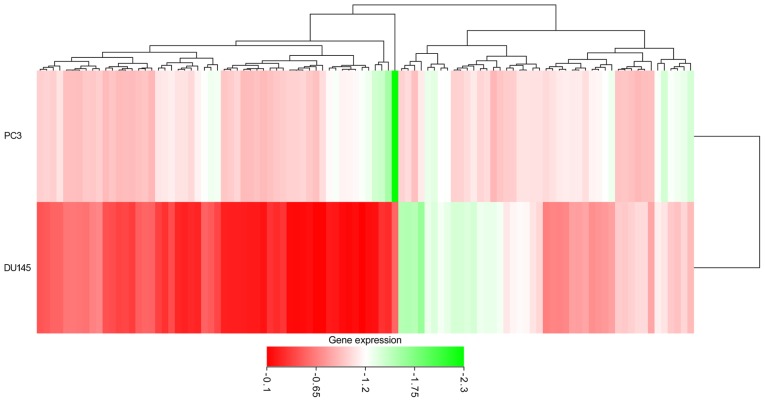
The prospective target genes of moR-224-5p in PCa were determined based on the results of TCGA, GEO and prediction tools. Based on TCGA data, 3,019 differentially expressed genes in PCa tissues were acquired from GEPIA, and the FC was used as a measure (log2FC>1.0). Finally, 687 overexpressed genes were obtained from the PCa tissues (Fig. 12). In addition, from GEO two cell lines were retrieved that were transfected with PC3 and DU145, and the microarrays of differentially expressed mRNA (GSE51053 and GSE56243) were examined. According to the unions in which the gene expression value was below −0.1, a total of 3,616 (GSE51053) and 3,326 (GSE56243) genes with low expression were obtained. Following combining the results of two microarrays and eliminating the duplicates, 6,038 genes with low expression that had been transfected with miR-224-5p were obtained. The top 100 genes are shown in the heat-map in Fig. 13. In addition, 102,240 potential target genes of miR-224-5p were accumulated via miRWalk2.0. Genes that appeared in at least three prediction tools were selected, with 9,876 genes acquired. A total of 82 overlapped mRNA genes qualified for further analysis following combining the results of TCGA, GEO and prediction tools (Fig. 11B).
Figure 12.
687 overexpressed gene positions based on GRCh38.p2 (NCBI).
Figure 13.
Heat-map of the top 100 of 6,038 genes. The depth of color represents the expression value of the gene.
In silico analysis
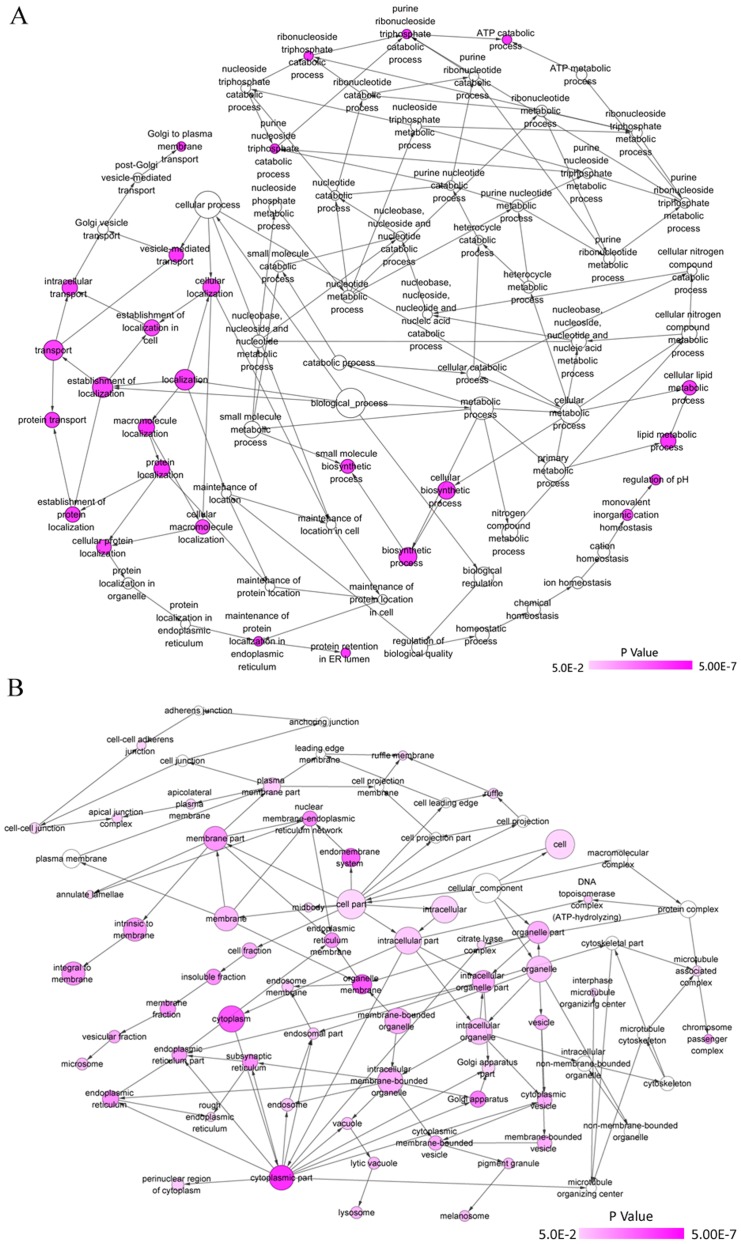
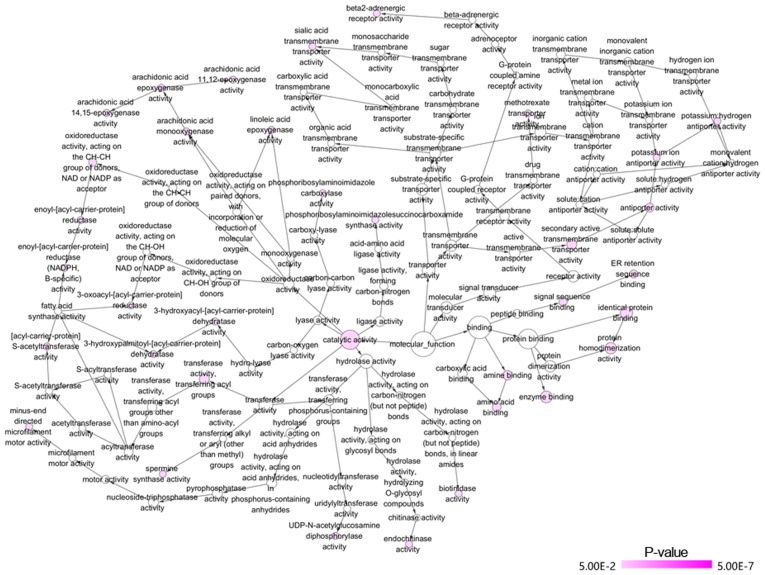
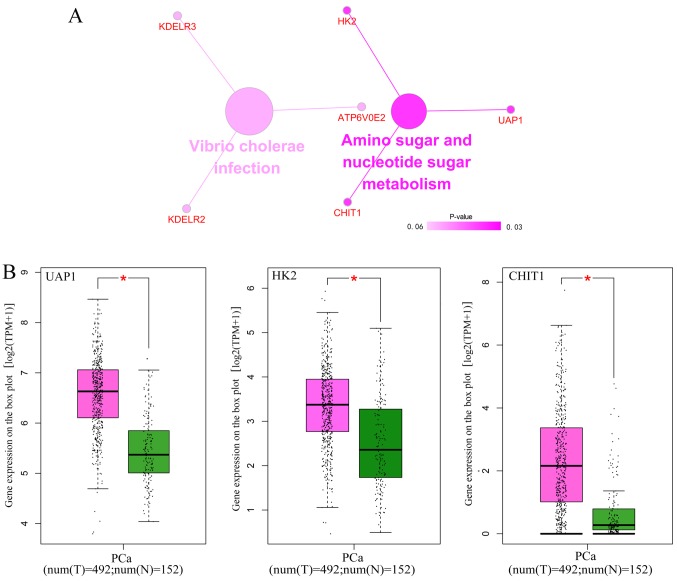
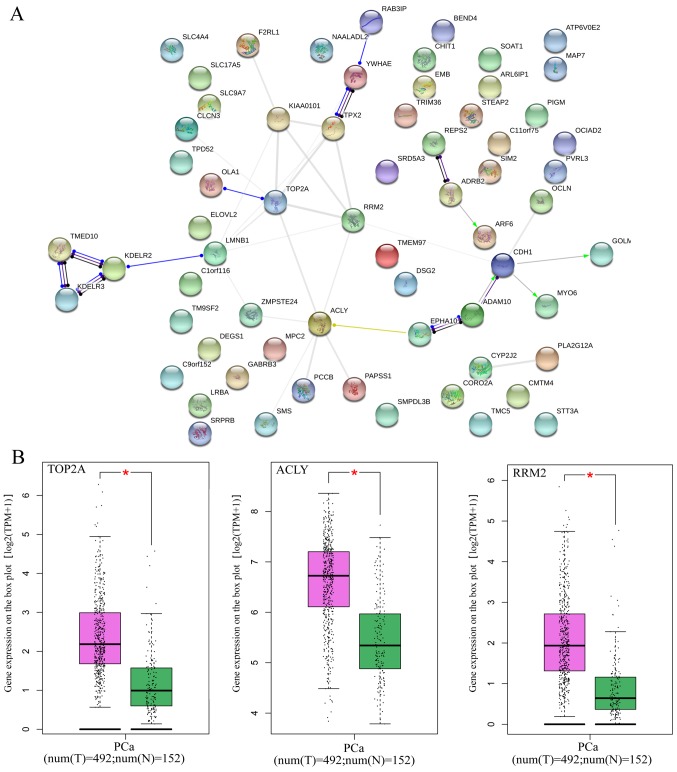
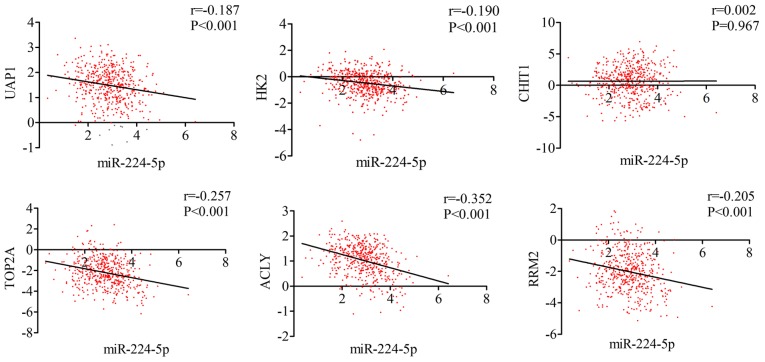
GO and KEGG analyses were performed for the 82 overlapped target genes using DAVID, which indicated that the GO terms were enriched in the following pathways: Intracellular transport, vesicle-mediated transport, protein transport of biological processes; Golgi apparatus, membrane fraction, insoluble fraction of cellular components; protein homodimerization activity, protein dimerization activity, identical protein binding of molecular functions (Table VI and Figs. 14 and 15). KEGG analysis demonstrated that these 82 genes were simply enriched in two pathways: Amino sugar and nucleotide sugar metabolism, and Vibrio cholerae infection. Significance was only found in the amino sugar and nucleotide sugar metabolism pathway (P<0.05; Table VI and Fig. 16A), in which UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1), hexokinase 2 (HK2) and chitinase 1 (CHIT1) were notably upregulated in PCa tissues (P<0.05; Fig. 16B). The PPI analysis suggested that DNA topoisomerase 2-α (TOP2A), ATP citrate lyase (ACLY) and ribonucleotide reductase regulatory subunit M2 (RRM2) were key genes for protein interaction, which had higher expression levels in PCa tissues (P<0.05; Table VII and Fig. 17). miR-224-5p may have negative axial associations with the expression of UAP1, HK2, CHIT1, TOP2A, ACLY and RRM2 in PCa, which requires further experiments for confirmation (Fig. 18).
Table VI.
Top 10 most significant GO terms of the potential target genes of microRNA-224-5-p in prostate adenocarcinoma.
| Category | Term | Count | P-value | FDR |
|---|---|---|---|---|
| Biological process | ||||
| GOTERM_BP_FAT | GO:0046907~intracellular transport | 10 | 0.002874 | 4.327707 |
| GOTERM_BP_FAT | GO:0016192~vesicle-mediated transport | 9 | 0.004584 | 6.818167 |
| GOTERM_BP_FAT | GO:0015031~protein transport | 10 | 0.007548 | 10.992314 |
| GOTERM_BP_FAT | GO:0045184~establishment of protein localization | 10 | 0.007995 | 11.607428 |
| GOTERM_BP_FAT | GO:0006163~purine nucleotide metabolic process | 5 | 0.010383 | 14.821117 |
| GOTERM_BP_FAT | GO:0006885~regulation of pH | 3 | 0.010551 | 15.043556 |
| GOTERM_BP_FAT | GO:0034613~cellular protein localization | 7 | 0.011114 | 15.783945 |
| GOTERM_BP_FAT | GO:0070727~cellular macromolecule localization | 7 | 0.011491 | 16.275859 |
| GOTERM_BP_FAT | GO:0008610~lipid biosynthetic process | 6 | 0.016047 | 22.014591 |
| GOTERM_BP_FAT | GO:0006605~protein targeting | 5 | 0.016883 | 23.026892 |
| Cellular component | ||||
| GOTERM_CC_FAT | GO:0005794~Golgi apparatus | 15 | 0.000193 | 0.241372 |
| GOTERM_CC_FAT | GO:0005624~membrane fraction | 14 | 0.000337 | 0.419916 |
| GOTERM_CC_FAT | GO:0005626~insoluble fraction | 14 | 0.000477 | 0.595260 |
| GOTERM_CC_FAT | GO:0005783~endoplasmic reticulum | 15 | 0.000517 | 0.644979 |
| GOTERM_CC_FAT | GO:0042598~vesicular fraction | 7 | 0.001999 | 2.470347 |
| GOTERM_CC_FAT | GO:0000267~cell fraction | 14 | 0.004767 | 5.799806 |
| GOTERM_CC_FAT | GO:0031090~organelle membrane | 14 | 0.005271 | 6.394020 |
| GOTERM_CC_FAT | GO:0005768~endosome | 7 | 0.006966 | 8.368649 |
| GOTERM_CC_FAT | GO:0016021~integral to membrane | 40 | 0.008019 | 9.576264 |
| GOTERM_CC_FAT | GO:0005792~microsome | 6 | 0.009016 | 10.705916 |
| Molecular function | ||||
| GOTERM_MF_FAT | GO:0042803~protein homodimerization activity | 8 | 0.000927 | 1.226713 |
| GOTERM_MF_FAT | GO:0046983~protein dimerization activity | 9 | 0.003674 | 4.779822 |
| GOTERM_MF_FAT | GO:0042802~identical protein binding | 9 | 0.009776 | 12.255478 |
| GOTERM_MF_FAT | GO:0043176~amine binding | 4 | 0.014922 | 18.132938 |
| GOTERM_MF_FAT | GO:0046982~protein heterodimerization activity | 5 | 0.016408 | 19.761183 |
| GOTERM_MF_FAT | GO:0046923~ER retention sequence binding | 2 | 0.018664 | 22.176377 |
| GOTERM_MF_FAT | GO:0031406~carboxylic acid binding | 4 | 0.030136 | 33.449456 |
| GOTERM_MF_FAT | GO:0016597~amino acid binding | 3 | 0.032286 | 35.386583 |
| GOTERM_MF_FAT | GO:0019899~enzyme binding | 7 | 0.035546 | 38.224143 |
| GOTERM_MF_FAT | GO:0015297~antiporter activity | 3 | 0.040576 | 42.376479 |
| KEGG category | Term | Count | P-value | Genes |
| KEGG_PATHWAY | hsa00520: Amino sugar and nucleotide sugar metabolism | 3 | 0.038278 | UAP1, HK2, CHIT1 |
| KEGG_PATHWAY | hsa05110: Vibrio cholerae infection | 3 | 0.059125 | KDELR3, KDELR2 ATP6V0E2 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate.
Figure 14.
(A) Network of biological processes of GO terms of miR-224-5p potential target genes in PCa was constructed with the Bingo plugin of Cytoscape v3.5.0. Each node represents a GO term, node color indicates the P-value of a GO term. (B) Network of cellular components of GO terms of miR-224-5p potential target genes in PCa was constructed with the Bingo plugin of Cytoscape v3.5.0. Each node represents a GO term, the node color indicates the P-value of a GO term. miR, microRNA; PCa, prostate adenocarcinoma; GO, Gene Ontology.
Figure 15.
Network of molecular function of GO terms of miR-224-5p potential target genes in PCa was constructed with the Bingo plugin of Cytoscape v3.5.0. Each node represents a GO term, the node color indicates the P-value of a GO term. miR, microRNA; PCa, prostate adenocarcinoma; GO, Gene Ontology.
Figure 16.
(A) Signaling pathways of Kyoto Encyclopedia of Genes and Genomes-enriched miR-224-5p potential targets genes in PCa constructed using the ClueGO plugin. The color of circles represents the degree of enrichment of the pathway (P-value). (B) Potential target genes of miR-224-5p between PCa tissue and normal prostate tissue. Pink indicates the PCa group, green indicates the normal group (*P<0.05). miR, microRNA; PCa, prostate adenocarcinoma; UAP1, UDP-N-acetylglucosamine pyrophosphorylase 1; HK2, hexokinase 2; CHIT1, chitinase 1.
Table VII.
Top 10 genes with combined scores in the protein-protein interaction network of potential target genes of microRNA-224-5p in prostate adenocarcinoma.
| Node 1 | Node 2 | Node 1 string internal ID | Co-expression | Experimentally determined interaction | Database annotated | Automated text mining | Combined score |
|---|---|---|---|---|---|---|---|
| TOP2A | RRM2 | 1860923 | 0.880 | 0.000 | 0.000 | 0.673 | 0.960 |
| MYO6 | CDH1 | 1855148 | 0.000 | 0.360 | 0.900 | 0.214 | 0.945 |
| CDH1 | ADAM10 | 1844988 | 0.050 | 0.091 | 0.900 | 0.409 | 0.942 |
| TMED10 | KDELR2 | 1848494 | 0.310 | 0.137 | 0.900 | 0.105 | 0.939 |
| KDELR3 | TMED10 | 1859221 | 0.270 | 0.137 | 0.900 | 0.105 | 0.936 |
| TOP2A | KIAA0101 | 1860923 | 0.926 | 0.000 | 0.000 | 0.145 | 0.934 |
| RAB3IP | YWHAE | 1862399 | 0.000 | 0.346 | 0.900 | 0.000 | 0.931 |
| TOP2A | TPX2 | 1860923 | 0.879 | 0.000 | 0.000 | 0.340 | 0.916 |
| EPHA10 | ADAM10 | 1855819 | 0.053 | 0.000 | 0.900 | 0.111 | 0.908 |
| ADRB2 | ARF6 | 1848724 | 0.000 | 0.000 | 0.900 | 0.112 | 0.907 |
| TOP2A | RRM2 | 1860923 | 0.880 | 0.000 | 0.000 | 0.673 | 0.960 |
| MYO6 | CDH1 | 1855148 | 0.000 | 0.360 | 0.900 | 0.214 | 0.945 |
| CDH1 | ADAM10 | 1844988 | 0.050 | 0.091 | 0.900 | 0.409 | 0.942 |
| TMED10 | KDELR2 | 1848494 | 0.310 | 0.137 | 0.900 | 0.105 | 0.939 |
| KDELR3 | TMED10 | 1859221 | 0.270 | 0.137 | 0.900 | 0.105 | 0.936 |
Figure 17.
(A) Protein-protein interaction network analysis of miR-224-5p potential target genes in PCa. (B) Hub genes of the expression of miR-224-5p between PCa tissue and normal prostate tissue. Red indicates the cancer group, green indicates the normal group (*P<0.05). miR, microRNA; PCa, prostate adenocarcinoma; TOP2A, DNA topoisomerase 2-α; ACLY, ATP citrate lyase; RRM2, ribonucleotide reductase regulatory subunit M2.
Figure 18.
Correlation between the targets (UAP1, HK2, CHIT1, TOP2A, ACLY, RRM2) and miR-224-5p in prostate adenocarcinoma. miR, microRNA; UAP1, UDP-N-acetylglucosamine pyrophosphorylase 1; HK2, hexokinase 2; CHIT1, chitinase 1; TOP2A, DNA topoisomerase 2-α; ACLY, ATP citrate lyase; RRM2, ribonucleotide reductase regulatory subunit M2.
Discussion
In the present study, referring to data in the TCGA, GEO, ArrayExpress and previous literature, it was confirmed that the expression of miR-224-5p was notably downregulated in PCa tissues, biofluids and cell lines, and its downregulated expression may be associated with the progression of PCa. In addition, prospective target genes of miR-224-5p in PCa were identified via prediction tools and microarrays of differentially expressed mRNAs in TCGA and GEO. Bioinformatics analysis was subsequently performed on these potential genes. GO and KEGG analyses revealed that miR-224-5p mediated the expression of UAP1, HK2 and CHIT1 in order to regulate the amino sugar and nucleotide sugar metabolism signaling pathway, thus exerting its effects in PCa. miR-224-5p may also be vital in PCa by being involved in the protein interaction through TOP2A, ACLY and RRM2.
Previous studies have demonstrated that the abnormal expression of miR-224-5p was a crucial factor in the initiation and progress of tumors. For example, the high expression of miR-224-5p was likely to be involved in the onset of digestive tract malignancy (38); the expression of miR-224-5p was significantly reduced in mucinous breast cancer (39); and cell experiments and histologic examination performed by Zheng et al revealed that miR-224-5p exhibited lower expression in uveal melanoma (40). However, studies on the expression of miR-224-5p in PCa have been limited. Only a small number of studies with small sample sizes have found that miR-224-5p was expressed at a low level in prostatic cancer (26–28), and this was not confirmed by a second study involving a larger sample size. Therefore, these findings may, to a certain extent, contain errors. The present study, using data from TCGA, GEO and ArrayExpress databases, and previous literature, investigated the expression of miR-224-5p in PCa from PCa tissues, biofluids and cell lines. Taking advantage of the public data, it was confirmed that the expression of miR-224-5p was low in PCa. The results of a study by Fu et al indicated that downregulated miR-224-5p was closely associated with terminal clinical staging and metastasis (27). The present study, based on analysis of TCGA data, revealed that the expression of pre-miR-224 was decreased only in the T stage and M stage of PCa, although additional investigations are required to confirm its expression in other stages. Furthermore, when investigating the association between miR-224-5p and prognosis, Lin et al found that the low expression of miR-224-5p was associated with a poorer RFS rate (41). Wan et al examined the recurrence rate of patients with PCa following surgery, and found that downregulated miR-224-5p was closely associated with RFS rate (42). Mavridis et al investigated the survival rate of 58 patients with PCa, and confirmed that the lower the expression of miR-224-5p, the poorer the RFS rate of the patients (26). Fu et al examined 20 patients of PCa and demonstrated that those patients with upregulated miR-224-5p tended to have improved OS rates (27). In the present study, data on the survival rate of 500 patients with PCa was acquired from TCGA. No significant association was found between the expression of pre-miR224 and the OS rates of patients, with additional follow-up investigations required to confirm this.
Bioinformatics analysis was used in the present study to examine the molecular mechanism of miR-224-5p in PCa. KEGG analysis indicated that potential target mRNAs were significantly enriched in the amino sugar and nucleotide sugar metabolism signaling pathway. Previous studies have demonstrated that this pathway may be involved in carbohydrate metabolism in tumor tissues, affecting the onset and development of cancer (43,44). Genes enriched in this pathway included UAP1, HK2 and CHIT1, all of which exhibited notably high expression in PCa tissues. It was demonstrated that higher expression of UAP1 in PCa tissues accelerated the growth of cancer cells (45). In addition, a high expression of HK2 in PCa is essential in cell proliferation, apoptosis and carbohydrate metabolism (46–48). Although experiments have shown that the differentially expressed CHIT1 is correlated with colon carcinoma (49), its association with PCa has not been elucidated. Additionally, analysis of three key genes in the PPI network, TOP2A, ACLY and RRM2, revealed that the expression levels of these three genes were markedly elevated. A study by Schaeferklein et al in 2015 showed that the increased expression of TOP2A in PCa stimulated the growth and proliferation of PCa cells via androgen receptor (50). Similarly, a study by Shah et al in 2016 further elucidated the correlations of ACLY with androgen receptor gene expression, and the proliferation and apoptosis of PCa cells. Xin et al found that ACLY inhibited the generation of fat in PCa tissues via a targeting association, which may suppress the growth and metastasis of PCA cells (51,52). RRM2 has been shown to convert ribonucleotide into deoxynucleotide and be involved in multiple biological processes, including the synthesis of DNA and cell growth. In vitro experiments have demonstrated that upregulated RRM2 promotes the proliferation and metastasis of tumor cells in PCa (53,54). Several studies have examined miR-224-5p targeting mRNA. Wan et al confirmed that miR-224-5p targeted and upregulated APLN, and these were involved in the onset and progression of PCa (42). Fu et al identified CAMKK2 as the target gene of miR-224-5p, and noted the negative axial regulatory associations between them; these 2 genes were also found to collaborate to promote the progression of PCa (27). According to the results of the present study, the target genes of miR-224-5p in PCa may include UAP1, HK2, CHIT1, TOP2A, ACLY and RRM2. miR-224-5p is likely to regulate the expression of these genes, therefore performing its functions in the initiation and development of PCa. However, these results based on theoretical analysis require additional in vivo and in vitro experiments for confirmation.
In conclusion, the present study, based on data from TCGA, GEO and ArrayExpress databases, and previous literature, examined the expression of miR-224-5p in PCa and its clinical significance. It was confirmed that the expression of miR-224-5p was low in PCa and was associated with its clinical progression. In addition, miR-224-5p may exert its effects in PCa by mediating UAP1, HK2, CHIT1, TOP2A, ACLY and RRM2. The present study provides a theoretical foundation for future investigations on the oncogenesis of PCa.
Acknowledgements
The authors would like to thank all members of the Molecular Oncology Group of the First Affiliated Hospital of Guangxi Medical University (Nanning, Guangxi Zhuang Autonomous Region 530021, China) for their professional suggestions. FCM, JM and JCZ reviewed and edited the manuscript.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
BLG and XHH and GC conceived and designed the study. BLG, LJZ, LG and RQH performed the experiments. BLG and GC wrote the paper. BLG, LJZ, LG, RQH, GC and XHH reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
This study does not involve animal or human related experiments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.An H, Tao N, Li J, Guan Y, Wang W, Wang Y, Wang F. detection of prostate cancer metastasis by whole body magnetic resonance imaging combined with bone scintigraphy and PSA Levels. Cell Physiol Biochem. 2016;40:1052–1062. doi: 10.1159/000453161. [DOI] [PubMed] [Google Scholar]
- 3.Chen SL, Wang SC, Ho CJ, Kao YL, Hsieh TY, Chen WJ, Wu PR, Ko JL, Lee H, Sung WW. Prostate cancer mortality-to-incidence ratios are associated with cancer care disparities in 35 countries. Sci Rep. 2017;7:400003. doi: 10.1038/srep40003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan XW, Gan SS, Ye JQ, Fan YH, Hong Υ, Chu CM, Gao Y, Li L, Liu X, Chen L, et al. SMC1A promotes growth and migration of prostate cancer in vitro and in vivo. Int J Oncol. 2016;49:1963–1972. doi: 10.3892/ijo.2016.3697. [DOI] [PubMed] [Google Scholar]
- 5.Ost P, Bossi A, Decaestecker K, De Meerleer G, Giannarini G, Karnes RJ, Roach M, III, Briganti A. Metastasis-directed therapy of regional and distant recurrences after curative treatment of prostate cancer: A systematic review of the literature. Eur Urol. 2015;67:852–863. doi: 10.1016/j.eururo.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Pettersson A, Robinson D, Garmo H, Holmberg L, Stattin P. Age at diagnosis and prostate cancer treatment and prognosis: A population-based cohort study. Ann Oncol. 2018;29:377–385. doi: 10.1093/annonc/mdx742. [DOI] [PubMed] [Google Scholar]
- 7.Jemal A, Fedewa SA, Ma J, Siegel R, Lin CC, Brawley O, Ward EM. prostate cancer incidence and PSA testing patterns in relation to USPSTF screening recommendations. JAMA. 2015;314:2054–2061. doi: 10.1001/jama.2015.14905. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Chen Z, Wang T, Liu L, Zhao L, Guo G, Wang D. influence of four radiotracers in PET/CT on diagnostic accuracy for prostate cancer: A Bivariate Random-Effects Meta-Analysis. Cell Physiol Biochem. 2016;39:467–480. doi: 10.1159/000445639. [DOI] [PubMed] [Google Scholar]
- 9.Barrington WE, Schenk JM, Etzioni R, Arnold KB, Neuhouser ML, Thompson IM, Jr, Lucia MS, Kristal AR. Difference in association of obesity with prostate cancer risk between US African American and Non-Hispanic White Men in the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA Oncol. 2015;1:342–349. doi: 10.1001/jamaoncol.2015.0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vidal AC, Howard LE, Sun SX, Cooperberg MR, Kane CJ, Aronson WJ, Terris MK, Amling CL, Freedland SJ. Obesity and prostate cancer-specific mortality after radical prostatectomy: Results from the Shared Equal Access Regional Cancer Hospital (SEARCH) database. Prostate Cancer Prostatic Dis. 2017;20:72–78. doi: 10.1038/pcan.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: Νew trends in the development of miRNA therapeutic strategies in oncology (Review) Int J Oncol. 2016;49:1–32. doi: 10.3892/ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y, Feng B, Han S, Lu L, Chen Y, Chu X, Wang R, Chen L. MicroRNA-129 in human cancers: From tumorigenesis to clinical treatment. Cell Physiol Biochem. 2016;39:2186–2202. doi: 10.1159/000447913. [DOI] [PubMed] [Google Scholar]
- 13.Paul P, Chakraborty A, Sarkar D, Langthasa M, Rahman M, Bari M, Singha RS, Malakar AK, Chakraborty S. Interplay between miRNAs and Human Diseases: A Review. J Cell Physiol. 2017;233:2007–2018. doi: 10.1002/jcp.25854. [DOI] [PubMed] [Google Scholar]
- 14.Xie T, Huang M, Wang Y, Wang L, Chen C, Chu X. MicroRNAs as regulators, biomarkers and therapeutic targets in the drug resistance of colorectal cancer. Cell Physiol Biochem. 2016;40:62–76. doi: 10.1159/000452525. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Tang W, Li R, He R, Gan T, Luo Y, Chen G, Rong M. Downregulation of microRNA-132 indicates progression in hepatocellular carcinoma. Exp Ther Med. 2016;12:2095–2101. doi: 10.3892/etm.2016.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang WT, Wang HL, Yang H, Ren FH, Luo YH, Huang CQ, Liang YY, Liang HW, Chen G, Dang YW. Lower expressed miR-198 and its potential targets in hepatocellular carcinoma: A clinicopathological and in silico study. OncoTargets Ther. 2016;9:5163–5180. doi: 10.2147/OTT.S108828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang C, Liu M, Fang W, Guo J, Zhang Z, Wu P, Zhang Y, Wang J. MiR-139-5p is increased in the peripheral blood of patients with prostate cancer. Cell Physiol Biochem. 2016;39:1111–1117. doi: 10.1159/000447819. [DOI] [PubMed] [Google Scholar]
- 18.van Beijnum JR, Giovannetti E, Poel D, Nowak-Sliwinska P, Griffioen AW. miRNAs: Micro-managers of anticancer combination therapies. Angiogenesis. 2017;20:269–285. doi: 10.1007/s10456-017-9564-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, Zhuang Y, Jiang S, Liu S, Zhou J, Wu J, Teng Y, Xia B, Wang R, Zou X. Interaction between Wnt/β-catenin pathway and microRNAs regulates epithelial-mesenchymal transition in gastric cancer (Review) Int J Oncol. 2016;48:2236–2246. doi: 10.3892/ijo.2016.3480. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Pang YY, He RQ, Lin P, Cen JM, Yang H, Ma J, Chen G. Diagnostic value of strand-specific miRNA-101-3p and miRNA-101-5p for hepatocellular carcinoma and a bioinformatic analysis of their possible mechanism of action. FEBS Open Bio. 2017;8:64–84. doi: 10.1002/2211-5463.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Q, Huang X, Urology DO. Expression of miRNA-32, −196a,-218,-128 and let7i in serum of patients with prostate cancer and its clinical significance. Guangdong Yixue. 2016 [Google Scholar]
- 22.Pan Y, Gan Q, Jun LU, Huang T, Mang KE. Serological determination and its clinical significance of miRNA-129 and miRNA-21 in prostate cancer patients. Chinese Journal of Health Laboratory Technology. 2016 [Google Scholar]
- 23.Yao CH, Yuan XC, Liu C, Deng JP. Expression and diagnostic value of miRNA-15a in prostate cancer. Chin J Immunol. 2016 [Google Scholar]
- 24.Lu S, Wang MS, Chen PJ, Ren Q, Bai P. miRNA-186 inhibits prostate cancer cell proliferation and tumor growth by targeting YY1 and CDK6. Exp Ther Med. 2017;13:3309–3314. doi: 10.3892/etm.2017.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian XM, Luo YZ, He P, Li J, Ma ZW, An Y. Inhibition of invasion and migration of prostate cancer cells by miRNA-509-5p via targeting MDM2. Genet Mol Res. 16 doi: 10.4238/gmr16019195. doi: 10.4238/gmr16019195. [DOI] [PubMed] [Google Scholar]
- 26.Mavridis K, Stravodimos K, Scorilas A. Downregulation and prognostic performance of microRNA 224 expression in prostate cancer. Clin Chem. 2013;59:261–269. doi: 10.1373/clinchem.2012.191502. [DOI] [PubMed] [Google Scholar]
- 27.Fu H, He HC, Han ZD, Wan YP, Luo HW, Huang YQ, Cai C, Liang YX, Dai QS, Jiang FN, et al. MicroRNA-224 and its target CAMKK2 synergistically influence tumor progression and patient prognosis in prostate cancer. Tumour Biol. 2015;36:1983–1991. doi: 10.1007/s13277-014-2805-0. [DOI] [PubMed] [Google Scholar]
- 28.Kristensen H, Haldrup C, Strand S, Mundbjerg K, Mortensen MM, Thorsen K, Ostenfeld MS, Wild PJ, Arsov C, Goering W, et al. Hypermethylation of the GABRE~miR-452~miR-224 promoter in prostate cancer predicts biochemical recurrence after radical prostatectomy. Clin Cancer Res. 2014;20:2169–2181. doi: 10.1158/1078-0432.CCR-13-2642. [DOI] [PubMed] [Google Scholar]
- 29.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen M, Wang J, Luo Y, Huang K, Shi X, Liu Y, Li J, Lai Z, Xue S, Gao H, et al. Identify Down syndrome transcriptome associations using integrative analysis of microarray database and correlation-interaction network. Hum Genomics. 2018;12:2. doi: 10.1186/s40246-018-0133-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He X, Zhang C, Shi C, Lu Q. Meta-analysis of mRNA expression profiles to identify differentially expressed genes in lung adenocarcinoma tissue from smokers and non-smokers. Oncol Rep. 2018;39:929–938. doi: 10.3892/or.2018.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Shi W, Wan Y, Wang Q, Feng W, Yan X, Wang J, Chai L, Zhang Q, Li M. Prediction of target genes for miR-140-5p in pulmonary arterial hypertension using bioinformatics methods. FEBS Open Bio. 2017;7:1880–1890. doi: 10.1002/2211-5463.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang L, Zeng JH, Wang JY, He RQ, Ma J, Chen G, Cai XY, Hu XH. Down-regulation of miR-26a-5p in hepatocellular carcinoma: A qRT-PCR and bioinformatics study. Pathol Res Pract. 2017;213:1494–1509. doi: 10.1016/j.prp.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Mi B, Liu G, Zhou W, Lv H, Liu Y, Liu J. Identification of genes and pathways in the synovia of women with osteoarthritis by bioinformatics analysis. Mol Med Rep. 2018;17:4467–4473. doi: 10.3892/mmr.2018.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Peng Z, Wang K, Qi Y, Yang Y, Zhang Y, An X, Luo S, Zheng J. NDUFA4L2 is associated with clear cell renal cell carcinoma malignancy and is regulated by ELK1. PeerJ. 2017;5:e4065. doi: 10.7717/peerj.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei D. A multigene support vector machine predictor for metastasis of cutaneous melanoma. Mol Med Rep. 2018;17:2907–2914. doi: 10.3892/mmr.2017.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Liu J, Fu X, Yang A. Identification of key genes and pathways in tongue squamous cell carcinoma using bioinformatics analysis. Med Sci Monit. 2017;23:5924–5932. doi: 10.12659/MSM.905035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Huang LS, Chen G, Feng ZB. Potential targets and clinical value of miR-224-5p in cancers of the digestive tract. Cell Physiol Biochem. 2017;44:682–700. doi: 10.1159/000485281. [DOI] [PubMed] [Google Scholar]
- 39.Zhou F, Li S, Meng HM, Qi LQ, Gu L. MicroRNA and histopathological characterization of pure mucinous breast carcinoma. Cancer Biol Med. 2013;10:22–27. doi: 10.7497/j.issn.2095-3941.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng X, Tang H, Zhao X, Sun Y, Jiang Y, Liu Y. Long non-coding RNA FTH1P3 facilitates uveal melanoma cell growth and invasion through miR-224-5p. PLoS One. 2017;12:e0184746. doi: 10.1371/journal.pone.0184746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Lin ZY, Huang YQ, Zhang YQ, Han ZD, He HC, Ling XH, Fu X, Dai QS, Cai C, Chen JH, et al. MicroRNA-224 inhibits progression of human prostate cancer by downregulating TRIB1. Int J Cancer. 2014;135:541–550. doi: 10.1002/ijc.28707. [DOI] [PubMed] [Google Scholar]
- 42.Wan Y, Zeng ZC, Xi M, Wan S, Hua W, Liu YL, Zhou YL, Luo HW, Jiang FN, Zhong WD. Dysregulated microRNA-224/apelin axis associated with aggressive progression and poor prognosis in patients with prostate cancer. Hum Pathol. 2015;46:295–303. doi: 10.1016/j.humpath.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 43.Cao J, Lu XX, Li Y, Zhu LQ, Yang C, Ou C, Tang YP. Applying gene set enrichment analysis and meta-analysis to screen key genes controlling the development and progression of hepatic carcinoma. World Chin J Digestology. 2012;20:754. doi: 10.11569/wcjd.v20.i9.754. (In Chinese) [DOI] [Google Scholar]
- 44.Li Z, Li BQ, Jiang M, Chen L, Zhang J, Liu L, Huang T. Prediction and analysis of retinoblastoma related genes through gene ontology and KEGG. Biomed Res Int. 2013;2013:304029. doi: 10.1155/2013/304029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itkonen HM, Engedal N, Babaie E, Luhr M, Guldvik IJ, Minner S, Hohloch J, Tsourlakis MC, Schlomm T, Mills IG. UAP1 is overexpressed in prostate cancer and is protective against inhibitors of N-linked glycosylation. Oncogene. 2015;34:3744–3750. doi: 10.1038/onc.2014.307. [DOI] [PubMed] [Google Scholar]
- 46.Kudryavtseva AV, Nyushko KM, Zaretsky AR, et al. Expression of HK2 gene is deregulated in Prostate cancer. Asian J Pharm. 2017;11:S158–S161. [Google Scholar]
- 47.Tao T, Chen M, Jiang R, Guan H, Huang Y, Su H, Hu Q, Han X, Xiao J. Involvement of EZH2 in aerobic glycolysis of prostate cancer through miR-181b/HK2 axis. Oncol Rep. 2017;37:1430–1436. doi: 10.3892/or.2017.5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tao T, Xiang P, Huang T. Expression of HK2 in prostate cancer and its effect on malignant phenotype of prostate cancer cells. Chinese Journal of Clinical & Experimental Pathology. 2017 [Google Scholar]
- 49.Li FF, Yan P, Zhao ZX, Liu Z, Song DW, Zhao XW, Wang XS, Wang GY, Liu SL. Polymorphisms in the CHIT1 gene: Associations with colorectal cancer. Oncotarget. 2016;7:39572–39581. doi: 10.18632/oncotarget.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaefer-Klein JL, Murphy SJ, Johnson SH, Vasmatzis G, Kovtun IV. Topoisomerase 2 alpha cooperates with androgen receptor to contribute to prostate cancer progression. PLoS One. 2015;10:e0142327. doi: 10.1371/journal.pone.0142327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah S, Carriveau WJ, Li J, Campbell SL, Kopinski PK, Lim HW, Daurio N, Trefely S, Won KJ, Wallace DC, et al. Targeting ACLY sensitizes castration-resistant prostate cancer cells to AR antagonism by impinging on an ACLY-AMPK-AR feedback mechanism. Oncotarget. 2016;7:43713–43730. doi: 10.18632/oncotarget.9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xin M, Qiao Z, Li J, Liu J, Song S, Zhao X, Miao P, Tang T, Wang L, Liu W, et al. miR-22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: Evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget. 2016;7:44252–44265. doi: 10.18632/oncotarget.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afrasiabi A, Fiuji F, Mirhafez R, Avan A. RRM2 (ribonucleotide reductase M2) Atlas Genet Cytogenet Oncol Haematol. 2015;19:p32–37. [Google Scholar]
- 54.Huang Y, Liu X, Wang YH, Yeh SD, Chen CL, Nelson RA, Chu P, Wilson T, Yen Y. The prognostic value of ribonucleotide reductase small subunit M2 in predicting recurrence for prostate cancers. Urol Oncol. 2014;32:51.e9–51.e19. doi: 10.1016/j.urolonc.2013.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.