Abstract
The interferon-induced transmembrane protein 3 (IFITM3, also called 1-8U) gene represents dysregulated expression in various tumors and is involved in tumorigenesis and progression. However, the role of IFITM3 and its underlying mechanism in hepatocellular carcinoma (HCC) are still far from elucidated. MicroRNAs (miRNAs), a class of endogenous (approximately 22 nucleotides) small noncoding RNAs, can post-transcriptionally regulate gene expression by repressing protein translation or silencing the expression of target genes that play critical roles in various cancers. miR-29a was identified as being aberrantly expressed in a significant proportion of HCC. However, the correlation between IFITM3 and miR-29a has not been reported to date. In this study, we investigated the expression of IFITM3 in HCC and its effect on the biological behavior of HCC cells as well as the association between IFITM3 and miR-29a. We determined that IFITM3 was upregulated and miR-29a downregulated in HCC tissues and that they were associated with HCC tumor size, tumor multifocal, and venous invasion. The expression of IFITM3 in HCC tissues was negatively correlated with miR-29a expression. Additionally, IFITM3 overexpression and miR-29a nonoverexpression were related to poor prognosis of HCC patients. Knockdown of IFITM3 inhibited migration, invasion, proliferation and promoted apoptosis of HCC cells, which are consistent with the effects of upregulated miR-29a. Additionally, after upregulation of IFITM3, the invasion, migration and proliferation abilities of HL-7702 cells were increased, but the apoptosis rate was decreased. Furthermore, using a Dual-Luciferase reporter gene assay, we identified IFITM3 as a new functional target gene of miR-29a. In conclusion, our findings demonstrated that the migration, invasion, proliferation and apoptosis features of HCC cells could be regulated by miR-29a via IFITM3. Thus, the present study indicated that miR-29a and IFITM3 play critical roles in the development and progression of HCC, revealing that miR-29a and IFITM3 may be novel potential therapeutic targets for patients with HCC.
Keywords: miR-29a, IFITM3, hepatocellular carcinoma, migration, invasion, proliferation, apoptosis
Introduction
Liver cancer, a fatal disease, is one of the most common cancers and the leading cause of cancer-related mortality worldwide (1). According to the latest statistics, in the United States in 2017, it is estimated that approximately 40,710 new cases of liver cancer will be diagnosed and approximately 28,920 cases, equivalent to 71% of these diagnosed cases, will eventually suucumb to this disease, and the 5-year relative survival rate for liver cancer is currently only 18% (2). Hepatocellular carcinoma (HCC), the most common primary liver malignancy (accounting for 70–90%), has an unsatisfactory clinical efficacy due to the high incidence of recurrence and metastasis even after curative resection (1,3,4). In addition, sorafenib, a multi-kinase inhibitor, is a drug that is available for the treatment of patients with advanced HCC, however it only modestly improves survival, while a range of side-effects commonly exist and most patients will eventually become drug-resistant (5,6). Therefore, its clinical application is restricted. The main reason for the unsatisfactory treatment for HCC is that the underlying molecular mechanism of HCC oncogenesis remains unclear. Hence, it is urgent to elucidate the mechanism involved in HCC progression and to identify novel therapeutic targets to improve the clinical outcome.
The interferon-induced transmembrane protein 3 (IFITM3, also called 1–8 U) gene (along with IFITM1 and IFITM2) belongs to the IFITM family of genes, which are clustered on chromosome 11 and have been identified as antiviral factors that interfere with viral entry following endocytosis (7–9). In recent years, an increasing number of studies have found dysregulated expression of IFITM3 in various tumors (including colon cancer, colorectal tumor and glioma), which is involved in cell migration and invasion, tumorigenesis and progression (10–12). However, the clinical significance and underlying mechanisms of aberrant IFITM3 expression in HCC have not been investigated and remain to be established.
MicroRNAs (miRNAs), a family of endogenous (approximately 22 nucleotides) small noncoding RNAs, are able to post-transcriptionally regulate gene expression by repressing protein translation or silencing the expression of target genes by directly binding to the 3′-untranslated region (3′-UTR) of messenger RNAs (mRNAs) (13–15). miRNAs play important roles in various biological and pathological processes of HCC, including migration, invasion, carcinogenesis, tumor growth and metastasis (16–19). Several studies have reported aberrant expression of miRNA-29a (miR-29a) in HCC. miR-29a is involved in multiple processes of tumor progression, including cell proliferation, growth, migration, and epithelial-mesenchymal transition (EMT) (20–22). Furthermore, low expression of miR-29a is a poor prognostic marker in HCC patients (23).
However, the expression and biological function of IFITM3 in HCC and the association between IFITM3 and miR-29a remain unclear. Previously, we performed a bioinformatics analysis and found that a potential binding site for miR-29a exists in the 3′UTR of IFITM3 mRNA. Thus, we presumed that miR-29a could affect HCC progression by regulating IFITM3. In the present study, we aimed to investigate the role of IFITM3 and miR-29a in HCC and to identify whether IFITM3 is a new direct target gene of miR-29a. In the addition, the association between IFITM3, miR-29a expression and the clinicopathological characteristics of HCC patients was investigated.
Materials and methods
Patients and tissue specimens
HCC and paracancerous tissue specimens (confirmed by histological diagnosis) were obtained from 52 patients (37 males and 15 females) who underwent hepatectomy at the Second Affiliated Hospital of Nanchang University (Nanchang, China) from January 2012 to December 2016. Their age ranged from 31 to 74 years, and the average age was 49 years. The sex, age, tumor size, TNM stage, alpha-fetoprotein (AFP) levels and other clinicopathological characteristics of patients were obtained from surgical and pathological records. Prior to liver resection, no treatments, including radiotherapy and chemotherapy, had been carried out in these patients. After the tissue specimens were resected during surgery, HCC and paracancerous tissue specimens were immediately collected and placed in liquid nitrogen and then stored. Informed consent was obtained from all patients for using their tissue specimens and clinicopathological data. The study protocol was approved by the Medical Ethics Committee of the Second Affiliated Hospital of Nanchang University. This study was performed in accordance with the ethical standards of the Declaration of Helsinki.
Cell lines and cell culture
The human normal liver cell line (HL-7702) and 3 HCC cell lines (Hep3B, HCCLM3 and Huh-7) were selected to conduct the assays, and they were purchased from the Shanghai Institute of Cell Biology (Shanghai, China). These cell lines were cultured in high-glucose Dulbeccos modified Eagles medium (DMEM; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) supplemented with 10% fetal bovine serum (FBS; Biological Industries, Kibbutz Beit-Haemek, Israel), 100 µg/ml streptomycin and 100 U/ml penicillin at 37°C, in a 5% CO2 and 95% humidity cell culture incubator.
Cell transfection
IFITM3 siRNA, pcDNA3.1(+)-IFITM3 plasmids and negative control (NC) were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). miR-29a mimics and NC miRNA were purchased from Guangzhou RiboBio Biotechnology, Co., Ltd. (Guangzhou, China). The HCCLM3 cell lines were eventually selected as experimental subjects and assigned to the NC and treatment groups. Transfections were performed using the Lipofectamine 3000 kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturers instructions. Two different IFITM3 siRNA sequences were transfected into the cells, and the sequences were as follows: IFITM3-s1 sense, 5′-CCAUUCUGCUCAUCGUCAUTT-3′ and antisense, 5′-AUGACGAUGAGCAGAAUGGTT-3′; IFITM3-s2 sense, 5′-GCUGAUCUUCCAGGCCUAUTT-3′ and antisense, 5′-AUAGGCCUGGAAGAUCAGCTT-3′. The sequences of pcDNA3.1(+)-IFITM3 plasmids were as follows: pcDNA3.1(+)-IFITM3 sense, 5′-AAGCTGGCTAGCGTTGCGGCCGCGCCACCATGAATCACACTGTCCAAACCTT-3′; pcDNA3.1(+)-IFITM3 antisence, 5′-AGAGTCGGTACCGTCGGATCCCTATCCATAGGCCTGGAAGATCAG-3′.
Hematoxylin and eosin (H&E) and immunohistochemistry (IHC)
After the HCC and adjacent tissues were fixed in 10% formalin solution, we embedded them in paraffin blocks and then cut the tissue into paraffin sections. After deparaffnization and hydration, some sections were stained with H&E, which was used to detect morphological changes, and some sections were used for immunostaining. Then, these sections were incubated with 0.3% hydrogen peroxide for 15 min to eliminate endogenous peroxidase, and microwave-heated at 100°C for 10 min in sodium citrate buffer (10 mmol/l, pH 6.0) for antigen retrieval and blocked with goat serum. Subsequently, the tissue sections were incubated with IFITM3 rabbit monoclonal antibody at a 1:200 dilution (cat. no. ab109429; Abcam, Cambridge, MA, USA) at 4°C overnight. Subsequently, the sections were washed with phosphate-buffered saline (PBS) 3 times at 5-min intervals. The next step was to add the secondary antibody biotinylated goat anti-rabbit serum IgG at a 1:2,000 dilution (cat. no. HS101-01; TransGen Biotech, Beijing, China) to the sections and incubate them for 30 min at 37°C. Then, the sections were stained with diaminobenzidine (DAB) and hematoxylin dyes and were sealed with neutral resins. Two pathologists blindly and randomly evaluated and semi-quantitatively scored the staining intensity and percentage of positive cells. The methods that we used to grade the staining intensity and score the percentage of staining have already been reported by Liu et al (24). For each immunostained section, the overall staining index was computed by multiplying the grades and scores to reach a value from 0 to 9, which was finally designated as follows: 0–1, IFITM3 nonoverexpression; 2–9, IFITM3 overexpression.
Protein extraction and western blot analysis
Western blot analysis was conducted to detect total protein expression in tissues and treated cells after 48 h of transfection. Total protein was extracted from tissues or cells, which were lysed in radioimmunoprecipitation assay (RIPA) buffer (cat. no. R0020; Solarbio Science & Technology Co., Ltd.) with 1% phenylmethanesulfonyl fluoride (PMSF), and then, the protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein determination was based on bicinchoninic acid (BCA) method. A total of 12 ml protein samples were loaded per lane. Protein samples were electrophoresed on a 15% sodium dodecyl sulfate-polyacrylamide gel and transferred onto polyvinylidene fluoride (PVDF) membranes. Afterwards, the membranes were blocked with 5% Difco skim milk at room temperature for 2 h. The membranes were then incubated with anti-IFITM3 antibody (1:5,000 dilution) (cat. no. ab109429; Abcam) at 4°C overnight. Tris-HCl buffer solution + Tween-20 (TBST) was used to wash the membranes 3 times for 10 min. Subsequently, they were incubated with horseradish peroxidase-conjugated secondary antibody at a 1:10,000 dilution (cat. no. HS101-01; TransGen Biotech) for 1 h at room temperature. Finally, the blots were detected by enhanced chemiluminescence (ECL) kit (cat. no. cw0049s; CWBIO, Beijing, China), and the intensity was measured by Quantity One software (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The reference protein in the study is GAPDH mouse monoclonal antibody with 1:6,000 dilution (cat. no. 60004-1-Ig; Proteintech Group Inc., Rosemont, IL, USA).
Isolation of mRNA and quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA of tissues and cells was isolated with an E.Z.N.A.™ Total RNA Kit II (Omega Bio-Tek, Inc., Norcross, GA, USA). Reverse transcription was performed with a PrimeScript RT reagent kit with gDNA Eraser (cat. no. RR047A; Takara Biotechnology, Co., Ltd., Dalian, China) following the protocols of the manufacturer. qRT-PCR was performed with SYBR Premix Ex Taq™ II (cat. no. RR820A; Takara Biotechnology, Co., Ltd.) according to the manufacturers instructions. The levels of IFITM3 and miR-29a were calculated with the 2−ΔΔCq method (25) and were normalized to those of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6, respectively. We randomly chose five non-tumor tissues of HCC patients and selected their average ΔCq of miR-29a as an internal control to calculate the ΔΔCq of each HCC tissue. In addition, we selected the mean level of miR-29a relative expression as a cut-off value according to the study of Li et al (26). The HCC patients with miR-29a expression less than the mean value were classified as the non-overexpression group, while the patients higher than the median value were considered as the overexpression group. The RNA relative expression levels of the experimental group were reported as the fold change compared to the corresponding NC group, which was defined as 1.0. All assays were performed in triplicate. The specific stem-loop primer for miR-29a and U6 primer were designed by Guangzhou RiboBio Biotechnology, Co., Ltd. (Guangzhou, China). The primer sequences of GAPDH and IFITM3 were purchased from GenScript Biotechnology (Nanjing, China) and were as follows: GAPDH forward, 5′-CAGGGCTGCTTTTAACTCTGGT-3′ and reverse, 5′-GATTTTGGAGGGATCTCGCT-3′; IFITM3 forward, 5′-ACTGTCCAAACCTTCTTCTCTCC-3′ and reverse, 5′-TCGCCAACCATCTTCCTGTC-3′.
Cell wound healing assays
A wound healing assay was performed to evaluate the cell migration capacity. Cells were seeded into 6-well plates and incubated in DMEM with 10% FBS at 37°C. After the cells reached 90–100% confluence, artificial wounds were generated by scraping a 200 µl pipette tip across the cell surface. Subsequently, the cells were gently washed with PBS 3 times to remove the detached cells, and then, they were incubated in fresh complete medium for 48 h. After 48 h, we observed and measured the cell migration distance from the edge of the scratch towards the center.
Cell migration and invasion assays
To detect cell invasion and migration, 8-µm pore size Corning Transwell chambers (Corning Inc., Corning, NY, USA) with and without Matrigel-coating were used, respectively. At 48 h after transfection, 4×104 cells in serum-free medium were placed into the upper chambers for the migration assay, and 8×104 cells were seeded for invasion assays, while medium with 10% FBS was added to the lower chamber. After 24 h of incubation, the cells remaining in the upper surface of the membrane were removed, and the cells that had migrated/invaded to the lower surface of the membrane were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Cells were counted in five random fields (magnification, ×200) taken by a light microscope (Olympus Corp., Tokyo, Japan).
Cell proliferation assays
The cell proliferation ability was assessed using the 5-ethynyl-2-deoxyuridine (EdU) proliferation assay. At 24 h after transfection, treated cells were resuspended and seeded in 96-well plates (1×104 cells/well) and were continually cultured for 24 h. Subsequently, after being washed 3 times with PBS, the cells were incubated for 4 h in serum-free DMEM supplemented with 50 µM EdU (Guangzhou RiboBio Biotechnology, Co., Ltd., Guangzhou, China). Then, the cells were fixed with 4% polyformaldehyde in PBS at room temperature for 30 min. Finally, the cells were subsequently incubated with Apollo staining solution and Hoechst 33342 for 30 min. The percentage of EdU-positive cells relative to the total number of cells was intended to represent the proliferation index. A fluorescence microscope (Olympus Corp.) was used to obtain images, and five random fields (magnification, ×200) were selected to evaluate the proliferation rate.
Cell apoptosis assays
Cell apoptosis was assessed by fluorescence activated cell sorting (FACS) analysis using the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (BD Biosciences, Franklin Lakes, NJ, USA). In brief, cells were firstly digested with trypsin and then collected and rinsed with pre-cooled PBS at 4°C; then, they were resuspended in 100 µl of 1X binding buffer and placed into a plastic 12×75 mm test tube, followed by the addition of 5 µl of Annexin V-FITC and 5 µl of PI solution in the dark at room temperature. After cells were gently vortexed and incubated for 15 min, 400 µl of 1X binding buffer was added to the cell suspension. Finally, the cell apoptosis rate was determined by a flow cytometry (BD Biosciences).
Dual luciferase reporter gene assay
The Dual-Luciferase reporter gene assay was performed to ascertain whether IFITM3 was the direct target gene of miR-29a. The IFITM3 3UTR dual luciferase reporter plasmids (WT and MUT) were constructed by Guangzhou RiboBio Biotechnology, Co., Ltd. They were co-transfected using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) with miR-29a mimics or miRNA NC into 293T cells, respectively (Shanghai Beinuo Biotech, Co., Ltd., Shanghai, China). After incubation for 48 h, the luciferase activities were detected using a Dual-Luciferase Assay System (Promega Corp., Madison, WI, USA).
Statistical analysis
GraphPad Prism 7.0 and SPSS 22.0 software were used for statistical analysis. Wilcoxons paired test was applied to compare the IFITM3 expression in HCC tissues and paracancerous tissues. The statistically significant positive correlation between IFITM3 and miR-29a expression levels was analyzed by Spearmans correlation analysis. Chi-square tests were used to examine the possible associations between the expression of IFITM3 or miR-29a and the clinicopathological characteristics of patients. The log-rank (Mantel-Cox) test was used to assess the differences in the overall survival rate. Comparisons between the two groups were analyzed with t-test or Students t-test. Multiple group comparisons were performed with the one-way analysis of variance (ANOVA), and the post hoc test was Student-Newman-Keuls (SNK) test. A P-value <0.05 was considered to indicate a statistically significant difference.
Results
IFITM3 is upregulated and miR-29a downregulated in HCC tissues and both are associated with HCC progression as well as poor prognosis
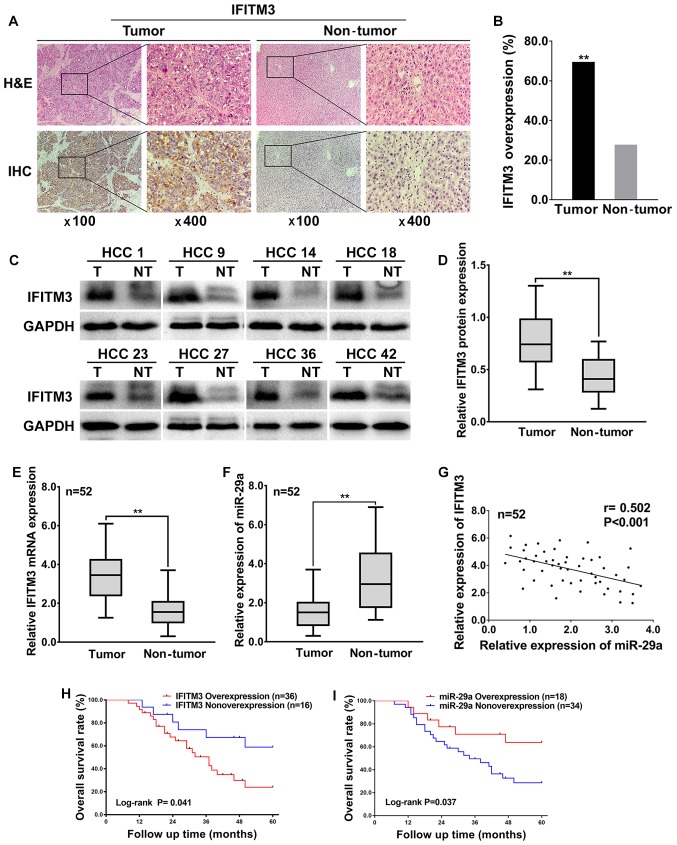
To investigate the expression levels and significance of IFITM3 in HCC tissues, a total of 52 HCC tissues and corresponding adjacent non-tumor tissues were examined using IHC and western blotting. The IHC results revealed that the IFITM3 protein was expressed in the cytoplasm as well as highly expressed in 69.23% (36 of 52) of HCC tissues and only 26.92% (14 of 52) of adjacent non-tumor tissues (Fig. 1A), which indicated that IFITM3 protein expression was significantly upregulated in HCC tissues compared with adjacent non-tumor tissues (P<0.01, Fig. 1B). In addition, the western blotting results revealed that the IFITM3 protein levels were significantly elevated in 73.08% (38 of 52) of HCC tissues, whereas the IFITM3 protein levels were elevated in only 21.15% (11 of 52) of adjacent non-tumor tissues (Fig. 1C), which was consistent with the IHC results, indicating that IFITM3 protein expression was significantly upregulated in HCC tissues (P<0.01; Fig. 1D).
Figure 1.
IFITM3 is upregulated and miR-29a downregulated in HCC tumor tissues and is associated with poor prognosis. (A) Representative IHC staining of IFITM3 in HCC tissues including tumor and non-tumor (magnification, left image ×100; right image ×400). (B) Comparison of overexpression rate among HCC tumor tissues and adjacent non-tumor tissues (**P<0.01 compared with the adjacent tissues). (C) Representative Western blotting analysis of IFITM3 protein expression (T, tumor; NT, non-tumorous tissues). (D) Quantification of IFITM3 protein expression using western blot analysis in 52 paired HCCs and their adjacent non-tumoral liver tissues. GAPDH protein expression was used as an internal control (**P<0.01). (E and F) qRT-PCR analysis of IFITM3 mRNA and miR-29a expression in 52 HCC tumors and peritumoral liver tissues (**P<0.01, paired Students t-test). (G) Scatter plots revealing a negative correlation between IFITM3 mRNA and miR-29a at the RNA level in 52 HCCs (r=−0.502, P<0.001). (H and I) HCC patients with IFITM3 and miR-29a high or low expression were both determined by log-rank (Mantel-Cox) test (P=0.041 and 0.037 respectively), revealing that overexpression of IFITM3 and non-overexpression of miR-29a was correlated to a poor overall survival for HCC patients. IFITM3, interferon-induced transmembrane protein 3; HCC, hepatocellular carcinoma.
To explore the expression and correlation of IFITM3 and miR-29a in HCC tissues, qRT-PCR was performed for 52 HCC tissues and adjacent non-tumor tissues. The qRT-PCR results revealed that the average fold change of IFITM3 mRNA expression in tumor tissues was significantly upregulated relative to that in paired non-tumor tissues (P<0.01; Fig. 1E). However, miR-29a was significantly decreased in tumor tissues compared with adjacent non-tumor tissues (P<0.01; Fig. 1F). Furthermore, the expression levels of IFITM3 in HCC tissues were negatively correlated with the miR-29a levels (two-tailed Spearmans correlation, r=−0.502, P<0.001; Fig. 1G).
The highest and lowest relative expression of miR-29a in 52 HCC tissues was 3.77 and 0.399, respectively. The mean level of miR-29a relative expression (mean value=1.521) was selected as a cut-off value. A total of 34 HCC tissues with miR-29a expression less than the mean value were considered to have non-overexpression of miR-29a, while the remaining 18 HCC tissues higher than the median value were considered overexpression. Moreover, the overall survival rate of HCC patients was both significantly decreased in IFITM3 overexpression patients [log-rank (Mantel-Cox) test, P=0.041; Fig. 1H] and miR-29a non-overexpression patients [log-rank (Mantel-Cox) test, P=0.037; Fig. 1I], indicating that IFITM3 overexpression and miR-29a non-overexpression were related to the poor prognosis of HCC patients. In addition, the highest and lowest survival time for HCC patients with IFITM3 overexpression or miR-29 non-overexpression were 8 and 60 months, respectively. The median survival time for HCC patients with IFITM3 overexpression was 37 months, and for the miR-29 non-overexpression it was 31 months.
Finally, we analyzed the associations between IFITM3 along with miR-29a expression in HCC and clinicopathological characteristics. The results revealed that IFITM3 overexpression and miR-29a non-overexpression were both closely associated with tumor size, tumor multifocal, and venous invasion (P<0.05 for all; Table I). These data indicated that IFITM3 overexpression and miR-29a non-overexpression were associated with HCC aggressive behavior and metastasis.
Table I.
Associations between miR-29a or IFITM3 and the clinicopathological characteristics in 52 HCC cases.
| miR-29a | IFITM3 | ||||||
|---|---|---|---|---|---|---|---|
| Clinicopathologic characteristics | (n) | Overexpression (n) | Nonoverexpression (n) | P-value | Overexpression (n) | Nonoverexpression (n) | P-value |
| Age (years) | 0.686 | 0.330 | |||||
| ≤51 | 24 | 9 | 15 | 15 | 9 | ||
| >51 | 28 | 9 | 19 | 21 | 7 | ||
| Sex | 0.245 | 0.683 | |||||
| Male | 37 | 11 | 26 | 25 | 12 | ||
| Female | 15 | 7 | 8 | 11 | 4 | ||
| Tumor size (cm) | 0.005 | 0.005 | |||||
| ≤5 | 21 | 12 | 9 | 10 | 11 | ||
| >5 | 31 | 6 | 25 | 26 | 5 | ||
| TNM stage | 0.703 | 0.309 | |||||
| I–II | 27 | 10 | 17 | 17 | 10 | ||
| III–IV | 25 | 8 | 17 | 19 | 6 | ||
| Tumor encapsulation | 0.115 | 0.115 | |||||
| Absent | 28 | 7 | 21 | 22 | 6 | ||
| Present | 24 | 11 | 13 | 14 | 10 | ||
| Tumor multifocal | 0.001 | 0.049 | |||||
| Absent | 22 | 13 | 9 | 12 | 10 | ||
| Present | 30 | 5 | 25 | 24 | 6 | ||
| Venous invasion | <0.001 | <0.001 | |||||
| Absent | 21 | 13 | 8 | 9 | 12 | ||
| Present | 31 | 5 | 26 | 27 | 4 | ||
| HBsAg | |||||||
| Negative | 13 | 6 | 7 | 0.313 | 7 | 6 | 0.165 |
| Positive | 39 | 12 | 27 | 29 | 10 | ||
| AFP (ng/ml) | 0.542 | 0.209 | |||||
| ≤400 | 29 | 9 | 20 | 18 | 11 | ||
| >400 | 23 | 9 | 14 | 18 | 5 | ||
| Cirrhosis | 0.278 | 0.734 | |||||
| Absent | 18 | 8 | 10 | 13 | 5 | ||
| Present | 34 | 10 | 24 | 23 | 11 | ||
The P-value represents probabilities for miR-29a or IFITM3 expression between variable subgroups determined by χ2 test. IFITM3, interferon-induced transmembrane protein 3; HCC, hepatocellular carcinoma; AFP, α-fetoprotein; HBsAg, hepatitis B surface antigen; TNM, tumor-node-metastasis; n, number of cases.
Knockdown of IFITM3 inhibits migration, invasion, proliferation and promotes apoptosis of HCC cells
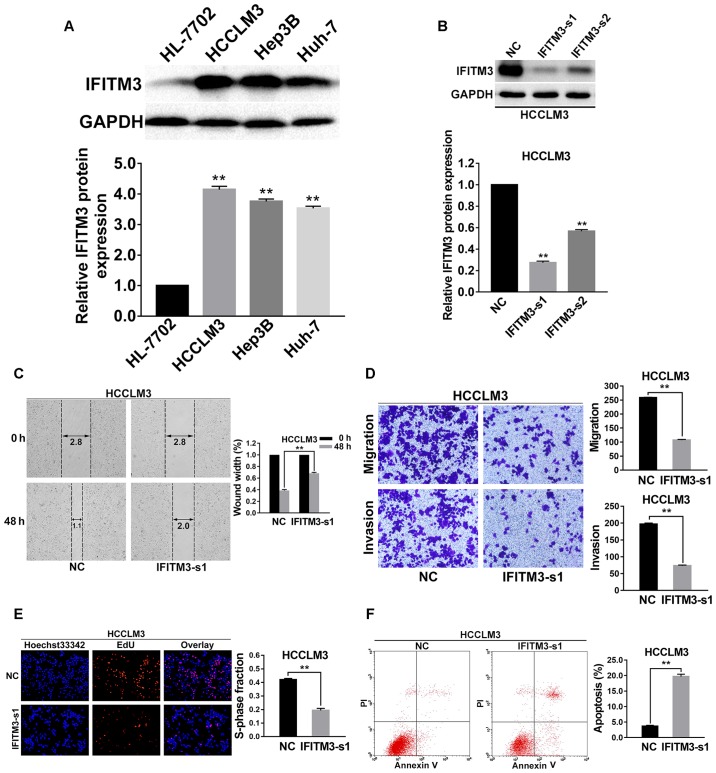
To investigate the effects of IFITM3 on metastasis, proliferation and apoptosis of HCC cells, we firstly examined the protein expression levels of IFITM3 in normal hepatocyte cells (HL-7702) and 3 HCC cell lines (Hep3B, HCCLM3 and Huh-7) by western blot analysis. The results revealed that expression of the IFITM3 protein in HCC cells was higher than that in the normal hepatocyte cell line. The HCCLM3 cells with the highest expression level of IFITM3 were finally selected for subsequent functional assays (P<0.01; Fig. 2A). Furthermore, two types of IFITM3 siRNA were used to transfect the HCCLM3 cell lines to detect the more effective sequence. At 48 h after transfection, IFITM3 protein expression was examined using western blotting, and the results revealed that IFITM3-s1 siRNA was more effective than IFITM3-s2 (P<0.01 for all; Fig. 2B). Thus, IFITM3-s1 siRNA was selected for subsequent RNA interference assays.
Figure 2.
Knockdown of IFITM3 inhibits invasion, migration, proliferation and promotes apoptosis of HCC cells. (A) Western blot analysis of IFITM3 expression in human normal hepatocyte cells (HL-7702) and 3 HCC cell lines (Hep3B, HCCLM3 and Huh-7) which indicated that IFITM3 protein expression in HCC cells was significantly higher than that in normal liver cells. **P<0.01 compared with the HL-7702 cells. (B) Two different IFITM3 siRNA sequences were transiently transfected into HCCLM3 cells to knockdown expression of IFITM3. After 48 h of transfection, western blot analysis revealed that the expression of IFITM3 was markedly downregulated with the IFITM3-siRNA groups. (C) Wound healing assay results revealed that wound closure was delayed with IFITM3 knockdown in HCCLM3 cells compared with NC groups at 48-h time-points after transfection. (D) Migration and invasion assays indicated that the migration and invasion abilities of IFITM3-s1 transfection HCCLM3 cells were decreased compared with the NC groups. (E) EdU proliferation assays revealed that the proliferation abilities were significantly reduced in IFITM3-s1-transfected HCCLM3 cells than in the NC groups. Blue, Hoechst 33342 staining of nuclei with all cells. Red, Apollo staining of EdU with proliferating cells. Overlay, the percentage of proliferating cells. (F) The apoptosis rate of HCCLM3 cells transfected with IFITM3-s1 was significantly higher than in the NC groups. **P<0.01 compared with the NC groups. NC, negative control; HCC, hepatocellular carcinoma; IFITM3, interferon-induced transmembrane protein 3.
Wound healing assays were performed to assess the variation of the cell migration ability for IFITM3 knockdown, and the results revealed that cell migration ability was significantly decreased in the IFITM3-s1 group (P<0.01; Fig. 2C). In addition, the results of the Transwell chamber assay without Matrigel for HCCLM3 cells were also consistent with the wound healing assay results, and using a Matrigel-coated Transwell chamber, we found that HCC cells with IFITM3 knockdown invaded through the matrix slower than the NC group cells (P<0.01 for all; Fig. 2D).
To explore the effects of IFITM3 knockdown on cell viability, EdU proliferation assays were conducted. The results revealed that growth of HCCLM3 cells was significantly decreased in the IFITM3-s1 group (P<0.01; Fig. 2E). To confirm whether IFITM3 was involved in the apoptosis of HCC cells, we used flow cytometry to examine the cell apoptosis rate of the treated HCC cells. The results indicated that the apoptosis rate of the IFITM3-s1 group was higher than that of the NC group (P<0.01; Fig. 2F).
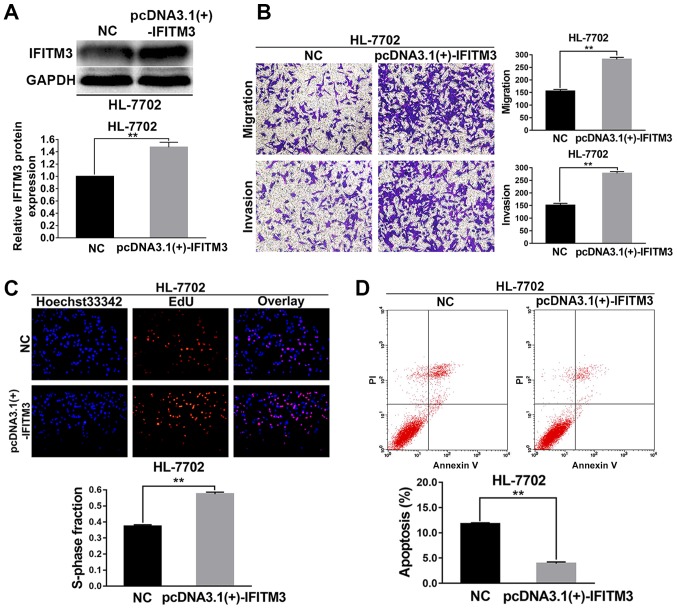
Upregulated IFITM3 promotes migration, invasion and proliferation as well as inhibits apoptosis of HL-7702 cells
The HL-7702 cells were selected for the IFITM3 upregulated assays. pcDNA3.1(+)-IFITM3 plasmids were transiently transfected into HL-7702 cells to enhance the expression of IFITM3. Firstly, western blot analysis revealed that the IFITM3 protein was increased after transfection with the pcDNA3.1(+)-IFITM3 plasmid, which indicated that the plasmid was effective (P<0.01; Fig. 3A). After IFITM3 was upregulated with pcDNA3.1(+)-IFITM3 plasmids, the results of the Transwell chamber assay revealed that the migration and invasion abilities of the HL-7702 cells were significantly increased (P<0.01; Fig. 3B). The results of the EdU proliferation assays revealed that growth ability of the HL-7702 cells was significantly increased after upregulation of IFITM3 (P<0.01; Fig. 3C). The results of flow cytometry indicated that the apoptosis rate of HL-7702 cells in the pcDNA3.1(+)-IFITM3 group was lower than the NC group (P<0.01; Fig. 3D).
Figure 3.
Upregulated IFITM3 promotes invasion, migration, proliferation and inhibits apoptosis of HL-7702 cells. (A) After 48 h of transfection, western blot analysis revealed that the expression of IFITM3 was markedly upregulated in the pcDNA3.1(+)-IFITM3 group. (B) Transwell chamber assay results revealed that the migration and invasion abilities of HL-7702 cells in the pcDNA3.1(+)-IFITM3 group were higher than in the NC group. (C) EdU proliferation assays revealed that the proliferation abilities of HL-7702 cells were increased in the pcDNA3.1(+)-IFITM3 group compared with the NC group. Blue, Hoechst 33342 staining of nuclei with all cells. Red, Apollo staining of EdU with proliferating cells. Overlay, the percentage of proliferating cells. (D) The apoptosis rate of HL-7702 cells transfected with pcDNA3.1(+)-IFITM3 plasmids was significantly lower than the NC group. **P<0.01 compared with the NC group. NC, negative control; HCC, hepatocellular carcinoma; IFITM3, interferon-induced transmembrane protein 3.
Upregulated miR-29a suppresses migration, invasion and proliferation as well as promotes apoptosis of HCC cells
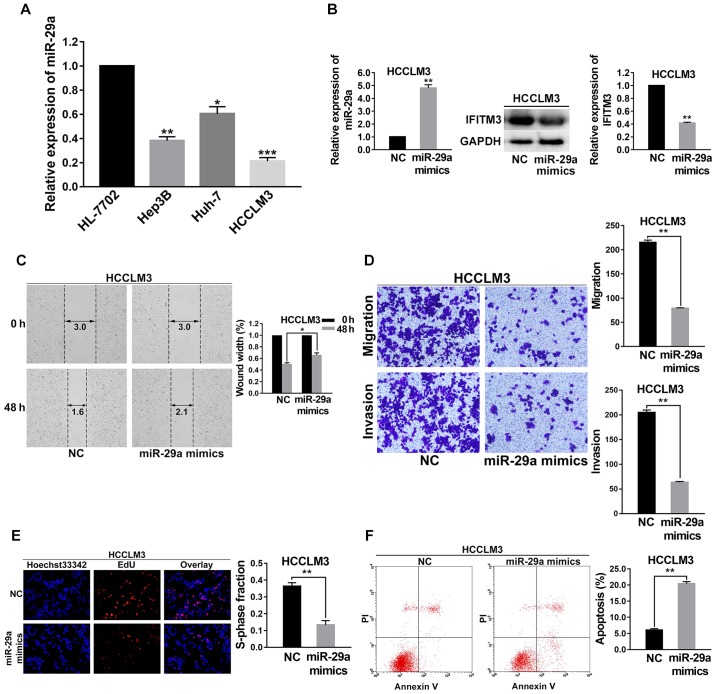
As shown in Fig. 4, the effects of upregulated miR-29a in HCC cells on the biological behaviors of those cells were consistent with the effects of IFITM3 knockdown. To investigate the function and mechanism of miR-29a in HCC, the expression level of miR-29a was first examined in 3 HCC cell lines (Hep3B, HCCLM3 and Huh-7) and normal hepatocyte cells (HL-7702) using qRT-PCR. As shown in Fig. 4A, the level of miR-29a in these HCC cell lines was decreased compared with the HL-7702 cells; and the HCCLM3 cell lines had the lowest expression levels of miR-29a (P<0.001; Fig. 4A). Hence, HCCLM3 cells were selected for subsequent assays and were transfected with miR-29a mimics. At 36 h post-transfection, a satisfactory transfection efficiency was observed by qRT-PCR, and using western blot analysis, we found that the IFITM3 protein expression was reduced after 48 h of transfection (P<0.01; Fig. 4B).
Figure 4.
Upregulated miR-29a suppresses migration, invasion, proliferation and promotes apoptosis of HCC cells. (A) The level of miR-29a in 3 HCC cell lines (Hep3B, HCCLM3 and Huh-7) and a normal liver cell line (HL-7702). (B) The satisfactory transfection efficiency of miR-29a mimics with HCCLM3 cells, and the IFITM3 protein expression in response to the transfection of miR-29a. (C) Wound healing assays indicated that the migration capacity of HCCLM3 cells was decreased after miR-29a upregulation. (D) The invasion and metastatic abilities of HCCLM3 cells were decreased after miR-29a upregulation. (E) EdU proliferation assays demonstrated that the proliferation abilities were significantly decreased in the miR-29a-upregulated HCCLM3 cells than that in the NC groups. (F) The apoptosis rate of HCCLM3 cells transfected with miR-29a mimics was significantly higher than in the NC groups. *P<0.05, **P<0.01, ***P<0.001 compared with the NC groups. NC, negative control; IFITM3, interferon-induced transmembrane protein 3.
Wound healing assays were performed to detect the variation of migration capacity of HCCLM3 cells after they were transfected with miR-29a mimics. The results revealed that the migration capacity of HCCLM3 cell lines was decreased after miR-29a was upregulated (P<0.05; Fig. 4C). Transwell chamber migration and invasion assays demonstrated that the invasion and metastatic abilities of HCCLM3 cells were reduced after miR-29a was upregulated (P<0.01 for all; Fig. 4D).
To investigate the effects of miR-29a upregulation on the cell proliferation capacity, EdU proliferation assays were carried out. The results indicated that the growth of HCCLM3 cells was significantly decreased in the miR-29a mimics group (P<0.01; Fig. 4E). Flow cytometry was performed to examine the cell apoptosis rate change of the miR-29a mimic-treated HCCLM3 cells. The results revealed that the apoptosis rate of the miR-29a mimic group was higher than that of the NC groups (P<0.01; Fig. 4F).
IFITM3 is the target gene of miR-29a
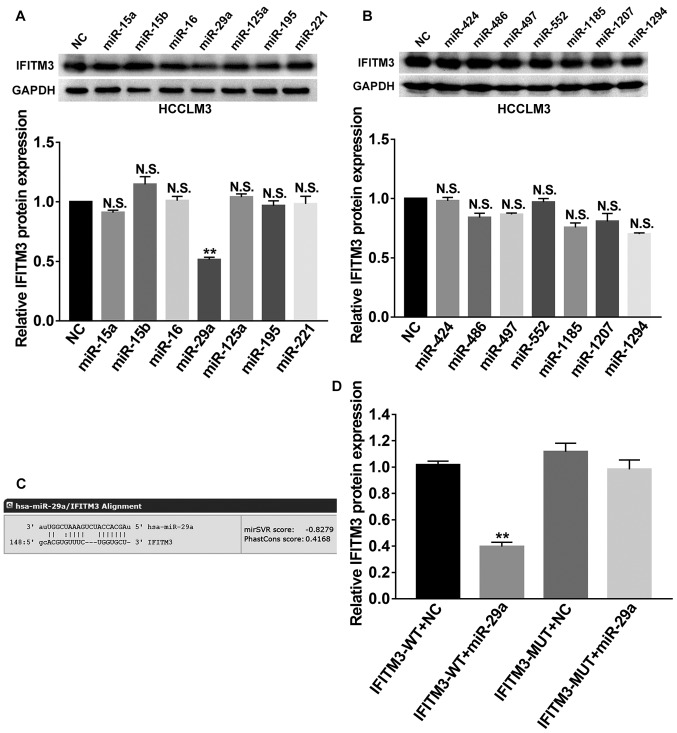
We focused on the post-transcriptional regulation of IFITM3 and investigated the effects of miRNAs on IFITM3. Next, we employed bioinformatics analysis to predict potential miRNAs that targeted the 3′UTR of IFITM3 mRNA by using 2 websites, including TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do). Then, a total of 14 candidate miRNAs (miR-15a, miR-15b, miR-16, miR-29a, miR-125a, miR-195, miR-221, miR-424, miR-486, miR-497, miR-552, miR-1185, miR-1207 and miR-1294) with good scores were selected for further screening and their corresponding mimics were respectively transfected into HCCLM3 cells. Finally, western blot analysis revealed that miR-29a could markedly suppress the protein expression of IFITM3 (P<0.01; Fig. 5A and B), which indicated that miR-29a was involved in the regulation of IFITM3. As shown in Fig. 5C, the 3′-UTR of IFITM3 contained a binding site for miR-29a. Subsequently, we performed a dual luciferase reporter gene assay to confirm the regulation relationship between miR-29a and IFITM3. The results revealed that the miR-29a mimics could reduce the luciferase activities of the wild-type IFITM3 reporter vector (IFITM3-WT) rather than the mutant-type IFITM3 reporter vector (IFITM3-MUT) (P<0.01; Fig. 5D), indicating that IFITM3 was the target gene of miR-29a.
Figure 5.
IFITM3 is the targeting gene of miR-29a. (A and B) Western blot analysis screened 14 candidate miRNAs and indicated that miR-29a could markedly suppress the protein expression of IFITM3 in HCCLM3 cells. (C) The 3′-UTR of IFITM3 contained a binding site for miR-29a. (D) Dual-Luciferase reporter gene assay revealed that miR-29a mimics could reduce the luciferase activities of IFITM3-WT reporter vector rather than IFITM3-MUT, indicating that IFITM3 was the targeting gene of miR-29a. **P<0.01. N.S., non-significant; WT, wild-type; MUT, mutant type; IFITM3, interferon-induced transmembrane protein 3.
Discussion
HCC is one of the most common and aggressive cancers. Although surgical resection, chemotherapy and radiation have been used to treat patients with HCC, the strong invasive and metastatic properties of HCC result in a high incidence of recurrence, metastasis and poor survival for patients (1–4,27). Thus, there is a pressing need to reveal the molecular mechanisms of HCC to develop novel therapeutic strategies for treating this disease.
Initiation and progression of HCC have been demonstrated to involve the dysregulation of various genes and miRNAs (28,29). The human IFITM protein family, including IFITM1 (also named 9–27), IFITM2 (also named 1–8D), and IFITM3 (also named 1-8U), initially received attention because human IFITM3 was found to limit influenza A virus (IAV) infection in cultured cells. Then, they were identified as antiviral factors and possess antiviral activities that limit the infection of cultured cells by many viruses (7,9,30). However, recently, accumulating evidence has shown that aberrant expression of IFITM3 is associated with the progression of various cancers. For example, Li et al (10) revealed that IFITM3 was expressed at higher levels in colon cancer and played a critical role in its progression and metastasis. The study of Andreu et al (11) confirmed that IFITM3 gene expression was significantly upregulated in colorectal tumors and could be rapidly induced after activation of the β-catenin signaling pathway. Bing et al (12) also demonstrated that IFITM3 played an important role in glioma cell growth and migration and was associated with tumorigenesis. In addition, overexpression of IFITM3 in gastric cancer has been observed and related to tumor metastasis (31). For patients with esophageal squamous cell carcinoma, overexpression of IFITM3 indicated a high risk of lymphatic metastatic recurrence after Ivor-Lewis esophagectomy (32). Breast cancer cell growth and colony formation could be suppressed through the downregulation of IFITM3 expression (33).
To date, however, the expression and potential biological function of IFITM3 in HCC have not been elucidated. The present study revealed that IFITM3 expression was significantly increased in human HCC tissues and HCC cells. In addition, knockdown of IFITM3 inhibited migration, invasion and proliferation as well as promoted apoptosis of HCC cells. Furthermore, after upregulation of IFITM3, the invasion, migration and proliferation abilities of HL-7702 cells were increased, but the apoptosis rate was decreased. These results indicated that IFITM3 plays important roles in the tumorigenesis and development of HCC. However, the upstream molecular mechanism remains unclear.
miRNAs are a class of endogenous ~22 nucleotide RNAs that have central roles in gene regulatory networks. Generally, miRNAs control gene expression by regulating the mRNA or translation of target genes (34). miRNAs always play important roles in various biological and pathological processes of HCC. Thus, we focused on the post-transcriptional regulation of IFITM3 and investigated the effects of miRNAs on IFITM3. Therefore, we employed two bioinformatics analysis websites to predict the potential miRNAs targeting the 3′UTR of IFITM3 mRNA. Initially, 14 candidate miRNAs (miR-15a, miR-15b, miR-16, miR-29a, miR-125a, miR-195, miR-221, miR-424, miR-486, miR-497, miR-552, miR-1185, miR-1207 and miR-1294) with good scores were selected for additional screening. Eventually, only miR-29a was identified as a functional target gene of IFITM3 by using dual luciferase reporter gene assay and western blot analysis. Previously, some studies have already reported aberrant expression of miR-29a in HCC that was associated with multiple processes involved in tumor progression. For example, the study of Zhu et al (20) noted that miR-29a was downregulated in HCC and could suppress cell proliferation by targeting SPARC. Mahati et al (21) also reported that miR-29a was downregulated in HCC and that it could suppress growth and migration of HCC by regulating CLDN1. Additionally, miR-29a could also regulate TGF-β-induced EMT in HCC (22). However, the effect of miR-29a on IFITM3 remains to be established, and the biological significance of miR-29a in HCC has not been fully elucidated.
In the present study, we investigated the effects of upregulated miR-29a in HCC cells on the IFITM3 expression levels and on the biological behaviors of HCC cells. We found that IFITM3 protein expression was reduced after cells were transfected with miR-29a mimics, indicating that miR-29a could downregulate the expression of IFITM3. The results also indicated that the invasion, migration and proliferation capabilities of HCC cells were suppressed and the apoptosis rate was increased after miR-29a upregulation, which were consistent with the effects of IFITM3 knockdown. The results provided further evidence that IFITM3 is the target of miR-29a.
Our results also revealed that the level of miR-29a was significantly upregulated in HCC tissues and was negatively correlated with IFITM3 expression. Furthermore, the overall survival rate of HCC patients was significantly decreased in patients with IFITM3 overexpression and miR-29a non-overexpression, indicating that they were related to poor prognosis. Moreover, IFITM3 overexpression and miR-29a non-overexpression were both closely correlated with tumor size, tumor multifocal and venous invasion, which are always associated with tumor growth and metastasis. In general, multifocal HCC can be caused by two distinct biological processes. One is intrahepatic metastasis (IM) which represents that a primary lesion occurs firstly and then spreads to additional locations in the liver. The other one is multicentric carcinogenesis (MC) which represents multiple lesions that occur independently due to underlying pathogenesis (35). The results indicated that miR-29a and IFITM3 may be involved in the progression of HCC carcinogenesis. Therefore, HCC with miR-29a non-overexpression and IFITM3 overexpression may be more invasive with higher malignancy, and patients may be more inclined to tumor multifocal and intrahepatic recurrence.
In summary, the results of the present study collectively indicated that miR-29a suppressed migration, invasion, and proliferation as well as promoted apoptosis of HCC via IFITM3. Thus, IFITM3 was identified as a novel target of miR-29a. These data laid the foundation for using miR-29a and IFITM3 regulatory pathways for the diagnosis and treatment of HCC patients.
Acknowledgements
The authors are especially grateful to ‘Jiangxi Provincial Key Laboratory of Molecular Medicine in the Second Affiliated Hospital of Nanchang University’ for providing the experimental facilities.
Funding
The present study was supported by grants from the Natural Science Foundation of Jiangxi Province, China (no. 20171BAB205063), the Youth Science Foundation of Jiangxi Province, China (no. 20171BAB215037) and the Graduate Innovation Special Foundation of Nanchang University (no. cx2016397).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YL, EL, WL, LW conceived and designed the study. YL, EL, CG, JG and JA contributed to the acquisition of the data. YL, EL, JM, WL and LW analyzed and interpreted the data. YL, EL, JM, CG, JG, JA, WL and LW wrote, reviewed, and/or revised the manuscript. WL and LW supervised the study. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The study protocol was approved by the Medical Ethics Committee of the Second Affiliated Hospital of Nanchang University. This study was performed in accordance with the ethical standards of the Declaration of Helsinki. Informed consent was obtained from all patients for using their tissue specimens and clinicopathological data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Ochiai T, Ikoma H, Okamoto K, Kokuba Y, Sonoyama T, Otsuji E. Clinicopathologic features and risk factors for extrahepatic recurrences of hepatocellular carcinoma after curative resection. World J Surg. 2012;36:136–143. doi: 10.1007/s00268-011-1317-y. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y, Nagano H, Ota H, Morimoto O, Nakamura M, Wada H, Noda T, Damdinsuren B, Marubashi S, Miyamoto A, et al. Patterns and clinicopathologic features of extrahepatic recurrence of hepatocellular carcinoma after curative resection. Surgery. 2007;141:196–202. doi: 10.1016/j.surg.2006.06.033. [DOI] [PubMed] [Google Scholar]
- 5.Gauthier A, Ho M. Role of sorafenib in the treatment of advanced hepatocellular carcinoma: An update. Hepatol Res. 2013;43:147–154. doi: 10.1111/j.1872-034X.2012.01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 7.Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-family proteins: The cells first line of antiviral defense. Annu Rev Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey CC, Kondur HR, Huang IC, Farzan M. Interferon-induced transmembrane protein 3 is a type II transmembrane protein. J Biol Chem. 2013;288:32184–32193. doi: 10.1074/jbc.M113.514356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen LM, Gaiha GD, Ryan BJ, Donis RO, Elledge SJ, Brass AL. IFITM3 inhibits influenza a virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, Peng Z, Tang H, Wei P, Kong X, Yan D, Huang F, Li Q, Le X, Li Q, Xie K. KLF4-mediated negative regulation of IFITM3 expression plays a critical role in colon cancer pathogenesis. Clin Cancer Res. 2011;17:3558–3568. doi: 10.1158/1078-0432.CCR-10-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreu P, Colnot S, Godard C, Laurent-Puig P, Lamarque D, Kahn A, Perret C, Romagnolo B. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res. 2006;66:1949–1955. doi: 10.1158/0008-5472.CAN-05-2731. [DOI] [PubMed] [Google Scholar]
- 12.Bing Z, Wang H, Gang Z, Ping L. The role of IFITM3 in the growth and migration of human glioma cells. BMC Neurol. 2013;13:210. doi: 10.1186/1471-2377-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Kota J, Chivukula RR, ODonnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852–1862. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 18.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 19.Wong CC, Wong CM, Tung EK, Au SL, Lee JM, Poon RT, Man K, Ng IO. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140:322–331. doi: 10.1053/j.gastro.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Zhu XC, Dong QZ, Zhang XF, Deng B, Jia HL, Ye QH, Qin LX, Wu XZ. microRNA-29a suppresses cell proliferation by targeting SPARC in hepatocellular carcinoma. Int J Mol Med. 2012;30:1321–1326. doi: 10.3892/ijmm.2012.1140. [DOI] [PubMed] [Google Scholar]
- 21.Mahati S, Xiao L, Yang Y, Mao R, Bao Y. miR-29a suppresses growth and migration of hepatocellular carcinoma by regulating CLDN1. Biochem Biophys Res Commun. 2017;486:732–737. doi: 10.1016/j.bbrc.2017.03.110. [DOI] [PubMed] [Google Scholar]
- 22.Kogure T, Kondo Y, Kakazu E, Ninomiya M, Kimura O, Shimosegawa T. Involvement of miRNA-29a in epigenetic regulation of transforming growth factor-beta-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Hepatol Res. 2014;44:907–919. doi: 10.1111/hepr.12188. [DOI] [PubMed] [Google Scholar]
- 23.Cho HJ, Kim SS, Nam JS, Kim JK, Lee JH, Kim B, Wang HJ, Kim BW, Lee JD, Kang DY, et al. Low levels of circulating microRNA-26a/29a as poor prognostic markers in patients with hepatocellular carcinoma who underwent curative treatment. Clin Res Hepatol Gastroenterol. 2017;41:181–189. doi: 10.1016/j.clinre.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Liu W, Liang B, Liu H, Huang Y, Yin X, Zhou F, Yu X, Feng Q, Li E, Zou Z, Wu L. Overexpression of nonSMC condensin I complex subunit G serves as a promising prognostic marker and therapeutic target for hepatocellular carcinoma. Int J Mol Med. 2017;40:731–738. doi: 10.3892/ijmm.2017.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Li C, Miao R, Liu S, Wan Y, Zhang S, Deng Y, Bi J, Qu K, Zhang J, Liu C. Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis. Oncotarget. 2017;8:28683–28695. doi: 10.18632/oncotarget.15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan R, Wang K, Hu J, Yan C, Li M, Yu X, Liu X, Lei J, Guo W, Wu L, et al. Ubiquitin-like protein FAT10 promotes the invasion and metastasis of hepatocellular carcinoma by modifying beta-catenin degradation. Cancer Res. 2014;74:5287–5300. doi: 10.1158/0008-5472.CAN-14-0284. [DOI] [PubMed] [Google Scholar]
- 28.Fabbri M, Calore F, Paone A, Galli R, Calin GA. Epigenetic regulation of miRNAs in cancer. Adv Exp Med Biol. 2013;754:137–148. doi: 10.1007/978-1-4419-9967-2_6. [DOI] [PubMed] [Google Scholar]
- 29.Yu G, Chen X, Chen S, Ye W, Hou K, Liang M. miR-19a, miR-122 and miR-223 are differentially regulated by hepatitis B virus X protein and involve in cell proliferation in hepatoma cells. J Transl Med. 2016;14:122. doi: 10.1186/s12967-016-0888-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewin AR, Reid LE, McMahon M, Stark GR, Kerr IM. Molecular analysis of a human interferon-inducible gene family. Eur J Biochem. 1991;199:417–423. doi: 10.1111/j.1432-1033.1991.tb16139.x. [DOI] [PubMed] [Google Scholar]
- 31.Hu J, Wang S, Zhao Y, Guo Q, Zhang D, Chen J, Li J, Fei Q, Sun Y. Mechanism and biological significance of the overexpression of IFITM3 in gastric cancer. Oncol Rep. 2014;32:2648–2656. doi: 10.3892/or.2014.3522. [DOI] [PubMed] [Google Scholar]
- 32.Jia Y, Zhang M, Jiang W, Zhang Z, Huang S, Wang Z. Overexpression of IFITM3 predicts the high risk of lymphatic metastatic recurrence in pN0 esophageal squamous cell carcinoma after Ivor-Lewis esophagectomy. PeerJ. 2015;3:e1355. doi: 10.7717/peerj.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang M, Gao H, Chen P, Jia J, Wu S. Knockdown of interferon-induced transmembrane protein 3 expression suppresses breast cancer cell growth and colony formation and affects the cell cycle. Oncol Rep. 2013;30:171–178. doi: 10.3892/or.2013.2428. [DOI] [PubMed] [Google Scholar]
- 34.Meister G. miRNAs get an early start on translational silencing. Cell. 2007;131:25–28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 35.Chianchiano P, Pezhouh MK, Kim A, Luchini C, Cameron A, Weiss MJ, He J, Voltaggio L, Oshima K, Anders RA, Wood LD. Distinction of intrahepatic metastasis from multicentric carcinogenesis in multifocal hepatocellular carcinoma using molecular alterations. Hum Pathol. 2018;72:127–134. doi: 10.1016/j.humpath.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.